Abstract
The human pathogen Trichomonas vaginalis lacks conventional mitochondria and instead contains divergent mitochondrial-related organelles. These double-membrane bound organelles, called hydrogenosomes, produce molecular hydrogen. Phylogenetic and biochemical analyses of hydrogenosomes indicate a common origin with mitochondria; however identification of hydrogenosomal proteins and studies on its metabolism have been limited. Here we provide a detailed proteomic analysis of the T. vaginalis hydrogenosome. The proteome of purified hydrogenosomes consists of 569 proteins, a number substantially lower than the 1,000 - 1,500 proteins reported for fungal and animal mitochondrial proteomes, yet considerably higher than proteins assigned to mitosomes. Pathways common to and distinct from both mitochondria and mitosomes were revealed by the hydrogenosome proteome. Proteins known to function in amino acid and energy metabolism, Fe-S cluster assembly, flavin-mediated catalysis, oxygen stress response, membrane translocation, chaperonin functions, proteolytic processing and ATP hydrolysis account for ∼30% of the hydrogenosome proteome. Of the 569 proteins in the hydrogenosome proteome, many appear to be associated with the external surface of hydrogenosomes, including large numbers of GTPases and ribosomal proteins. Glycolytic proteins were also found to be associated with the hydrogenosome proteome, similar to that previously observed for mitochondrial proteomes. Approximately 18% of the hydrogenosomal proteome is composed of hypothetical proteins of unknown function, predictive of multiple activities and properties yet to be uncovered for these highly adapted organelles.
Keywords: Hydrogenosome, Mitochondria, Mitosome, Organelle evolution, Parasite, Trichomonas
Graphical abstract
1. Introduction
Trichomonas vaginalis, a pathogenic protist, causes the most common non-viral sexually transmitted human infection worldwide, with ∼170 million cases reported annually (WHO, 2001; Johnston and Mabey, 2008). This parasite belongs to a group of microaerophilic and anaerobic unicellular eukaryotes that lack conventional mitochondria and instead contain related specialized double-membrane organelles called hydrogenosomes (Lindmark et al., 1975; Shiflett and Johnson, 2010). These organelles, which are also found in specific fungi (chytrids) and ciliates (Boxma et al., 2004, 2005), are defined by the ability to produce molecular hydrogen. Hydrogenosomes are polyphylogenetic and have arisen independently in several eukaryotic lineages (Embley and Hirt, 1998).
Some eukaryotes lack either hydrogenosomes or mitochondria and instead contain highly reduced, double-membrane bound organelles called mitosomes (Mai et al., 1999; Tovar et al., 1999; Williams et al., 2002; Putignani et al., 2004; Regoes et al., 2005; Shiflett and Johnson, 2010). Studies demonstrating the presence of mitochondrial-type proteins in hydrogenosomes and mitosomes, together with similarities in the biogenesis of hydrogenosomes and mitochondria, support the hypothesis that these organelles evolved from a single α-proteobacterial endosymbiont (reviewed in Shiflett and Johnson, 2010). The relationship between hydrogenosomes, mitosomes and mitochondria has generated much debate and raises the question whether the acquisition of the endosymbiont that gave raise to mitochondria may have been present in the earliest eukaryotic cell (reviewed in Martin et al., 2001). Efforts to trace the origin of the T. vaginalis hydrogenosome have relied on phylogenetic analyses of nuclear-encoded hydrogenosomal proteins as there is no genome to allow analyses of organellar genes (Clemens and Johnson, 2000). Hypotheses put forth differ primarily in whether a single endosymbiotic event gave rise to both hydrogenosomes and mitochondria through divergent evolution or whether an additional second endosymbiont contributed to the formation of hydrogenosomes in T. vaginalis (Dyall et al., 2004a; Embley, 2006; Shiflett and Johnson, 2010). As the list of analysed proteins has grown, a consensus has emerged that hydrogenosomes, mitosomes and other mitochondrion-like organelles evolved from a single endosymbiont that also gave rise to mitochondria (reviewed in Shiflett and Johnson, 2010).
Until recent years, the only characterized function for T. vaginalis hydrogenosomes was carbohydrate metabolism, specifically in the conversion of pyruvate and malate to the end products of ATP, acetate, CO2 and hydrogen (Muller, 1993). Hydrogenosomes are also the target and site of activation of the 5-nitroimidazole drugs used to treat trichomoniasis (Narcisi and Secor, 1996). Recently, enzymes responsible for iron-sulfur (Fe-S) cluster assembly typically found in mitochondria have been localized to the T. vaginalis hydrogenosome (Tachezy et al., 2001; Carlton et al., 2007; Dolezal et al., 2007). Similarly, mitosomes have also been shown to be the site of Fe-S biogenesis (Tachezy et al., 2001; Regoes et al., 2005; Goldberg et al., 2008). This supports the theory that the only required function for mitochondria is Fe-S biogenesis and may be why these organelles have been preserved throughout evolution (Lill and Kispal, 2000). In Entamoeba histolytica, the machinery for Fe-S cluster assembly has been reported in both the cytosol and the mitosome (Maralikova et al., 2010). This mitosome also houses proteins involved in a sulfate activation pathway, indicating an additional potential function (Mi-Ichi et al., 2009).
Both hydrogenosomes and mitochondria contain many more metabolic pathways than mitosomes (Shiflett and Johnson, 2010). Mitosomes appear to be extremely reduced in complexity and none have been demonstrated to generate ATP (Shiflett and Johnson, 2010). Hydrogenosomes can generate ATP by substrate level phosphorylation but not via oxidative phosphorylation as mitochondria do. Hydrogenosomes also lack a trichloroacetic acid (TCA) cycle, cytochromes and members of complex I-IV, with the exception of NADH dehydrogenase 51 kDa (Ndh51) and 24 kDa (Ndh24) subunits (Dyall and Johnson, 2000; Dyall et al., 2004b; Hrdy et al., 2004). To mediate reduction of reactive oxygen species (ROS), T. vaginalis contains a bacterial-type thioredoxin reduction system within the hydrogenosome (Coombs et al., 2004; Putz et al., 2005). The organism, however, lacks peroxisomes and the glutathione reducing pathway normally present in mitochondria.
Being devoid of a genome, all T. vaginalis hydrogenosomal proteins are nuclearly encoded, synthesized in the cytosol and subsequently targeted and translocated into the organelle. Many hydrogenosomal matrix proteins contain conserved N-terminal presequences that are similar to sequences known to target proteins to the mitochondrial matrix (Bradley et al., 1997; Hausler et al., 1997). In the case of the ferredoxin protein, the presequence has been shown to be necessary for targeting and translocation of the protein into hydrogenosomes in vitro (Bradley et al., 1997). Homologs of several proteins known to be involved in protein import and the biogenesis of yeast mitochondria are present in the hydrogenosome. These include mitochondrial-like chaperones Hsp70, Hsp60 and Hsp10, a processing peptidase and putative members of the translocation machinery (Pam18 and Tim17/22/23 orthologs) (Bui et al., 1996; Dolezal et al., 2006; Brown et al., 2007; Carlton et al., 2007; Smid et al., 2008; Shiflett and Johnson, 2010).
The sequencing of the T. vaginalis genome allowed bioinformatic identification of putative hydrogenosomal proteins through screening for the conserved N-terminal presequence motif. We originally identified 138 putative hydrogenosomal presequences in the genome using this approach (Carlton et al., 2007). Subsequently, using a less strict consensus sequence, 222 putative hydrogenosomal precursor proteins were found (Smid et al., 2008). These data support the presence of additional metabolic pathways in the organelle but suffer from the weakness of the inability to detect proteins that contain divergent targeting signals or lack an N-terminal presequence and may mistakenly identify a non-hydrogenosomal protein (Mentel et al., 2008).
In this study, we conducted a proteomics analysis of the T. vaginalis hydrogenosome to gain a better understanding of the metabolic processes of this organelle. These studies allow the comparison of a hydrogenosome proteome with the mitochondrial proteomes of yeast, protistan and human mitochondria, and the mitosome proteomes of E. histolytica and Giardia lamblia, further defining similarities and differences between these organelles (Mi-Ichi et al., 2009; Jedelsky et al., 2011). Using multiple fractionation techniques, 569 proteins were identified in the T. vaginalis hydrogenosome, uncovering new members of known hydrogenosomal pathways and revealing new metabolic pathways present in this unique organelle.
2. Materials and methods
2.1. Parasite culture
Trichomonas vaginalis strain T1 were grown in Diamond's medium supplemented with 10% (v/v) horse serum and iron as described previously (Diamond, 1957). Transformed T. vaginalis T1 cultures were grown as described (Delgadillo et al., 1997).
2.2. Isolation of hydrogenosomes
Hydrogenosomes of T. vaginalis strain T1 were purified by collecting cells grown as described in Section 2.1 by centrifugation. All procedures were done at 4°C. Cell pellets were washed twice in SMD (0.25 M sucrose, 0.01 M morpholine propane sulfonic acid (MOPS) pH 8.0, 0.01 M DTT) and resuspended in SMDI (SMD plus 25 μg/ml N-p-tosyl-L-lysine chloromethylketone (TLCK) and 10 μg/ml leupeptin). Cells were lysed in a Stansted cell disruptor with 30 psi front pressure and 12 psi back pressure. Unbroken cells were pelleted by centrifugation at 1,000 g for 10 min and the supernatant was then centrifuged at 5,000 g to pellet the organelles. This pellet was resuspended in 45% Percoll in SMDI and subjected to o centrifugation at 68,000 g at 4°C for 1 h. The fraction containing hydrogenosomes (Bradley et al., 1997) was collected from the resulting Percoll gradient, washed twice with 10 vol. of SMDI and reisolated by centrifugation at 7,500 g for 10 min. Hydrogenosomes were resuspended in freezing buffer (0.25 M sucrose, 0.01 M morpholine propane sulfonic acid pH 8.0, 0.5% BSA, 8% glycerol) and stored at -80 °C.
2.3. Fractionation of hydrogenosomal proteins by sodium carbonate or sodium hydroxide extraction
Isolated hydrogenosomes were alkaline extracted at 1 mg/ml in 0.1 M Na2CO3, pH 11.5 or in 0.5 M NaOH, for 30 min at 4 °C. The insoluble pellet and soluble supernatant were subsequently obtained by centrifugation at 100,000 g for 30 min. The proteins found in the supernatant were TCA-precipitated and resuspended in sample buffer. The proteins from the insoluble pellet were resuspended in 0.1 M Na2CO3 and subjected to a second centrifugation step at 100,000 g for 20 min. The insoluble pellet from the second centrifugation step was resuspended in SDS-PAGE sample buffer. Approximately 100 μg of total protein from the soluble fraction and approximately 10 μg of total protein from the membrane fraction were resolved on 15% SDS-PAGE gels and stained with 0.04% Coomassie Brilliant Blue G-250/3.5% perchloric acid. Proteins from both the soluble and insoluble membrane fractions from the sodium carbonate extraction and only the insoluble membrane fraction from the sodium hydroxide extraction were excised and prepared for mass spectrometry analysis as described in Section 2.6.
