Abstract
Expression of the metastasis suppressor NME1 in melanoma is associated with reduced cellular motility and invasion in vitro and metastasis in vivo, but the underlying molecular mechanisms are not completely understood. Herein, we report a novel mechanism through which NME1 controls melanoma cell morphology via upregulation of the extracellular matrix (ECM) protein fibronectin. Expression of NME1 strongly suppressed cell motility in melanoma cell lines 1205LU and M14. The resulting sedentary phenotype was associated with a more flattened appearance and marked increases in actin stress fibre and focal adhesion formation. NME1‐induced focal adhesions were colocalized with dense deposits of fibronectin, which were absent or minimal in the corresponding NME1‐deficient parental lines. NME1 was a strong inducer of fibronectin mRNA and protein expression, shown with reciprocal approaches of forced NME1 expression and shRNA‐mediated knock‐down. Increased synthesis and ECM deposition of fibronectin was necessary for NME1‐induced cell spreading, as knock‐down of fibronectin opposed the effects of NME1 on cell morphology. Fibronectin knock‐down also reversed the ability of NME1 to promote aggregation when cells were plated on a non‐adherent substratum. Similarly, inhibiting activation of the fibronectin receptor integrin α4β1 with an anti‐α4 antibody reversed the motility‐suppressing effect of NME1. A positive correlation was observed between NME1 and fibronectin mRNA in clinical biopsies of normal skin, benign nevi and primary melanomas, but not in metastatic forms, suggesting the NME1/fibronectin axis represents a barrier to melanoma progression. In summary, these findings indicate fibronectin is an important effector of the motility‐suppressing function of NME1 in melanoma cells.
Keywords: fibronectin, melanoma, motility, NME1
Introduction
Melanoma arises from the melanocyte, the melanin‐producing cell of skin. Interestingly, melanotic tumors exhibit much higher incidence of metastasis and decreased survival, relative to their amelanotic counterparts 1. While 5‐year survival rates are relatively favourable for Stage I and Stage II melanoma patients (98.3 and 64.2%, respectively), the prognosis for patients with metastatic disease is extremely poor (16.1%) 2. An inverse correlation has been established between expression of the metastasis suppressor NME1 and poor survival or tumor grade in a number of human cancers, including melanoma as well as carcinomas of breast, stomach, lung non‐small cell, hepatocellular and oral squamous cell origins 3, 4, 5, 6, 7, 8. Moreover, NME1 exerts metastasis suppressor activity in HGF‐overexpressing transgenic mice 9, a well‐characterized model of UV‐inducible melanoma 9, 10. NME1 exhibits multiple biochemical activities in vitro, including nucleoside diphosphate kinase, histidine kinase and a 3′‐5′ exonuclease function 9, 10, 11. While site‐directed mutagenesis analysis of the NME1 molecule strongly suggests both the kinase and exonuclease activities contribute to its metastasis suppressor function 3, 12, the underlying molecular mechanisms of how NME1 suppresses melanoma metastasis have yet to be determined.
Cell motility requires establishment of physical contacts between the cell and its microenvironment, a process mediated by the integrin superfamily 4. Integrin‐mediated adhesion is regulated both by intra‐cellular signalling networks and in response to composition and organization of the extracellular matrix (ECM) 5, 6. During tumor progression, melanoma and other solid tumors show reduced expression of the ECM protein fibronectin, and similar to the loss of NME1, this phenomenon is linked to increased proliferation, migration, invasion and metastasis 7, 8, 13. Moreover, melanoma cells of low metastatic potential deposit an elaborate fibronectin matrix that supports formation of large focal adhesions and associated actin stress fibres, which in turn promote adhesion and can inhibit motility 14.
In this study, we provide evidence for a novel mechanism through which NME1 suppresses cell motility via stabilization of cell‐substrate adhesions, at least in part, through upregulation of the ECM protein fibronectin. Moreover, a positive correlation between the expression of NME1 and fibronectin is associated with earlier stages of melanoma progression. These findings offer a new mechanistic perspective on the role of NME1 in suppression of metastasis in melanoma.
Materials and methods
Cell lines and cell culture
Melanoma cell lines 1205LU, WM278 and WM793 were obtained from the Wistar Institute (Philadelphia, PA, USA), and their authenticity was confirmed by STR DNA typing. MDA‐MB‐435s/M14 cells (referred to as M14 in this text) were obtained from R. Plattner (University of Kentucky) 15. NME1 was stably expressed in 1205LU as described previously 3. For stable forced expression in M14, NME1 cDNA was cloned into pEGFP‐N1 vector (Clontech, Mountain View, CA, USA), and the construct or an empty vector was then transfected to M14 using Lipofectamine (Life Technologies, Carlsbad, CA, USA). Stable transfectants were obtained by selection with G418 (Life Technologies). Knock‐down of NME1 expression was achieved by lentiviral delivery of Mission shRNA (Sigma‐Aldrich, St. Louis, MO, USA) TRCN0000010062 (shNME1#1) or TRCN0000010055 (shNME1#2), while fibronectin was targeted with shRNA TRCN0000064828 (shFN#1) or TRCN0000064832 (shFN#2). Non‐target shRNA pLK0.1 (SHC002, Sigma‐Aldrich, St. Louis, MO, USA)) was used as a negative control. Production of viral vectors and transduction were performed according to manufacturer's instructions. All melanoma cell lines with the exception of M14 were cultured in Tu 2% growth medium, consisting of a 1:4 mixture of Leibovitz's L‐15 and MCDB 153 medium, and supplemented with 2% FBS (Life Technologies) and 5 μg/ml bovine insulin (Sigma) and maintained at 37°C with 5% CO2. M14 cell lines derived cultured in DMEM (Life Technologies) supplemented with 10% FBS and maintained at 37°C in 10% CO2. For assessment of focal adhesions, 1205LU and M14 cell lines were transduced with a lentiviral vector encoding a DsRed‐paxillin fusion protein (plasmid provided by Cai Huang, University of Kentucky). Human umbilical vein endothelial cells (HUVECs) were cultured in EGM2 according to manufacturer's instructions (Lonza, Walkersville, MD, USA).
