Abstract
The earliest steps of embryonic development involve important changes in chromatin and transcription factor networks, which are orchestrated to establish pluripotent cells that will form the embryo. DNA methylation, histone modifications, the pluripotency regulatory network of transcription factors, maternal factors and newly translated proteins all contribute to these transitions in dynamic ways. Moreover, these dynamics are linked to the onset of zygotic transcription. We will review recent progress in our understanding of chromatin state and regulation of gene expression in the context of embryonic development in vertebrates, in particular mouse, Xenopus and zebrafish. We include work on mouse embryonic stem cells and highlight work that illustrates how early embryonic dynamics establish gene regulatory networks and the state of pluripotency.
Keywords: pluripotency, embryo, chromatin, methylation, zygotic genome activation
1. Introduction: Routing fertilized eggs to pluripotency
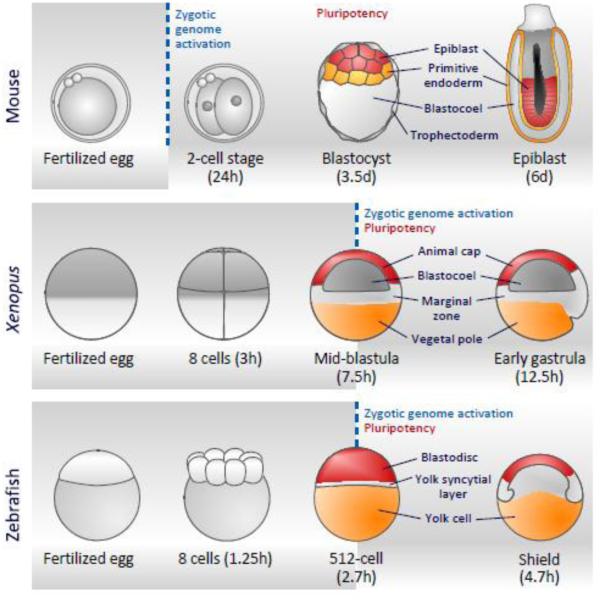
At the very beginning of embryonic development two specialized and highly differentiated cells, the gametes, fuse to form the zygote which in turn produces all cell types of the organism (as well as extra-embryonic tissue in the case of mammals, see below). The parental genomes show different histone modification patterns and are subject to dramatic chromatin reorganization, DNA demethylation and remethylation after fertilization and during early development, in order to reprogram the sperm and oocyte epigenomes [1-7]. Early in development a maternal to zygotic transition (MZT) is triggered, which passes regulatory control of development from maternal to newly synthesized components [8, 9]; this regulatory event can be defined as the period of time encompassing the initial degradation of maternal transcripts, zygotic genome activation (ZGA, the onset of transcription), until the first major morphological requirement for zygotic transcripts in embryonic development [9]. Following the ZGA, pluripotent cells emerge which will give rise to the three germ layers of the embryo, ectoderm, mesoderm and endoderm, however the relationship between ZGA and pluripotency is different between mammals and non-mammalian vertebrates (Fig. 1, Table 1).
Figure 1.
Embryonic development in relation to zygotic gene activation (ZGA) and pluripotency in mouse (top panel), Xenopus (middle panel) and zebrafish (bottom panel). ZGA occurs at the late 1-cell stage in mouse, but does not happen until the 12th and 10th cell cycle in Xenopus and zebrafish respectively. In mouse, before pluripotency is established two other lineages, trophectoderm and primitive endoderm, need to be formed. The inner cell mass (ICM) epiblast cells of the pre-implantation blastocyst (mouse, 3.5 days, red cells) represent the ground state of pluripotency in vivo, whereas in day 6 embryos (still before gastrulation) these cells have been primed towards differentiation. In Xenopus, vegetal pole cells have been maternally specified to form endoderm and secrete signals to induce mesoderm in the marginal zone. Animal pole cells (red) correspond to the pluripotent cells of the mid-blastula embryo. Times post fertilization are indicated for X. laevis at 22 °C. In zebrafish, the mid-blastula transition (MBT) starts two cell cycles earlier. Zebrafish has one large yolk cell which does not divide. Nuclei from blastodisc cells closest to the yolk cell form a syncytium with yolk cell. This region has a mesendodermal fate. Zebrafish times post fertilization are temperature-dependent and indicated at 28.5 °C.
Table 1.
Overview of early development and plurlpotency In Mouse, Xenopus and zebrafish. Note the interspecific differences in the relationship between (1) zygotic genome activation (ZGA) and plurlpotency and (2) DNA methylation and pluripotency. Abbreviations: hypoM, hypomethylated; hyperM, hypermethylated; deM, demethylated; ND, not determined.
| Mouse | Xenopus | Zebrafish | |
|---|---|---|---|
| ZGA stage | 2 cell | blastula | 512 cell |
|
| |||
| Pluripotent stage | blastocyst (E3.5) | blastula | 512 cell |
|
| |||
|
Global DNA
methylation |
hypoM at ZGA and pluripotency |
hyperM at ZGA and pluripotency |
hyperM at ZGA and pluripotency |
| Paternal DNA
methylation |
hyperM, active deM
after fertilization |
ND | hyperM, maintained |
| Maternal DNA
methylation |
relatively hypoM,
passive deM |
ND | relatively hypoM, hyperM
after 16-cell stage |
|
| |||
|
Histone H3
methylation |
ZGA and later | ZGA and later | ZGA and later |
|
| |||
|
Pluripotency
transcription factors |
OCT4(POU5Fl) S0X2 >NANOG |
Oct91, −25, −60 (Pou5f3) Sox2 Ventxl, −2 |
Oct4 (Pou5f3) Sox2, Soxl9b Nanog |
In amniotic species such as the mammals, the zygote starts transcribing its own genes just before the two-cell stage. Subsequently, trophectoderm and primitive endoderm, cell lineages that contribute to placental development, are set up in addition to the pluripotent cells of the inner cell mass of the blastocyst that will form the organism (Fig. 1). Therefore the mammalian zygote is referred to as totipotent, being able to produce all cell types of both the embryo proper and embryonic placental tissues [10].
Although early embryonic development is strikingly different in Xenopus and zebrafish compared to mouse, pluripotency and subsequent germ layer commitment, patterning and convergent extension are functionally highly analogous in these species. In Xenopus and zebrafish early cleavage development produces a blastula embryo, which undergoes ZGA (Fig. 1). This is also referred to as the mid-blastula transition (MBT) [8, 9, 11]. Cell cycle lengthening and the acquisition of cell motility coincide with the onset of embryonic transcription at the MBT [11] and at this stage, at and immediately after the MBT, cells at the animal pole of the embryo are pluripotent (Fig. 1). These cells are normally fated to give rise to ectoderm (epidermal ectoderm and neural ectoderm) but can also give rise to mesoderm and endoderm derivatives when exposed to specific factors [12].
Pluripotency, as it emerges from the zygote, represents a functional cellular state, much in the same way differentiated cells have a defined set of biochemical and cellular properties. These properties emerge from a cell type-specific reading of the genomic sequence information, which is part of what is referred to as epigenetic regulation. At the molecular level this involves chemical modifications of either the DNA itself (for example methylation of cytosine residues) or the chromosomal proteins associated with genomic DNA (chromatin). The profiles of epigenetic modifications can vary between cells and developmental stages and form a molecular regulatory intermediate between genomic sequence information and biochemical and cellular properties of cells. Epigenetics therefore constitutes a developmental stage- and cell type-specific filter of genomic sequence information.
In this review we compare vertebrate studies on the pluripotent chromatin state and how it emerges during embryonic development. We will discuss DNA methylation, histone modifications, cis-regulatory elements and transcription factors that prime the zygote for pluripotency. We will illustrate the findings from mouse, frog and fish embryo models but also discuss findings in cellular models of pluripotency such as mouse embryonic stem (ES) cells where warranted. ES cells represent stable pluripotency in vitro, whereas embryonic pluripotency is transitory. However much has been learned from these systems that also is highly relevant for pluripotency in vivo. The reader is referred to excellent reviews for other aspects of pluripotent chromatin and embryogenesis, including more detailed discussions of the MZT [8, 9], chromatin interactions and complexes [13-17], and naive and primed pluripotency in cell culture [18, 19].
2. Global DNA methylation dynamics in relation to pluripotency
2.1 DNA methylation dynamics in mammalian development
DNA methylation is an important epigenetic modification with a role in a variety of processes, including tissue-specific gene expression, development and cellular differentiation, carcinogenesis and aging, and specifically for mammals, genomic imprinting and X chromosome inactivation [20-22]. Methylation at the 5-position of cytosine (5mC) occurs mainly at CG dinucleotides (commonly referred to as CpG dinucleotides) in vertebrates and is a prerequisite for normal embryogenesis; the DNA methyltransferases DNMT1, DNMT3a and DNMT3b are all essential for early mouse development [23, 24]. DNA methylation can be reversed passively (lack of maintenance during DNA replication) and by members of the Ten-eleven translocation (TET) family of 2OG-Fe(II) dioxygenases which catalyze the hydroxylation of 5mC to generate 5-hydroxymethylcytosine (5hmC), which can be further modified to 5-formylcytosine and 5-carboxylcytosine and subsequently can be removed by base excision repair [13, 25-27].
DNA methylation in somatic cells portrays a bimodal configuration, in which the majority of CpG sites are methylated; unmethylated CpGs are primarily found in clusters, known as CpG islands (CGIs), which are frequently associated with gene promoters [28, 29]. Two-thirds of gene promoters in mammals are associated with CGIs. 5hmC signatures are enriched at sites of DNaseI hypersensitivity, which are indicative of genomic regions bound by regulatory proteins, whereas 5mC is generally much less abundant at these locations. 5hmC is particularly enriched near transcription start sites and at active and poised enhancers.
In mouse the paternal genome derived from sperm shows 80 to 90% overall CpG methylation but is almost completely demethylated shortly after fertilization (Table 1) [30, 31]. TET3, which is abundant in oocytes and zygotes, plays an important role in this active demethylation process by oxidizing 5mC, which is then passively lost during subsequent rounds of replication [32-34]. TET1 is not required for pluripotency and development, although its loss causes a decrease of 5hmC levels in ES cells and a skewed differentiation towards trophectoderm in vitro, suggesting a role in inner cell mass specification [35-37]. In addition, loss of TET1 causes skewed differentiation and allows ES cells to colonize the placenta in embryo chimeras [38]. TET2 is important for maintaining active chromatin at the Hoxa cluster during differentiation [39]. Knockdown of Tet1 and Tet2 reduces the expression of pluripotency-related genes including Esrrb, Prdm14 and Klf2 [40].
Whereas the paternal genome is actively demethylated in a TET3-dependent fashion, the maternal genome shows lower global methylation levels (40%) and undergoes replication-dependent (passive) demethylation, leading to the observed epigenetic asymmetry [41]. In this way the early mouse totipotent zygote is devoid of DNA methylation except at imprinted regions. Erasure of gametic methylation patterns and genomic remethylation are not required for pluripotency. Although the genome of DNMT3a, DNMT3b, and DNMT1 triple-knockout mouse ES cells is hypomethylated, these cells retain self-renewal and pluripotency but exhibit a host of differentiation defects [42]. The global demethylation may contribute to a relatively open and accessible pluripotent chromatin state.
2.2 DNA methylation dynamics in non-mammalian vertebrates
Promoters in non-mammalian species tend to be much less CpG-dense and many do not meet the general sequence criteria of CGIs, but like mammalian CGI promoters they contain a cluster of unmethylated CpG dinucleotides and they also have a similar core promoter architecture [43-45].
In zebrafish the oocyte methylome is significantly hypomethylated compared to sperm, similar to mammals [46, 47]. The zygote shows an average of the DNA methylation levels of sperm and oocytes immediately after fertilization, which gradually increases to the level observed in sperm by the MBT (Table 1). Genes which, against the general trend, are hypermethylated in oocytes, are demethylated during early development [47], showing that both the generally hypomethylated oocyte genome as well as specific hypermethylated oocyte genes are remodeled to a sperm-like profile during early embryogenesis. Importantly, maternal haploid embryos remodel their epigenome to a sperm-like pattern, showing that the paternal methylome is not instructive for maternal methylome remodeling [47]. By the time of ZGA and the emergence of pluripotent cells the zebrafish embryonic genome is relatively hypermethylated with no major changes before and after the onset of transcription. This is in striking contrast to the mouse genome which is globally hypomethylated by the time of ZGA, to be remethylated during pre-implantation development [7]. Another difference is the extent of methylome remodeling. Early reports indicated an absence of global demethylation in both fish and frogs [48, 49] and the remodeling of the maternal methylome identified in recent zebrafish studies indeed does not lead to global demethylation but affects specific sequences [46, 47].
In Xenopus, genomic DNA in sperm as well as embryos of early blastula and later stages are globally hypermethylated [44, 49]. Depletion of maternal dnmt1 by antisense RNA during cleavage stages is associated with a decrease in the genomic 5mC content and leads to the activation of zygotic transcription approximately two cell cycles earlier than normal [50]. Hypomethylation allows the early expression of mesodermal and organizer genes such as t (brachyury), cerberus, and otx2, which are subsequently down-regulated during gastrulation of the dnmt1-depleted embryos. However, Dnmt1-dependent gene repression was subsequently found to be independent of its catalytic activity, suggesting Dnmt1 can also act as a potent transcriptional repressor independent of DNA methylation [51].
Of the three TET family members in mammals, only two (tet2 and tet3) are present in the Xenopus genome [52]. In contrast to mouse, in which Tet3 is abundant in oocytes and zygotes and contributes to active demethylation of sperm DNA, frog tet3 is extremely low in oocytes and cleavage stage embryos, consistent with an absence of global demethylation in Xenopus early development. Tet3 expression increases after the MBT and peaks at neurula and neural tube stages. Knockdown experiments showed that Tet3 and its catalytic activity are important for neural development and the expression of neural and eye genes [52].
In summary, these data point to a major difference in global DNA methylation dynamics between vertebrates (Table 1). Whereas the mouse genome is globally demethylated from ZGA through the pluripotency at the blastocyst stage, the zebrafish genome is not globally demethylated in early development and is hypermethylated during ZGA and pluripotency. Data on DNA methylation is more limited in Xenopus, but it is clear that genomic DNA is also globally hypermethylated during ZGA and blastula stages, similar to the situation in fish.
2.3 Direct and indirect reading of DNA methylation
The studies cited above indicate that the temporal paths and the extent of DNA methylation remodeling are different between mammals and fish, and that global hypomethylation is not a conserved aspect of ZGA or pluripotency among vertebrates. This may suggest that, generally, not the demethylated state, but the act of remodeling is significant for early development. DNA methylation is known to recruit methyl CpG binding proteins, which cause repression of transcription [53]. However, contrary to this general paradigm of DNA methylation-associated repression, methylated DNA can also recruit many other proteins including transcriptional activators, and methylated promoters can drive active transcription in some cases [53-57]. This diversity in methylation readout is probably highly regulated. Notable differences have been observed in DNA methylation readers in mouse ES cells, neural progenitor cells (NPCs) and brain tissue. For example, the MBD2-containing NuRD complex is a reader of 5mC in NPCs and brain, but not in ES cells [56]. Moreover, in both oocytes and late stage embryos of Xenopus methylated promoter templates are strongly repressed in cis, however this is not observed in early blastula and gastrula embryos. In early embryos CpG-dense methylated promoters drive active transcription while DNA methylation is maintained. These results point to a temporal uncoupling of DNA methylation and repression of transcription during development [44].
Interestingly, in mouse ES cells, global DNA methylation status depends on the culture conditions. When grown in 2i media, the genome is globally hypomethylated in much the same way as the ICM cells of the blastocyst, whereas ES cells grown in serum plus LIF are also pluripotent but relatively hypermethylated and in this respect more similar to the mouse epiblast [58, 59]. Strikingly, mouse ES cells lacking all three DNA methyl transferases, DNMT1, DNMT3a and DNMT3b, are viable but cannot differentiate [42]. These data suggest that DNA methylation is dispensable for pluripotency but not differentiation.
Interestingly, DNA methylation patterns in fish, frogs and mammals can be predicted on the basis of conserved DNA sequence content [60] and inserting DNA sequences in a defined locus determines their DNA methylation state in a fashion that depends on CpG density and other sequence content, including transcription factor motifs [61]. Moreover, histone modifications such as the promoter mark H3K4me3 (histone H3 lysine 4 trimethylation) and the Polycomb mark H3K27me3 (histone H3 lysine 27 trimethylation) are preferentially targeted to unmethylated regions. The locations of each of these histone modifications can, like DNA methylation itself, be predicted based on DNA sequence content [60]. This interaction between the DNA methylome and histone modifications raises the possibility that early embryonic remodeling of the methylome serves other functions than altering the patterns of methylation-dependent transcriptional repression. Indeed, remodeling of DNA methylation has been found to influence active and repressive histone modifications in the process of reprogramming fibroblasts to induced pluripotent stem (iPS) cells [62]. As such, “indirect reading” of DNA methylation by influencing other epigenetic modifications may have a very strong impact on pluripotency independent of the classical function of DNA methylation in repression of transcription. In addition, remodeling of DNA methylation in early developmental stages may influence gene expression at later stages by a more direct readout of interfering with DNA binding of transcription factors and targeting transcriptional repression. This may be very important during differentiation which requires the DNA methyltransferases [42].
3. Histones and histone modifications
The major proteins in chromatin are the core histone proteins H2A, H2B, H3, and H4, which form nucleosome particles. The N-terminal tails undergo a diverse array of posttranslational modifications and regulate the accessibility to the underlying DNA. These modifications are deposited and removed by enzymes (‘writers’ and ‘erasers’) and are recognized by specific proteins (‘readers’) which causes a variety of functional outcomes such as transcriptional activation or repression of genes (reviewed in refs. [20, 63-65]). These chromatin states can be transmitted from mother to daughter cells. Lysine acetylation correlates with chromatin accessibility, recruitment of specific proteins and transcriptional activity, whereas lysine methylation can have different effects depending on which residue is modified. Both ES cells and early embryos have a relatively open chromatin with histones showing relatively high levels of modifications that are involved in gene activation such as histone H3 lysine 4 trimethylation (H3K4me3), acetylation of lysines 9 and 14 (H3K9ac, H3K14ac), and histone H4 acetylation (reviewed in [65, 66]). H3K4me3 is found at active promoters while methylation of another lysine, H3K36me3, is associated with transcribed chromatin and reflects RNAPII activity. In contrast, H3K9me3, H3K27me3 and H4 lysine 20 (H4K20me3) reflect repressive chromatin states. In ES cells relatively low levels are observed of modifications associated with gene repression like histone H3 lysine 9 methylation (H3K9me2 and H3K9me3) compared with those in differentiated cells. For example large, megabase scale chromosomal regions marked with H3K9me2, referred to as large organized chromatin K9 modifications (LOCKs) are decreased markedly in ES cells: LOCKs decorate 4% of the genome in ES cells, compared to 10, 31 and 46% respectively in brain, differentiated ES cells and liver [67]. These domains depend on the H3K9 methyltransferase EHMT2 (G9A). Tissue-specific methylated K9 domains are associated with tissue-specific repression of transcription. These findings illustrate the relatively open nature of pluripotent chromatin relative to that of differentiated cells.
3.1 Histone H3 K4 and K27 methylation
Genome-wide epigenome maps of mouse ES cells, neural progenitor cells and embryonic fibroblasts revealed that H3K4me3 and H3K27me3 can be used to uncover expressed, poised or repressed genes, allowing the profiling of cells for lineage potential [68, 69]. When these two modifications, activating H3K4me3 and repressing H3K27me3, decorate the same genomic location the chromatin is referred to as bivalent [16]. H3K27 methylation is catalyzed by the Polycomb Repressive Complex (PRC) 2. PRC1 and PRC2 are among the most extensively studied histone modifying complexes involved in gene repression. PRC2 contains SUZ12, EZH2, EED, and RBAP46/48 as core components and catalyzes H3K27 methylation through the methyltransferase activity of EZH2. H3K27me2 and H3K27me3 can recruit PRC1, which induces monoubiquitylation of H2AK119 with the ubiquitin ligases RING1A and RING1B in PRC1. PRC1 and PRC2, however, also have independent functions. H3K27me3 is a marker for developmentally repressed genes, typically decorating the promoters and transcription initiation sites of these genes. Many of its targets encode transcription factors important for differentiation. The Polycomb group of proteins are classical antagonists of the Trithorax group of proteins, which include the H3K4 methyltransferases [15, 70].
The remarkable phenomenon of bivalency with opposing H3K4me3 and H3K27me3 modifications was described first in ES cells but was subsequently found in many other systems [69]. Bivalent domains have garnered attention because the H3K27me3 mark might help repress cell lineage-regulatory genes during pluripotency while the H3K4me3 mark could poise these genes for activation upon differentiation. In serum-grown ES cells, most bivalent promoters are transcribed at very low levels (with some exceptions, such as Klf4, Klf5 and Rex1) and are considered to be poised for activation by developmental signals.
In mammals six methyltransferase proteins, SET1A, SET1B and MLL1 to MLL4, found in COMPASS-like complexes, are capable of methylating H3K4 [17]. In fibroblasts, MLL1 is required for the H3K4 trimethylation of a small subset of promoters, including a subset of Hox genes [71]. Loss of H3K4 methylation at these loci causes reduced levels of transcription initiation by RNA polymerase II. MENIN, a shared subunit of both MLL1 and MLL2 complexes, is required for most of H3K4 methylation at Hox loci, whereas this does not depend on the MLL3, MLL4 or SET1 complexes. In line with these findings, in mouse ES cells MLL2 is required for H3K4 methylation of bivalent genes that are also regulated by H3K27me3, including many of the Hox genes [72, 73]. Loss of MLL2, however, has very little effect on gene activation during ES cell differentiation.
The H3K4me3 and H3K27me3 modifications are unlikely to occur on the same histone H3 tail [74]. Two copies of H3 are present in the nucleosome octamer, however, which could accommodate the two modifications. This would correspond to an asymmetrically modified nucleosome or asymmetric bivalency, rather than intra-molecular bivalency (Fig. 2). In heat maps of the genomic distribution of histone modifications of bivalent chromatin around transcription start sites, it is apparent that the H3K4me3-positive center of bivalent promoter regions contains less H3K27me3 signal compared to flanking regions [75], suggesting the presence of poly-nucleosomal bivalency (Fig. 2). In this type of bivalency adjacent nucleosomes contain the opposing histone modifications. In some cases cell populations in which bivalency is observed are heterogeneous. In this case the opposing histone modifications may be present on the same locus in different cells [76]. This represents pseudo-bivalency among cells with different patterns of epigenetic modifications (Fig. 2). Pseudo-bivalency however, cannot explain the co-occurrence of H3K4 and H3K27 methylation in highly homogenous cells such as naïve pluripotent stem cells [75]. In addition, mass spectrometry-based analyses of ES cells have indicated that 15% of mononucleosomes modified with H3K4me3 also carry H3K27me3 on the second copy of histone H3 within the nucleosome [77]. Together these studies suggest that asymmetric, poly-nucleosomal and pseudo-bivalency all contribute to the observations of co-occurring H3K4me3 and H3K27me3 in chromatin in ES cells.
Figure 2.
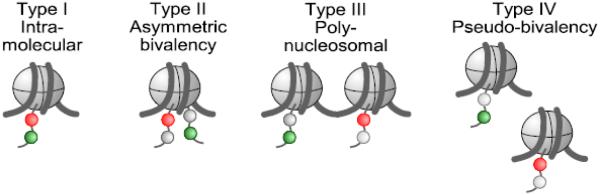
Different types of bivalent chromatin. Bivalent chromatin is defined by the co-occurrence of histone H3 lysine 4 and lysine 27 trimethylation (H3K4me3 and H3K27me3). In the case of Intra-molecular bivalency they would co-occur on the same H3 tail; however this is unlikely to happen in vivo (see text). Asymmetric bivalency: Nucleosomes can carry the two histone modifications at two different H3 tails within the same nucleosome. Poly-nucleosomal bivalency: Adjacent nucleosomes can carry the opposing histone modifications. Pseudo-bivalency: Different cells within a population (or alleles within a cell) carry different histone modifications. This type is highly relevant in vivo where pluripotency is a transient and unstable state, leading to lineage commitment in different cells. H3K4me3 and H3K27me3 are indicated in green and red respectively.
During mouse early development, both H3K4me3 and H3K27me3 appear from the 2-cell stage onwards (Table 1). In the pre-implantation blastocyst, the inner cell mass (ICM) contains the pluripotent cells that will give rise to the embryo proper, whereas trophectoderm and primitive endoderm cells will contribute to extra-embryonic tissue. Trophoblast stem cells and endoderm stem cells (cultured cells derived from the embryo) show very little H3K27 methylation in contrast to ES cells, whereas these cells have similar levels of H3K4me3 [78]. Similarly, ICM and trophectoderm show different H3K27me3 levels [79]. Therefore, there is significant H3K27 methylation in mouse ICM cells. The H3K27me3 modification however is not universally abundant in pluripotent cells in vitro. When mouse ES cells are grown in 2i medium, which causes the cells to be in a ground state of pluripotency that is less poised to differentiation, H3K27me3 is much reduced in bivalent domains compared to cells grown in serum plus LIF [75].
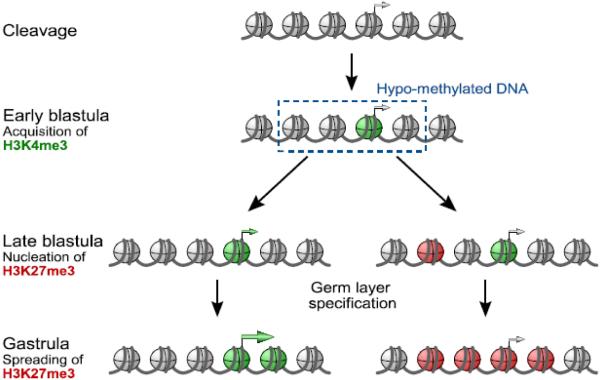
In Xenopus early embryos, bivalent chromatin was observed, but not as a quantitatively dominant feature [80]. Instead, the marks accumulate by and large with different spatio-temporal kinetics during the time between the MBT and the end of gastrulation. H3K4me3 and H3K27me3 segregate spatially within the embryo (pseudo-bivalency, Figs. 2 and 3), reflecting localized gene expression in different cells. Moreover H3K4me3 precedes accumulation of H3K27me3 and classical bivalency is present at low levels but is not a dominant feature of the pluripotent chromatin state in Xenopus. Interestingly, H3K27me3 nucleates first in promoter-distal locations in blastula embryos, before accumulating in larger domains (Fig. 3), highlighting the spatio-temporal hierarchy of H3K4 and H3K27 methylation during pluripotency and germ layer induction [60, 80]. Consistent with these findings, Geminin promotes Polycomb binding and H3K27 methylation and contributes to the transition from pluripotency to lineage commitment by preventing multi-lineage commitment [81]. The abundance of histone modifications has also been determined by quantitative mass spectrometry; the results showed relatively abundant H3K4me3 from early stages on, whereas H3K27me3 accumulated over time and was much less abundant at any stage compared to mouse ES cells [82].
Figure 3.
Histone modifications and DNA methylation in relation to pluripotency and germ layer induction in Xenopus. Overall the genome is hypermethylated, but both H4K4me3 and H3K27me3 emerge in unmethylated CpG islands with different kinetics.
Bivalent chromatin has been reported in early zebrafish embryos, immediately after the MBT [83]. Also in zebrafish embryos K4 accumulates first, being very abundant at the MBT, with K27 methylation accumulating significantly between the MBT and 50% epiboly (mid-gastrulation) [84]. These studies have also highlighted the accumulation of H3K4me3 before the MBT and transcriptional activation, suggesting a role for maternal factors in setting a permissive mark for transcription in preparation for the pluripotent stage of development.
3.2 Enhancer histone modification signatures
Enhancers are distinguished from promoters by robust levels of H3K4me1 while H3K4me3 is absent or very low [85]. In addition they may be decorated with H3K27 acetylation and feature the recruitment of RNAPII and the EP300 / CBP histone acetyltransferase which catalyzes H3K27 acetylation. Several hundred thousand enhancers have been identified in the human genome by profiling these histone modifications and the transcriptional coactivator EP300 [86-90]. Many of these enhancers drive cell type-specific gene expression, for example in ES cells. Enhancers decorated with both H3K4me1 and H3K27ac are considered active enhancers, as opposed to primed enhancers only marked with H3K4me1, and the active enhancer subset is more predictive of developmental state than all enhancers [86, 90]. In addition, H3K4me1-positive enhancers may be enriched for H3K27me3 rather than H3K27ac, which are referred to as poised enhancers. Profiling these histone modifications in ES cells has uncovered human enhancers that are poised in ES cells but become active during differentiation [89]. Validation of the activity of these human enhancers in zebrafish embryos showed that they are able to direct cell-type and stage-specific expression even in the absence of sequence conservation in the fish genome. During differentiation a subset of poised enhancers acquires H3K27 acetylation, RNAPII association and enhancer RNAs. These data show that developmental enhancers are epigenetically pre-marked and kept in a poised state during pluripotency. There are indications that the H3K4me1 mark is important not just for poised or primed enhancers, but also for active enhancers. For example, the Drosophila Trr complex and its mammalian MLL3 and MLL4 counterparts, are involved in H3K4 monomethylation at enhancers [91-93]. These complexes also contain UTX (KDM6A), an H3K27 demethylase which counteracts PRC2-mediated H3K27 methylation. This is significant as the lysine cannot accommodate both acetylation and methylation simultaneously. Indeed, loss of Trr/MLL4 leads to global reduction of H3K27acetylation, implicating the MLL3/4 complexes in the function of both poised and active enhancers.
Profiling of enhancers in Xenopus blastula embryos using H3K4me1 uncovered many enhancers, a subset of which recruited RNAPII, and another partially overlapping subset recruited the PRC2 catalytic subunit Ezh2 and the PRC2-associated factor Jarid2 [60], illustrating the dynamic balance of opposing activities present on enhancers during pluripotency. In zebrafish, changes in H3K27 acetylation enrichment accompany a shift from pluripotent to tissue-specific gene expression [94]. A marked increase in the number of enhancers was observed from the dome stage (blastula) to 80% epiboly (gastrula). This illustrates the high complexity of gene expression associated with germ layer specification and patterning. Enhancers that lose H3K27 acetylation at blastula and gastrula stages are enriched for pluripotency factor Pou5f1 (Oct4) and Sox2 binding sites, and the majority of them overlap with genomic peaks of the Nanog-like factor at the dome stage [94]. Like the mammalian pluripotency factors, the zebrafish Nanog-like protein likely contributes to the pluripotent program and its down regulation or eviction could facilitate developmental progression.
3.3 Histone variants and linker histones
The histone proteins are highly conserved; however there are nonallelic variants of the major histones that show a divergence ranging from almost no amino acid differences to significant changes. Localized replacement of a canonical histone with a variant can alter chromatin state. Among the core histones, H2A has the largest number of variants, including H2A.Z, MacroH2A, H2A-Bbd, H2AvD, and H2A.X [95-97]. H2A-H2B dimers can be replaced by several different H2A variants independently of replication. Histone variant incorporation is one of the mechanisms to create different states through a combination of altered chromatin structure or trans-acting factors. For instance, depletion of H2A.Z leads to reduced chromatin accessibility with a decrease in the binding of both active and repressive complexes and a reduction in the binding of the pluripotency transcription factor OCT4 to its binding sites at pluripotency genes. As a consequence the cells showed a reduction in the efficiency of self-renewal [98]. A study investigating the reprogramming of mammalian nuclei transplanted to Xenopus oocytes showed that the HIRA-dependent deposition of H3.3 at the oct4 locus was required for transcriptional reprogramming to a pluripotent state [99]. Another study in Xenopus investigated the role of H3.3 during embryonic development. Partial depletion of H3.3 results in abnormal development, whereas substantial depletion of both H3.1 and H3.3 showed a distinct gastrulation arrest phenotype. Using lineage marker analyses, the defect was attributed to a loss of competence to respond to mesoderm inducing cues and this defect was attributed to perturbation in chromatin organization brought about by loss of H3 and abnormal incorporation of linker histone H1A [100]. Replacement of an oocyte-specific linker histone by somatic linker histones has been shown to contribute to a loss of mesoderm competence[101]. The H1 linker histone contributes to silencing of pluripotency factors and participates in mediating changes in DNA methylation and histone marks necessary for silencing of pluripotency genes during differentiation [102]. These findings illustrate that the pluripotency gene network requires both the incorporation and loss of specific histone variants, and that remodeling chromatin state plays a role in both establishing and the exit of pluripotency.
4. Chromatin remodeling
As outlined above, chromatin state is affected by myriad proteins, including enzymes that catalyze the particular chemical modifications, proteins that recognize and bind the modifications, and the enzymes that remove this modification. In addition, chromatin state is affected by chromatin remodeling complexes that enable context-dependent access to packaged DNA. Chromatin remodelers act on nucleosomes, the basic repeating unit of packaged DNA. They use the energy of ATP hydrolysis to move, destabilize, eject, or restructure nucleosomes and thereby increase the fluidity of chromatin. There are four families of nucleosome remodelers [103, 104]: (1) SWI/SNF [switching defective/sucrose nonfermenting], also called BAF [Brg/Brahma-associated factor]; (2) ISWI [imitation switch]; (3) CHD [chromodomain, helicase, and DNA binding]; and (4) INO80 [inositol requiring 80]). All these families of nucleosome remodelers are involved in diverse activities, including transcription, DNA repair, and DNA replication. As such many subunits of the complexes are essential for the maintenance of pluripotency and proliferation of ES cells.
The SWI/SNF (BAF) complexes of mammals contain either BRM or BRG1 as nucleosomal ATPase subunits. Whereas BRM is dispensable for mouse development, BRG1 is essential for inner cell mass and trophectoderm survival and preimplantation development [105]. esBAF is an ES cell-specific SWI/SNF complex, characterized by subunits BRG1, BAF155 and BAF60A, and the absence of BRM, BAF170, and BAF60C. esBAF is required for the maintenance of pluripotency. Like BRG1, BAF155 is important for preimplantation development, maintenance of Oct4 expression and proliferation of ES cells [106]. In ES cells, knockdown of BRG1 has been shown to downregulate pluripotency genes and upregulate differentiation genes. BRG1 not only occupied the promoters of pluripotency genes but also occupied a subset of OCT4, SOX2, NANOG and PcG protein target genes [107]. esBAF is characterized by the presence and absence of specific subunits and this complex composition has been shown to direct its specific role in the core pluripotency network [106]. In murine ES cells, esBAF has been shown to be involved in a complex regulatory circuitry. BRG1-esBAF functions positively with PcG to repress differentiation genes (all four Hox loci), but prevents PcG mediated repression of pluripotency genes activated by ES cells transcription factors like OCT4, SOX2 and STAT3 [108].
CHD11 is necessary for the maintenance of open chromatin in ES cells [109]. Knockdown of Chd1 increases heterochromatin in ES cells and reduced Oct4 expression. Surprisingly this knockdown decreased the expression of only 25 genes, suggesting a functional redundancy with other chromatin remodeling complexes. A much larger group of genes was upregulated upon Chd1 knockdown, including genes involved in neurogenesis. Although ES cells with Chd1 knockdown can maintain an undifferentiated state, the cells lose the ability to differentiate into primitive endoderm and tend to differentiate into neural lineages, indicating that CHD1 is necessary for the maintenance of pluripotency [109]. More recently CHD1 has been specifically implicated in maintaining high levels of RNAPI and II in transcribing protein-coding genes and ribosomal DNA, thereby sustaining the transcriptional competence of pluripotent cells [110].
Nucleosome remodeling deacetylase (NuRD) complexes contain Mi2 (Chd3 / Chd4) nucleosomal ATPase, as well as methyl-CpG-binding proteins MBD2 or MBD3, histone deacetylase and a number of other subunits. This complex is the most abundant source of histone deacetylase in Xenopus eggs and early embryos [111]. Mouse ES cells lacking MBD3 are viable but cannot differentiate properly and show LIF-independent self-renewal [112]. A functional NuRD complex has been implicated in maintaining transcriptional heterogeneity in ES cell populations by inhibiting pluripotent gene expression and promoting exit from pluripotency [113]. Several hundred genes show increased expression in MBD3 knockdown, decreased expression in BRG1 knockdown and a wild-type like expression in double knockdowns, uncovering their opposing effects [114]. Unlike ES cells, MBD3-deficient blastocyst ICM cells grown ex vivo fail to maintain a population of Oct4-positive pluripotent cells [115]. On a similar note, MBD3 is required for reprogramming and the derivation of induced pluripotent stem (iPS) cells [116]. Depleting MBD3/NuRD complex in somatic cells, impairs somatic cell reprogramming by establishing heterochromatic features and repression of key pluripotency genes [117].
Like esBAF, the INO80 nucleosome remodeling complex is found at pluripotency genes along with Oct4, Sox2 and Nanog [118]. It mediates the recruitment of RNAPII and maintains open chromatin structure in ES cells as shown by micrococcal nuclease and DNAseI hypersensitivity assays. Knockdown of Ino80 causes a decreased expression of pluripotency factors and a loss of ES cell morphology. Expression of Ino80 is important for iPS cell colony formation. Knockdown of Ino80 in early embryos caused impaired blastocyst formation and reduced expression of pluripotency factors in the ICM [118]. The role of chromatin remodeling in Xenopus and zebrafish pluripotency has yet to be determined. In mouse, the results discussed above illustrate how mobilization of nucleosomes and/or nucleosome eviction at gene-regulatory regions is required for development and maintenance of pluripotency.
5. The pluripotency transcription factor network
The concomitant actions of cis- and trans-acting elements orchestrate the early developmental programs, influencing the cascade of events that leads to functional pluripotency. In this section we will highlight how transcription factors control the emergence of pluripotency.
5.1 The core pluripotency network in ES cells
OCT4, SOX2 and NANOG form a “core” network of pluripotency transcription factors [119, 120]. They are co-recruited to regulatory elements of target genes and form auto-regulatory and feedforward loops that provide stability to pluripotency gene expression and suppress lineage-specific gene expression. These factors regulate each other and process signals to direct self-renewal and block differentiation by maintaining the expression of the pluripotency network. The binding of these transcription factors is transduced by the mediator complex to recruit general transcription factors to the promoter, which promote the recruitment of RNAPII complex. The Mediator subunit MED12 interacts with Nanog and these factors work together in ES cell gene regulation [121]. In addition to the well-characterized core network of OCT4, SOX2 and NANOG, an additional set of transcription factors, including ESRRB, TFCP2L1, KLF2, and KLF4 make important contributions to the pluripotent regulatory circuitry and can reset human ES cells to ground-state pluripotency [122, 123]. Three KLF transcription factors, KLF2, KLF4 and KLF5, share many transcriptional targets with NANOG and regulate Nanog and other pluripotency genes [124].
Mouse ES cells can not only be grown in 2i or serum plus LIF conditions, conditions that correspond to naïve (ground-state) and primed pluripotent states, they can also be primed by exposure to FGF2 and activin, leading to the formation of a transient epiblast-like cells (EpiLCs). ES cells and EpiLCs share the core transcription network of OCT4, SOX2, and NANOG. The transition between these two pluripotent states, however, is associated with widespread OCT4 relocalization accompanied by global rearrangement of enhancer chromatin landscapes [125]. Motif analyses suggested OTX2 and ZIC2/3 as mediators of primed pluripotency-specific OCT4 binding. Blocking differentiation signals and overexpressing OTX2 was sufficient to drive exit from the naive state and induce transcription of several primed pluripotency genes. OTX2 drives the early stage of ES cell differentiation in collaboration with OCT4 at enhancers [126]. ZIC3 is an activator of NANOG and contributes to maintenance of pluripotency in mouse ES cells [127, 128], but in zebrafish and Xenopus embryos it functions in neural and neural crest gene expression [129, 130]. Epiblast stem cells (EpiSCs) are derived directly from developing mouse epiblast embryos; a subset of these cells express OCT4 in addition to SOX2. However EpiSCs also express post-implantation markers and show heterogeneous expression of lineage specific genes of mesendoderm, definitive endoderm and the primitive streak [131, 132].
The transcription initiation factor TBP and many of its associated factors (TAFs) of the TFIID complex are expressed at higher levels in mouse ES cells than in somatic cells. Knockdown of these factors affected the pluripotent circuitry leading to inhibition of reprogramming of fibroblasts. Transient expression of TFIID subunits greatly enhanced reprogramming showing that TFIID is critical for transcription factor-mediated reprogramming and the maintenance of pluripotency [133].
5.2 Embryonic pluripotency transcription factors in non-mammalian vertebrates
The oct4 gene (pou5f1) has been duplicated early in the vertebrate lineage, giving rise to pou5f1 and pou5f3 [134]. Some vertebrates, for example marsupials and sharks, have retained both copies, whereas mammals have lost pou5f3 (Table 1). Both teleost fish and frogs have lost the other copy, pou5f1, but retained pou5f3. Additionally, there are three tandem-duplicated oct4-like pou5f3 genes in Xenopus: oct91 (pou5f3.1) which is not expressed in the oocyte but is induced at the MBT, oct25 (pou5f3.2) which is maternal but not translated until after fertilization, and oct60 (pou5f3.3) which is exclusively maternally expressed [135]. Sox2 is present in the Xenopus genome, but there is no clear orthologue of nanog. Its activity may be taken over by ventx1 and ventx2, which are closely related and structurally and functionally very similar to nanog. Loss of these factors leads to down-regulation of oct91 and premature differentiation, suggesting these factors play a Nanog-like role in amphibian development [136].
In Xenopus tropicalis, the zygotic genome activates in a broad wave of new transcription extending from the 7th cleavage division (early blastula) to beyond the 12th cleavage division (MBT) [137]. The Oct25 protein levels increase dramatically in early development and Oct25 is with 400 million copies per embryo among the most abundant transcription factors at the early gastrula stage [138]. Oct25 represses Nodal and Wnt signaling and antagonizes the T-box transcription factor Vegt, thereby antagonizing mesoderm induction [139, 140]. Nodal-responsive enhancers are marked with H3K4me1 and H3K27ac at the blastula stage, independent of zygotic Nodal signaling [141]. Foxh1 mediates Nodal signaling together with Smad2/3, but also functions independently and co-occupies Oct25 targets, which may modulate the transition between pluripotency and mesendoderm induction [142]. In both mouse ES cells and Xenopus embryos neural differentiation requires Suv4-20h (Kmt5b/c) to repress Oct25, illustrating the necessity to down-regulate the pluripotency network upon exit of pluripotency [143].
Two studies in zebrafish have highlighted the role of the pluripotency network in early development and zygotic genome activation (ZGA). In zebrafish ZGA coincides with the stage of embryonic pluripotency. Using ribosome profiling it was found that zebrafish Nanog, Pou5f3 (a homolog of the mammalian pluripotency transcription factor OCT4, also referred to as Pou5f1) and SoxB1 (related to Sox2) are the most highly translated transcription factors just before zygotic genome activation sets in [144]. Pou5f3 is recruited to Sox2-POU binding sites before the MBT, and similarly Nanog binds to promoters of first wave expression genes. Combined loss of these factors resulted in developmental arrest before gastrulation and a failure to activate many embryonic genes [144, 145]. These data link ZGA to the pluripotency network in zebrafish. This may be different from the situation in mouse, as genetic removal of OCT4 from mouse oocytes does not affect ZGA, totipotency-pluripotency and developmental competence [146, 147].
6. Long non-coding RNA
RNA is an integral component of chromatin and plays a role in the pluripotent regulatory circuitry. Large intergenic non-coding RNAs (lincRNAs) have been identified in ES cells [148], and shRNA loss of function experiments showed that 137 of 147 lincRNAs affected the expression of an average of 175 protein-coding genes in trans [149]. Of these, 15 lincRNAs were found to be important for the expression of multiple pluripotency markers and ES cell morphology. In addition lincRNAs were found to repress gene expression related to endoderm, (neuro-) ectoderm, mesoderm and trophectoderm differentiation, thereby maintaining pluripotency. Many regulatory sequences of these lincRNAs are bound by Oct4, Nanog and Sox2. Moreover, many of the lincRNAs interact with chromatin modifying enzymes, interacting with chromatin readers, writers and erasers, including the Polycomb complexes PRC1 and PRC2 [149]. PRC2 has also been found to interact with cis-acting RNA; at loci where these RNAs are made by RNAPII. Although this contributes to targeting of the complex, enzymatic activity seems to require the PRC2-associated factor Jarid2 [150, 151].
One lincRNA named TUNA was shown to be highly expressed when mouse ES cells differentiated toward the neural lineages. TUNA depletion inhibited neural differentiation. TUNA was shown to occupy promoter regions of Nanog, Sox2, and Fgf4, genes that are important for pluripotency and neural lineage commitment [152].
The DNA methyltransferase Dnmt1 also interacts with many cellular transcripts, including the extra-coding CEBPA lincRNA [153]. This lincRNA adopts a stem-loop structure critical for interaction with Dnmt1 and when transcribed, acts to shield the Cebpa locus from DNA methylation. This study provides another line of evidence establishing how cells employ RNAs to modulate the deposition of repressive epigenetic marks in a genome-wide manner [153]. The WDR5 subunit of MLL complexes also interacts with RNA, including many lincRNAs important for ES cell pluripotency. WDR5 RNA binding activity is specifically required for H3K4 methylation and maintenance of pluripotency in ES cells [154]. These data highlight that non-coding RNAs, some of which are transcribed under the control of the pluripotency regulatory network, can act in cis or in trans to maintain pluripotency and repress lineage specific gene expression, and they do so, at least in part, by interactions with chromatin-modifying complexes. Developmental stage-specific lincRNAs have been identified in Xenopus and zebrafish embryos [155-159], but their role in establishing pluripotency has not been determined.
7. Perspective
By its very nature embryonic development is highly dynamic, and it is no surprise that many of the underlying molecular processes are dynamic as well. At its many different levels of organization, DNA methylation, chromatin composition, histone modifications acting at diverse genomic elements, and transcription factor networks, are all acting within their own molecular logic to orchestrate development. As illustrated above, there are integral links between zygotic genome activation and the cellular state of pluripotency. In a simple scenario, exemplified by zebrafish, the core pluripotency network is instrumental for the onset of embryonic transcription while establishing the pluripotent state at the same time. In mouse, the zygote activates its genome, but first needs to produce the extra-embryonic lineages as well as the pluripotent cells of the inner cell mass of the blastocyst. Despite these differences, accumulating evidence summarized in this review shows that a great deal of functional conservation is apparent in the pluripotency network.
One of the major current challenges is to elucidate the interplay between different layers of regulation that together instruct vertebrate development. In recent years, techniques like ChIP-sequencing, RNA-sequencing, and proteomics have been applied to ES cells, their differentiated derivatives, and embryos of mouse, Xenopus, zebrafish and many others. Ongoing efforts in these areas should allow elucidating systems level views of the regulatory networks in each of these model systems. These highly fruitful endeavors will provide further insight in the differences and similarities of pluripotency in the embryo, in culture and in different species.
Acknowledgements
The authors thank Dr. Hendrik Marks for critical reading. This work has been supported by grants of the US National Institutes of Health (NICHD, grant R01HD069344) and the Netherlands Organization of Scientific Research (NWO-CW, grant 700.58.007). The authors apologize to those whose work could not be cited due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46(3):317–20. [PubMed] [Google Scholar]
- 2.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, Jin X, Shi X, Liu P, Wang X, Wang W, Wei Y, Li X, Guo F, Wu X, Fan X, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–10. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 3.Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, van Lohuizen M, Peters AH. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40(4):411–20. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 4.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280(1):225–36. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43(8):811–4. doi: 10.1038/ng.864. PMCID 3146050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511(7511):611–5. doi: 10.1038/nature13581. PMCID 4178976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484(7394):339–44. doi: 10.1038/nature10960. PMCID 3331945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–42. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 10.Solter D. From teratocarcinomas to embryonic stem cells and beyond: a history of embryonic stem cell research. Nat Rev Genet. 2006;7(4):319–27. doi: 10.1038/nrg1827. [DOI] [PubMed] [Google Scholar]
- 11.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30(3):675–86. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 12.Borchers A, Pieler T. Programming pluripotent precursor cells derived from Xenopus embryos to generate specific tissues and organs. Genes (Basel) 2010;1(3):413–26. doi: 10.3390/genes1030413. PMCID 3966229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–9. doi: 10.1038/nature12750. PMCID 4046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luna-Zurita L, Bruneau BG. Chromatin modulators as facilitating factors in cellular reprogramming. Curr Opin Genet Dev. 2013;23(5):556–61. doi: 10.1016/j.gde.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 16.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27(12):1318–38. doi: 10.1101/gad.219626.113. PMCID 3701188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. PMCID 4010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marks H, Stunnenberg HG. Transcription regulation and chromatin structure in the pluripotent ground state. Biochim Biophys Acta. 2014;1839(3):129–37. doi: 10.1016/j.bbagrm.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Welling M, Geijsen N. Uncovering the true identity of naive pluripotent stem cells. Trends Cell Biol. 2013;23(9):442–8. doi: 10.1016/j.tcb.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 22.Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–65. doi: 10.1007/s00412-009-0221-9. PMCID 2729420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 24.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 25.Piccolo FM, Fisher AG. Getting rid of DNA methylation. Trends Cell Biol. 2014;24(2):136–43. doi: 10.1016/j.tcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 26.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–7. doi: 10.1126/science.1210944. PMCID 3462231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–3. doi: 10.1126/science.1210597. PMCID 3495246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22. doi: 10.1101/gad.2037511. PMCID 3093116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28(8):812–28. doi: 10.1101/gad.234294.113. PMCID 4003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 31.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10(8):475–8. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 32.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–10. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 33.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108(9):3642–7. doi: 10.1073/pnas.1014033108. PMCID 3048122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334(6053):194. doi: 10.1126/science.1212483. PMCID 3799877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–75. doi: 10.1016/j.stem.2011.07.010. PMCID 3154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–33. doi: 10.1038/nature09303. PMCID 3491567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–8. doi: 10.1038/nature10066. PMCID Pmc3408592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–13. doi: 10.1016/j.stem.2011.01.008. PMCID 3134318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bocker MT, Tuorto F, Raddatz G, Musch T, Yang FC, Xu M, Lyko F, Breiling A. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nat Commun. 2012;3:818. doi: 10.1038/ncomms1826. PMCID 3576573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 41.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 42.Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, Nakayama J, Okano M. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11(7):805–14. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 43.Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grutzner F, Odom DT, Patient R, Ponting CP, Klose RJ. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. 2013;2:e00348. doi: 10.7554/eLife.00348. PMCID 3583005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogdanovic O, Long SW, van Heeringen SJ, Brinkman AB, Gomez-Skarmeta JL, Stunnenberg HG, Jones PL, Veenstra GJ. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res. 2011;21(8):1313–27. doi: 10.1101/gr.114843.110. PMCID 3149498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Heeringen SJ, Akhtar W, Jacobi UG, Akkers RC, Suzuki Y, Veenstra GJ. Nucleotide composition-linked divergence of vertebrate core promoter architecture. Genome Res. 2011;21(3):410–21. doi: 10.1101/gr.111724.110. PMCID 3044855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, Zhang J, Huang X, Yu M, Wang X, Liu F, Wu CI, He C, Zhang B, Ci W, Liu J. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153(4):773–84. doi: 10.1016/j.cell.2013.04.041. PMCID 4081501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153(4):759–72. doi: 10.1016/j.cell.2013.04.030. PMCID 4030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macleod D, Clark VH, Bird A. Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat Genet. 1999;23(2):139–40. doi: 10.1038/13767. [DOI] [PubMed] [Google Scholar]
- 49.Veenstra GJ, Wolffe AP. Constitutive genomic methylation during embryonic development of Xenopus. Biochim Biophys Acta. 2001;1521(1-3):39–44. doi: 10.1016/s0167-4781(01)00280-9. [DOI] [PubMed] [Google Scholar]
- 50.Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 2000;14(3):313–27. PMCID 316362. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development. 2008;135(7):1295–302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He HH, Cui Q, Clark E, Ma C, Barbara A, Veenstra GJ, Xu G, Kaiser UB, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151(6):1200–13. doi: 10.1016/j.cell.2012.11.014. PMCID 3705565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spruijt CG, Vermeulen M. DNA methylation: old dog, new tricks? Nat Struct Mol Biol. 2014;21(11):949–54. doi: 10.1038/nsmb.2910. [DOI] [PubMed] [Google Scholar]
- 54.Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, Cairns BR. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15(2):239–53. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107(47):20311–6. doi: 10.1073/pnas.1008688107. PMCID 2996703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2(2):160–9. doi: 10.1016/j.stem.2007.12.011. PMCID 3070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, Benedetti R, Altucci L, Jansen JH, Walter J, Gut IG, Marks H, Stunnenberg HG. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13(3):360–9. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, Surani MA, Hajkova P. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20(3):311–6. doi: 10.1038/nsmb.2510. PMCID 3591483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Heeringen SJ, Akkers RC, van Kruijsbergen I, Arif MA, Hanssen LL, Sharifi N, Veenstra GJ. Principles of nucleation of H3K27 methylation during embryonic development. Genome Res. 2014;24(3):401–10. doi: 10.1101/gr.159608.113. PMCID 3941105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43(11):1091–7. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 62.Lee DS, Shin JY, Tonge PD, Puri MC, Lee S, Park H, Lee WC, Hussein SM, Bleazard T, Yun JY, Kim J, Li M, Cloonan N, Wood D, Clancy JL, Mosbergen R, Yi JH, Yang KS, Kim H, Rhee H, et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat Commun. 2014;5:5619. doi: 10.1038/ncomms6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95. doi: 10.1038/cr.2011.22. PMCID 3193420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11(4):285–96. doi: 10.1038/nrg2752. PMCID 3760772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bogdanovic O, van Heeringen SJ, Veenstra GJ. The epigenome in early vertebrate development. Genesis. 2012;50(3):192–206. doi: 10.1002/dvg.20831. PMCID 3294079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–32. doi: 10.1146/annurev-cellbio-051809-102012. PMCID 3085914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41(2):246–50. doi: 10.1038/ng.297. PMCID 2632725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. PMCID 2921165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 70.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29(22):6074–85. doi: 10.1128/MCB.00924-09. PMCID 2772563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1093–7. doi: 10.1038/nsmb.2653. PMCID 3805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141(3):526–37. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- 74.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42(3):330–41. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 75.Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149(3):590–604. doi: 10.1016/j.cell.2012.03.026. PMCID 3398752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong SH, Rampalli S, Lee JB, McNicol J, Collins T, Draper JS, Bhatia M. Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell Stem Cell. 2011;9(1):24–36. doi: 10.1016/j.stem.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Voigt P, LeRoy G, Drury WJ, 3rd, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. Asymmetrically modified nucleosomes. Cell. 2012;151(1):181–93. doi: 10.1016/j.cell.2012.09.002. PMCID 3498816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci U S A. 2010;107(24):10783–90. doi: 10.1073/pnas.0914507107. PMCID 2890770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P. Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS One. 2010;5(2):e9150. doi: 10.1371/journal.pone.0009150. PMCID 2818844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17(3):425–34. doi: 10.1016/j.devcel.2009.08.005. PMCID 2746918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim JW, Hummert P, Mills JC, Kroll KL. Geminin cooperates with Polycomb to restrain multi-lineage commitment in the early embryo. Development. 2011;138(1):33–44. doi: 10.1242/dev.059824. PMCID 2998164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider TD, Arteaga-Salas JM, Mentele E, David R, Nicetto D, Imhof A, Rupp RA. Stage-specific histone modification profiles reveal global transitions in the Xenopus embryonic epigenome. PLoS One. 2011;6(7):e22548. doi: 10.1371/journal.pone.0022548. PMCID 3142184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464(7290):922–6. doi: 10.1038/nature08866. PMCID 2874748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Ostrup O, Winata C, Mathavan S, Muller F, Alestrom P, Collas P. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell. 2011;21(6):993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 85.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 86.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. PMCID 3003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–12. doi: 10.1038/nature07829. PMCID 2910248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–8. doi: 10.1038/nature07730. PMCID 2745234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–83. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–37. doi: 10.1016/j.molcel.2013.01.038. PMCID 3857148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26(23):2604–20. doi: 10.1101/gad.201327.112. PMCID 3521626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33(23):4745–54. doi: 10.1128/MCB.01181-13. PMCID 3838007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. PMCID 3869375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ, Gomez-Skarmeta JL. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 2012;22(10):2043–53. doi: 10.1101/gr.134833.111. PMCID 3460198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19(5):662–74. doi: 10.1016/j.devcel.2010.10.014. PMCID 3033583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19(3):295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 97.Skene PJ, Henikoff S. Histone variants in pluripotency and disease. Development. 2013;140(12):2513–24. doi: 10.1242/dev.091439. [DOI] [PubMed] [Google Scholar]
- 98.Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12(2):180–92. doi: 10.1016/j.stem.2012.11.003. PMCID 3570599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin. 2012;5(1):17. doi: 10.1186/1756-8935-5-17. PMCID 3538669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lim CY, Reversade B, Knowles BB, Solter D. Optimal histone H3 to linker histone H1 chromatin ratio is vital for mesodermal competence in Xenopus. Development. 2013;140(4):853–60. doi: 10.1242/dev.086611. [DOI] [PubMed] [Google Scholar]
