A Sec14-nodulin protein model is used to identify the nodulin domain as a novel phosphoinositide effector module with a role in controlling lateral organization of phosphoinositide. The domain organization of Sec14-nodulin proteins suggests a versatile principle for the bit mapping of membrane surfaces into high-definition lipid-signaling screens.
Abstract
Polarized membrane morphogenesis is a fundamental activity of eukaryotic cells. This process is essential for the biology of cells and tissues, and its execution demands exquisite temporal coordination of functionally diverse membrane signaling reactions with high spatial resolution. Moreover, mechanisms must exist to establish and preserve such organization in the face of randomizing forces that would diffuse it. Here we identify the conserved AtSfh1 Sec14-nodulin protein as a novel effector of phosphoinositide signaling in the extreme polarized membrane growth program exhibited by growing Arabidopsis root hairs. The data are consistent with Sec14-nodulin proteins controlling the lateral organization of phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) landmarks for polarized membrane morphogenesis in plants. This patterning activity requires both the PtdIns(4,5)P2 binding and homo-oligomerization activities of the AtSfh1 nodulin domain and is an essential aspect of the polarity signaling program in root hairs. Finally, the data suggest a general principle for how the phosphoinositide signaling landscape is physically bit mapped so that eukaryotic cells are able to convert a membrane surface into a high-definition lipid-signaling screen.
INTRODUCTION
A cardinal feature of eukaryotic cells is their ability to register lipid-signaling reactions with high spatial and temporal precision on large membrane surfaces. Membrane morphogenesis is a fundamental process that relies on this feature and is the foundation upon which cell shape control, tissue formation, and organogenesis are built (Hepler et al., 2001; Affolter et al., 2009; Cáceres et al., 2012). Polarized membrane growth is developmentally controlled, and the process is also induced by environmental factors, as exemplified by dimorphic switch of fungal pathogens from budding to mycelial growth modes (Whiteway and Bachewich, 2007; Richter et al., 2011). Both modes of membrane morphogenic control are on display in the bacteria–plant symbiosis required for nitrogen fixation (Oldroyd and Downie, 2008; Oldroyd, 2013).
Biological nitrogen fixation is a prokaryotic activity of planetary significance that captures atmospheric N2 by reducing the gas into “combined” nitrogen forms suitable for assimilation by plants (Vance, 2001). The primary source for symbiotic N2 fixation relies on an intimate partnership between highly polarized membrane structures (root hairs) of leguminous plants and N2-fixing rhizobia bacteria in soil (Oldroyd and Downie, 2008; Oldroyd, 2013). This symbiosis involves a bidirectional chemical dialogue between the prokaryotic and eukaryotic partners, culminating in bacterial colonization and infection of growing root hairs. Subsequent formation of specialized microenvironments, termed nodules, provides the anaerobic niche essential for the fixing of N2 by bacterial nitrogenase (Long 2001; Brewin 2002; Debrosses et al., 2011; Suzaki et al., 2014). Bacteria foster establishment of the symbiotic state by secreting a battery of nodulation factors to which the plant responds by deforming growing tips of root hairs (Irving et al., 2000; Oldroyd et al., 2013). The resulting structures entrap the rhizobia and initiate the infection process through a local invagination of the root hair plasma membrane and establishment of a polarized growing infection structure, termed the infection thread, into underlying cortical cells (van Spronsen et al., 2001; Monahan-Giovanelli et al., 2006). Activation of meristematic activity in cortical cells subsequently initiates formation of a nodule primordium (Oldroyd, 2013). Organ-specific plant proteins expressed only during symbiotic nitrogen fixation are called nodulins, and these polypeptides define the plant's contribution to symbiosis (van Kammen 1984; Mylona et al., 1995). Here we focus on Nlj16-like nodulins. The founding member of this family, Nlj16, was first described in the legume Lotus japonicus as a 15-kDa protein of unknown function specifically expressed late in the nodulation program (Kapranov et al., 1997).
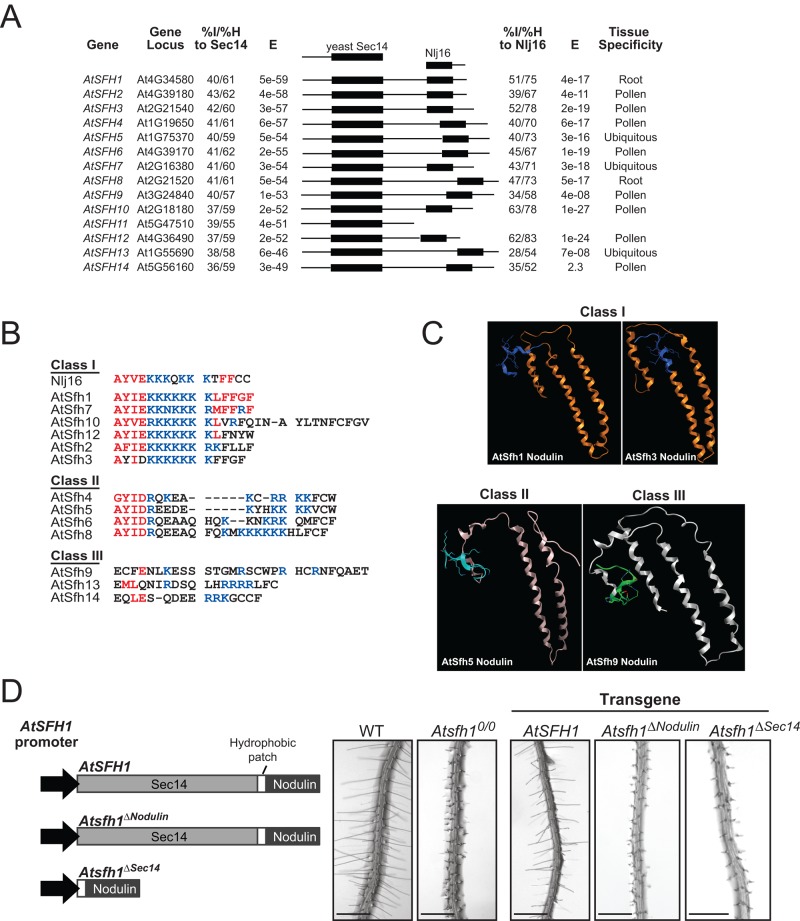
Sec14-like phosphatidylinositol transfer proteins (PITPs) integrate stimulated phosphatidylinositol 4-phosphate (PtdIns(4)P) production with multiple aspects of intracellular lipid metabolism and diversify biological outcomes for phosphoinositide signaling (Schaaf et al., 2008; Bankaitis et al., 2010). It is from this perspective that discovery of a conserved family of Sec14-Nlj16–like nodulin proteins in Lotus and other plants brings together root hair development, lipid signaling, and nodulation in unexpected ways (Kapranov et al., 2001; Vincent et al., 2005; Huang et al., 2013). Arabidopsis produces 31 Sec14-like proteins. Of the 14 Arabidopsis Sec14-like proteins with highest homology to yeast Sec14, 13 exhibit C-terminal Nlj16-like nodulin domains (Figure 1A). Moreover, these Arabidopsis Sec14-nodulin proteins are expressed predominantly in tip-growing cells that execute developmental programs of extreme polarized membrane growth (pollen, root hairs; Figure 1A). Arabidopsis mutants lacking the AtSfh1 Sec14-nodulin elaborate short, distorted root hairs characterized by loss of tip-directed phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) gradients, disorganized cytoskeleton networks, and delocalized Ca2+ signaling (Vincent et al., 2005). Whereas Sec14-domains are well characterized, the roles of the nodulin domains remain mysterious.
FIGURE 1:
The Arabidopsis Sec14-nodulin protein family. (A) Alignment of the 14 highest-scoring Sec14 homologues of the Arabidopsis Sec14-like PITP family. All Arabidopsis proteins with homology to the L. japonicus Nlj16 nodulin are also shown. Percentage identities (% I) and similarities (% H) and corresponding E-values are indicated (Sec14, left; nodulin, right), as are tissue expression profiles (www.ncbi.nlm.nih.gov/geo/). (B) Alignments of the C-termini of class I, II, and III nodulin domains. Conserved residues and basic amino acids are in red and blue, respectively. (C) Homology models for nodulin domains. Models were generated by structural templating using nodulin homology to a region of a DNA topoisomerase. (D) Diagram of Atsfh1 constructs used to transform Atsfh10/0 plants. All transgenes were expressed under native AtSFH1 promoter control and encoded epitope-tagged myc-AtSfh1–hemagglutinin (HA), myc-AtSfh1ΔNodulin-HA, and AtSfh1ΔSec14-HA, respectively. Bright-field images of root hairs of transgenic seedling. Scale bars, 1 mm.
The nodulin domains of all known Sec14-nodulins belong to the Nlj16 family, and the known Nlj16-like nodulins are genetically encoded as C-terminal domains of Sec14-like proteins (Kapranov et al., 2001; Vincent et al., 2005). In Lotus, nodulation-specific production of free-standing Nlj16 results from developmentally controlled reconfiguration of LjPLP-IV (encoding Sec14-Nlj16) gene transcription. This reprogramming drives robust expression of the Nlj16 nodulin as a free-standing domain at the expense of the full-length Sec14-Nlj16 (Kapranov et al., 2001). Whereas the joining of Sec14 modules with Nlj16-nodulins in leguminous and nonleguminous plants forecasts that these units execute coordinated activities, the developmental expression profiles indicate these activities are differentially used in root hair biogenesis versus nodulation (Kapranov et al., 2001).
Here we report the first mechanistic insights into the activities of Nlj6-like nodulin domains. We identify the AtSfh1 nodulin as a novel PtdIns(4,5)P2-binding module whose lipid-binding and self-assembly activities are essential for root hair morphogenesis when the nodulin domain is produced in the context of a full-length AtSfh1 protein. By contrast, the stand-alone AtSfh1 nodulin domain exhibits properties of a potent PtdIns(4,5)P2 sink that antagonizes PtdIns(4,5)P2 signaling by sequestering the lipid from active signaling pools. The data describe the AtSfh1 Sec14-nodulin as a novel polarity regulator that organizes PtdIns(4,5)P2 landmarks for root hair morphogenesis. The results further highlight the nodulin domain as a versatile PtdIns(4,5)P2 clamp whose activities can be developmentally modulated so as to promote or, as in late stages of nodulation, help subvert polarized morphogenetic programs in developing plant tissues.
RESULTS
Arabidopsis Sec14-nodulin proteins
Nlj16-like nodulin domains share extensive primary sequence homology along their length of ∼120 amino acids (Ile et al., 2006) and fall into three classes distinguished by their extreme C-terminal sequences. Class I nodulins (including AtSfh1 nodulin) are characterized by an uninterrupted stretch of seven or more basic C-terminal amino acids with vicinal aromatic residues. The class I AtSfh10 nodulin domain additionally harbors a Cys residue that represents a potential palmitoylation site. Class II and class III nodulins exhibit C-terminal stretches of more than seven contiguous basic residues. All class II modules show penultimate Cys residues. Class III modules exhibit the least basic C-termini (Figure 1B).
Templated and ab initio modeling simulations, although generating a number of potential structural models, nonetheless consistently predicted the AtSfh1 nodulin adopts an α-helical coiled-coil structure. For example, one templated model predicts the AtSfh1 nodulin domain assumes an elongated antiparallel three-helix coiled-coil (Figure 1C). Templated modeling of AtSfh3, AtSfh5, and AtSfh9 nodulin domains similarly arrived at elongated three-helix coiled-coil folds. Although these simulations do not confidently generate precise structural details, these experiments nonetheless forecast that a common structural feature of Nlj16-like nodulin domains is an α-helical coiled-coil fold. As described later, the biochemical properties of these nodulin domains are consistent with this inference. The conserved joining of a Sec14 domain with an Nlj16-like nodulin suggests that both domains contribute to AtSfh1 function. Indeed, expression of neither the isolated Sec14 nor nodulin domain complemented short–root hair phenotypes of Atsfh10/0 plants (Figure 1D).