2.4. Fractionation of hydrogenosomal proteins by zinc chelating column chromatography
Isolated hydrogenosomes (0.5 mg/ml) were solubilized in 1% Triton X-100, 0.3 M NaCl, 20 mM Tris pH 7.5, with a protease inhibitor cocktail (Sigma) for 30 min and insoluble material removed by centrifugation at 100,000 g at 4 °C for 20 min. The solubilized material was fractionated with the Amersham Chelating Sepharose Fast Flow column, which had been charged with Zn2+ and equilibrated with the solubilization buffer. Fractions from the flow-through and the subsequent washes were collected. Wash steps included the solubilisation buffer supplemented with the following: 1 M NaCl for the first wash, 50 mM NaCl for the second wash, 20 mM imidazole/300 mM NaCl for the third wash, 250 mM imidazole/300 mM NaCl for the fourth wash and 50 mM EDTA/1 M NaCl to strip the remaining proteins from the column. The collected fractions were precipitated using TCA and resuspended in SDS-PAGE sample buffer. Samples from the third and fourth washes were resolved by 8-16% Tris SDS-PAGE (Criterion, USA) and stained with 0.04% Coomassie Brilliant Blue G-250/3.5% perchloric acid. Protein bands were excised and prepared for mass spectrometry analysis as described in Section 2.6.
2.5. Fractionation of hydrogenosomal proteins using sodium carbonate extraction and sucrose gradient
Isolated hydrogenosomes (1.3 mg/ml) were alkaline extracted in 0.1 M Na2CO3, pH 11.5, for 30 min at 4 °C. Then 2.4 M sucrose/0.1 M Na2CO3 was added to adjust the final concentration to 1.5 M sucrose/0.1 M Na2CO3 in a final volume of 4 ml. A sucrose gradient was established by overlaying the extracted protein solution with 3.75 ml of 1.4 M sucrose/0.1 M Na2CO3 and then 3 ml of 0.25 M sucrose/0.1 M Na2CO3. The gradient was subjected to centrifugation for 4 h in a SW41 rotor (Beckman, USA) at 274,000 g at 4 °C. Aliquots from the interface of the top and middle layer were precipitated using TCA, resuspended and resolved on 8-16% SDS PAGE gels, then analyzed by western blot, Coomassie blue stain or silver stain. Interface proteins were excised and prepared for analysis by mass spectrometry. Western blot analysis was also used to compare the protein collected from the interface layer with proteins from the middle and bottom layers of the sucrose gradient. Protein bands were excised and prepared for mass spectrometry analysis as described in Section 2.6.
2.6. Protein gel extraction in preparation for LC-MS/MS analysis
Following gel electrophoresis, gel slices containing the target protein(s) were excised from the gel and incubated for 10 min with 0.1 M NH4HCO3 followed by a 1:1 solution of NH4HCO3 and acetonitrile. These two wash steps were repeated three times in succession and then gel slices were dried in a Speedvac. The gel slices were then incubated in 10 mM DTT in 0.1 M NH4HCO3 for 1 h at 60 °C, followed by incubation in 50 mM iodoacetimide in 0.1 M NH4HCO3 for 45 min at 45 °C. The gel slices were then washed as described above and dried in a Speedvac. Gel slices were then incubated with 83 ng/μl of sequencing-grade trypsin (Promega, USA) in 0.1 M NH4HCO3 and incubated for 45 min at 4 °C. NH4HCO3 (0.1 M) was added to the gel slices, which were then incubated overnight at 37 °C. Water was added and the supernatant was subsequently removed. A solution of 50% acetonitrile with 1% trifluoroacetic acid (TFA) was then added to the gel slices and incubated for 10 min. The supernatant was removed and added to the pool of peptides. The last step was repeated three to five times. The peptides were then dried in a Speedvac and analyzed by LC-MS/MS.
2.6.1. Protein identification and analysis of protein sequences
Protein identification was accomplished by reversed-phase high performance LC-MS/MS using a hybrid quadrupole time-of-flight (QTOF) mass spectrometer. A QSTAR Pulsar XL QTOF mass spectrometer (Sciex/Applied Biosystems, Toronto, ON, Canada) was equipped with a nanoelectrospray (nanoESI) interface (Protana, Odense, Denmark) and a Dionex/LC Packings (Sunnyvale, CA, USA) nano-HPLC system. The nano-HPLC system was equipped with a homemade precolumn (150 μm × 5 mm) and analytical column (75 μm × 150 mm) packed with Jupiter Proteo C12 resin (particle size 4 μm; Phenomenex, USA). The dried peptides were resuspended in 1% (v/v) formic acid (FA) solution; 6 μL of sample solution was loaded onto the precolumn for each LC-MS/MS run. The precolumn was washed with the loading solvent (0.1% FA) before the sample was injected onto the LC column. The eluants used for the LC were 0.1% FA (solvent A) and 95% acetonitrile containing 0.1% FA (solvent B). The flow rate was 200 nL/min and the following gradient was used: 3% B to 6% B in 6 s, 6% B to 24% B in 18 min, 24% B to 36% B in 6 min, 36% B to 80% B in 2 min, and maintained at 80% B for 7.9 min. The column was finally equilibrated with 3% B for 15 min before the next run. Electrospray ionization was performed using a 30 μm (inner diameter) nano-bore stainless steel online emitter and a voltage set at 1900 V.
Peptides identified by LC-MS/MS were analysed using the MASCOT sequence database searching program (Matrix Science, London, UK) against the T. vaginalis peptide sequence database (www.trichdb.org v1.0) (Perkins et al., 1999). Derived from the size of the T. vaginalis genome and the protein query database, all searches were performed allowing for phosphorylated Ser, Thr and Tyr, oxidized Met, carbamidomethylated Cys and N-terminal pyro-Gly, using a mass tolerance of 0.3 Da. A MASCOT score of 42 corresponds to a P value < 0.05; thus, peptides with scores < 42 were excluded from the dataset. This dataset was further analysed by the Basic Local Alignment Search Tool (BLAST) using the BLOSUM62 scoring matrix (Altschul et al., 1990). Individual examination of the program output included analysis of sequences with the use of programs available from the Swiss Institute of Bioinformatics (SIB) (ftp://ftp.expasy.org/databases/uniprot) and ClustalX or MUSCLE alignment programs, identification of motif or domains, and identification of any predicted transmembrane domains using the TMpred or HMMTOP prediction algorithm. In addition, the KEGG Orthology-Based Annotation System (KOBAS) program was used to compare the sequences with protein orthologs found in other organisms and predict putative associated metabolic pathways. Putative hydrogenosomal targeting sequences were also identified with a perl script regular expression corresponding to the consensus sequences ML(S/T/A) X(1..15)R (N/F/E/XF), MSLX(1..15)R(N/F/XF) or MLR(S/N)F.
To compare the hydrogenosomal proteome with mitochondrial protein sequences from Homo sapiens and Saccharomyces cerevisiae, the mitochondrial proteomes were obtained from the MitoMiner database (http://mitominer.mrc-mbu.cam.ac.uk) (Smith and Robinson, 2009). The Trypanosoma brucei mitochondrial proteome (Panigrahi et al., 2009) was obtained from the TriTrypDB (http://tritrypdb.org/tritrypdb/), and the mitochondrial Tetrahymena thermophila protein sequences (Smith et al., 2007) were obtained from GenBank.
2.7. Immunofluorescent microscopy
Trichomonas vaginalis T1 transfectants were fixed in 3.5% formaldehyde/0.2% Triton X-100/PBS solution. Cells were preincubated in blocking buffer (PBS/3% BSA) prior to incubation with mouse anti-HA monoclonal antibody (Covance) and/or rabbit anti-Hsp70 polyclonal antibody diluted 1:5,000 (v/v) in blocking buffer. Secondary Alexa Fluor-488 anti-mouse and Alexa Fluor-594 anti-rabbit antibodies (Invitrogen), diluted 1:5,000 (v/v) were added for 1 h. Cells were mounted in Prolong Gold antifade mounting medium (Invitrogen). Images were taken on a Zeiss Axioscope 2 microscope and analyzed using Axiovision LE software (Zeiss).
3. Results and Discussion
3.1. Identification of hydrogenosomal proteins by mass spectrometry
We determined the proteome of the T. vaginalis hydrogensome to better define its metabolic pathways and to allow comparison with the proteomes of mitochondria and mitosomes. Hydrogenosomes were purified using gradient density centrifugation (Bradley et al., 1997). We showed that this method yields highly purified hydrogenosomes as judged by electron microscopy and the absence of activities of cytosolic enzymes (Bradley et al., 1997). Hydrogenosomal proteins were extracted using a combination of sodium hydroxide, sodium carbonate extractions, sucrose gradients and zinc chromatography. Multiple fractionation strategies were employed to optimize inclusion of proteins in the proteome. Proteins were then subjected to trypsin digestion and the resulting peptides were identified by mass spectrometry. The complete proteome dataset was found to contain 569 proteins (Supplementary Tables S1 - S3). Present in the proteome were 66 previously characterized hydrogenosomal proteins and their paralogs, confirming both their localization and our methodology (Table 1). Of the 569 proteins identified, 175 proteins can be placed in pathways previously associated with the hydrogenosome such as energy metabolism, Fe-S cluster assembly, oxygen stress response and amino acid metabolism, better defining the organelle's metabolic capacity (Fig. 1, Supplementary Table S1). Many of these proteins are also candidates for playing a role in biogenesis or protein and solute membrane translocation. In addition, we identified 101 hypothetical proteins which are mostly unique to the T. vaginalis genome (Fig. 1, Supplementary Table S1). Our proteome also contains 137 small GTPases and related proteins as well as 123 proteins that are likely associated with the external surface of the organelle (Fig. 1, Supplementary Table S2). Thirty-three proteins were classified as contaminants as they are homologues of proteins known to be localized in the endoplasmic reticulum, nucleus or vesicles in other organisms (Fig. 1, Supplementary Table S3). Definitive classification as a contaminant, however, awaits future experiments to directly test whether these proteins localize to hydrogenosomes. To independently confirm the hydrogenosomal localization of proteins assigned to the protein, 13 randomly selected proteins were epitope-tagged and expressed in T. vaginalis transfectants and localized by immunofluorescent microscopy using an antibody against hydrogenosomal Hsp70 as the organelle marker. Twelve of the 13 proteins were seen to co-localize with Hsp70, consistent with the low level of assigned contaminants (data not shown). Nevertheless, based on the association of hydrogenosome with other subcellular structures and the difficulty of obtaining purity of any subcellular fraction, our preparations are likely to contain contaminating, non-hydrogenosomal proteins.
Table 1.
Known Trichomonas vaginalis hydrogenosomal proteins and their representation in the proteome.
| Protein | # Paralogs (Proteome) | # Paralogs (Genome) |
|---|---|---|
| Energy Metabolism | ||
| Hydrogenosome Malic Enzyme | 7 | 15 |
| Hydrogenase | 5 | 14 |
| Ferredoxin | 3 | 7 |
| Pyruvate:ferredoxin oxidoreductase (PFO) | 6 | 7 |
| Adenylate Kinase | 1 | 8 |
| Acetate:succinate CoA-transferases | 3 | 4 |
| Succinate Thiokinase (α subunit) | 3 | 3 |
| Succinate Thiokinase (β subunit) | 3 | 3 |
| NADH dehydrogenase (24 kD subunit) | 1 | 1 |
| NADH dehydrogenase (51 kD subunit) | 1 | 2 |
| Oxygen Scavenging System | ||
| Thioredoxin (Trx) | 1 | 32 |
| Thioredoxin peroxidase (TrxP), Type 1 | 1 | 3 |
| Thioredoxin peroxidase (TrxP) Type 2 | 1a | 7 |
| Rubrerythrin | 1 | 6 |
| Iron Superoxide Dismutase (SOD) | 2 | 7 |
| Amino Acid Metabolism | ||
| H-protein | 2 | 2 |
| L-protein | 1 | 1 |
| Serine Hydroxymethyltransferase | 1 | 1 |
| Fe-S cluster assembly/hydrogenase maturation | ||
| IscS | 1 | 2 |
| IscU | 1 | 1 |
| IscA | 2 | 3 |
| HydG | 1 | 2 |
| Chaperones | ||
| Hydrogenosomal Hsp10 | 4 | 4 |
| Hydrogenosomal Hsp60 | 3 | 3 |
| Hydrogenosomal Hsp70 | 3 | 3 |
| Peptidases | ||
| Hydrogenosome Processing Peptidase (α subunit) | 1 | 1 |
| Hydrogenosome Processing Peptidase (β subunit) | 1 | 1 |
| Membrane Proteins | ||
| Hydrogenosome Membrane Protein (Hmp35) | 3 | 3 |
| Hydrogenosome Membrane Protein (Hmp31 MCF) | 3 | 3 |
Multiple protein sequences that cannot differentiate between individual genes based on spectra.
