Highlight
The characterization of Medicago truncatula R108 mutants affected in strigolactone synthesis or perception supports a role for strigolactones in shoot elongation and in the formation of leaf margin serrations.
Key words: Medicago truncatula, elongation, leaf, mutant, phytohormone, shoot, strigolactone.
Abstract
Strigolactones were recently identified as a new class of plant hormones involved in the control of shoot branching. The characterization of strigolactone mutants in several species has progressively revealed their contribution to several other aspects of development in roots and shoots. In this article, we characterize strigolactone-deficient and strigolactone-insensitive mutants of the model legume Medicago truncatula for aerial developmental traits. The most striking mutant phenotype observed was compact shoot architecture. In contrast with what was reported in other species, this could not be attributed to enhanced shoot branching, but was instead due to reduced shoot elongation. Another notable feature was the modified leaf shape in strigolactone mutants: serrations at the leaf margin were smaller in the mutants than in wild-type plants. This phenotype could be rescued in a dose-dependent manner by exogenous strigolactone treatments of strigolactone-deficient mutants, but not of strigolactone-insensitive mutants. Treatment with the auxin transport inhibitor N-1-naphthylphtalamic acid resulted in smooth leaf margins, opposite to the effect of strigolactone treatment. The contribution of strigolactones to the formation of leaf serrations in M. truncatula R108 line represents a novel function of these hormones, which has not been revealed by the analysis of strigolactone mutants in other species.
Introduction
Strigolactones (SL) are carotenoid-derived metabolites long known for their ability to trigger the germination of parasitic plant seeds (Cook et al., 1966). They also play a role in plant–microbe symbiotic interactions, as stimulants of the growth and metabolism of arbuscular mycorrhizal fungi (Akiyama et al., 2005; Besserer et al., 2006, 2008). In addition to these effects in the rhizosphere, we and others have proposed that SL or related compounds also act in planta as phytohormones, contributing to the control of shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008). This discovery stemmed both from the elucidation of the biosynthetic origin of SL (Matusova et al., 2005) and from long-standing studies of the control of shoot architecture in several plant species (reviewed in Xie et al., 2010; Ruyter-Spira et al., 2013). Central to these pioneer studies are a series of mutants in Arabidopsis thaliana (max—more axillary growth), pea (rms—ramosus), rice (d—dwarf) and Petunia (dad—decreased apical dominance), isolated in forward genetic screens on the basis of their enhanced shoot branching phenotypes. The physiological characterization of these mutants including, in particular, a series of graft experiments, revealed that they lack or are insensitive to an unknown mobile signal able to suppress shoot branching. Cloning of the mutated genes in Arabidopsis showed that two carotenoid cleavage dioxygenase (CCD) isoforms, CCD7 and CCD8, are necessary for the synthesis of this signal (Sorefan et al., 2003; Booker et al., 2004). As carotenoid cleavage was proposed to be necessary for SL production (Matusova et al., 2005), the hypothesis that the unknown signal could be SL was investigated. Indeed, pea and rice shoot branching mutants are either SL-deficient or SL-insensitive, and SL exhibits all the expected properties of the long sought-after signal (Gomez-Roldan et al., 2008; Umehara et al., 2008). The SL biosynthetic pathway has since been further elucidated with the discovery of additional enzymes and intermediates (Alder et al., 2012). Studies of SL perception and signalling have identified two key actors, an α/β hydrolase probably acting as the SL receptor (Hamiaux et al., 2012) and an F-box protein essential for signal transduction and integration (Stirnberg et al., 2007).
Early analyses of SL-deficient and SL-insensitive mutants have mainly focused on shoot branching or tillering enhancement, although other phenotypes including dwarfism, altered root growth, reduced shoot diameter, delayed leaf senescence, decreased flower size, and modifications of leaf shape parameters have also been documented. In the last few years, more extensive characterization of the SL mutants in Arabidopsis, pea, and rice has established a role for SL in root architecture and root hair development (Kohlen et al., 2011; Kapulnik et al., 2011), mesocotyl elongation (Hu et al., 2010), vascular secondary growth (Agusti et al., 2011), adventitious rooting (Rasmussen et al., 2012), seedling response to light (Tsuchiya et al., 2010), and seed germination (Toh et al., 2012). Furthermore, the characterization of SL-deficient mutants or transgenic lines in other plant species has revealed the contribution of SL to fruit development (Kohlen et al., 2012), tuber differentiation (Roumeliotis et al., 2012), and nodule formation (Foo and Davies, 2011). It therefore seems that SL, like other plant hormones, exert a wide range of effects in various physiological contexts. This is fully consistent with observations that SL signalling crosstalks with several other plant hormonal pathways (Vanstraelen and Benkova, 2012). It is likely that other effects of SL remain to be discovered, and the study of additional plant species should contribute to a more comprehensive assessment of SL functions.
