Abstract
The forebrain is the seat of higher order brain functions, and many human neuropsychiatric disorders are due to genetic defects affecting forebrain development, making it imperative to understand the underlying genetic circuitry. Recent progress now makes it possible to begin fully elucidating the genomic regulatory mechanisms that control forebrain gene expression. Herein, we discuss the current knowledge of how transcription factors drive gene expression programs through their interactions with cis-acting genomic elements, such as enhancers; how analyses of chromatin and DNA modifications provide insights into gene expression states; and how these approaches yield insights into the evolution of the human brain.
Keywords: forebrain, telencephalon, enhancer, transcription factor, genome, non-coding, gene regulation, distant-acting
Overview
Working at the turn of the twentieth century, Santiago Ramón y Cajal demonstrated the variety of neuronal cell types and provided insights into the network of connections within the brain using simple histological stains and light microscopes (Cajal, 1899). Over one hundred years later, despite the availability of advanced imaging, molecular and functional analysis tools, much remains unknown about the genetic factors controlling the development, structure, and function of the intricate features that, in their entirety, form the human central nervous system. The forebrain houses the neural structures that control higher order brain functions, including the pallium (cortex and hippocampus), subpallium (striatum, pallidum, preoptic area and septum), hypothalamus and thalamus. Understanding the development, evolution, function, and dysfunction of the forebrain requires a deep understanding of the genetic control of how its components are assembled and interconnected.
At the core of the processes that regulate forebrain development and function is the transcriptional circuitry. Over the last 25 years, numerous regional and cell type-specific transcription factors (TFs) have been identified and characterized. We are now aware of some components of TF networks that control regionalization within the embryonic brain. While general paradigms for identifying TFs and defining their cellular functions have been established, studies are now needed to explore and understand the molecular and genomic mechanisms through which networks of such TFs function during development. Of paramount importance is elucidating the cis-regulatory genomic elements where sets of TFs interact to control forebrain gene expression.
Recent technological advances, highlighted by the invention and application of chromatin immunoprecipitation (ChIP) -based and high-throughput sequencing assays, have enabled large-scale efforts to functionally annotate the genome. The results from studies that have applied these technologies to the developing brain reveal the essential role of neuronal TFs, the dynamic gene expression landscapes in brain development, and the extensive role of non-coding regulatory elements. These findings point to complex regulatory systems underlying the diversification of neuronal cell types and structural connectivity that Cajal drew in his formative illustrations. While many of the details of the emerging regulatory landscape of the brain remain to be explored, recent advances highlight the role of transcriptional control in normal development and in neurological disorders and disease. In this review, we describe the interplay between TFs, distal transcriptional enhancers, chromatin structure and epigenomic features, and DNA-binding and chromatin remodeling proteins in establishing the regulatory circuitry underlying transcriptional control and development of the forebrain. In addition, we describe approaches for the identification and characterization of enhancers and other regulatory elements and provide a perspective on emerging and exciting research on the genomic and regulatory control of forebrain development, evolution, and disease.
Annotating regulatory elements active in forebrain development
Metazoan gene regulation via cis-regulatory elements
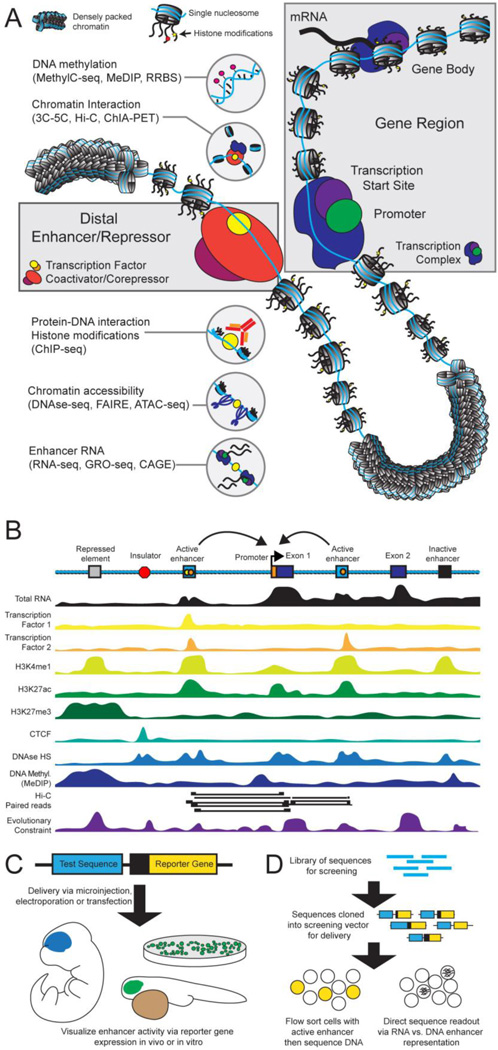
cis-regulatory control of gene expression during development is a complex process, dependent on distal sequences, spatial organization of the chromosome, and chromatin or epigenetic state [Figure 1A]. For some genes, notably housekeeping genes, the proximal regulatory sequence is sufficient for correctly activating transcription (Lenhard et al., 2012). In contrast, genes with complex expression patterns can be acted on by many distal transcriptional enhancers located in intronic, intergenic, and even exonic sequence, with enhancers potentially located far from target genes. Enhancers appear to be the most numerous regulatory elements in mammalian genomes and exhibit extensive tissue and stage specificity, suggesting that distal enhancers are required for the precise control of gene expression (ENCODE Project Consortium et al., 2012; Mouse ENCODE Consortium et al., 2012; Nord et al., 2013; Shen et al., 2012). With the binding of activating TFs, enhancers are brought within spatial proximity of target promoters through the formation of loops in DNA (Kulaeva et al., 2012), with structural proteins such as mediator and cohesin involved in this process (Kagey et al., 2010). It is now clear that enhancers may interact with multiple promoters, and clusters of co-regulated genes may exhibit promoter interactions, with the spatial aspects of gene regulation just now beginning to be characterized (Zhang et al., 2013). Many of the details regarding the mechanisms and timing of transcriptional control via regulatory sequences remain uncertain. There is evidence that enhancer-promoter looping may actually be very stable and not reflect activation (DeMare et al., 2013; Kieffer-Kwon et al., 2013). Additional classes of regulatory sequences are also important in gene regulation, such as insulators and silencers (Cuddapah et al., 2009). Significant progress in characterizing the complexity of non-coding regulatory circuitry has been made in model systems (Lagha et al., 2012), and in specific lineages and developmental loci (Montavon et al., 2011; Stamatoyannopoulos, 2005). It is increasingly recognized that transcriptional regulation is achieved endogenously through a complex interplay of cues that involve the binding of activating and repressive TFs, chromatin state and structure, epigenomic modifications and chromatin remodeling proteins, the activity of long non-coding RNAs, and cis-regulatory elements that can have activating or repressing function depending on developmental context. Despite this complexity, new technologies have enabled the identification and characterization of hundreds of thousands of candidate regulatory elements in the human and mouse genomes.
Figure 1. Types of gene regulatory sequences and methods for their discovery and characterization.
A) Schematic view of different chromatin states in genic and intergenic regions, with characteristic classes of epigenomic features and an overview of selected methods for their genome-wide mapping. B) Track-style view of features commonly associated with different types of regulatory sequences. C) Transgenic reporter assays enable the validation and detailed characterization of enhancer activity patterns in vitro and in vivo. D) Massive-parallel reporter assays enable medium- and large-scale function-based enhancer discovery screens.
Early approaches to enhancer identification in the brain
Before the publication of the human and mouse reference genome sequences, regulatory regions were typically found via trial-and-error, such as through deconstructing BACs or other large sequence fragments to determine subregions that controlled expression patterns of target genes. The first genome-wide predictions of regulatory elements were based on the presence of evolutionary sequence conservation (homology across species) or constraint (relative local sequence conservation across evolution) (Cooper et al., 2005; Frazer et al., 2004; Ovcharenko et al., 2004; Prabhakar et al., 2006b; Schwartz et al., 2000; Siepel et al., 2005) coupled to functional screening via reporter assays (Kothary et al., 1989; Nobrega et al., 2003; Pennacchio et al., 2006). The combination of trial-and-error and comparative genomics-guided screens, when applied to individual loci of interest, led to the identification of regulatory elements with activity in the developing forebrain near genes including Arx, Dach1, Dlx1/2, Dlx5/6, Emx2, Fezf2, Meis1, Otx1/2, Pax6, and Sox2 (Ahituv et al., 2007; Colasante et al., 2008; Ghanem et al., 2007; Kammandel et al., 1999; Kurokawa et al., 2004; 2014; Machon et al., 2002; Mariani et al., 2012; Royo et al., 2012; Shim et al., 2012; Suda et al., 2010; Theil et al., 2002; Zerucha et al., 2000) These elegant and labor-intensive studies provided first insights into the regulatory architecture of these key developmental loci. However, due to the required effort they were limited in the scope of genomic regions covered and likely missed additional regulatory elements, particularly those far from the genes of interest or lacking strong cross-species conservation. Many additional embryonic brain enhancers have been identified via large-scale unguided genome-wide screens of extremely conserved non-regions for sequences that drive reporter gene expression at specific embryonic time points, with the results available in the VISTA enhancer database (Visel et al., 2007). This database contains over 2100 tested human and mouse sequences, over 1100 of which function as enhancers in vivo in embryonic mouse tissues with whole mount staining images available. The VISTA enhancer set includes over 350 annotated to drive expression in the forebrain at e11.5, and 147 of these enhancers additionally include high resolution images of developmental brain sections that can be used to map the spatial activity of forebrain enhancers (Visel et al., 2013).
Epigenomic approaches to study gene regulation
Two parallel developments have resulted in rapid expansion of the catalogue of regulatory elements in mammalian genomes and in annotation of their function. The first is the availability of next-generation sequencing technologies that cost-effectively generate enough sequence coverage to enable genome-wide enrichment maps in a single experiment. The second is the knowledge about interpreting epigenomic marks that emerged from early studies in the area of cellular and chromatin biology, with additional traction from ENCODE pilot studies (ENCODE Project Consortium et al., 2007). Current proxy signatures of regulatory element activity and chromatin state include co-activator binding (e.g. p300), histone modifications, binding of TFs or other DNA-associated proteins, chromatin accessibility, DNA methylation, and non-genic RNA transcription [Figure 1A/B]. Using approaches to assay these signals, it is possible to identify and differentiate classes of regulatory elements and thus to identify enhancers that are active in particular cell lines or tissues. There are also emerging genome-scale tools to map interactions between regulatory sequences and their target genes (e.g. ChIA-PET (Fullwood et al., 2009)) and for generating genome-wide interaction maps (e.g. chromosome capture assays such as HiC (Lieberman-Aiden et al., 2009). There is support for sequential chromatin modifications that are associated with repressed, poised, and active enhancers (Creyghton et al., 2010; Rada-Iglesias et al., 2011). For example, the histone modification H3K27me3, shown in Figure 2b, can be indicative of a repressed region while H3K27ac can indicate active enhancers (Creyghton et al., 2010; Rada-Iglesias et al., 2011). Despite the general correlation between specific chromatin modification patterns and activity states, no specific signatures that have been reported appear to capture function exactly (Cotney et al., 2012; Visel et al., 2009b).