Fig. 1.
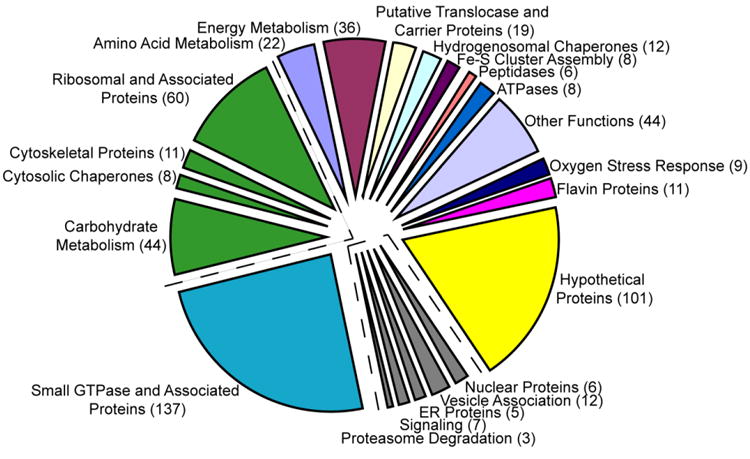
Functional distribution of the 569 proteins identified in the Trichomonas vaginalis hydrogenosome proteome. Proteins involved in amino acid or energy metabolism, membrane functions, oxygen stress response and other functions are depicted on the top right in multiple colors. Hypothetical proteins are indicated in yellow and small GTPase and associated proteins are shown in blue. Proteins predicted to be associated with the external surface of the organelle are depicted in green and contaminants are indicated in gray. Numbers of proteins in each category are listed in parentheses.
The peptides used to identity these proteins had MASCOT scores ranging from 42-1,843 in a skewed bell curve distribution pattern (Fig. 2A). The majority of the proteins (66%) correspond to MASCOT scores ranging from 50–200 and approximately 63% of the proteins were found to have > 3 matched peptides (Fig. 2A, B). The proteome consists of 468 (82%) unique proteins and 101 (18%) proteins that correspond to protein families with multiple sequences containing such high identity at the amino acid level that the expressed genes could not be distinguished by the derived peptides (Supplementary Table S4). In comparison, most mitochondrial proteomes have been shown to contain ∼1,000 proteins (Sickmann et al., 2003; Reinders et al., 2006; Smith et al., 2007), whereas the recently published G. lamblia mitosomal proteome contains 139 proteins (Jedelsky et al., 2011), and the E. histolytica mitosomal proteome contains only 95 proteins (Mi-Ichi et al., 2009).
Fig. 2.
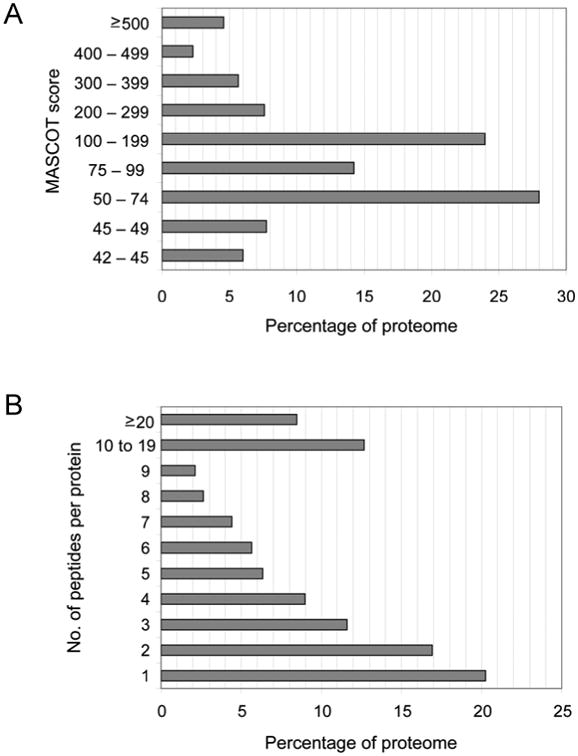
Statistical analysis of the Trichomonas vaginalis hydrogenosome proteome. A) Distribution of MASCOT scores. MASCOT scores ranged from 42- 1,843. The top MASCOT score is reported for those proteins identified in multiple fractions. MASCOT Scores > 42 correspond to a P value < 0.05 in this dataset. B) Number of peptides per protein. Peptides per protein ranged from 1- 128.
The number of proteins (101/569; ∼18%) annotated as hypothetical in the hydrogenosome proteome (Supplementary Table S1) is comparable to that observed for mitochondrial proteomes. For example, 25% of the 750 proteins found in the S. cerevisiae mitochondrial proteome do not have a known function (Sickmann et al., 2003). This is also true for mitosomes; approximately 28% of the G. lamblia mitosome proteome consists of hypothetical proteins (Jedelsky et al., 2011), however ∼67% of the E. histolytica mitosome proteome is hypothetical (Mi-Ichi et al., 2009). Twenty-six percent of the hypothetical proteins in the hydrogenosomal proteome are predicted to have at least one transmembrane domain (Supplementary Table S5) using the prediction algorithms TMHMM 2.0 (Krogh et al., 2001), HMMTOP 2.0 (Tusnady and Simon, 2001) or SPOCTOPUS (Viklund et al., 2008). Further investigation of these hypothetical proteins may reveal proteins that function as additional members of the protein translocase machinery, carriers or proteins involved in biogenesis and homeostasis. These hypothetical proteins likely also represent novel functions associated with each type of organelle.
3.2. The highly redundant nature of the hydrogenosomal proteome
The T. vaginalis genome is highly redundant, containing many multi-copy gene families (Carlton et al., 2007). This is reflected in the hydrogenosomal proteome where multiple paralogs of protein families are present (Table 1, Supplementary Table S4). For example, the genome contains 15 genes encoding malic enzymes and seven of these distinct proteins are found in the proteome; similarly six of seven predicted pyruvate ferredoxin oxidoreductases (PFOs) are present and three of seven ferredoxin proteins are also found in the organelle (Table 1). Although it is not known whether the three ferredoxin paralogs are functionally redundant, their presence could explain the lack of an observable phenotype when a single ferredoxin gene was deleted by homologous recombination (Land et al., 2004). Multiple genes encoding four types of hydrogenases, as well as two hydrogenase-related proteins, are present in the genome (Carlton et al., 2007). The proteome contains four hydrogenase proteins, two type I and two type II, differing in the number of Fe-S domains, together with a previously documented but uncharacterized type IV hydrogenase containing a hydrogenase domain and a C-terminal electron-supplying reductase region (Supplementary Table S1). Determining whether the different hydrogenases present in the hydrogenosome have overlapping or distinct catalytic activities awaits further analyses.
3.3. Comparative analysis of the proteome and expressed sequence tag (EST) databases
The observation that in some cases every member of a hydrogenosomal protein family is found in the proteome while other families lack the full complement of proteins (Table 1, Supplementary Table S4) may be the result of regulated expression of multigene families under different conditions. To address this possibility, we compared the expression profile of proteins found in the hydrogenosome proteome with a T. vaginalis EST database generated from cells grown under the same normal growth conditions used to prepare organelles for proteomic analyses (http://tvxpress.cgu.edu.tw/). Thirteen protein families comprised of 3-10 paralogs that had at least one paralog absent from the proteome were compared. From this dataset of 71 proteins, 34 are present in the proteome and 29 of these are expressed under normal growth conditions (Fig. 3). Of the remaining 37 proteins absent from the proteome, 31 are only expressed under specialized conditions or lack ESTs under all conditions tested. This observation, while based on a small dataset, raises the possibility that T. vaginalis has used gene expansion and differential gene expression as a way of adapting quickly to changing environmental pressures.
Fig. 3.
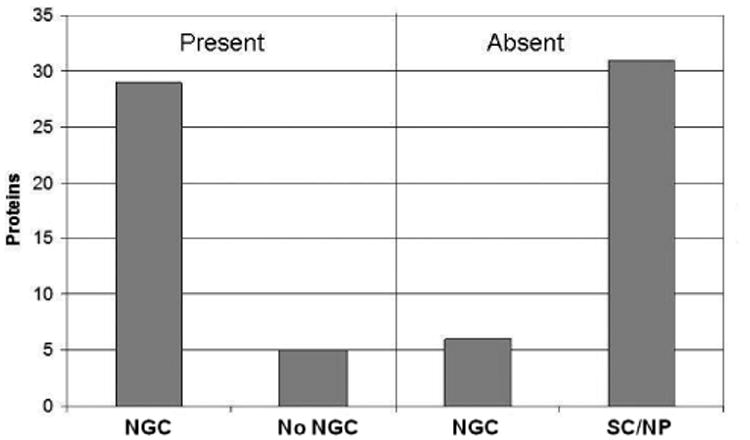
Comparison of expressed sequence tags (ESTs) for the presence or absence of the corresponding protein in the Trichomonas vaginalis hydrogenosome proteome. Seventy-one proteins comprising 13 gene families were compared. Twenty-nine of the 34 proteins represented in ESTs from cells grown under the conditions used to isolate hydrogenosomes for proteomic analysis (NGC) are present in the proteome, whereas 31 of the 37 proteins not represented in the proteome are either expressed under specialized conditions (SC) or not presence (NP) in EST databases.
3.4. Functional categorization of the proteome
The proteins composing the hydrogenosome proteome were analysed using BLAST to assign putative functions based on sequence similarity to previously identified proteins in the NCBI database (Supplementary Table S1-S3; Altschul et al., 1990). Proteins were further examined using KOBAS to search the KEGG database and build putative metabolic pathways (Kanehisa et al., 2008). These analyses led to the discovery of several new pathways and putative functions for the trichomonad hydrogenosome, as well as providing additional information on pathways previously assigned to the organelle.
As shown in Fig. 1, ∼1/3 of the proteome is involved in metabolic processes including sugar (44 proteins), amino acid (22 proteins) and energy metabolism (36 proteins), protein membrane translocation (14 proteins), chaperonin functions (12 proteins), Fe-S cluster assembly (eight proteins), proteolytic processing (peptidases) (six proteins), ATP hydrolysis (eight proteins), oxygen stress response (nine proteins), flavin-mediated catalysis (11 proteins) and other known functions. Although a limited role in amino acid metabolism has been previously implicated for the hydrogenosome, these data greatly expand the role of this organelle in this process (Mukherjee et al., 2006a, b). The identification of additional proteins with activities previously described in T. vaginalis hydrogenosomes, such as Fe-S cluster assembly or maturation, peptidases and flavin proteins, further broaden our understanding of metabolic activities compartmentalized within this organelle.