The mechanisms by which SL affect these various aspects of plant development have not been fully elucidated. For example, two models have been put forward to explain the respective roles of auxin and SL in the control of shoot branching. One model proposes that SL dampens polar auxin transport in the main stem in a systemic manner (Bennett et al., 2006). This would prevent auxin export from the buds that is necessary for bud growth (Crawford et al., 2010). The other model favours a local action of SL in buds as second messengers of auxin (Dun et al., 2013). These divergent views are not mutually exclusive and may reflect, to some extent, differences in the plant species and physiological contexts in these independent studies (Dun et al., 2006). Among other known functions of SL, the modulation of primary root growth has been associated with modifications of the auxin gradient at the root tip (Ruyter-Spira et al., 2011), whereas stimulation of vascular secondary growth seems to occur independently or downstream of auxin transport (Agusti et al., 2011) and enhanced root hair elongation is at least partly independent from auxin (Kapulnik et al., 2011). The emerging picture is that a universal mode of SL action is not to be discovered, and that SL can display opposite properties depending on the target cells and physiological context (de Saint-Germain et al., 2013).
Among legumes, Medicago truncatula Gaertn. has emerged as an attractive model species for the study of aerial development. Many mutants with shoot or leaf phenotypes have been identified through forward genetics screens, and extensive reverse genetic resources have been generated. The aim of the present study was to investigate the role of SL in the regulation of aerial development in M. truncatula. We first identified transposon insertional mutants affected in SL biosynthesis or response genes. Both types of mutants displayed a bushy phenotype, which could be attributed to reduced internode elongation rather than to enhanced shoot branching. In addition, mutant phenotypic characterization in combination with SL treatments revealed a novel function for SL in leaf margin development.
Materials and methods
Identification of M. truncatula SL-related genes
A BLAST search was performed with the amino acid sequences of pea CCD7 and CCD8, and Arabidopsis D14, against the NCBI database. For phylogenetic analysis, sequences were aligned using the MUSCLE program (Edgar, 2004). Maximum-likelihood trees were built with MEGA6 (Tamura et al., 2013), using Jones-Taylor-Thornton (JTT) as the amino acid substitution model and the nearest-neighbour-interchange (NNI) heuristic method. The partial deletion (95%) mode was used to treat gaps and missing data. Accession numbers of all genes used in this analysis are listed in Supplementary Table S1.
Plant material and growth conditions
All mutants were obtained from the Noble Foundation Tnt1 insertion library (Tadege et al., 2008) by PCR screening (Cheng et al., 2014). Plants homozygous for the presence of a Tnt1 insertion in the target gene were selected by PCR. The position of Tnt1 insertions was confirmed by PCR and sequencing. Mutant lines ccd7-1 and ccd8-1 were backcrossed twice to the R108 wild type. In the F2 progeny of the second backcross, homozygous lines carrying a Tnt1 insertion in the CCD gene were selected. Wild-type siblings identified in this progeny were used for comparison.
Plants were grown on an inert substrate (OilDri, Brenntag, France) and fertilized with a modified Long Ashton nutrient solution (Balzergue et al., 2011). Plants were kept in a growth chamber under a 16h photoperiod at 24 °C. For the analysis of shoot architecture, 40-day-old plants were photographed and shoot length was measured with a ruler.
Treatments
The synthetic strigolactone analogue GR24 was obtained from Chiralix (The Netherlands) and NPA was purchased from Sigma. Stock solutions were prepared in acetone for GR24, and in DMSO for NPA, and diluted 1000-fold in water supplemented with 0.05% Tween 20 to reach the indicated concentrations. Control treatments were performed with the solvent(s) alone, diluted 1000-fold in water supplemented with 0.05% Tween 20.
Plants were treated five times a week throughout the culture by application of 100 µl of solution directly to the primary shoot apex with a pipette.
Leaf shape analysis
After four to five weeks of growth, several fully expanded leaves were collected. The three leaflets of each leaf were carefully separated from the rachis and petiole, and leaflets were scanned at 600 dpi resolution using an Epson scanner. Images were analysed using ImageJ software to obtain solidity values.
Statistical analyses
All analyses were carried out using Statgraphics Centurion software (SigmaPlus). Two-sample comparisons were performed using the unequal variance t-test. Multiple comparisons were performed by one-way ANOVA followed by Fisher’s LSD test, or using non-parametric tests when normality or homoscedasticity criteria were not satisfied. In that case datasets were analysed using the Kruskal-Wallis test followed by pairwise comparisons with the Mann-Whitney test. A Bonferroni correction was applied to take into account multiple testing, so that differences are reported at a 0.05 significance level.