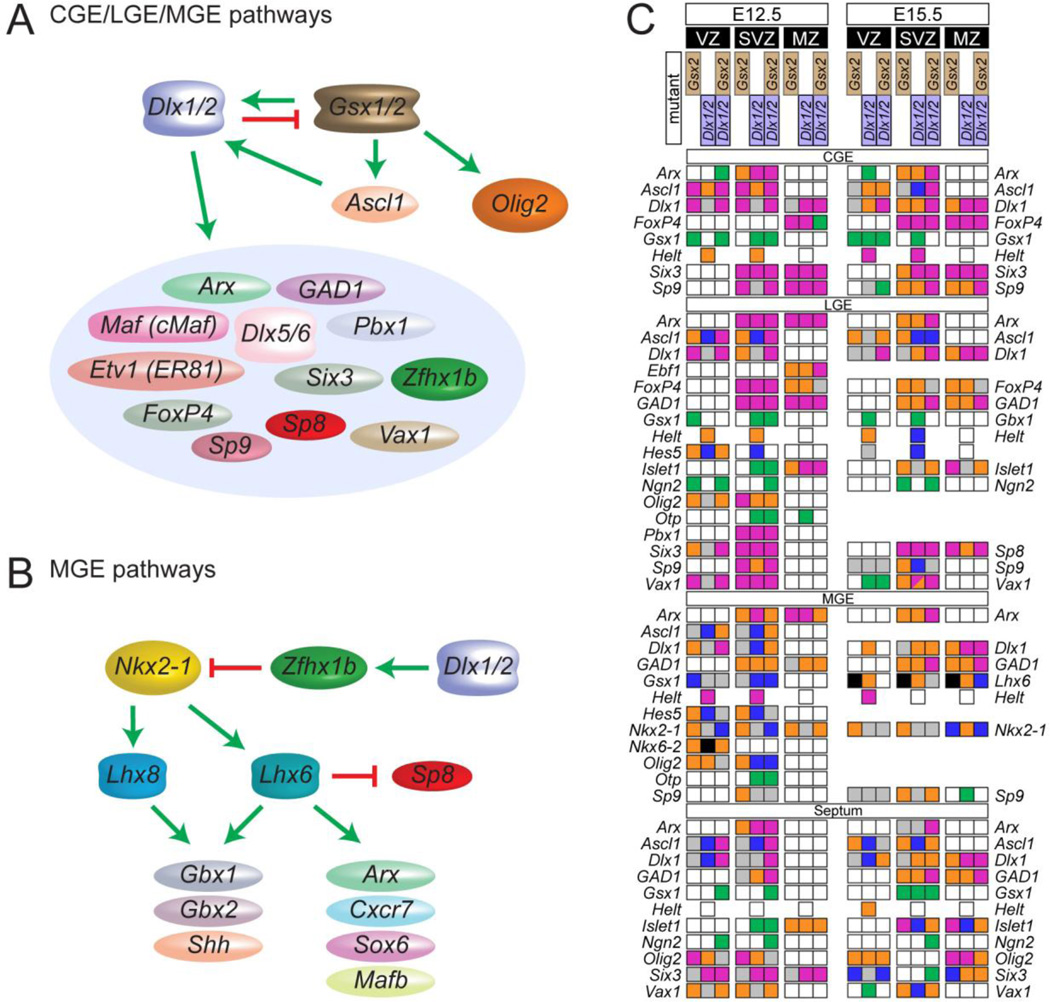
Figure 2. TFs with known roles in forebrain development.
A/B) Models of transcriptional pathways in the developing mouse basal ganglia based on RNA expression analyses in the embryonic brain of loss of function TF mutants. Green arrows: activation; red stop signal: repression. A) Pathways in the caudal, lateral and medial ganglionic eminences (CGE, LGE, MGE) based on data in references in main text B) Pathways in the medial ganglionic eminence (MGE) based on data in references in main text:. C) Expression of TFs in the basal ganglia of Gsx2−/−, Dlx1−/−Dlx2−/−, and Gsx2−/−Dlx1−/−Dlx2−/− mutant mice. Expression changes are reported separately for two different developmental stages (E12.5 and E15.5) in the ventricular zone (VZ), subventricular zone (SVZ) and mantle zone (MZ) of the CGE, LGE, MGE and Septum. Colors indicate the effect of each mutation on TF expression: Black or no square, not analyzed; gray, no obvious expression change in mutant; white, no detectable expression; magenta, severe reduction in expression; orange, moderate/mild reduction in expression; green, ectopic expression; blue, increased expression. In diagonally divided boxes, the top part represents the dorsal region and the bottom the ventral region. Modified from (Wang et al., 2013).
While the resources generated by large scale centralized efforts, such as the ENCODE and Epigenomics Roadmap initiatives, have significantly advanced our understanding of cis-regulatory elements, there are limitations in utilizing these datasets to characterize regulatory elements active in the developing brain due to the incomplete representation of representative cell lines and tissue types. Nonetheless, application of functional genomics to identify regulatory circuits controlling brain function has already produced major insights. There are now many publicly available genomic datasets relevant to the developing brain (see Table 1 for a partial list). These resources can be of tremendous value to researchers. For example, the Brainspan expression datasets have been instrumental in mapping the expression networks of genes perturbed in autism and schizophrenia (Gulsuner et al., 2013; Parikshak et al., 2013; Willsey et al., 2013).
Table 1.
Functional genomics datasets relevant to gene regulation in the developing brain
| Resource name | URL | Description |
|---|---|---|
| Genomic datasets focusing on the brain | ||
| Allen Brain Atlas | http://www.brain-map.org/ | Developmental transcriptomes, |
| Brainspan | http://www.brainspan.org/ | Mouse and human developmental |
| MethylomeDB | http://www.neuroepigenomics.org/methylomedb/ | Mouse and human |
| Nord et al. 2013 | http://enhancer.lbl.gov/mouse_timecourse/ | Mouse forebrain developmental |
| Brain expression resources (in situ, enhancer, BAC) | ||
| VISTA | http://enhancer.lbl.gov/ | Whole mount e11.5 mouse activity |
| Gensat | http://www.gensat.org/index.html | BAC-driven reporter |
| Large-scale centralized functional genomics initiatives | ||
| ENCODE | http://www.genome.gov/encode/ | Many data types. Expanding |
| Roadmap Epigenomics | http://www.roadmapepigenomics.org/ | Many data types, |
| Blueprint Epigenome | http://www.blueprint-epigenome.eu/ | Population level epigenomic data, |
| FANTOM | http://fantom.gsc.riken.jp/ | CAGE expression data for limted |
| Shen et al. 2012 | http://chromosome.ucsd.edu/mouse/download.html | Mouse ENCODE project |
Functional characterization of enhancer activity in the brain
While a number of epigenomic markers, signatures and assays are now available to predict the genomic location of enhancers, a continued major challenge is validation of functional predictions and determination of the exact activity of non-coding sequences. Reporter-based enhancer assays (Kothary et al., 1989) have been used extensively to map enhancer activity of both human and mouse regulatory sequences in vivo, and have been extended via library-based screening to examine hundreds to thousands of sequences in parallel. In comparison to assays performed in cell lines, in vivo functional enhancer analyses reveal specific cell types, tissue sub-regions, and developmental stages at which an enhancer drives expression (Pennacchio et al., 2006; Visel et al., 2013). Enhancer assays further differ in how the DNA is delivered, the size of fragment that is introduced, the number of copies or constructs per cell, stable or transient integration, and whether the readout is image or sequence based, as reviewed recently (Shlyueva et al., 2014). Beyond assaying in vivo activity, reporter assays have been used to document the functional autonomy of distal enhancers (Visel et al., 2009a), changes in enhancer activity due to sequence variation associated with evolution and human disease (Poitras et al., 2010; Prabhakar et al., 2008), and to label cells in fate mapping experiments (Chen et al., 2013; Pattabiraman et al., 2014; Visel et al., 2013). Reporter-based approaches have been further adapted to discover new enhancers in the genome (e.g. enhancer trapping methods (Hill and Wurst, 1993)), generate molecular reagents for regional/cell-type specific expression in the brain (Gong et al., 2003), and been employed in high throughput function-based identification and characterization of enhancers (Arnold et al., 2013; Dickel et al., 2014; Kheradpour et al., 2013; Murtha et al., 2014; Patwardhan et al., 2012; White et al., 2013). Similar methods have been used in simpler and more distant model organisms, which offer the trade-off of lower cost but larger differences in brain anatomy and function compared to humans (Ariza-Cosano et al., 2012). The combination of single-enhancer studies that produce detailed spatial and temporal maps of in vivo enhancer activity complemented by sequence-based assays of function have the potential to greatly expand understanding of regulatory element activity and the effects of regulatory sequence variation on brain development. Considering the results from traditional single gene studies alongside genome-wide or high-throughput approaches, we next attempt to synthesize current understanding of how transcriptional regulation orchestrates forebrain development, evolution, and function.