The majority of identified proteins in our proteome were not previously assigned to the organelle, however 66 characterized hydrogenosomal proteins and their paralogs were found (Table 1). As discussed in Section 3.1, a substantial number of hypothetical proteins (101) were found, pointing to the presence of multiple activities and properties yet to be revealed (Fig. 1, Supplementary Table S1). Further studies will be necessary to determine what percentage of these hypothetical proteins are localized to the hydrogenosome; nevertheless their high frequency is predictive of as-yet uncovered hydrogenosomal activities. We classified 123 proteins as organelle-associated proteins that are not predicted to compartmentalize within the organelles, but instead associate with the external surface of hydrogenosomes (Supplementary Table S2, Fig. 1). These include proteins involved in carbohydrate metabolism (glycolysis, gluconeogenesis and the pentose phosphate pathway, 44 proteins), cytosolic chaperones (eight proteins), cytoskeletal proteins (11 proteins) and ribosomal proteins (60 proteins). Additionally, an exceptionally large number of GTPase proteins (137 proteins) were found, consistent with the massive expansion of this protein family in the T. vaginalis genome (Supplementary Table S1, Figure 1; Lal et al., 2005; Carlton et al., 2007). It is probable that the GTPases are primarily components of cytoskeletal structures that have been shown to associate with the external surface of the hydrogenosome (Benchimol, 2009), however some may function within the organelle. Due to this ambiguity we did not classify the GTPases as contaminants; although many are likely to be associated with the outer surface of the organelle. Confirmatory localization using immunomicroscopy of parasites transfected with tagged candidate hydrogenosomal proteins will be required to definitively classify them as hydrogenosomal.
3.5. Metabolic pathways of the hydrogenosome
The T. vaginalis hydrogenosome has previously been shown to be a site of ATP production, Fe-S cluster biosynthesis, and oxygen stress response (Muller, 1993; Tachezy et al., 2001; Coombs et al., 2004; Sutak et al., 2004; Putz et al., 2005). Molecular hydrogen, acetate and CO2 are by-products of ATP production via substrate level phosphorylation. Proteins known to be involved in energy metabolism include malic enzyme, PFO, [2Fe-2S] ferredoxin (Fdx), succinate thiokinase (SCS), adenylate kinase (AK), [Fe] hydrogenase (Hyd), Ndh51 and 24kDa Ndh24 subunits and acetyl:succinate CoA-tranferase (ASCT) (Johnson et al., 1990; Lahti et al., 1992; Lahti et al., 1994; Hrdy and Muller, 1995b; Bui and Johnson, 1996; Horner et al., 2000; Dyall et al., 2004b; Hrdy et al., 2004; van Grinsven et al., 2008). All of these proteins were represented in the proteome, in several instances by multiple paralogs (Table 1, Supplementary Tables S1 - S4).
3.5.1. Fe-S Cluster Pathway
Multiple proteins involved in energy metabolism contain Fe-S clusters and are dependent on enzymes required for Fe-S biosynthesis for their maturation (Lill, 2009). Similar to mitochondria and mitosomes, enzymes involved in the Iron-Sulfur Cluster (ISC) machinery are localized to the hydrogenosome (Tachezy et al., 2001; Tovar et al., 2003; Regoes et al., 2005; Dolezal et al., 2007). Twelve genes encoding proteins similar to components of the ISC assembly or protein maturation pathways are present in the T. vaginalis genome (Carlton et al., 2007). We identified eight of these proteins in the proteome: IscU, IscA, IscS, Fdx, Hsc20 (HscB), GrpE (Mge1), HydF and HydG (Fig. 4A, Supplementary Table S1). Three proteins of the ISC pathway missing from the proteome are frataxin, HydE and an Isd11-like protein that contains a putative N-terminal presequence (Richards and van der Giezen, 2006). Of these, frataxin and HydE have been localized in the T. vaginalis hydrogenosome (Dolezal et al., 2007; Putz et al., 2006; Dolezal et al., 2007). Two additional proteins found in mitochondria that compose the Fe-S export machinery, Atm1 and Erv1, are absent from the proteome and genes encoding these proteins are also not readily identifiable in the genome (Lill and Muhlenhoff, 2008).
Fig. 4.
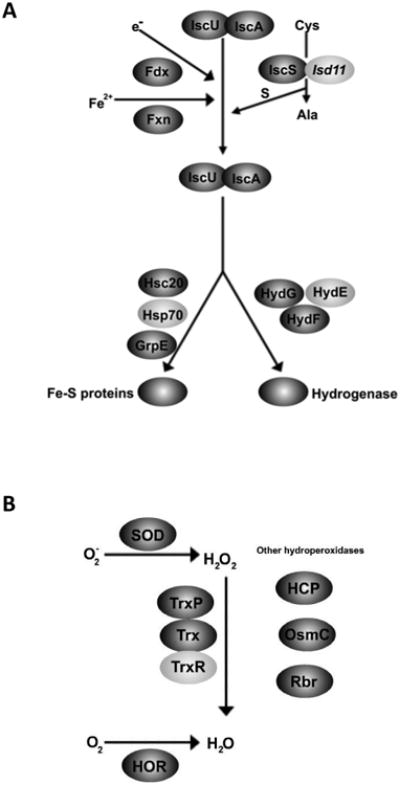
Proteins from known pathways localized to the Trichomonas vaginalis hydrogensome. A) Iron-Sulfur (Fe-S) cluster assembly pathway. Proteins involved in Fe-S cluster assembly were identified in the hydrogenosome proteome (dark grey); some proteins not present in the proteome have been identified in the genome (light grey). B) Proteins involved in oxygen stress response were also identified in the hydrogensome proteome (dark grey). Only one protein, TrxR (light grey), known to be involved in this pathway, was not found in the proteome.
The mitosome of G. lamblia contains only eight proteins known to be involved in ICS transfer and assembly: IscS, IscU, Nfu, IscA, Glutaredoxin5, Hsp70, HscB, and GrpE (Jedelsky et al., 2011). During reductive evolution, E. histolytica has lost the ISC machinery entirely and instead gained the Nif system, likely through a lateral gene transfer from a ε-proteobacterium (Ali et al., 2004; van der Giezen et al., 2004). This system has been described in only one other eukaryotic organism, Mastigamoeba balamuthi, where it is predicted to be cytosolic (Gill et al., 2007; Maralikova et al., 2010). In E. histolytica the Nif machinery has been demonstrated to possess a dual localization in both the cytosol and mitosome (Maralikova et al., 2010).
3.5.2. Oxygen stress response
Inhibition of specific oxygen sensitive hydrogenosomal proteins, such as PFO and [Fe]-hyd, can block energy metabolism in the organelle (Hrdy and Muller, 1995a; Page-Sharp et al., 1996). Iron-dependant superoxide dismutase (SOD) proteins that convert reactive oxygen superoxide species (O2-) to hydrogen peroxide (H2O2) have been characterized but not previously localized in T. vaginalis (Viscogliosi et al., 1998). SOD activity has been reported in both the cytosol and the hydrogenosomes of the related parasite Tritrichomonas foetus (Lindmark and Muller, 1974). Proteins encoded by two of the seven genomic copies of SOD were identified in the T. vaginalis hydrogenosome proteome (Fig. 4B, Table 1). Mitochondria typically utilize a glutathione system and catalase to remove H2O2, proteins that are not present in trichomonads (Muller, 1993; Coombs et al., 2004). Instead proteins of the thioredoxin pathway (Trx, TrxP, TrxR) that convert H2O2 into H2O have been localized to hydrogenosomes, as has peroxidase rubrerythrin (Rbr) (Coombs et al., 2004; Putz et al., 2005). Three of these four proteins, Trx, TrxP and Rbr, were also identified in our proteomic data (Fig. 4B, Supplementar Table S1). In addition a protein with weak similarity to the OsmC/Ohr hydroperoxidase protein of bacteria was present, which may represent another pathway for the metabolism of H2O2 (Fig. 4B, Supplementaryl Table S1; Rehse et al., 2004). A protein that is up-regulated in bacteria by H2O2, the hybrid cluster protein (HCP) is also present in the proteome (Fig. 4B, Supplementaryl Table S1; Almeida et al., 2006).
To confirm the localization of this protein in hydrogenosomes, a C-terminally HA-tagged HCP protein was expressed in T. vaginalis. Immunostaining of the transformants showed that HCP-HA colocalized with the hydrogenosomal marker protein Hsp70 (Fig. 5B). Another protein that plays a role in oxygen stress response that is also present in the hydrogenosome proteome is the recently identified flavo-diiron protein called the hydrogenosome oxygen reductase (HOR) (Fig. 4B). This protein has been shown to metabolize another ROS, O2 (Smutna et al., 2009). Several of the proteins involved in these pathways appear to be the result of lateral gene transfer based on phylogenetic analyses, including HCP, Trx, TrxP, TrxR, Rbr and quite possibly OsmC (Coombs et al., 2004; Rehse et al., 2004; Putz et al., 2005; Andersson et al., 2006).
Fig. 5.
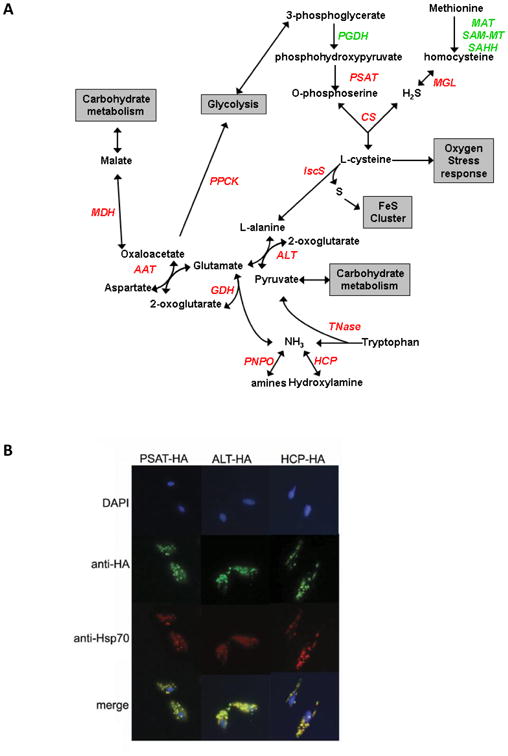
Proteins involved in amino acid metabolism pathways identified in the Trichomonas vaginalis hydrogenosome proteome. A) Proteins involved in amino acid metabolism pathways found in the hydrogenosome proteome are labeled red and proteins found in the genome are labeled green. Grey boxes list intersecting pathways. PGDH, phosphoglycerate dehydrogenase; PSAT. phosphoserine aminotransferase; CS, cysteine synthase; MGL, methionine-g-lyase; MAT, SAM-MT, SAHH, methionine adenosyltransferase, S-adenosyl-methioinine-dependent methyltransferase, S-adenosylhomocysteine hydrolase, respectively; IscS, iron sulphur cluster C protein; ALT, alanine aminotransferase; Tnase, tryptophanase; HCP, hybrid cluster protein; PNPO, pyridoxamine 5′-phophate oxidase; GDH, glutamate dehydrogenase; AAT, aspartate aminotransferase; MDH, malate dehydrogenase; PPCK, phosphoenolpyruvate carboxykinase. B) Subcellular localization of PSAT, ALT and HCP was confirmed using immunofluorescence. Trichomonas vaginalis cells were transformed with C-terminally HA-tagged PSAT, ALT or HCP and analysed as described in Section 2.7 (green). The hydrogenosome was stained with anti-Hsp70 (red). Merged images show co-localization of PSAT-HA, ALT-HA and HCP-HA with Hsp70 in the hydrogenosome. Nuclei are stained with DAPI.