Results and discussion
Identification of Tnt1-insertion SL mutants
Previous studies have demonstrated that SL biosynthesis involves the cleavage of a carotenoid substrate (Matusova et al., 2005; Alder et al., 2012). Enzymes able to carry out such a cleavage are classified into two main groups (Vallabhaneni et al., 2010). While 9-cis-epoxycarotenoid dioxygenases (NCEDs) are specialized isoforms associated with the biosynthesis of abscisic acid, the carotenoid cleavage dioxygenases (CCDs) collectively use a wider range of substrates and contribute to varied physiological functions. CCDs can be further divided into four clades represented by the Arabidopsis isoforms CCD1, CCD4, CCD7, and CCD8 (Auldridge et al., 2006). The involvement of CCD7 and CCD8 in SL synthesis is now firmly established in several species including pea (Pisum sativum), a close relative of M. truncatula. CCD7 and CCD8 act successively in the SL biosynthetic pathway (Alder et al., 2012), so that a loss-of-function mutant of either of the corresponding genes is SL-deficient as demonstrated by biochemical analyses (e.g. Gomez-Roldan et al., 2008 for pea; Umehara et al., 2008 for rice).
We searched for M. truncatula orthologues of pea CCD7 and CCD8 genes. For CCD7, one strong homologous gene and four less closely related genes were identified. A search with the pea CCD8 sequence yielded the same five genes, which therefore probably comprised the whole set of M. truncatula CCD genes. Phylogenetic analysis (Supplementary Fig. S1A) revealed that one of these genes (hereafter named MtCCD7) fell into the CCD7 clade, and another into the CCD8 clade (MtCCD8). The remaining three genes were more closely related to CCD1 or CCD4. At the amino acid level, MtCCD7 and MtCCD8 respectively share 88% and 91% identity (93% and 95% similarity) with their pea orthologues.
Mutants of MtCCD7 and MtCCD8 were identified by PCR-based reverse screening from a collection of M. truncatula Tnt1 retrotransposon insertion lines (Tadege et al., 2008; Cheng et al., 2014). Tnt1 insertion lines were generated using the R108-1(c3) line, which is highly embryogenic and more amenable to genetic transformation than the widely used Jemalong A17 ecotype (Trinh et al., 1998). Two mutant alleles (ccd7-1 and ccd7-2) of MtCCD7, and one for MtCCD8 (ccd8-1) were isolated. They all harbour a Tnt1 insertion in the coding sequence (Supplementary Fig. S2) leading to a premature STOP codon, and can therefore be considered KO mutants.
The α/β hydrolase D14/DAD2 recently emerged as a strong candidate for the SL receptor (Hamiaux et al., 2012; Nakamura et al., 2013). The binding of SL to D14 triggers downstream responses and d14 mutants in several species are insensitive to SL (Arite et al., 2009; Hamiaux et al., 2012; Waters et al., 2012). D14 belongs to a multigene family in which close relatives called D14-like can be identified (Delaux et al., 2013). Only D14 seems to be essential for SL perception as indicated by binding capacities and phenotypes of mutants of D14-like genes (Waters et al., 2012; Kagiyama et al., 2013).
Five genes homologous to Arabidopsis D14 could be found in the M. truncatula genome. Only one of these, hereafter called MtD14, falls into the D14 clade (Supplementary Fig. S1B). The remaining four can be classified as D14-like1 or D14-like2. A mutant harbouring a Tnt1 insertion in the second exon of MtD14 (d14-1, Supplementary Fig. S2) was identified by PCR-based reverse screening (Cheng et al., 2014).
Aerial architecture of SL mutants
Shoot development in M. truncatula follows a complex but ordered pattern (Bucciarelli et al., 2006; Moreau et al., 2006). In brief, the main shoot axis produces growth units called metamers comprising an internode, a leaf, and an axillary bud. Under our growth conditions, internodes on the main axis remain very short in the early stages of growth. Axillary buds from metamers 1 to 4 (m1–m4) grow out to form axillary shoots, that elongate substantially and adopt a prostrate growth habit. The main axis then elongates vertically and produces additional metamers, from which new axillary shoots can later emerge. Overall, one axillary bud is present at each leaf axil and eventually grows out into a shoot of higher order.
After several weeks of growth, both ccd7-1 and ccd8-1 mutants appeared smaller and more compact than the wild type (Fig. 1A). While M. truncatula wild-type plants adopted a trailing growth habit as axillary shoots became longer and heavier, mutant plants did not. SL-insensitive M. truncatula d14-1 mutants displayed an aerial phenotype very similar to that of ccd7-1 and ccd8-1 mutants (Fig. 1B), establishing a firm link between this phenotype and the SL pathway.
Fig. 1.
