Summary
The enzyme flavin reductase 1 (FR1) from Trichomonas vaginalis, formerly known as NADPH oxidase, was isolated and identified. Flavin reductase is part of the antioxidative defense in T. vaginalis and indirectly reduces molecular oxygen to hydrogen peroxide via free flavins. Importantly, a reduced or absent flavin reductase activity has been reported in metronidazole-resistant T. vaginalis, resulting in elevated intracellular oxygen levels and futile cycling of metronidazole. Interestingly, FR1 has no close homologue in any other sequenced genome, but seven full-length and three truncated isoforms exist in the T. vaginalis genome. However, out of these, only FR1 has an affinity for flavins, i.e. FMN, FAD, and riboflavin, which is high enough to be of physiological relevance. Although there are no relevant changes in the gene sequence or any alterations of the predicted FR1-mRNA structure in any of the strains studied, FR1 is not expressed in highly metronidazole-resistant strains. Transfection of a metronidazole-resistant clinical isolate (B7268), which does not express any detectable amounts of FR, with a plasmid bearing a functional FR1 gene nearly completely restored metronidazole sensitivity. Our results indicate that FR1 has a significant role in the emergence of metronidazole resistance in T. vaginalis.
Keywords: Trichomonas vaginalis, flavin reduction, oxygen scavenging, metronidazole resistance
Introduction
The microaerophilic protist Trichomonas vaginalis is a ubiquitously occurring human parasite which can cause severe vaginitis or urethritis (Petrin et al., 1998). In some cases, T. vaginalis infection can lead to preterm deliveries or even miscarriages in infected pregnant women (Petrin et al., 1998). Since trichomoniasis is the most frequent non-viral sexually-transmitted disease, T. vaginalis can be defined as a major human pathogen. In practice, treatment is exclusively based on 5-nitroimidazole drugs, i.e. either on metronidazole or tinidazole (Upcroft and Upcroft, 2001). Although metronidazole was introduced for the treatment of T. vaginalis infections already more than 50 years ago, it has remained a highly reliable drug up to the present day and drug resistance is still comparably rare (Schwebke and Barrientes, 2006). However, in some parts of the world, such as Papua New Guinea, resistance rates can exceed 15% (Upcroft et al., 2009). In most cases, drug resistance can be overcome by higher doses of metronidazole or by prescribing tinidazole instead (Sobel et al., 2001), but not always (Goldman et al., 2009). Therefore, metronidazole resistance in T. vaginalis constitutes a serious problem because alternative treatments regimens are rather ineffective.
Metronidazole and other 5-nitroimidazoles are prodrugs and need to be reduced at their nitro groups in order to become toxic (Moreno and Docampo, 1985). Oxygen, however, interferes with nitroimidazole reduction by reoxidizing the nitroradical anion, i.e. the single electron transfer product, resulting in the restoration of the parent drug and formation of superoxide radical anions. Since this “futile cycle” (Mason and Holtzman, 1975) only occurs quantitatively in aerobic cells, 5-nitroimidazoles are comparably safe in humans but highly toxic to microaerophilic and anerobic organisms. However, it was shown that clinical metronidazole-resistant T. vaginalis isolates have a reduced oxygen scavenging activity (Yarlett et al., 1986a), resulting in elevated intracellular oxygen levels. As these strains are normally susceptible to the drug in the absence of oxygen (Meingassner and Thurner, 1979), this type of resistance has been termed “aerobic resistance”.
Two enzymatic pathways for the removal of oxygen have been described in T. vaginalis (Tanabe et al., 1979; Linstead and Bradley, 1988): an NADH-dependent oxidase which reduces oxygen to water and an NADPH-dependent oxidase, a flavin reductase, which reduces oxygen to hydrogen peroxide via FMN as cofactor. Interestingly, only flavin reductase enzyme activity has been found to be diminished or even absent in clinical metronidazole-resistant isolates (Ellis and Lloyd, 1992; Leitsch et al., 2012). Flavin reductase activity is also missing in a T. vaginalis C1 cell line (C1res) with high level metronidazole resistance induced in vitro (Leitsch et al., 2009). This strain displays “anaerobic metronidazole resistance” because the presence of oxygen is not a prerequisite for this type of resistance. Anaerobic resistance to metronidazole has been suggested to be based on defective metronidazole-activating pathways, including the pyruvate:ferredoxin oxidoreductase (PFOR) – ferredoxin couple (Yarlett et al., 1985; Yarlett et al., 1986b; Kulda et al., 1993) and thioredoxin reductase (Leitsch et al., 2009). Thus, aerobic and anaerobic metronidazole resistance to metronidazole seem to differ fundamentally but have in common that flavin reductase activity is diminished or lost, respectively.
The potentially very high importance of flavin reductase in the development of metronidazole resistance in T. vaginalis prompted us to isolate and identify this enzyme, and to characterize it in detail at the genetic and enzymatic level.
Results
Isolation and identification of flavin reductase
In our previous work (Leitsch et al., 2009, Leitsch et al., 2012), we had identified flavin reductase activity in T. vaginalis as a distinctive feature between metronidazole-sensitive and metronidazole-resistant strains (Table 1). In cell extracts of metronidazole-resistant strains, either diminished reduction or no reduction at all of free flavins (i.e. riboflavin, FAD, FMN) could be observed (Leitsch et al., 2012). We aimed at isolating the enzyme responsible for this activity and to perform a thorough genetic and enzymological analysis. A gel-based strategy for the isolation of flavin reductase was devised, based on the assumption that flavin reduction would not only result in the formation of fully reduced flavin (e.g. FMNH2) but also in the formation of flavin semiquinones (e.g. FMNH·) which can transfer an electron to oxygen to generate superoxide radical anions. The generation of superoxide, in turn, can be visualized with nitroblue tetrazolium (NBT) staining in polyacrylamide (PAA) gels. Gel extracts of nine T. vaginalis strains and one highly-metronidazole resistant cell line derived from strain C1 were prepared and aliquots of 40 μg per lane were loaded on native 12.5% PAA gels. After gel gel electrophoresis, NBT staining was performed and staining patterns were analyzed (Figure 1). Three major bands were discernible of which the lowest was either weak or missing altogether in metronidazole-resistant strains (Figure 1). Most conclusive was the absence of the lowest band in C1res, a highly metronidazole-resistant cell line derived from strain C1 in our laboratory several years ago (Leitsch et al., 2009). We hypothesized that the band contained flavin reductase, excised it from the lane corresponding to G3 cell extract, and submitted it for analysis and identification by reversed-phase liquid chromatography tandem mass spectrometry (RP-LC/MS/MS). In total, 12 proteins were positively identified in two separate runs in the NBT positive band (Supplementary Table 1), including a flavodoxin-like fold family protein of about 27 kDa size (GI:123365845 or TVAG_517010) with unknown function. The flavodoxin-like fold of TVAG_517010 suggested that this enzyme could be flavin reductase, so we expressed it recombinantly in E. coli, and assayed its function. Indeed, TVAG_517010 proved to be a potent flavin reductase (data not shown) when added to reaction buffer (100 mM Tris pH 7.5 buffer containing 200 μM NADPH and 10 μM FMN). Flavin reduction was measured as rate of NADPH oxidation as described before (Leitsch et al., 2009, Leitsch et al. 2012).
Table 1.
T. vaginalis strains used in this study. Summary of resistance status and FR activity, as determined previously (Leitsch et al., 2012).
| Strain | Resistance status | FR activity |
|---|---|---|
| C1 | Sensitive | High |
| C1res | Highly resistant (anaerobic) | None |
| G3 | Sensitive | Very High |
| JH31A | Sensitive | High |
| Tv2 | Reduced sensitivity | Low |
| IR78 | Intermediately resistant | Low |
| Fall River | Intermediately resistant | Low |
| CDC085 | Highly resistant (aerobic) | None |
| LA1 | Highly resistant (aerobic) | None |
| B7268 | Highly resistant (aerobic) | None |
Figure 1.