Class I nodulin domains are PtdIns(4,5)P2-binding modules
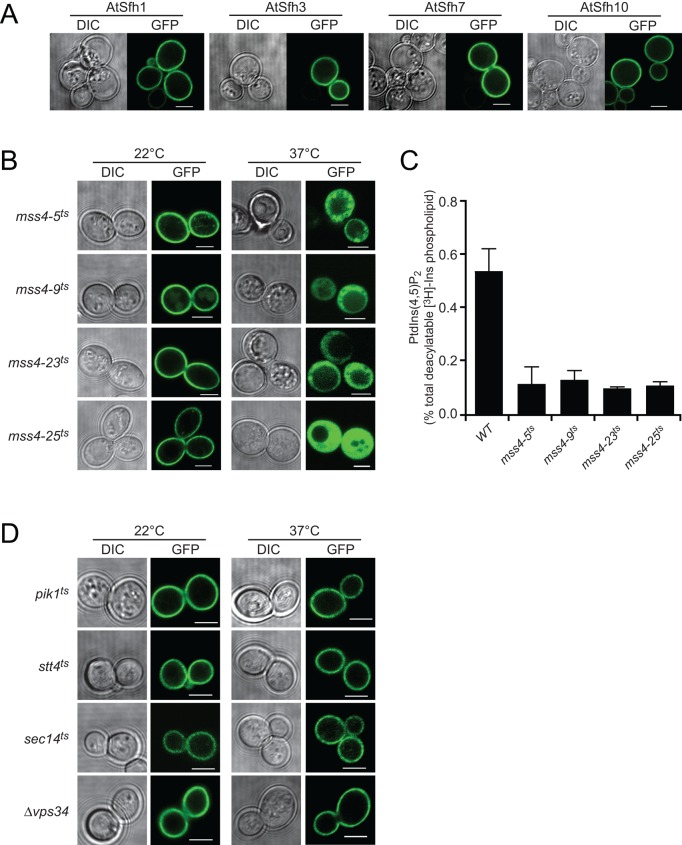
The signature basic patch/aromatic residue motifs of class I nodulin C-termini resemble mammalian plasma membrane (PM) targeting motifs that bind acidic lipids, including phosphoinositides (McLaughlin and Aderem, 1995; McLaughlin and Murray, 2005; Murray et al., 2002). Phosphoinositides, particularly PtdIns(4,5)P2, are well-established regulators of polarized membrane growth in various unicellular and multicellular organisms, including plant root hairs (Braun et al., 1999; Devreotes and Janetopoulos, 2003; Gervais et al., 2008; Yang, 2008; Heilmann, 2009; Yakir-Tamang and Gerst, 2009; Krahn and Wodarz, 2012). We therefore considered the possibility that nodulin domains play important roles in execution of PtdIns(4,5)P2 signaling in tip-growing root hairs and exploited yeast (a system in which the composition of a biological membrane can be manipulated) to examine whether nodulin domains display specific lipid interactions. AtSfh1, AtSfh3, AtSfh7, and AtSfh10 class I GFP-nodulins targeted to the PM, whereas class II and class III GFP-nodulins did not (Figure 2A and Supplemental Figure S1, A and B). Membrane targeting of class I nodulins was PtdIns(4,5)P2 dependent, as inactivation of a temperature-sensitive version of yeast PtdIns-4-phosphate 5-OH kinase (Mss4) released class I GFP-nodulins from the PM (Figure 2B and Supplemental Figure S1C). PtdIns(4,5)P2 levels were reduced in all mss4ts mutants at 37°C (Figure 2C).
FIGURE 2:
Class I nodulins localize to yeast PM in a PtdIns(4,5)P2-dependent manner. (A) GFP-tagged class I chimeras localize to PM when expressed in WT yeast. Images are representative of 158, 261, 352, and 216 cells expressing AtSfh1, AtSfh3, AtSfh7, and AtSfh10 nodulins, respectively. All cells showed exclusive PM localization. (B) GFP-tagged class I nodulins are released from PM in four independently isolated mss4ts mutants when Mss4 is inactivated at 37°C. Images are representative of an aggregate of 901 and 1094 cells imaged at 22 and 37°C, respectively, and 107–355 cells were scored for each nodulin at each temperature. In all cases, >95% of the cells imaged at 22°C showed PM localization of the GFP-nodulin reporter, whereas, in all cases, >93% of the cells imaged at 37°C showed exclusively cytoplasmic localization for the indicated GFP-nodulin reporter. GFP-nodulin profiles were also imaged in WT yeast at 22°C (aggregate of 275 cells) and 37°C (aggregate of 184 cells). As expected, at both temperatures, 100% of the cells showed exclusively PM localization profiles. (C) Quantification of PtdIns(4,5)P2 in WT and mss4ts strains at 37°C. Analyses involved steady-state radiolabeling of cells with [3H]inositol at 22°C and shift of cells to 37°C for 2 h; total deacylated 3H-labeled inositol glycerolipids were quantified by anion-exchange HPLC. PtdIns(4,5)P2 values are expressed as percentage of total deacylatable [3H]inositol lipid. The unpaired t test p value (mutant compared with WT) is <0.012. (D) GFP-tagged class I nodulins localize to PM when expressed in yeast with temperature sensitive PtdIns-4-OH kinase (stt4ts and pik1ts), PtdIns-3-OH kinase (vps34ts), and Sec14 (sec14ts) incubated at 22 and 37°C. Differential interference contrast (DIC) and GFP confocal images are identified, and 107–170 cells were imaged for each mutant at each temperature. For each mutant and condition, >91% of the cells imaged showed exclusive PM localization for the indicated GFP-nodulin reporter. Scale bars, 2 μm.
Bulk PtdIns(4)P levels were not depressed upon shift of mss4ts mutants to 37°C, suggesting that PtdIns(4)P did not contribute to class I nodulin association with yeast PM. Indeed, reduction of bulk PtdIns(4)P by inactivation of the Pik1 or Stt4 yeast PtdIns 4-OH kinases failed to compromise class I nodulin PM targeting (Figure 2D and Supplemental Figure S1D). PtdIns-3OH phosphoinositides did not contribute to PM targeting of the AtSfh1 nodulin domain either. Challenge of cells with 1.4 M NaCl for 5–15 min to elevate PtdIns(3,5)P2 levels in the cytosolic leaflets of vacuolar membranes (Dove et al., 1997) failed to redistribute class I nodulins from PM to vacuoles. Moreover, class I nodulins targeted to PM in vps34Δ mutants devoid of all yeast PtdIns 3OH phosphoinositides (PtdIns(3)P and PtdIns(3,5)P2; Figure 2D and Supplemental Figure S1D).
Nodulin peptide binds PtdIns(4,5)P2
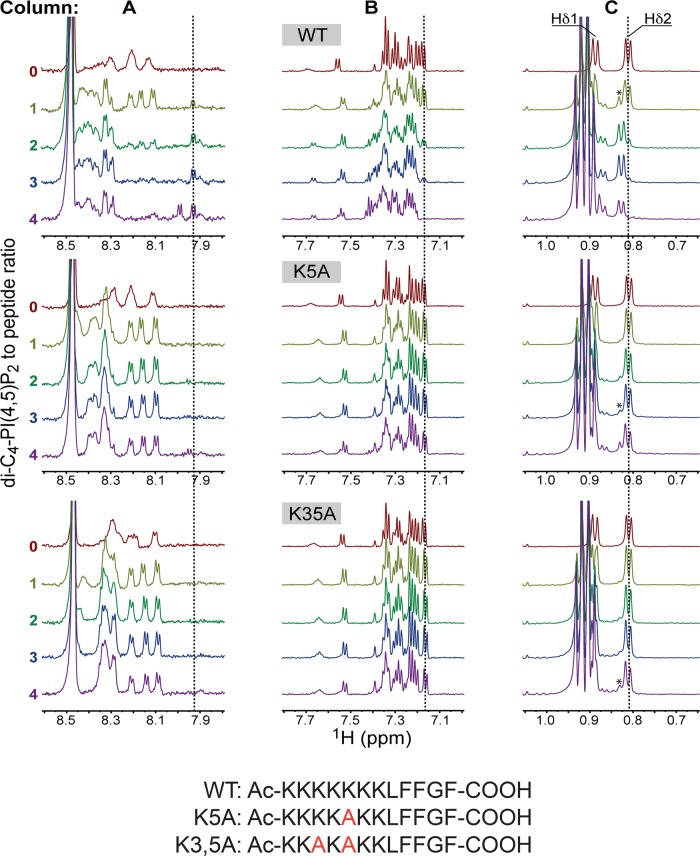
Nuclear magnetic resonance (NMR) titration experiments using short-chain PtdIns(4,5)P2 (di-C4-PtdIns(4,5)P2) as ligand confirmed that the C-terminal region of AtSfh1 nodulin interacts with PtdIns(4,5)P2. Binding assays were carried out with di-C4-PtdIns(4,5)P2 below the critical micellar concentration. Addition of di-C4-PtdIns(4,5)P2 to the wild-type AtSfh1 nodulin peptide (WT; Ac-KKKKKKKLFFGFCOOH) resulted in significant changes in the 1H NMR spectrum. In the presence of equimolar ligand, a new set of 1H peaks appeared in the amide region between 7.9 and 8.0 ppm and in the aromatic region that contains 1H peaks of all three Phe side chains (Figure 3). The two –CH3 groups of Leu-8 resonated at 0.81 (Hδ1) and 0.89 ppm (Hδ2). Although Hδ1 was obscured by the –CH3 protons of di-C4-PtdIns(4,5)P2, a shifted Leu Hδ2 peak also appeared at equimolar peptide::ligand concentrations. Increasing di-C4-PtdIns(4,5)P2 concentrations drove full conversion of ligand-free peptide to the ligand-bound species (Figure 3).
FIGURE 3:
NMR analyses of AtSfh1 nodulin peptide binding to PtdIns(4,5)P2. The 1H NMR spectra of three AtSfh1 nodulin peptide variants (peptide sequences given at bottom; Lys → Ala highlighted in red) are stacked and color coded according to di-C4-PtdIns(4,5)P2:peptide molar ratio. Three 1H spectral regions are shown: amide (A), aromatic (B), and upfield methyl (0.75–1.05 ppm; C). Significant chemical shift changes resulting from di-C4-PtdIns(4,5)P2 binding to nodulin peptide are marked (vertical lines). Peaks centered at 0.91 ppm correspond to methyl protons of di-C4-PtdIns(4,5)P2 acyl chains. The Leu Hδ peak of di-C4-PtdIns(4,5)P2-bound peptides is marked by an asterisk.
Two mutant nodulin peptides, K5A (Ac-KKKKAKKLFFGFCOOH) and K3,5A (Ac-KKAKAKKLFFGFCOOH), were also analyzed. The 1H NMR spectra of the K5A and K3,5A peptides showed few chemical shifts in the presence of equimolar di-C4-PtdIns(4,5)P2 (Figure 3). Addition of greater than twofold to threefold molar excess of the ligand was required to evoke mutant peptide chemical shift perturbations similar to those observed for WT peptide. Using Leu Hδ chemical shifts as reporters of peptide binding to di-C4-PtdIns(4,5)P2, we qualitatively ranked the relative peptide affinities for di-C4-PtdIns(4,5)P2 as WT > K5A > K3,5A.
Modeling nodulin peptide interactions with PtdIns(4,5)P2
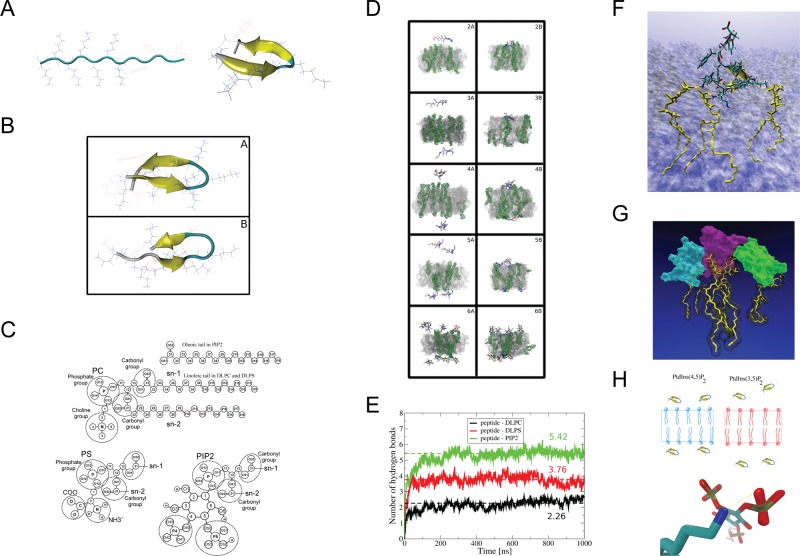
Two-stage atomistic molecular dynamics simulations (MDSs) modeled how the AtSfh1 nodulin peptide binds PtdIns(4,5)P2. In the first stage, WT nodulin peptide folding was simulated in aqueous solution (Figure 4A). The second stage simulated interactions of structured and unstructured peptides (systems 5 and 6 and systems 2–4, respectively; Figure 4B and Supplemental Table S1A) to bilayers composed of dilineoylphosphatidylcholine (DLPC), dilineoylphosphatidylserine (DLPS), and PtdIns(4,5)P2 at 297 K (Figure 4C). Membrane simulations were initiated with one, two, four, or eight AtSfh1 peptides positioned randomly in water (Figure 4D; details in Materials and Methods). Nodulin peptide bound firmly to the membrane after 300 ns via H-bond interactions with PtdIns(4,5)P2 (Figure 4E), and the residues engaged in peptide binding to PtdIns(4,5)P2 were K1–K7 (Supplemental Table S1B and Figure 4F). MDS further predicted that a single peptide bound one, two, or three PtdIns(4,5)P2 with similar probabilities (∼30%), and could engage four PtdIns(4,5)P2 simultaneously (Supplemental Table S1C and Figure 4G). Of interest, aggregations of two and three peptides were observed when binding of multiple nodulin peptides was simulated on membrane surfaces containing PtdIns(4,5)P2 (Supplemental Table S1A and Figure 4G).
