Abstract
Recent studies showed that cyclophilin A (CypA) promotes NF-κB/p65 nuclear translocation, resulting in enhanced NF-κB activity and altered expression of its target genes, such as the Sox9 transcriptional factor, which plays a critical role in chondrogenic differentiation and endochondral ossification. In this report, we unveil the role of CypA in signal-induced chondrogenic differentiation and endochondral ossification. Expression levels of the chondrogenic differentiation markers and transcriptional regulators Sox9 and Runx2 were all significantly lower in CypA knockdown chondrogenic cells than in wild-type cells, indicating that CypA plays a functional role in chondrogenic differentiation. In vitro differentiation studies using micromass cultures of mouse limb bud cells further supported the conclusion that CypA is needed for chondrogenic differentiation. Newborn CypA-deficient pups double stained with alcian blue and alizarin red exhibited generalized, pronounced skeletal defects, while high-resolution micro-computed tomography (microCT) analyses of the femurs and lumbar vertebrae revealed delayed or incomplete endochondral ossification. Comparative histology and immunohistochemistry (IHC) analyses further verified the effects of CypA deficiency on chondrogenic differentiation. Our results provide evidence for the important contribution of CypA as a pertinent component acting through NF-κB–Sox9 in regulation of chondrogenesis signaling. These findings are important to better understand signal-induced chondrogenesis of chondrogenic progenitors in physiological and pathophysiological contexts.
INTRODUCTION
Chondrogenesis is an essential process in vertebrates. It leads to the formation of cartilage growth plates, thereby driving bodily growth while providing structural templates and induction signals for the formation of long bones through endochondral ossification (1). On the other hand, defects in chondrogenesis cause various chondrodysostoses and chondrodysplasias, with such skeletal malformations accounting for a significant proportion of human birth defects that often result in embryonic and perinatal lethality (2). To identify the molecular mechanisms that drive chondrocyte differentiation and impact underlying cartilage diseases, the transcriptional mechanisms governing their cartilage-specific expression have been intensely studied. As a general overview, the chondrocyte differentiation pathway corresponds to a succession of major genetic program switches that are likely controlled by a specific set of transcriptional activators, repressors, and associated factors. While some of these factors play essential roles in determining cell fate and differentiation, other factors are found to be mutated in severe diseases of cartilage and bone malformation (3). The framework of the cartilage matrix is a collagen fiber network comprised primarily of type II collagen (Col2) (encoded by the Col2α1 gene) and secondarily of type IX collagen (encoded by Col9α1, Col9α2, and Col9α3). Collagen type X (Col10) (encoded by Col10α1) is also produced in abundance, but exclusively by prehypertrophic and hypertrophic chondrocytes (2).
Importantly, chondrocyte differentiation requires Sox family transcription factors in the early stages and Runx2 in the late stages (4, 5). In particular, Sox9 has an essential, nonredundant role in specifying the commitment and differentiation of mesenchymal cells toward the chondrogenic lineage in all developing skeletal elements. While Sox9 is turned on in chondrogenic and osteogenic mesenchymal cells prior to condensation and remains highly expressed in prechondrocytes and chondroblasts, it is turned off when cells undergo prehypertrophy (6). Moreover, Sox9 not only promotes chondrocyte differentiation but also promotes the expression of cartilage-specific extracellular matrix genes, including Col2α1, Col11α2, and aggrecan (5). Two other members of the Sox family, L-Sox5 and Sox6, are also critical effectors of chondroblast differentiation. Moreover, both in vivo and in vitro experiments suggest that Sox9 and L-Sox5/Sox6 cooperate directly to activate Col2α1 (7).
The Runt domain transcriptional activator Runx2, also referred to as core-binding factor α1 (Cbfa1), and its close relative, Runx3, promote chondroblast proliferation and its organization into columns. Specifically, Runx2 and Runx3 have essential roles in inducing chondrocyte prehypertrophy and hypertrophy, with Runx2−/− Runx3−/− mice displaying a complete absence of prehypertrophic and hypertrophic chondrocytes (8). While both activators are upregulated in chondroblasts that reach prehypertrophy, Runx2 remains expressed throughout hypertrophy and terminal differentiation. Importantly, Runx2−/− mice lack bones, demonstrating that Runx2 is required for osteoblast differentiation (9). Furthermore, Runx2-deficient mice also exhibit a disturbance in chondrocyte maturation, suggesting that Runx2 is a direct transcriptional activator of chondrocyte maturation markers (10). In fact, Runx2 has been found to bind multiple recognition sites in the Col10α1 promoter in vivo and to activate Col10α1 reporter constructs through these elements in vitro (11).
Recently, several reports have described the effects of cyclophilin B (CypB, or PPIB) mutants on type I collagen modification and components of the prolyl 3-hydroxylation complex, where CypB-deficient mice present with severe osteogenesis imperfecta-like phenotypes (12). Similarly, the molecular chaperone Hsp47 was recently reported to play an essential role in cartilage and endochondral bone formation (13). However, the role of a closely related protein, cyclophilin A (CypA), in the formation of cartilage and endochondral bone remains to be elucidated. CypA, the Ppia gene product, is a member of the peptidyl-prolyl isomerase (PPIase) family, catalyzing not only the cis-trans isomerization of peptidyl-prolyl bonds during protein folding but also conformational changes (14). CypA was first identified as the primary intracellular target of the immunosuppressive drug cyclosporine (CsA) (15). The immunosuppressive activity of CsA is thought to result from the engagement of calcineurin by the CsA-CypA complex (16).
Several lines of research have since revealed that PPIases, such as CypA, may function as molecular signaling “switches” that can act as novel molecular timers to help control the amplitude and duration of cellular processes (17). In addition, the role of CypA in the activation of other factors and their nuclear translocation has an impact on various cellular functions by acting as a signal transducer and activator of transcription (18, 19). One report demonstrated that the knockdown of CypA inhibits Stat3 interleukin-6-induced tyrosine phosphorylation and nuclear translocation, resulting in altered gene expression in myeloma cell lines (20). Additional data also suggest that CypA may contribute to the pathology of certain human malignancies (21). Therefore, targeting CypA and inhibiting its PPIase activity have been proposed as novel therapeutic strategies for the treatment of human cancers.
In this study, we found that the prolyl isomerase CypA is required for chondrogenic differentiation and endochondral ossification. CypA functions as a novel NF-κB/p65-interacting protein (22). It acts in concert with p65 to mediate BMP-2-induced Sox9 production and hence regulation of chondrogenesis signaling. Either knockdown of CypA or pharmacological inhibition of CypA or NF-κB reduces or delays BMP-2-induced chondrogenic differentiation. CypA-deficient mice display chondrodysplasis and defective endochondral bone formation. Micromass cultures of primary mesenchymal limb bud cells confirmed delayed hypertrophic differentiation in CypA-deficient mice. High-resolution micro-computed tomography (microCT) analyses of femurs and lumbar vertebrae also revealed delayed or incomplete endochondral ossification. Comparative histology and immunohistochemistry (IHC) analyses further verified the effects of CypA deficiency on chondrogenic differentiation. These molecular details are important to better understand BMP-2-induced chondrogenesis signaling during vertebrate development and will help to develop better strategies for tissue repair and treatment of cartilage-related diseases.
MATERIALS AND METHODS
Cell culture and generation of stable CypA knockdown cell lines.
ATDC5 and C3H10T1/2 cells were obtained from the American Type Culture Collection (ATCC). ATDC5 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–F-12 (Invitrogen, Carlsbad, CA) containing 5% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin, and C3H10T1/2 cells were cultured in DMEM with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. For chondrogenic differentiation, ATDC5 cells were cultured in a maintenance medium supplemented with insulin-transferring sodium selenite medium supplement (ITS) (Sigma-Aldrich, St. Louis, MO); C3H10T1/2 cells were cultured in DMEM–F-12 containing 5% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 ng/ml BMP-2 (Sigma-Aldrich).
To knock down CypA in ATDC5 and C3H10T1/2 cells, the cells were transfected with the pSilencer-CypARNAi vector for 48 h, followed by screening in 500 mg/ml G418 for 48 h. Twelve single clones of each cell line were selected and maintained in medium with 200 mg/ml G418 for 2 weeks. Western blotting was performed to test the knockdown efficiency. 2T7 and 2T3 single clones (ATDC5 cells) and cd-1 and cd-2 single clones (C3H10T1/2) were selected for comparison to scrambled clones (SC) and wild-type (WT) parental cells.