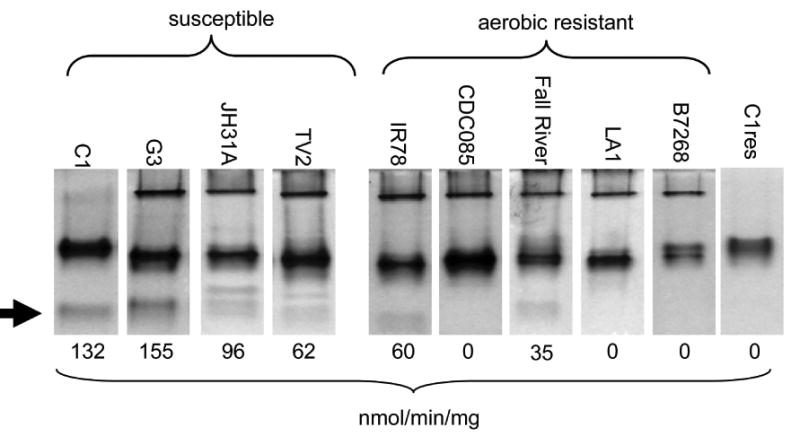
Flavin reductase activity in T. vaginalis gel extracts as visualized by NBT staining in native polyacrylamide gels (12.5% PAA). The lowest band, marked by an arrow, was missing or at least weaker in all metronidazole-resistant clinical isolates and the highly metronidazole-resistant C1 cell line, C1res. Fourty μg of protein were loaded per isolate. Flavin reductase activites as measured and published before (Leitsch et al., 2012) are given below the gels for comparison.
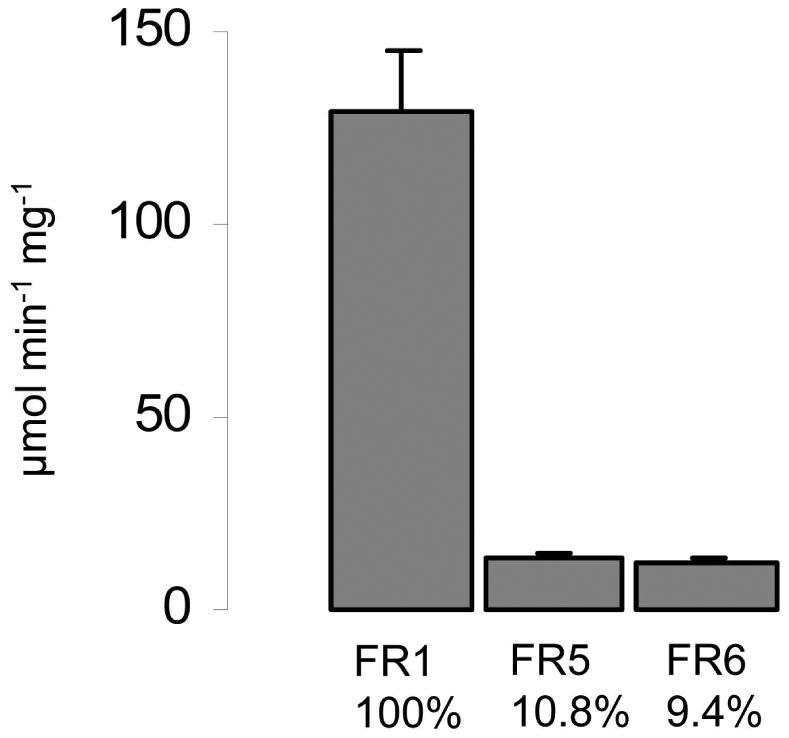
When we performed a BLAST search with the protein sequence of TVAG_517010, we found seven additional isoforms (Table 2) encoded in the T. vaginalis genome (Carlton et al., 2007). All isoforms were given the designation “flavin reductase” (FR). TVAG_517010 was named flavin reductase 1 (FR1), with the others designated FR 2-7 relative to their amino acid similarity to FR1 (Table 2). Further, three truncated isoforms are present in the genome (TVAG_445900, TVAG_502470, TVAG_502480). Interestingly, there are no close FR homologues in any other sequenced genome. The closest homologue in another organism is an NAD(P)H dehydrogenase from Streptomyces acidiscabies (WP_010360348) with a 28% amino acid sequence identity and 45% sequence similarity to FR1. Because of the abundance of flavin reductase homologs in the T. vaginalis genome, we sought to identify which protein(s) were possibly responsible for flavin reduction observed in the PAA/NBT gels. All full-length isoforms of TVAG_517010 were recombinantly expressed in E. coli for further characterization, with the exception of TVAG_144100 which was not taken into further consideration because it differs from TVAG_144070 only by one missense mutation, leading to the exchange of Ser124 to Gly124. Only FR5 and FR6 displayed relevant flavin reductase activity when 50 μM FMN were added as substrate, whereas FR2, 3, 4, and 7 displayed only marginal activity (Figure 2A) and were not further characterized. At a lower concentration of FMN (10 μM), FR5 and FR6 displayed a proportionately lower activity as compared to FR1 than observed with 50 μM, indicating that FR5 and FR6 have a weaker affinity to FMN than FR1 (Figure 2B).
Table 2.
Flavin reductases in T. vaginalis as revealed by a BLAST search with TVAG_517010. Indicated is percentage of amino acid (aa) identity when compared to the FR1 sequence.
| ID | Size (aa) | aa identity to FR1 | |
|---|---|---|---|
| FR1 | TVAG_517010 | 239 | 100% |
| FR2 | TVAG_311580 | 240 | 75% |
| FR3 | TVAG_407030 | 240 | 73% |
| FR4 | TVAG_406950 | 238 | 66% |
| FR5 | TVAG_144070 TVAG_144100 |
235 | 65% |
| FR6 | TVAG_009980 | 235 | 64% |
| FR7 | TVAG_127310 | 235 | 62% |
Figure 2.
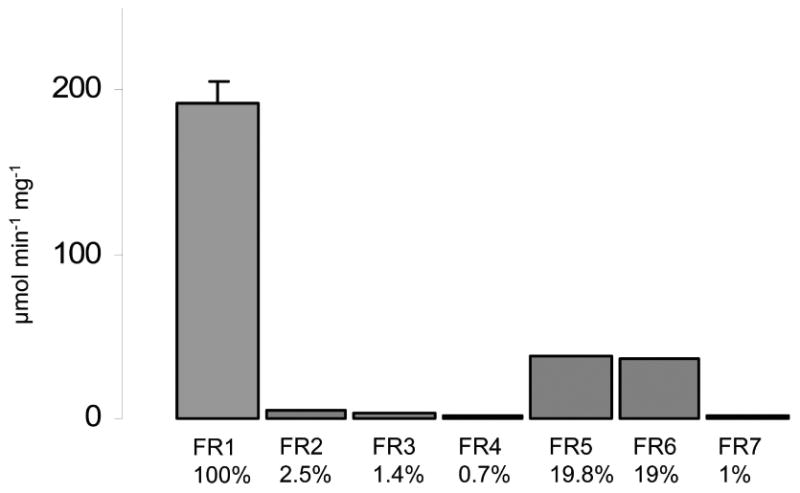
A, Activities of recombinantly expressed FR1-7. Activities in relation to FR1 are given below the bars in %. All measurements were performed with 1 μg ml-1 enzyme and 50 μM FMN in 100 mM potassium phosphate buffer, pH 5.5. All measurements with FR1, FR5 and FR6 were performed at least three times. Error bars indicate SEM.
B, Activities with of FR1, FR5, FR6 with 10 μM FMN. Activities in relation to FR1 are given below the bars in %. All measurements were performed with 1 μg ml-1 enzyme in 100 mM potassium phosphate buffer, pH 5.5. All measurements were performed at least three times. Error bars indicate SEM.