Immunofluorescence and TIRF microscopy
For fluorescence and total internal reflection fluorescence (TIRF) microscopy, cells were plated onto glass bottom chamber slides, cultured for 2 days in complete media, then serum‐starved for 24 h. Cells were then fixed and stained with antifibronectin antibody (clone FN‐15, Sigma). Actin cytoskeleton was stained with fluorescent phalloidin (Cytoskeleton, Denver, CO, USA) in permeabilized cells. DsRed‐paxillin was imaged in fixed cells or, for TIRF time lapse, in living, serum‐starved cells. Nuclei of fixed cells were counterstained with 1 μg/ml DAPI (Sigma). All fluorescence microscopy was conducted with a Nikon Ti series inverted microscope equipped with a 40× or 60× objective (regular fluorescence) or 60× APO TIRF objective.
Cell migration measurement
Transwell migration assays were performed using 8‐μm pore size transwell inserts (BD Biocoat; BD Biosciences, Bedford, MA, USA) as previously described 3, with migration assessed at 18 h postplating. For time‐lapse microscopy and motility measurements, cells were grown for 2 days in complete growth media and serum‐starved for 24 h prior to experiments and maintained in 0.1% FBS during imaging. Phase‐contrast time‐lapse videomicroscopy was conducted with a Nikon Ti series microscope equipped with an environmental chamber. Time‐lapse videos were captured over 18 h with image acquisition every 20 min. Measurements of migration path length, velocity and cell area were calculated using NIS Elements AR 3.0 software (Nikon Instruments, Melville, NY, USA).
Transendothelial migration assays were performed on 5‐μm pore polycarbonate transwell inserts coated with Matrigel (BD Biosciences), with HUVECs seeded onto the coated inserts at 105 cells/well and cultured for 2 days to form a confluent monolayer (Lonza). Once the HUVEC monolayer was established, 1205LU cells were pretreated with complete growth medium supplemented with 10 ng/ml CXCL12 or 0.1% BSA control for 6 h before labelling with CellTracker (Life Technologies) and seeded on top of HUVEC monolayers at 2 × 105 cells/well. Five hundred ul of the invasion medium containing 10 nm CXCL12 attractant or vehicle control was added to the bottom chamber of the transwell plate. Transendothelial migration assays were conducted at 37°C in 5% CO2 for 18 h. At the end of the assay, cells from the top surface of the insert were removed with a cotton swab and transmigrated cells on the bottom were fixed in 4% paraformaldehyde. Fluorescent transmigrated cells were counted from one 4× field per membrane.
Slow aggregation assay
To study the effect of fibronectin loss on cell–cell adhesion, cells stably expressing control (shCON) or fibronectin‐specific shRNA (shFN#1 or shFN#2) were seeded onto 96‐well plates coated with 0.75% agarose to prevent cell‐substrate adhesion. A total of 10 000 cells per well were plated in a final volume of 100 μl complete media (i.e. serum containing). Aggregate diameter was then measured 24 h later using ImageJ (National Institutes of Health). Viability of cells within aggregates was also measured 24 h postplating. Aggregates were dissociated with 0.05% trypsin–EDTA, and cell viability was assessed by trypan blue exclusion.
Western blots
For preparation of whole‐cell lysates, cells were cultured for 2 days in complete media and then serum‐starved for 24 h prior to harvesting, unless indicated otherwise. Cells were lysed in RIPA buffer, supplemented with EDTA‐free HALT protease inhibitors (Thermo Scientific, Rockford, IL, USA), and 10 mm sodium orthovanadate (Sigma). Equal amounts of cell lysate, as measured by Lowry DC reagent kit (Bio‐Rad, Hercules, CA, USA), were subjected to SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose. Proteins of interest were detected via chemiluminescence after exposure to primary antibodies against NME1 (610247; BD Biosciences, San Jose, CA, USA), fibronectin (FN‐15, Sigma) and beta‐tubulin (H‐235, Santa Cruz Biotechnology, Santa Cruz, CA, USA) followed by isotype‐specific HRP‐conjugated secondary antibodies.