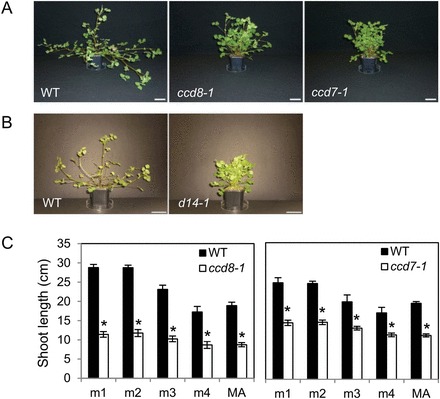
Shoot architecture of strigolactone mutants. (A, B). Photographs of 40-day-old plants. Bar=5cm. Mutants ccd7-1 and ccd8-1 (A) and d14-1 (B) were examined in separate experiments. (C) Shoot lengths of the first axes (emerging from metamers m1 to m4) and of the main axis (MA) measured after 40 days of growth. Mutants were compared with their respective wild-type siblings. Values correspond to the mean±SEM of 5–6 plants per genotype. Asterisks indicate statistically significant differences between the mutant and wild type for each axis (unequal variance t-test, P<0.05). (This figure is available in colour at JXB online.)
The bushy appearance of SL-deficient M. truncatula mutants could not be attributed to enhanced shoot branching as indicated by the observation that in the wild type as well as the mutants, most axillary buds eventually developed into shoots. For example, 40-day-old wild-type and ccd8-1 mutant plants, respectively, harboured an average of 8.38±0.6 and 8.38±0.26 first-order shoots, and 4±0.76 and 3.86±1.6 second-order shoots. Thus, the mutant and wild type could not be distinguished on the basis of bud outgrowth patterns. This situation is in contrast with other species studied for the role of SL in development: wild-type plants usually exhibit limited shoot branching and the number of outgrowing lateral buds increases in SL mutants. An exception is the legume Lotus japonicus, which exhibits profuse basal shoot branching at the cotyledonary node. Still, in this species, many buds in the aerial stem remain dormant, which allows for enhanced aerial branching in LjCCD7 RNAi knock-down lines, in addition to an increased number of basal shoots (Liu et al., 2013). To reveal a putative function for SL in the control of shoot branching in M. truncatula, it would be necessary to define growth conditions that limit the development of axillary shoots in the wild type. A starting point could be the observation that growing M. truncatula in the absence of a nitrogen source results in a complete inhibition of axillary shoot development, accompanied by a marked reduction of aerial organ growth (Bucciarelli et al., 2006). One might be able to define an intermediate nitrogen supply level that does not impair plant growth too severely, while limiting axillary shoot development in the wild type, to investigate whether SL-deficient mutants exhibit enhanced shoot branching in such conditions.
The compact phenotype of the SL-deficient mutants could be attributed to reduced shoot elongation, as documented by the shoot length of the first four metamers and the main axis (Fig. 1C). Similar results were obtained with the ccd7-2 mutant allele (Supplementary Fig. S3A). This phenotype persisted throughout the plants’ life cycle, and therefore did not reflect a mere growth delay. Dwarfism has been reported in SL-deficient mutants of several species, and has been suggested to be an indirect consequence of enhanced branching, through reduced resource allocation to a larger number of shoots (Kohlen et al., 2012) or tillers (Zou et al., 2006). Our observations indicate that it is not the case in M. truncatula, as reduced shoot length in SL-deficient mutants is observed in the absence of any effect on shoot branching. This conclusion is consistent with the recent report of de Saint Germain et al. (2013), where it is proposed that in pea plants the effects of SL on shoot branching and internode elongation are independent. Exogenous application of the synthetic SL analogue GR24 (Zwanenburg et al., 2009) at the primary shoot apex partly rescued the reduced shoot elongation observed in SL-deficient mutants of M. truncatula (Supplementary Fig. S3). This contrasts with the observations of de Saint Germain et al. (2013) in pea, where application of GR24 to the shoot tip did not stimulate shoot elongation. One hypothesis in that report was that SLs are unstable and therefore a single application might not be sufficient to obtain visible effects. In our case, GR24 was applied repeatedly over several weeks, which could account for the observed effect on shoot elongation. In any case, the positive effect of GR24 on shoot elongation of M. truncatula further supports our conclusion that SL deficiency is the cause of dwarfism in the Mtccd7 and Mtccd8 mutants.
Leaf shape alterations
When wild-type and SL mutant plants were grown side by side for the analysis of shoot architecture, we noticed differences in leaf shape. Apart from the first unifoliate leaf, M. truncatula bears compound leaves with three leaflets. Leaflets of R108 plants display an overall circular shape with serrations on the leaf margin. In both SL-deficient mutants serrations seemed shallower than in the wild type, as illustrated in Fig. 2A, B. To investigate whether this novel phenotype was due to SL deficiency, mutant plants were treated with the synthetic SL analogue GR24 at different concentrations applied directly at the primary shoot apex. These treatments clearly modified the leaf shape: GR24-treated leaflets displayed fewer, deeper, and wider serrations than mock-treated leaflets (Fig. 2A, B).
Fig. 2.