Kinetic parameters of flavin reductases 1, 5, and 6
To further characterise the flavin reductase activity of FR1, 5, and 6 we determined the kinetic parameters with the substrates riboflavin, FAD and FMN (Table 3). A relatively low enzyme concentration of 0.25 μg/ml was chosen because of the very high activity of the enzymes and because reduced flavins had to be re-oxidized in the buffer by oxygen in order to guarantee stable substrate levels. When enzyme concentrations of more than 1 μg/ml were applied, flavins in the enzyme buffer were not re-oxidized quickly enough and the assay buffer turned from yellow to colourless, indicating that no more substrate was present. Interestingly, maximum reduction rates (Vmax) for FMN were rather similar with all three isoforms (Table 3) but FR1 displayed a 10 to 20-fold higher affinity to FMN than FR5 and FR6. Also the affinity of FR1 to FAD was at least 10-fold higher than observed with FR5 and FR6 but the Vmax was considerably lower (roughly 33% of the Vmax for FMN). In contrast, FR5 and FR6 displayed higher affinity for riboflavin (Table 3) than FR1. Moreover, FR1 was inhibited by riboflavin (possibly either by the semiquinone or the fully reduced form) as reactions almost stopped after 2 min. This was not the case with FR5 and FR6. However, when small amounts of FMN (400 nM) were added to reactions, riboflavin was efficiently and rapidly reduced by FR1 suggesting that FMN is a loosely-bound, canonical cofactor of FR1. The same concentration of FAD did not have this effect on the riboflavin reduction rate of FR1 (not shown) and reduction rates of riboflavin by FR5 and FR6 were unaffected by 400 nM FMN (not shown). Interestingly, FR1 was found to be much more specific for NADPH than FR5 and FR6. Whereas FR1 displayed only 3% activity when NADH was added instead of NADPH (200 μM NADPH or NADH, 50 μM FMN, 1 μg/ml enzyme, 100 mM potassium phosphate pH 5.5, 37°C), FR5 and FR6 only showed a decrease in activity of 40 to 50% (data not shown). The Km of FR1 for NADPH (89 μM) was, nevertheless, rather high (Table 3).
Table 3.
Kinetic parameters of recombinant FR1, FR5, and FR6. ND…not determined.
| FR1 | FR5 | FR6 | |
|---|---|---|---|
| Km FMN [μM] | 10 ± 1.4 | 186.9 ± 30.3 | 160 ± 23.9 |
| Km FAD [μM] | 16.5 ± 2.5 | 267.4 ± 21.6 | 192.1 ± 12.2 |
|
Km riboflavin/(+ 400 nM FMN) [μM] |
148.6 ± 29.4 (22.5 ± 4.6) |
94.4 ± 11.5 | 97.5 ± 28.8 |
| Km NADPH [μM] | 89.4 ± 12.7 | ND | ND |
| Vmax FMN [μmol sec-1 mg-1] | 236. 4 ± 9,1 | 185.2 ± 12.2 | 170.8 ± 11.3 |
| Vmax FAD [μmol sec-1 mg-1] | 73.5 ± 3.8 | 163.8 ± 7 | 154.4 ± 4.6 |
|
Vmax riboflavin/(+ 400 nM FMN) [μmol sec-1 mg-1] |
136.3 ± 14.6 (129.3 ± 9.7) |
144.9 ± 8.2 | 158.7 ± 22 |
Taken together, it can be concluded that FR1 is responsible for the flavin reductase activity as observed before in T. vaginalis cell extracts (Leitsch et al., 2009; Leitsch et al., 2012) because FR5 and 6 have a Km for FMN too high in order to be relevant under the assay conditions which had been applied (10 μM FMN, Tris pH 7.5). Further, FR1 is far more specialized with regard to its function as an NADPH-dependent reductase of FMN than FR5 and FR6.
Determination of end products of FR1-mediated flavin reduction
Flavin reductase, originally designated as NADPH oxidase (Linstead and Bradley, 1988), had been found to produce hydrogen peroxide as an end product. In fact, this enzyme activity was identified as the major source of hydrogen peroxide in T. vaginalis (Chapman et al., 1999). We wanted to confirm these previous results with purified recombinant FR1 and measured hydrogen peroxide formation after addition of NADPH in the presence of 10 μM FMN via formation of iron-thiocyanate complexes in a colorimetric assay. Read-outs were compared with standards of known hydrogen peroxide concentrations. Hydrogen peroxide concentrations measured proved to be virtually equimolar to the NADPH concentrations applied (Figure 3), indicating that hydrogen peroxide is the main if not the single product of FR1. However, as NBT staining in gels (Figure 1) also suggested superoxide to be a product of FR1, we measured superoxide formation in a cytochrome c-coupled assay. Indeed, superoxide formation could be observed (30.7 ± 10.1 nmol mg-1 sec-1) when small amounts of enzyme (0.025 μg/ml) and low concentrations of FMN (400 nM) were applied. Superoxide dismutase was added to the enzyme buffer in order to distinguish between direct reduction of cytochrome c by the semiquinone FMNH· and reduction by superoxide which had been generated by the oxidation of FMNH· by oxygen. Our results are consistent with the previous observation (Chapman et al., 1999) that FR is a relevant source of hydrogen peroxide but also show that superoxide radical anions are a minor by-product of the reaction which allows visualization of the enzyme in PAA gels by NBT staining.
Figure 3.
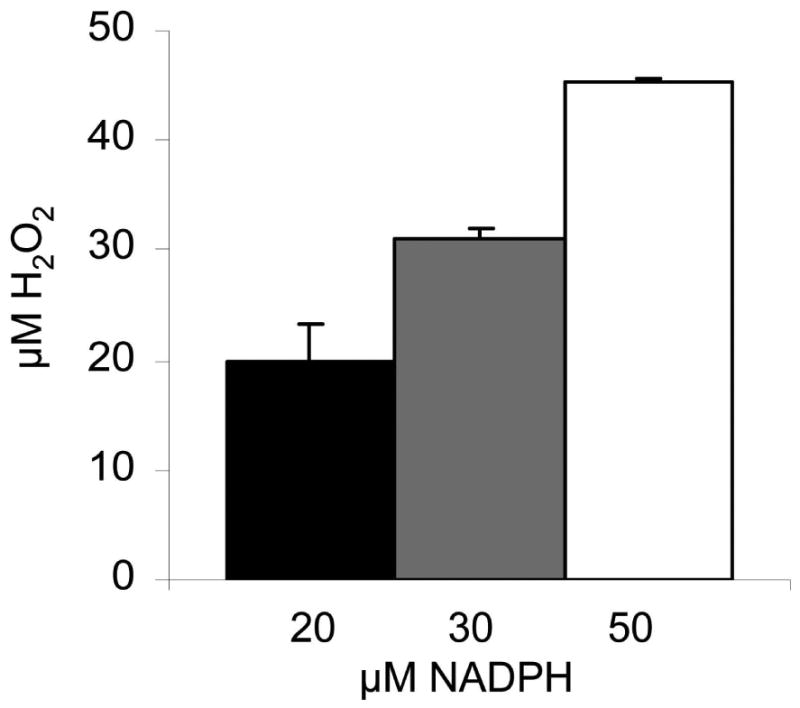
Twenty, 30, and 50 μM NADPH were oxidized by recombinant flavin reductase. Afterwards generated hydrogen peroxide was measured and compared to standards. Hydrogen peroxide is the only quantitative product of FR1, and NADPH is oxidized in amounts equimolar to the hydrogen peroxide generated. All measurements were performed three times. Error bars indicate SEM.
Measuring the activity and the expression of flavin reductases in T. vaginalis strains
In a previous study (Leitsch et al., 2012), we had found flavin reductase activity absent in highly-metronidazole resistant isolates. Interestingly, however, this finding was not fully supported by an earlier study by others (Ellis and Lloyd, 1992) in which one of the highly metronidazole-resistant strains, CDC085, had shown some residual flavin reductase activity when very high concentrations of FMN (100 μM) were used. Having tested all FRs for activity (Figure 2A), we speculated that this residual activity might be exerted by FRs different from FR1 and argued that under optimal assay conditions (as determined in this study, i.e. 100 mM potassium phosphate buffer, pH 5.5, as opposed to 100 mM Tris, pH 7.5) the much lower activities of other FR isoforms could be measurable. In analogy to our measurements with the purified recombinant FR isoforms (Figure 2), FR activity in strains G3, C1, Fall River, CDC085, LA1, B7268 and C1res was measured at two concentrations of FMN (10 and 100 μM) in order to obtain information on the affinity of the active FR(s) to the substrate in the respective strains. In accordance with the measured Km of FR1 to FMN (Table 3), reduction rate was slightly less than twice as high in the strains G3, C1 and Fall River when 100 μM instead of 10 μM FMN were used in the assay (Figure 4). Under the new assay conditions applied, low levels of FR activity could be indeed measured in CDC085 and LA1 (Figure 4), but not in C1res and B7268. In CDC085 and LA1, activity was three times higher with 100 μM FMN than with 10 μM FMN (Figure 4), indicating a higher Km for FMN than observed with FR1. Thus, these measurements strongly suggested that, in contrast to G3, C1, and Fall River, the FR isoforms expressed in CDC085 and LA1 are different from FR1 and that in B7268 and C1res FRs are not expressed at all.