TFs controlling forebrain development
Elucidating the transcriptional networks in the developing forebrain requires the marriage of defining the cellular and developmental functions of individual TFs with their genome-wide molecular functions. In the telencephalic region of the forebrain (cerebral hemispheres), this has begun through the identification of TFs with region and cell-type specific expression patterns. For instance, telencephalic expression of TFs such as Emx1, Emx2, Fezf2, Ngn1, Ngn2, Pax6, Satb2, Tbr1 and Tbr2 are largely restricted to cortically derived glutamatergic progenitors and neurons (Lai et al., 2013; MacDonald et al., 2013), whereas Dlx1, Dlx2, Dlx5, Dlx6, Gsx1, Gsx2, Lhx6, Lhx8 and Nkx2-1 are largely restricted to subcortically derived GABAergic and cholinergic progenitors and neurons (Rubenstein and Campbell, 2013). Note that interneurons of cortical structures (neocortex, hippocampus and olfactory bulb) are believed to be largely generated by subcortical progenitors (Batista-Brito and Fishell, 2013; Rubenstein and Campbell, 2013). Other telencephalic TFs are jointly expressed by cortical and subcortical regions, such as Arx, Ascl1 (Mash1), Brn1, Brn2, COUPTFI, COUPTFII, CTIP1, CTIP2, Cux1, Cux2, Foxg1, Lhx2, Pbx1, Pbx2, Satb1, Sox5, Sox6, Sp8 and Zfhx1b (Sip1; Zeb2) (Lai et al., 2013; MacDonald et al., 2013; Rubenstein and Campbell, 2013; Rubenstein and Rakic, 2013; Thompson et al., 2014) suggesting that they may share similar functions in both cortical and basal ganglia development. Many other TFs are likely to be centrally involved in forebrain development and defining these factors and their roles is an active area of research. Below we highlight two signaling pathways active in the developing subcortical forebrain as examples of how understanding of these transcription factor networks reveals the transcriptional control of neurodevelopment.
Analysis of forebrain development using TFs mutants
Many of the TFs involved in forebrain development have been studied in loss-of-function mouse mutants with the goal to define their individual and combined in vivo functions (Figure 2). For instance, analysis of Dlx1−/−, Dlx2−/−, Dlx5−/−, and Dlx6−/− single mutants demonstrated distinct selective phenotypes in subsets of neurons in lineages that express the Dlx genes (Cobos et al., 2005; Long et al., 2009a; 2009b; Qiu et al., 1995; Rubenstein and Campbell, 2013; Wang et al., 2011; 2010), such as defects in dendrite innervating cortical interneurons (Cobos et al., 2005; Howard et al., 2014; Mao et al., 2009; Seybold et al., 2012). On the other hand, the double mutants have earlier and more pervasive phenotypes that affect regional and cell type identity, differentiation and cell migration (Anderson et al., 1997a; 1997b; Cobos et al., 2007; Wang et al., 2010; Yun et al., 2002).
Dlx1&2-associated network
Systematic analysis by RNA expression arrays and RNA in situ-hybridization showed that Dlx1−/−Dlx2−/− mice have altered expression of tens of TFs in the embryonic subpallium (Cobos et al., 2007; Long et al., 2009a; 2009b). Dlx1−/−Dlx2−/− mice have overexpression of Ascl1, Gsx1, Gsx2 and Olig2, suggesting that some of the Dlx1&2 mutant phenotype is due to overexpression of these TFs (Figure 2A,C). This hypothesis has been tested by generating Dlx1&2 triple mutants with Ascl1 (Mash1), Gsx1, Gsx2 and Olig2. For instance, Dlx1&2 promote neuronogenesis and repress oligodendrogenesis; Dlx1−/−Dlx2−/− mutants generate excessive oligodendrocytes; this phenotype is reversed in the Dlx1−/−Dlx2−/−Olig2−/− triple mutant, and exacerbated in the Dlx1−/−Dlx2−/−Ascl1−/−mutant (Petryniak et al., 2007). Like Olig2, Olig1 represses Dlx-mediated neurogenesis (Silbereis et al., 2014). Triple mutant analyses also provided evidence that Dlx1/2, together with Ascl1 and Gsx2, promote neurogenesis and fundamental subcortical properties, such as expression of GAD1, the gene encoding the GABA synthesizing enzyme (Long et al., 2009a; 2009b; Wang et al., 2013) (see Figure 2C for an approach to annotate TF expression changes in Dlx1/2, Gsx2, and Dlx1/2;Gsx2 mutants).
Dlx1−/−Dlx2−/− mutants have reduced expression of a distinct set of TFs, including Arx, Sp8 and Zfhx1b (Figure 2A,C) (Long et al., 2009b; McKinsey et al., 2013). Analysis of Arx, Sp8 and Zfhx1b single mutants has given insights into their individual contributions to the Dlx1−/−Dlx2−/− phenotype. Arx mutants have a time-dependent block in the maturation and migration of neurons from basal ganglia progenitor zones (Colombo et al., 2007), similar to Dlx1−/−Dlx2−/− mutants, which have a progressive accumulation of non-migrated immature cells in the subventricular zone (Anderson et al., 1997b; Yun et al., 2002). Sp8 mutants (Waclaw et al., 2006), as well as Dlx single and compound mutants (Long et al., 2007; 2003; Qiu et al., 1995), have olfactory bulb interneuron differentiation defects. Zfhx1b mutants have abnormal migration of interneurons generated by the MGE subcortical progenitor region. Zfhx1b and Dlx1−/−Dlx2−/− mutants both fail to repress expression of Nkx2-1 (Figure 2B) (McKinsey et al., 2013; van den Berghe et al., 2013), a TF that is essential for MGE identity (Sussel et al., 1999). Nkx2-1 expression needs to be turned off during maturation of interneurons that migrate to the cortex (Nóbrega-Pereira et al., 2008). In Zfhx1b mutants Nkx2-1 expression persists, leading to an accumulation of MGE-derived interneurons in the striatum, suggesting that Zfhx1b regulates the switch between the generation of cortical and striatal interneurons, and is required for the expression of Maf (cMaf) in migrating cortical interneurons (Figure 2A) (McKinsey et al., 2013). Furthermore, there is evidence that Zfhx1b regulates the expression of the Unc5b receptor, which contributes to the migration defect (van den Berghe et al., 2013).
Nkx2-1-associated network
Nkx2-1 functions in part parallel to the Dlx-driven TF hierarchy (Figure 2B). Nkx2-1 has a fundamental role in specification of MGE progenitor cell identity – in its absence, the MGE changes fate, taking on the more dorsal properties of the LGE and CGE (Butt et al., 2008; Flandin et al., 2010; Sussel et al., 1999). Nkx2-1 drives the expression of Lhx6 and Lhx8 (Du et al., 2008; Sussel et al., 1999), which together with their cofactor Ldb1 (Zhao et al., 2014) are required for the differentiation of GABAergic (Lhx6) and cholinergic (Lhx8) neurons (Fragkouli et al., 2009; 2005; Liodis et al., 2007; Zhao et al., 2008; 2003). Interneurons lacking Lhx6, have reduced expression of the Arx, MafB, Npas1 and Sox6 TFs (Figure 2B) (Denaxa et al., 2012; Zhao et al., 2008) Satb1 function has been linked to activity-dependent differentiation of cortical interneurons (Close et al., 2012; Denaxa et al., 2012). Arx function was noted above; Npas1 represses the generation of a specific set of cortical interneurons (Stanco and Rubenstein); Sox6 is required in the MGE to repress pallial proneural gene expression and to promote interneuron development (Azim et al., 2009; Batista-Brito and Fishell, 2013; Batista-Brito et al., 2009). MafB function in the telencephalon is currently under study. To identify the role of individual molecules that are downregulated in the Lhx6 mutant, we have employed a novel complementation approach (Vogt et al., 2014). For instance, to test whether reduced Arx expression contributes to the Lhx6 mutant phenotype, lentiviral transduction introduces Arx downstream of a Dlx1&2 enhancer into dissociated MGE cells in vitro; these are then transplanted into a wild type cortex, and the derived interneurons are phenotyped. In this case, restoring Arx expression partially restored interneuron expression of parvalbumin (Vogt et al., 2014).
Mapping TF function, combinatorial activity and genomic binding in the forebrain
Thus, even examining these two networks, loss of function analyses of over 20 TFs have enabled the field to perform detailed histological, cellular and molecular analyses of the mutant developing subpallium and its derivatives, including cortical and olfactory bulb interneurons. Similar progress has been made in defining TF function during pallial development, including its regionalization and generation of projection neuron subtypes, although due to space constraints we will not amplify upon this important subject (MacDonald et al., 2013; O'Leary et al., 2013). However, very little is known about how these TFs fit into the transcriptional circuitry orchestrating such processes, including the combinatorial activity of TFs. Models based on co-transcription (Ravasi et al., 2010) and DNAse I footprinting (Neph et al., 2012) to predict co-occurrence of transcription factor binding sites show that there are robust combinatorial sets of transcription factors that are associated with specific lineages and tissues, including in the brain. Understanding this combinatorial functionality of TFs will likely be essential to understanding neurodevelopment. In a functional study of odorant receptor (OR) regulation in sensory neurons in Drosophila, it was shown via systematic RNAi-based TF knockdown that combinations of seven TFs accomplish OR regulation via transcriptional activation and repression to prevent ectopic expression (Jafari et al., 2012). This study highlights how combinatorial TF activity via differential expression and binding can drive neurodevelopmental processes and suggests that integrative models of TF activity will be necessary to understand the complex developmental architecture of the forebrain. Several additional critical gaps of knowledge remain to be filled, including the identification of all of the TFs that may participate, identification of the gene regulatory elements (enhancers), and identifying the genomic regions where TFs bind.
As noted above, there are now expression databases that define the spatial and temporal expression patterns of most TFs (Table 1). Careful annotation of this information, and additional analyses using RNA expression arrays and RNA-seq in specific cell types, will greatly facilitate defining the sets of TFs that are candidates for contributing to TF circuitries. What remains perhaps the largest unmet gap, is the mapping of TFs to their in vivo genomic binding sites. One method to obtain this information is ChIP-seq, however, performing TF ChIP-seq on embryonic brain tissue has lagged, in part because the identification of high-affinity antibodies that specifically bind to a given TF that is bound to fixed chromatin has been problematic. Nonetheless, there are examples where inroads have been made. For instance, there is ChIP-qPCR evidence that DLX2 directly binds to, and regulates the Dlx1&2, Dlx5&6 and Zfhx1b loci (Colasante et al., 2008; McKinsey et al., 2013; Potter et al., 2009; Zhou et al., 2004), that NKX2-1 directly binds to, and regulates the Lhx6 locus (Du et al., 2008), and that LHX6 directly binds to, and regulates the Arx locus (Vogt et al., 2014). Genome-wide analyses of ASCL1 promoter binding in the embryonic brain and in neural stem cell cultures demonstrated that ASCL1 is bound to promoters of genes regulating cell cycle progression (Castro et al., 2011). Ongoing studies are now making progress using ChIP-seq to define the genome-wide landscape of TF binding in the developing forebrain.