Real‐time PCR
Total RNA was isolated from cells using the RNeasy kit as per manufacturer's instructions (Qiagen, Valencia, CA, USA). Complementary DNA was generated with random hexamer primers and the TaqMan RT reagents (Applied Biosystems, Carlsbad, CA, USA). SYBR green‐based quantitative real‐time quantitative PCR (qRT‐PCR) was conducted for human fibronectin (forward 5′‐CCACCCCCATAAGGCATAGG‐3′; reverse 5′‐GTAGGGGTCAAAGCACGAGTCATC‐3′) and normalized to beta‐2‐microglobulin (forward 5′‐CTCGCTCCGTGGCCTTAG‐3′; reverse 5′‐ATCTTCAAACCTCCATGATG‐3′) using the 2−ΔΔCT method.
Gene expression profiling
Gene expression profiles of 17 normal skin and benign nevi, 31 primary melanoma and 73 metastatic melanoma biopsies published by Kabbarah et al. 16 were obtained from Gene Expression Omnibus (accession number GSE46517). Log2 expression values of NME1 and fibronectin (FN1) were evaluated for linear correlation using the Pearson product moment.
Statistical analysis
Results were analysed with SigmaStat v3.5 statistical software (Systat Software, Chicago, IL, USA). Unless stated otherwise in the figure legends, all data are representative of n ≥ 2 independent experiments. Additional details for individual statistical analyses can be found in figure legends.
Additional Methods can be found in Data S1.
Results
NME1 suppresses random and directed migration of metastatic melanoma cells
Consistent with prior studies 3, 17, stable forced expression of NME1 in the human metastatic melanoma‐derived cell lines M14 and 1205LU significantly reduced directionality and distance of random cell migration, as measured by time‐lapse microscopy (Fig. 1a,b, respectively). Control 1205LU and M14 cells were motile even under conditions of low mitogen concentration (0.1% serum), with average migration path lengths of approximately 400 μm and 300 μm, respectively (Fig. 1b). Forced NME1 expression inhibited migration path lengths by twofold in both cell lines and also significantly reduced migration velocity (Fig. 1c). Velocity of cell migration is a function of the strength of cell adhesion to the substrate 18, suggesting a role for NME1 in regulation of cell adhesion.
Figure 1.
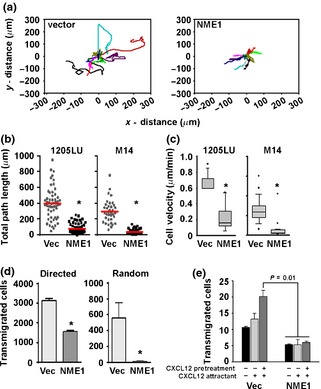
NME1 suppresses migration in the metastatic melanoma‐derived cell lines 1205LU and M14. (a) Representative traces of individual 1205LU cells stably expressing the pCI vector or NME1 over 24 h. Total path lengths from 1205LU and M14 cells are shown in (b), while individual cell velocities are quantitated in (c). *P ≤ 0.006 as determined by Mann–Whitney rank sum test, with horizontal bars indicating means in (b) and error bars represent standard error of the mean for (c). (d) Serum‐starved M14 cells stably expressing NME1 or vector were subjected to transwell (8 μm pore) migration assays towards a 2% FBS gradient (directed migration, left) or in the absence of chemoattractant (random migration, right) for 18 h. Error bars represent standard deviation with *P ≤ 0.05 as determined by Student's t‐test. (e) Serum‐starved 1205LU cells stably expressing NME1 or vector were subjected to transendothelial migration assays. Cells were exposed to a 10 nm CXCL12 attractant or pretreated with 10 ng/ml CXCL12 for 6 h to stimulate migration. Error bars represent standard deviation with *P ≤ 0.05 as determined by one‐way ANOVA.
As NME1‐expressing cells changed direction less frequently than vector control cells when allowed to migrate randomly on tissue culture plastic (Fig. 1a), we next measured the extent to which NME1 suppressed directed migration towards growth factors. M14 cells expressing NME1 were significantly less efficient than vector cells at migrating towards a gradient of 2% FBS in a Boyden chamber assay (Fig. 1d). NME1 expression also suppressed migration of 1205LU cells through a monolayer of human umbilical endothelial cells (HUVEC) in transendothelial migration (TEM) assays, which model the intravasation step of metastasis. Stable expression of NME1 in 1205LU cells inhibited TEM towards a CXCL12 attractant (Fig. 1e). This effect was even more pronounced in cells pretreated for 6 h with 10 ng/ml CXCL12, which stimulates migration in melanoma 19. Together, these results demonstrate that NME1 suppresses multiple features of melanoma cell motility and invasion.
NME1 expression promotes cell spreading and focal adhesion formation
In addition to suppressing cell migration, NME1 induced marked changes in cell morphology and focal adhesion formation, while cell lines transfected with empty vector displayed the narrow, bipolar spindle‐like shape associated with a motile phenotype in melanoma cells 20. NME1‐expressing 1205LU and M14 cells were more spread out and polygonal in appearance (Fig. 2a), with average cell surface area increasing nearly twofold (Fig 2b).