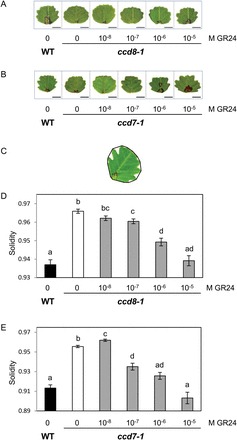
Exogenous SL can rescue the leaflet serration phenotype of SL-deficient mutants. GR24 was applied to the shoot tip at the indicated concentrations. (A, B) Scanned images of one representative leaflet in each condition. Representative leaflets with a solidity value equal to the average solidity of all leaflets in this condition were selected. Bar=5mm. (C) Illustration of solidity as a shape descriptor. The convex hull (black line) delimits the convex area of each leaflet. Solidity is calculated as the ratio of leaflet area/convex area. (D, E) Solidity values for leaflets of ccd8-1 and ccd7-1 mutants and their respective wild-type siblings, treated or not with GR24. Values correspond to the mean±SEM (n=49–85 leaflets for each condition). Different letters indicate statistically significant differences according to Mann-Whitney’s test (P<0.05 after Bonferroni adjustment). (This figure is available in colour at JXB online.)
A morphometric analysis was undertaken to quantify these effects. We chose to focus on serration size as serration number is difficult to determine accurately, especially for the smaller ones. Among commonly used shape descriptors, solidity (Neal and Russ, 2012) was the most appropriate to quantify differences in serration size. Solidity corresponds to the ratio of leaflet area over convex area, i.e. the proportion of pixels in the convex area that are also in the leaflet (Fig. 2C): the smaller the value, the larger the indentations. Solidity is affected only by irregularities at the surface perimeter, and not by overall form. Solidity was significantly higher in both SL-deficient mutants as compared with the wild type (Fig. 2D, E), confirming our initial visual observations that serrations were shallower in the SL-deficient mutants. Furthermore, solidity decreased as the applied concentration of GR24 increased. Concentrations of 10–7 M and higher significantly affected solidity, and application of 10–6 –10–5 M GR24 could restore this shape descriptor back to wild-type levels (Fig. 2D, E). The analysis of leaf shape in the second ccd7 mutant allele gave similar results (Supplementary Fig. S4). Finally, SL-insensitive d14-1 mutants displayed the same leaf serration phenotype as the SL-deficient ccd7 and ccd8 mutants, but the shape of their leaflets was not modified by application of 10–5 M GR24 (Fig. 3). Together, these results demonstrate a close link between the canonical SL pathway and leaflet margin morphology in M. truncatula R108. It is noteworthy that the compound leaf structure does not seem to be disturbed in SL mutants. This is consistent with the hypothesis that partly distinct mechanisms account for the formation of compound leaves and leaf margin serrations in M. truncatula (Zhou et al., 2011), and suggests that SL affects a process specific to leaf margins.
Fig. 3.
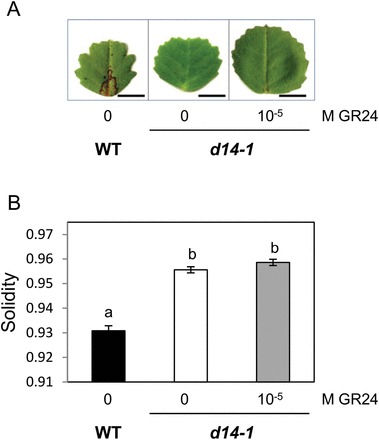
Exogenous SL does not affect the leaflet shape phenotype of SL-insensitive mutants. Mutant plants were treated with 10–5 M GR24 applied to the shoot tip. (A) Scanned images of one representative leaflet under each condition. Bar=5mm. (B) Leaflet solidity values for control WT plants and d14-1 mutants treated or not with GR24. Values correspond to the mean±SEM (n=53–127 leaflets for each condition). Different letters indicate statistically significant differences according to Mann-Whitney’s test (P<0.05 after Bonferroni adjustment). (This figure is available in colour at JXB online.)
Genetic studies in Arabidopsis have shed light on the mechanisms that drive the formation of leaf serrations. In the model proposed by Bilsborough et al. (2011), the growth-repressing transcription factor CUC2 allows the formation of convergence points of polar auxin transport. This process, together with auxin’s ability to stimulate its own polar transport, results in the accumulation of auxin at discrete points along the leaf margin. As auxin inhibits the expression of CUC2, a pattern of interspersed auxin and CUC2 maxima is created along the leaf margin, leading to differential growth and to the formation of serrations. In agreement with this model, impairment of auxin transport in Atpin1 mutants or following treatment with the auxin transport inhibitor N-1-naphthylphtalamic acid (NPA) results in the loss of leaf serrations (Hay et al., 2006). The smooth leaf margin phenotype of M. truncatula R108 mutants of MtPIN10 suggests that auxin transport is also required for the formation of leaf serrations in this species (Zhou et al., 2011; Peng and Chen, 2011).