FIGURE 4:
Molecular dynamics simulations of AtSfh1 nodulin peptide on membrane bilayers. (A) Unfolded extended structure of the AtSfh1 nodulin peptide (left) and folded β-hairpin structure after 4 μs of MDS in water (right). (B) Two most-populated clustered structures from REMD simulation at 297 K: 10.85 (A) and 8% (B) probability. Both structures adopt β-hairpins. (C) Chemical structures of lipids used in the simulations. (D) Snapshots of systems simulated with a lipid bilayer. Beginning (A, C, E, G, I) and end of the simulation (B, D, F, H, J) of systems 2 (A, B), 3 (C, D), 4 (E, F), 5 (G, H), and 6 (I, J) in Supplemental Table S1A. PtdIns(4,5)P2 molecules are in green (H2O not shown). (E) Time development of peptide::lipid H bonds (DLPC, black; DLPS, red; and PtdIns(4,5)P2, green). Data averaged over simulations 2–6 in Supplemental Table S1A. Dashed lines show average level of H bonds with each lipid type. (F) Snapshot of the peptide (licorice representation together with “new cartoon”) bound to three PtdIns(4,5)P2 molecules (yellow licorice) by H bonds (orange dots). Membrane shown as a transparent surface (H2O not shown). (G) Snapshot of three aggregated nodulin peptides (cyan, violet, and green surface) bound to 3 PtdIns(4,5)P2 (yellow licorice). Picture generated by VMD (Humphrey et al., 1996). (H) Schematic representation of AtSfh1 nodulin peptide bound to PtdIns(4,5)P2 (blue) and PtdIns(3,5)P2 (red). Bottom, H bonds established between nodulin peptide Lys side chain (blue) and PtdIns(4,5)P2 (red).
Additional 500-ns MDSs were run to examine how the AtSfh1 nodulin peptide might discriminate between PtdIns(4,5)P2 and PtdIns(3,5)P2. Indeed, binding of peptide to PtdIns(3,5)P2 was projected to be weaker than to PtdIns(4,5)P2. The number of H bonds established between peptide and phosphoinositide within the 200- to 500-ns simulation window differed significantly between these isomers (Supplemental Table S1D). Whereas PtdIns(4,5)P2 presented a conformation suitable for establishment of two H bonds between a coordinating Lys and an individual headgroup phosphate, PtdIns(3,5)P2 did not (Figure 4H).
C-terminal Lys mediates PtdIns(4,5)P2 binding
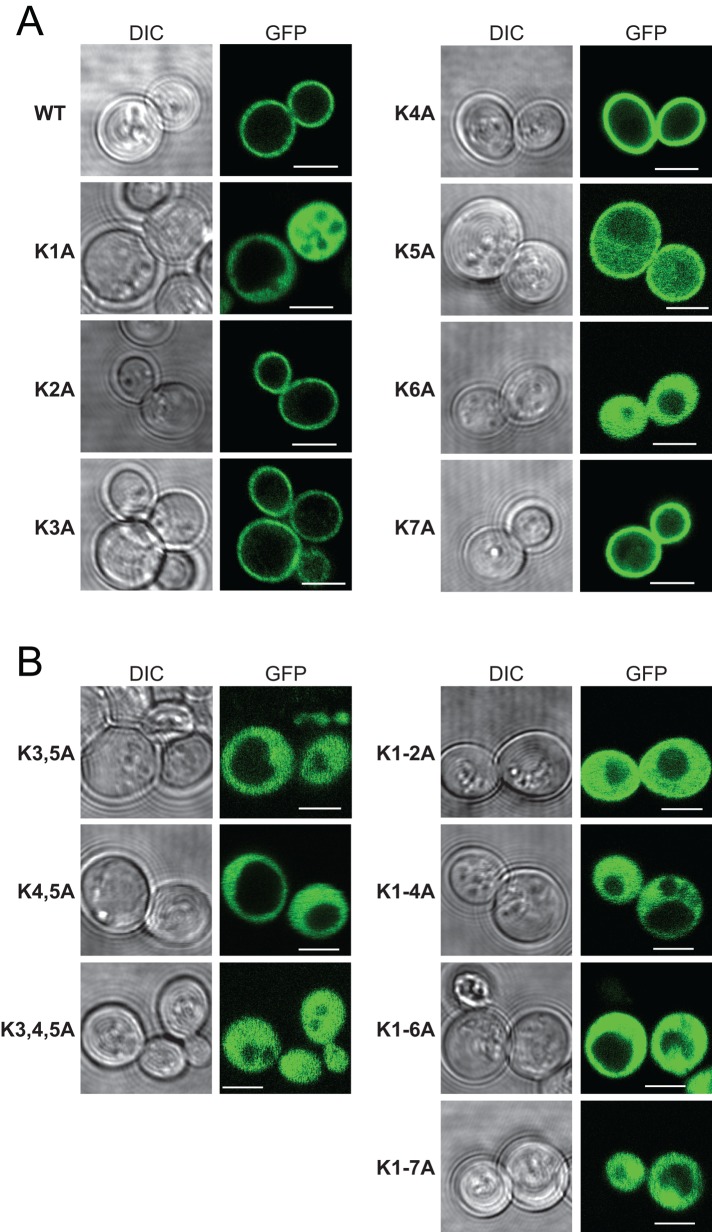
To examine the functional importance of AtSfh1 nodulin peptide::PtdIns(4,5)P2 interactions, we constructed an allelic series of K → A mutants. Yeast expression experiments demonstrated that PM localization of the AtSfh1 nodulin was dependent on integrity of the C-terminal basic motif and that individual Lys residues displayed differential contributions to PtdIns(4,5)P2 binding (Figure 5, A and B). Two single substitutions (K1A and K6A) and all multiple substitutions tested (K3,5A, K4,5A, K3,4,5A, K1-2A, K1-4A, K1-6A, and K1-7A) compromised nodulin association with PM. By contrast, the AtSfh1K2A, AtSfh1K3A, AtSfh1K4A, AtSfh1K5A, and AtSfh1K7A nodulins retained significant PM localization (Figure 5, A and B), indicating that those residues did not play critical individual roles in PtdIns(4,5)P2 binding, although collective contributions were important. The mutant nodulin localization properties, as scored using the yeast system, were recapitulated in tobacco leaf cells. Whereas AtSfh1 and mutant versions harboring single K → A substitutions targeted to the PM, none of the multiply substituted nodulins did (Supplemental Figure S2, A and B).
FIGURE 5:
Localization of GFP-tagged WT and the indicated single (A) and multiple (B) K → A substituted nodulin chimeras in WT yeast. Corresponding DIC and GFP confocal image panels. Scale bars, 2 μm. For all mutants, 100–235 cells were imaged and scored. All cells expressing the single K1A and K6A mutant reporters displayed exclusive localization of reporter to the cytoplasm, whereas all cells expressing the K5A reporter showed both PM and cytoplasmic localization for the reporter. Otherwise, >99% of the cells expressing single-mutant K → A derivatives showed exclusively PM localization for the reporter.
Bulky aromatic residues, such as the C-terminal Phe residues of the class I nodulin peptides, are a common feature of polybasic motifs, and these residues often stabilize protein::membrane interactions (McLaughlin and Murray, 2005; Gerlach et al., 2010; Li et al., 2014). Therefore the contributions of these C-terminal Phe residues to nodulin recruitment to membranes were also analyzed in yeast. Whereas single F → A substitutions did not affect PM targeting, the triple F → A substitution abolishes it (unpublished data).
C-terminal Lys is required for root hair biogenesis
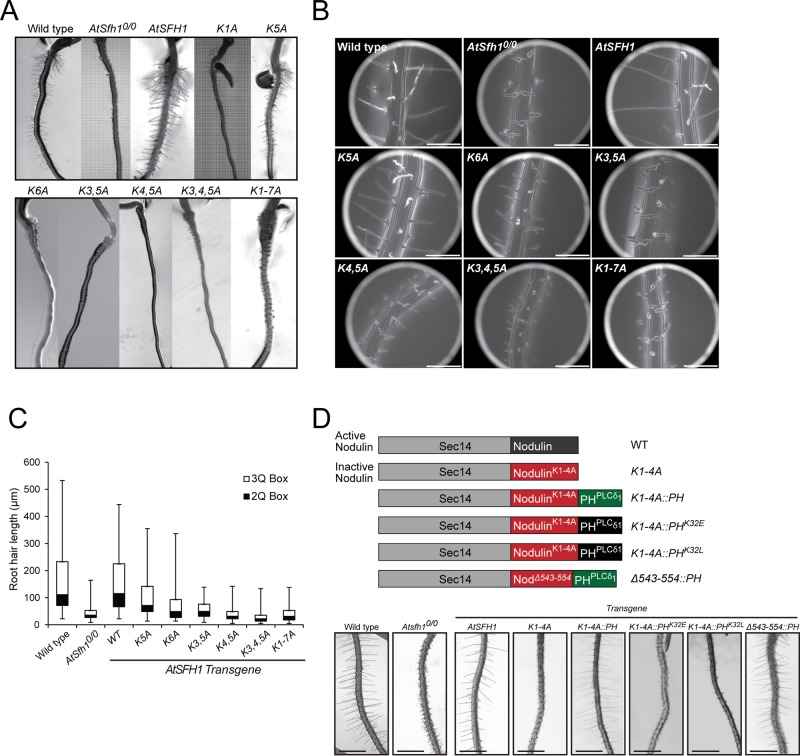
Root hair growth in Arabidopsis requires PtdIns(4,5)P2 synthesis catalyzed by the AtPIP5K3 PtdIns-4-phosphate 5-OH kinase (Kusano et al., 2008; Stenzel et al., 2008; Munnik and Nielsen, 2011). Therefore the PtdIns(4,5)P2-binding properties of the AtSfh1 nodulin domain suggested that AtSfh1 may itself be a novel PtdIns(4,5)P2 effector. To examine the biological significance of PtdIns(4,5)P2 binding by the AtSfh1 nodulin, we incorporated the K → A substitution series into an AtSFH1 transgene. The allelic series included conversion of all seven Lys residues to Ala (K1-7A) and the K1A, K5A, and K6A single, the K3,5A and K4,5A double, and K3,4,5A triple substitutions. An Atsfh10/0 Arabidopsis line was reconstituted with either WT or mutant AtSFH1 transgenes, T3 homozygous transgenic plant lines were generated, and those lines were analyzed for correction of Atsfh10/0 short–root hair phenotypes. Whereas AtSFH1+ complemented Atsfh10/0 phenotypes, variable efficiencies of rescue were recorded for Atsfh1 nodulin peptide mutants. Stable expression of Atsfh1K5A and Atsfh1K6A gene products partially rescued Atsfh10/0 root hair morphogenetic defects, as exemplified by most Atsfh1K5A and Atsfh1K6A root hairs presenting single growing tips and exhibiting intermediate lengths relative to AtSFH1+/+ and Atsfh10/0 root hairs (Figure 6, A–C, and Supplemental Table S1E). By contrast, Atsfh1K3,5A, AtsfhK4,5A, Atsfh1K3,4,5A, and Atsfh1K1-7A plants phenocopied Atsfh10/0 mutants (Figure 6, A–C, and Supplemental Table S1E).
FIGURE 6:
AtSfh1 C-terminal polybasic motif is required for biological function. (A) Bright- field images of root hairs of 3-d-old WT, Atsfh10/0, and T3 seedlings of Atsfh10/0 plants expressing AtSFH1 and indicated Lys → Ala transgenes. (B) Environmental scanning electron micrograph images of T3 seedlings of the same plant lines as in A. Single and double tip-root hairs, along with notched root hairs, are obvious. Scale bars, 200 μm. (C) Box-and-whisker representation of root hair length distributions measured for 3-d-old seedlings of transgenic plant lines represented in A and B. The 2Q and 3Q boxes represent second and third quartiles of the data set, respectively. Whiskers span the first quartile, from the second quartile box down to minimum, and the fourth quartile from the third quartile box up to the maximum. (D) Bright-field images of root hairs of 3-d-old T3 seedlings of WT, Atsfh10/0, and Atsfh10/0 plants expressing epitope-tagged AtSfh1, Atsfh1K1,4A, AtSfh1K1,4A::PHPLCδ1, AtSfh1K1,4A::PHPLCδ1K32E, AtSfh1K1,4A::PHPLCδ1K32L, and AtSfh1Δ543-554::PHPLCδ1 proteins. Proteins encoded by the respective transgenes are at the top. All constructs were expressed from the native AtSFH1 promoter.
The morphological defects of Atsfh10/0 root hairs reflect collapse of tip-directed PtdIns(4,5)P2 gradients (Vincent et al., 2005). Whereas Atsfh10/0 root hairs reconstituted for AtSfh1 expression recovered a strong tip-focused PtdIns(4,5)P2 gradient, expression of the hypomorphic AtSfh1K5A and AtSfh1K6A restored weak tip-directed PtdIns(4,5)P2 profiles that were reduced ∼50% in magnitude relative to WT (Supplemental Figure S2D). Tip-directed PtdIns(4,5)P2 gradients were completely ablated in root hairs expressing nonfunctional mutants as sole AtSfh1 species (AtSfh1K3,5A, AtSfh1K4,5A, AtSfh1K3,4,5A, AtSfh1K1-7A; Supplemental Figure S2D).
An unrelated PtdIns(4,5)P2-binding unit replaces the nodulin peptide
The severities of AtSfh1 nodulin peptide mutant phenotypes were proportional to the corresponding PtdIns(4,5)P2 binding defects, suggesting that PtdIns(4,5)P2 binding is the key activity of that motif. As a test, a PtdIns(4,5)P2-specific binding unit (the PHPLCδ1 domain) was appended to a mutant AtSfh1 with a Lys-deficient nodulin peptide (Figure 6D). The consequences of reconstituting PtdIns(4,5)P2-binding capacity in this way were then determined. Whereas the “parental” AtSfh1K1,4A was nonfunctional, AtSfh1K1,4A::PHPLCδ1 expression revived root hair development in Atsfh10/0 plants (Figure 6D). Similar results were obtained when the PHPLCδ1 domain was appended to the AtSfh1Δ543-554 C-terminus (lacks the entire polybasic motif, including the three Phe residues). Rescue was dependent on PHPLCδ1 binding to PtdIns(4,5)P2, since appending binding mutants (K32E and K32L; Yagisawa et al., 1998) failed to rescue Atsfh10/0 root hair defects (Figure 6D). Thus PtdIns(4,5)P2 binding is the crucial activity of the nodulin peptide, and binding of one PtdIns(4,5)P2 per nodulin meets the threshold for biological function.
AtSfh1 nodulin binds PtdIns(4,5)P2 with high affinity
Two lines of evidence demonstrated that class I nodulins bind PtdIns(4,5)P2 with high affinity. First, inducible expression of AtSfh1 nodulin interfered with yeast endocytosis, a PtdIns(4,5)P2-dependent process (Sun et al., 2007). This interference was manifested by delayed internalization of FM4-64 tracer from PM into endosomes when cells expressed AtSfh1 nodulin but not mutant versions unable to bind PtdIns(4,5)P2 (Supplemental Figure S3A). Second, whereas incubation of an mss4-102ts strain at 26°C released a green fluorescent protein (GFP)–tagged tandem PH domain from the PM, monomeric red fluorescent protein (mRFP)–nodulin association with PM was not compromised (Supplemental Figure S3B). These data indicate that AtSfh1 nodulin binds PtdIns(4,5)P2 more tightly than does a tandemized PHPLCδ1 and report a high affinity, as monomeric PHPLCδ1 binds PtdIns(4,5)P2 with KD ≈ 1–5 μM (Lemmon et al., 1995).
Nodulin domains homo-oligomerize
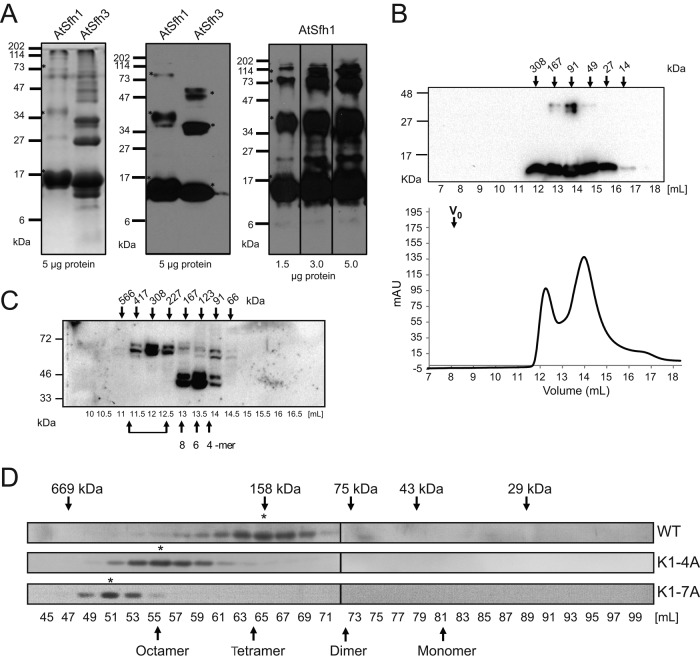
Isothermal titration calorimetry reported Ka values for WT, K5A, and K3,5A nodulin peptides for PtdIns(4,5)P2 of 5.4 × 104, 2.4 × 104, and 1.7 × 104, respectively (KD values of ∼19, 42, and 59 μM; Supplemental Figure S3, C–E). The low affinity of WT nodulin peptide for phosphoinositide could not account for the high affinity of AtSfh1 nodulin for PtdIns(4,5)P2 in vivo, however, and suggested that Nlj16-like nodulins self-assemble with accompanying enhancements in PtdIns(4,5)P2 binding avidity. All 13 Arabidopsis nodulin domains were expressed in Escherichia coli, and all but 3 (AtSfh3, AtSfh5, and AtSfh8 nodulins) distributed quantitatively into inclusion bodies. AtSfh1 nodulin was solubilized with 8 M urea and, consistent with formation of homo-oligomers, resolved as a ladder in SDS–PAGE. The ladder ranged from 14 to ∼112 kDa (octamer?) in increments of the monomeric mass of 14 kDa (Figure 7A). Soluble fractions of class I AtSfh3 nodulin (13 kDa) behaved similarly in SDS–PAGE (Figure 7A). AtSfh3 nodulin and the soluble class II AtSfh5 and AtSfh8 nodulins filtered at apparent molecular masses (Mr) of ∼91 kDa, suggesting assembly into hexamers (Figure 7B and Supplemental Figure S3, F and G).
FIGURE 7:
Homo-oligomerization of class I nodulins. (A) Purified recombinant histidine (His)-tagged AtSfh1 (solubilized in 8 M urea) and soluble AtSfh3 nodulins (5 μg) were resolved by SDS–PAGE and visualized by Coomassie blue staining (left) and immunoblotting with anti-His tag antibody (middle). Right, immunoblotting of purified AtSfh1 nodulin with polyclonal AtSfh1 nodulin antibody (load at bottom). SDS-resistant homo-oligomeric forms are identified in all panels (asterisks). (B) Gel filtration chromatogram for purified native AtSfh3 nodulin. Top, immunoblot profile across the elution, shown above the chromatogram; elution properties of the indicated standards are identified by arrows. (C) Immunoblots of gel filtration column fractions of total lysates from yeast expressing myc-tagged AtSfh1 nodulin. Bottom, elution volume (milliliters); top, corresponding apparent molecular masses. Presumptive oligomeric states are identified at bottom. Brackets denote higher-order complexes. (D) Immunoblot profiles of gel filtration column fractions of lysates from tobacco leaf tissues expressing mRFP-tagged AtSfh1 WT, AtSfh1K1-4A, and AtSfh1K1-7A nodulins using anti-mRFP antibodies to visualize nodulin chimeras. Peak fractions of homo-oligomeric forms are identified (asterisks). Monomeric and presumed homo-oligomeric forms of the mRFP-tagged proteins are identified (arrows).
The biochemical properties of the AtSfh1 nodulin were examined under native conditions by expressing the isolated domain in yeast and tobacco leaf cells. Size exclusion chromatography of cytosol prepared from yeast expressing myc-tagged AtSfh1 nodulin reported that this domain assembled into presumptive hexamers, octamers, and even higher-order complexes (Figure 7C). When expressed in tobacco leaf cells, mRFP-AtSfh1 nodulin filtered at an Mr consistent with assembly into homo-tetramers (Figure 7D). Of interest, native K1-4A and K1-7A nodulins assembled into homo-octamers and even higher-order complexes, respectively (Figure 7D). Thus charge neutralization of the nodulin peptide potentiates homo-oligomerization of the nodulin, an attractive feature, as such charge neutralization accompanies PtdIns(4,5)P2 binding.
Oligomerization-defective nodulins are impaired in membrane targeting
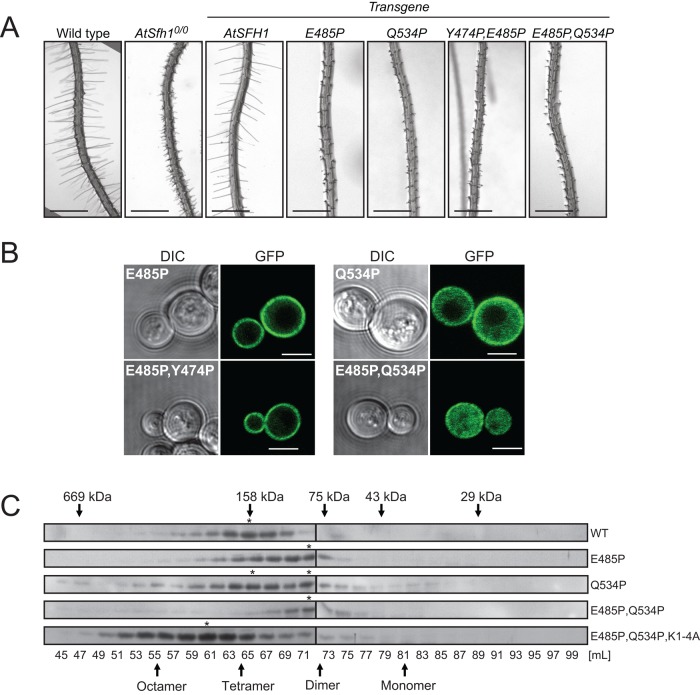
Residues E485 and Q534 are conserved in all Nlj16-like nodulins, whereas Y474 is conserved only in class I versions. All three residues lie outside the motif responsible for PtdIns(4,5)P2 binding, and each was converted to Pro in attempts to generate new classes of mutant nodulins. Whereas AtSfh1E485P, AtSfh1Q534P, AtSfh1E485P,Q534P, and AtSfh1E485P,Y474P nodulins were all stably expressed in yeast and plants, each failed to complement Atsfh10/0 root hair defects (Figure 8A). Although functionally compromised, the AtSfh1E485P and AtSfh1E485P,Y474P nodulins targeted to PM in a PtdIns(4,5)P2-dependent manner (Figure 8B and Supplemental Figure S4A). Thus the biological defects of AtSfh1E485P,Y474P nodulin revealed a critical activity independent of PtdIns(4,5)P2 binding. PtdIns(4)P or 3-OH phosphoinositide deficiency was without effect (Supplemental Figure S4B).
FIGURE 8:
Polybasic motif–independent functions of AtSfh1 nodulin. (A) Bright-field images of root hairs from 3-day-old WT, Atsfh10/0, and T3 seedlings of transgenic Atsfh10/0 plants expressing myc-AtSFH1-HA and myc-Atsfh1-HA with indicated Pro substitutions. Scale bars, 1 mm. (B) Confocal images of WT yeast expressing GFP-tagged AtSfh1E485P, AtSfh1Q534P, AtSfh1E485P,Q534P, and AtSfh1E485P,Y474P nodulins. These images are representative of 105, 187, 114, and 142 cells imaged, respectively. Ninety-eight percent and 92% of the cells expressing GFP-tagged AtSfh1E485P or AtSfh1E485P,Y474P nodulins showed exclusively PM localization of the reporter, respectively. All cells expressing GFP-tagged AtSfh1E485P,Q534P nodulin showed a cytoplasmic localization profile exclusively. For cells expressing GFP-tagged AtSfh1Q534P nodulin, 5% of the cells showed PM localization, 60% showed both PM and cytoplasmic staining, and 35% showed only a cytoplasmic localization of the reporter. Scale bars, 2 μm. (C) Immunoblots of gel filtration column fractions of lysates from tobacco leaf cells expressing mRFP-AtSfh1, AtSfh1E485P, AtSfh1Q534P, AtSfh1E485P,Q534P, and AtSfh1E485P,Q534P, K1-4A nodulins. Peak fractions of presumptive homo-oligomeric forms are identified (asterisks). Apparent molecular masses and fraction volumes in C are indicated.