Figure 2.
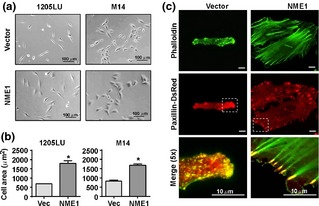
NME1 expression promotes cell spreading and discrete focal adhesion formation. (a) Representative phase‐contrast images of 1205LU and M14 stably expressing vector or NME1. (b) Quantitation of cell area in response to NME1 expression. Error bars represent standard deviation with *P = 0.002 as determined by Student's t‐test. (c) Representative total internal reflection fluorescence (TIRF) micrographs of 1205LU cells after 24 h serum starvation. Actin cytoskeleton was visualized by fluorescent phalloidin, and cell adhesions were visualized by stable expression of paxillin‐DsRed. Brackets denote magnified area shown in the bottom panel.
Cell morphology is determined in large part by the actin cytoskeleton, which provides mechanical and scaffolding functions essential for cell migration 21. We examined the impact of NME1 expression on organization of the actin cytoskeleton using fluorescent phalloidin to visualize filamentous actin. In control 1205LU cells, filamentous actin was finely punctate or diffuse and confined predominantly to cellular protrusions (Fig. 2c, top left panel). In contrast, forced NME1 expression was associated with formation of abundant actin stress fibres that originated from prominent plaques at the cell periphery and extended across the cell body (Fig. 2c, top right panel). Strong actin stress fibres are physically linked to the cell exterior via focal adhesions and are typically found in non‐migrating or slowly migrating cells 22. To visualize the effect of NME1 on formation of cell adhesions, we co‐expressed DsRed‐labelled paxillin, a marker of focal adhesions 23, 24. In control 1205LU cells, paxillin localized into small, dot‐like structures that were located at opposite ends of the spindle‐shaped cells (Fig. 2c, middle left). Stable expression of NME1, however, resulted in larger oval‐shaped focal adhesions distributed radially around the periphery of cells (Fig. 2c, middle right). Phalloidin and paxillin fluorescence were partially colocalized within small adhesive structures enriched at the protrusive end of the cell membrane in control 1205LU cells (Fig. 2c, lower left). In contrast, large, discrete focal adhesions anchored the prominent actin stress fibres in the NME1‐expressing line (Fig. 2c, lower right).
NME1 has been reported to suppress activation of the small GTPases Rac1 and Cdc42 25, 26, which facilitate formation of actin filaments and cell migration [reviewed in 27]. However, stable expression of NME1 in 1205LU cells failed to alter total cellular levels of active Rac1 or Cdc42, nor did it affect the response of either GTPase to the chemokine CXCL12 (Figure S1a). Stable expression of NME1 also had negligible effects of the activation of RhoA in response to serum (Figure S1b), which is known to promote the formation of focal adhesions and stress fibres 28, 29.
NME1 facilitates changes in cell morphology via deposition of a dense fibronectin matrix
As NME1 expression did not affect activation of cellular GTPases associated with cytoskeletal rearrangements (Figure S1), it was hypothesized that NME1 regulates cell morphology and motility via an ‘outside‐in’ mechanism. Deposition of fibronectin matrix has been associated with reorganization of actin into stress fibres, assembly of prominent focal adhesions and, ultimately, reduction in cell motility 18, 30. To determine whether NME1 expression modulated the presence and organization of fibronectin in the ECM, we employed TIRF (total internal reflection fluorescence) microscopy to visualize fibronectin deposition at the cell surface–substrate interface in 1205LU cells. Fibronectin was detected at very low levels in the ECM of control 1205LU cells and appeared only as fine, diffuse puncta below the cell body (Fig. 3a, top left panel). In contrast, NME1 expression induced the appearance of prominent fibronectin‐containing fibrils and plaques (Fig. 3a, top right panel). Fibronectin matrix was distributed across the cell surface, but enriched around the cell periphery. The majority of DsRed‐paxillin positive focal adhesions in NME1 overexpressing cells were found in contact with the dense fibronectin plaques and fibrils (Fig. 3a, bottom panels, arrows). Together, these results suggest that NME1 promotes focal adhesion formation by providing a fibronectin‐enriched ECM for cell attachment.
Figure 3.