3.6. Amino acid metabolism pathways identified in the proteome
The first evidence that hydrogenosomes, like mitochondria, may be involved in amino acid metabolism came when the H and L proteins of the Glycine Cleavage System (GCS), and the serine hydroxymethyltransferase protein (SHMT) were identified and characterized as hydrogenosomal (Mukherjee et al., 2006a, b). However the apparent absence of other proteins required in these pathways for glycine and serine metabolism left the role of these proteins undefined. Our proteomic data now show that in addition to these proteins, 10 other protein families involved in amino acid metabolism are present in the hydrogenosome (Fig. 5A, Supplementary Table S1).
Three of these proteins, phosphoserine aminotransferase (PSAT), methionine-γ-lyase (MGL) and cysteine synthase (CS), have been shown to be involved in the conversion of phosphohydroxypyruvate and homocysteine to cysteine in T. vaginalis (Westrop et al., 2006). CS localizes to mitochondria and plastids in Arabidopsis thaliana (Hesse et al., 1999) and PSAT is found in mitochondria in spinach nodules (Hoa le et al., 2004). However, in T. vaginalis localization of the pathway was predicted to be cytosolic based on the lack of classic presequences on these proteins. To further address whether this pathway exists in hydrogenosomes, C-terminally HA-tagged PSAT (PSAT-HA) was expressed in T. vaginalis. The resulting immunostaining showed that PSAT-HA colocalized with hydrogenosomal Hsp70, confirming its localization in the organelle (Fig. 5B).
The cysteine generated by this pathway (Fig. 5A) is a substrate for IscS to generate alanine and molecular sulfur used in ISC formation (Tachezy et al., 2001; Sutak et al., 2004). Alanine can then be converted to glutamate by the four distinct alanine aminotransferases (ALT) present in the proteome. To confirm the presence of this enzyme in the organelle, a C-terminally HA-tagged protein (ALT-HA) was shown to co-localize with Hsp70 in transfected T. vaginalis (Fig. 5B). These data indicate that enzymes that provide a source of cysteine for the hydrogenosomal Fe-S biogenesis pathway are also localized in the organelle.
The T. vaginalis CS has been shown to utilize O-phosphoserine as well as the more common substrate, O-acetylserine, in the synthesis of cysteine in vitro (Westrop et al., 2006). However the genes required for the synthesis of O-acetylserine are absent from the T. vaginalis genome. The use of O-phosphoserine by the T. vaginalis CS is surprising as this was previously observed exclusively in mycobacteria and Archeae (Westrop et al., 2006; Agren et al., 2008). Additionally, CS has only been identified in a few mitochondria, whereas MGL is absent and PSAT has only been previously documented in one organism's mitochondrion (Hoa le et al., 2004).
Two additional enzymes involved in amino acid metabolism, aspartate aminotransferase (AAT) and glutamate dehydrogenase (GDH), that catalyse the reversible metabolism of aspartate, glutamate, oxoglutarate and oxaloacetate, were also present in the hydrogenosomal proteome (Fig. 5A, Supplementary Table S1). GDH would also allow for the conversion of NH3, which is a by-product of a number of enzymes identified in the hydrogenosome proteome.
Compared with hydrogenosomes, human mitochondria harbor a larger number of amino acid metabolic pathways: 11 amino acids can be synthesized in human mitochondria and 17 can be metabolized (Guda et al., 2007). Very little, however, is known about amino acid metabolism in mitochondria of other organisms. In an effort to gain a better understanding of similarities and differences between T. vaginalis hydrogenosomes and mitochondria we compared our proteome and the mitochondrial proteomes of H. sapiens, S. cerevisiae, T. brucei and T. thermophila using KOBAS (Mao et al., 2005; Wu et al., 2006). We identified metabolic pathways for 16 amino acids in H. sapiens, 12 in S. cerevisiae, eight in T. brucei, nine in T. thermophila and six in T. vaginalis (Supplementary Table S6).
Comparison of the KOBAS results showed that the Ala, Asp, Glu pathway and the Gly, Ser pathway are conserved in the hydrogenosome and the majority of the mitochondria proteomes (Supplementary Table S6). Additionally, the mitochondrial proteomes of H. sapiens and S. cerevisiae reveal the presence of enzymes involved in cysteine metabolism, a feature shared with the hydrogenosome, albeit using different enzymes (Supplementary Table S6). In contrast, pathways that are present in the mitochondrial proteomes but absent from the hydrogenosome include the Val, Ile and Leu biosynthesis and degradation pathway; proline metabolism is also present in all of the mitochondrial proteomes but not the hydrogenosomal proteome (Supplementary Table S6). However, it is unlikely that T. vaginalis has lost these pathways as enzymes such as branched chain aminotransferase, known to play a major role in Val, Ile and Leu degradation, is present in the T. vaginalis genome. The differences in conservation of the Val, Ile, Leu and Pro pathways in the analyzed mitochondrial proteomes but not the T. vaginalis hydrogenosomes suggests that these pathways play an essential role within mitochondria.
Conservation of the Ala, Asp, Glu, Gly and Ser pathways in both hydrogenosomes and mitochondria emphasizes key similarities between the organelles, as does the presence of all or the majority of proteins involved in the GCS pathway (H-,L-, T-, P-proteins and SHMT proteins) in the hydrogenosome and mitochondrial proteomes. None of the current mitosome proteomes appear to contain amino acid metabolism pathways (Mi-Ichi et al., 2009; Jedelsky et al., 2011). A more in-depth analysis of the evolution of these pathways should provide a better understanding of the similarities and differences between these organelles.
3.7. Identification of hydrogenosomal membrane proteins
Eighty-nine proteins in the proteome were identified as potential membrane proteins based on TMHMM (Krogh et al., 2001), HMMTOP (Tusnady and Simon, 2001) and SPOCTOPUS (Viklund et al., 2008) analyses (Supplementary Table S7). Membrane proteins are difficult to identify by proteomic analyses due to hydrophobicity and solubility properties, therefore this subset of proteins is often under-represented (Santoni et al., 2000) and it is likely that many hydrogenosomal membrane proteins were not detected. The membrane proteins we have identified include both predicted translocases and carrier family proteins, as well as several hypothetical proteins (Supplementaryl Table S7).
3.7.1. Translocases
Only a few proteins found in the two membranes that surround the hydrogenosome have been characterized. Lacking a genome, all proteins involved in the metabolic pathways housed in the hydrogenosome must be imported. Proteins must traverse the outer membrane, the intermembrane space (IMS) and the inner membrane, and complexes containing multiple proteins have evolved to serve in these roles. As biochemical analyses of the translocation machinery of the T. vaginalis hydrogenosome are limited, we and others have adopted bioinformatics approaches to identify putative mitochondrial-like translocase proteins in the T. vaginalis genome (Table 2).
Table 2.
Comparative analyses of mitochondrial translocase proteins found in Saccharomyces cerevisiae and Tetrahymena thermophilia mitochondria, the Trichomonas vaginalis hydrogenosome and mitosomes of Giardia lamblia or Entamoeba histolytica.
| Translocase Complex Member | S. cerevisiae mitochondria | T. thermophilia mitochondria | T. vaginalis hydrogenosome | G. lamblia & E. histolytica mitosome |
|---|---|---|---|---|
| Outer Membrane Proteins | ||||
| Tom70 | X | |||
| Tom40 | X | X | Xa | Xa |
| Tom22 | X | X | ||
| Tom20 | X | |||
| Tom7 | X | X | ||
| Tom6 | X | |||
| Tom5 | X | |||
| Sam50 | X | X | X | Xb |
| Inner Membrane Proteins | ||||
| Tim50 | X | X | ||
| Tim44 | X | X | X | |
| Tim21 | X | |||
| Tim17/22/23 | X | X | X | |
| Motor Complex Proteins | ||||
| Pam18 | X | X | X | X |
| Pam16 | X | X | X | X |
X indicates the presence of a homolog.
These proteins have a porin-3 domain found in both Tom40 and VDAC. They have not been shown to function as a Tom40 biochemically.
Not present in the G. lamblia and E. histolytica proteomes; however, this protein localizes to the E. histolytica mitosome.
Saccharomyces cerevisiae mitochondrial proteins are initially recognized and transported across or into the outer membrane by the Translocase of the Outer Membrane (TOM) complex (Neupert and Herrmann, 2007; reviewed in Lithgow and Schneider, 2010).
The import of outer membrane β-barrel proteins also requires an additional complex called the Sorting and Assembly Machinery (SAM) (Neupert and Herrmann, 2007; Lithgow and Schneider, 2010). Of the seven proteins that form the S. cerevisiae mitochondrial TOM complex, four related proteins similar to one of these were present in the hydrogenosome proteome (Table 2, Supplementary Table S1). These proteins share the porin-3 domain with Tom40 based on Pfam domain predictions (http://pfam.sanger.ac.uk). Caution is required in interpreting this, however, as the Tom40 protein is a member of the Pfam PF01459 porin-3 superfamily. The presence of this domain in a membrane protein does not necessarily imply that it is involved in protein import, as these domains are also found in mitochondrial voltage-dependent anion channels (VDAC). Functional data will be necessary to demonstrate its identity. A single porin-3-domain containing protein was identified in both mitosome proteomes (Mi-Ichi et al., 2009; Jedelsky et al., 2011). In G. lamblia it is postulated to act as both a general import pore for proteins as well as serving in ion exchange (Jedelsky et al., 2011). In trypanosome mitochondria, a single porin-3 protein is also present, but has been demonstrated functionally to be a VDAC (Pusnik et al., 2009).
In addition to the porin-3 domain containing proteins, the hydrogenosome proteome contains a protein that was previously identified as Sam50 by Hidden Markov Modeling (Table 2, Supplementary Table S1; Dolezal et al., 2006). No Sam50 was identified in the G. lamblia or E. histolytica mitosome proteomes (Mi-Ichi et al., 2009; Jedelsky et al., 2011), however a Sam50-like gene is present in the E. histolytica genome (Loftus et al., 2005) and the protein it encodes has been localized to the mitosome in E. histolytica via immunofluoresence (Dolezal et al., 2010). Unlike that observed for the T. vaginalis hydrogenosome and the E. histolytica mitosome, it appears that the G. lamblia mitosome lacks a Sam50 as the gene is absent from its genome (http://giardiadb.org/giardiadb/).
The presence of proteins with weak similarity to Tom40 raises the possibility that a very divergent TOM complex exists; however, screening for additional members of the TOM complex failed to identify any candidates (Table 2). The absence of TOM complex members suggests that the translocase machinery in the hydrogenosome may be distinct from that found in mitochondria or that similarity is insufficient for identification by bioinformatics. Biochemical analyses will therefore be critical in defining these complexes. We have previously described an abundant hydrogenosomal integral membrane protein of approximately 35 kDa, Hmp35 (TVAG_104250), found in a stable ∼300 kDa complex, and with characteristics of a translocation pore (Dyall et al., 2003). This protein and two similar proteins, TVAG_031860 and TVAG_216170, were identified in the hydrogenosome proteome (Supplementary Table S1). The Hmp35 complex may function as an outer membrane protein translocase, acting in addition to or in place of Tom40 in these organelles.
In mitochondria, proteins are transferred from the TOM complex and across the IMS by soluble chaperone complexes (small translocase of the inner membrane (TIM) proteins), which include the Tim9-Tim10 and Tim8-Tim13 complexes (Neupert and Herrmann, 2007; Lithgow and Schneider, 2010). No small Tim proteins similar to those found in mitochondria IMS were found in the hydrogenosome proteome, nor have any been identified in the mitosome proteomes. The absence of these proteins in the hydrogenosome may reflect the paucity of inner membrane proteins found in hydrogenosomes, large sequence divergence in the these small Tims, or they may simply be low abundance in hydrogenosomes. Their absence in mitosomes likely reflects the extreme reduction that has occurred in these organelles.