By contrast, PM association of the AtSfh1Q534P and AtSfh1E485P,Q534P nodulins was strongly compromised (Figure 8B), suggesting that these mutant domains had diminished affinities for PtdIns(4,5)P2 and identifying these as candidates for oligomerization-defective mutants. Indeed, gel filtration of leaf extracts confirmed that E485P,Q534P missense substitutions collapsed AtSfh1 nodulin tetramers into dimers and interfered with AtSfh1K1-4A nodulin assembly into its typical octameric form (Figure 8C). These substitutions did not compromise assembly into dimers, indicating that AtSfh1E485P,Q534P nodulin was selectively deficient in its polymerization into tetramers and higher-order homo-oligomers.
Nodulin interaction with PtdIns(4,5)P2 is resistant to Ca2+ influx
A hallmark feature of root hairs engaged in active tip growth is the coupling of phosphoinositide and Ca2+ signaling (Braun et al., 1999; Ryan et al., 2001; Vincent et al., 2005; Cole and Fowler, 2006; Bibikova and Gilroy, 2008; Thole and Nielsen, 2008). Because elongating root hair tips are preferential sites of Ca2+ influx and direct interaction of Ca2+ with PtdIns(4,5)P2 is known to compete with protein binding to this phosphoinositide (Levental et al., 2009; Slochower et al., 2013; Shi et al., 2013; Li et al., 2014), we again exploited a well-characterized yeast model to test whether large local Ca++ influxes modulated nodulin::PtdIns(4,5)P2 interactions. Specifically, we took advantage of previous demonstrations that, relative to wild-type yeast, vps33 mutants experience rapid, large, and sustained elevations in cytosolic Ca2+ when challenged with high extracellular concentrations of this cation (Miseta et al., 1999).
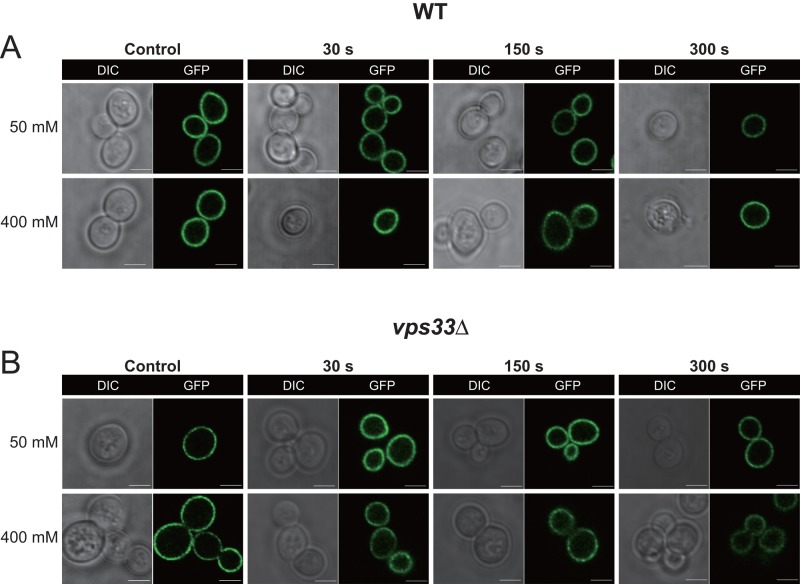
Wild-type yeast are able to reestablish cytoplasmic Ca2+ homeostasis rapidly when subjected to Ca2+ shock. This is exemplified by the fact that challenge of cells with 50 mM extracellular Ca2+ raises cytoplasmic Ca2+ from a resting concentration of ∼75 to 300 nM within seconds, and this elevation is resolved within 90 s to near resting levels (Miseta et al., 1999). As expected, the AtSfh1 GFP-nodulin reporter remained associated with yeast PM under these conditions and even when cells were subjected to a 400 mM Ca2+ shock (Figure 9A). The isogenic vps33Δ partner strain is defective in maintaining Ca2+ homeostasis. Challenge of the mutant with 50 mM extracellular Ca2+ raises cytosolic Ca2+ from a resting concentration of 165 nM to ∼1.75 μM, and these levels remain elevated (∼470 nM) for prolonged periods (Miseta et al., 1999). Even throughout these conditions of elevated cytoplasmic Ca2+, however, localization of the GFP-nodulin reporter to the PM was not disturbed in the vps33Δ strain (Figure 9B). Moreover, subjecting vps33Δ cells to an intense 400 mM Ca2+ shock also failed to chase the GFP-nodulin reporter from the PM (Figure 9B). These data suggest that elevating cytosolic Ca2+ to micromolar concentrations is by itself insufficient to break AtSfh1-nodulin interactions with PtdIns(4,5)P2.
FIGURE 9:
AtSfh1 nodulin domain interactions with phosphoinositide under conditions of Ca2+ influx. Confocal images of WT yeast cells expressing GFP-AtSfh1 nodulin in wild-type (A) and vps33Δ (B) strains, respectively, challenged with 50 or 400 mM CaCl2 as indicated. For both A and B, the control images were taken immediately before Ca2+ challenge. Images were collected every 30 s during a 30- to 300-s post–Ca2+-challenge window of analysis. The time point at which each image was taken is indicated. The vps33Δ mutant strain accumulates high levels of cytosolic Ca2+ under these conditions and sustains these elevated levels throughout the time period the cells were imaged (Miseta et al., 1999). The GFP-AtSfh1 nodulin remained bound to PM in all cells observed for both yeast strains, under both Ca2+ challenge conditions, and at all times imaged. The data are representative of three independent experiments, and 160–326 individual cells were analyzed for each strain, under each condition, for each time point. Scale bar, 2 μm.
DISCUSSION
Sec14-nodulin proteins are novel cell polarity regulators whose activities are critical for morphogenesis of highly polarized plant structures such as root hairs. The conserved genetic fusion of Sec14- and Nlj16-like nodulin modules is a biologically striking arrangement that sits at the interface of root hair development, lipid signaling, and symbiotic N2 fixation. Using the nonleguminous plant Arabidopsis thaliana as experimental model, we now demonstrate the nodulin domain is required for coherent organization of polarized root hair growth signaling and identify the AtSfh1 and related class I nodulins as functionally versatile PtdIns(4,5)P2-binding modules. These units can either support or subvert polarized cell growth programs, as dictated by developmental or environmental demands. Collectively the data identify Sec14-nodulin proteins as novel scaffolds for templating the patterning of PtdIns(4,5)P2 signaling in plants. This organization of PtdIns(4,5)P2 landmarks is an essential aspect of the polarity-signaling program in root hairs, as the system fails without it, even in the face of uncompromised PtdIns(4,5)P2 biosynthetic capability. Taken together, these data reveal new principles for how lipid signaling is functionally diversified, with exquisite spatial and temporal precision, on membrane surfaces.
The three Nlj16-like nodulin classes are distinguished by their C-terminal peptide sequences, and this motif endows class I nodulins their signature properties as PtdIns(4,5)P2-binding units. Membrane targeting of class I nodulins to a model eukaryotic (yeast) PM was dependent on PtdIns(4,5)P2 and no other yeast phosphoinositide, including the positional isomer PtdIns(3,5)P2. This selectivity was further highlighted by the fact that yeast PM is rich in acidic phospholipids such as PtdIns and PtdSer. Given that PtdIns and PtdSer mass in yeast exceeds that of PtdIns(4,5)P2 by ∼100- and 50-fold, respectively (Zinser et al., 1991), these anionic lipids (as well as the monophosphorylated phosphoinositides) would efficiently compete for class I nodulin binding if the module were to harbor even weak affinities for them. Class I nodulin specificity for PtdIns(4,5)P2 comes with high affinity. Thus AtSfh1 nodulin is an avid electrostatic sink for PtdIns(4,5)P2, whose avidity and capacity are governed by homo-oligomerization into higher-order structures (e.g., hexamers/octamers and higher). Although an electrostatic sink of the sort presented by the AtSfh1 nodulin is expected to select for the most anionic lipid molecules available—for example, PtdIns(4,5)P2 and PtdIns(3,5)P2—the positional specificity of this sink is a remarkable property.
The Lys-rich C-terminal motif of class I nodulins prosecutes PtdIns(4,5)P2 binding by these modules, and PtdIns(4,5)P2 binding is essential for the biological function of AtSfh1 as polarity regulator in developing root hairs. Evidence to this effect includes 1) the direct correspondence between strength of lipid-binding defect of a mutant nodulin with severity of loss-of-function phenotype when the defect is incorporated into a full-length AtSfh1, and 2) the ability of a PtdIns(4,5)P2-specific binding unit (PHPLCδ1) to act as functional surrogate for the nodulin peptide in the context of a full-length AtSfh1. Physical appendage of a Sec14 domain that stimulates PtdIns(4)P production to a unit that both laterally sequesters PtdIns(4,5)P2 and assembles into higher-order structures recommends AtSfh1-like proteins as novel scaffolds for coupling phosphoinositide production and organization. That is, Sec14-nodulins are well designed to arrange PtdIns(4,5)P2 pools so that the nodulin-bound phosphoinositide pool is appropriately prepatterned to support spatially coherent signaling upon register of physiological trigger. In this model, PtdIns(4,5)P2-independent mechanisms of AtSfh1 organization template the spatial patterning of PtdIns(4,5)P2 pools essential for root hair morphogenesis. Supporting evidence comes from demonstrations that the intricate tip-directed distributions of AtSfh1 and PtdIns(4,5)P2 in Arabidopsis root hairs show similar patterns (Vincent et al., 2005) and that both the Sec14 and nodulin domains contribute to a functional AtSfh1.
It remains to be determined how AtSfh1-bound PtdIns(4,5)P2 pools are made available to other effectors. The Ca2+-calmodulins and related calmodulin-like activities (of which plants express many) remain attractive candidates for PtdIns(4,5)P2 “displacement factors,” as these proteins have high affinities for basic peptides (O'Neil and Degrado, 1990; McLaughlin and Murray, 2005). In this scenario, calmodulins release PtdIns(4,5)P2 molecules from the AtSfh1 nodulin peptide upon register of a Ca2+ trigger. Also plausible are Ca2+-regulated posttranslational modifications of the nodulin domains. Such designs hold the appealing feature of spatially and temporally coupling Ca2+- and PtdIns(4,5)P2-regulated signaling reactions, a functional coordination on stark display in tip-growing cells. Our experiments in yeast do suggest, however, that direct Ca2+::PtdIns(4,5)P2 interactions of the sort that would occur at sites of robust Ca2+ influx into cells (e.g., growing root hair tips) are insufficient to break the nodulin::phosphoinositide interaction.
The Lotus LjPLP-IV gene, which encodes the AtSfh1 orthologue in this leguminous plant, is subjected to a peculiar transcriptional control involving a bidirectional promoter that is activated in mature nitrogen-fixing nodules. This bidirectional promoter resides in intron 10 of the LjPLP-IV gene and drives expression of both the stand-alone Nlj16 nodulin and antisense transcripts directed against Sec14-domain sequences (Kapranov et al., 2001). The biochemical properties of class I nodulins now suggest a molecular rationale for why this major reprogramming of Sec14-nodulin gene transcription occurs. Late stages of nodulation demand that polarized morphogenetic programs, such as root hair formation, root hair curling, or growth of the infection thread, be subverted to establish the anaerobic microenvironment suitable for nitrogenase activity (Gage, 2004; Oldroyd, 2013). We posit that the nodulation program terminates polarized morphogenetic programs by silencing expression of a master polarity regulator (Sec14-nodulin). This effect is reinforced by production of a potent antagonist of polarized membrane growth (stand-alone nodulin). The basis for the antagonism lies with the class I nodulin's ability to laterally sequester PtdIns(4,5)P2 in the absence of auxiliary activities supplied by full-length protein. By physically uncoupling the Sec14 and nodulin domains and shifting relative stoichiometries in favor of the PtdIns(4,5)P2 clamp (Nlj16 is highly expressed, whereas expression of full-length protein is silenced; Kapranov et al., 1997, 2001), the phosphoinositide signaling circuit required for polarized membrane trafficking is quenched. We interpret the legume/bacterial symbiosis as a biological context in which the nodulin's developmental versatility as PtdIns(4,5)P2 clamp is on display.