The regulatory circuitry underlying forebrain development
Forebrain development is characterized by dynamic transcription
Analysis of TF expression and effects of loss-of-function in neuronal stem cell proliferation, differentiation, and migration, reviewed elsewhere (Hébert and Fishell, 2008; Kohwi and Doe, 2013; Molyneaux et al., 2007), have led to models of sequential or combinatorial expression of TFs in the regulation of these processes. The dynamic expression profiles of key TFs are mirrored by genome-wide transcription patterns generated from microarrays and more recently RNA-seq approaches. Transcriptome analysis of multiple brain regions across multiple developmental stages revealed extensive regional and temporal differences (Kang et al., 2011; Miller et al., 2014; Pletikos et al., 2013). These expression differences coalesce into transcriptional modules of co-expressed genes that are involved in stage-specific processes in specific structures, such as opposing expression patterns of genes associated with neuronal differentiation (at early stages) and ion channels (at later stages) in the developing neocortex and hippocampus. To achieve tightly regulated dynamic expression profiles, it is likely that cis-regulatory elements represent a substrate for TF binding, enabling precise control via localization of transcriptional machinery to target promoters, activate transcription, and recruit chromatin remodeling proteins that together direct specific spatiotemporal expression of target genes.
Enhancers drive sub-regional expression patterns in the developing forebrain
Gene-centric studies demonstrated that some genes, such as Arx (Ahituv et al., 2007; Colasante et al., 2008) and Dlx1&2 and Dlx5&6 (Ghanem et al., 2007; Zerucha et al., 2000), have multiple enhancers with similar activity patterns, raising the possibility of enhancer redundancy. In addition, some loci (e.g. in the Dlx family) had multiple enhancers with overlapping, yet distinct activity patterns, suggesting that a given gene may be regulated by many enhancers with distinct temporal and spatial activities. A recent study examined in vivo activity patterns of 145 human enhancer sequences at high spatial resolution in the mouse telencephalon at e11.5, using reporter assays and serial histological sectioning to generate a digital atlas of enhancer function in the developing brain (Figure 3A–D) (Visel et al., 2013).
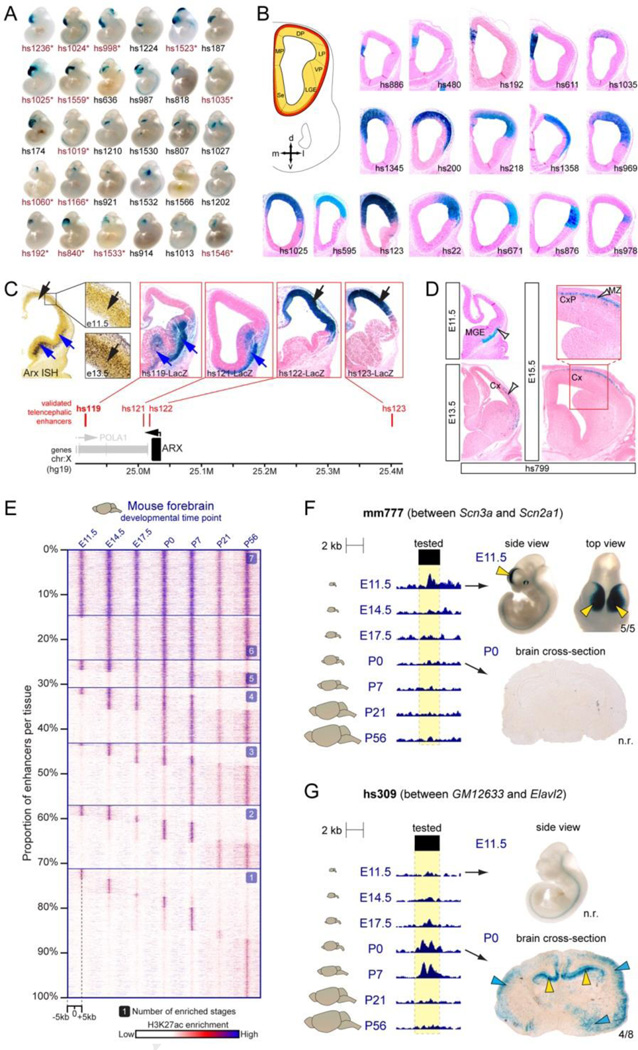
Figure 3. Spatial and temporal specificity of enhancers active in the developing forebrain.
A) Subset of forebrain enhancers with a spectrum of subregional specificities at whole-mount resolution. B) Examples of enhancers with restricted pallial activity in the mouse telencephalon at E11.5. C) Multiple enhancers in the larger region surrounding the Arx gene show subregional forebrain activity patterns that recapitulate endogenous Arx mRNA expression in the mouse forebrain. Notably, enhancer activities show partial spatial redundancy. D) Example of an enhancer with activity across multiple developmental stages, labeling cell populations whose location is consistent with migration from the MGE, through the LGE, to the cortex (white arrows). E) Developmental dynamics of enhancer-associated histone mark H3K27ac at candidate forebrain enhancers analyzed by ChIP-seq across seven stages of brain development. Most sites show temporally restricted H3K27ac marks. F,G) Examples of in vivo validated temporally dynamic enhancer activity in the forebrain, as predicted by temporally dynamic H3K27ac signatures. CP, choroid plexus; Cx, cortex; CxP, cortical plate; DP, dorsal pallium; LGE, lateral ganglionic eminence; LP, lateral pallium; MGE, medial ganglionic eminence; MP, medial pallium; MZ, marginal zone; VP, ventral pallium. A-D modified from Visel et al., 2013. E-G modified from Nord et al., 2013.
In addition to generating activity maps for enhancers that are located nearby genes with key functions in brain development and neurological disorders, this analysis identified several recurrent characteristic features of enhancers active in the developmental brain. First, individual enhancers drove highly reproducible, spatially restricted enhancer patterns, with large variation in patterns of expression observed across the sampled enhancers that represent all the major subregions of the e11.5 telencephalon (Figure 3A,B,D). Generation of stable transgenic lines for 15 pallial enhancers enabled fate mapping analysis, showing that enhancers with activity in distinct pallial progenitor domains generate distinct cortical regions (Pattabiraman et al., 2014), consistent with the protomap hypothesis (Rakic, 1988).
Second, when combined to form a composite pattern, the expression patterns of individual enhancers recapitulate gene expression patterns as mapped via in situ hybridization, consistent with observations in other organ systems (e.g. (Schwartz and Olson, 1999)) that the combined activity of discrete enhancer sequences drive complex endogenous gene expression patterns. For example, four distant-acting enhancer sequences were described located in the extended locus containing Arx, a gene that regulates pallial and subpallial development and is associated with mental deficiency and epilepsy (Colasante et al., 2008; Friocourt and Parnavelas, 2010; Kitamura et al., 2009; 2002; Marsh et al., 2009; Olivetti and Noebels, 2012). Together, these enhancers recapitulate endogenous Arx expression (Figure 3C). Third, enhancers that activated gene expression in the same anatomical structures were enriched for shared sequence motifs, indicating that highly specific enhancer activity is, at least in part, due to the presence of binding sites for particular transcriptional regulators active in the brain. This and other in vivo studies of enhancer activity in the developing forebrain establish that the cis-regulatory landscapes guiding tissue-specific expression patterns are complex, with the combined action of multiple, sometimes redundant, enhancers likely involved in regulation of specific loci.
Functional genomics reveal regulatory circuits in the brain
Experiments that characterize neuronal cis-regulation one element at a time have revealed many aspects of the regulatory control of forebrain development, yet these studies fail to capture global patterns of enhancer usage and chromatin state during forebrain development. Early attempts to fill this gap involved ChIP-seq experiments targeting p300-binding sites in the developing mouse brain and targeting various histone modifications in neuronal cell lineages (Creyghton et al., 2010; Visel et al., 2009b). More recent genome-wide studies in human and mouse tissues, including across brain regions, assaying p300 and histone modifications, DNase hypersensitivity, enhancer [e]RNA, and DNA hypomethylation are in line with results from in vivo transgenic assays showing that enhancers are highly tissue-specific (Andersson et al., 2014; Shen et al., 2012; Stergachis et al., 2013; Visel et al., 2007; Zhu et al., 2013)
Two recent studies have directly interrogated temporal changes in enhancer and epigenomic landscapes by profiling mouse and human forebrain tissues across developmental stages. In one approach, ChIP-seq targeting H3K27ac, a histone modification strongly associated with active enhancers, was performed on mouse forebrain collected across fetal and postnatal development (Nord et al., 2013) (Figure 3E–G). This study generated in depth maps of enhancer activity in vivo across development in the forebrain, identifying over 50,000 candidate enhancers active across development in the forebrain, with the majority of enhancers predicted to be transiently active. The predicted activity of a set of enhancers was validated in vivo using transgenic assays (Figure 3F–G). Dynamic enhancer activity was associated with genes expressed in the control of stage-specific biological processes such as neuronal proliferation at early embryonic time points and synapse development and plasticity later. In another time course study, Methyl-seq was performed on cortical tissues from the human and mouse at three time points to profile dynamic methylation at cytosine residues, with additional examination of 5-hydroxymethylcytosine in selected samples (Lister et al., 2013). Hypomethylation was indicative of enhancer activity, and differentially methylated regions were detected that indicate extensive turnover in enhancer activity consistent with the H3K27ac signatures observed in the developing mouse brain. As further evidence of the dynamic activity of enhancers in the developing brain, comparison of embryonic stem cells, neural stem cells, and neural progenitors showed that enhancer-promoter interactions are specific to each cell population (Zhang et al., 2013). In a finding suggestive of how some non-coding regions act to control lineage specification, it was recently observed that there are large domains exhibiting chromatin modifications consistent with regulatory sequence that are particularly responsive to essential lineage-specification or pluripotency factors. These loci, referred to as “super-enhancers” (Chapuy et al., 2013; Hnisz et al., 2013; Whyte et al., 2013) or “stretch enhancers” (Parker et al., 2013) have been predicted to exist in brain tissues as well, with initial analysis suggesting that TFs such as NKX2-2, OLIG1, BRN2, SOX10, and SOX2 are master regulators that interact with these regulatory regions in the brain (Hnisz et al., 2013). It is unclear whether these larger domains represent regulatory elements that act in a cooperative way that is qualitatively different from enhancers as previously described, or if they instead are simply regions that are densely packed with regulatory sequences.