To investigate the relationship between SL, auxin transport, and serrations in M. truncatula, we compared the effects of SL and NPA on wild-type leaflets (Fig. 4). Consistent with the report of Zhou et al. (2011), NPA treatment resulted in smoother leaf margins with few and shallow serrations, or even no serrations at all except the terminal one (Fig. 4A). Similar results were obtained with ccd7-1 mutants (Fig. 4B). Morphometric analysis confirmed the opposite effects of GR24 and NPA on the formation of serrations (Fig. 4C, D). These results do not lend support to an impact of SL on leaflet serrations through reduced auxin transport, as has been proposed for their effect on shoot branching (Crawford et al., 2010). They do not rule out this mechanism either, as SL and NPA probably affect auxin transport in different ways, both qualitatively and quantitatively. Indeed, SL and NPA seem to target distinct auxin transporters (Shinohara et al., 2013; Kim et al., 2010), and they reduce auxin transport to different extents (Crawford et al., 2010). It is also possible that their spatial or temporal range of action is different. A more detailed examination of the consequences of GR24 treatment on PIN turnover in leaf cells and on auxin distribution along the leaflet margin would be needed to determine whether SL control the formation of serrations by altering auxin transport. Alternatively, similar to some of their other functions in plant development, SL may act on leaf serrations either downstream of auxin or via a separate pathway.
Fig. 4.
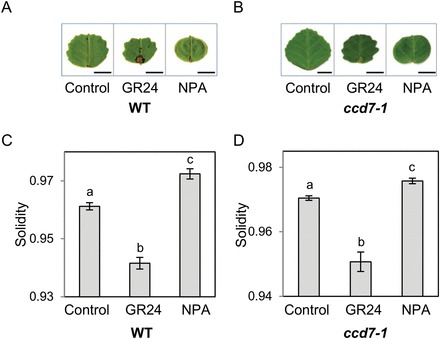
GR24 and NPA effects on leaflet serrations. Wild-type (A, C) and ccd7-1 mutant plants (B, D) were treated with 5×10–6 M GR24 or NPA, or the solvents alone (control). (A, B) Scanned images of one representative leaflet in each condition. Bar=5mm. (C, D) Solidity values. Values correspond to the mean±SEM (n=37–72 leaflets for each condition). Different letters indicate statistically significant differences according to Mann-Whitney’s test (P<0.05 after Bonferroni adjustment). (This figure is available in colour at JXB online.)
Other M. truncatula mutants with a leaf serration phenotype have been described, such as lol1/ago7 (Zhou et al., 2013) and mtphan (Ge et al., 2014). In these cases, however, the serration shape was distinct from the SL mutants, and was accompanied by additional phenotypes that we did not observe. This suggests that different mechanisms account for the alteration of leaf serrations in these different mutant backgrounds. Moderate alterations of leaf shape have been reported previously for SL-deficient mutants in other species, notably a lower length/width ratio and other modifications related to lamina overall shape and position on the petiole (Beveridge et al., 1997; Stirnberg et al., 2002; Challis et al., 2013). Nonetheless, the present article is to our knowledge the first report of a role for SL in the formation of leaf margin serrations. As leaf shape has been examined in great detail in Arabidopsis SL mutants (Hepworth, 2012), a serration phenotype is unlikely to have gone unnoticed in this species. The so far unreported effect of SL on leaf serrations may reflect particularities in leaf morphogenesis in M. truncatula. The inverted repeat-lacking clade (IRLC) of Fabaceae comprising Medicago spp is already known to use a different mechanism to control compound leaf development compared with the rest of the vascular plants (Champagne et al., 2007). Other authors have also underlined the strong context-dependency of the mechanisms and pathways governing leaf shape (Zhou et al., 2013). When primary shoot apices of the commonly used M. truncatula Jemalong A17 ecotype were treated with SL, no effect on the formation of leaf serrations could be observed (Supplementary Fig. S5), although Jemalong is able to respond to SL application to the root system by a decreased formation of lateral roots (De Cuyper et al., 2014). This discrepancy indicates that the leaf SL response observed in R108 is not a general property in the Medicago genus. The R108 ecotype is morphologically different from other M. truncatula accessions. Its increased stem elongation (Schnurr et al., 2007) and deeper leaf serrations may reflect an enhanced sensitivity to SL, at least in aerial organs. It will be interesting to determine whether these characteristics were already present in the original R108 ecotype, or appeared during the in vitro selection of the highly embryogenic R108-1(c3) line (Hoffmann et al., 1997). In the latter case, a putative contribution of enhanced strigolactone sensitivity to embryogenic potential would be worth investigating.