Figure 4.
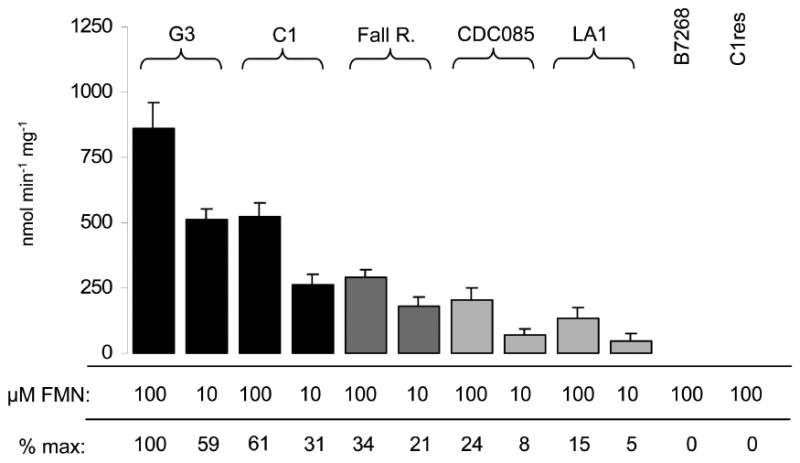
Flavin reductase activity in the fully metronidazole-suscpetible isolates G3 and C1 (black bars), the intermediately metronidazole-resistant isolate Fall River (dark grey bars), and the highly metronidazole-resistant isolates CDC085, LA1, B7268, and C1res (light grey bars) was measured at an FMN concentration of 100 μM or 10 μM, respectively. Percentages below the bars indicate FR activity in relation to FR activity in G3 extract with 100 μM FMN. Note that FR activity is approximately halved in G3, C1, and Fall River if 10 μM FMN are used instead of 100 μM. In CDC085 and LA1, FR activities are reduced to a third if 10 μM FMN instead of 100 μM are used, indicating a weaker affinity of the respective FR to FMN than observed in G3 and C1. B7268 and C1res do not display any FR activity. All measurements were performed at least three times with 20 μg ml-1 (mg protein)-1 extract and in 100 mM potassium phosphate buffer, pH 5.5. Error bars indicate SEM.
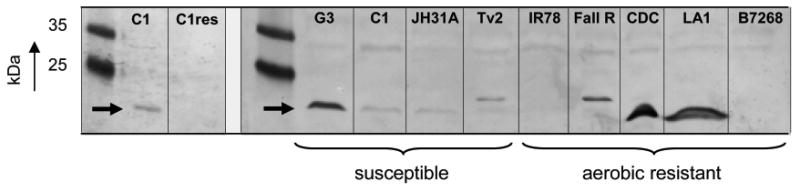
To confirm this result, polyclonal antiserum from rabbit was raised and used for the detection of recombinant FR1 in immunoblot analyses. However, due to the high similarity to the other FRs, the antiserum cross-reacted with all FRs (Figure 5A) which enabled us to check overall FR expression in the strains studied. It is important to note that the FR proteins migrated faster in the gel than suggested by their theoretical size (approximately 27 kDa). FR1 displayed a narrow double band, presumably caused by binding of FMN (molecular weight 456 Da), originating from E. coli during recombinant expression of the protein. Moreover, FR4-7 migrated slower in the gels than FR1 although being slightly smaller (Table 2). The reason for this is unclear but may be caused by structural differences of the proteins. When the anti-FR antiserum was tested on T. vaginalis cell extracts, the highly metronidazole-resistant cell line of strain C1, C1res (Figure 5B, left panel), and the clinical isolate B7268 were found to lack detectable FRs (Figure 5B, right panel). This corroborated our previous measurements of FR activity in these two strains as shown in Figure 4. Moreover, LA1 and CDC085, appear to express very high amounts of FRs which by far exceed the FR levels as observed in the metronidazole-susceptible isolates C1, G3, JH31A and Tv2 (Figure 5B). Thus, the residual FR activity found in CDC085 and LA1likely results from alternative FRs that are highly expressed. Since FR5 and FR6 have a Vmax for FMN that is quite similar to that of FR1 (Table 3), it is likely that either one of the remaining FRs, which have very low activities (Figure 2A), is expressed in CDC085 and LA1. Further, the location of the FR bands in CDC085 and LA1 suggests that FR2 and/or FR3 are expressed in these strains (Figure 5B) because FR4-7 show a different migration behaviour in PAA gels (Figure 5A). Tv2 and Fall River, both displaying diminished flavin reductase activity (Leitsch et al., 2012) (Figure 4), also expressed an alternative FR which, according to the positions of the recombinant FRs on the immunoblots (Figure 5A), are possibly either one of FR4-7 (Figure 5B). Another metronidazole-resistant isolate, IR78, expressed low but detectable amounts of FRs (Figure 5B). To summarize, expression of FR isoforms overall corresponded well to FR activities under optimal assay conditions (Figure 4), as neither the presence nor the activity of FRs was detectable in C1res and B7268. In the case of C1res this constitutes especially strong evidence for a correlation of FR1 expression and metronidazole resistance as C1res, a highly metronidazole-resistant cell line, is derived directly from the metronidazole-sensitive C1 which expresses FR1. As to strain IR78, the reduced FR activity observed (Leitsch et al., 2012) could be reconciled with a weaker band appearing in the immunoblots (Figure 5B). Also RT-qPCR was performed to quantify FR expression in all strains and to identify the alternative FRs expressed in strains Tv2, Fall River, CDC085 and LA1. However, due to the considerable biological variation of FR mRNA levels in the strains (Supplementary Figure 1), the experiment was considered inconclusive and, therefore, discontinued.
Figure 5.
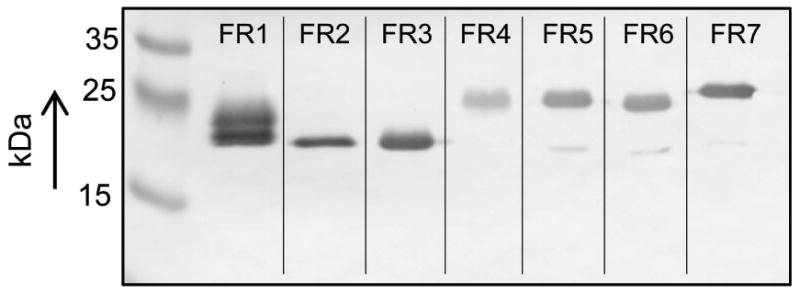
A, Anti-FR1 polyclonal antiserum (1:1000) was tested on 1 μg of purified recombinant FR1-7. All FRs cross-reacted with anti-FR1 antiserum. All FRs migrate faster in the gel than predicted by their nominal mass (approx. 27 kDa), FR1-3 were migrating faster in the gel than FR4-7.
B, Anti-FR1 polyclonal antiserum was used to detect FRs in gel extracts of T. vaginalis (100 μg protein/lane). As shown in the left panel, C1res does not express any FR. In the right panel, the other strains were tested: 1, G3; 2, C1; 3, JH31A; 4, Tv2; 5, IR78; 6, Fall River; 7, CDC085; 8, LA1; 9, B7268.
To check whether differences in FR activity and expression could be caused by mutations in the FR1 gene, the gene was sequenced in all strains and searched for mutations possibly causing reduced FR activity or even loss of FR activity in metronidazole-resistant strains. However, the FR1 gene sequences of all strains were almost identical, with only strains C1 and B7268 showing a single nucleotide exchange of G609 to T609, leading to the replacement of alanine to valine. As C1 displays distinct FR1 activity (Leitsch et al., 2012) (Figure 1 and Figure 4) this amino acid exchange does not substantially alter enzyme activity. Strain LA1 showed a single nucleotide exchange of T95 to A95 that does not affect the amino acid sequence. Further, 3′ RACE and 5′ RACE analyses were performed with FR1 mRNA from all strains to identify changes that could affect transcript stability. The 3′ UTR proved to be 47 bp long (without stop codon) and to be perfectly identical with regard to sequence in all strains (Supplementary Figure 2). The 5′ UTR was found to be four bases long (GTTC) in all strains (Supplementary Figure 2), with the exception of C1res and B7268 in which FR1 transcript numbers were so low that 5′ RACE was unsuccessful even after three rounds of PCR with nested primers.