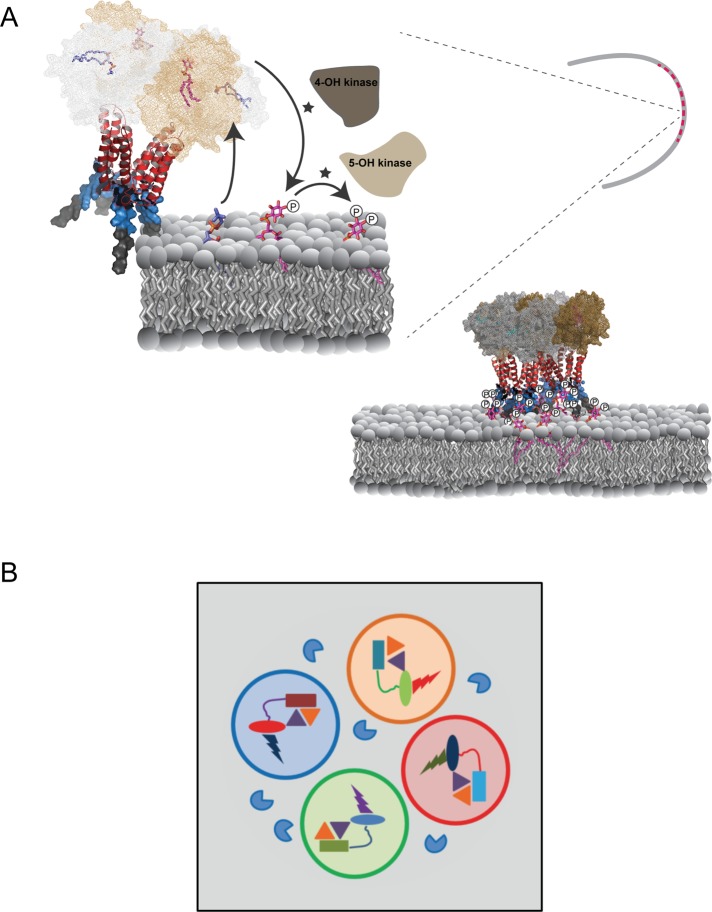
Finally, the biochemical properties of class I Sec14-nodulins suggest an interesting mechanism for scaffolding assembly of PtdIns kinases and/or appropriate effectors to support exquisite spatial and temporal control of functionally privileged phosphoinositide signaling foci (Figure 10A). These foci could operate on scales as small as single macromolecular complexes, thereby coupling synthesis of a specific phosphoinositide with its channeling to distinct cohorts of privileged effectors (Figure 10B). Higher-order arrangement of such highly integrated signaling “bits” by homo-oligomeric interactions would have the capacity to organize phosphoinositide signaling into macroscale patterns. Such a design enables differential functional channeling of even chemically identical phosphoinositide molecules produced in immediate proximities on the membrane surface. When superimposed upon the phosphoinositide chemical code, this design imprints what is effectively “point” resolution to the lipid-signaling landscape. Point resolution not only provides a facile mechanism for diversifying biological outcomes for phosphoinositide signaling, but it also codifies a versatile principle for the intricate bit mapping of a large membrane surface for production of a high-definition lipid-signaling screen.
FIGURE 10:
Patterning of phosphoinositide signaling by Sec14-nodulins. (A) AtSfh1-dependent patterning of PtdIns(4,5)P2 in root hair tips. On the basis of the mechanism for how yeast Sec14 stimulates PtdIns 4-OH kinase activity (Schaaf et al., 2008) and our data with the AtSfh1 Sec14 domain (unpublished data), we propose that Sec14 domains (mesh) of an AtSfh1 tetramer promote phosphoinositide synthesis by presentation of PtdIns (magenta) to PtdIns 4-OH kinases during heterotypic lipid exchange with amino phospholipid (blue). Specific interaction of C-terminal lysines (blue surface mode) with PtdIns(4,5)P2, in conjunction with the weak association of the AtSfh1 C-terminal aromatic motif (LFFGF, gray surface mode), stabilizes interaction of the nodulin domain with membranes. Charge neutralization of Lys residues by PtdIns(4,5)P2 promotes assembly of AtSfh1 into higher-order oligomers by interaction of the helical coiled-coil motifs (red ribbon diagram). (B) Engineering phosphoinositide signaling with point resolution. Distinct classes of Sec14-nodulins (Sec14 domains, rectangles; nodulin domains, ovals) scaffold PtdIns and PtdIns-phosphate kinase assemblies (gold and purple triangles, respectively) with distinct phosphoinositide effectors (bolts). Classes of individual complexes that prosecute distinct biological outcomes for phosphoinositide signaling are organized into signaling pixels denoted by open circles of different color. Phosphoinositide phosphatases (blue PacMan) hydrolyze phosphoinositides that escape pixel boundaries.
MATERIALS AND METHODS
Yeast strains and methods
Standard reagents/kits were purchased from Sigma-Aldrich (St. Louis, MO), Fisher Scientific (Pittsburgh, PA), Invitrogen (Grand Island, NY), or Promega (Madison, WI). Lipids were purchased from Echelon Biosciences (Salt Lake City, UT) and Sigma-Aldrich. Peptides were purchased from Eton Bioscienes (San Diego, CA). Strains used are as follows: wild-type yeast CTY182 (MATα ura3-52 lys2-801 his3Δ-200), CTY1568 (MATα leu2 ura3 his3 trp lys suc2-Δg stt4Δ::HIS, YCp(URA3, stt4ts)), CTY1537 (MATa ura3-52 leu2 Gal+ pik1ts), and CTY1-1A (MATα ura3-52 lys2-801 his3Δ-200 sec14-1ts). Lipid kinase mutant strains included RG1 (pRS315-mss4-5ts; mss4Δ::kanR,ura3-52, his3-Δ200, ade−), RG2 (pRS315-mss4-9ts; mss4Δ::kanR, ura3-52, his3-Δ200, ade−), RG3 (pRS315-mss4-23ts; mss4Δ::kanR, ura3-52, his3-Δ200, ade−), RG4 (pRS315-mss4-25ts; mss4Δ::kanR, ura3-52, his3-Δ200, ade−), and PtdIns-3-OH kinase mutant strain RG5 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vps34Δ::kanMX). The Vps33-deficient strain LBY317 (MATα ura3-52 leu2-3 112 his3Δ-200 trp1-Δ901 lys2-801 suc2-Δ9 vps33Δ::HIS3) and its isogenic wild-type partner SEY6210 (MATα ura3-52 leu2-3 112 his3Δ-200 trp1-Δ901 lys2-801 suc2-Δ9) were previously described (Miseta et al., 1999). Yeast media, genetic techniques, [3H]inositol labeling, and phosphoinositide determinations by anion exchange chromatography of deacylated inositol lipids were described (Rivas et al., 1999; Guo et al., 1999; Phillips et al., 1999; Li et al., 2000). Jeremy Thorner (University of California, Berkeley, CA) provided pik1ts and stt4ts strains, Scott Emr (Cornell University, Ithaca, NY) the mss4-102ts strain (Stefan et al., 2002), and Tsuyoshi Nakagawa (Shimane University, Nishikawatsu, Japan) the Gateway binary vectors that contain the bar gene (Nakamura et al., 2010).
Isolation of mss4ts alleles
The mss4ts strains were generated by John Moskow in the laboratory of Daniel Lew (Duke University, Durham, NC) by gap repair mutagenesis as described (Moskow et al., 2000). Briefly, the entire MSS4 open reading frame (ORF) and flanking sequences (1180 base pairs upstream and 400 base pairs downstream) were PCR amplified under conditions of relaxed stringency. The PCR products were cotransformed directly into yeast along with an MSS4-containing pRS315 plasmid digested with NheI (to remove the 3.47-kb restriction fragment containing the MSS4 ORF plus 930 and 200 base pairs of upstream and downstream flanking sequences, respectively). The yeast host for the transformations was an mss4Δ::kanR-null strain in which the normally lethal mss4Δ::kanR was complemented by an ectopic PGAL1-MSS4 cassette. KanR transformants were selected on glucose medium (PGAL1 inactive) at 24°C. The strains were replica plated at 37°C to screen for mss4ts alleles (identified by lack of growth at this temperature in glucose, but not galactose, medium). Ts isolates were cured of the PGAL1-MSS4 cassette and characterized.
Plant materials and cDNA isolation
A. thaliana Columbia-0 (Col-0) was used as the wild-type strain (WT). The Atsfh10/0 mutant refers to the Col-0 AtSFH1::T-DNA insertion line described (Vincent et al., 2005). Total mRNA (∼100 μg) was prepared from 100 mg of leaves, flowers, or roots using the RNeasy Plant Mini Kit (Qiagen). The 300– to 400–base pair AtSfh mRNAs encoding nodulin domains were amplified by reverse transcriptase-PCR (Superscript FirstStrand Synthesis System; Invitrogen), and cDNAs were cloned into the pGEM vector using the pGEMTeasy cloning kit (Promega) and subcloned into the doxycycline-controlled yeast expression vector pCM189. A sequence encoding a translational mRFP-AtSFH1 nodulin fusion was amplified from pGWB655-AtSFH1 nodulin and subcloned into the yeast expression vector pDR199 (Schaaf et al., 2011).
Templated homology modeling of nodulin domains
Homology models were generated using Schrödinger's Prime Homology Modeling module (Prime, version 3.1; Schrödinger, New York, NY). DNA topoisomerase (Protein Data Bank ID 1A36) was used as a structural template to build the model. The templating exploited sequence similarity between the topoisomerase linker region and the AtSfh1 nodulin domain. The topoisomerase linker region was pruned and used as a template after sequence realignment. A position-specific substitutional matrix for the query sequence, derived from PSI-BLAST, was used to match the template sequence.
To minimize the inaccuracy in a secondary structure prediction, a composite secondary structure was predicted for query sequence by iterative optimization and was aligned to the secondary structure alignment of the template. The models were build using “full-build” option, which builds insertions, closes gaps, and predicts side-chain conformations of nonconserved residues to produce models with no unphysical clashes. Extensive conformational sampling further refined the structures by particularly refining the regions where insertions were made and gaps closed and repredicting side-chain conformations.
Several (intermediate) main-chain models were generated as a result of permutational selection of different loop candidates and side-chain rotamers and were refined using AMBER99 and generalized Born/volume integral methodology (MOE-2011.10; Chemical Computing Group, Montreal, Canada). The model, which scored best according to the selected force-field (AMBER99), was chosen as the final model. Homology models of Atsfh1 mutants were similarly built using the same computational protocol.
NMR of AtSfh1 nodulin peptides
Three C-terminal AtSfh1 nodulin peptides were investigated: the WT peptide, Ac-KKKKKKKLFFGF-COOH; the K5A variant, Ac-KKKKAKKLFFGF-COOH; and the K3,5A variant, Ac-KKAKAKKLFFGF-COOH. Peptides of >95% purity were purchased from Eton Bioscience. Stock solutions of peptides were prepared in HPLC-grade water (Fisher Scientific) and adjusted to pH 6.5 with ammonium hydroxide (EM Science). Stock solution concentrations were determined by measuring absorption at 205 nm. di-C4-PtdIns(4,5)P2 was purchased from Echelon Bioscience. The stock solutions of phosphoinositides were prepared in the NMR buffer containing 5 mM [U-2H4, 98%]imidazole at pH 6.5 and 8% D2O. For NMR measurements, the peptide stocks were diluted with NMR buffer to a final concentration of 0.15 or 0.25 mM. Binding experiments were conducted by adding aliquots of di-C4-PtdIns(4,5)P2 stock solutions to the peptide samples.
NMR experiments were carried out at 298.15 K on Varian Inova NMR instruments operating at 1H Larmor frequencies of 500 and 600 MHz. One-dimensional 1H NMR spectra of all PtdIns(4,5)P2 titration points were collected using the “water flip-back” technique (Grzesiek and Bax, 1993) to suppress water signal and processed with MestReNova. Spectra were referenced externally with the chemical shift standard 4,4-dimethyl-4-silapentane-1-sulfonic acid.
Atomistic simulations of the AtSfh1 nodulin peptide
Atomistic 4000-ns MDS was performed for AtSfh1 nodulin peptide (N-terminal acetyl cap-KKKKKKKLFFGF) solvated in bulk water. In addition, five MDSs were run (>1000 ns each) of peptide interacting with a lipid bilayer comprising DLPC, DLPS, and PtdIns(4,5)P2 lipids. In membrane simulations, one, two, four, or eight AtSfh1 peptides were positioned randomly in water, and the simulations were performed at physiological salt concentration of 140 mM NaCl. In all cases, counterions were included to neutralize the systems.
The Optimized Potential for Liquid Simulations (OPLS) all-atom force field (Jorgensen et al., 1988) was used to parameterize all molecules. For lipids, we used an extension of the OPLS all-atom force field (Maciejewski et al., 2014). For water, we used the TIP3P model, which is compatible with the OPLS parameterization (Jorgensen et al., 1983). The system setup used in this study is identical to that used in our previous simulations of lipid bilayers with OPLS all-atom parameterization (Kaiser et al., 2011; Orłowski et al., 2011). Periodic boundary conditions with the usual minimum image convention were used in all three directions. The LINCS algorithm (Hess et al., 1997) was used to preserve the length of each hydrogen atom covalent bond. The time step was set to 2 fs, and the simulations were carried out at constant pressure (1 bar) and temperature (298 K). The temperature and pressure were controlled by the Parrinello–Rahman and v-rescale methods, respectively (Parrinello et al., 1981; Bussi et al., 2007). The temperatures of the solute and solvent were controlled independently. For pressure, we used a semi-isotropic control for systems with a lipid bilayer and an isotropic one for the peptide in water. The Lennard-Jones interactions were cut off at 1.0 nm. For the electrostatic interactions, we used the particle mesh Ewald method (Essmann et al.,1995) with a real-space cutoff of 1.0 nm, β-spline interpolation (order of 6), and direct sum tolerance of 10−6.