In parallel to gene activation, cis-regulatory sequences and chromatin state are also involved in the repression of transcription. Specific TFs can have activating or repressing effects depending on context. For example, REST/NRSF is a master regulator of neurogenesis that acts to repress transcription of neuronal genes in non-neuronal lineages by recruiting chromatin remodeling proteins (Ballas et al., 2005). Major markers of repressive remodeling in the brain include H3K27me3 and DNA hypermethylation at gene bodies and distal enhancers (Lister et al., 2013; Zhu et al., 2013). Likely due to extensive repressive chromatin in intergenic regions and non-expressed genes in mature neuronal lineages, the majority of enhancers active in the adult brain were found within gene bodies of neuronal genes or near the transcription start site in two independent studies (Nord et al., 2013; Zhu et al., 2013). These studies indicate that chromatin state dynamics are an important factor in brain development, a finding paralleled by recent studies of neurodevelopmental disorders discussed below.
Enhancers as tools for analysis of forebrain development
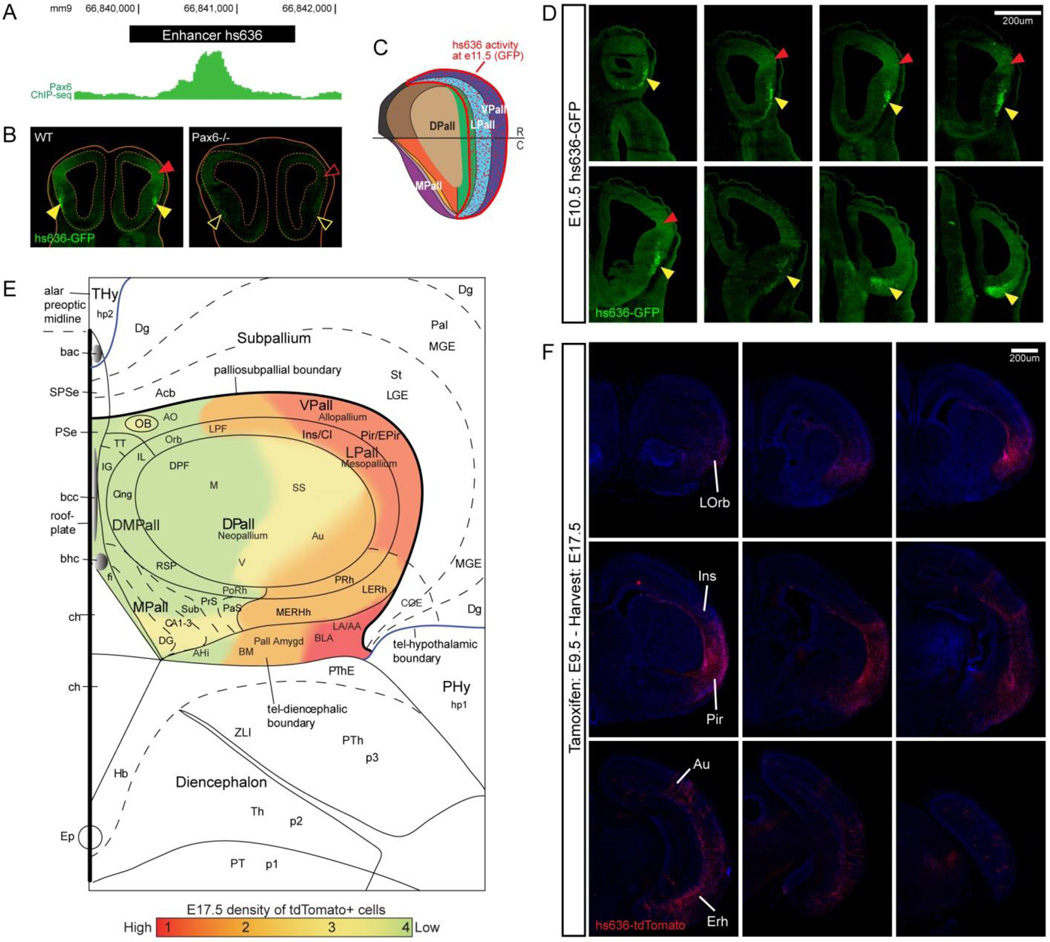
Enhancer elements generally maintain correct temporal-spatial control when ectopically positioned in the genome (e.g. (Ghanem et al., 2007; Visel et al., 2013; Zerucha et al., 2000)). Thus, these elements can be used experimentally to drive spatiotemporally restricted gene expression in specific brain regions and cell types. For instance, enhancers driving Cre expression have been used for fate mapping experiments of the subpallium (Potter et al., 2009; Waclaw et al., 2010) or pallium (Pattabiraman et al., 2014). Figure 4 illustrates the use of an enhancer (hs636) with activity in ventral parts of the pallial primordium (GFP expression in Figure 4D, schematically summarized in Figure 4C) to fate map its derivatives (Cre-mediated induction of tdTomato expression in Figure 4F, schematically summarized in Figure 4E). Enhancer hs636 is bound by PAX6 in vivo (Figure 4A), and depends on Pax6 function for its expression (Figure 4B). Enhancers driving markers such as GFP can be used in stem cell differentiation experiments to indicate when particular telencephalic cell states differentiate and for cell purification using FACS (Chen et al., 2013). Given their small size, they can also be used in viral vectors to confer cell type-specific gene expression, and have recently been used to drive expression in cortical interneurons (Lee et al., 2014; Vogt et al., 2014).
Figure 4. Enhancers as tools for analyzing forebrain development.
A) PAX6 ChIP-seq analysis from E12.5 cortex showing a peak directly over endogenous enhancer 636 (black bar). B) GFP pallial expression driven by enhancer hs636 in E11 cortex and reduced pallial GFP expression in Pax6−/−. C) Schema showing approximate position of GFP expression (red) within flattened view of E11.5 pallial progenitor zones. D) Enhancer hs636 activity in E10.5 telencephalon in stable transgenics (yellow arrowheads: ventrolateral pallial neurons; red arrowheads: ventrolateral pallial progenitors 24127591). E,F) Fate mapping using enhancer hs636. Cre recombination was tamoxifen-induced at E9.5 and brains were analyzed at E17.5 for tdTomato staining (F). Results are summarized in a schematic map showing dtTomato expression within a flattened view of E17.5 pallial subdivisions, color coded according to approximate density of tdTomato+ cells (E). A-F modified from (Pattabiraman et al., 2014). Abbreviations according to region: Ventral Pallium (VPall, allopallium); AO: anterior olfactory nuclei; OB: olfactory bulb; Pir/EPir; piriform and ectopiriform; LERh: lateral entorhinal; MERh: medial entorhinal. Lateral Pallium (LPall, mesopallium): Ins/Cl: insula/claustrum; LO: lateral orbital; PRh: perirhinal; Orb: orbitofrontal. Dorsal Pallium (DPall; neopallium): AU (A); auditory; DPF: dorsal prefrontal; F: frontal; LPF: lateral prefrontal; M: motor; SS: somatosensory; V: visual. Dorsomedial Pallium (DMPall): Cing (C): cingulate gyrus; IL: infralimbic (and PrL: prelimbic); MOrb: medial orbital; RSP: retrosplenial; PoRh: postrhinal. Medial Pallium (MPall): CA1–3: CA fields 1–3; DG: dentate gyrus; fi (F); fimbria; IG: indusium griseum; Sub (S): subiculum; PaS: parasubiculm; PrS: presubiculum; TT: tenia tecta. Dorsal Midline: bac: brachium of the anterior commissure; bcc: brachium of the corpus callosum; bhc: brachium of the hippocampal commissure; ch; choroid plexus; PSe (PS): pallial septum. Pallial Amygdala (Pall Amygd): AA: anterior amygdala; Ahi: amygdalohippocampal area; BM: basomedial; BLA; basolateral; LA: lateral. Subpallium: Acb: accumbens; CGE: caudal ganglionic eminence; Dg: Diagonal area; LGE: lateral ganglionic eminence; MGE: medial ganglionic eminence; Pal: pallidum; SPSe: subpallial septum; St: striatum. Hypothalamus: hp1, 2: hypothalamic prosomere 1 and 2; PHy: peduncular; Thy: hypothalamus. Diencephalon: Hb; habenula; p2, p3: prosomeres 2 and 3; Thy: terminal hypothalamus; PThE: prethalamic eminence; Th: thalamus.