Total nuclear protein extraction and Western blot analysis.
The cell nuclear protein fraction was isolated using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific, Hudson, NH). The primary antibodies anti-p65 (sc-109; Santa Cruz Biotechnology, Santa Cruz, CA), anti-p50 (H119; Santa Cruz Biotechnology), anti-p84 (ab487; Abcam, Cambridge, MA), anti-CypA (sc-133494; Santa Cruz Biotechnology), and anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) (sc-20357; Santa Cruz Biotechnology) were used according to the manufacturers' instructions.
Animals.
We conducted all mouse experiments in accordance with experimental protocols approved by the Institutional Animal Care and Use Committee at the University of California, Los Angeles, Los Angeles, CA. We purchased Ppia+/− mice from the Jackson Laboratory and backcrossed the Ppia+/− mice with C57BL/6 mice for 8 generations prior to the experiments.
Real-time PCR.
Total mRNAs were extracted with TRIzol (Invitrogen) according to the manufacturer's instructions. First-strand cDNA synthesis and amplification were performed using the Superscript III reverse transcription system (Invitrogen). Quantitative reverse transcription (qRT)-PCRs were performed in triplicate in a total volume of 25 μl containing 2 μM primers and 12.5 μl of Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA). The sequences of the primers were as follows: Sox9 forward, AGACTCACATCTCTCCTAATGCT, and reverse, ACGTCGGTTTTGGGAGTGG; Col2α1 forward, ACACTGGTAAGTGGGGCAAG, and reverse, GCGTCTGACTCACACCAGAT; Col10α1 forward, CATAAAGGGCCCACTTGCTA, and reverse, TGGCTGATATTCCTGGTGGT; Runx2 forward, CCGCACGACAACCGCACCAT, and reverse, CGCTCCGGCCCACAAATCTC; Gapdh forward, TGCACCACCAACTGCTTAGC, and reverse, CCACCACCCTGTTGCTGTAG.
Alcian blue and alizarin red staining.
Alcian blue staining was performed using alcian blue solution (0.1% alcian blue 8GX in 0.1 N HCl) for 30 min at room temperature. Counterstaining was conducted using 1% nuclear fast red solution. For alizarin red staining, cells were fixed in 4% paraformaldehyde (PFA) for 20 min and stained with alizarin red solution (Lifeline Cell Technology, Frederick, MD) for 5 min.
Micromass culture of mouse limb bud cells.
Micromass pellet culture was prepared as described with some modifications (23). Briefly, the anterior and posterior limb buds of embryonic day 12.5 (E12.5) mouse embryos were digested with 10 mg/ml dispase (Sigma-Aldrich) solution containing 10% FBS (Invitrogen)-Puck's saline A buffer (PSA) and shaken gently at 37°C for 1.5 h. The isolated single-cell suspensions were resuspended in growth medium at a concentration of 107 cells/ml and spotted in 10-μl droplets in each 1.5-ml centrifuge tube. The culture medium was DMEM–F-12 (1:1) plus 10% FBS supplemented with 0.5 mM glutamine, 0.25 mM ascorbic acid, and 1 mM β-glycerol phosphate. Growth media were replaced every 3 days.
Skeleton staining.
Newborn Ppia+/+ and Ppia−/− mice were collected, and their skin was carefully peeled off with forceps. The mice were then fixed overnight with 95% ethanol, followed by acetone incubation overnight at room temperature. The specimens were stained with alcian blue solution containing 0.1% alcian blue 8GX (Sigma-Aldrich), 20% acetic acid, and 80% ethanol. After 24 h, the skeleton samples were rinsed with 95% ethanol and 95% ethanol-2% KOH (1:1 [vol/vol]) and counterstained overnight with 0.03% alizarin red S (Sigma-Aldrich) in 1% KOH. Clearing of the samples was conducted by placing them in 1% KOH-20% glycerol for at least 2 days before transfer into glycerol-95% ethanol (1:1 [vol/vol]). Images were taken using an Olympus SZX12 microscope and a MicroFire digital microscope camera with PictureFrame software (Optronics, Goleta, CA).
Immunohistochemical staining.
Four CypA WT and two knockout (KO) 13-week-old adult mice were euthanized and fixed in 10% formalin. Femurs were isolated and embedded in paraffin. Paraffin sections (5 μm thick) were dewaxed in xylene and rehydrated in ethanol baths. Endogenous peroxidases were blocked by incubating the sections in 3% hydrogen peroxide for 20 min at room temperature. The sections were incubated with anti-Col II (II-II6B3-c; Development Studies Hybridoma Bank), anti-Col X (X-AC9), anti-p65 (sc-109; Santa Cruz Biotechnology, Santa Cruz, CA), or anti-CypA (sc-133494; Santa Cruz Biotechnology) primary antibody at room temperature for 1 h and overnight at 4°C and then incubated with a biotinylated secondary antibody (Dako, Carpinteria, CA) for 1 h at room temperature. Positive immune reactivity was detected using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA) and AEC chromogenic substrate (Dako) according to the manufacturer's instructions. A negative control was generated by replacing primary antibody solutions with phosphate-buffered saline (PBS). The sections were counterstained with hematoxylin for 30 s and then rinsed for 5 min in running water. Photomicrographs were acquired using an Olympus BX51 microscope and a MicroFire digital microscope camera with PictureFrame software.
Three-dimensional (3D) microCT analysis.
Three WT and three KO neonatal mice were processed postmortem for microCT analysis. The whole bodies were fixed in 4% PFA for 48 h and stored in 70% ethanol. Samples were then scanned using high-resolution microCT (SkyScan 1172; SkyScan N.V., Belgium) at an image resolution of 15.9 μm (39 kV and 246 μA radiation source, using a 0.5-mm aluminum filter) and analyzed using DataViewer, NRecon, CTAn, and CTVol software provided by the manufacturer. The quantitative and structural parameters examined follow the nomenclature described by the American Society for Bone and Mineral Research Nomenclature Committee (24, 25). After examining the overall skeleton, samples were reoriented using DataViewer for detailed analysis of the most commonly examined bone sites, femurs and individual lumbar vertebral bodies at levels L3 (the third lumbar vertebra) through L6, and were saved as separate data sets. For femurs, the trabecular bone was analyzed between the proximal growth plate and the distal growth plate, spanning 1.91 mm of the bone (or 82% of the whole length), and the cortical bone was analyzed in 0.45 mm at the center of the shaft. For the lumbar vertebral body, the trabecular bone was analyzed throughout the height of each bone (average, 0.59 mm at L3, 0.54 mm at L4, and 0.53 mm at L5). For the analysis of trabecular bone, regions of interest (ROI) were drawn to discretely select the trabecular bone only, maintaining a 0.025-mm clearance from the endosteal bone surface. For the analysis of cortical bone, ROI were drawn to exactly fit the outer border on the periosteal surface and the inner border on the endosteal surface.
To determine the optimal threshold, two representative (median) femurs were selected (based on radiographic findings) from each group. The threshold ranges for trabecular and cortical bones were determined to be 70 to 255 and 100 to 255 (equivalent to 1.22 to 4.95 g hydroxyapatite (HA)/cm3 and 1.82 to 4.95 g HA/cm3), respectively, to optimally represent the normal structures in the WT samples.
Chondrogenic differentiation in hydrogel.
Glycol chitosan (molecular mass, ∼500 kDa), glycidyl methacrylate, and riboflavin (RF) sodium salt were purchased from Sigma-Aldrich (St. Louis, MO) and used as received.
3D culture of ATDC5 cells.
Photopolymerizable methacrylated glycol chitosan (MeGC) was prepared as previously described (26, 27). ATDC5 cells were harvested and resuspended in 40 μl of MeGC solution (2% [wt/vol]) at a density of 1 × 107 cells/ml. The cell suspension was exposed to visible blue light (400 to 500 nm; 500 mW/cm2; Bisco Inc., Schaumburg, IL) for 40 s in the presence of 6 μM RF as a photoinitiator. The cross-linked hydrogel was cultured in 1 ml of chondrogenic medium at 37°C and 5% CO2 in humidified incubators for up to 3 weeks. The 3D cultured samples were then analyzed for viability and chondrogenic differentiation.
LIVE/DEAD assay.