The simulations were performed using the GROMACS 4.5.5 software package (Hess et al., 2008), resulting in ∼10 μs of trajectories. Nodulin peptide folding was examined in long (4 μs) MDS, as well as in 200-ns atomistic replica exchange molecular dynamics (REMD) simulations (system 1; Supplemental Table S1A). During the 4-μs MDS, the peptide folded from extended to common β-hairpin structures in ∼1.3 μs and remained stable until the end of the simulation, with only two brief (∼300 ns) episodes of partial unfolding. In all MDS runs (systems 2–6; Figure 4D and Supplemental Table S1A), the nodulin peptide secondary structures were monitored throughout. These simulations consumed 400,000 core-hours of computing time.
REMD Simulations of the C-terminal AtSfh1 nodulin peptide
Atomistic REMD simulations for a linear extended structure of AtSfh1 used GROMACS 4.5.5 (Hess et al., 2008). REMD is the technique of choice to sample the peptide conformational space. The sequence (see earlier description) was constructed using the VMD (Humphrey et al., 1996) program and solvated with 2423 water molecules and 6 Cl− ions using a simulation box of size 4.29 × 3.62 × 4.87 nm. These dimensions are large enough to prevent interactions between the peptide and its periodic images. The energy of the system was minimized with the steepest descent and conjugate gradients algorithms, and 1-ns simulation in 297 K was performed for equilibration. This system was then used as a starting configuration for the REMD simulations. Simulation parameters were the same as in the foregoing MDSs, except for temperature, for which we used 48 replicas at 275.00, 276.81, 278.64, 280.47, 282.31, 284.16, 286.02, 287.89, 289.77, 291.66, 293.56, 295.46, 297.38, 299.31, 301.24, 303.19, 305.15, 307.11, 309.09, 311.07, 313.07, 315.08, 317.09, 319.12, 321.16, 323.20, 325.26, 327.33, 329.41, 331.50, 333.60, 335.71, 337.83, 339.96, 342.10, 344.26, 346.42, 348.60, 350.82, 353.01, 355.23, 357.45, 359.68, 361.92, 364.18, 366.45, 368.72, and 371.02 K. These temperatures were generated using a temperature predictor for parallel tempering simulations (Patriksson et al., 2008) with a desired exchange probability set to 0.2. Simulations were 200 ns each, and collectively summed to 9.6 μs of simulation time. Continuous trajectory at 297 K was used to perform cluster analyses with the pairwise method of Daura et al. (1999). Cluster analyses of the REMD trajectory at 297 K showed that the two most-populated structures (10.85 and 8%) agreed with 4-μs MDS at the same temperature (Figure 4B), suggesting that the nodulin peptide adopted a β-hairpin conformation in which two β-strands (residues 1–5 and 7–9) were separated by a turn at residue 6 (Figure 4B). The REMD simulations consumed 70,000 core-hours of computing time.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) was performed using a VP-ITC high-sensitivity titration calorimeter (MicroCal, Northampton, MA) with nodulin peptide and lipid vesicle concentrations clamped at 9 and 2 mM, respectively. The calorimetric cell was filled with nodulin peptide solution at 9 μM. Lipid vesicles at 2 mM concentration (as phosphate) were injected into the cell (1.43 ml) in 10-μl steps, leading to a 100- to 200-fold dilution of lipid vesicles. To minimize the contribution of dilution to the heat of partitioning, all samples were prepared in the same buffer and degassed under vacuum immediately before use. Typically, the injections were made at 10-min intervals and 2 s/μl. Constant stirring speed of 290 rpm was maintained during the experiment to ensure proper mixing after each injection. Titration experiments were performed at 25°C in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer. Dilution heats of vesicles in buffer were determined separately and subtracted from experimental heats of binding. At each lipid injection, free peptide partitioned into the bilayer membrane, and the corresponding heat of reaction was measured. The heat of reaction became smaller as less peptide remained free in solution. The integration of each calorimetric peak yielded a heat of reaction. These heats were plotted versus lipid concentration.
The reaction heat for each injection is related to the calorimetric enthalpy of binding, ΔH. The binding isotherms, ΔH versus molar ratio, were analyzed using MicroCal Origin. The fit of the binding curve yields the binding constant Ka (Kd = 1/Ka) and the enthalpy ΔH of the binding reaction. The Gibbs free energy of binding (ΔG) and the entropy (ΔS) are determined from the basic thermodynamic expression ΔG = −RT ln Ka = ΔH − TΔS, where R and T are the gas constant and the absolute temperature, respectively.
Protein purification and size exclusion chromatography
Recombinant proteins were expressed in E. coli BL21 and expression induced with 100 μM IPTG (see the Supplemental Methods). AtSfh1 nodulin was purified from 25 mM sodium phosphate, 300 mM NaCl (pH 7.5), and 8 M urea, and AtSfh3 nodulin was purified from 25 mM potassium phosphate and 300 mM KCl (pH 7.5). Histidine-tagged proteins were purified by TALON affinity chromatography. The AtSfh3, AtSfh5, and AtSfh8 nodulin domains were further purified by S200 gel filtration chromatography. mRFP-tagged nodulin proteins expressed in Nicotiana benthamiana leaves were isolated with 50 mM Na2HPO4, 300 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and 5 mM β-mercaptoethanol (pH 7.5) and separated on a HiLoad 16/600 Superdex 200 column.
Microscopy and imaging
Light microscopy experiments used an Olympus MVX-10 Macroview with charge-coupled device camera (Hamamatsu ORCA). Confocal imaging was done using a NikonA1R. Images were processed in Photoshop 7.0 (Adobe). Confocal and Nomarski microscopy was performed on 3-d-old seedlings mounted in water and covered with number 1.5 coverslips. Fluorescence was scanned with an inverted spinning-disk confocal microscope (Olympus DSU; 60×/1.2 numerical aperture oil immersion lens) or a Leica TCS SP8 confocal laser scanning microscope. For each experiment, seven seedlings were analyzed and one or two root hair images were generated for each seedling. Yeast cells were imaged on a confocal spinning disk microscope (Olympus DSU; 100× oil immersion lens). FM4-64 staining traced the endocytic pathway as described (Vida and Emr, 1995). For GFP, yellow fluorescent protein (YFP), and FM4-64 fluorescence, fluorescein isothiocyanate, YFP, and tetramethylrhodamine isothiocyanate filter settings were used. Live 3-d-old seedlings mounted on 0.8% (wt/vol) top agar were imaged by environmental scanning electron microscopy using an FEI Quanta 200 Field Emission Gun. Images were taken at 95% humidity and 15 kV at a magnification of 200×. Images were processed in Photoshop 7.0.
Plant growth and transformation
Seeds were plated in 0.8% (wt/vol) top agar (low-melt agar in 1× Murashige and Skoog salt and vitamin mixture medium [MS; Life Technologies, Grand Island, NY]). Seeds were stratified at 4ºC for 2 d and then grown vertically under constant light (90 μM/m2 s) at 22ºC for 3 d before being imaged for root hair size. Agrobacterium tumefaciens (Rhizobium radiobacter)–mediated transformation by the floral dip method was used to generate transgenic plant lines (Clough and Bent, 1998). Agrobacterium strain GV3101 strain was used for all transformations. Transformed plants were propagated to the T3 generation to obtain homozygous lines.
Transient expression in tobacco leaves
Agrobacteria were transformed with pGWB655-AtSFH1 nodulin and mutant versions (encoding mRFP-AtSfh1 nodulin chimera), pH7WGF2-BRI1, encoding a Bri1-GFP translational fusion used as a plasma membrane marker (kindly donated by Klaus Harter, University of Tübingen, Tübingen, Germany), and pBIN61-p19, encoding the silencing suppressor p19 (Voinnet et al., 2003). Transformants were then used for cotransfection of N. benthamiana leaves. For this, Agrobacteria transformants were grown at 28°C in Luria–Bertani (LB) medium supplemented with 100 μg/ml rifampicin (Genaxxon Bioscience, Ulm, Germany), 30 μg/ml gentamicin (Duchefa Biochemie B.V., Haarlem, The Netherlands), and appropriate antibiotic for plasmid selection to stationary phase. Subsequently, 1 ml of saturated culture was added to 4 ml of fresh medium and grown for an additional 4 h. Transformants were then sedimented at 4000 × g for 20 min at 4°C and resuspended in 10 mM 2-(N-morpholino)ethanesulfonic acid (MES; Carl Roth GmbH, Karlsruhe, Germany), 10 mM MgCl2, and 150 μM 3′,5′-dimethoxy-4′-hydroxyacetophenone (acetosyringone; Sigma-Aldrich) and infiltrated into the abaxial air spaces of 2-wk-old tobacco plants. Leaves were imaged 5 d after infiltration using a confocal laser scanning microscope (Leica TCS SP8).
Ca2+-influx experiments
Rapid imposition of large Ca2+ influx into yeast cell cytoplasm exploited vps33 mutant strains conducted as described previously (Miseta et al., 1999). Isogenic wild-type (SEY6210) and vps33 (LBY317) yeast strains expressing GFP-AtSfh1 nodulin were grown to mid exponential phase in uracil-free minimal medium buffered with MES-Tris to pH 5.5. Yeast cells from 5-ml cultures were harvested and washed twice with 1 ml of test medium (uracil-free minimal medium, 2% dextrose, 2 mM ethylene glycol tetraacetic acid [EGTA], 40 mM MES-Tris, pH 6.5). Cells were resuspended in 500 μl of test medium and allowed to equilibrate for 30 min at room temperature. CaCl2 was then added to the medium to a final concentration of 50 or 400 mM. The yeast cells were then immediately and continuously imaged by confocal microscopy in the window of 30–300 s, with images collected at 30-s intervals during this time window.
Immunoblotting of root extracts
Proteins were extracted from root hairs in extraction buffer (5 mM HEPES, pH 7.5, 10 mM magnesium acetate, 2 mM EGTA, 2 mM phenylmethylsulfonyl fluoride, 15 mM β-mercaptoethanol, protease cocktail inhibitors [Roche]), resolved by SDS–PAGE, and blotted with AtSfh1 antibodies raised against the linker joining the Sec14 and nodulin domains. Total proteins from myc-AtSfh1 nodulin proline substitution mutant lines were blotted with anti-myc antibodies.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM44530) and the Robert A. Welch Foundation (BE-0017) to V.A.B. M.K.F.C., M.D., H.C.L., H.S., and G.S. were supported by Emmy Noether Grant SCHA 1274/2-1, Grant SCHA 1274/3-1 and SFB 1101/TP A05 from the Deutsche Forschungsgemeinschaft (G.S.). A.O., T.R., and I.V. were supported by grants from the Academy of Finland, the Academy of Finland Center of Excellence Program (I.V.), the European Research Council Advanced Grant CROWDED-PRO-LIPIDS (I.V.), and the Sigrid Juselius Foundation (I.V.). Y.Y. and T.I.I. were supported by Welch Research Foundation Grant A-1784 (T.I.I.), and A.A., M.G.L., and H.A. by grants from the Spanish Ministerio de Economia (BFU2011-28566) and the Basque Government (IT838-13; A.A.). M.G.L. was a predoctoral student supported by the Basque Government. The Laboratory for Molecular Simulation at Texas A&M University (College Station, TX) and CSC-IT Center for Science (Espoo, Finland) provided software, support, and computer time. We thank Daniel J. Lew (Duke University, Durham, NC) for permission to use and report the mss4ts mutant strains constructed in his laboratory by John J. Moskow (supported by National Institutes of Health Grant GM62300 to D. J. Lew), David Bedwell (University of Alabama–Birmingham Medical Center, Birmingham, AL) for helpful discussions regarding calcium experiments and vps33 mutant strains, and Mark McDermott for assistance in plant maintenance.
Abbreviations used:
- AtPIP5K3
Arabidopsis phosphatidylinositol 4-phosphate 5-OH kinase
- C-terminal
carboxy-terminal
- DIC
differential interference contrast
- DLPC
dilineoylphosphatidylcholine
- DLPS
dilineoylphosphatidylserine
- GFP
green fluorescent protein
- ITC
isothermal titration calorimetry
- MDS
molecular dynamics simulation
- mRFP
monomeric red fluorescent protein
- NMR
nuclear magnetic resonance
- OPLS
optimized potential for liquid simulations
- ORF
open reading frame
- PH domain
pleckstrin homology domain
- PITP
phosphatidylinositol transfer protein
- PLC
phospholipase C
- PM
plasma membrane
- PtdIns
phosphatidylinositol
- PtdIns(3)P
phosphatidylinositol 3-phosphate
- PtdIns(3,5)P2
phosphatidylinositol 3,5-bisphosphate
- PtdIns(4)P
phosphatidylinositol 4-phosphate
- PtdIns(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- REMD
replica exchange molecular dynamics
- VMD
visual molecular dynamics
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1475) on March 4, 2015.