Two TIM complexes, the Tim23 complex and the Tim22 complex, exist in yeast mitochondria (Neupert and Herrmann, 2007; Lithgow and Schneider, 2010). The Tim23 complex mediates the translocation or membrane insertion of preproteins containing an N-terminal presequence (Neupert and Herrmann, 2007; Lithgow and Schneider, 2010). Matrix preprotein translocation is further assisted by the presequence translocase-associated motor (PAM) complex, which is localized on the matrix side of the Tim23 complex (Neupert and Herrmann, 2007; Lithgow and Schneider, 2010). The Tim22 complex mediates the insertion of inner membrane proteins containing internal signal sequences (Neupert and Herrmann, 2007; Lithgow and Schneider, 2010). Tim22, Tim17 and Tim23 are evolutionarily related and are grouped into the Tim17/22/23 protein family (Pfam PF02466).
Several putative homologues of the TIM and PAM complexes were identified in the hydrogenosomal proteome (Table 2, Supplementary Tables S1 and S5). Three putative homologues of the Tim 17/22/23 family are present in the T. vaginalis genome and all three were found in the hydrogenosome proteome, together with a Tim44 homolog and Pam16 (Dolezal et al., 2006). Together with the previously identified Pam18 like protein (Dolezal et al., 2005), many of the members of the Tim23 inner membrane complex and PAM complex have been identified via sequence similarity in T. vaginalis, although the similarity is very low (Dolezal et al., 2006). Again, further investigation will determine whether these putative hydrogenosomal translocase proteins are functionally equivalent to their mitochondrial counterparts, including whether they form separate complexes with different roles in substrate import.
This is in sharp contrast to G. lamblia mitosomes, which contain no Tim17/22/23 homologs (Table 2; Jedelsky et al., 2011). These organelles do however contain conserved Pam16 and Pam 18 proteins, as well as Hsp70, indicating that the motor complex is well-conserved evolutionarily across phyla and organelle types, despite considerable diversification (Table 2; Jedelsky et al., 2011). The mitosome of E. histoytica has only Hsp70 and entirely lacks any proteins with homology to Tim17/22/23 or Pam 16/18 (Table 2; Mi-Ichi et al., 2009).
3.7.2. Carrier family proteins
Another abundant membrane protein, Hmp31 (TVAG_237680), which is phylogenetically related to the mitochondrial carrier protein family (MCF), and two Hmp31 paralogs were present in the proteome (Table 1; Dyall et al., 2000). BLAST analyses indicate that these proteins, TVAG_051820 and TVAG_262210, share 49 and 41% sequence identity at the amino acid level and 69 and 62% sequence similarity, respectively, with Hmp31, although their function has not been assessed. Three additional MCF proteins were also found. This protein family is responsible for transporting a variety of molecules, including ADP/ATP, glutamate, thiamine pyrophosphate and succinate/fumurate, across the mitochondrial inner membrane (Kunji, 2004). Compared with the MCFs in Arabidopsis thaliana (45) and yeast (35), there are relatively few MCFs in T. vaginalis, most likely due to the fewer metabolites produced by a reduced number of metabolic pathways in the hydrogenosome. Further investigation will be required to determine whether these proteins can function as MCFs, as possible aspartate/glutamate or oxodicarboxylate carriers or as a malate-aspartate shuttle system.
Both the G. lamblia and E. histolytica mitosomes contain transporters. Giardia lamblia contains seven; three members of the major facilitator superfamily and four ABC transporter family proteins (Jedelsky et al., 2011). The E. histolytica mitosome is further reduced, containing only four distinct transporter proteins; an ADP/ATP transporter, a sodium/sulfate symporter, an ABC transporter and a phosphate transporter (Chan et al., 2005; Mi-Ichi et al., 2009; Dolezal et al., 2010).
3.8. Characterization of the hydrogenosomal presequence
Many hydrogenosomal matrix proteins contain cleavable N-terminal presequences thought to be necessary for targeting proteins to the organelle (Johnson et al., 1990, 1993; Lahti et al., 1992, 1994; Lange et al., 1994; Hrdy and Muller, 1995a; Bui and Johnson, 1996; Bradley et al., 1997; Mentel et al., 2008). Analyses of a limited subset of presequences revealed that they typically contain hydroxylated amino acids (Ser and Thr) and Leu residues, and a conservation of an Arg residue at the -2 or -3 position relative to the cleavage site; these features are also common in mitochondrial presequences, although hydrogenosomal presequences are typically much shorter than those of mitochondrial targeted proteins (Dyall and Johnson, 2000). Several mitosome-targeted proteins also contain presequences; these are even shorter than those found on hydrogenosome-bound proteins but also contain hydrophobic amino acids (Burri and Keeling, 2007).
A search of the T. vaginalis genome for genes encoding putative hydrogenosomal matrix proteins identified 138 proteins with a predicted presequence, 53 of which encode hypothetical proteins (Carlton et al., 2007). Putative sequence motifs compiled from these data identified the motifs M(L/I)(S/T/A/C/G/Q/K/N) or M(S/T)(L/I) at the N-terminus and R(N/F/XF/S/A/I/E/G/Q/Y/XY) within the first 20 amino acid residues of the protein. These motifs were used to search the hydrogenosome proteome. Ninety-five of the 569 proteins (81 known and 14 hypothetical) had matching N-terminal sequences (Supplementary Table S8). These data indicate that either the majority of hydrogenosomal proteins do not possess a presequence or that the parameters used to define the presequence are too stringent. Proteins have been demonstrated to be targeted to the T. vaginalis hydrogenosome in the absence of an N-terminal presequence and only a few putative presequences have been experimentally validated as targeting signals, leaving the role of a presequence unclear (Mentel et al., 2008). It is notable that ∼ 1/3 of mitochondrial matrix proteins lack an identifiable targeting signal and many mitosome targeting signals appear to be dispensable (Gakh et al., 2002; Burri and Keeling, 2007). An explanation for the absence of presequences is the presence of yet-to-be-defined internal signal sequences for matrix-targeted proteins. The combination of bioinformatics and proteomics will be helpful in teasing out the requirements of protein translocation, possibly redefining the presequence motif and identifying putative internal targeting sequences.
A comparison of the predicted presequences of paralogous proteins in the proteome revealed that while some contain the predicted Arg at the -2 or -3 position, others have a Lys residue at this position. This exchange was found in IscS, Hsc20, flavodoxin protein (Wrba-like), MRP protein and in ALT paralogs (Supplementary Table S8). A presequence search substituting a Lys for Arg in the motif above identified 18 additional proteins with putative presequences. The binding pocket of the β subunit of the hydrogenosomal processing peptidase (HPP) which binds presequence substrates is large enough to accommodate varied presequences and should permit a Lys residue to be exchanged for an Arg residue (Brown et al., 2007; Smid et al., 2008). Experimental data testing targeting and cleavage of such a presequence will be necessary to determine whether a Lys at the -2 or -3 position is permitted.
To determine an average length and amino acid composition of predicted hydrogenosomal presequences and the residues immediately downstream of the cleavage site we performed a LOGOs analysis on the first 25 amino acids of these proteins containing a predicted presequence with either Arg or Lys at the -2 or -3 position (Supplementary Fig. S1 A) (http://weblogo.berkeley.edu/logo.cgi). The average length of the putative hydrogenosomal presequence was found to be 8-10 amino acids, with 84% of the presequences being ≤14 residues in length (Supplementary Fig. S1A). Hydroxylated residues, specifically Ser, and hydrophobic residues such as Ala, Phe, Ile and Leu were common in the first seven residues. The majority of the N-terminal presequences also started with MLS, which is strikingly similar to the consensus N-terminus of targeting signals in T. thermophila mitochondrial matrix protein presequences (MLSK) (Smith et al., 2007). The aligned predicted cleavage sites reveal that Phe and Asn are preferred at the -1 position relative to the cleavage site and hydrophobic and hydroxylated residues are preferred at positions +1 and +2 (Supplementary Fig. S1B).
3.9. Identification of externally associated proteins
The purified hydrogenosome fractions used to determine the proteome were not subjected to mild protease treatment in order to remove proteins associated with the outer surface of the organelle, thus allowing for the identification of hydrogenosome-associated proteins. Approximately 22% of the proteome (124 proteins) are likely to be externally associated based on the assigned localization and properties of these proteins in other organisms (Supplementary Table S2). This group includes proteins that share sequence similarity with glycolytic enzymes, ribosomal proteins, cytosolic chaperones and cytoskeletal proteins (Fig. 1). Similar externally associated proteins are also found in mitochondrial proteomes (Giege et al., 2003; MacKenzie and Payne, 2004; Ohlmeier et al., 2004; Anesti and Scorrano, 2006; Brandina et al., 2006; Graham et al., 2007). Moreover, the presence of cytoskeletal proteins in the hydrogenosomal proteome is expected as hydrogenosomes can be seen closely associated with components of the cytoskeleton in electron micrographs (Benchimol, 2009).
Although the presence of glycolytic enzymes may have been expected, the presence of the entire glycolytic pathway in this proteome is unprecedented (Supplementary Table S2). Glycolytic enzymes were reported in multiple mitochondrial proteomes including those of A. thaliana (Heazlewood et al., 2004), S. cerevisiae (Reinders et al., 2006), human heart (Taylor et al., 2003) and T. thermophila (Smith et al., 2007), although typically only a few members of the pathway were found. In contrast, four of the eight glycolytic enzymes in the hydrogenosomal proteome are found in multiple isoforms (Pi-dependent fructose 6-P 1-phosphotransferase, pyruvate dikinase, phosphofructokinase and pyruvate kinase) (Supplementary Table S2). Trichomonas vaginalis genes encoding both pyrophosphate and ATP-dependent glycolytic enzymes have been previously documented (Slamovits and Keeling, 2006) and the presence of enzymes of both types in the proteome indicate their co-localization within the cell. The presence of glycolytic proteins in mitochondrial proteomes was originally thought to be contamination, however recent studies have shown that glycolytic proteins form complexes on the surface of these organelles (Giege et al., 2003; Graham et al., 2007). Our data indicate that this may also be the case for the hydrogenosome.
Ribosome and ribosome-associated proteins were found to constitute 11% of the proteome (Fig. 1, Supplementary Table S2). The association of ribosomal proteins with the hydrogenosome has not been previously noted, however mammalian mitochondria are known to associate with cytosolic ribosome populations (MacKenzie and Payne, 2004). It is notable that the T.vaginalis genome contains unusually large ribosomal protein gene families, the expression of which may contribute to the large number of ribosomal proteins associated with hydrogenosomes (Lal et al., 2005).
Another group of proteins that appear to associate with the hydrogenosome are small GTPase proteins (Fig. 1, Supplementary Table S1). GTPase and associated proteins represent 24% of the hydrogenosomal proteome, which is a significantly higher proportion than the 6% found in the yeast mitochondrial proteome (Sickmann et al., 2003). In mitochondria GTPases and associated proteins are known to be involved in fusion, fission, motility and maintenance of inner membrane architecture (Rube and van der Bliek, 2004; McBride et al., 2006). For example, the mitochondrial Rab protein, Ypt11p, has been shown to be involved in mitochondrial retention and inheritance and dynamin related proteins (DRPs) are involved in fission (Frederick et al., 2004; Rube and van der Bliek, 2004; Boldogh and Pon, 2007). It is possible that these hydrogenosomal proteins carry out similar functions. However, with the sheer number of GTPases identified it is also likely that they have other unknown roles or are merely organelle-associated and do not function within the hydrogenosome.