NADH-oxidase activity is impaired in metronidazole-treated T. vaginalis
Trichomonas vaginalis has two different enzymatic pathways to remove intracellular oxygen, i.e. NADH oxidase (Tanabe, 1979; Linstead and Bradley, 1988) and flavin reductase. Since it is likely that clinical metronidazole resistance is caused by impaired oxygen scavenging leading to the reoxidation of activated metronidazole by intracellular oxygen (Mason and Holtzman, 1975), it remained an open question why metronidazole-resistant clinical isolates display impaired flavin reductase activity (Leitsch et al., 2012) but normal NADH oxidase activity (Müller and Gorrell, 1983; Rasoloson et al., 2001). We hypothesized that NADH oxidase might be impaired or deactivated by metronidazole, rendering T. vaginalis solely dependent on flavin reductase for the removal of intracellular oxygen. In order to test this hypothesis we treated the metronidazole-sensitive strain G3 and in cysteine-free growth medium with 200 μM metronidazole for 2h. The omission of cysteine, a potent oxygen scavenger, leads to a higher oxygen level in the medium which emulates “aerobic” test conditions. After this treatment, the parasites had been rendered immotile but were still intact (as judged by trypan blue staining) and could be used for assaying NADH oxidase and flavin reductase activities. Flavin reductase activity was hardly, if at all, affected. It should be noted that the measured flavin reductase activity in G3 was much higher than determined before (Leitsch et al., 2012) because the enzyme assays were performed under optimal buffer conditions (100 mM potassium phosphate, pH 5.5 as opposed to 100 mM Tris, pH 7.5). In contrast, NADH oxidase activity had been reduced to about 50% (Figure 6). Also in the metronidazole-resistant isolate CDC085, NADH oxidase activity was approximately halved after treatment with 200 μM metronidazole for 2h (Supplementary Figure 3). These results suggest that NADH oxidase is negatively affected by metronidazole and that in metronidazole-treated T. vaginalis, flavin reductase might be the only remaining major pathway for the removal of intracellular oxygen.
Figure 6.
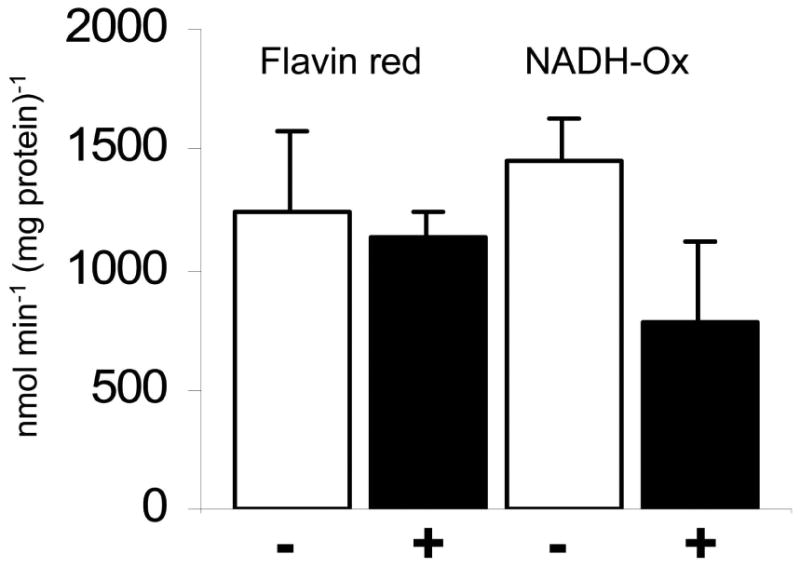
NADH-oxidase activity was measured in T. vaginalis G3 (metronidazole-sensitive), either untreated (-) or treated (+) with 200 μM metronidazole for two hours. Cells had been incubated in cysteine-free medium in order to guarantee elevated oxygen levels. For comparison, also flavin reductase activity was measured, either in absence (-) or presence (+) of metronidazole (200 μM, 2h). All measurements were repeated at least twice. Bars indicate SEM.
Transfection of the highly metronidazole-resistant isolate B7268 with the FR1 gene restores metronidazole susceptibility
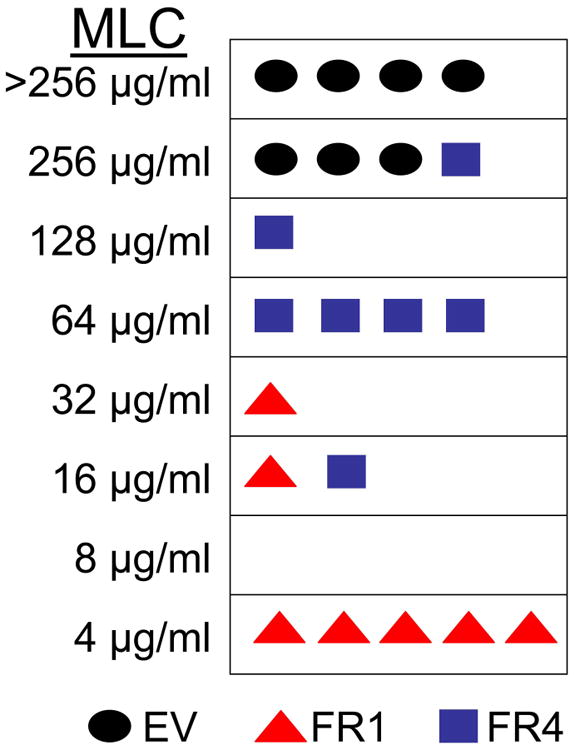
We hypothesized that re-introduction of FR1 into an aerobically-resistant strain would lead to restoration of metronidazole sensitivity. To test this, the highly-metronidazole resistant strain B7268 was transfected with a plasmid bearing either the FR1 or the FR4 gene or an empty vector control. B7268 had been found to not express any FR at the protein level (Figure 5B), rendering it an ideal candidate for the transfection experiment. Flavin reductase 4 was chosen as a control because it is all but inactive in vitro (Figure 2A). After electroporation, transfectants were selected with geneticin, assayed by western blot analysis for expression (Figure 7A), and used in metronidazole susceptibility assays as described before (Leitsch et al., 2012). The episomal expression of FR1 in B7268 almost restored full susceptibility to metronidazole whereas FR4 only had a slight effect on metronidazole sensitivity of B7268 (Figure 7B). This observation strongly argues that the lack of FR1 activity in B7268 leads to aerobic resistance.
Figure 7.
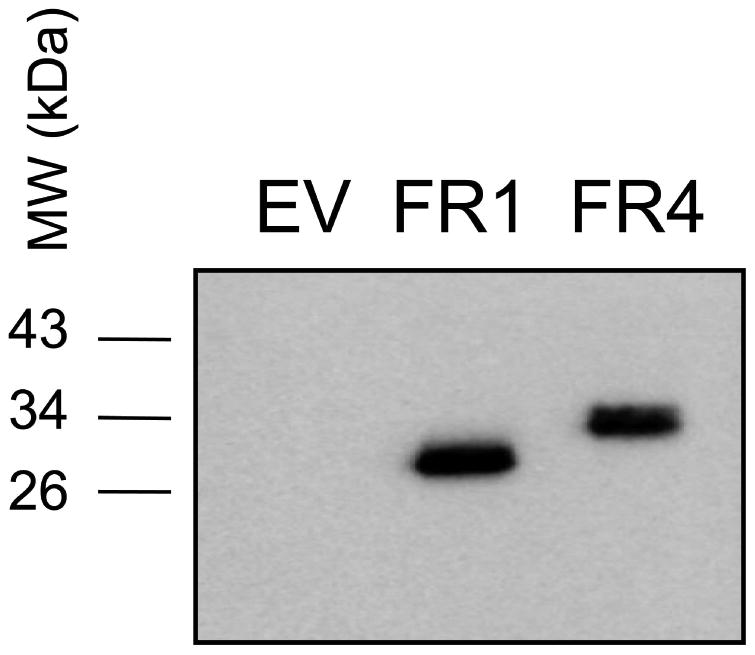
A, Western blot against T. vaginalis B7268 cell preparations with an anti-HA antibody (1:5000) which recognizes the HA-tag fused C-terminally to FR1 and FR4. Preparations of 1 × 104 parasites each were loaded on 17.5% SDS PAGE gels. EV, empty vector.
B, Summary of the metronidazole-susceptibility assays conducted with B7268 transfectants bearing the empty vector (EV, black circles), FR4 (blue squares), and FR1 (red triangles). Each shape represents one assay performed over 48 h. Seven independent experiments were performed for each of the transfectants. Each shape in the summary represents the minimum lethal concentration (MLC) determined for the transfectant in one of the experiments. The MLC was determined for each transfectant after 48 hours of incubation with a series of concentrations (0-256 μg/ml) of metronidzaole for each experiment. The range of MLCs is given alongside the column.
Discussion
In our pursuit of factors contributing to metronidazole resistance in T. vaginalis, we isolated flavin reductase 1 (FR1) and identified it as the enzyme responsible for the activity of reducing free flavins observed earlier in T. vaginalis cell extracts (Linstead and Bradley, 1988). This enzyme activity had attracted considerable interest because it is negatively affected in metronidazole-resistant T. vaginalis strains (Ellis and Lloyd, 1992; Leitsch et al., 2009; 2012). We found additional seven full-length (Table 2) and three truncated isoforms of the enzyme in the T. vaginalis genome. Highly redundant gene families are a common feature in the T. vaginalis genome (Carlton et al., 2007; Cui et al., 2007). Notably, only FR1 was found to have a Km for FMN and FAD which is low enough to likely be of physiological importance (Table 3). Flavin reductases 5 and 6 proved to be equally active (Table 3) but showed a Km for FMN and FAD one to two orders of magnitude above that observed with FR1. Given the presence of multiple FR genes in the T. vaginalis genome, with three being truncated, we speculate that FR2-7 are degenerative isoforms of FR1 which have lost their original function. It is presently impossible to determine whether FR1 is derived from a less specialized enzyme and whether the other isoforms are indeed degenerative forms of FR1, as closely related enzymes are absent from the sequenced genomes of other organisms. This is the more surprising as an according activity was also found in G. lamblia extracts (Ellis et al., 1993; Leitsch et al. 2011).
Our sequence analyses of the FR1 gene and its mRNA revealed no relevant mutations or shortened UTRs in any of the strains studied. This indicates that the loss or the diminished expression of FR1 in metronidazole-resistant strains is not lost due to deleterious mutations but, rather, down-regulation of expression. Consistent with this observation, FR activity is low or absent in resistant strains compared to sensitive strains (Figure 4). However, the underlying mechanisms remain to be studied.
The results of our transfection experiment (Figure 7B) provide strong support for the notion that the loss of FR1 activity is a major step in the development of metronidazole resistance (Leitsch et al., 2012). Two properties of FR1 activity could play a role in metronidazole resistance. First, FR1 removes intracellular oxygen by reducing it to hydrogen peroxide utilizing NADPH, and thus its loss may lead to increased intracellular oxygen concentrations, which in turn results in enforced redox-cycling, i.e. detoxification of activated metronidazole (reviewed in Kulda, 1999). Secondly, upon loss of FR1 hydrogen peroxide levels will strongly decrease as FR1 is probably the main source of hydrogen peroxide in T. vaginalis (Chapman et al., 1999). This is highly favourable because hydrogen peroxide is detrimental to T. vaginalis (Davis and Lushbaugh, 1993). The toxicity of hydrogen peroxide is further exacerbated by metronidazole which counteracts removal of hydrogen peroxide by diminishing the activity of thioredoxin reductase (TrxR) (Leitsch et al., 2009), an enzyme that activates thioredoxin-dependent peroxidases (Coombs et al., 2004; Leitsch et al., 2009). Thus, when FR1 is inactive in metronidazole-treated T. vaginalis, then inhibition of TrxR activity by metronidazole has a reduced effect on viability.
The restoration of metronidazole sensitivity demonstrated by transfection (Figure 7) cannot distinguish which aspect of the loss of FR1 activity is of greater importance for the development of metronidazole resistance. However, it was shown by others that the metronidazole-resistant clinical isolate IR78 takes up far less metronidazole in the presence of high oxygen concentrations than C1, a highly metronidazole susceptible strain (Müller and Gorrell, 1983). In contrast, at a lower oxygen concentration, metronidazole uptake was nearly equal. These observations favour the view that enhanced redox cycling of metronidazole nitroradical anions is of decisive importance for clinical metronidazole resistance.
It is an unresolved issue, how the normal activity levels of NADH oxidase in metronidazole-resistant T. vaginalis isolates can be reconciled with the observed elevated oxygen levels in these strains (Yarlett et al., 1986a), because this enzyme reduces oxygen to water very efficiently (Tanabe, 1979; Linstead and Bradley, 1988). However, our results with metronidazole-treated G3 (susceptible) and CDC085 (resistant) suggest that NADH oxidase activity is impaired by metronidazole, whereas FR1 activity remains unaffected (Figure 6 and Supplementary Figure 3). Thus, it is possible that NADH oxidase is rendered inactive during metronidazole treatment and that metronidazole-resistant isolates cannot rely on a second line enzymatic pathway for the removal of oxygen, i.e. by FR1. The previously made observation that NADH oxidase is highly vulnerable to oxygen (Linstead and Bradley, 1988) further strengthens this hypothesis. Together, this study suggests that the inactivation of FR1 acts as a mechanism underlying metronidazole resistance not only in anaerobically resistant laboratory strains but also in many clinical isolates of T. vaginalis.
Experimental Procedures
Chemicals
Riboflavin, FMN, FAD, NADPH, and NADH were purchased from Sigma-Aldrich. Acrylamide and bis-acrylamide were purchased from Bio-Rad.
Strains and cell culture
The T. vaginalis strains used were already described before (Leitsch et al., 2012). All strains were routinely grown in trypticase, yeast extract, maltose medium (TYM) as described (Leitsch et al., 2012). Parasites were subcultured either every day or every second day. The highly metronidazole-resistant cell line of C1, C1res, was routinely grown in presence of 1mM metronidazole.
Preparation of cell extracts, gel electrophoresis and nitroblue-tetrazolium blue (NBT) staining
Fully grown cultures were harvested by centrifugation at 800 × g for 5 min, followed by a washing step in 1 × PBS. Afterwards, cells were resuspended in 100 mM Tris pH 7.5 and disrupted in a Dounce homogenizer. Cell debris and large organelles were removed by centrifugation at 20,000 × g for 10 min in a cryocentrifuge (4° C). Supernatants were used for native gel electrophoresis after determination of protein concentrations using Bradford assay. Appropriate amounts of cell extract were loaded on polyacrylamide gels lacking SDS (stacking gel: 5% polyacrylamide, separation gel: 12.5% polyacrylamide) and gel runs were performed at 100 V. After gel electrophoresis, gels were rinsed twice with ultrapure water and subsequently immersed into 100 mM Tris pH 7.5 buffer containing 0.5 mM NADPH and 0.2% NBT. Staining was allowed to proceed until bands were distinctly visible. Afterwards, gels were rinsed thoroughly with water and scanned using an Epson Perfection V750 PRO scanner.
Protein identification using Mass Spectrometry
Gel bands of interest were excised from the gel, digested with trypsin and analysed using reversed phase LC ESI ion trap tandem mass spectrometry using an Ultimate 3000 UHPLC system (Dionex, part of Thermo Fisher) coupled to an amazon speed ETD ion trap (Bruker Daltonics, Bremen, Germany) as described in detail previously (Kolarich et al., 2012; Williams et al., 2012). MS/MS spectra were extracted and processed using Data Analysis 4.0 (Bruker Daltonics) and searched against the NCBI protein database (Version 28/08/2011, containing 15148518 sequences) using the in house MascotServer 2.3 (Matrixscience, UK) and ProteinScape 3.1 (Bruker Daltonics, Bremen, Germany). The following search parameters were used: taxonomy was limited to “other Eukaryotes”, Carbamidomethyl was set as a fixed Cysteine modification, Asparagine/Glutamine deamidation and Oxidation of Methionine were set as variable modifications; trypsin allowing one missed cleavage was defined as the proteolytic enzyme; MS and MS/MS tolerance parameters were 0.15 and 0.25 Da, respectively. Identification of two independent peptide sequences per protein and a mascot score of > 85 were considered a minimum requirement. All analyses were performed in duplicate.