Calcein-AM and propidium iodide (PI) dyes (1 mg/ml; Molecular Probes, Invitrogen) were used to perform the LIVE/DEAD assay. Briefly, at each time point, the culture medium was removed and 1 ml of PBS containing 2 μl of calcein-AM and 1 μl of PI was added to each well. Samples were then incubated at 37°C for 10 min protected from light. Afterward, the samples were washed three times with PBS and immediately visualized in the dark by fluorescence microscopy (BX51; Olympus, Tokyo, Japan).
Statistics.
In vitro experiments were performed in triplicate and repeated at least twice; a representative experiment (means ± standard errors [SE]) was selected for the figures. Statistical analysis was performed using the appropriate analysis of variance (ANOVA) to analyze more than two groups, followed by post hoc Tukey's test analysis between specific groups. P values of <0.05 and <0.01 were considered to be significant.
RESULTS
Effect of CypA deficiency on chondrocyte differentiation in vitro.
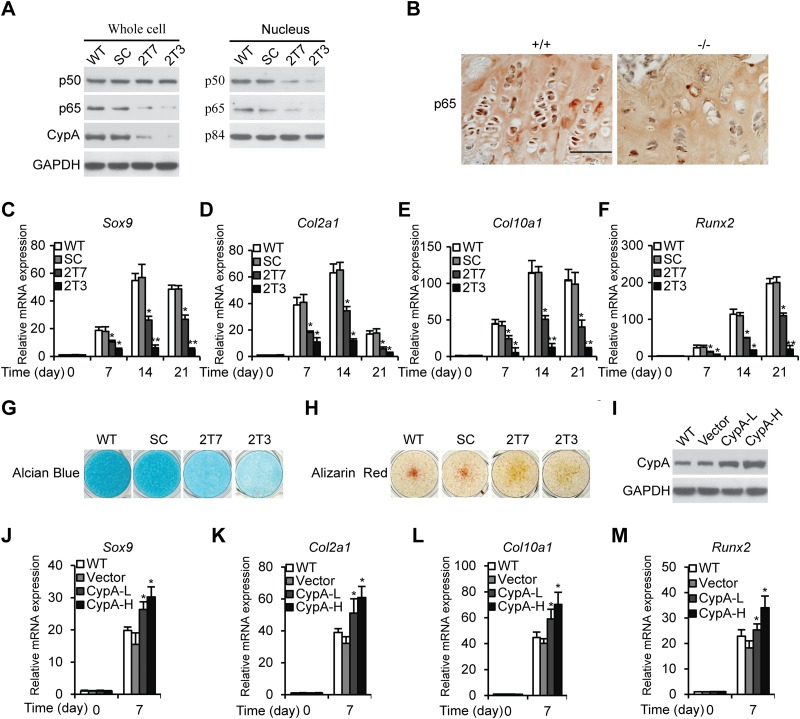
We had previously demonstrated that CypA knockdown (Kd) results in the downregulation of NF-κB/p65 (RelA, a subunit of NF-κB) in glioblastoma cell lines and a murine myoblast line, C2C12 (22, 28). Decreased NF-κB/p65 expression levels were also observed in CypA Kd mouse chondrogenic ATDC5 stable clones, 2T7 and 2T3, as well as in CypA KO mouse proximal tibia growth plates (Fig. 1A and B). As reported previously, activation of NF-κB/p65 facilitates early chondrogenic differentiation (29). To study whether CypA Kd ATDC5 cells lead to downregulated NF-κB/p65 and subsequently affect chondrogenic differentiation, we performed chondrogenic differentiation of CypA Kd cells, 2T7 and 2T3, and compared them to WT ATDC5 cells and SC in the presence of ITS (Sigma-Aldrich, St. Louis, MO) for 3 weeks. Then, at various time points, we measured the mRNA levels of genes for chondrogenic differentiation markers, Col2α1- and Col10α1 collagen, and transcription factors, Sox9 and Runx2, by qRT-PCR. In the CypA Kd stable clones, chondrogenic differentiation markers and transcriptional factors were both significantly reduced (Fig. 1C to F). In WT cells, the expression levels of Col10α1 and Runx2 mRNAs increased from day 14 on, indicating late-stage differentiation of chondrocytes (Fig. 1E and F). Similar results were observed in a murine embryonic mesenchymal cell line, C3H10T1/2 (see Fig. S1 in the supplemental material). Decreased CypA expression levels in CypA Kd ATDC5 cells (2T7 and 2T3) and C3H10T1/2 cells (Cd-1 and Cd-2) were compared with the WT and SC by Western blotting (Fig. 1A; see Fig. S1A in the supplemental material). To verify the effects of CypA deficiency on chondrogenic differentiation, alcian blue and alizarin red staining were used to determine the amounts of matrix proteoglycan and calcium deposition, respectively. Matrix proteoglycan and calcium depositions were found to be markedly reduced in the CypA Kd ATDC5 stable clones compared to controls (Fig. 1G and H). These data clearly showed that CypA Kd cells result in reduction of NF-κB/p65 activation and decreased Sox9 transcriptional activity, suggesting CypA acts through the NF-κB–Sox9 pathway in regulation of chondrogenic differentiation. To examine whether overexpression of CypA influences chondrocyte differentiation, we transfected CypA expression plasmids into wild-type ATDC5 cells, followed by Western blot analyses of CypA expression (Fig. 1I), and determined the relative expression levels of Sox9, Col2α1, Col10α1, and Runx2 (Fig. 1J to M). Our results showed that overexpression of CypA enhanced ATDC5 differentiation in a dose-dependent manner, further suggesting that CypA has a functional role in chondrogenic differentiation.
FIG 1.
Knockdown of CypA attenuates chondrogenic differentiation of ATDC5 cells. (A) Western blot analyses. Whole-cell lysates and nuclear extracts prepared from ATDC5 WT, SC, and CypA Kd stable clones 2T7and 2T3 were immunoblotted to detect CypA, p65, and p50 expression levels. GAPDH and p84 were used as loading controls. (B) Immunohistochemical staining of p65 in a dewaxed section prepared from the distal femoral growth plates of 3-week-old WT and CypA KO mice (scale bar, 50 μm). (C to F) Chondrogenic differentiation of ATDC5 WT, SC, and CypA Kd clones 2T7 and 2T3. The relative mRNA levels of Sox9 (C), Col2α1 (D), Col10α1 (E), and Runx2 (F) were measured by RT-quantitative PCR at several time points. Mean values (n = 3) and standard deviations (SD) are shown. *, P < 0.05, and **, P < 0.01 compared to WT cells at the same time points. (G and H) Alcian blue (G) and alizarin red (H) staining of cell surface proteoglycans and calcium deposits from 21-day chondrogenic differentiated ATDC5 WT, SC, and CypAKd clones. (I) Western blot analyses of CypA expression levels. WT, wild-type ATDC5 cells; Vector, empty-vector-transfected WT cells as a control; CypA-L, WT cells transfected with low concentrations of CypA plasmids (1.25 μg/well) in 6-well plates; CypA-H, WT cells transfected with high concentrations of CypA plasmids (2.5 μg/well) in 6-well plates. (J to M) Relative mRNA expression levels of Sox9 (J), Col2α1 (K), Col10α1 (L), and Runx2 (M) in ATDC5 WT, vector-transfected, and CypA-L-transfected or CypA-H-transfected ATDC5 cells cultured in chondrogenic differentiation medium for 21 days. The data are shown as means and SD of three experiments. *, P < 0.05 compared to WT cells at the same time points.
CypA Kd stable clones exhibit diminished chondrocyte proliferation, reduced chondrogenic differentiation, and reduced matrix proteoglycans in a three-dimensional hydrogel.