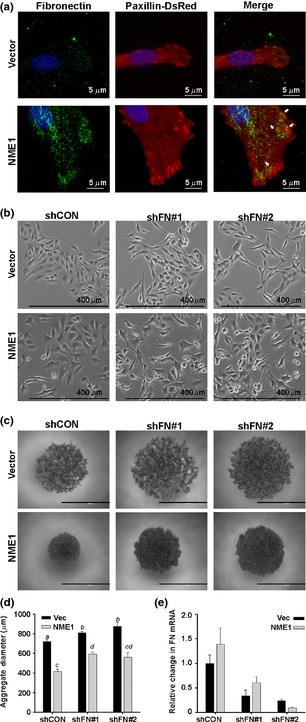
NME1 facilitates cell spreading and aggregation via deposition of fibronectin. (a) Representative total internal reflection fluorescence (TIRF) micrographs of 1205LU cells stably expressing vector or NME1 after staining for fibronectin (FN) plaques in the extracellular matrix. Cell‐substrate adhesions were visualized by stable expression of paxillin‐DsRed (middle panel). Colocalization of cell adhesions with FN deposits is seen in yellow and depicted by white arrows (right panel). (b) Representative phase‐contrast images of M14 cells stably expressing control (shCON) or fibronectin‐specific shRNAs (shFN). (c) Representative phase‐contrast images of aggregates formed by M14 cells stably expressing control (shCON) or fibronectin‐specific shRNAs (shFN) 24 h after seeding under non‐adherent conditions. Scale bar equals 1 mm. (d) Quantitation of the aggregate size (n = 2 independent experiments). Groups not sharing a common superscript are statistically different (P < 0.05) by ANOVA with Holm–Sidak post hoc testing. (e) Efficiency of fibronectin knock‐down was quantitated by qRT‐PCR in M14 cells.
To determine whether increased deposition of fibronectin and its association with focal adhesions contributed to the flattened morphology induced by exogenous NME1, we inhibited endogenous fibronectin synthesis with shRNA. Phase‐contrast micrographs demonstrate that the knock‐down of fibronectin led to an elongated, spindle‐like morphology that was more pronounced in the presence of NME1 overexpression (Fig. 3b). Loss of fibronectin also impaired cell–cell adhesion of M14 cells in a slow aggregation assay (Fig. 3c). The ability of cells to form aggregates in vitro is commonly associated with less invasive cell types 31. Similarly, M14 cells expressing NME1 formed significantly tighter aggregates than vector cells after seeding onto a semisolid agar substratum that prevented cell attachment (Fig. 3c, quantified 3e). Fibronectin knock‐down resulted in less compact aggregates in both vector and NME1 expressing cells, suggesting fibronectin promotes formation of cell–cell contacts. Importantly, fibronectin knock‐down inhibited aggregate formation more strongly in cells with forced NME1 expression than vector cells, with a 37.25% increase in aggregate diameter compared to a 16.75% increase in vector cells (Fig. 3e). Similar but more modest effects of fibronectin knock‐down on aggregate formation were seen in the VGP‐derived melanoma cell line WM793 (Figure S2). Transduction with fibronectin‐specific shRNAs resulted in 50% knock‐down of fibronectin mRNA (Fig. 3e). Sustained knock‐down of >90% could not be employed for these studies, as it induced cell death. However, the modest fibronectin knock‐down used had no effect on cell viability over the course of the aggregation assay in either cell line (data not shown). Together, these findings demonstrate fibronectin expression contributes to NME1‐associated changes in cell morphology and cell–cell adhesion.
Knock‐down of fibronectin increased migration of M14 cells in transwell assays, consistent with its role as a motility‐suppressing effector of NME1 (Figure S3). Fibronectin knock‐down had no significant effect on motility in the context of forced NME1 expression, possibly due to incomplete knock‐down (Figure S3). In an alternative approach, we assessed the impact of antibodies that blocked outside‐in signalling via the fibronectin receptors, integrins α4β1 and α5β1. Treatment with anti‐α4 integrin antibody ablated the motility‐suppressing effects of NME1 as assessed by time‐lapse microscopy, with no effect seen in untreated or IgG‐treated controls (Figure S4). Integrin α5‐blocking antibody did not significantly alter cell motility, which may have been secondary to low α5β1 expression or poor blocking activity. Nevertheless, the motility‐enhancing effect of anti‐α4 integrin antibody strongly suggests fibronectin deposition and its association with α4β1 integrins mediates NME1‐induced changes in morphology and motility in melanoma cells.
NME1 upregulates fibronectin mRNA in human melanoma cell lines and NME1 mRNA expression is positively correlated with fibronectin mRNA in human melanoma tissues
As expression of NME1 and fibronectin mRNAs was positively correlated in M14 melanoma cells (Fig. 3e), steady‐state levels of cellular fibronectin and the corresponding mRNA were measured in other melanoma cell lines. Forced expression of NME1 also upregulated fibronectin protein in 1205LU cells (Fig. 4a, left panel), while, conversely, suppression of endogenous NME1 expression with two separate shRNAs in WM278 cells led to a concomitant decrease in fibronectin protein (Fig. 4a, right panel). qRT‐PCR analysis revealed upregulation of fibronectin mRNA in 1205LU transfected with NME1 (Fig. 4b, left panel) and a corresponding decrease in fibronectin mRNA when NME1 was silenced in WM278 cells (Fig. 4b, right panel). Together, these results suggest that NME1 induces expression of fibronectin mRNA via a transcriptional or post‐transcriptional mechanism.
Figure 4.