Evolutionary conservation and novelty of gene regulation in the brain
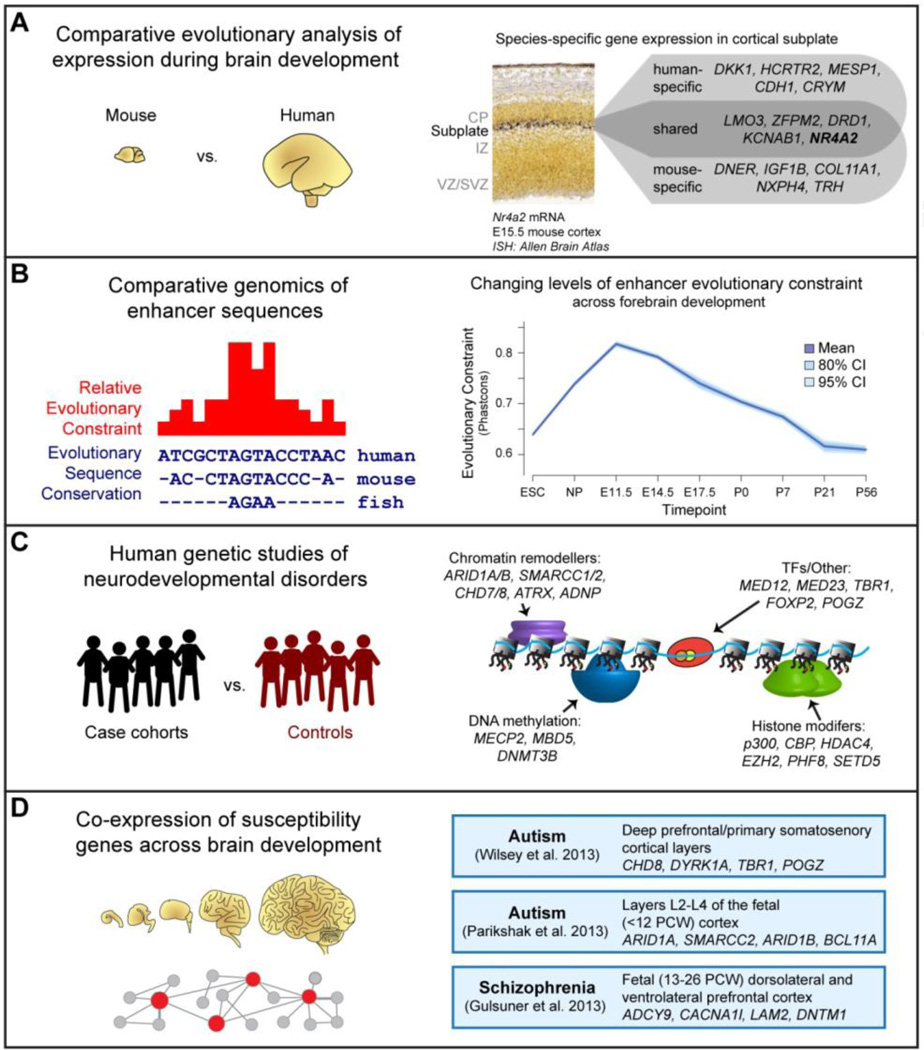
Ongoing studies of various vertebrate species, including human, are defining the transcription factors and enhancers that control cortical development (Miller et al., 2014; Shim et al., 2012). These analyses are expected to shed light on the genetic mechanisms that have contributed to cortical evolution and disease (Willsey et al., 2013). Comparative transcriptomic analyses have demonstrated differences in expression of genes in the developing brain across primates (Khaitovich et al., 2004; Konopka et al., 2009; Nowick et al., 2009) and between mice and humans (Miller et al., 2014) (Figure 5A). Evolutionary differences in the structure and representative cell types and connectivity of the forebrain are observed across vertebrates, with the six-layered laminar structure arising in mammals and the expansion in cortical surface area producing complex involutions observed in a number of species, including humans. These differences may be associated with changes in the expression patterns of and the interactions between specific transcription factors in the developing brain. Studies on the evolution of regulatory control systems and cis-regulatory elements have produced paradoxical findings highlighting both extreme evolutionary constraint and human-specific regulatory changes as strong forces in shaping the forebrain. King and Wilson demonstrated high levels of coding sequence conservation between human and chimpanzee and postulated that non-coding changes account for the majority of sequence level differences between the species (King and Wilson, 1975). The availability of sequenced genomes has enabled comparisons of non-coding sequence homology across the vertebrate evolutionary tree and recently, between humans and extinct hominins (Green et al., 2010). Perhaps surprisingly, very high levels of sequence conservation have been observed at regulatory sequences active in the developing forebrain across the vertebrate lineage, with ultraconserved regulatory elements enriched for forebrain activity in transgenic assays compared to other tissues in e11.5 mice and increased constraint levels observed genome-wide for forebrain enhancers relative to other tissues (Blow et al., 2010; Nord et al., 2013; Pennacchio et al., 2006). Interestingly, the level of constraint on stage-specific forebrain enhancers decreases significantly from mid-gestation to adult (Nord et al., 2013) (Figure 5B), a pattern also observed across various developmental lineages (Bogdanović et al., 2012; Stergachis et al., 2013). This pattern is observable in the constraint of gene expression patterns and general morphology (Domazet-Lošo and Tautz, 2010; Kalinka et al., 2010; Quint et al., 2012), in line with the hourglass model of evolutionary constraint on development (Casci, 2011). Further supporting the conservation of neuronal regulatory networks, a study of transcription factor co-expression and interaction comparing human and mouse identified a number of transcription factor networks that appear conserved between mouse and human in the developing brain (Ravasi et al., 2010). These findings suggest that cis-regulatory landscapes controlling early forebrain development are generally relatively ancient.
Figure 5. Insights and Challenges in Deciphering the Regulatory Architecture of Forebrain Development.
A) Comparative functional genomic studies, such as large-scale studies of gene expression patterns in the developing brain by RNA in situ hybridization, provide insight into general and human-specific aspects of molecular pathways involved in brain development. B) Comparative genomic studies reveal a deeply conserved regulatory framework associated with brain development, but can also identify specific changes in regulatory sequences that underlie structural and functional innovations in the brain observed in vertebrate evolution. Remarkably, regulatory sequences active during early stages of brain development (mid-gestation in mouse) tend to be under higher evolutionary constraint than those active later in development and in the adult brain. C) Genome-wide association, exome sequencing, copy number variation, and whole-genome sequencing studies of patient cohorts are powerful tools for identifying genes and non-coding sequences associated with neurodevelopmental disorders. These studies have revealed a major role for proteins involved in chromatin remodeling, DNA methylation processes, histone modification, and other transcriptional regulatory pathways and processes, with individual genes reported in human genetic studies offered as examples. D) Systems-level analysis integrating expression, genetic, epigenomic and functional data has the potential to elucidate genetic and functional networks required for normal brain development and function, which are thought to be disrupted in neurodevelopmental and neuropsychiatric disorders.
Despite the observation of strong evolutionary constraint at brain enhancers, other studies examining sequence and functional conservation of enhancers suggest that human regulatory elements exhibit high levels of evolutionary innovation both in sequence and function. One locus that has been examined in detail is AUTS2, a gene implicated in human evolution and in neurological disorders. Sequence comparisons at this locus between human and Neanderthal genomes identified one of the strongest signals for human-specific non-coding sequence changes (Green et al., 2010), and non-coding sequences at this locus exhibiting human-specific changes drove expression in the developing brain (Oksenberg et al., 2013). A second approach has been to look for regulatory regions that have high levels of evolutionary constraint but where there are human-specific changes, also known as human-accelerated regions (HARs) or accelerated conserved non-coding sequences (aCNSs), or where there is no mappable human homolog (McLean et al., 2011; Pollard et al., 2006; Prabhakar et al., 2006a). HARs and aCNSs are enriched near TFs and near genes associated with neuronal cell adhesion and human-specific deletions are similarly associated with neural functions, findings suggestive of sequence-level changes that drive human-specific aspects of brain development. Genome-wide comparison of functional conservation of enhancers as assayed by p300 interaction revealed evidence that sequences exhibiting significant homology between human and mouse can show functional differences in the developing brain. This comparison only included a single developmental time point at which mouse and human brains should be relatively stage matched (Clancy et al., 2007) and candidate enhancers identified using only a single epigenomic mark (Visel et al., 2013), and further experiments are necessary to elucidate functional conservation across tissues and developmental stages. While not in a neuronal tissue, recent work comparing functional conservation using epigenomic assays in hepatocytes (Odom et al., 2007) and developmental limb tissue (Cotney et al., 2013) demonstrate significant lack of functional conservation between humans and other vertebrate lineages. These findings are supported by functional enhancer assays comparing activity in Drosophila lineages (Arnold et al., 2013). Finding of limited functional conservation of non-coding elements in the brain is in line with studies directly comparing TF binding and H3K27ac across evolution in other tissues, which suggest substantial turnover in TF binding sites and evolution of new enhancers via functional modification or co-option of enhancers active in other tissues (Lettice et al., 2011; Schmidt et al., 2010; Stefflova et al., 2013). Annotated maps of human forebrain enhancers will be required to determine the balance between evolutionary conservation and innovation in cis-regulatory control of human forebrain development.
Gene regulation and neurodevelopmental disorders
Loss of function of regulatory genes in forebrain development
Mouse knockout models are now available for thousands of genes, including many genes known or proposed to be involved in transcriptional regulation during brain development. Studies using these models have revealed severe neuroanatomical consequences and frequently embryonic lethal phenotypes associated with loss of function of neuronal transcriptional regulators, as described above. In parallel, some of the first successful efforts to map human monogenic neurodevelopmental disorders implicate transcriptional regulation and chromatin remodeling in synapse development and plasticity, such as MECP2 in Rett Syndrome and FMR1 in Fragile X Syndrome (Amir et al., 1999; Verkerk et al., 1991). A growing body of evidence supports that multiple human neurodevelopmental disorders are associated with mutations in TFs, including Arx, Pax6, Six3 and Tbr1 (Bhatia et al., 2013; Hehr et al., 2010; Olivetti and Noebels, 2012; van Heyningen and Williamson, 2002; Willsey et al., 2013).
Human genetic studies of forebrain transcriptional regulation
There has been a leap forward in understanding the genetic components of complex neurodevelopmental disorders in recent years (McCarroll and Hyman, 2013). While the relative contribution to these traits across the spectrum of allele frequency remains an active discussion (McClellan and King, 2010), replicated findings at specific loci have established a role for both common and rare variants via genome-wide association studies (GWAS), rare or de novo CNV screening, and exome sequencing, as reviewed recently (Krystal and State, 2014; McCarroll and Hyman, 2013). The findings from human genetics studies point to polygenicity and a significant role for genes involved in transcriptional regulation and chromatin remodeling in the developing forebrain in the etiology of disorders such as autism, schizophrenia, and intellectual disability (Figure 5C). Studies of autism and schizophrenia, have highlighted pathways required for transcriptional regulation and chromatin remodeling, as well as other functional classes, such as calcium channels and synaptic plasticity genes (Krystal and State, 2014; McCarroll and Hyman, 2013). Evidence is mounting for dosage effects or haploinsufficiency of developmental TFs, such as TBR1, and chromatin remodeling proteins, such as CHD8, as a mechanism in these disorders (O'Roak et al., 2012a; 2011; 2012b). It is also likely that regulatory sequence variation contributes to these traits, as regions around many of the replicated GWAS signals as well as rare disease-associated CNVs do not contain coding sequence variants and disease-linked variants have been identified within enhancers active in the developing brain that result in changes in enhancer activity (Poitras et al., 2010).
Transcriptional regulation and chromatin remodeling in autism and schizophrenia
While there are now many genes identified via rare and de novo CNV screening and exome sequencing, it will take larger screening efforts and functional follow-up to establish the causality and pathophysiology of these mutations. An approach to learn more about these networks that has yielded success for autism and schizophrenia is to identify brain sub-regions and developmental stages at which the implicated gene networks are co-expressed using publicly available brain transcriptome data (Kang et al., 2011) (Figure 5d). In autism, co-expression networks present in midfetal layer 5/6 cortical projection neurons were identified that were organized around high-confidence “seed” genes that are mutated in autism (Willsey et al., 2013). A similar study linked rare de novo coding mutations in autism to co-regulated highly expressed genes during early neuronal fate determination, migration, and establishment of the cortical layers, providing suggestive evidence of a gene set that contributes to autism via haploinsufficiency (Parikshak et al., 2013). The same study identified glutamatergic neurons in superficial cortical layers (layer 2–4) as a potential cell type where autism-associated expression networks may be critical. These two studies describe different specific cortical layers as relevant, but the developmental regulatory processes affected by both sets of gene network analyses are highly convergent. An earlier study of gene expression in autism-associated cortical regions from autistic subjects versus controls identified differentially expressed genes and expression networks in autism, with a number of genes that overlap the two recent studies (Voineagu et al., 2011). A study examining de novo mutations in schizophrenia linked genes with predicted deleterious coding mutations in cases to expression networks in the fetal dorsolateral and ventrolateral prefrontal cortex (Gulsuner et al., 2013). While a complex combination of genetic and environmental factors is likely to influence these phenotypes, these studies illustrate how human genetic studies in tandem with functional genomics data can reveal the pathophysiology of neurodevelopmental disorders and the causal role that loss of endogenous gene expression and transcriptional regulatory circuits during brain development plays.
Knockout models of enhancer function
Non-coding sequence variation is predicted to represent a substantial proportion of disease-associated variants (ENCODE Project Consortium et al., 2012), yet the requirement for specific enhancers during development remains largely unclear. There are examples of sequence variation at enhancers that are linked to developmental phenotypes, with one of the most well-known instances being mutations in the ZRS enhancer of Shh that lead to limb malformations (Lettice et al., 2003). There are a few examples of enhancers whose deletion was shown to cause clear neurodevelopmental phenotypes. For instance, deletion of an enhancer regulating Fezf2 resulted in loss of Fezf2 expression and anatomical changes in the brain characterized by loss of specification of corticospinal neurons (Shim et al., 2012). On the other hand, a study that targeted four ultraconserved enhancers, which included enhancers predicted to regulate brain-expressed genes that produced severe phenotypes when knocked out, found that mice homozygous for the enhancer deletion allele for each of the four enhancers appeared did not have gross neurodevelopmental or neurological phenotypes (Ahituv et al., 2007), indicating that even the evolutionarily most conserved enhancers may not necessarily have functions at the organism level that that are required for viability.
Two related explanations may account for the mixed consequences of enhancer loss of function in these mouse models. The first is that there is a high level of functional redundancy in enhancers that regulate a specific locus. In this model, loss of function of a single enhancer is masked by the activity of another enhancer that drive a similar expression pattern, as has been observed for “shadow” enhancers in Drosophila (Lagha et al., 2012). In a similar model, enhancers may not be functionally redundant, but they act in a combinatorial manner where each enhancer acts to fine tune the expression of a gene. In this model, careful phenotyping would be necessary to identify the changes caused by loss of enhancer function. Outside the brain, this general paradigm was recently demonstrated in a study where deletions of craniofacial enhancers produced subtle morphological changes (Attanasio et al., 2013). While there remain a small number of regulatory loss-of-function models in mouse, results from human genetic studies, especially GWAS, suggest a significant role for regulatory variation across phenotypes, including in neurodevelopmental disorders (Ripke et al., 2013). It is quite possible that each regulatory circuit will be different, with strong effects possible for loss of function at some enhancers and weak or no observable phenotypes seen when other enhancers are deleted or when common variation changes enhancer activity.
Conclusions and major outstanding questions
The combination of detailed functional dissection of the roles of individual TFs with genome-wide approaches to mapping gene expression, enhancer activity, and chromatin dynamics reveal an emerging picture of the role of transcriptional regulation in forebrain development. These studies link cell-specific expression of TFs during processes such as proliferation, differentiation, migration, and synapse development to target genes via binding at TF binding motifs found in promoter-proximal sequences and distal enhancers. These regulatory circuits are emerging as tightly linked to the evolution of the human brain and to human neurodevelopmental disorders, with initial indications of specific developmental cortical sub-regions and neuronal classes that may be affected by mutations that contribute to disorders such as autism and schizophrenia. Figure 5 summarizes findings from recent studies highlighting the role of gene regulation in evolution and neurodevelopmental disorders.
With the availability of new technologies for epigenomic and functional profiling of regulatory sequences, availability of patient-specific models such as induced pluripotent stem cells and precision-engineered animal models, where is the field headed? We now discuss major challenges for three areas: basic biological mechanisms underlying gene regulation, the role of regulatory elements and chromatin structure in brain development, and linking findings from human genetic studies to biological mechanisms focusing on gene regulation.
Challenge 1: Basic mechanisms
General models of gene regulation via TF binding and activation or repression through co-factor recruitment, changes in chromatin accessibility, and DNA looping are becoming clearer, yet many of the basic mechanisms required for this process are still poorly defined. TF binding sites can be predicted based on various experimental methods and can be validated using direct measures of interaction, however it appears that a large proportion of the validated binding sites of many TFs may not be functionally relevant (White et al., 2013), and that combinatorial binding of TFs may govern function (Teng et al., 2014). Consistent with these observations, levels of individual TF expression do not have extensive global effects as measured by TF knockdown and expression profiling (Cusanovich et al., 2014). There are additionally unresolved questions regarding regulatory function and the balance of conservation versus binding site turnover within enhancers across evolution (Villar et al., 2014), the order and spacing of binding sites within enhancers (Smith et al., 2013), the requirement for specific TF binding interactions within enhancers (Teng et al., 2014), and the relationship between structural contact through looping and transcriptional activation (DeMare et al., 2013; Kieffer-Kwon et al., 2013).
A second major mechanistic question is how enhancers drive expression of a specific gene or set of target genes given the observation that many enhancers appear to skip nearby transcription start sites to interact with more distal targets (Zhang et al., 2013). While recent advances mapping chromatin interactions enable mapping of enhancer-promoter interactions, the mechanism for establishing specificity has been studied only for a very small number of loci. It is clear that enhancers vastly outnumber their transcribed targets. The model of specific enhancers driving cell-type or tissue-specific expression patterns is attractive and is true at a genomic scale, yet it appears that many genes are controlled by complex landscapes of potentially redundant regulatory elements and that loss of function of even highly-conserved enhancers near critical genes may not produce significant phenotypic effects (Ahituv et al., 2007). Recent studies suggest that enhancer redundancy is required to maintain robust expression patterns (Lagha et al., 2012) and that arrays of regulatory elements with correlated activity may enable tighter quantitative or qualitative regulation of expression (Whyte et al., 2013), however, this remains a relatively unexplored area especially in mammalian systems.
At this time, no single known epigenomic signature is sufficient to identify all enhancers and distinguish enhancers that are active with perfect specificity (Cotney et al., 2012), and an active area of research is honing prediction algorithms that are based on regulatory sequence composition, epigenomic signatures, and genomic structure and in developing methods to enable function-based screening of regulatory element activity as described above. A central assumption to the current models is that the census of TFs present combined with the relevant regulatory sequence substrate represents the information necessary for interpreting endogenous enhancer function, yet the field remains a long way from determining the rules that govern the processes involved in the establishment and function of mammalian regulatory circuitry and in predicting the effects of perturbations to these systems.
Challenge 2: Gene regulation in forebrain development
The framework and major actors in many TF signaling networks active in control of differentiation across a variety of neuronal subtypes are becoming clearer, with emerging Dlx1/2 and Nkx2-1 networks in the LGE/CGE/MGE detailed above. However, these existing pictures are likely to represent a gross simplification of the intrinsic and external factors directing developmental processes in the brain. Moving from single genes to networks will require systems level analysis of brain development. Efforts are already underway to map the transcriptome (Kang et al., 2011; Miller et al., 2014) and connectome (Oh et al., 2014) of developing brains, and significant resources will be developed towards developing new technologies. Genomic analysis has led the way, with major accomplishments using RNA-seq to understand transcriptional differences underlying normal and pathogenic brain development (Kang et al., 2008; Miller et al., 2014; Voineagu et al., 2011). In the future, the direct and indirect regulatory interactions that drive brain development will be revealed at a systems level with the level of rigor and detail that is currently only possible via in depth examination of individual circuits.
In contrast to the substantial effort dedicated to mapping TF activity in the developing brain, other aspects of the regulatory circuitry orchestrating normal and pathogenic neurodevelopmental processes remain underexplored. While genome-wide studies have established that the dynamic activity of tens of thousands of enhancers orchestrate spatiotemporal gene expression in forebrain development, mapping and epigenomic and functional characterization of these elements in the human genome remains a priority. Furthermore, technology exists to establish enhancer-promoter interactions (Fullwood and Ruan, 2009; Lieberman-Aiden et al., 2009), which will be critical to understanding the control of loci involved in specific processes or implicated in specific disorders. Out of the tens of thousands of enhancers predicted in functional genomics studies of brain development, for only a fraction have the target genes been predicted with experimental confirmation of required regulatory activity available for only a small subset of these predictions (e.g. (Shim et al., 2012)). It is unlikely that current centralized efforts will have the bandwidth to profile all of the multidimensional axes of brain sub-regions, temporal stages across differentiation and development, and the different lineages represented in the brain. Nonetheless, the technology required to map and characterize regulatory elements is accessible and individual research groups can now profile specific systems of interest. Through these combined efforts, a more complete picture of the location, function, and targets of neural regulatory elements should emerge.
Complementary to characterizing the activity of individual regulatory elements genome-wide across the multidimensional space of brain development is the less studied role of chromatin remodeling and epigenetic marking in brain development. Early studies in this area suggest a central role for chromatin remodeling factors in forebrain development, substantial transcriptional control via chromatin restriction as lineage specification proceeds in the brain, and widespread changes in DNA methylation patterns across brain development (Lister et al., 2013; Ronan et al., 2013; Zhu et al., 2013). Further studies are necessary to understand the mechanism and requirement for these processes in neurodevelopment. Finally, the availability of genomic datasets, functional genomic assays, and efficient genome modification technology will enable deeper exploration of the regulatory circuits and activities of specific TFs in brain development. With new technologies for precision genome engineering emerging (Bedell et al., 2012; Christian et al., 2010; Jinek et al., 2012; Mali et al., 2013), we foresee rapid expansion in the understanding of the interplay of specific genes that control neural lineage specification, regionalization, and function. Through the results of these studies, it will be possible to ask more detailed questions regarding the role of regulatory sequences and chromatin remodeling processes in the development and function of the brain and potentially in the role of non-coding sequence in the evolution of the human brain and cognitive capacity.
Challenge 3: Gene regulation in neurodevelopmental disorders
As described above, neurodevelopmental disorders appear to be driven in part by changes in gene expression levels during development, highlighted by the dosage sensitivity observed in genes identified by CNV and exome screening. The compelling evidence generated by recent attempts to intersect neurodevelopmental transcriptome data with genes linked to autism and schizophrenia is, as of now, more suggestive than conclusive regarding the pathophysiology of these disorders (Parikshak et al., 2013; Voineagu et al., 2011; Willsey et al., 2013). Further work is required to map the affected cell types and document the molecular changes and cell biology associated with changes in expression of susceptibility genes. Additionally, there are two major unexplored potential driver mechanisms behind neurodevelopmental disorders. First is the effect of non-coding sequence variation. As regulatory elements are predicted to direct robust expression patterns in the developing brain, it is likely that sequence variation that changes the function of these elements has the potential to drive changes in gene expression and downstream dosage-sensitivity driven effects. While many GWAS have identified non-coding regions as associated to phenotypes such as autism and schizophrenia, the causal variants have yet to be characterized. In a recent example of a large effect non-coding variant with severe phenotypic consequences in the brain, a short deletion in regulatory sequence was identified in subjects exhibiting gyral abnormalities (Bae et al., 2014). The deletion variant disrupted the regulatory function, leading to a change of expression of isoforms of GPR56 and resulting in changes in the cortical expression pattern of this gene, which resulted in restricted polymicrogyria surrounding the Sylvan fissure that is linked to intellectual and language difficulty and seizures. Another suggestive finding is the duplication of the region containing VIPR2 in schizophrenia, where a minimal duplication of non-coding sequence was observed in cases (Vacic et al., 2011).
The current list of characterized causal variants found in non-coding regions appears exceptionally small when viewed in light of the current prominence and success of screening coding sequence (e.g. exome sequencing). In the future, whole genome analysis will enable unbiased examination of non-coding sequences as well, but there will be major challenges connected to the expected large number of variants and the difficulty of predicting functionality of affected non-coding sequence (Jiang et al., 2013). The expansion of regulatory element mapping and characterization described above will enable screening of non-coding regions. In the meantime, targeted examination of non-coding regions may be a viable intermediate approach, where predicted functional or conserved non-coding elements around susceptibility genes could be screened at a volume sufficient to start to dissect differences between case and control population variation at these loci.
An area poised for large-scale growth in the near future is the characterization of epigenomic or epigenetic changes in the brain (or in induced pluripotent stem cell-derived differentiated neurons) of individuals afflicted with neurological and neurodevelopmental disorders. These studies are confounded by the difficulty of assigning causal relationships for observed differences in driving original pathophysiological processes or in contributing to later manifestations of the traits. Nonetheless, the emergence of consistent patterns associated with specific phenotypes has the potential to provide evidence for a role of stable epigenetic changes that will ultimately lead to better understanding of the forces contributing to neurodevelopmental disorders and could provide a link between environmental factors and long-term changes in gene regulation.
The identification of specific regulatory elements or of widespread epigenetic changes will additionally generate novel targets for therapeutic intervention. Already, major changes are underway in the molecular diagnosis of neurodevelopmental disorders that enable better clinical care (Krystal and State, 2014), as well as the development of promising targeted therapeutic approaches (Gross et al., 2012). The ability to target the CRISPR/Cas9 or TALE systems to specific sequences has been harnessed to build synthetic regulatory proteins that can act to change gene expression or epigenomic markings at specific regulatory elements (Mendenhall et al., 2013). In the not-too-distant future, it may be possible to use synthetic regulators to modify gene dosage of target genes involved in key processes in specific neuronal cell populations implicated via studies of transcriptional regulation in normal and pathogenic brain development. With the combination of new technologies and the rapidly growing understanding of the role of transcriptional regulation in neurodevelopment, this is an exciting and rapidly changing field that has the potential to transform our understanding of the evolution, development and function of the human brain.
Acknowledgments
A.S.N. was supported by NIH/NIGMS NRSA F32 fellowship GM105202. A.V. was supported by National Institute of Neurological Disorders and Stroke grant R01NS062859A and by National Human Genome Research Institute grant R01HG003988. J.L.R.R. was supported by the Nina Ireland, Weston Havens Foundation, NINDS grant R01NS34661, NIMH grant R01MH081880, and NIMH grant R37MH049428. K.P. was supported by NIH NIGMS MSTP Grant T32 GMO7618. Work at the E.O. Lawrence Berkeley National Laboratory was performed under Department of Energy Contract DE-AC02-05CH11231, University of California.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariza-Cosano A, Visel A, Pennacchio LA, Fraser HB, Gómez-Skarmeta J, Irimia M, Bessa J. Differences in enhancer activity in mouse and zebrafish reporter assays are often associated with changes in gene expression. Dev. Biol. 2012;13:713–531. doi: 10.1186/1471-2164-13-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342:1241006–1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim E, Jabaudon D, Fame RM, Macklis JD. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat. Neurosci. 2009;12:1238–1247. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae B-I, Tietjen I, Atabay KD, Evrony GD, Johnson MB, Asare E, Wang PP, Murayama AY, Im K, Lisgo SN, et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 2014;343:764–768. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Fishell G. The Generation of Cortical Interneurons. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing CNS and PNS. Elsevier Inc.; 2013. pp. 503–518. [Google Scholar]
- Batista-Brito R, Rossignol E, Hjerling-Leffler J, Denaxa M, Wegner M, Lefebvre V, Pachnis V, Fishell G. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63:466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, Tan W, Penheiter SG, Ma AC, Leung AYH, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Bengani H, Fish M, Brown A, Divizia MT, de Marco R, Damante G, Grainger R, van Heyningen V, Kleinjan DA. Disruption of autoregulatory feedback by a mutation in a remote, ultraconserved PAX6 enhancer causes aniridia. Am. J. Hum. Genet. 2013;93:1126–1134. doi: 10.1016/j.ajhg.2013.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanović O, Fernández Miñán A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJC, Gómez-Skarmeta JL. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Research. 2012;22:2043–2053. doi: 10.1101/gr.134833.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJB, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008;59:722–732. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SRY. Textura del sistema nervioso del hombre y de los vertebrados. Madrid: Imprenta N. Moya; 1899. [Google Scholar]
- Casci T. Development: Hourglass theory gets molecular approval. Nat. Rev. Genet. 2011;12:76–76. doi: 10.1038/nrg2940. [DOI] [PubMed] [Google Scholar]
- Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MGM, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-JJ, Vogt D, Wang Y, Visel A, Silberberg SN, Nicholas CR, Danjo T, Pollack JL, Pennacchio LA, Anderson S, et al. Use of “MGE Enhancers” for Labeling and Selection of Embryonic Stem Cell-Derived Medial Ganglionic Eminence (MGE) Progenitors and Neurons. PLoS ONE. 2013;8:e61956. doi: 10.1371/journal.pone.0061956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy B, Finlay BL, Darlington RB, Anand KJS. Extrapolating brain development from experimental species to humans. Neurotoxicology. 2007;28:931–937. doi: 10.1016/j.neuro.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close J, Xu H, De Marco García N, Batista-Brito R, Rossignol E, Rudy B, Fishell G. Satb1 is an activity-modulated transcription factor required for the terminal differentiation and connectivity of medial ganglionic eminence-derived cortical interneurons. J. Neurosci. 2012;32:17690–17705. doi: 10.1523/JNEUROSCI.3583-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JLR. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JLR. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat. Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Colasante G, Collombat P, Raimondi V, Bonanomi D, Ferrai C, Maira M, Yoshikawa K, Mansouri A, Valtorta F, Rubenstein JLR, et al. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J. Neurosci. 2008;28:10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Collombat P, Colasante G, Bianchi M, Long J, Mansouri A, Rubenstein JLR, Broccoli V. Inactivation of Arx the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J. Neurosci. 2007;27:4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A NISC Comparative Sequencing Program. Distribution and intensity of constraint in mammalian genomic sequence. Genome Research. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, Gerstein MB, Noonan JP. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Research. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh T-Y, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Research. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Pavlovic B, Pritchard JK, Gilad Y. The functional consequences of variation in transcription factor binding. PLoS Genet. 2014;10:e1004226. doi: 10.1371/journal.pgen.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP. The genomic landscape of cohesin-associated chromatin interactions. Genome Research. 2013;23:1224–1234. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denaxa M, Kalaitzidou M, Garefalaki A, Achimastou A, Lasrado R, Maes T, Pachnis V. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Reports. 2012;2:1351–1362. doi: 10.1016/j.celrep.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Zhu Y, Nord AS, Wylie JN, Akiyama JA, Afzal V, Plajzer-Frick I, Kirkpatrick A, Göttgens B, Bruneau BG, et al. Function-based identification of mammalian enhancers using site-specific integration. Nat. Methods. 2014;11:566–571. doi: 10.1038/nmeth.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 2008;135:1559–1567. doi: 10.1242/dev.015123. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, et al. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JLR. The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus but few neocortical interneurons. J. Neurosci. 2010;30:2812–2823. doi: 10.1523/JNEUROSCI.4228-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development. 2009;136:3841–3851. doi: 10.1242/dev.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Hearn C, Errington M, Cooke S, Grigoriou M, Bliss T, Stylianopoulou F, Pachnis V. Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur. J. Neurosci. 2005;21:2923–2938. doi: 10.1111/j.1460-9568.2005.04141.x. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G, Parnavelas JG. Mutations in ARX Result in Several Defects Involving GABAergic Neurons. Front Cell Neurosci. 2010;4:4. doi: 10.3389/fncel.2010.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J. Cell. Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long J, Hatch G, Rubenstein JLR, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J. Neurosci. 2007;27:5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Berry-Kravis EM, Bassell GJ. Therapeutic Strategies in Fragile X Syndrome: Dysregulated mGluR Signaling and Beyond. Neuropsychopharmacology. 2012;37:178–195. doi: 10.1038/npp.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, et al. Consortium on the Genetics of Schizophrenia (COGS), PAARTNERS Study Group. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehr U, Pineda-Alvarez DE, Uyanik G, Hu P, Zhou N, Hehr A, Schell-Apacik C, Altus C, Daumer-Haas C, Meiner A, et al. Heterozygous mutations in SIX3 and SHH are associated with schizencephaly and further expand the clinical spectrum of holoprosencephaly. Hum. Genet. 2010;127:555–561. doi: 10.1007/s00439-010-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat. Rev. Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DP, Wurst W. Gene and enhancer trapping: mutagenic strategies for developmental studies. Curr. Top. Dev. Biol. 1993;28:181–206. doi: 10.1016/s0070-2153(08)60213-6. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MA, Rubenstein JLR, Baraban SC. Bidirectional homeostatic plasticity induced by interneuron cell death and transplantation in vivo. Proc. Natl. Acad. Sci. U.S.A. 2014;111:492–497. doi: 10.1073/pnas.1307784111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari S, Alkhori L, Schleiffer A, Brochtrup A, Hummel T, