Abstract
We examined the association between posttraumatic stress disorder (PTSD) and gene expression using whole blood samples from a cohort of trauma-exposed white non-Hispanic male veterans (115 cases and 28 controls). 10,264 probes of genes and gene transcripts were analyzed. We found 41 that were differentially expressed in PTSD cases versus controls (multiple-testing corrected p<0.05). The most significant was DSCAM, a neurological gene expressed widely in the developing brain and in the amygdala and hippocampus of the adult brain. We then examined the 41 differentially expressed genes in a meta-analysis using two replication cohorts and found significant associations with PTSD for 7 of the 41 (p<0.05), one of which (ATP6AP1L) survived multiple-testing correction. There was also broad evidence of overlap across the discovery and replication samples for the entire set of genes implicated in the discovery data based on the direction of effect and an enrichment of p<0.05 significant probes beyond what would be expected under the null. Finally, we found that the set of differentially expressed genes from the discovery sample was enriched for genes responsive to glucocorticoid signaling with most showing reduced expression in PTSD cases compared to controls.
Keywords: PTSD, gene expression, whole blood, DSCAM, ATP6AP1L, glucocorticoid responsive
1. Introduction
The past decade of research on the genetics of PTSD has led to important discoveries through twin studies, candidate gene studies, genome-wide association studies (GWASs), methylation, and expression studies (see e.g. Guffanti et al., 2013; Logue et al., 2013; Mehta et al., 2013; Ressler et al., 2011; Sarapas et al., 2011; Segman et al., 2005; Xie et al., 2013; Yehuda et al., 2009; Zieker et al., 2007). Gene expression studies permit measurement of the downstream effects of genetic and epigenetic variation and can potentially lead to the identification of biomarkers not only for PTSD risk, (as with DNA loci), but potentially also of the actual manifestation of the syndrome. However, gene expression studies conducted to date have been limited by small samples and/or narrow scopes of analysis focused on a limited number of candidate genes. Other studies have used a transcriptome-wide approach to study PTSD expression. Here we use transcriptome-wide to indicate an examination of the expression of genes throughout the genome without prior hypothesis about the function of any associated genes, analogous to the way “genome-wide” is used to indicate an agnostic investigation of variants throughout the genome in a genetic association study. Four of the five published studies that have used expression arrays for transcriptome-wide investigations of RNA from peripheral blood were based on samples with less than 30 cases and 30 controls (Sarapas et al., 2011; Segman et al., 2005; Yehuda et al., 2009). The exception was a study by Mehta et al. (2013) who examined PTSD-associated gene expression in 61 PTSD cases vs. 108 trauma-exposed controls. They found that genes and biological pathways implicated by comparing PTSD cases with childhood trauma to controls differed from those implicated when PTSD cases with only adult trauma were compared to controls. The investigators concluded from this that PTSD related gene expression changes might vary as a function of population (e.g. military vs. epidemiological samples) due to differences in type and/or developmental timing of trauma exposure.
Both transcriptome-wide and candidate gene expression studies of PTSD have implicated loci related to functioning of the hypothalamic-pituitary-adrenal axis and glucocorticoid signaling. For example, using a sample of 24 cases and 40 controls Ressler et al. (2011) found that expression levels of pituitary adenylate cyclase-activating polypeptide (PACAP)—a gene involved in stress response—and its 38 residue peptide, PACAP38 (encoded by ADCYAP1), were associated with PTSD symptom severity and acoustic startle reflex in women. They then replicated this finding in an all-female sample of 30 cases and 45 controls. Other candidate-gene expression studies have linked PTSD to expression of the brain-derived neurotrophic factor (BDNF) gene (see e.g. Dell'Osso et al., 2009; Matsuoka et al., 2013), which plays a role in glucocorticoid receptor (GR) regulation. Lambert et al. 2013, Yehuda et al. 2009, and Sarapas et al. 2011 reported that the FK506 Binding Protein 5 (FKBP5) gene, which is also implicated in moderating activity of the GR, was differentially expressed among participants with PTSD secondary to exposure to the World Trade Center attacks (n=15 PTSD cases and 20 controls in Yehuda et al., n=20 cases and 20 controls in Sarapas et al.). These two studies also showed that expression of the signal transducer and activator of transcription 5B (STAT5B) gene, which encodes a protein that inhibits the GR, was differentially expressed among participants with PTSD. FKBP5 and STAT5B were further implicated by Mehta et al (2011) in a candidate-gene study examining expression as a function of an FKBP5 polymorphism and PTSD severity (n=41 cases and 70 controls, FKBP5 interaction confirmed in an additional n=98 subjects; Mehta et al., 2011).
Building on previous work, the primary aim of this study was to perform a transcriptome-wide examination of gene expression in PTSD. To increase power, we initially focused our attention on a homogeneous (in sex and ancestry) discovery sample of 143 trauma-exposed white non-Hispanic male veterans. Our second aim was to examine the robustness of the findings from the discovery sample and to look for evidence of replication in two additional (more diverse) samples: women and men of other ancestral groups from the veteran cohort (n=73) and an independent civilian cohort from the Detroit Neighborhood Health Study (DNHS; n=129). To our knowledge, when considered together (N = 345), these samples constituted the largest PTSD expression study conducted to date. A final aim was to examine the discovery sample for association between lifetime PTSD and expression of candidate genes previously identified as glucocorticoid responsive.
2. Materials and Methods
2.1 Samples
We examined expression in a clinical sample of 216 veterans (final n after quality filters and removal of subjects with missing data, see supplementary materials for details) enrolled in a study on the PTSD and its comorbidities. The recruitment methods and sample composition have been described in detail elsewhere (Miller et al., 2012; Wolf et al., 2012). This is a subset of a larger cohort previously used in a genome-wide association study of PTSD (Logue et al., 2013). The research was approved and conducted under the oversight of the appropriate institutional review boards. All subjects were assessed with the Clinician Administered PTSD Scale for DSM-IV (CAPS-IV; Blake et al., 1990; Blake et al., 1995), which ascertains PTSD diagnosis per the DSM-IV diagnostic criteria. CAPS inter-rater reliability for the lifetime PTSD diagnosis, as determined through independent ratings of approximately one-third of the diagnostic interviews using videotape recordings, was excellent (kappa=0.87). The Traumatic Life Events Questionnaire (TLEQ; Kubany et al., 2000) was used to assess exposure to different types of traumatic events in both childhood and adulthood. Ancestral population was determined by self-report and analysis of genome-wide genotype data (described in detail in Logue et al., 2013).
The details of sample preparation, storage, and RNA extraction, RNA processing and data cleaning, including quality control (QC) and normalization, are presented in the supplementary materials. Expression was measured using the Illumina (San Diego, CA, USA) HumanHT12_v4 BeadChip which assesses 47,323 probes. Each probe measures the expression of a particular transcript (i.e. variant) of a particular gene. Approximately 40,000 of these map to coding gene transcripts, 4,000 map to non-coding gene transcripts, while the remaining 3,000 probes map to confirmed RNA sequences that align to express sequence tag (EST) clusters. After QC filters based on the annotation from the Bioconductor illuminaHumanv4.db annotation (Barbosa-Morais et al., 2010; Dunning et al.) and removal of probes that were unexpressed in blood, 10,264 probes remained for analysis. The proportion of monocytes and lymphocytes were estimated for each individual using expression data from GEO (GSE50008) data using a linear model similar to those proposed elsewhere for cell-count estimation (Houseman et al., 2012; described in the Supplementary Methods). The sample was then split into a discovery cohort of white non-Hispanic (WNH) males (n=143, with 115 cases and 28 controls) and a replication cohort of the remaining subjects (n=73, with 51 cases and 22 controls), which included WNH females (n=17) as well as men and women of other ancestries or undetermined descent (n=56).
Replication was also examined in an independent sample from the Detroit Neighborhood Health Study (DNHS, n=129 with 64 cases and 65 controls; Goldmann et al., 2011; Uddin et al., 2010). Fifty-eight of these participants (45%) were male; the majority (n=108, or 84%) were African American. Expression was assessed using the same HumanHT12_v4 BeadChip and analyzed using similar methods (see supplementary materials). Descriptive characteristics for each sample are presented in Supplementary Table 1.
2.2 Study Design and Analysis
Technical details of the analysis are in the Supplementary Methods. Briefly, Bioconductor’s limma package (Smyth, 2005) was used to test for association between lifetime PTSD and gene expression. Limma fits a linear regression model of gene expression as a function of PTSD and the included covariates. Age and estimated proportion of monocytes and lymphocytes were included as covariates in all analyses. Multiple-testing adjusted significance (pcor) was computed using a Benjamini–Hochberg False Discovery Rate procedure (Benjamini and Hochberg, 1995).
We first examined all 10,264 blood-expressed probes that passed QC for association with PTSD in the discovery sample. Probes with pcor<0.05 were then examined in the two replication cohorts and in a meta-analysis across the two samples performed using the METAL (Willer et al., 2010) software. The p-value method of meta-analysis was used, which incorporates the p-value and direction of effect. We also performed a series of post-hoc analyses examining the potential influences of genomic variation (single nucleotide polymorphisms; SNPs), comorbid depression, medication and substance abuse, and childhood vs. adult trauma. These analyses are described in the supplementary materials. We additionally examined the effect of PTSD severity (rather than the dichotomous diagnosis) in the model of gene expression.
INGENUITY’s (IPA; www.ingenuity.com) network analysis was used to identify biological links between a “target set” of 90 genes associated with PTSD in the discovery sample at the pcor<0.10 (Supplementary Table 3) level. IPA’s network analysis accepts an input of a target list of genes and creates a number of local networks of up to 35 genes which both maximize the number of target genes and the number of connections between them, adding other non-target genes/molecules as necessary. The network analysis optimizes the connectivity of the genes in the list, but it should be stressed that the network is not necessarily restricted to or representative of a single biological pathway.
The possible involvement of the differentially expressed genes in particular biological process or disease pathways was examined using the Gene-Set Enrichment Analysis (GSEA) software (Mootha et al., 2003; Subramanian et al., 2005) and INGENUITY’s Canonical Pathway and Disease/Biological Function analyses. These tools are similar, in that they search for evidence of enrichment of genes in particular gene-set lists. GSEA looks for enrichment corresponding to GO categories and KEGG pathways, and the INGENUITY tools look for enrichment of curated gene-sets created according to INGENUITY’s knowledge base.
In addition to these curated lists, we compared our “target set” of 90 top genes to a list of probes for GR-regulated genes derived from results of an in vivo dexamethasone stimulation experiment that identified GR-regulated transcripts in peripheral blood. In that study, gene expression levels in peripheral blood were compared pre- and 3 hours post- dexamethasone administration in 160 Caucasian males, (93 healthy controls and 71 male in-patients with depressive disorders). Recruitment strategies, participant characteristics and blood sample collection, processing and analysis was described in (Menke et al., 2012) and (Hennings et al., 2009). Total RNA was extracted from PaxGene RNA tubes using standard procedures and hybridized to Illumina HumanHT-12 v3.0 Expression BeadChips (Illumina, Inc., San Diego, CA, USA). Quality control and data analysis was performed in R as described in (Menke et al., 2012). The position of the probes and possible SNPs within these sequences were annotated using ReMOAT version August 2009 (http://remoat.sysbiol.cam.ac.uk/, Barbosa-Morais et al., 2010). These expression data have been deposited in the NCBI Gene expression omnibus (GSE46743). In these data, 4,375 expression probes that showed an absolute fold change of 1.3 or greater from pre- to post-dexamethasone administration in at least 20% of all samples and were classified therefore as “glucocorticoid responsive”. See Supplementary Table 5 for a complete listing. We then used this list to evaluate if the set of PTSD-associated genes in this study was enriched for glucocorticoid-responsive genes using both Fisher’s exact test and a permutation test (with 10,000 replicates).
Finally, we examined the discovery dataset for association between lifetime PTSD and expression of previously implicated candidate genes. We focused on GWAS-implicated genes (Guffanti et al., 2013; Logue et al., 2013; Xie et al., 2013), published candidate genes (based on Skelton et al. 2012 review), and other loci which had been implicated in expression studies of PTSD (Ressler et al., 2011; Uddin et al., 2011; Yehuda et al., 2009; Zieker et al., 2007). This yielded 14 probes of interest: one each for COMT, ADCYAP1, ADCYAP1R1, RORA, STAT5B, and BDNF, and two probes each for TXNRD1, IL16, S1PR1, and NR3C1 (see Supplementary Table 2 for details). Probes were not available for several of the genes of interest either because they were not included on the microarray (AC068718.1), because probes for these genes failed to pass quality filters based on the illuminaHumanv4.db annotation (FKBP5- poor mapping quality and secondary matches, NPY-poor mapping quality, DBH-poor mapping quality, DAT1-poor mapping quality and secondary matches, 5-HTR2A -poor mapping quality and secondary matches, RGS2-secondary matches, MAN2C1-secondary matches, IL18-poor mapping quality and secondary matches, and SOD1-secondary matches), or because they were not expressed in blood at sufficient levels (TLL1, DRD2, GABARA2, SLC6A4, CASP2, and TET1). For this portion of the analysis, the FDR-corrected p-value (pcor) was adjusted for the examination of these 14 probes.
3. Results
3.1 Transcriptome-wide Association Analyses
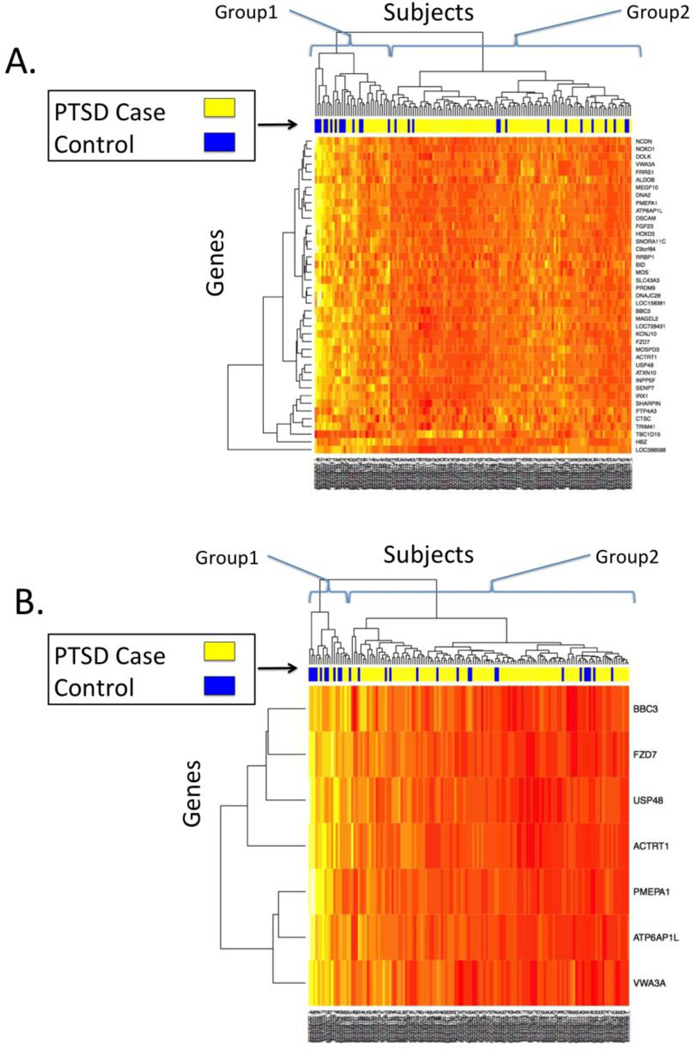
The transcriptome-wide analysis of 10,264 probes yielded 41 probes with multiple testing-corrected associations with PTSD (Table 1; pcor<0.05). All but one of the implicated genes (TBC1D15) had lower expression in PTSD cases than controls (t<0 in Table 1). The most significant association was with the Down syndrome cell adhesion molecule (DSCAM) gene (ILMN_1658420; p=2.77×10−6, pcor=0.010). Expression of DSCAM was lower in cases than controls (Figure 1). A heatmap of expression for the 41 PTSD-associated genes is presented in Figure 2a. The bar between the heatmap and the dendogram represents PTSD status, with yellow signifying PTSD cases and blue signifying controls. The dendogram at the top of the figure represents an unsupervised (i.e. without including information about PTSD status) hierarchical clustering of the subjects based on expression values. Subjects are divided into two groups based on the first split in the dendogram. Group 1 consisted of 34 subjects (58.8% PTSD) who showed higher expression of most PTSD-associated genes. Group 2 consisted of 109 subjects with lower expression levels and a greater proportion of cases with PTSD (95 of 109 or 87.2%, OR=4.68, p=0.00082). Results of the transcriptome-wide analysis of PTSD severity were less significant; no probes were significantly associated with PTSD severity (pcor>.05).
Table 1.
Probes significantly associated (pcor<0.05) with lifetime PTSD in the white non-Hispanic male discovery cohort of n=143 cases and 28 controls, and meta-analysis of two diverse replication cohorts: a VA cohort of 51 cases and 22 controls and the DNHS cohort of 64 cases and 65 controls.
| VA Discovery Sample | VA Rep. Sample | DNHS Rep. Sample | META analysis of Rep. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IlluminaID | Gene** | t | p | pcor | t | p | pcor | t | p | pcor | ED*** | pmeta | pcor |
| ILMN_1658420 | DSCAM | −4.88 | 2.77E-06* | 0.010* | −2.14 | 0.036* | 0.32 | 0.17 | 0.87 | 0.89 | -+ | 0.27 | 0.51 |
| ILMN_1729495 | TRIM41 | −4.87 | 2.92E-06* | 0.010* | −0.069 | 0.95 | 0.97 | −0.53 | 0.59 | 0.86 | -- | 0.64 | 0.77 |
| ILMN_1756873 | USP48 | −4.87 | 2.94E-06* | 0.010* | −0.84 | 0.40 | 0.67 | −1.93 | 0.055 | 0.41 | -- | 0.041* | 0.24 |
| ILMN_1755990 | ATP6AP1L | −4.77 | 4.61E-06* | 0.012* | −1.45 | 0.15 | 0.49 | −3.67 | 0.00036* | 0.015* | -- | 0.00019* | 0.0078* |
| ILMN_1652306 | MEGF10 | −4.67 | 6.92E-06* | 0.012* | −0.53 | 0.60 | 0.72 | −0.49 | 0.62 | 0.86 | -- | 0.48 | 0.61 |
| ILMN_1735353 | IRX1 | −4.67 | 6.92E-06* | 0.012* | −1.50 | 0.14 | 0.49 | −0.39 | 0.70 | 0.86 | -- | 0.24 | 0.49 |
| ILMN_2309156 | PMEPA1 | −4.64 | 7.98E-06* | 0.012* | −1.31 | 0.20 | 0.49 | −2.28 | 0.024* | 0.33 | -- | 0.010* | 0.10 |
| ILMN_1708337 | NOXO1 | −4.45 | 1.75E-05* | 0.023* | −0.54 | 0.59 | 0.72 | −1.00 | 0.32 | 0.86 | -- | 0.26 | 0.51 |
| ILMN_1729645 | BBC3 | −4.32 | 2.93E-05* | 0.030* | −2.28 | 0.026* | 0.32 | −1.78 | 0.078 | 0.41 | -- | 0.0062* | 0.085 |
| ILMN_1805345 | MOSPD3 | −4.26 | 3.73E-05* | 0.030* | −1.57 | 0.12 | 0.49 | −0.52 | 0.60 | 0.86 | -- | 0.18 | 0.47 |
| ILMN_2108823 | FZD7 | −4.24 | 4.06E-05* | 0.030* | −2.10 | 0.039* | 0.32 | −1.13 | 0.26 | 0.86 | -- | 0.034* | 0.23 |
| ILMN_1705900 | ATXN10 | −4.24 | 4.07E-05* | 0.030* | −0.46 | 0.65 | 0.74 | 0.49 | 0.63 | 0.86 | -+ | 0.90 | 0.97 |
| ILMN_2282959 | DNA2 | −4.24 | 4.08E-05* | 0.030* | −1.76 | 0.082 | 0.48 | −0.19 | 0.85 | 0.89 | -- | 0.24 | 0.49 |
| ILMN_3240883 | SNORA11C | −4.21 | 4.55E-05* | 0.030* | −0.50 | 0.62 | 0.73 | −0.67 | 0.51 | 0.86 | -- | 0.41 | 0.60 |
| ILMN_1760479 | MOS | −4.20 | 4.71E-05* | 0.030* | −1.19 | 0.24 | 0.51 | −0.98 | 0.33 | 0.86 | -- | 0.14 | 0.43 |
| ILMN_1657478 | MAGEL2 | −4.19 | 4.97E-05* | 0.030* | −0.99 | 0.33 | 0.64 | −1.39 | 0.17 | 0.68 | -- | 0.089 | 0.37 |
| ILMN_1755920 | LOC158381 | −4.19 | 4.98E-05* | 0.030* | −0.76 | 0.45 | 0.67 | −1.63 | 0.10 | 0.48 | -- | 0.079 | 0.36 |
| ILMN_1815392 | ACTRT1 | −4.15 | 5.77E-05* | 0.032* | −1.41 | 0.16 | 0.49 | −1.76 | 0.080 | 0.41 | -- | 0.025* | 0.21 |
| ILMN_1747716 | ALDOB | −4.14 | 5.85E-05* | 0.032* | −1.29 | 0.20 | 0.49 | −0.60 | 0.55 | 0.86 | -- | 0.22 | 0.49 |
| ILMN_1761598 | PRDM9 | −4.08 | 7.53E-05* | 0.039* | −1.88 | 0.064 | 0.44 | −0.52 | 0.61 | 0.86 | -- | 0.13 | 0.43 |
| ILMN_2253246 | DNAJC28 | −4.06 | 8.02E-05* | 0.039* | −1.15 | 0.25 | 0.52 | −0.34 | 0.73 | 0.86 | -- | 0.34 | 0.54 |
| ILMN_2359710 | PTP4A3 | −4.04 | 8.78E-05* | 0.040* | −0.30 | 0.77 | 0.81 | 0.40 | 0.69 | 0.86 | -+ | 0.88 | 0.97 |
| ILMN_1798076 | HOXD3 | −4.03 | 8.93E-05* | 0.040* | −0.33 | 0.74 | 0.80 | −0.68 | 0.50 | 0.86 | -- | 0.46 | 0.61 |
| ILMN_1808272 | KCNJ10 | −4.02 | 9.44E-05* | 0.040* | −1.31 | 0.19 | 0.49 | −0.90 | 0.37 | 0.86 | -- | 0.14 | 0.43 |
| ILMN_2268381 | RRBP1 | −4.01 | 9.93E-05* | 0.041* | −1.19 | 0.24 | 0.51 | 1.77 | 0.079 | 0.41 | -+ | 0.47 | 0.61 |
| ILMN_1696488 | FGF23 | −3.97 | 0.00011* | 0.045* | −0.96 | 0.34 | 0.64 | 0.19 | 0.85 | 0.89 | -+ | 0.68 | 0.80 |
| ILMN_1792885 | CTSC | −3.94 | 0.00013* | 0.047* | −0.036 | 0.97 | 0.97 | −1.15 | 0.25 | 0.86 | -- | 0.34 | 0.54 |
| ILMN_2214734 | FRRS1 | −3.94 | 0.00013* | 0.047* | −2.23 | 0.029* | 0.32 | −0.18 | 0.86 | 0.89 | -- | 0.15 | 0.43 |
| ILMN_3305849 | LOC728431 | −3.93 | 0.00013* | 0.047* | −0.70 | 0.49 | 0.67 | −0.70 | 0.49 | 0.86 | -- | 0.33 | 0.54 |
| ILMN_2387328 | C9orf84 | −3.92 | 0.00014* | 0.047* | −0.34 | 0.73 | 0.80 | −2.00 | 0.047* | 0.41 | -- | 0.071 | 0.36 |
| ILMN_1749834 | SMIM1 | −3.91 | 0.00014* | 0.047* | −3.00 | 0.0037* | 0.15 | 0.36 | 0.72 | 0.86 | -+ | 0.16 | 0.43 |
| ILMN_1794780 | SHARPIN | −3.89 | 0.00016* | 0.047* | −0.76 | 0.45 | 0.67 | −0.39 | 0.70 | 0.86 | -- | 0.45 | 0.61 |
| ILMN_1762319 | SLC43A3 | −3.88 | 0.00016* | 0.047* | −1.51 | 0.13 | 0.49 | −0.50 | 0.62 | 0.86 | -- | 0.20 | 0.48 |
| ILMN_2259495 | BID | −3.88 | 0.00016* | 0.048* | 0.74 | 0.46 | 0.67 | −0.43 | 0.67 | 0.86 | +- | 0.93 | 0.97 |
| ILMN_1807556 | VWA3A | −3.86 | 0.00017* | 0.048* | −1.29 | 0.20 | 0.49 | −2.79 | 0.0061* | 0.13 | -- | 0.0030* | 0.061 |
| ILMN_1778294 | SENP7 | −3.86 | 0.00017* | 0.048* | −0.77 | 0.45 | 0.67 | 0.73 | 0.46 | 0.86 | -+ | 0.88 | 0.97 |
| ILMN_1713458 | HBZ | −3.85 | 0.00018* | 0.048* | −0.71 | 0.48 | 0.67 | 0.43 | 0.67 | 0.86 | -+ | 0.95 | 0.97 |
| ILMN_2359500 | NCDN | −3.85 | 0.00018* | 0.048* | −0.59 | 0.55 | 0.71 | −0.76 | 0.45 | 0.86 | -- | 0.34 | 0.54 |
| ILMN_1803941 | TBC1D15 | 3.83 | 0.00019* | 0.0498* | −0.63 | 0.53 | 0.71 | −0.27 | 0.79 | 0.89 | -- | 0.56 | 0.69 |
| ILMN_1753627 | DOLK | −3.82 | 0.00020* | 0.0499* | −0.69 | 0.49 | 0.67 | 0.54 | 0.59 | 0.86 | -+ | 0.98 | 0.98 |
| ILMN_1813650 | INPP5F | −3.82 | 0.00020* | 0.0499* | −1.49 | 0.14 | 0.49 | 0.053 | 0.96 | 0.96 | -+ | 0.41 | 0.60 |
indicates significant at the .05 level.
Symbol from the updated annotations in illuminaHumanv4.db.;
ED= Effect Direction in the replication cohorts.
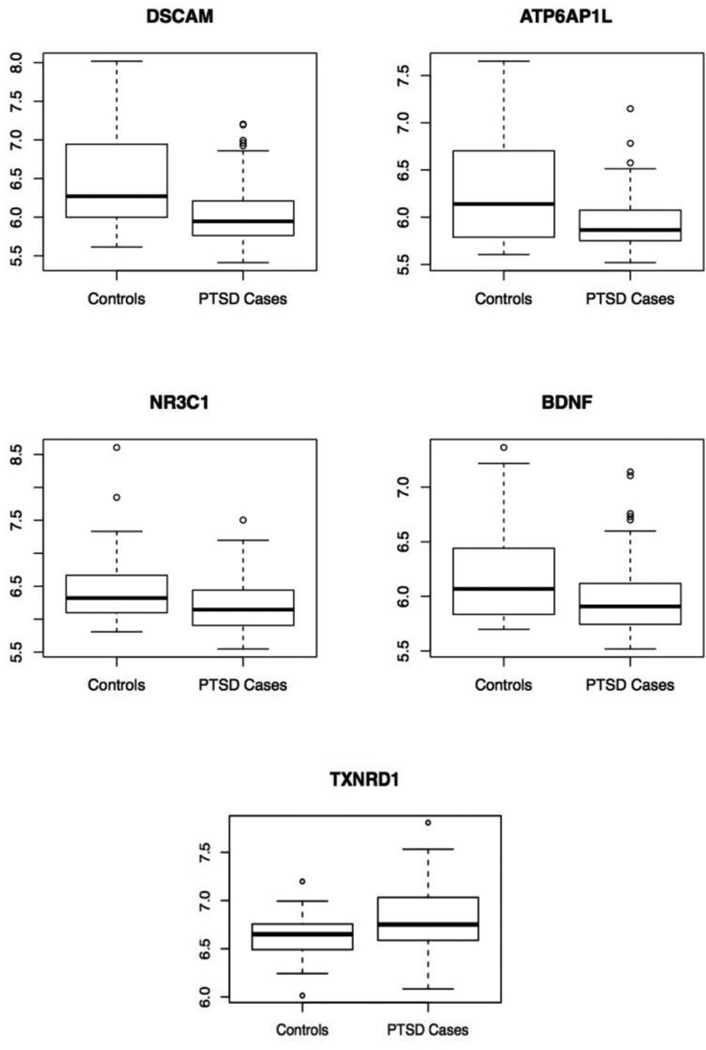
Figure 1.
Boxplots of DSCAM, ATP6AP1L, NR3C1, BDNF, and TXNRD1 expression in the white non-Hispanic male discovery cohort of n=143 cases and 28 controls as a function of PTSD status.
Figure 2.
Heatmap of expression in the white non-Hispanic male discovery cohort of n=143 cases and 28 controls, for A) the 41 significant (pcor<0.05) probes in the discovery sample and B) the 7 genes with pmeta<0.05 in the replication meta-analysis. Rows represent genes, columns represent subjects. The degree of expression from high to low is represented by the colors white and yellow on the high end, and orange and red on the low end. The colored bar above the heatmap represents PTSD status, with blue=no PTSD, yellow=Lifetime PTSD.
We then examined these 41 probes for association with PTSD in both the VA replication sample and the DNHS cohorts. In the VA sample, expression of DSCAM and four other genes were significant (p from 0.0037 to 0.039), but none survived correction for multiple testing (pcor>0.05). However, the direction of association was consistent across the VA discovery and VA replication samples, agreeing for 39 of the 41 probes, which is more than would be expected by chance (p=3.9×10−10). In the DNHS replication sample, 4 genes (9.8%) were associated with PTSD (p from 0.00036 to 0.047), and expression of the ATPase, H+ transporting, lysosomal accessory protein 1-like (ATP6AP1L) gene remained associated after multiple-testing correction (pcor=0.015). Again, the direction of the associations in the DNHS sample was more consistent with the discovery data than would be expected by chance (30 out of 41, p=0.0022).
Meta-analysis of the replication cohorts showed that the association between ATP6AP1L expression and PTSD was strengthened compared to what had been observed in DNHS alone (p=0.00019), and ATP6AP1L remained significant after multiple testing correction. (pcor=0.0078). In addition, 7 genes, more than would be expected by chance (17%, p=0.0039), were associated with PTSD at the pmeta<0.05 level (Table 1). All 7 genes replicating at the p<0.05 level had lower expression in PTSD cases compared to controls. A heatmap of expression for these 7 genes is presented in Figure 2B and shows clearer distinction between PTSD cases and controls compared with the set of 41 probes that were identified using only the discovery sample. Using this set of replicating genes, there is still a high (n=19) and a low (n=124) expression group. The high expression group (Group 1 in Figure 2B) contains fewer PTSD cases (42%) than the low expression group (Group 2; 86%). Hence, clustering based on expression of these 7 genes is strongly associated with PTSD (OR=8.46, p=6.72×10−5).
3.2 Pathway and Network Analyses
INGENUITY’s canonical pathway analyses of the top 90 (pcor<0.10) “target list” genes from our discovery-sample identified no pathways that were significantly enriched for the PTSD-associated genes at the multiple-testing corrected significant level. Similarly, analysis of the log fold-change values in GSEA did not identify any multiple-testing correction significant GO terms or KEGG pathways. However, INGENUITY’s Disease and Biological Function tool identified several pathways enriched for our target genes, including pathways involved in Neurological Disease, (i.e., Ataxia and Neurodegeneration; see Table 3).
Table 3.
Top disease and biological function gene categories according to INGENUITY analysis of the 90 “target” genes* with pcor<0.10 in the association analysis of the white non-Hispanic male discovery cohort of n=143 PTSD cases and 28 controls.
| Category | Disease/Function Annotation |
p | pcor** | Genes |
|---|---|---|---|---|
| Neurological Disease |
Dysmetria | 4.5×10−5 | 0.025 | PRNP, SPRN |
| Ataxia | 7.4×10−5 | 0.025 |
ATXN10,BID,KCNJ10 MOS, SLC4A7 |
|
| Neurodegeneration of nervous tissue. |
3.78×10−3 | 0.046 | KCNJ10, MOS, SLC4A7 | |
| Tissue Morphology |
Degeneration of organ of Corti |
7.22×10−5 | 0.025 | CLRN1,KCNJ10,SLC4A7 |
| Quality of Chondrocytes |
8.33×10−5 | 0.025 | FGF18, FGF23, FOSL2 | |
| Neurodegeneration of nervous tissue |
3.78×10−3 | 0.046 | KCNJ10, MOS, SLC4A7 | |
| Connective Tissue Development and Function |
Quality of Chondrocytes |
8.33×10−5 | 0.025 | FGF18, FGF23, FOSL2 |
| Proliferation of Chondrocytes |
2.59×10−3 | 0.046 | FGF18, FGF23, FOSL2 | |
| Abnormal morphology of atlas. |
3.01×10−3 | 0.046 | HOXD3, SKI | |
| Auditory Disease |
Collapse of Reissner’s Membrane |
1.49×10−3 | 0.036 | KCNJ10, SLC4A7 |
| Degeneration of supporting cell layer of inner ear |
5.23×10−4 | 0.046 | CLRN1, SLC4A7 | |
| Usher syndrome 3A | 3.91×10−3 | 0.046 | CLRN1 |
The complete list of target genes is presented in Supplementary Table 3.
pcor is the Benjamini Hochberg FDR corrected p-value.
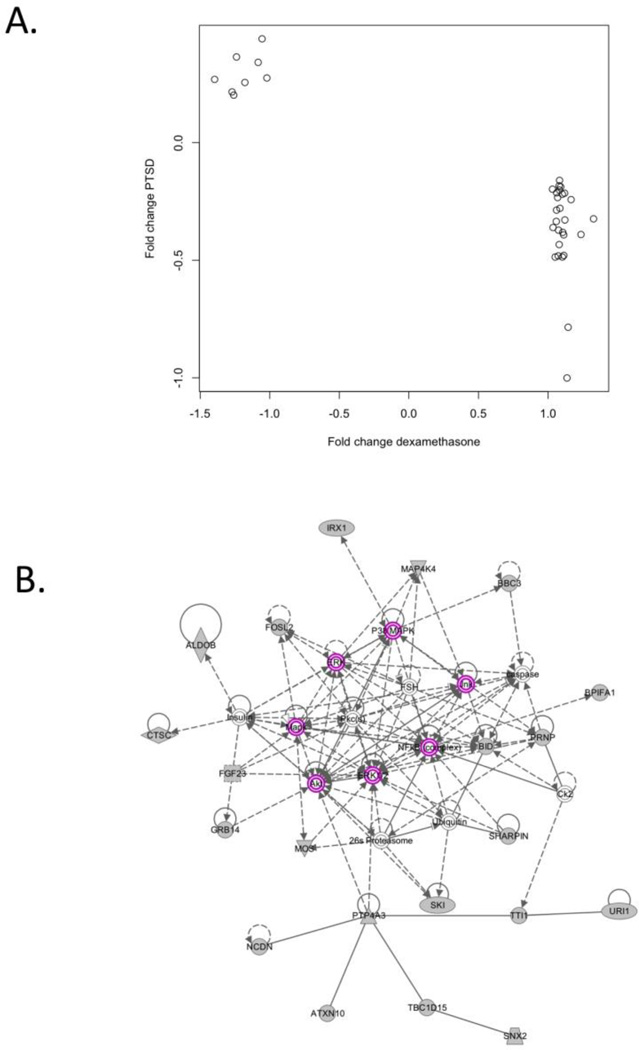
We then evaluated whether the target list genes were enriched for genes related to the GR signaling pathway by comparing the target list to the experimentally derived set of glucocorticoid-responsive probes. Of the 4,375 GR-responsive probes, 2,703 passed quality filters and were analyzed in the VA sample. Hence, 26% of the 10,264 analyzed probes were identified as “glucocorticoid responsive”. In comparison, 35 (39%) of our top 90 probes were on the list (see Supplementary Table 3) suggesting that our list of differentially expressed genes contained a greater proportion of glucocorticoid-responsive genes than would be expected by chance (Fisher's exact p=0.0017, permutation test p=0.0059). Of the 35 target-list genes that were also glucocorticoid-responsive, 27 (77%) had lower expression in cases compared to controls and 8 (23%) had higher expression in cases compared to controls. When we compared these effects to the fold-change estimates from the dexamethasone stimulation experiment, we found perfect concordance. That is, All 27 genes with lower expression in PTSD cases had higher expression after dexamethasone administration and all 8 genes with higher expression in PTSD cases had lower expression after dexamethasone administration (Figure 3A), which is highly unlikely to be due to chance (sign-test significance based on Fisher’s exact p=4.25×10−8).
Figure 3.
Analyses of the “target list” of 90 genes implicated in PTSD at the pcor<0.10 level which implicate glucocorticoid receptor signaling. A) A plot of the fold-change estimates for the 35 PTSD-associated “target list” genes identified as GR regulated in the dexamethasone stimulation experiment. Fold change estimates from the dexamethasone experiment are presented on the x axis and the fold-change estimates from the comparison of male PTSD cases and controls in the white non-Hispanic discovery sample are presented on the y axis. B) INGENUITY-generated network of interacting genes with glucocorticoid receptor signaling genes/molecules highlighted in purple. The grey shapes indicate genes and molecules from the target list, while open shapes indicate that a gene was added by INGENUITY to increase network connectivity.
We then explored the relationship between the PTSD-associated genes and the GR pathway using the INGENUITY network analysis to identify possible biological links between the 90 target-list genes. INGENUITY’s top network, that is, the network with the highest proportion of target genes, is presented in Figure 3B. This network included 21 genes from our target list and 14 genes/molecules to increase connectivity. Of the added genes/molecules, half (7) were related to GR signaling. The GR signaling molecules are highlighted in purple in Figure 3B.
3.3 Candidate Genes and Covariates
Analysis of probes for the 14 candidate genes implicated in previous PTSD studies showed that the GR gene (NR3C1), BDNF, and the thioredoxin reductase 1 (TXNRD1) gene were significantly associated with PTSD in our discovery cohort (pcor<0.05 when adjusted for 14 candidate genes examined; Table 2). For both BDNF and NR3C1, expression was lower in PTSD cases than controls while the opposite was true for TXNRD1 (Figure 1).
Table 2.
Previously implicated PTSD genes which are differentially expressed in the white non-Hispanic male discovery cohort of n=143 cases and 28 controls. (Pcor<.05).*,**
| Gene | IlluminaID | t | p | pcor*** |
|---|---|---|---|---|
| BDNF | ILMN_1761910 | −3.25 | 0.0014 | 0.010 |
| NR3C1 | ILMN_1668525 | −3.31 | 0.0012 | 0.010 |
| TXNRD1 | ILMN_2324421 | 2.83 | 0.0053 | 0.025 |
No probes were significant for RORA, COMT, ADCYAP1, ADCYAP1R1, NPSR1, STAT5B, IL16, or S1PR1.
The following genes did not have probes which passed quality checks or were not expressed at sufficient levels to be included in the analysis: TLL1, AC068718.1, FKBP5, NPY, DBH, DRD2, DAT1, SLC6A4, 5-HTR2A, GABRA2, RGS2, MAN2C1, IL18, CASP2, SOD1, and TET1.
Here, pcor represents correction for 14 probes targeting previously implicated PTSD genes.
Finally, examination of potential confounders (i.e., depression, medication use, and substance abuse, as described in the Supplementary Materials) revealed no evidence of significant effects of these factors. We also examined the potential role of genomic (SNP) variation might play and identified several expression quantitative trait loci (eQTLs), for SMIM1, ATP6AP1L, and BDNF (see Supplementary Results and Supplementary Table 4). However, none of these eQTLs were associated with PTSD, and the associations between PTSD and expression of these genes remained significant when these SNPs were included as predictors in the same analysis.
4. Discussion
Transcriptome-wide analysis of gene expression levels in peripheral blood drawn from a sample of white non-Hispanic veterans (115 PTSD cases and 28 controls) revealed that expression levels for 41 of 10,264 probes differed significantly as a function of PTSD diagnosis. A replication meta-analysis showed that 7 of these 41 genes were associated with the disorder at the p<0.05 level, with one of them surviving multiple-testing correction. Despite differences in the discovery and replication samples, including sex differences, recruitment differences, and the ratio of cases to controls (see Supplementary Materials for details), we found broad evidence of overlap across the discovery and replication samples for the set of genes implicated in the discovery sample based on the direction of effect and enrichment of p<0.05 associations beyond what would be expected by chance. If the discovery and replication samples were more closely matched, this agreement might have been greater.
Several findings suggested that differential expression of genes observed in PTSD cases is (at least in part) related to glucocorticoid signaling. When we compared our 90 most differentially expressed genes to a list of genes whose expression is altered by dexamethasone administration (Hennings et al., 2009; Menke et al., 2012), we found that a greater proportion of our differentially expressed genes were on this list than would be expected by chance, as were two of our most differentially expressed genes: DSCAM (differentially expressed in the discovery sample) and ATP6AP1L (differentially expressed in both the discovery and replication analyses). For the genes on our top list which where altered by dexamethasone, genes with lower expression in PTSD cases had increased expression after administration of dexamethasone and vice versa (Figure 3A). INGENUITY network analysis independently confirmed that many of the PTSD-associated genes in this study, though not classified as GR signaling genes themselves, interact with elements of the GR signaling pathway (Figure 3B). Finally, our candidate-gene analysis of loci previously implicated in PTSD also revealed reduced expression of the GR gene (NR3C1).
Together, these findings contribute to a vast body of research implicating abnormalities in HPA-axis reactivity and GR sensitivity and signaling in PTSD. NR3C1 regulation has been linked to the presence of developmental adversity in both mouse experiments and humans (see e.g. Champagne and Curley, 2009; Perroud et al., 2011). Mouse models of GR deficiency have also implicated Nr3c1 in behavioral traits and emotional stability (e.g. Boyle et al., 2006; Pepin et al., 1992; Tronche et al., 1999). A transcriptome-wide study of the differences between stress vulnerable and resilient rats (Daskalakis et al., 2014) found that transcription factors predicted to regulate their differentially expressed genes were consistently related to the GR signaling network and NR3C1 was a predicted transcription factor for expression changes observed in blood and brain tissue from the amygdala and hippocampus. The latter raises the intriguing possibility that the NR3C1 expression differences observed in this study might reflect NR3C1 regulated alterations in the brain.
The most significant differentially expressed gene in our discovery sample, DSCAM, is known to play a role in neural development, including axon guidance and branching and synapse maturation (Schmucker and Chen, 2009) and it is likely responsible for some of the neurological features of Down syndrome (Yamakawa et al., 1998). The DSCAM association should be considered preliminary, however, as it was only associated at the p<0.05 level in the VA replication cohort and not significantly associated with the disorder in the DNHS replication sample.
One gene survived multiple-testing correction in our replication meta-analysis: ATP6AP1L. ATP6AP1L is the largely uncharacterized paralogue of ATP6AP1. ATP6AP1 is part of an enzyme that assists in the acidification of organelles and may play a role in regulation of neuroendocrine responses through the acidification of neuroendocrine secretory granules (Supek et al., 1994).
It is interesting to note that of the genes whose expression levels were associated with PTSD in this study, one is known to be imprinted (MAGEL2; Boccaccio et al., 1999), one has an imprinted isoform (INPP5F; Choi et al., 2005), and two more (IRX1 and PTP4A3) are predicted to be imprinted (Luedi et al., 2007). Imprinted genes are regulated epigenetically with many involved in embryonic development and neurobehavioral traits (Tycko and Morison, 2002). While it is not likely that the mechanism of imprinting is itself is related to the expression differences that we found between PTSD cases and controls, expression differences in these genes may point to a role for DNA methylation or other epigenetic mechanisms in PTSD and its expression signature.
Analysis of candidate genes previously implicated in PTSD revealed differential expression of BDNF, TXNRD1 and, as noted above, the GR gene (NR3C1). BDNF plays a role in learning, memory, and fear extinction (Andero and Ressler, 2012). Lower BDNF expression in serum has been linked to untreated depression (Molendijk et al., 2013). Studies have also linked BDNF expression levels to PTSD, but findings have been inconsistent with some reporting increased BDNF expression in PTSD cases and others showing the opposite (see e.g. Dell'Osso et al., 2009; Matsuoka et al., 2013). In this study, we found that PTSD cases had lower BDNF expression than trauma-exposed controls (probe ILMN_1761910, logFC= −0.24, p= 0.0014; Figure 1). The inconsistency across studies of BDNF expression may be a result of the different tissues studied, differences in study population, or differences in the duration of symptoms since the trauma.
The other candidate gene that was differentially expressed, TXNRD1, encodes a protein, thioredexin, which plays an important role in responding to oxidative stress by mounting an antioxidant response. Levels of the protein are thought to index oxidative stress (Nakamura, 2005) and so the elevated levels observed in PTSD cases versus controls in this study is consistent with growing evidence for the role of oxidative stress in disorders of the trauma and stress-related spectrum (Miller & Sadeh, 2014). TXNRD1, is also associated with neuroprotection in individuals with neurodegenerative disorders and has been found to index neurocognitive impairment among individuals with schizophrenia (Zhang et al., 2013).
There are several limitations that should be kept in mind when interpreting our results. As these data were cross sectional, we cannot determine whether the expression differences represent expression signature of past or current PTSD, or if it is it is marker of a pre-trauma vulnerability to the development of PTSD. We examined expression in whole blood, and not all genes and transcripts are expressed in blood. Additionally, whole blood is itself contains a heterogeneous mixture of cell types, each of which may have unique expression patterns. We recognize that differences in cell proportions may be caused by differences in the proportion of cell types if not properly controlled. To address this issue, we estimated the proportion of different cell types and included these estimates as covariates in our analysis models. Additionally, we cannot determine specificity of expression changes to any particular cell type. Studies in other tissues, or a specific cell type isolated from whole blood may not show the same effects. Our statistical adjustment for different cell type proportions included an adjustment for white blood cell proportions but did not adjust for RNA from reticulocytes. This leaves open the possibility that the proportion of reticulocytes (i.e., immature red blood cells) in the sample could explain observed associations. We find this unlikely, as broad agreement was observed between the VA discovery and replication samples and the DNHS replication sample, and reticulocyte RNA was removed from the DNHS-study samples. While we have evidence of broad agreement across the discovery and replication samples, only one specific gene replicated at a multiple-testing corrected level. This may be due to the noted difference in sample preparation, or some of the differentially expressed genes may be population-specific and not universally generalizable. A final limitation was that we did not have qPCR validation of the results in our discovery sample.
To conclude, this study identified a set of genes whose expression was associated with PTSD. We obtained evidence of partial replication in results from two additional samples. Unsupervised clustering of the expression levels of replicating genes yielded a classification that was strongly associated with PTSD (OR > 8). This suggests that as expression study sample sizes increase, it may be possible in the future to develop reliable biomarkers of PTSD risk and/or diagnostic status using expression probes. We also found that many of our PTSD-associated effects involved lower expression levels for genes related to the GR pathway, possibly reflecting HPA axis dysregulation. Prior studies have indicated that lymphocyte GRs are increased in PTSD cases (Yehuda et al., 1996a; Yehuda et al., 1991; Yehuda et al., 1993; Yehuda et al., 1996b) resulting in enhanced suppression of the HPA axis in response to GR activation. The suppressed expression of our top genes may be a reflection of these alterations, although the genes we identified here, including DSCAM and ATP6AP1 have not been previously linked to PTSD. Examining the function of these peripheral and downstream affiliates of the GR signaling pathway may lead to identification of novel biomarkers of PTSD and potential targets for treatment.
Supplementary Material
Acknowledgements
This research was supported by U.S. Department of Veterans Affairs VA CSR&D and BLR&D grants and NIH grant R21MH102834 to MWM. This work was also supported by a Career Development Award to EJW from the United States (U.S.) Department of Veterans Affairs, Clinical Sciences Research and Development Program. Data collection and analysis of GSE46743 was supported within the 7th framework programme by an ERC starting grant to EB, GxEmolmech – 281338.
Footnotes
Disclosures.
MWL, AKS, CB, EJW, GG, AR, AS, SS, DH, EBB, JA AM, MU, DW, SG AEA, and KCK have no conflicts of interest to report. MWM owns stock in Illumina, Inc. All authors have approved the final article.
References
- Andero R, Ressler KJ. Fear extinction and BDNF: translating animal models of PTSD to the clinic. Genes Brain Behav. 2012;11:503–512. doi: 10.1111/j.1601-183X.2012.00801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, Darot JF, Ritchie ME, Lynch AG, Tavare S. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Charney DS, Keane TM. The Clinician-Administered PTSD Scale-IV. Boston: National Center for Posttraumatic Stress Disorder, Behavioral Sciences Division; 1990. [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM. The development of a Clinician-Administered PTSD Scale. J Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- Boccaccio I, Glatt-Deeley H, Watrin F, Roeckel N, Lalande M, Muscatelli F. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum Mol Genet. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- Boyle MP, Kolber BJ, Vogt SK, Wozniak DF, Muglia LJ. Forebrain glucocorticoid receptors modulate anxiety-associated locomotor activation and adrenal responsiveness. J Neurosci. 2006;26:1971–1978. doi: 10.1523/JNEUROSCI.2173-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Curley JP. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neurosci Biobehav Rev. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Choi JD, Underkoffler LA, Wood AJ, Collins JN, Williams PT, Golden JA, Schuster EF, Jr, Loomes KM, Oakey RJ. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol Cell Biol. 2005;25:5514–5522. doi: 10.1128/MCB.25.13.5514-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis NP, Cohen H, Cai G, Buxbaum JD, Yehuda R. Expression profiling associates blood and brain glucocorticoid receptor signaling with trauma-related individual differences in both sexes. Proc Natl Acad Sci U S A. 2014;111:13529–13534. doi: 10.1073/pnas.1401660111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Osso L, Carmassi C, Del Debbio A, Catena Dell'Osso M, Bianchi C, da Pozzo E, Origlia N, Domenici L, Massimetti G, Marazziti D, Piccinni A. Brain-derived neurotrophic factor plasma levels in patients suffering from post-traumatic stress disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33:899–902. doi: 10.1016/j.pnpbp.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Dunning M, Lynch A, Eldridge M. illuminaHumanv4.db: Illumina HumanHT12v4 annotation data (chip illuminaHumanv4) [Google Scholar]
- Guffanti G, Galea S, Yan L, Roberts AL, Solovieff N, Aiello AE, Smoller JW, De Vivo I, Ranu H, Uddin M, Wildman DE, Purcell S, Koenen KC. Genome-wide association study implicates a novel RNA gene, the lincRNA AC068718.1, as a risk factor for post-traumatic stress disorder in women. Psychoneuroendocrinology. 2013;38:3029–3038. doi: 10.1016/j.psyneuen.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings JM, Owashi T, Binder EB, Horstmann S, Menke A, Kloiber S, Dose T, Wollweber B, Spieler D, Messer T, Lutz R, Kunzel H, Bierner T, Pollmacher T, Pfister H, Nickel T, Sonntag A, Uhr M, Ising M, Holsboer F, Lucae S. Clinical characteristics and treatment outcome in a representative sample of depressed inpatients - findings from the Munich Antidepressant Response Signature (MARS) project. J Psychiatr Res. 2009;43:215–229. doi: 10.1016/j.jpsychires.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubany ES, Haynes SN, Leisen MB, Owens JA, Kaplan AS, Watson SB, Burns K. Development and preliminary validation of a brief broad-spectrum measure of trauma exposure: the Traumatic Life Events Questionnaire. Psychol Assess. 2000;12:210–224. doi: 10.1037//1040-3590.12.2.210. [DOI] [PubMed] [Google Scholar]
- Lambert WM, Xu CF, Neubert TA, Chao MV, Garabedian MJ, Jeanneteau FD. Brain-derived neurotrophic factor signaling rewrites the glucocorticoid transcriptome via glucocorticoid receptor phosphorylation. Mol Cell Biol. 2013;33:3700–3714. doi: 10.1128/MCB.00150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2013;18:937–942. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Nishi D, Noguchi H, Kim Y, Hashimoto K. Longitudinal changes in serum brain-derived neurotrophic factor in accident survivors with posttraumatic stress disorder. Neuropsychobiology. 2013;68:44–50. doi: 10.1159/000350950. [DOI] [PubMed] [Google Scholar]
- Mehta D, Gonik M, Klengel T, Rex-Haffner M, Menke A, Rubel J, Mercer KB, Putz B, Bradley B, Holsboer F, Ressler KJ, Muller-Myhsok B, Binder EB. Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: evidence from endocrine and gene expression studies. Arch Gen Psychiatry. 2011;68:901–910. doi: 10.1001/archgenpsychiatry.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, Loeschner A, Gonik M, Mercer KB, Bradley B, Muller-Myhsok B, Ressler KJ, Binder EB. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke A, Arloth J, Putz B, Weber P, Klengel T, Mehta D, Gonik M, Rex-Haffner M, Rubel J, Uhr M, Lucae S, Deussing JM, Muller-Myhsok B, Holsboer F, Binder EB. Dexamethasone stimulated gene expression in peripheral blood is a sensitive marker for glucocorticoid receptor resistance in depressed patients. Neuropsychopharmacology. 2012;37:1455–1464. doi: 10.1038/npp.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW, Wolf EJ, Reardon A, Greene A, Ofrat S, McInerney S. Personality and the latent structure of PTSD comorbidity. J Anxiety Disord. 2012;26:599–607. doi: 10.1016/j.janxdis.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk ML, Spinhoven P, Polak M, Bus BA, Penninx BW, Elzinga BM. Serum BDNF concentrations as peripheral manifestations of depression: evidence from a systematic review and meta-analyses on 179 associations (N=9484) Mol Psychiatry. 2013 doi: 10.1038/mp.2013.105. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nakamura H. Thioredoxin and its related molecules: update 2005. Antioxid Redox Signal. 2005;7:823–828. doi: 10.1089/ars.2005.7.823. [DOI] [PubMed] [Google Scholar]
- Pepin MC, Pothier F, Barden N. Impaired type II glucocorticoid-receptor function in mice bearing antisense RNA transgene. Nature. 1992;355:725–728. doi: 10.1038/355725a0. [DOI] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Olie E, Salzmann A, Nicastro R, Guillaume S, Mouthon D, Stouder C, Dieben K, Huguelet P, Courtet P, Malafosse A. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry. 2011;1:e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, Norrholm SD, Kilaru V, Smith AK, Myers AJ, Ramirez M, Engel A, Hammack SE, Toufexis D, Braas KM, Binder EB, May V. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarapas C, Cai G, Bierer LM, Golier JA, Galea S, Ising M, Rein T, Schmeidler J, Muller-Myhsok B, Uhr M, Holsboer F, Buxbaum JD, Yehuda R. Genetic markers for PTSD risk and resilience among survivors of the World Trade Center attacks. Dis Markers. 2011;30:101–110. doi: 10.3233/DMA-2011-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23:147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- Segman RH, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev AY. Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry. 2005;10:500–513. 425. doi: 10.1038/sj.mp.4001636. [DOI] [PubMed] [Google Scholar]
- Skelton K, Ressler KJ, Norrholm SD, Jovanovic T, Bradley-Davino B. PTSD and gene variants: new pathways and new thinking. Neuropharmacology. 2012;62:628–637. doi: 10.1016/j.neuropharm.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Supekova L, Mandiyan S, Pan YC, Nelson H, Nelson N. A novel accessory subunit for vacuolar H(+)-ATPase from chromaffin granules. J Biol Chem. 1994;269:24102–24106. [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- Uddin M, Galea S, Chang SC, Aiello AE, Wildman DE, de los Santos R, Koenen KC. Gene expression and methylation signatures of MAN2C1 are associated with PTSD. Dis Markers. 2011;30:111–121. doi: 10.3233/DMA-2011-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf EJ, Miller MW, Harrington KM, Reardon A. Personality-based latent classes of posttraumatic psychopathology: personality disorders and the internalizing/externalizing model. J Abnorm Psychol. 2012;121:256–262. doi: 10.1037/a0023237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa K, Huot YK, Haendelt MA, Hubert R, Chen XN, Lyons GE, Korenberg JR. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet. 1998;7:227–237. doi: 10.1093/hmg/7.2.227. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Cai G, Golier JA, Sarapas C, Galea S, Ising M, Rein T, Schmeidler J, Muller-Myhsok B, Holsboer F, Buxbaum JD. Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biol Psychiatry. 2009;66:708–711. doi: 10.1016/j.biopsych.2009.02.034. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Levengood RA, Schmeidler J, Wilson S, Guo LS, Gerber D. Increased pituitary activation following metyrapone administration in post-traumatic stress disorder. Psychoneuroendocrinology. 1996a;21:1–16. doi: 10.1016/0306-4530(95)00055-0. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Lowy MT, Southwick SM, Shaffer D, Giller EL., Jr Lymphocyte glucocorticoid receptor number in posttraumatic stress disorder. Am J Psychiatry. 1991;148:499–504. doi: 10.1176/ajp.148.4.499. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Southwick SM, Krystal JH, Bremner D, Charney DS, Mason JW. Enhanced suppression of cortisol following dexamethasone administration in posttraumatic stress disorder. Am J Psychiatry. 1993;150:83–86. doi: 10.1176/ajp.150.1.83. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Teicher MH, Trestman RL, Levengood RA, Siever LJ. Cortisol regulation in posttraumatic stress disorder and major depression: a chronobiological analysis. Biol Psychiatry. 1996b;40:79–88. doi: 10.1016/0006-3223(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Chen da C, Xiu MH, Yang FD, Tan YL, He S, Kosten TA, Kosten TR. Thioredoxin, a novel oxidative stress marker and cognitive performance in chronic and medicated schizophrenia versus healthy controls. Schizophr Res. 2013;143:301–306. doi: 10.1016/j.schres.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Zieker J, Zieker D, Jatzko A, Dietzsch J, Nieselt K, Schmitt A, Bertsch T, Fassbender K, Spanagel R, Northoff H, Gebicke-Haerter PJ. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Mol Psychiatry. 2007;12:116–118. doi: 10.1038/sj.mp.4001905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.