3.10. Conclusions
The work described here has greatly added to our knowledge of the hydrogenosome of T. vaginalis. Previously thought to play a role in only a few metabolic pathways, these analyses indicate the T. vaginalis hydrogenosome to be a far more complex organelle. In-depth phylogenetic analyses of pathways identified in the proteome should allow for a better understanding of the evolution of this organelle. Determining whether these proteins and pathways also exist in the hydrogenosomes of fungi and ciliates will provide a better understanding of the evolution of these divergent organelles. Currently, proteomes for the mitosomes of both G. lamblia and E. histolytica are available (Mi-Ichi et al., 2009; Jedelsky et al., 2011). These proteomes indicate that the mitosome family of mitochondrial-related organelles are substantially more reduced than the hydrogenosome. Further studies made possible by comparison of organellar proteomes will serve to broaden our knowledge of the various functions and biogenetic properties that both unify and differentiate eukaryotic organelles.
Supplementary Material
Supplementary Fig S1. Analysis of N-terminal presequences identified from the Trichomonas vaginalis hydrogenosome proteome. A) LOGOs analysis of hydrogenosomal presequences. A dataset comprising the first 25 amino acid residues of 113 putative presequences from proteins in the hydrogenosome proteome were compared by LOGOs analysis. B) LOGOs analysis of the cleavage site. A dataset including predicted cleavage sites of 113 putative presequence-containing proteins from the proteome were compared by LOGOs analysis. LOGOs program is available at http://weblogo.berkeley.edu/logo.cgi.
Acknowledgments
We thank members of our laboratory for helpful discussions. This work was funded by the National Institutes of Health (NIH), USA grant (R37 AI027587) to PJJ, a NIH Microbial Pathogenesis Training Grant (2-T32-AI-007323) to AMS and RDH and a NIH Kirschstein-NRSA Fellowship (F32-AI080084) to AMS. The UCLA Mass Spectrometry and Proteomics Technology Center, USA was established with a grant from the W.M. Keck Foundation, USA.
Footnotes
Note: Supplementary data is associated with this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agren D, Schnell R, Oehlmann W, Singh M, Schneider G. Cysteine synthase (CysM) of Mycobacterium tuberculosis is an O-phosphoserine sulfhydrylase: evidence for an alternative cysteine biosynthesis pathway in mycobacteria. J Biol Chem. 2008;283:31567–31574. doi: 10.1074/jbc.M804877200. [DOI] [PubMed] [Google Scholar]
- Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J Biol Chem. 2004;279:16863–16874. doi: 10.1074/jbc.M313314200. [DOI] [PubMed] [Google Scholar]
- Almeida CC, Romao CV, Lindley PF, Teixeira M, Saraiva LM. The role of the hybrid cluster protein in oxidative stress defense. J Biol Chem. 2006;281:32445–32450. doi: 10.1074/jbc.M605888200. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Hirt RP, Foster PG, Roger AJ. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol Biol. 2006;6:27. doi: 10.1186/1471-2148-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesti V, Scorrano L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2006;1757:692–699. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Benchimol M. Hydrogenosomes under microscopy. Tissue Cell. 2009;41:151–168. doi: 10.1016/j.tice.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Mitochondria on the move. Trends in Cell Biology. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Boxma B, de Graaf RM, van der Staay GWM, van Alen TA, Ricard G, Gabaldon T, van Hoek A, Moon-van der Staay SY, Koopman WJH, van Hellemond JJ, Tielens AGM, Friedrich T, Veenhuis M, Huynen MA, Hackstein JHP. An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- Boxma B, Voncken F, Jannink S, van Alen T, Akhmanova A, van Weelden SW, van Hellemond JJ, Ricard G, Huynen M, Tielens AG, Hackstein JH. The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase E. Mol Microbiol. 2004;51:1389–1399. doi: 10.1046/j.1365-2958.2003.03912.x. [DOI] [PubMed] [Google Scholar]
- Bradley PJ, Lahti CJ, Plumper E, Johnson PJ. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 1997;16:3484–3493. doi: 10.1093/emboj/16.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochimica et Biophysica Acta (BBA) -Bioenergetics. 2006;1757:1217–1228. doi: 10.1016/j.bbabio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Brown MT, Goldstone HMH, Bastida-Corcuera F, Delgadillo-Correa MG, McArthur AG, Johnson PJ. A functionally divergent hydrogenosomal peptidase with protomitochondrial ancestry. Mol Microbiol. 2007;64:1154–1163. doi: 10.1111/j.1365-2958.2007.05719.x. [DOI] [PubMed] [Google Scholar]
- Bui ET, Bradley PJ, Johnson PJ. A common evolutionary origin for mitochondria and hydrogenosomes. Proc Natl Acad Sci U S A. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui ET, Johnson PJ. Identification and characterization of [Fe]-hydrogenases in the hydrogenosome of Trichomonas vaginalis. Mol Biochem Parasitol. 1996;76:305–310. doi: 10.1016/0166-6851(96)02567-4. [DOI] [PubMed] [Google Scholar]
- Burri L, Keeling PJ. Protein targeting in parasites with cryptic mitochondria. Int J Parasitol. 2007;37:265–272. doi: 10.1016/j.ijpara.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UCM, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Muller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM, Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. Draft Genome Sequence of the Sexually Transmitted Pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KW, Slotboom DJ, Cox S, Embley TM, Fabre O, van der Giezen M, Harding M, Horner DS, Kunji ER, Leon-Avila G, Tovar J. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr Biol. 2005;15:737–742. doi: 10.1016/j.cub.2005.02.068. [DOI] [PubMed] [Google Scholar]
- Clemens DL, Johnson PJ. Failure to detect DNA in hydrogenosomes of Trichomonas vaginalis by nick translation and immunomicroscopy. Mol Biochem Parasitol. 2000;106:307–313. doi: 10.1016/s0166-6851(99)00220-0. [DOI] [PubMed] [Google Scholar]
- Coombs GH, Westrop GD, Suchan P, Puzova G, Hirt RP, Embley TM, Mottram JC, Muller S. The Amitochondriate Eukaryote Trichomonas vaginalis Contains a Divergent Thioredoxin-linked Peroxiredoxin Antioxidant System. J Biol Chem. 2004;279:5249–5256. doi: 10.1074/jbc.M304359200. [DOI] [PubMed] [Google Scholar]
- Delgadillo MG, Liston DR, Niazi K, Johnson PJ. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc Natl Acad Sci U S A. 1997;94:4716–4720. doi: 10.1073/pnas.94.9.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LS. The establishment of various trichomonads of animals and man in axenic cultures. J Parasitol. 1957;43:488–490. [PubMed] [Google Scholar]
- Dolezal P, Dagley MJ, Kono M, Wolynec P, Likic VA, Foo JH, Sedinova M, Tachezy J, Bachmann A, Bruchhaus I, Lithgow T. The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog. 2010;6:e1000812. doi: 10.1371/journal.ppat.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Dancis A, Lesuisse E, Sutak R, Hrdy I, Embley TM, Tachezy J. Frataxin, a conserved mitochondrial protein, in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2007;6:1431–1438. doi: 10.1128/EC.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Smid O, Rada P, Zubacova Z, Bursac D, Sutak R, Nebesarova J, Lithgow T, Tachezy J. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc Natl Acad Sci U S A. 2005;102:10924–10929. doi: 10.1073/pnas.0500349102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004a;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Dyall SD, Johnson PJ. Origins of hydrogenosomes and mitochondria: evolution and organelle biogenesis. Curr Opin Microbiol. 2000;3:404–411. doi: 10.1016/s1369-5274(00)00112-0. [DOI] [PubMed] [Google Scholar]
- Dyall SD, Koehler CM, Delgadillo-Correa MG, Bradley PJ, Plumper E, Leuenberger D, Turck CW, Johnson PJ. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol Cell Biol. 2000;20:2488–2497. doi: 10.1128/mcb.20.7.2488-2497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Lester DC, Schneider RE, Delgadillo-Correa MG, Plumper E, Martinez A, Koehler CM, Johnson PJ. Trichomonas vaginalis Hmp35, a putative pore-forming hydrogenosomal membrane protein, can form a complex in yeast mitochondria. J Biol Chem. 2003;278:30548–30561. doi: 10.1074/jbc.M304032200. [DOI] [PubMed] [Google Scholar]
- Dyall SD, Yan W, Delgadillo-Correa MG, Lunceford A, Loo JA, Clarke CF, Johnson PJ. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature. 2004b;431:1103–1107. doi: 10.1038/nature02990. [DOI] [PubMed] [Google Scholar]
- Embley TM. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos Trans R Soc B-Biol Sci. 2006;361:1055–1067. doi: 10.1098/rstb.2006.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Hirt RP. Early branching eukaryotes? Curr Opin Genet Dev. 1998;8:624–629. doi: 10.1016/s0959-437x(98)80029-4. [DOI] [PubMed] [Google Scholar]
- Frederick RL, McCaffery JM, Cunningham KW, Okamoto K, Shaw JM. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J Cell Biol. 2004;167:87–98. doi: 10.1083/jcb.200405100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ. Enzymes of Glycolysis Are Functionally Associated with the Mitochondrion in Arabidopsis Cells. Plant Cell. 2003;15:2140–2151. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill EE, Diaz-Trivino S, Barbera MJ, Silberman JD, Stechmann A, Gaston D, Tamas I, Roger AJ. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol Microbiol. 2007;66:1306–1320. doi: 10.1111/j.1365-2958.2007.05979.x. [DOI] [PubMed] [Google Scholar]
- Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, Delbac F, Vivares CP, Hirt RP, Lill R, Embley TM. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–628. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- Graham JWA, Williams TCR, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ. Glycolytic Enzymes Associate Dynamically with Mitochondria in Response to Respiratory Demand and Support Substrate Channeling. Plant Cell. 2007;19:3723–3738. doi: 10.1105/tpc.107.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda P, Guda C, Subramaniam S. Reconstruction of pathways associated with amino acid metabolism in human mitochondria. Genomics Proteomics Bioinformatics. 2007;5:166–176. doi: 10.1016/S1672-0229(08)60004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausler T, Stierhof YD, Blattner J, Clayton C. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur J Cell Biol. 1997;73:240–251. [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse H, Lipke J, Altmann T, Hofgen R. Molecular cloning and expression analyses of mitochondrial and plastidic isoforms of cysteine synthase (O-acetylserine(thiol)lyase) from Arabidopsis thaliana. Amino Acids. 1999;16:113–131. doi: 10.1007/BF01321531. [DOI] [PubMed] [Google Scholar]
- Hoa le TP, Nomura M, Kajiwara H, Day DA, Tajima S. Proteomic analysis on symbiotic differentiation of mitochondria in soybean nodules. Plant Cell Physiol. 2004;45:300–308. doi: 10.1093/pcp/pch035. [DOI] [PubMed] [Google Scholar]
- Horner DS, Foster PG, Embley TM. Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol Biol Evol. 2000;17:1695–1709. doi: 10.1093/oxfordjournals.molbev.a026268. [DOI] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Embley TM. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Hrdy I, Muller M. Primary structure and eubacterial relationships of the pyruvate:ferredoxin oxidoreductase of the amitochondriate eukaryote Trichomonas vaginalis. J Mol Evol. 1995a;41:388–396. [PubMed] [Google Scholar]