*These authors contributed equally to this work.
The authors declare no financial conflicts.
REFERENCES
- Affolter M, Zeller R, Caussinus E. Tissue remodelling through branching morphogenesis. Nat Rev Mol Cell Biol. 2009;10:831–842. doi: 10.1038/nrm2797. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova T, Gilroy S. Calcium in root hair growth. In: Emons AMC, Ketelaar T, editors. In: Root Hairs: Excellent Tools for the Study of Plant Molecular Cell Biology. Berlin: Springer; 2008. pp. 1–26. [Google Scholar]
- Braun M, Baluska F, von Witsch M, Menzel D. Redistribution of actin, profilin and phosphatidylinositol-4,5-bisphosphate in growing and maturing root hairs. Planta. 1999;209:435–443. doi: 10.1007/s004250050746. [DOI] [PubMed] [Google Scholar]
- Brewin NJ. Pods and nods: a new look at symbiotic nitrogen fixing. Biologist. 2002;49:1–5. [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Cáceres A, Ye B, Dotti CG. Neuronal polarity: demarcation, growth and commitment. Curr Opin Cell Biol. 2012;24:547–553. doi: 10.1016/j.ceb.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cole RA, Fowler JE. Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol. 2006;9:579–588. doi: 10.1016/j.pbi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Peptide folding: when simulation meets experiment. Angew Chem Int Ed. 1999;38:236–240. [Google Scholar]
- Desbrosses GJ, Stougaard J. Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe. 2011;10:348–358. doi: 10.1016/j.chom.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach H, Laumann V, Martens S, Becker CFW, Goody RS, Geyer M. HIV-1 NEF membrane association depends on charge, curvature, composition and sequence. Nat Chem Biol. 2010;6:46–53. doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- Gervais L, Claret S, Januschke J, Roth S, Guichet A. PIP5K-dependent production of PIP2 sustains microtubule organization to establish polarized transport in the Drosophila oocyte. Development. 2008;135:3829–3838. doi: 10.1242/dev.029009. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Bax A. The importance of not saturating H2O in protein nmr—application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and human synaptojanin encode phosphoinotide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Heilmann I. Using genetic tools to understand plant phosphoinositide signalling. Trends Plant Sci. 2009;14:171–179. doi: 10.1016/j.tplants.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Hepler PK, Luis Vidali L, Cheung AY. Polarized cell growth in higher plants. Annu Rev Cell Dev Biol. 2001;17:159–187. doi: 10.1146/annurev.cellbio.17.1.159. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Huang J, Kim CM, Xuan Y-H, Park SJ, Piao HL, Je BI, Liu J, Kim TH, Kim B-K, Han C-D. OsSNDP1, a Sec14-nodulin domain-containing protein, plays a critical role in root hair elongation in rice. Plant Mol Biol. 2013;82:39–50. doi: 10.1007/s11103-013-0033-4. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ile KE, Schaaf G, Bankaitis VA. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat Chem Biol. 2006;2:576–583. doi: 10.1038/nchembio835. [DOI] [PubMed] [Google Scholar]
- Irving HI, Boukli NM, Kelly MN, Broughton WJ. Nod-factors in symbiotic development of root-hairs. In: Ridge RW, Emons AM, editors. In: Root Hairs Cell and Molecular Biology. Tokyo: Springer; 2000. pp. 241–265. [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- Kaiser H-J, Orlowski A, Rog T, Nyholm TK, Chai W, Feizi T, Lingwood D, Vattulainen I, Simons K. Lateral sorting in model membranes by cholesterol-mediated hydrophobic matching. Proc Natl Acad Sci USA. 2011;108:16628–16633. doi: 10.1073/pnas.1103742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, de Bruijn FJ, Szczyglowski K. Novel, highly expressed late nodulin gene (LjNOD16) from Lotus japonicus. Plant Physiol. 1997;113:1081–1090. doi: 10.1104/pp.113.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Routt SM, Bankaitis VA, de Bruijn FJ, Szczglowski K. Nodule-specific regulation of phosphatidylinositol transfer protein expression in Lotus japonicus. Plant Cell. 2001;13:1369–1382. doi: 10.1105/tpc.13.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn MP, Wodarz A. Phosphoinositide lipids and cell polarity: linking the plasma membrane to the cytocortex. Essays Biochem. 2012;53:15–27. doi: 10.1042/bse0530015. [DOI] [PubMed] [Google Scholar]
- Kusano H, Testerink C, Vermeer JE, Tsuge T, Shimada H, Oka A, Munnik T, Aoyama T. The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell. 2008;20:367–380. doi: 10.1105/tpc.107.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, O'Brien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I, Christian DA, Wang YH, Madera JJ, Discher DE, Janmey PA. Calcium-dependent lateral organization in phosphatidylinositol 4,5 bisphosphate (PIP2) and cholesterol-containing monolayers. Biochemistry. 2009;48:8241–8248. doi: 10.1021/bi9007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shi X, Guo X, Li H, Xu C. Ionic protein-lipid interaction at the plasma membrane: what can the charge do. Trends Biochem Sci. 2014;39:130–140. doi: 10.1016/j.tibs.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Li X, Routt SM, Xie Z, Cui X, Fang M, Kearns MA, Bard M, Kirsch DR, Bankaitis VA. Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol Biol Cell. 2000;11:1989–2005. doi: 10.1091/mbc.11.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SR. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 2001;125:69–72. doi: 10.1104/pp.125.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewski A, Pasenkiewicz-Gierula M, Cramariuc O, Vattulainen I, Rog T. Refined OPLS all-atom force field for saturated phosphatidylcholine bilayers at full hydration. J Phys Chem. 2014;B 118:4571–4581. doi: 10.1021/jp5016627. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. PM phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Miseta A, Fu L, Kellermayer R, Buckley J, Bedwell D. The Golgi apparatus plays a significant role in the maintenance of Ca2+ homeostasis in the vps33D vacuolar biogenesis mutant of Saccharomyces cerevisiae. J Biol Chem. 1999;274:5939–5947. doi: 10.1074/jbc.274.9.5939. [DOI] [PubMed] [Google Scholar]
- Monahan-Giovanelli H, Pinedo CA, Gage DJ. Architecture of infection thread networks in developing root nodules induced by the symbiotic bacterium Sinorhizobium meliloti on Medicago truncatula. Plant Physiol. 2006;140:661–670. doi: 10.1104/pp.105.072876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskow JJ, Gladfelter AS, Lamson RE, Pryciak PM, Lew DJ. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. J Biol Chem. 2000;20:7559–7571. doi: 10.1128/mcb.20.20.7559-7571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol. 2011;14:489–497. doi: 10.1016/j.pbi.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Murray D, Arbuzova A, Honig B, McLaughlin S. The role of electrostatic and nonpolar interactions in the association of peripheral proteins with membranes. Curr Top Membr. 2002;52:271–301. [Google Scholar]
- Mylona P, Pawlowski K, Bisseling T. Symbiotic nitrogen fixation. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Mano S, Tanaka Y, Ohnishi M, Nakamori C, Araki M, Niwa T, Nishimura M, Kaminaka H, Nakagawa T, et al. Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci Biotech Biochem. 2010;74:1315–1319. doi: 10.1271/bbb.100184. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- O'Neil KT, DeGrado WF. How calmodulin binds its targets: sequence independent recognition of amphiphilic α-helices. Trends Biochemi Sci. 1990;15:59–64. doi: 10.1016/0968-0004(90)90177-d. [DOI] [PubMed] [Google Scholar]
- Orłowski A, St-Pierre J-F, Magarkar A, Bunker A, Pasenkiewicz-Gierula M, Vattulainen I, Róg T. Properties of the membrane binding component of catechol-O-methyltransferase revealed by atomistic molecular dynamics simulations. J Phys Chem. 2011;B115:13541–13550. doi: 10.1021/jp207177p. [DOI] [PubMed] [Google Scholar]
- Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- Patriksson A, van der Spoel D. A temperature predictor for parallel tempering simulations. Phys Chem Chem Phys. 2008;10:2073–2077. doi: 10.1039/b716554d. [DOI] [PubMed] [Google Scholar]
- Phillips SE, Sha B, Topalof L, Xie Z, Alb JG, Klenchin VA, Swigart P, Cockcroft S, Martin TF, Luo M, Bankaitis VA. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol Cell. 1999;4:187–197. doi: 10.1016/s1097-2765(00)80366-4. [DOI] [PubMed] [Google Scholar]
- Richter S, Müller LM, Stierhof Y-D, Mayer U, Takada N, Kost B, Vieten A, Geldner N, Koncz C, Jürgens G. Polarized cell growth in Arabidopsis requires endosomal recycling mediated by GBF1-related ARF exchange factors. Nat Cell Biol. 2011;14:80–86. doi: 10.1038/ncb2389. [DOI] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xi Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E, Steer M, Dolan L. Cell biology and genetics of root hair formation in Arabidopsis thaliana. Protoplasma. 2001;215:140–149. doi: 10.1007/BF01280310. [DOI] [PubMed] [Google Scholar]
- Schaaf G, Dynowski M, Mousley CJ, Shah SD, Yuan P, Winklbauer EM, Kaphan Freitas de Campos F, Trettin K, Quinones MC, Smirnova T, et al. Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol Biol Cell. 2011;22:892–905. doi: 10.1091/mbc.E10-11-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Ortlund E, Tyeryar K, Mousley CJ, Ile K, Woolls M, Garrett T, Raetz CRH, Redinbo M, Bankaitis VA. The functional anatomy of PL binding and regulation of PIP homeostasis by proteins of the Sec14-superfamily. Mol Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Bi Y, Yang W, Guo X, Jiang Y, Wan C, Li L, Bai Y, Guo J, Wang Y, Chen X, et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature. 2013;493:111–115. doi: 10.1038/nature11699. [DOI] [PubMed] [Google Scholar]
- Slochower DR, Huwe PJ, Radhakrishnan R, Janmey PA. Quantum and all-atom simulations of protonation and divalent ion binding to phosphatidylinositol 4,5-bisphosphate (PIP2) J Phys Chem. 2013;117:8322–8329. doi: 10.1021/jp401414y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Audhya A, Emr SD. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol Biol Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Ischebeck T, König S, Hołubowska A, Sporysz M, Hause B, Heilmann I. The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell. 2008;20:124–141. doi: 10.1105/tpc.107.052852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4, 5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Sun Y, Carroll S, Kakosnen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin and actin dependent endocytic internalization. J Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Kawaguchi M. Root nodulation: a developmental program involving cell fate conversion triggered by symbiotic bacterial infection. Curr Opin Plant Biol. 2014;21C:16–22. doi: 10.1016/j.pbi.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Thole JM, Nielsen E. Phosphoinositides in plants: novel functions in membrane trafficking. Curr Opin Plant Biol. 2008;11:620–631. doi: 10.1016/j.pbi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Vance C. Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol. 2001;127:391–397. [PMC free article] [PubMed] [Google Scholar]
- van Kammen A. Suggested nomenclature for plant genes involved in nodulation and symbiosis. Plant Mol Biol Rep. 1984;2:4345. [Google Scholar]
- van Spronsen PC, Grønlund M, Bras CP, Spaink HP, Kijne JW. Cell biological changes of outer cortical root cells in early determinate nodulation. Mol Plant Microbe Interact. 2001;14:839–847. doi: 10.1094/MPMI.2001.14.7.839. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Biol Chem. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol. 2005;168:801–812. doi: 10.1083/jcb.200412074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagisawa H, Sakuma K, Paterson HF, Cheung R, Allen V, Hirata H, Watanabe Y, Hirata M, Williams RL, Katan M. Replacement of single basic amino acids in the pleckstrin homology domain of phospholipase C-δ1 alter the ligand binding, phospholipase activity, and interaction with the plasma membrane. J Biol Chem. 1998;273:417–424. doi: 10.1074/jbc.273.1.417. [DOI] [PubMed] [Google Scholar]
- Yakir-Tamang L, Gerst JE. Phosphoinositides, exocytosis and polarity in yeast: all about actin. Trends Cell Biol. 2009;19:677–684. doi: 10.1016/j.tcb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Yang Z. Cell polarity signaling in Arabidopsis. Annu Rev Cell Dev Biol. 2008;24:551–575. doi: 10.1146/annurev.cellbio.23.090506.123233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Gottlieb SDM, Fasch E-V, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote S.cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.