In conclusion, we show here that both SL-deficient and SL-insensitive mutants of M. truncatula plants displayed a compact architecture that was attributed to reduced shoot elongation rather than enhanced shoot branching. In addition, the SL-deficient and SL-insensitive mutants displayed a modified leaf shape, with reduced serrations at the leaf margin. Exogenously applied SL could rescue the serration phenotype of the SL-deficient mutants in a dose-dependent manner, but had no effect on SL-insensitive mutants. These observations demonstrate the importance of SL in the formation of leaf serrations in M. truncatula R108. This novel function of SLs represents another example of the versatility of SL action depending on the target tissue and species context (de Saint-Germain et al., 2013), and highlights the usefulness of investigating SL functions in a wide range of plant species. It remains to be investigated whether the effect of SL on leaf margin serrations is widespread and could have contributed to the evolutionary diversification of leaf shape.
Supplementary data
Figure S1. Phylogeny of strigolactone biosynthesis and response genes.
Figure S2. Position of Tnt1 insertions in the different mutant alleles.
Figure S3. Shoot elongation phenotype of the ccd7-2 mutant allele; partial rescue of the shoot elongation phenotype by exogenous SL application.
Figure S4. Leaflet serration phenotype of ccd7-2 mutants.
Figure S5. Effect of GR24 on leaflet serrations in M. truncatula ecotype A17.
Table S1. Sequence accession numbers.
Acknowledgements
This work was supported by the French Agence Nationale de la Recherche (grant number ANR-09-BLAN-0241-0 ‘MycSignalling’), and by the French Laboratory of Excellence project ‘TULIP’ (grant numbers ANR-10-LABX-41, ANR-11-IDEX-0002-02). The Medicago truncatula plants used in this research project, which are jointly owned by the Centre National de la Recherche Scientifique, were obtained from the Samuel Roberts Noble Foundation, Inc. and were created through research funded, in part, by a grant from the National Science Foundation (grant number NSF-0703285).
Glossary
Abbreviations:
- SL
strigolactone
- CCD
carotenoid cleavage dioxygenase
- NPA
N-1-naphthylphtalamic acid.
References
- Agusti J, Herold S, Schwarz M, et al. 2011. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences, USA 108, 20242–20247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. 2012. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology 50, 1416–1424. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, McCarty DR, Klee HJ. 2006. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Current Opinion in Plant Biology 9, 315–321. [DOI] [PubMed] [Google Scholar]
- Balzergue C, Puech-Pagès V, Bécard G, Rochange SF. 2011. The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. Journal of Experimental Botany 62, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. 2006. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Current Biology 16, 553–563. [DOI] [PubMed] [Google Scholar]
- Besserer A, Puech-Pages V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. 2006. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biology 4:e226. 10.1371/journal.pbio.0040226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Bécard G, Jauneau A, Roux C, Séjalon-Delmas N. 2008. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiology 148, 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C. 1997. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiology 115, 1251–1258. [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proceedings of the National Academy of Sciences, USA 108, 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. 2004. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Current Biology 14, 1232–1238. [DOI] [PubMed] [Google Scholar]
- Bucciarelli B, Hanan J, Palmquist D, Vance CP. 2006. A standardized method for analysis of Medicago truncatula phenotypic development. Plant Physiology 142, 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis RJ, Hepworth J, Mouchel C, Waites R, Leyser O. 2013. A role for more axillary growth1 (MAX1) in evolutionary diversity in strigolactone signaling upstream of MAX2. Plant Physiology 161, 1885–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne CE, Goliber TE, Wojciechowski MF, Mei RW, Townsley BT, Wang K, Paz MM, Geeta R, Sinha NR. 2007. Compound leaf development and evolution in the legumes. The Plant Cell 19, 3369–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee HK, Tadege M, Ratet P, Udvardi M, Mysore KS, Wen J. 2014. An efficient reverse genetics platform in the model legume Medicago truncatula . New Phytologist 201, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. 1966. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190. [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137, 2905–2913. [DOI] [PubMed] [Google Scholar]
- De Cuyper C, Fromentin J, Endah Yocgo R, De Keyser A, Guillotin B, Kunert K, Boyer FD, Goormachtig S. 2014. From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula . Journal of Experimental Botany , doi: 10.1093/jxb/eru404. [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Ligerot Y, Dun EA, Pillot JP, Ross JJ, Beveridge C, Rameau C. 2013. Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiology 163, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, Ferguson BJ, Beveridge CA. 2006. Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiology 142, 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. 2013. Dynamics of strigolactone function and shoot branching responses in Pisum sativum . Molecular Plant 6, 128–140. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Davies NW. 2011. Strigolactones promote nodulation in pea. Planta 234, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Ge L, Peng J, Berbel A, Madueño F, Chen R. 2014. Regulation of compound leaf development by PHANTASTICA in Medicago truncatula . Plant Physiology 164, 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–194. [DOI] [PubMed] [Google Scholar]
- Hay A, Barkoulas M, Tsiantis M. 2006. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis . Development 133, 3955–3961. [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. 2012. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology 22, 2032–2036. [DOI] [PubMed] [Google Scholar]