- Hrdy I, Muller M. Primary structure of the hydrogenosomal malic enzyme of Trichomonas vaginalis and its relationship to homologous enzymes. J Eukaryot Microbiol. 1995b;42:593–603. doi: 10.1111/j.1550-7408.1995.tb05913.x. [DOI] [PubMed] [Google Scholar]
- Jedelsky PL, Dolezal P, Rada P, Pyrih J, Smid O, Hrdy I, Sedinova M, Marcincikova M, Voleman L, Perry AJ, Beltran NC, Lithgow T, Tachezy J. The Minimal Proteome in the Reduced Mitochondrion of the Parasitic Protist Giardia intestinalis. PLoS ONE. 2011;6:e17285. doi: 10.1371/journal.pone.0017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PJ, d'Oliveira CE, Gorrell TE, Muller M. Molecular analysis of the hydrogenosomal ferredoxin of the anaerobic protist Trichomonas vaginalis. Proc Natl Acad Sci U S A. 1990;87:6097–6101. doi: 10.1073/pnas.87.16.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PJ, Lahti CJ, Bradley PJ. Biogenesis of the hydrogenosome in the anaerobic protist Trichomonas vaginalis. J Parasitol. 1993;79:664–670. [PubMed] [Google Scholar]
- Johnston VJ, Mabey DC. Global epidemiology and control of Trichomonas vaginalis. Curr Opin Infect Dis. 2008;21:56–64. doi: 10.1097/QCO.0b013e3282f3d999. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kunji ER. The role and structure of mitochondrial carriers. FEBS Lett. 2004;564:239–244. doi: 10.1016/S0014-5793(04)00242-X. [DOI] [PubMed] [Google Scholar]
- Lahti CJ, Bradley PJ, Johnson PJ. Molecular characterization of the alpha-subunit of Trichomonas vaginalis hydrogenosomal succinyl CoA synthetase. Mol Biochem Parasitol. 1994;66:309–318. doi: 10.1016/0166-6851(94)90157-0. [DOI] [PubMed] [Google Scholar]
- Lahti CJ, d'Oliveira CE, Johnson PJ. Beta-succinyl-coenzyme A synthetase from Trichomonas vaginalis is a soluble hydrogenosomal protein with an amino-terminal sequence that resembles mitochondrial presequences. J Bacteriol. 1992;174:6822–6830. doi: 10.1128/jb.174.21.6822-6830.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal K, Field MC, Carlton JM, Warwicker J, Hirt RP. Identification of a very large Rab GTPase family in the parasitic protozoan Trichomonas vaginalis. Mol Biochem Parasitol. 2005;143:226–235. doi: 10.1016/j.molbiopara.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Land KM, Delgadillo-Correa MG, Tachezy J, Vanacova S, Hsieh CL, Sutak R, Johnson PJ. Targeted gene replacement of a ferredoxin gene in Trichomonas vaginalis does not lead to metronidazole resistance. Mol Microbiol. 2004;51:115–122. doi: 10.1046/j.1365-2958.2003.03791.x. [DOI] [PubMed] [Google Scholar]
- Lange S, Rozario C, Muller M. Primary structure of the hydrogenosomal adenylate kinase of Trichomonas vaginalis and its phylogenetic relationships. Mol Biochem Parasitol. 1994;66:297–308. doi: 10.1016/0166-6851(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- Lindmark DG, Muller M. Superoxide dismutase in the anaerobic flagellates, Tritrichomonas foetus and Monocercomonas sp. J Biol Chem. 1974;249:4634–4637. [PubMed] [Google Scholar]
- Lindmark DG, Muller M, Shio H. Hydrogenosomes in Trichomonas vaginalis. J Parasitol. 1975;61:552–554. [Google Scholar]
- Lithgow T, Schneider A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos Trans R Soc Lond B Biol Sci. 2010;365:799–817. doi: 10.1098/rstb.2009.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillen N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri WA, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- MacKenzie JA, Payne RM. Ribosomes Specifically Bind to Mammalian Mitochondria via Protease-sensitive Proteins on the Outer Membrane. J Biol Chem. 2004;279:9803–9810. doi: 10.1074/jbc.M307167200. [DOI] [PubMed] [Google Scholar]
- Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2010;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 2001;382:1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mentel M, Zimorski V, Haferkamp P, Martin W, Henze K. Protein import into hydrogenosomes of Trichomonas vaginalis involves both N-terminal and internal targeting signals: a case study of thioredoxin reductases. Eukaryot Cell. 2008;7:1750–1757. doi: 10.1128/EC.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-Ichi F, Yousuf MA, Nakada-Tsukui K, Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A. 2009;106:21731–21736. doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Brown MT, McArthur AG, Johnson PJ. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2006a;5:2062–2071. doi: 10.1128/EC.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Sievers SA, Brown MT, Johnson PJ. Identification and biochemical characterization of serine hydroxymethyl transferase in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2006b;5:2072–2078. doi: 10.1128/EC.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. The hydrogenosome. J Gen Microbiol. 1993;139:2879–2889. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- Narcisi EM, Secor WE. In vitro effect of tinidazole and furazolidone on metronidazole-resistant Trichomonas vaginalis. Antimicrob Agents Chemother. 1996;40:1121–1125. doi: 10.1128/aac.40.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Ohlmeier S, Kastaniotis AJ, Hiltunen JK, Bergmann U. The Yeast Mitochondrial Proteome, a Study of Fermentative and Respiratory Growth. J Biol Chem. 2004;279:3956–3979. doi: 10.1074/jbc.M310160200. [DOI] [PubMed] [Google Scholar]
- Page-Sharp M, Behm CA, Smith GD. Tritrichomonas foetus and Trichomonas vaginalis: the pattern of inactivation of hydrogenase activity by oxygen and activities of catalase and ascorbate peroxidase. Microbiology. 1996;142:207–211. doi: 10.1099/13500872-142-1-207. [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Charriere F, Maser P, Waller RF, Dagley MJ, Lithgow T, Schneider A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- Putignani L, Tait A, Smith HV, Horner D, Tovar J, Tetley L, Wastling JM. Characterization of a mitochondrion-like organelle in Cryptosporidium parvum. Parasitology. 2004;129:1–18. doi: 10.1017/s003118200400527x. [DOI] [PubMed] [Google Scholar]
- Putz S, Dolezal P, Gelius-Dietrich G, Bohacova L, Tachezy J, Henze K. Fe-hydrogenase maturases in the hydrogenosomes of Trichomonas vaginalis. Eukaryot Cell. 2006;5:579–586. doi: 10.1128/EC.5.3.579-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz S, Gelius-Dietrich G, Piotrowski M, Henze K. Rubrerythrin and peroxiredoxin: two novel putative peroxidases in the hydrogenosomes of the microaerophilic protozoon Trichomonas vaginalis. Mol Biochem Parasitol. 2005;142:212–223. doi: 10.1016/j.molbiopara.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Regoes A, Zourmpanou D, Leon-Avila G, van der Giezen M, Tovar J, Hehl AB. Protein import, replication, and inheritance of a vestigial mitochondrion. J Biol Chem. 2005;280:30557–30563. doi: 10.1074/jbc.M500787200. [DOI] [PubMed] [Google Scholar]
- Rehse PH, Ohshima N, Nodake Y, Tahirov TH. Crystallographic structure and biochemical analysis of the Thermus thermophilus osmotically inducible protein C. J Mol Biol. 2004;338:959–968. doi: 10.1016/j.jmb.2004.03.050. [DOI] [PubMed] [Google Scholar]
- Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- Richards TA, van der Giezen M. Evolution of the Isd11-IscS complex reveals a single alpha-proteobacterial endosymbiosis for all eukaryotes. Mol Biol Evol. 2006;23:1341–1344. doi: 10.1093/molbev/msl001. [DOI] [PubMed] [Google Scholar]
- Rube DA, van der Bliek AM. Mitochondrial morphology is dynamic and varied. Mol Cell Biochem. 2004;256-257:331–339. doi: 10.1023/b:mcbi.0000009879.01256.f6. [DOI] [PubMed] [Google Scholar]
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Shiflett AM, Johnson PJ. Mitochondrion-related organelles in eukaryotic protists. Annu Rev Microbiol. 2010;64:409–429. doi: 10.1146/annurev.micro.62.081307.162826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamovits CH, Keeling PJ. Pyruvate-Phosphate Dikinase of Oxymonads and Parabasalia and the Evolution of Pyrophosphate-Dependent Glycolysis in Anaerobic Eukaryotes. Eukaryotic Cell. 2006;5:148–154. doi: 10.1128/EC.5.1.148-154.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smid O, Matuskova A, Harris SR, Kucera T, Novotny M, Horvathova L, Hrdy I, Kutejova E, Hirt RP, Embley TM, Janata J, Tachezy J. Reductive evolution of the mitochondrial processing peptidases of the unicellular parasites Trichomonas vaginalis and Giardia intestinalis. PLoS Pathog. 2008;4:e1000243. doi: 10.1371/journal.ppat.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Robinson AJ. MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data. Mol Cell Proteomics. 2009;8:1324–1337. doi: 10.1074/mcp.M800373-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Gawryluk RM, Spencer DF, Pearlman RE, Siu KW, Gray MW. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J Mol Biol. 2007;374:837–863. doi: 10.1016/j.jmb.2007.09.051. [DOI] [PubMed] [Google Scholar]
- Smutna T, Goncalves VL, Saraiva LM, Tachezy J, Teixeira M, Hrdy I. Flavodiiron protein from Trichomonas vaginalis hydrogenosomes: the terminal oxygen reductase. Eukaryot Cell. 2009;8:47–55. doi: 10.1128/EC.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutak R, Dolezal P, Fiumera HL, Hrdy I, Dancis A, Delgadillo-Correa M, Johnson PJ, Muller M, Tachezy J. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc Natl Acad Sci U S A. 2004;101:10368–10373. doi: 10.1073/pnas.0401319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy J, Sanchez LB, Muller M. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol Biol Evol. 2001;18:1919–1928. doi: 10.1093/oxfordjournals.molbev.a003732. [DOI] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- Tovar J, Leon-Avila G, Sanchez LB, Sutak R, Tachezy J, van der Giezen M, Hernandez M, Muller M, Lucocq JM. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Cox S, Tovar J. The iron-sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol Biol. 2004;4:7. doi: 10.1186/1471-2148-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grinsven KW, Rosnowsky S, van Weelden SW, Putz S, van der Giezen M, Martin W, van Hellemond JJ, Tielens AG, Henze K. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J Biol Chem. 2008;283:1411–1418. doi: 10.1074/jbc.M702528200. [DOI] [PubMed] [Google Scholar]
- Viklund H, Bernsel A, Skwark M, Elofsson A. SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology. Bioinformatics. 2008;24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- Viscogliosi E, Delgado-Viscogliosi P, D G, Dauchez M, Gratepanche S, Alix AJ, Dive D. Cloning and expression of an iron-containing superoxide dismutase in the parasitic protist, Trichomonas vaginalis. FEMS Microbiol Lett. 1998;161:115–123. doi: 10.1111/j.1574-6968.1998.tb12936.x. [DOI] [PubMed] [Google Scholar]
- Westrop GD, Goodall G, Mottram JC, Coombs GH. Cysteine biosynthesis in Trichomonas vaginalis involves cysteine synthase utilizing O-phosphoserine. J Biol Chem. 2006;281:25062–25075. doi: 10.1074/jbc.M600688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Health Organization, Global Prevalence and Incidence of Selected Curable Sexually Transmitted Infections. 2001 www.who.int/docstore/hiv/GRSTI/006.htm. [PubMed]
- Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig S1. Analysis of N-terminal presequences identified from the Trichomonas vaginalis hydrogenosome proteome. A) LOGOs analysis of hydrogenosomal presequences. A dataset comprising the first 25 amino acid residues of 113 putative presequences from proteins in the hydrogenosome proteome were compared by LOGOs analysis. B) LOGOs analysis of the cleavage site. A dataset including predicted cleavage sites of 113 putative presequence-containing proteins from the proteome were compared by LOGOs analysis. LOGOs program is available at http://weblogo.berkeley.edu/logo.cgi.