Recombinant expression of flavin reductases
Flavin reductase (FR) genes 1 – 7 were amplified from T. vaginalis G3 genomic DNA using the primers as listed in Supplementary Table 2. All genes were cloned into the pET-17b expression vector. The FR1 gene was ligated into the vector via NdeI and EcoRI restriction sites in the forward and reverse primers, respectively, and the FR2 – 6 genes were ligated via NdeI and XhoI restriction sites. As the gene of FR7 contains an NdeI restriction site, the gene was first cloned into pET-17b via XbaI and EcoRI and subsequently mutagenized. The T residue in the NdeI recognition sequence was exchanged for a C residue using a QuikChange II site-directed mutagenesis kit (Agilent Technologies) and two complementary primers bearing the mutation (Supplementary Table 2). Site-directed mutagenesis was performed according to the manufacturer's instructions. Subsequently, the mutagenized FR7 gene was amplified from the plasmid using a new forward primer bearing an NdeI restriction site and cloned into pET-17b via NdeI and EcoRI restriction sites. Expression plasmids were introduced into E. coli BL21 AI and protein expression in batch cultures was induced by addition of 0.2% L-arabinose at OD600= 0.3. All recombinant flavin reductases were purified via their 6 × His tags in Ni-NTA spin columns (Qiagen) according to the manufacturer's protocol.
Enzymological characterization of recombinant flavin reductases
For the determination of kinetic constants of FR1-7, measurements were performed in 100 mM potassium phosphate buffer pH 5.5 at 37°C using a Perkin Elmer Lambda 25 spectrophotometer and a PTP A Peltier element (Perkin Elmer). Flavin reduction activity was measured by determining oxidation of NADPH or NADH at λ= 340 nm. Riboflavin, FMN, FAD, NADPH and NADH were added in the amounts indicated in the text. The enzyme concentrations applied were 0.25 μg/ml in case of FR1, 5, and 6 and 1 μg/ml in case of FR2, 3, 4, and 7.
For the determination of end products of FR1, superoxide and hydrogen peroxide formation were measured in appropriate assays. Superoxide formation was measured at 37°C and λ= 550 nm in the presence of cytochrome c (100 mM potassium phosphate pH 5.5, 50 μM cytochrome c, 200 μM NADPH, 400 nM FMN, 0.025 μg/ml FR1) which received electrons either directly from reduced FMN (the semiquinone FMNH·) or from superoxide radical anions which are the reaction product of oxygen and reduced FMN. Specific reduction of cytochrome c by superoxide was determined by adding 1 μM bovine superoxide dismutase (Sigma-Aldrich) and subsequent subtraction of the resulting diminished reduction rate of cytochrome c from the overall reduction rate. Measurements were performed three times.
For the measurement of generated hydrogen peroxide, flavin reduction by FR1 was allowed to take place in 700 μl of 100 mM Tris pH 7.5, containing 1 μg FR1 and 10 μM FMN, in the presence of varying concentrations of NADPH (20 to 50 μM). The amount of hydrogen peroxide generated was quantified as described before (Wassmann et al., 1999) by measuring [FeSCN]2+ complex formation after Fe(II) had been oxidized by hydrogen peroxide to Fe(III). Briefly, after all NADPH had been oxidized, 300 μl of 30% trichloroacetic acid were added, followed by 200 μl 10 mM Fe(II) ammonium sulphate and 100 μl of 2.5 M potassium thiocyanate. Colorimetric quantification of [FeSCN]2+ was performed at λ= 480 nm. Hydrogen peroxide standards ranged from 5 to 50 μM.
Measurement of NADH-oxidase and flavin reductase activity in cell extracts
Cell extracts were prepared as described earlier. Flavin reductase and NADH-oxidase activites were measured at 37° C in 100 mM potassium phosphate pH 5.5 (200 μM NADPH or NADH) using 20 μg protein/ml cell extract. FMN was added at concentrations as indicated in the text.
Western Blotting
Either 1 μg of purified recombinant flavin reductases or 100 μg protein T. vaginalis cell extracts were loaded on 12.5% PAA gels containing 0.1% SDS. After gel electrophoresis, proteins were blotted (300 mA, 1h) on a nitrocellulose membrane (Protran from Whatman) using Mini-PROTEAN Tetra System equipment (Bio-Rad). Subsequently, blots were exposed at 4° C overnight to anti-FR1 antibody raised in rabbit (David's Biotechnologie) at a concentration of 1:1000 in 1 × TBST buffer containing 1% milk powder. After washing (5 times in 1% milk powder in 1 × TBST), alkaline phosphatase (AP)-coupled anti-rabbit antibody was added and blots were incubated at RT for 1 h. After five additional washing steps in 1 × TBST buffer containing 1% milk powder, blots were equilibrated in AP buffer (100 mM Tris pH 9.5, 100 mM NaCl, 5 mM MgCl2) for 15 min and AP staining was started by adding AP buffer containing 5 μg/ml NBT and 0.83 μg/ml BCIP.
5′ RACE and 3′ RACE
Amplification of the 5′ and 3′ ends of FR1 mRNA was performed with the 5′/3′ RACE kit (2nd generation) from Roche according to the provider's manual. For 5′ RACE the following primers were used: first primer, GGAGAATTAGCGCCGAAG; first nested primer, GTAAGCATAGAAGCAGGCTG; second nested primer, TTGGCTGAATCAGCGAAACG. For 3′ RACE following primers were used: first primer, ATGTCTCACATGCACG; nested primer, GTTGCTCACCCAGACCC.
Transfection of B7268 with the FR1 and FR4 genes and determination of sensitivity to metronidazole
Flavin reductase genes 1 (TVAG_517010) and 4 (TVAG_406950) were PCR amplified from T. vaginalis G3 genomic DNA using the primers For-517010-Nde (5′ - TTC ATT TTC CAT ATG TCT CAC ATG CAC GTC TTG) and Rev-517010-Kpn (5′ - CAC GGT ACC CAA GAG GAT GTT TTC TGA ATT GAC) for FR1 and For-406950-Nde (5′ - TAT AAA TAA CAT ATG CGT GTC TTA ATA TTG GTC G) and Rev-406950-Kpn (5′ - AAA GGT ACC TTC ACT AAC GAG AAT GTT ATC GG) for FR4. Underlined indicates the restriction site in each primer. PCR products were digested with NdeI and KpnI and ligated to plasmid MasterNeo-(HA)2 (Dyall et al., 2000) using standard procedures. Resulting constructs produce flavin reductase proteins with a 2 x HA epitope at the C-terminus. T. vaginalis strain B7268 was electroporated with the constructs and an empty vector control (EV) as described previously (Delgadillo et al, 1997). Transfectants were selected with 200 μg/ml G418 (Sigma).
Protein samples for western blot analysis were taken from lysates of parasites boiled in 2% SDS – 10 mM Tris-HCl (pH 8.0) and 1x HALT protease inhibitor (Thermo Scientific). Preparations of 1 x 104 parasites each were resolved on SDS-PAGE gels (5% stacking, 17.5% resolving – 35 mAmps for 70 min) then electroblotted (60 V for 60 min) onto PVDF membranes (Bio Rad). Membranes were then incubated in 5% milk – 1 x TBST solution for 30 min. Membranes were then incubated with a 1:5000 dilution of mouse anti-HA antibody (Covance) overnight at 4°C. After washing (3 x with 1 x TBST) membranes were incubated with a 1:25000 dilution of donkey anti-mouse HRP (Jackson ImmunoResearch) at room temperature for one hour. After three additional washes with 1 x TBST, blots were incubated with ECL reagent and exposed to film (Genemate).
Metronidazole sensitivity was determined as described previously (Leitsch et al., 2012) with the modification that 200 μg/ml G418 was included in the media at all times.
Supplementary Material
Acknowledgments
This work was supported by the Austrian Science Fund (grant P22546), the Max Planck Society (Daniel Kolarich), and NIH grants R01AI103182 and T32AI007323 (Brian D. Janssen and Patricia J. Johnson).