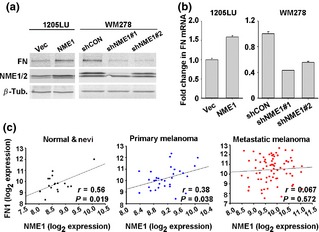
NME1 induces fibronectin mRNA in vitro and is positively correlated with fibronectin mRNA in vivo. (a) Western blot analysis for the indicated proteins after stable overexpression of NME1 in 1205LU cells or stable knock‐down of NME1 in WM278 cells. (b) qRT‐PCR analysis of FN mRNA levels after overexpression of NME1 in 1205LU cells and after stable knock‐down of NME1 down WM278 cells. Error bars represent standard deviation with *P ≤ 0.02 as determined by Student's t‐test. (c) NME1 and FN mRNA levels were extracted from GSE46517 and subjected to Pearson product moment analysis. The microarray consisted of 17 normal skin and benign nevi, 31 primary melanoma and 73 metastatic melanoma biopsies.
To determine whether induction of fibronectin mRNA by NME1 observed in cultured melanoma cell lines translates to human melanoma patients, we analysed the gene expression profiles from 17 biopsies of normal skin and benign nevi, 31 biopsies of primary melanoma and 73 biopsies of metastatic melanoma from the publically available GEO data set GSE46517 originally prepared by Kabbarah et al. 16. A significant positive correlation was observed between expression of NME1 and fibronectin (FN1) mRNAs in normal and benign nevi biopsies (Fig. 4c, left panel), as well as in primary melanoma lesions (Fig. 4c, left panel). Interestingly, this correlation was not seen in metastatic lesions, suggesting introduction of additional complexity during later stages of malignant progression [e.g. destabilization of NME1 protein 32]. The loss of correlation between NME1 and FN1 within metastatic melanoma samples was not due to changes in FN1 expression as no significant difference was observed in FN1 mRNA in primary versus metastatic melanomas (Figure S5).
Discussion
Migration is critical for metastasis during both early and late stages of dissemination of malignant cells from the primary tumor. Previous studies addressing the antimotility function of NME1 were conducted predominantly in tumors of epithelial origin 33, 34, 35, which are biologically distinct from melanoma. The current study identifies a novel mechanism by which NME1 suppresses migration of metastatic melanoma cells through upregulation of fibronectin which drives formation of stable, immobilizing cell‐substrate adhesions.
Migration of metastatic melanoma cells relies on optimally coordinated functions of the actin cytoskeleton and cell adhesion. The organization and function of the actin cytoskeleton is controlled to a large extent by intra‐cellular signalling pathways, specifically by the spatial and temporal activity of Rho GTPases, such as RhoA, Rac1 and Cdc42 27. In kidney and Burkitt's lymphoma cell lines, NME1 has been reported to suppress activity of these GTPases through interactions with their respective guanine exchange factors, TIAM1 25 and Dbl1 36. In our current studies of melanoma cells, however, NME1 overexpression effectively inhibited cell migration (Fig. 1) and promoted cell spreading and formation of actin stress fibres (Fig. 2) without altering the activation status of Rac1 or Cdc42 (Figure S1). Another recent study in breast cancer cells demonstrated that NME1 inhibited motility by preventing turnover of actin stress fibres by directly binding the actin‐severing protein gelsolin 37. Further study will be required to determine whether the extent to which these discrepancies are secondary to the lineage differences among the cancer cell lines under study and the methodologies employed.
Our observations in metastatic melanoma cell lines prompted us to examine whether NME1 blocks cell motility by regulating cell adhesion via outside‐in signalling as an alternative avenue to its functions in intra‐cellular signalling. The studies show that NME1 upregulates expression of fibronectin, which is deposited into the ECM of tumor cells to influence cell morphology (Fig. 3b), cell–cell adhesion (Fig. 3c) and suppress migration (Fig. S2). This finding is in line with a number of previous reports, where reduced or diminished expression of fibronectin in solid tumors, including melanoma, has been linked to increased migration, invasion and metastasis, while fibronectin overexpression suppressed multiple features of the transformed phenotype 7, 8, 13, 38. Consistent with our data, melanoma cell lines of low metastatic potential deposit elaborate fibronectin matrix that enhances formation of large focal adhesions with actin stress fibres, resulting in an increased substrate adhesion and reduced motility 14. Nevertheless, many studies have reported fibronectin overexpression in cancerous lesions 39, 40, including melanoma 41. Fibronectin fibrils also promote adhesion‐dependent growth of tumor cells 42, as well as formation of new blood vessels in primary and metastatic lesions 43. In addition to its synthesis within tumor cells, fibronectin is elaborated by tumor stroma or drawn from pools of soluble plasma fibronectin. The latter lacks specific domains that support stable cell adhesion and actually promotes cell migration in vitro 44. The context‐dependent nature of fibronectin function is evidenced in the work of Soikkeli et al. 42 in metastatic melanoma, which showed that cellular fibronectin deposited into the ECM supports cell migration in the presence of the anti‐adhesive protein periostin. In the absence of periostin, however, cellular fibronectin did not increase the migration of either melanoma or microvascular endothelial cells.
Fibronectin is likely to be only one of a spectrum of NME1‐regulated genes that affect cell adhesion. This is supported by the observation that fibronectin knock‐down was only able to impair, but not completely reverse the effects of NME1 on cell–cell adhesion (Fig. 3c). Additionally, blocking antibodies to the fibronectin receptor, α4β1 integrin, reversed the effect of NME1 on cell motility, suggesting receptor‐specific ‘outside‐in’ signalling from fibronectin contributes to the motility‐suppressing function of NME1. In fact, NME1 has previously been reported to induce expression of E‐cadherin 35 as well as regulate the expression and glycosylation of integrin β1 45, 46. Thus, fibronectin induction alone may be necessary, but not sufficient, for NME1‐mediated suppression of cell motility.