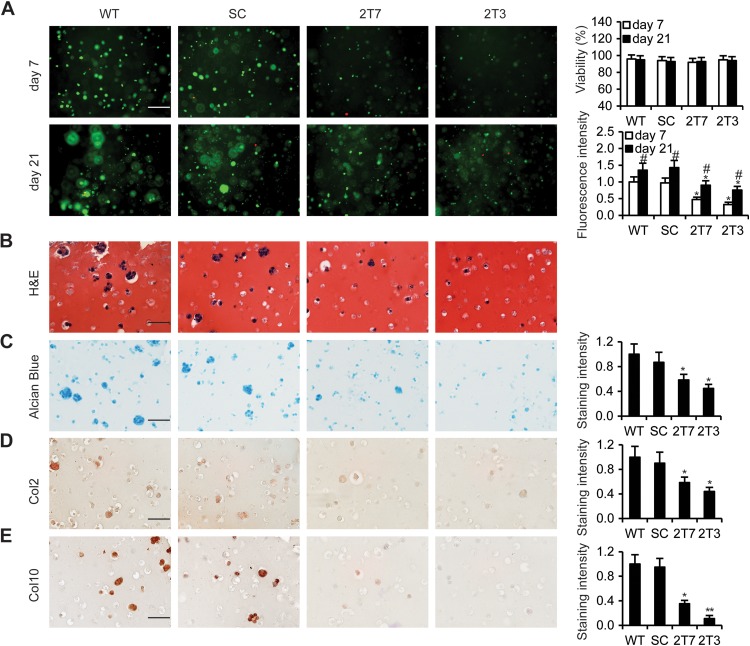
To evaluate chondrocyte proliferation and extracellular matrix deposition in a 3D hydrogel that closely mimics an in vivo microenvironment, we encapsulated CypA Kd stable clones, 2T7 and 2T3, and control cells in a photo-cross-linked hydrogel and cultured them for up to 21 days. In LIVE/DEAD assays, CypA Kd stable clones showed markedly reduced proliferation at 7 days and 21 days in comparison to the controls (Fig. 2A). Histological examination revealed round shapes characteristic of chondrocytes and lacuna formation in the hydrogels at 21 days in the controls but not in the CypA Kd stable clones (Fig. 2B). The levels of alcian blue staining to evaluate the deposition of glycosaminoglycans and immunochemical staining to evaluate the expression of the typical chondrogenic differentiation markers Col2 and Col10 were significantly lower in CypA Kd clones than in the WT and scrambled clones (Fig. 2C, D, and E). Collectively, CypA Kd mouse chondrogenic ATDC5 cells showed diminished chondrocyte proliferation, as well as reduced chondrogenic differentiation and cartilage formation.
FIG 2.
Knockdown of CypA inhibited chondrogenic differentiation in hydrogel. (A) LIVE/DEAD fluorescent staining of encapsulated WT, SC, 2T7, and 2T3 ATDC5 cells undergoing chondrogenic differentiation on days 7 and 21 of culture. The fluorescence intensity was determined using Image Pro 6 software. The percent viability was determined by calculating the number of live cells (green) normalized to the total number of cells (green and red). Scale bar, 50 μm; n = 5 in each group. *, P < 0.05 compared to WT cells; #, P < 0.05 (day 21 compared to day 7). (B and C) Cells were stained with H&E for histological examination of the characteristics of chondrocytes and lacuna formation (B) and with alcian blue to show synthesis of sulfated glycosaminoglycans (C). (D and E) Encapsulated CypA Kd clones 2T7 and 2T3 and control cells in a photo-cross-linked hydrogel were incubated with chondrogenic differentiation medium for 21 days, and the cells were immunohistochemically stained to detect the chondrogenic differentiation markers Col2 (D) and Col10 (E). Scale bars, 50 μm. Quantification of the staining intensity was conducted using Image Pro 6.0 software. n = 5 in each group. *, P < 0.05, and **, P < 0.01 compared to WT cells.
CypA isomerase activity and NF-κB/p65 are indispensable for normal chondrocyte differentiation in vitro.
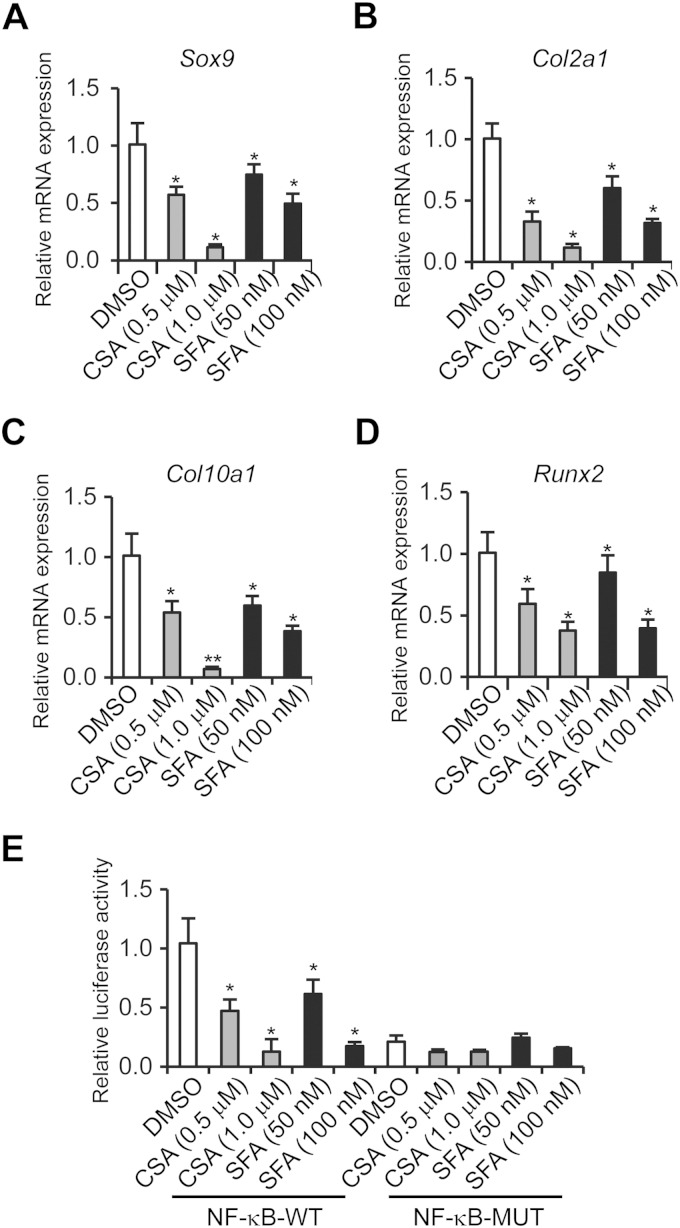
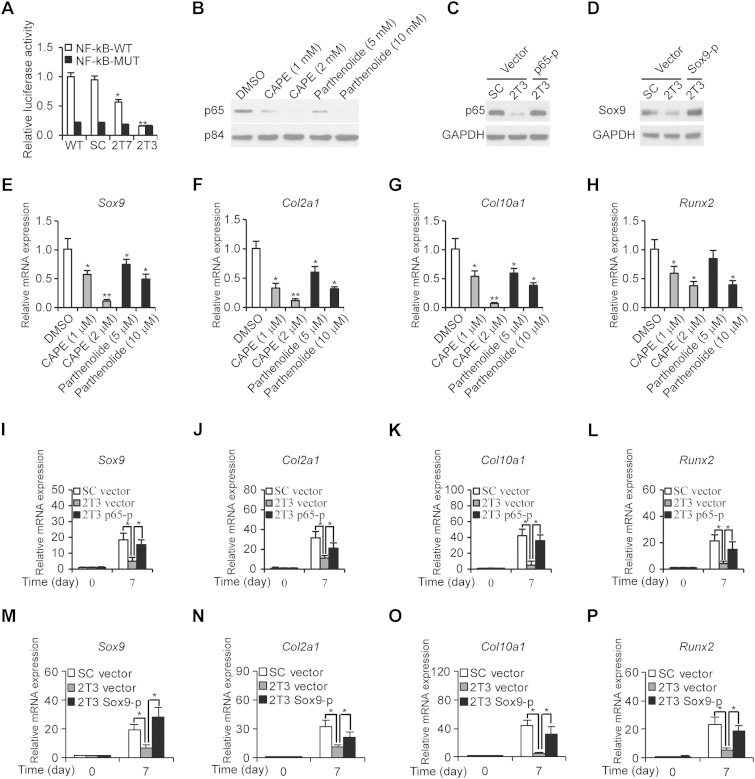
To ensure that the reduced chondrogenic differentiation was due to the lack of CypA isomerase activity and downregulated NF-κB, we treated WT ATDC5 cells with CypA isomerase inhibitors, CsA and sanglifehrin A (SFA), and with NF-κB inhibitors, caffeic acid phenethyl ester (CAPE) and pathenolide. The relative mRNA levels of Sox9, Col2α1, Col10α1, and Runx2 were then determined after 14 days of culture in the presence of ITS. The results clearly demonstrated that either isomerase binding compounds or NF-κB inhibitors led to decreased relative expression levels of chondrogenic differentiation markers (Fig. 3A to D and 4E to H). Meanwhile, decreased κB-luciferase activities were observed in the CypA isomerase inhibitor-treated ATDC5 cells (Fig. 3E), as well as in the CypA Kd stable clones, 2T7 and 2T3 (Fig. 4A). In addition, NF-κB inhibitor-treated ATDC5 cells exhibited decreased levels of p65, a subunit of NF-κB (Fig. 4B). Furthermore, the ectopic expression of p65 and Sox9 plasmids (Fig. 4C and D) were found to restore differentiation markers of CypA Kd chondrogenic cells after 7 days of culture with ITS (Fig. 4I to P). These results suggest that CypA, NF-κB/p65, and Sox9 are required for normal chondrocyte differentiation in vitro and further implied that CypA regulates chondrogenic differentiation through transcriptional activation of Sox9.
FIG 3.
PPIase activity of CypA is required for chondrogenic differentiation. (A to D) ATDC5 cells were treated with DMSO (as a control) and PPIase inhibitor, CSA or SFA, at the indicated concentrations in chondrogenic differentiation medium for 14 days, and the relative mRNA expression levels of Sox9 (A), Col2α1 (B), Col10α1 (C), and Runx2 (D) were quantitated by qRT-PCR. Mean values (n = 3) and standard deviations are shown. (E) NF-κB luciferase assay in ATDC5 cells treated with DMSO, CSA, or SFA for 24 h. Luciferase reporter plasmids, pGL4 nf-κb-Luc with the consensus NF-κB binding sequence GGGACTTTCC and its mutant with a mutated sequence (CTCACTTTCC), were used. *, P < 0.05, and **, P < 0.01 compared to the DMSO treatment group. The data are shown as means and SD of three experiments.
FIG 4.
CypA regulates ATDC5 differentiation through the NF-κB pathway. (A) Relative luciferase activities of NF-κB were determined in WT, SC, 2T7, and 2T3 ATDC5 cells 2 days posttransfection with κB-Luc plasmids. n = 4. *, P < 0.05, and **, P < 0.01 compared to the WT. (B) Western blot analysis of nuclear p65 expression levels. Nuclear extracts prepared from ATDC5 and NF-κB inhibitor-treated ATDC5 cells were subjected to immunoblotting to detect p65 expression levels. p84 was used as a loading control. (C and D) Western blot analyses of ectopic expressed levels of p65 and Sox9 compared to controls. (E to H) The relative mRNA expression levels of Sox9 (E), Col2α1 (F), Col10α1 (G), and Runx2 (H) were determined by qRT-PCR. ATDC5 cells were treated with NF-κB inhibitors at the indicated concentrations and cultured in chondrogenic differentiation medium for 7 days, followed by determination of their relative mRNA expression levels. Cells treated with DMSO were used as controls. n = 4. *, P < 0.05, and **, P < 0.01 compared to the DMSO-treated group. (I to L) Restoration of relative expression levels of Sox9 (I), Col2α1 (J), Col10α1 (K), and Runx2 (L) by transfection of p65 plasmids (p65-p) into CypA Kd ATDC5 2T3 cells. (M to P) Restoration of relative expression levels of Sox9 (M), Col2α1 (N), Col10α1 (O), and Runx2 (P) by transfection of Sox9 plasmids (Sox9-p) into CypA Kd ATDC 2T3 cells. *, P < 0.05 compared to vector-transfected CypA Kd 2T3 cells at the same time points. The data are shown as means and SD of three experiments.
Double staining reveals several skeletal defects in Ppia−/− animals.
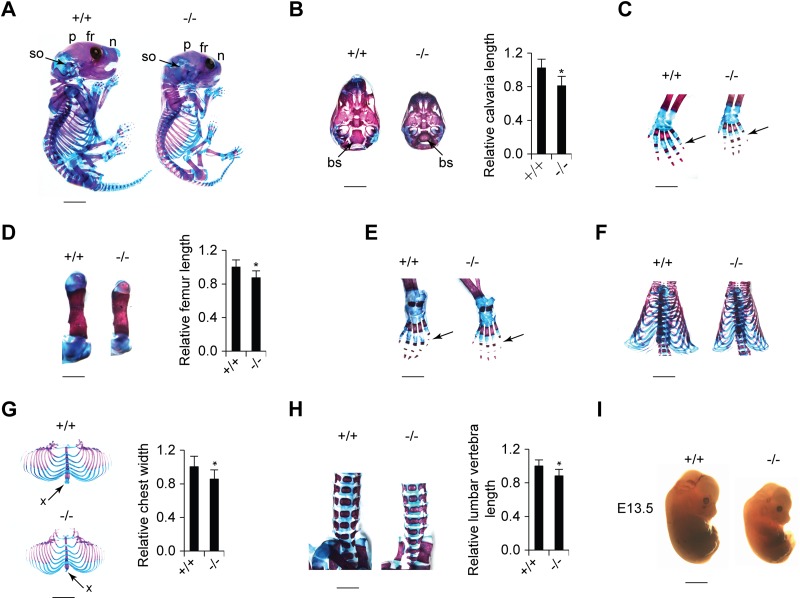
To analyze the physiological role of CypA in skeletal growth, we compared the skeletal phenotypes of wild-type and CypA KO (Ppia−/−) mice. One-day-old (P1) Ppia−/− mice exhibited shorter statures than their WT littermates (Fig. 5A). In addition, Ppia−/− mice were found to have considerably reduced body weights throughout embryonic (E13.5) and postnatal (P1) life (Fig. 5A and I). Next, WT and Ppia−/− skeletons at P1 were prepared, and the cartilage and bone samples were visualized by alcian blue and alizarin red double staining. From a lateral view, the skulls of Ppia−/− CypA-deficient mice exhibited delayed ossification of the frontal, parietal, nasal, and supraoccipital bones in comparison to their WT littermates (Fig. 5A). From a ventral view, the caudal part of the skulls of CypA-deficient mice exhibited a smaller and developmentally delayed basisphenoid base (Fig. 5B). In the trunk region, CypA-deficient mice were found to have shorter vertebrae, smaller rib cages, and underdeveloped xiphoid processes (Fig. 5G and H). In addition, the same mice were found to have shorter long bones (Fig. 5D) and incomplete ossification centers on the distal phalanges of the fifth forepaws and hind paws (Fig. 5C and E). Taken together, these results suggest that CypA plays a functional role in skeletal development.
FIG 5.
Ppia−/− mice exhibit small body size and defects in skeletal development. (A) Alcian blue/alizarin red staining of skeletons of newborn Ppia+/+ and Ppia−/− mice (scale bar, 3 mm). A lateral view of the skull exhibits differences in the development of the frontal (fr), parietal (p), nasal (n), and supraoccipital (so) bones. (B) (Left) Ventral view of the caudal part of the skull showing that the basisphenoid bone (bs) is smaller in the Ppia−/− newborn mouse (scale bar, 2 mm). (Right) Quantification of skull length. (C) Left forelimbs of newborn mice (scale bar, 1 mm). The arrows indicate the distal phalanges of the fifth digit of the forepaws. (D) Comparison of left femurs of Ppia+/+ and Ppia−/− mice with double staining (left) (scale bar, 2 mm) and length quantification (right). (E) Double staining of left hind limbs (scale bar, 1 mm). The arrows indicate the distal phalanges of the fifth digit of the hind paws. (F) Ventral view of backbones and rib cages (scale bar, 3 mm). (G) Rib cages (left) (scale bar, 3 mm) and width quantification (right). The arrows indicate xiphoid processes. (H) Frontal view of the lumbar vertebrae with alcian blue and alizarin red staining (left) (scale bar, 2 mm) and length quantification (right). (I) Comparison of the sizes of Ppia+/+ and Ppia−/− embryos at E13.5 (scale bar, 1 mm).
High-resolution microCT analysis of skeletal phenotype.
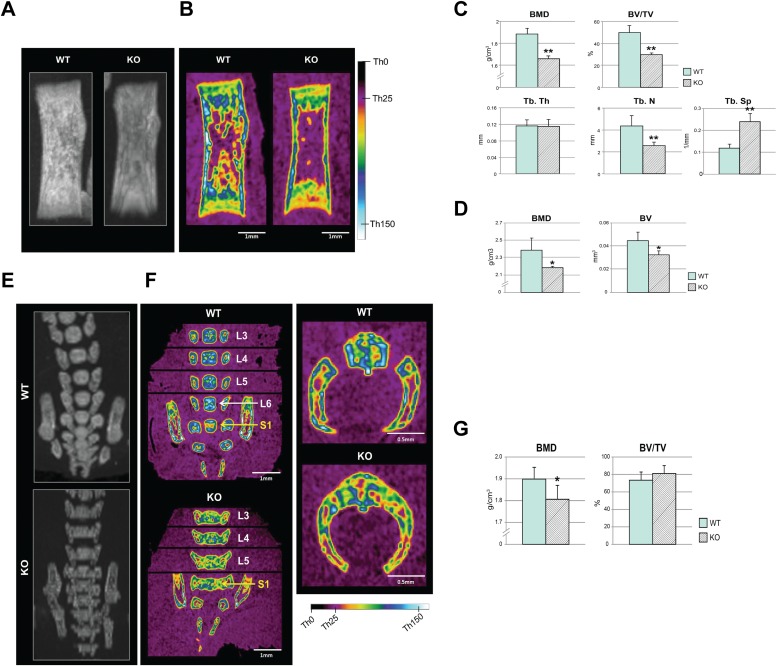
MicroCT analysis of the whole body revealed a 25% average decrease in body height in the CypA KO group, with the femurs showing a 10% average reduction in length. Detailed microCT analysis of the femurs revealed a statistically significant reduction in the bone mineral density (BMD), bone volume (BV), and percent bone volume (BV/total volume [TV]) in trabecular and cortical bones, both qualitatively (Fig. 6A and B) and quantitatively (Fig. 6C and D). The structural measurements of the trabecular bone also exhibited diminished microstructure in the KO mice (Fig. 6A). Interestingly, the CypA KO lumbar vertebrae showed a decrease in height by 5 to 10% (variable by the vertebral level, from L3 to L5) while presenting an increase in width by 4 to 7.5% (variable by the vertebral level) in comparison to the WT, attributable to bilaterally fused vertebral bodies and dorsal arches (Fig. 6F and G).
FIG 6.
High-resolution microCT analysis of femurs and lumbar vertebrae. (A and B) Overview radiographic maximum intensity projection (MIP) (A) and color-coded midcoronal sectional images (B) of femurs generated from a microCT data set, with a color scale marked with reference threshold values. (C) Quantification of BMD, BV/TV, and trabecular structures (Tb.Th, Tb.N, and Tb.Sp) in the trabecular bone. (D) Quantification of BMD and BV in the cortical bone. (E and F) MIP (E) and color-coded midcoronal sectional images (F) of vertebral bodies at L3 to S1 (the first sacral vertebra) levels showing abnormal ossification patterns in KO mice (the exact midcoronal image of each vertebral body was captured separately and then stitched together, as marked by black lines in panel F). (F) (Left) Overall mineral density was visibly reduced in KO mice, as shown by colors corresponding to lower threshold values. (Right) Bilaterally fused vertebral body and dorsal arches. (G) Quantification of the lumbar vertebrae at levels L3 to L5 presented significantly reduced BMD but slightly increased BV/TV in KO mice. n = 4 in each group. *, P < 0.05, and **, P < 0.01 compared to the WT. The error bars indicate means and SD.
The microCT analysis of the lumbar vertebrae further revealed a missing L6 in KO mice that was present in WT littermates (Fig. 6E and F). The quantitative analysis of the lumbar vertebrae at the L3 to L5 levels was in accordance with the visual analysis and presented a significantly reduced BMD but a slightly increased BV/TV in KO mice (Fig. 6G). This is due to the abnormally fused and widened vertebral bodies in the CypA KO group, adding more bone tissue in the ROI.
Micromass cultures show delayed hypertrophic differentiation of chondrocytes in CypA KO mice.
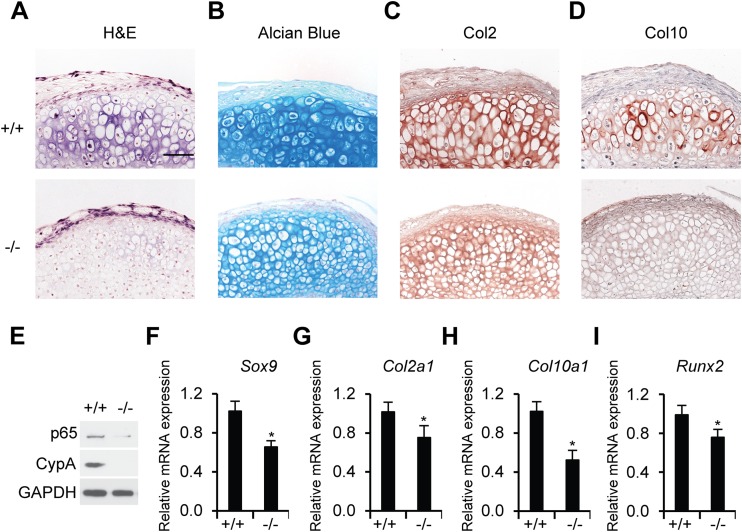
To analyze the role of CypA in coordinating the proliferation and hypertrophic differentiation of growth plate chondrocytes, we established a micromass system to achieve reproducible hypertrophic differentiation in mouse mesenchymal limb bud cells. Upon treatment with ascorbic acid and β-glycerol phosphate in the micromass culture, mesenchymal limb bud cells isolated from Ppia−/− mice showed decreased chondrogenesis in comparison to those from Ppia+/+ mice. Hematoxylin and eosin (H&E) staining of the micromass cultures showed a marked delay in hypertrophic differentiation of CypA-deficient mesenchymal cells (Fig. 7A). Decreased chondrogenesis was further reflected in significant reduction in alcian blue stain and Col2 and Col10 expression levels in the CypA-deficient micromass cultures in comparison to a WT control (Fig. 7B to D). Relative mRNA levels of the transcriptional factor genes Sox9 and Runx2 and the chondrogenic differentiation marker genes Col2α1 and Col10α1 were reduced in CypA-deficient cells in comparison to WT controls (Fig. 7F to I). The delay in hypertrophic differentiation of CypA-deficient mesenchymal cells was correlated with downregulated NF-κB/p65 and undetectable levels of CypA expression, suggesting that CypA and NF-κB/p65 play key functional roles in chondrocyte differentiation (Fig. 7A to E).
FIG 7.
CypA deficiency suppresses chondrogenic differentiation of primary mesenchymal limb bud cells. (A) H&E staining of sections of micromass pellets. (B) Alcian blue staining of sections of micromass pellets. (C and D) Immunohistochemical detection of Col2 (C) and Col10 (D) in micromass pellet cultures of mesenchymal limb bud cells from Ppia+/+ and Ppia−/− mice (E12.5). (A to D) Scale bar, 50 μm. (E) Western blot analyses of CypA and p65 expression levels in mesenchymal limb bud cells (E12.5) isolated from Ppia+/+ and Ppia−/− mice. GAPDH was used as a loading control. (F to I) Relative mRNA expression levels of Sox9 (F), Col2α1 (G), Col10α1 (H), and Runx2 (I) in 21-day-cultured micromass pellets. The data are shown as means and SD of three experiments. n = 4 in each group. *, P < 0.05 compared to chondrogenic differentiated limb bud cells from WT mice.
Comparative histology and IHC of mouse proximal tibias confirm decreased growth plate activity and reduced trabeculation in Ppia−/− mice.
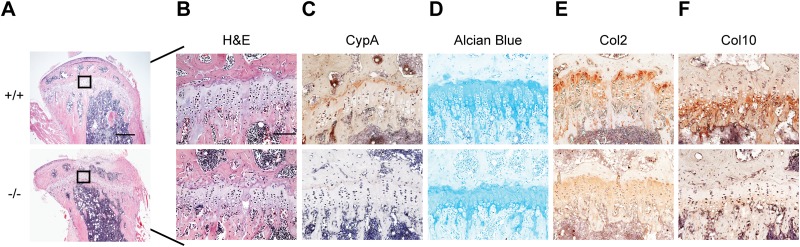
Histological analysis of the H&E-stained tibia sections prepared from 13-week-old mice showed substantially greater trabeculation in WT mice than in Ppia−/− mice (Fig. 8A). To verify whether CypA deficiency affects chondrogenic differentiation in vivo, alcian blue and immunochemical staining were performed. H&E and alcian blue staining of growth plate sections revealed a wider growth plate and a longer chondrocyte column in the proximal tibia in WT mice than in Ppia−/− mice (Fig. 8B and D). Immunochemical staining of proximal tibia sections with an anti-CypA antibody was used to measure the expression levels of CypA (Fig. 8C). In addition, alcian blue staining of proximal tibia sections revealed a substantially stronger positive stain in the growth plate areas of WT mice than in Ppia−/− mice, suggesting reduced cartilage formation in Ppia−/− mice (Fig. 8D). Moreover, immunochemical staining revealed that Col2 and Col10 collagen were more highly expressed in WT mice, further suggesting Ppia−/− mice had delayed chondrocyte differentiation and decreased growth plate activity during chondrogenesis (Fig. 8E and F).
FIG 8.
H&E, alcian blue, and immunohistochemical staining of proximal tibia prepared from Ppia+/+ and Ppia−/− adult mice. (A) H&E staining of proximal tibia sections prepared from 13-week-old Ppia+/+ and Ppia−/− mice at low magnification (scale bar, 200 μm). (B) H&E staining of proximal tibia sections at high magnification. (C) Immunohistochemical staining of CypA in a dewaxed section of the proximal tibia prepared from Ppia+/+ and Ppia−/− mice. (D) Alcian blue staining to detect cartilage matrix in the growth plate. (E and F) Immunohistochemical staining of Col2 (E) and Col10 (F) expression levels in dewaxed proximal tibia sections prepared from Ppia+/+ and Ppia−/− mice. (B to F) Scale bar, 50 μm.
DISCUSSION
Knockdown of CypA by RNA interference in the chondrogenic cell line ATDC5 and the murine mesenchymal stem cell line C3H10T1/2 significantly suppressed not only chondrogenic differentiation but also matrix proteoglycan accumulation and calcium deposition. These results were further confirmed by encapsulating CypA Kd stable clones of ATDC5 cells in a 3D hydrogel to evaluate chondrocyte proliferation and extracellular matrix deposition. Our study of the loss of function of CypA in the mouse model also indicated the following. (i) In micromass cultures, mesenchymal limb bud cells isolated from Ppia−/− mice had markedly delayed hypertrophic differentiation and significantly decreased expression of chondrogenic differentiation markers in comparison to cells isolated from Ppia+/+ mice. (ii) Based on skeleton staining, the Ppia−/− mice exhibited short stature, the frontal and parietal bones were delayed in growth, the vertebrae and rib cages were smaller, and the long bones were shorter than in the WT littermates. (iii) As demonstrated by high-resolution computed tomography analyses, CypA KO mice exhibited significant reductions in bone mineral density and bone volume in trabecular and cortical bones (Fig. 6C and D). (iv) As revealed by H&E and IHC staining, CypA is required for normal growth plate activity and expression of the chondrogenic differentiation markers type II and type X collagen. Collectively, these results demonstrate that CypA has a pivotal physiological role in skeletal growth with particular implications for chondrogenesis.
CypB (PPIB)-deficient mice were found to possess severe osteogenesis imperfecta-like phenotypes (12). However, the functional role of the closely related PPIase family member CypA (PPIA) in skeletal development remains elusive. In previous reports, we demonstrated a close relationship between CypA and NF-κB signaling (22, 28). NF-κB was found to regulate certain aspects of endochondral ossification during embryogenesis (29, 30). In this report, we clearly showed that CypA Kd cells result in reduction of NF-κB/p65 activity and decreased Sox9 transcriptional activity, suggesting that CypA acts through an NF-κB–Sox9 pathway in the regulation of chondrogenic differentiation. A different approach for inactivation of NF-κB is to use a dominant-negative IκB(AA) superrepressor, which prevents phosphorylation and degradation of IκBα, thereby retaining the NF-κB dimer in an inactive form in cytoplasm (31, 32). Therefore, the IκB(AA) mutant transgenic mice may develop defects similar to those observed in Ppia−/− mice.
Sox9 is another key molecule for chondrogenic differentiation and cartilage formation (5), and its activity is dependent on NF-κB activation (33). Sox9 plays a critical role in the commitment and differentiation of mesenchymal stem cells into chondrocytes in the early stages of chondrocyte differentiation (34). The molecular mechanism of CypA involved in chondrogenesis is clearly different from the function of CypB in osteogenesis. Sox9 also promotes the expression of cartilage-specific extracellular matrix genes, including Col2α1, Col11, and Aggrecan genes (4, 5). Heterozygous Sox9 mutant mice and Sox9-deficient mice exhibit severe impairment in endochondral ossification (34). Our studies of CypA-deficient cultured cells showed significantly suppressed chondrogenic differentiation, matrix proteoglycan accumulation, and calcium deposition. These findings from CypA-deficient animals correspond to the in vitro data and provide further evidence for the effect of CypA on markedly delayed hypertrophic differentiation and significantly decreased chondrogenic differentiation markers, cartilage matrix, and collagen type II and type X expression. This chondrodysplasia-like phenotype of CypA-deficient mice is similar to the phenotype of Sox9-null mutants, which exhibited severe generalized chondrodysplasia (34).
Importantly, CypA-deficient animals expired at various times within 5 weeks after birth, with the rare exception of survival up to 13 weeks, as presented in this study. These observations resulted after backcrossing 129s6/SvEV Ppia+/− mice with C57BL/6 mice for 8 generations, suggesting that the penetration of the abnormal phenotype is dependent upon the genetic background. Early perinatal death could be attributable to respiratory distress or malnutrition, based on morphological findings from double staining and microCT imaging. Respiratory distress and delayed failure were suspected because of the reduced size and constricted morphology of the rib cage, leading to a considerably reduced pleural cavity that could restrict the expansion of the lungs. Moreover, the xiphoid process was consistently missing in CypA-deficient animals, which could have further contributed to respiratory distress. However, the complete absence of a xiphoid process has not always led to death in past mouse knockout models (35). As a result, malnutrition was also suspected because of an invariably foreshortened nose and reduced oral cavity that could impede suckling.
In addition, the CypA-deficient animals were found to be significantly lighter in weight than the WT animals, with both approximately 20% shorter height and individual bone size. These phenotypic changes are similar to the Runx2 KO phenotype, which also exhibits a 20% decrease in weight between WT and KO animals (36). Furthermore, the middle phalanges of the fifth digit of the paws, the xiphoid process of the sternum, and the sixth lumbar vertebra (L6) were consistently absent in the CypA KO animals. L6, however, was always present in WT littermates. MicroCT-based examination further confirmed that there was neither full nor partial sacralization of the terminal lumbar vertebrae in KO animals. This finding was particularly interesting because the abnormalities in the phalanges, xiphoid process, and lumbar vertebrae are all due to a delay in endochondral ossification. Similar findings were reported in mice lacking the transcription factor Sp3, which is required for proper ossification (35).
A detailed analysis of the lumbar vertebrae showed bilaterally fused vertebral bodies and arches in CypA KO animals, while the WT animals exhibited a normal cartilaginous joint connection between the vertebral bodies and arches. The vertebral body and arches must fuse, and this fusion normally occurs in mammals only after birth (37). This is presumably due to the connected primary ossification centers in the centrum and in the vertebral arches bilaterally, as shown in the double staining. The fusion between the vertebral bodies and arches was a distinct finding specifically at the L3 to L5 vertebral levels, as L6 was absent in the CypA KO mice (Fig. 5H and 6F). Another suspected cause is the disruption of early apoptosis of chondrocytes that failed to separate the bones, thereby allowing ossification and fusion to occur. Nevertheless, these observations suggest that fusion of the vertebral bodies and arches is due to abnormal endochondral ossification. Early fusion could lead to restriction of the spinal canal and could also impede the normal development of the spinal nerves.
In the tibias of WT animals, histological analyses of the H&E- and alcian blue-stained growth plate sections revealed a wider growth plate and a longer chondrocyte column in the proximal tibia than in those of Ppia−/− mice (Fig. 8B and D). Immunochemical staining revealed that Col2 and Col10 collagen were more highly expressed in WT mice but not in the CypA-deficient mice (Fig. 8E and F), further suggesting Ppia−/− mice had delayed chondrocyte differentiation and decreased growth plate activity during chondrogenesis. The reduced or delayed hypertrophic chondrocytes in the tibias of Ppia−/− mice suggested that the reduction in limb length was coupled with the inhibition of chondrocyte terminal maturation. Double staining of the skulls of CypA-deficient mice also revealed delayed ossification of the frontal, parietal, and nasal bones. These results suggested that CypA deficiency could influence intramembranous ossification, in addition to endochondral ossification, as described here. The potential effect of CypA deficiency in intramembranous ossification is under investigation, and we expect that CypA may also mediate different pathways in the regulation of skeletogenesis.
Our molecular findings provide a basis for explaining the important role of CypA in regulation of chondrogenic differentiation and endochondral ossification. The novel in vitro data presented here advance the current understanding of how CypA influences cellular signals via the NF-κB–Sox9 pathway in the regulation of chondrogenesis. The molecular mechanism identified here is also important for mesenchymal progenitor cells that respond to cellular signals in vivo during embryonic development. With these novel molecular findings, our future goal is the application and translation of this knowledge to the fields of developmental biology and tissue repair for cartilage-related diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sotirios Tetradis (UCLA School of Dentistry) and Aaron James (David Geffen School of Medicine at UCLA) for critical review of the manuscript. We are grateful to Juyoung Park for excellent technical assistance and to Novartis Pharmaceuticals (Basel, Switzerland) and Karen Lyones for kindly providing SFA and Sox9 plasmids, respectively. Eric Chen and Albert Ha are gratefully acknowledged for critically reading the manuscript.
This work was supported by a CIRM Early Translational Research Award (TR2-0182) and by NIH-NIAMS (R01 AR061399).
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01414-14.
REFERENCES
- 1.Olsen BR, Reginato AM, Wang W. 2000. Bone development. Annu Rev Cell Dev Biol 16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre V, Smits P. 2005. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today 75:200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- 3.Mundlos S, Olsen BR. 1997. Heritable diseases of the skeleton. Part I. Molecular insights into skeletal development-transcription factors and signaling pathways. FASEB J 11:125–132. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol 17:2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Crombrugghe B, Lefebvre V, Nakashima K. 2001. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol 13:721–727. doi: 10.1016/S0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- 6.Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. 1995. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet 9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre V, Li P, de Crombrugghe B. 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J 17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T. 2004. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev 18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 10.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, Ochi T, Endo N, Kitamura Y, Kishimoto T, Komori T. 1999. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn 214:279–290. doi:. [DOI] [PubMed] [Google Scholar]
- 11.Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. 2003. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol 162:833–842. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dijk FS, Nesbitt IM, Zwikstra EH, Nikkels PG, Piersma SR, Fratantoni SA, Jimenez CR, Huizer M, Morsman AC, Cobben JM, van Roij MH, Elting MW, Verbeke JI, Wijnaendts LC, Shaw NJ, Hogler W, McKeown C, Sistermans EA, Dalton A, Meijers-Heijboer H, Pals G. 2009. PPIB mutations cause severe osteogenesis imperfecta. Am J Hum Genet 85:521–527. doi: 10.1016/j.ajhg.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masago Y, Hosoya A, Kawasaki K, Kawano S, Nasu A, Toguchida J, Fujita K, Nakamura H, Kondoh G, Nagata K. 2012. The molecular chaperone Hsp47 is essential for cartilage and endochondral bone formation. J Cell Sci 125:1118–1128. doi: 10.1242/jcs.089748. [DOI] [PubMed] [Google Scholar]
- 14.Fischer G, Aumuller T. 2003. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev Physiol Biochem Pharmacol 148:105–150. doi: 10.1007/s10254-003-0011-3. [DOI] [PubMed] [Google Scholar]
- 15.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 16.Bram RJ, Hung DT, Martin PK, Schreiber SL, Crabtree GR. 1993. Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporin A and FK506: roles of calcineurin binding and cellular location. Mol Cell Biol 13:4760–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu KP, Finn G, Lee TH, Nicholson LK. 2007. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol 3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 18.Colgan J, Asmal M, Neagu M, Yu B, Schneidkraut J, Lee Y, Sokolskaja E, Andreotti A, Luban J. 2004. Cyclophilin A regulates TCR signal strength in CD4+ T cells via a proline-directed conformational switch in Itk. Immunity 21:189–201. doi: 10.1016/j.immuni.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Lu YC, Yokoyama K, Rossi J, Chiu R. 2004. Cyclophilin A is required for retinoic acid-induced neuronal differentiation in p19 cells. J Biol Chem 279:24414–24419. doi: 10.1074/jbc.M311406200. [DOI] [PubMed] [Google Scholar]
- 20.Bauer K, Kretzschmar AK, Cvijic H, Blumert C, Loffler D, Brocke-Heidrich K, Schiene-Fischer C, Fischer G, Sinz A, Clevenger CV, Horn F. 2009. Cyclophilins contribute to Stat3 signaling and survival of multiple myeloma cells. Oncogene 28:2784–2795. doi: 10.1038/onc.2009.142. [DOI] [PubMed] [Google Scholar]
- 21.Obchoei S, Wongkhan S, Wongkham C, Li M, Yao Q, Chen C. 2009. Cyclophilin A: potential functions and therapeutic target for human cancer. Med Sci Monit 15:RA221–RA232. [PubMed] [Google Scholar]
- 22.Sun S, Guo M, Zhang JB, Ha A, Yokoyama KK, Chiu RH. 2014. Cyclophilin A (CypA) interacts with NF-kappaB subunit, p65/RelA, and contributes to NF-kappaB activation signaling. PLoS One 9:e96211. doi: 10.1371/journal.pone.0096211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James CG, Appleton CT, Ulici V, Underhill TM, Beier F. 2005. Microarray analyses of gene expression during chondrocyte differentiation identifies novel regulators of hypertrophy. Mol Biol Cell 16:5316–5333. doi: 10.1091/mbc.E05-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR. 1987. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2:595–610. [DOI] [PubMed] [Google Scholar]
- 25.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 26.Park H, Choi B, Hu J, Lee M. 2013. Injectable chitosan hyaluronic acid hydrogels for cartilage tissue engineering. Acta Biomater 9:4779–4786. doi: 10.1016/j.actbio.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Hu J, Hou Y, Park H, Choi B, Hou S, Chung A, Lee M. 2012. Visible light crosslinkable chitosan hydrogels for tissue engineering. Acta Biomater 8:1730–1738. doi: 10.1016/j.actbio.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 28.Sun S, Wang Q, Giang A, Cheng C, Soo C, Wang CY, Liau LM, Chiu R. 2011. Knockdown of CypA inhibits interleukin-8 (IL-8) and IL-8-mediated proliferation and tumor growth of glioblastoma cells through down-regulated NF-kappaB. J Neurooncol 101:1–14. doi: 10.1007/s11060-010-0220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caron MM, Emans PJ, Surtel DA, Cremers A, Voncken JW, Welting TJ, van Rhijn LW. 2012. Activation of NF-kappaB/p65 facilitates early chondrogenic differentiation during endochondral ossification. PLoS One 7:e33467. doi: 10.1371/journal.pone.0033467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S, Flint JK, Rezvani G, De Luca F. 2007. Nuclear factor-kappaB p65 facilitates longitudinal bone growth by inducing growth plate chondrocyte proliferation and differentiation and by preventing apoptosis. J Biol Chem 282:33698–33706. doi: 10.1074/jbc.M702991200. [DOI] [PubMed] [Google Scholar]
- 31.Attar RM, Macdonald-Bravo H, Raventos-Suarez C, Durham SK, Bravo R. 1998. Expression of constitutively active IkappaB beta in T cells of transgenic mice: persistent NF-kappaB activity is required for T-cell immune responses. Mol Cell Biol 18:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hettmann T, DiDonato J, Karin M, Leiden JM. 1999. An essential role for nuclear factor kappaB in promoting double positive thymocyte apoptosis. J Exp Med 189:145–158. doi: 10.1084/jem.189.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ushita M, Saito T, Ikeda T, Yano F, Higashikawa A, Ogata N, Chung U, Nakamura K, Kawaguchi H. 2009. Transcriptional induction of SOX9 by NF-kappaB family member RelA in chondrogenic cells. Osteoarthritis Cartilage 17:1065–1075. doi: 10.1016/j.joca.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gollner H, Dani C, Phillips B, Philipsen S, Suske G. 2001. Impaired ossification in mice lacking the transcription factor Sp3. Mech Dev 106:77–83. doi: 10.1016/S0925-4773(01)00420-8. [DOI] [PubMed] [Google Scholar]
- 36.Takarada T, Hinoi E, Nakazato R, Ochi H, Xu C, Tsuchikane A, Takeda S, Karsenty G, Abe T, Kiyonari H, Yoneda Y. 2013. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J Bone Mineral Res 28:2064–2069. doi: 10.1002/jbmr.1945. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan KM, Spivak JM, Bendo JA. 2005. Embryology of the spine and associated congenital abnormalities. Spine J 5:564–576. doi: 10.1016/j.spinee.2004.10.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.