References
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A, Cammack R, Linstead R, Lloyd D. The generation of metronidazole radicals in hydrogenosomes isolated from Trichomonas vaginalis. J Gen Microbiol. 1985;131:2141–2144. doi: 10.1099/00221287-131-9-2141. [DOI] [PubMed] [Google Scholar]
- Coombs GH, Westrop GD, Suchan P, Puzova G, Hirt RP, Embley TM, et al. The amitochondriate eukaryote Trichomonas vaginalis contains a divergent thioredoxin-linked peroxiredoxin antioxidant system. J Biol Chem. 2004;279:5249–5256. doi: 10.1074/jbc.M304359200. [DOI] [PubMed] [Google Scholar]
- Cui J, Smith TF, Samuelson J. Gene expansion in Trichomonas vaginalis: a case study on transmembrane cyclases. Genome Inform. 2007;18:35–43. [PubMed] [Google Scholar]
- Davis SR, Lushbaugh WB. Oxidative stress and Trichomonas vaginalis: the effect of hydrogen peroxide in vitro. Am J Trop Med Hyg. 1993;48:480–7. doi: 10.4269/ajtmh.1993.48.480. [DOI] [PubMed] [Google Scholar]
- Delgadillo MG, Liston DR, Niazi K, Johnson PJ. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc Natl Acad Sci U S A. 1997;94:4716–20. doi: 10.1073/pnas.94.9.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Koehler CM, Delgadillo-Correa MG, Bradley PJ, Plümper E, Leuenberger D, et al. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol Cell Biol. 2000;20:2488–97. doi: 10.1128/mcb.20.7.2488-2497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JE, Cole D, Lloyd D. Influence of oxygen on the fermentative metabolism of metronidazole-sensitive and resistant strains of Trichomonas vaginalis. Mol Biochem Parasitol. 1992;56:79–88. doi: 10.1016/0166-6851(92)90156-e. [DOI] [PubMed] [Google Scholar]
- Ellis JE, Wingfield JM, Cole D, Boreham PF, Lloyd D. Oxygen affinities of metronidazole-resistant and -sensitive stocks of Giardia intestinalis. Int J Parasitol. 1993;23:35–39. doi: 10.1016/0020-7519(93)90095-g. [DOI] [PubMed] [Google Scholar]
- Goldman LM, Upcroft JA, Workowski K, Rapkin A. Treatment of metronidazole-resistant Trichomonas vaginalis. Sex Health. 2009;6:345–347. doi: 10.1071/SH09064. [DOI] [PubMed] [Google Scholar]
- Kolarich D, Jensen PH, Altmann F, Packer NH. Determination of site-specific glycan heterogeneity on glycoproteins. Nat Protoc. 2012;7:1285–98. doi: 10.1038/nprot.2012.062. [DOI] [PubMed] [Google Scholar]
- Kulda J, Tachezy J, Cerkasovová A. In vitro induced anaerobic resistance to metronidazole in Trichomonas vaginalis. J Euk Microbiol. 1993;40:262–269. doi: 10.1111/j.1550-7408.1993.tb04915.x. [DOI] [PubMed] [Google Scholar]
- Kulda J. Trichomonads, hydrogenosomes and drug resistance. Int J Parasitol. 1999;29:199–212. doi: 10.1016/s0020-7519(98)00155-6. [DOI] [PubMed] [Google Scholar]
- Leitsch D, Kolarich D, Binder M, Stadlmann J, Altmann F, Duchêne M. Trichomonas vaginalis: metronidazole and other nitroimidazole drugs are reduced by the flavin enzyme thioredoxin reductase and disrupt the cellular redox system. Implications for nitroimidazole toxicity and resistance. Mol Microbiol. 2009;72:518–536. doi: 10.1111/j.1365-2958.2009.06675.x. [DOI] [PubMed] [Google Scholar]
- Leitsch D, Burgess AG, Dunn LA, Krauer KG, Tan K, Duchêne M, et al. Pyruvate:ferredoxin oxidoreductase and thioredoxin reductase are involved in 5-nitroimidazole activation while flavin metabolism is linked to 5-nitroimidazole resistance in Giardia lamblia. J Antimicrob Chemother. 2011;66:1756–1766. doi: 10.1093/jac/dkr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D, Drinić M, Duchêne M. Down-regulation of flavin reductase and alcohol dehydrogenase-1 (ADH-1) in metronidazole-resistant isolates of Trichomonas vaginalis. Mol Biochem Parasitol. 2012;183:177–183. doi: 10.1016/j.molbiopara.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstead DJ, Bradley S. The purification and properties of two soluble reduced nicotinamide:acceptor oxidoreductases from Trichomonas vaginalis. Mol Biochem Parasitol. 1988;27:125–133. doi: 10.1016/0166-6851(88)90032-1. [DOI] [PubMed] [Google Scholar]
- Mason RP, Holtzman JL. The role of catalytic superoxide formation in the O2 inhibition of nitroreductase. Biochem Biophys Res Commun. 1975;67:1267–1274. doi: 10.1016/0006-291x(75)90163-1. [DOI] [PubMed] [Google Scholar]
- Meingassner JG, Thurner J. Strain of Trichomonas vaginalis resistant to metronidazole and other 5-nitroimidazoles. Antimicrob Agents Chemother. 1979;15:254–257. doi: 10.1128/aac.15.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SNJ, Docampo R. Mechanism of toxicity of nitro compounds used in chemotherapy of trichomoniasis. Environ Health Perspect. 1985;64:199–208. doi: 10.1289/ehp.8564199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Gorrell TE. Metabolism and metronidazole uptake in Trichomonas vaginalis isolates with different metronidazole susceptibilities. Antimicrob Agents Chemother. 1983;24:667–673. doi: 10.1128/aac.24.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrin D, Delgaty K, Bhatt R, Garber G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin Microbiol Rev. 1998;11:300–317. doi: 10.1128/cmr.11.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasoloson D, Tomková E, Cammack R, Kulda J, Tachezy J. Metronidazole-resistant strains of Trichomonas vaginalis display increased susceptibility to oxygen. Parasitology. 2001;123:45–56. doi: 10.1017/s0031182001008022. [DOI] [PubMed] [Google Scholar]
- Schwebke JR, Barrientes FJ. Prevalence of Trichomonas vaginalis isolates with resistance to metronidazole and tinidazole. Antimicrob Agents Chemother. 2006;50:4209–4210. doi: 10.1128/AAC.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JD, Nyiresy P, Brown W. Tinidazole therapy for metronidazole-resistant vaginal trichomonosis. Clin Infect Dis. 2001;33:1341–1346. doi: 10.1086/323034. [DOI] [PubMed] [Google Scholar]
- Tanabe M. Trichomonas vaginalis: NADH oxidase activity. Exp Parasitol. 1979;48:145–143. doi: 10.1016/0014-4894(79)90063-8. [DOI] [PubMed] [Google Scholar]
- Upcroft P, Upcroft JA. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin Microbio Rev. 2001;14:150–164. doi: 10.1128/CMR.14.1.150-164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upcroft JA, Dunn L, Wal T, Tabrizi S, Delgadillo-Correa MG, Johnson PJ, et al. Metronidazole resistance in Trichomonas vaginalis from highland women in Papua New Guinea. Sex Health. 2009;6:334–338. doi: 10.1071/SH09011. [DOI] [PubMed] [Google Scholar]
- Wassman C, Hellberg A, Tannich E, Bruchhaus I. Metronidazole resistance in the protozoan parasite Entamoeba histolytica is associated with increased expression of iron-containing superoxide dismutase and peroxiredoxin and decreased expression of ferredoxin 1 and flavin reductase. J Biol Chem. 1999;274:26051–26056. doi: 10.1074/jbc.274.37.26051. [DOI] [PubMed] [Google Scholar]
- Williams CF, Lloyd D, Kolarich D, Alagesan K, Duchêne M, Cable J, Williams D, Leitsch D. Disrupted intracellular redox balance of the diplomonad fish parasite Spironucleus vortens by 5-nitroimidazoles and garlic-derived compounds. Vet Parasitol. 2012;190:62–73. doi: 10.1016/j.vetpar.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Gorell TE, Marczak R, Müller M. Reduction of nitroimidazole derivatives by hydrogenosomal extracts of Trichomonas vaginalis. Mol Biochem Parasitol. 1985;14:29–40. doi: 10.1016/0166-6851(85)90103-3. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Yarlett NC, Lloyd D. Metronidazole-resistant clinical isolates of Trichomonas vaginalis have lowered oxygen affinities. Mol Biochem Parasitol. 1986a;19:111–116. doi: 10.1016/0166-6851(86)90115-5. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Yarlett NC, Lloyd D. Ferredoxin-dependent reduction of nitroimidazole derivatives in drug-resistant and susceptible strains of Trichomonas vaginalis. Biochem Pharmacol. 1986b;35:1703–1708. doi: 10.1016/0006-2952(86)90327-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