- Hepworth JA. 2012. Comparative analysis of the MAX pathway. PhD thesis, University of York. [Google Scholar]
- Hoffmann B, Trinh TH, Leung J, Adam Kondorosi K, Kondorosi E. 1997. A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Molecular Plant–Microbe Interactions 10, 307–315. [Google Scholar]
- Hu Z, Yan H, Yang J, Yamaguchi S, Maekawa M, Takamure I, Tsutsumi N, Kyozuka J, Nakazono M. 2010. Strigolactones negatively regulate mesocotyl elongation in rice during germination and growth in darkness. Plant and Cell Physiology 51, 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. 2013. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells 18, 147–160. [DOI] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N, et al. 2011. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis . Planta 233, 209–216. [DOI] [PubMed] [Google Scholar]
- Kim JY, Henrichs S, Bailly A, Vincenzetti V, Sovero V, Mancuso S, Pollmann S, Kim D, Geisler M, Nam HG. 2010. Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. Journal of Biological Chemistry 285, 23309–23317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C. 2011. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis . Plant Physiology 155, 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Lammers M, et al. 2012. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytologist 19, 535–547. [DOI] [PubMed] [Google Scholar]
- Liu J, Novero M, Charnikhova T, Ferrandino A, Schubert A, Ruyter-Spira C, Bonfante P, Lovisolo C, Bouwmeester HJ, Cardinale F. 2013. Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus . Journal of Experimental Botany 64, 1967–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusova R, Rani K, Verstappen FW, Franssen MC, Beale MH, Bouwmeester HJ. 2005. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiology 139, 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau D, Salon C, Munier-Jolain N. 2006. Using a standard framework for the phenotypic analysis of Medicago truncatula: an effective method for characterizing the plant material used for functional genomics approaches. Plant, Cell and Environment 29, 1087–1098. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Xue YL, Miyakawa T, et al. 2013. Molecular mechanism of strigolactone perception by DWARF14. Nature Communications 4, 2613. [DOI] [PubMed] [Google Scholar]
- Neal FB, Russ JC. 2012. Measuring shape. Boca Raton: CRC Press. [Google Scholar]
- Peng J, Chen R. 2011. Auxin efflux transporter MtPIN10 regulates compound leaf and flower development in Medicago truncatula . Plant Signaling and Behaviour 6, 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, et al. 2012. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiology 158, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumeliotis E, Kloosterman B, Oortwijn M, Kohlen W, Bouwmeester HJ, Visser RG, Bachem CW. 2012. The effects of auxin and strigolactones on tuber initiation and stolon architecture in potato. Journal of Experimental Botany 63, 4539–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, et al. 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiology 155, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Al-Babili S, van der Krol S, Bouwmeester H. 2013. The biology of strigolactones. Trends in Plant Science 18, 72–83. [DOI] [PubMed] [Google Scholar]
- Schnurr JA, Jung HJG, Samac DA. 2007. A comparative study of Alfalfa and Medicago truncatula stem traits: Morphology, chemical composition, and ruminal digestibility. Crop Science 47, 1672–1680. [Google Scholar]
- Shinohara N, Taylor C, Leyser O. 2013. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biology 11, e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, et al. 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes and Development 17, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. 2002. MAX1 and MAX2 control shoot lateral branching in Arabidopsis . Development 129, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Leyser OHM. 2007. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. The Plant Journal 50, 80–94. [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, et al. 2008. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula . The Plant Journal 54, 335–347. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y. 2012. Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant and Cell Physiology 53, 107–117. [DOI] [PubMed] [Google Scholar]
- Trinh TH, Ratet P, Kondorosi E, Durand P, Kamaté K, Bauer P, Kondorosi A. 1998. Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Reports 17, 345–355. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P. 2010. A small-molecule screen identifies new functions for the plant hormone strigolactone. Nature Chemical Biology 6, 741–749. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni R, Bradbury LM, Wurtzel ET. 2010. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Archives of Biochemistry and Biophysics 504, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E. 2012. Hormonal interactions in the regulation of plant development. Annual Review of Cell and Developmental Biology 28, 463–487. [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. 2012. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis . Development 139, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K. 2010. The strigolactone story. Annual Review of Phytopathology 48, 93–117. [DOI] [PubMed] [Google Scholar]
- Zhou C, Han L, Hou C, Metelli A, Qi L, Tadege M, Mysore KS, Wang ZY. 2011. Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. The Plant Cell 23, 2106–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Fu C, et al. 2013. The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula . The Plant Cell 25, 4845–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. 2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. The Plant Journal 48, 687–698. [DOI] [PubMed] [Google Scholar]
- Zwanenburg B, Mwakaboko AS, Reizelman A, Anilkumar G, Sethumadhavan D. 2009. Structure and function of natural and synthetic signalling molecules in parasitic weed germination. Pest Management Science 65, 478–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