Fibronectin undergoes extensive post‐transcriptional modifications resulting in splice variants with different localization and activities in tumors 47, 48, and it still remains to be determined which splice variants of fibronectin are induced by NME1. Extracellular processing of fibronectin fibrils is critical for its tumor‐promoting effects 49 and may also be regulated by NME1. Many of the matrix metalloproteases responsible for the processing of fibronectin, such as MMP2 and MT1‐MMP, have already been shown to be inhibited by NME1 35, 50. Thus, NME1 may act as a ‘gatekeeper’, inhibiting pericellular proteolysis and turnover of fibronectin and thereby suppressing metastasis, while the loss of NME1 during melanoma progression could elicit increased fibronectin degradation and promotion of metastasis.
Supporting the notion that NME1 suppresses metastasis via induction of fibronectin, we observed that expression of NME1 and fibronectin mRNAs was strongly correlated in normal skin, benign nevi and primary melanoma lesions (Fig. 4c). The lack of a correlation between NME1 and fibronectin mRNA in metastatic biopsies is consistent with previous work showing downregulation of NME1 protein expression rather than the cognate mRNA in metastases 32. Collectively, our studies strongly suggest NME1 plays a prominent role in the metastatic cascade at least in part by promoting cell adhesion to the ECM via fibronectin deposition, thereby anchoring melanoma cells at the primary site.
Conflict of interests
The authors have declared no conflicting interests.
Supporting information
Figure S1. Forced expression of NME1 in 1205LU does not alter the activation status of RhoA, Rac1 or Cdc42 in 1205LU cells.
Figure S2. Knockdown of fibronectin impairs cell‐cell adhesion in WM793 cells.
Figure S3. Knockdown of fibronectin is insufficient to reverse NME1‐mediated effects on cell motility.
Figure S4. Blockade of the fibronectin receptor α4β1 impairs the suppressive effect of NME1 on cell motility.
Figure S5. Fibronectin mRNA expression in GEO dataset GSE46517.
Data S1. Methods.
Acknowledgements
This work was supported by the United States National Institutes of Health, National Cancer Institute grants CA83237 and CA159871 (DMK), and training grant T32CA15474 from the National Cancer Institute (MKL). MN and MKL performed the research, analysed the data and wrote the paper. XHY and AMB contributed to interpretation of the data. All authors contributed to the design of the research and approval of the manuscript.
References
- 1. Brozyna A A, Jozwicki W, Carlson J A et al Hum Pathol 2013: 44: 2071–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kosary C L, Altekruse S F, Ruhl J et al Cancer 2014: 120(Suppl 23): 3807–3814. [DOI] [PubMed] [Google Scholar]
- 3. Wang L S, Chow K C, Lien Y C et al Eur J Cardiothorac Surg 2004: 26: 419–424. [DOI] [PubMed] [Google Scholar]
- 4. Hartsough M, TSteeg P S. J Bioenerg Biomembr 2000: 32: 301–308. [DOI] [PubMed] [Google Scholar]
- 5. Guan‐Zhen Y, Ying C, Can‐Rong N et al Int J Exp Pathol 2007: 88: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Katakura H, Tanaka F, Oyanagi H et al Ann Thorac Surg 2002: 73: 1060–1064. [DOI] [PubMed] [Google Scholar]
- 7. Terasaki‐Fukuzawa Y, Kijima H, Suto A et al Int J Mol Med 2002: 9: 25–29. [PubMed] [Google Scholar]
- 8. Scambia G, Ferrandina G, Marone M et al J Clin Oncol 1996: 14: 334–342. [DOI] [PubMed] [Google Scholar]
- 9. Jarrett et al Clin Exp Metastasis 2013: 30: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Freije J M, Blay P, MacDonald N J et al J Biol Chem 1997: 272: 5525–5532. [DOI] [PubMed] [Google Scholar]
- 11. Agarwal R P, Robison B, Parks R E Jr. Methods Enzymol 1978: 51: 376–386. [DOI] [PubMed] [Google Scholar]
- 12. Chambers A F, Naumov G N, Varghese H J et al Surg Oncol Clin N Am 2001: 10: 243–255, vii. [PubMed] [Google Scholar]
- 13. Akamatsu H, Ichihara‐Tanaka K, Ozono K et al Cancer Res 1996: 56: 4541–4546. [PubMed] [Google Scholar]
- 14. Volk T, Geiger B, Raz A. Cancer Res 1984: 44: 811–824. [PubMed] [Google Scholar]
- 15. Ganguly S S, Fiore L S, Sims J T et al Oncogene 2012: 31: 1804–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kabbarah O, Nogueira C, Feng B et al PLoS One 2010: 5: e10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. MacDonald N J, Freije J M, Stracke M L et al J Biol Chem 1996: 271: 25107–25116. [DOI] [PubMed] [Google Scholar]