- 101.Steinbach OC, Wolffe AP, Rupp RA. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389(6649):395–9. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B, Ho PY, Medrzycki M, Berhe DT, Pan C, McDevitt TC, Fan Y. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012;8(5):e1002691. doi: 10.1371/journal.pgen.1002691. PMCID 3349736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21(3):396–420. doi: 10.1038/cr.2011.32. PMCID 3110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saladi SV, de la Serna IL. ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev. 2010;6(1):62–73. doi: 10.1007/s12015-010-9120-y. PMCID 2862992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 106.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106(13):5181–6. doi: 10.1073/pnas.0812889106. PMCID 2654396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27(2):317–28. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 108.Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13(8):903–13. doi: 10.1038/ncb2285. PMCID Pmc3155811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460(7257):863–8. doi: 10.1038/nature08212. PMCID 3891576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guzman-Ayala M, Sachs M, Koh FM, Onodera C, Bulut-Karslioglu A, Lin CJ, Wong P, Nitta R, Song JS, Ramalho-Santos M. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development. 2015;142(1):118–27. doi: 10.1242/dev.114843. PMCID Pmc4299150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8(14):843–6. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 112.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8(3):285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 113.Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O'Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, Costello I, Humphreys P, Mansfield W, Nakagawa K, Strouboulis J, Behrens A, Bertone P, Hendrich B. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10(5):583–94. doi: 10.1016/j.stem.2012.02.020. PMCID Pmc3402183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147(7):1498–510. doi: 10.1016/j.cell.2011.11.054. PMCID Pmc3252821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134(6):1123–32. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 116.dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JC. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15(1):102–10. doi: 10.1016/j.stem.2014.04.019. PMCID 4082719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, Shen M, Zong L, Lyu G, Zhao Y, Ye T, Gu J, Tao W, Lu Z, Grummt I. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31(7):1278–86. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 118.Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao YL, Zhou B, Han L, Fargo DC, Jothi R, Williams CJ, Wade PA, Hu G. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14(5):575–91. doi: 10.1016/j.stem.2014.02.013. PMCID 4154226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–56. doi: 10.1016/j.cell.2005.08.020. PMCID 3006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 121.Tutter AV, Kowalski MP, Baltus GA, Iourgenko V, Labow M, Li E, Kadam S. Role for Med12 in regulation of Nanog and Nanog target genes. J Biol Chem. 2009;284(6):3709–18. doi: 10.1074/jbc.M805677200. [DOI] [PubMed] [Google Scholar]
- 122.Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naive pluripotency. Science. 2014;344(6188):1156–60. doi: 10.1126/science.1248882. PMCID 4257066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, Reik W, Bertone P, Smith A. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–69. doi: 10.1016/j.cell.2014.08.029. PMCID 4162745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10(3):353–60. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 125.Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14(6):838–53. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang SH, Kalkan T, Morissroe C, Marks H, Stunnenberg H, Smith A, Sharrocks AD. Otx2 and Oct4 drive early enhancer activation during embryonic stem cell transition from naive pluripotency. Cell Rep. 2014;7(6):1968–81. doi: 10.1016/j.celrep.2014.05.037. PMCID 4074343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lim LS, Hong FH, Kunarso G, Stanton LW. The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. Stem Cells. 2010;28(11):1961–9. doi: 10.1002/stem.527. [DOI] [PubMed] [Google Scholar]
- 128.Lim LS, Loh YH, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng HH, Stanton LW. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol Biol Cell. 2007;18(4):1348–58. doi: 10.1091/mbc.E06-07-0624. PMCID 1838990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winata CL, Kondrychyn I, Kumar V, Srinivasan KG, Orlov Y, Ravishankar A, Prabhakar S, Stanton LW, Korzh V, Mathavan S. Genome wide analysis reveals Zic3 interaction with distal regulatory elements of stage specific developmental genes in zebrafish. PLoS Genet. 2013;9(10):e1003852. doi: 10.1371/journal.pgen.1003852. PMCID 3814314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc Natl Acad Sci U S A. 1997;94(22):11980–5. doi: 10.1073/pnas.94.22.11980. PMCID 23676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Han DW, Tapia N, Joo JY, Greber B, Arauzo-Bravo MJ, Bernemann C, Ko K, Wu G, Stehling M, Do JT, Scholer HR. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143(4):617–27. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 132.Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA, Power MD, Loebel DA, Jones V, Hor A, de Alencastro G, Logan GJ, Teber ET, Tam OH, Stutz MD, Alexander IE, Pickett HA, Tam PP. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14(1):107–20. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 133.Pijnappel WW, Esch D, Baltissen MP, Wu G, Mischerikow N, Bergsma AJ, van der Wal E, Han DW, Bruch H, Moritz S, Lijnzaad P, Altelaar AF, Sameith K, Zaehres H, Heck AJ, Holstege FC, Scholer HR, Timmers HT. A central role for TFIID in the pluripotent transcription circuitry. Nature. 2013;495(7442):516–9. doi: 10.1038/nature11970. [DOI] [PubMed] [Google Scholar]
- 134.Frankenberg SR, Frank D, Harland R, Johnson AD, Nichols J, Niwa H, Scholer HR, Tanaka E, Wylie C, Brickman JM. The POU-er of gene nomenclature. Development. 2014;141(15):2921–3. doi: 10.1242/dev.108407. [DOI] [PubMed] [Google Scholar]
- 135.Hinkley CS, Martin JF, Leibham D, Perry M. Sequential expression of multiple POU proteins during amphibian early development. Mol Cell Biol. 1992;12(2):638–49. doi: 10.1128/mcb.12.2.638. PMCID 364253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Scerbo P, Girardot F, Vivien C, Markov GV, Luxardi G, Demeneix B, Kodjabachian L, Coen L. Ventx factors function as Nanog-like guardians of developmental potential in Xenopus. PLoS One. 2012;7(5):e36855. doi: 10.1371/journal.pone.0036855. PMCID 3351468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, Gilchrist MJ. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141(9):1927–39. doi: 10.1242/dev.102012. PMCID 3994770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Smits AH, Lindeboom RG, Perino M, van Heeringen SJ, Veenstra GJ, Vermeulen M. Global absolute quantification reveals tight regulation of protein expression in single Xenopus eggs. Nucleic Acids Res. 2014;42(15):9880–91. doi: 10.1093/nar/gku661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cao Y, Siegel D, Donow C, Knochel S, Yuan L, Knochel W. POU-V factors antagonize maternal VegT activity and beta-Catenin signaling in Xenopus embryos. EMBO J. 2007;26(12):2942–54. doi: 10.1038/sj.emboj.7601736. PMCID 1894774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cao Y, Siegel D, Oswald F, Knochel W. Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. J Biol Chem. 2008;283(49):34168–77. doi: 10.1074/jbc.M803532200. PMCID 2662234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gupta R, Wills A, Ucar D, Baker J. Developmental enhancers are marked independently of zygotic Nodal signals in Xenopus. Dev Biol. 2014;395(1):38–49. doi: 10.1016/j.ydbio.2014.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chiu WT, Charney Le R, Blitz IL, Fish MB, Li Y, Biesinger J, Xie X, Cho KW. Genome-wide view of TGFbeta/Foxh1 regulation of the early mesendoderm program. Development. 2014;141(23):4537–47. doi: 10.1242/dev.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nicetto D, Hahn M, Jung J, Schneider TD, Straub T, David R, Schotta G, Rupp RA. Suv4-20h histone methyltransferases promote neuroectodermal differentiation by silencing the pluripotency-associated Oct-25 gene. PLoS Genet. 2013;9(1):e1003188. doi: 10.1371/journal.pgen.1003188. PMCID 3561085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503(7476):360–4. doi: 10.1038/nature12632. PMCID 3925760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leichsenring M, Maes J, Mossner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341(6149):1005–9. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- 146.Wu G, Han D, Gong Y, Sebastiano V, Gentile L, Singhal N, Adachi K, Fischedick G, Ortmeier C, Sinn M, Radstaak M, Tomilin A, Scholer HR. Establishment of totipotency does not depend on Oct4A. Nat Cell Biol. 2013;15(9):1089–97. doi: 10.1038/ncb2816. PMCID 3845671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu G, Scholer HR. Role of Oct4 in the early embryo development. Cell Regen (Lond) 2014;3(1):7. doi: 10.1186/2045-9769-3-7. PMCID 4230828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. PMCID 2754849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. PMCID 3175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55(2):171–85. doi: 10.1016/j.molcel.2014.05.009. PMCID 4107928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20(11):1250–7. doi: 10.1038/nsmb.2679. PMCID 3823624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, Head SR, Duester G, Dong PD, Rana TM. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53(6):1005–19. doi: 10.1016/j.molcel.2014.01.021. PMCID 4010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D'Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–6. doi: 10.1038/nature12598. PMCID 3870304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife. 2014;3:e02046. doi: 10.7554/eLife.02046. PMCID 3921674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJ. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics. 2013;14:762. doi: 10.1186/1471-2164-14-762. PMCID 3907017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–91. doi: 10.1101/gr.133009.111. PMCID 3290793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, Gong Z, Korzh V, Alestrom P, Mathavan S. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21(8):1328–38. doi: 10.1101/gr.116012.110. PMCID 3149499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23(1):201–16. doi: 10.1101/gr.141424.112. PMCID 3530680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vesterlund L, Jiao H, Unneberg P, Hovatta O, Kere J. The zebrafish transcriptome during early development. BMC Dev Biol. 2011;11:30. doi: 10.1186/1471-213X-11-30. PMCID 3118190. [DOI] [PMC free article] [PubMed] [Google Scholar]