- 18. Gupton S L, Waterman‐Storer C M. Cell 2006: 125: 1361–1374. [DOI] [PubMed] [Google Scholar]
- 19. Bartolome R A, Galvez B G, Longo N et al Cancer Res 2004: 64: 2534–2543. [DOI] [PubMed] [Google Scholar]
- 20. Bai S W, Herrera‐Abreu M T, Rohn J L et al BMC Biol 2011: 9: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lazaro‐Dieguez F, Colonna C, Cortegano M et al FEBS Lett 2007: 581: 3875–3881. [DOI] [PubMed] [Google Scholar]
- 22. Ridley A J, Schwartz M A, Burridge K et al Science 2003: 302: 1704–1709. [DOI] [PubMed] [Google Scholar]
- 23. Mitra S K, Hanson D A, Schlaepfer D D. Nat Rev Mol Cell Biol 2005: 6: 56–68. [DOI] [PubMed] [Google Scholar]
- 24. Deakin N O, Turner C E. J Cell Sci 2008: 121: 2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otsuki Y, Tanaka M, Yoshii S et al Proc Natl Acad Sci U S A 2001: 98: 4385–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murakami M, Meneses P I, Knight J S et al Int J Cancer 2008: 123: 500–510. [DOI] [PubMed] [Google Scholar]
- 27. Reymond N, Riou P, Ridley A J. Methods Mol Biol 2012: 827: 123–142. [DOI] [PubMed] [Google Scholar]
- 28. Schoenwaelder S M, Burridge K. Curr Opin Cell Biol 1999: 11: 274–286. [DOI] [PubMed] [Google Scholar]
- 29. Madura T, Yamashita T, Kubo T et al EMBO Rep 2004: 5: 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sechler J L, Schwarzbauer J E. Cell Adhes Commun 1997: 4: 413–424. [DOI] [PubMed] [Google Scholar]
- 31. Debruyne D, Boterberg T, Bracke M E. Methods Mol Biol 2014: 1070: 77–92. [DOI] [PubMed] [Google Scholar]
- 32. Fiore L S, Ganguly S S, Sledziona J et al Oncogene 2013: 33: 4508–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung S, Paek Y W, Moon K S et al Anticancer Res 2006: 26: 249–258. [PubMed] [Google Scholar]
- 34. Horak C E, Mendoza A, Vega‐Valle E et al Cancer Res 2007: 67: 11751–11759. [DOI] [PubMed] [Google Scholar]
- 35. Boissan M, De Wever O, Lizarraga F et al Cancer Res 2010: 70: 7710–7722. [DOI] [PubMed] [Google Scholar]
- 36. Murakami M, Meneses P I, Lan K et al Cancer Biol Ther 2008: 7: 677–688. [DOI] [PubMed] [Google Scholar]
- 37. Marino N, Marshall J C, Collins J W et al Cancer Res 2013: 73: 5949–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orgaz J L, Benguria A, Sanchez‐Martinez C et al Melanoma Res 2011: 21: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandez‐Garcia B, Eiro N, Marin L et al Histopathology 2014: 64: 512–522. [DOI] [PubMed] [Google Scholar]
- 40. Waalkes S, Atschekzei F, Kramer M W et al BMC Cancer 2010: 10: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gaggioli C, Robert G, Bertolotto C et al J Invest Dermatol 2007: 127: 400–410. [DOI] [PubMed] [Google Scholar]
- 42. Soikkeli J, Podlasz P, Yin M et al Am J Pathol 2010: 177: 387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Castellani P, Viale G, Dorcaratto A et al Int J Cancer 1994: 59: 612–618. [DOI] [PubMed] [Google Scholar]
- 44. Pankov R, Yamada K M. J Cell Sci 2002: 115: 3861–3863. [DOI] [PubMed] [Google Scholar]
- 45. Li M Q, Shao J, Meng Y H et al Hum Reprod 2013: 28: 2822–2831. [DOI] [PubMed] [Google Scholar]
- 46. She S, Xu B, He M et al J Exp Clin Cancer Res 2010: 29: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaczmarek J, Castellani P, Nicolo G et al Int J Cancer 1994: 59: 11–16. [DOI] [PubMed] [Google Scholar]
- 48. Carnemolla B, Balza E, Siri A et al J Cell Biol 1989: 108: 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiao Y, Feng X, Zhan Y et al PLoS One 2012: 7: e41591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Horak C E, Lee J H, Elkahloun A G et al Cancer Res 2007: 67: 7238–7246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Forced expression of NME1 in 1205LU does not alter the activation status of RhoA, Rac1 or Cdc42 in 1205LU cells.
Figure S2. Knockdown of fibronectin impairs cell‐cell adhesion in WM793 cells.
Figure S3. Knockdown of fibronectin is insufficient to reverse NME1‐mediated effects on cell motility.
Figure S4. Blockade of the fibronectin receptor α4β1 impairs the suppressive effect of NME1 on cell motility.
Figure S5. Fibronectin mRNA expression in GEO dataset GSE46517.
Data S1. Methods.


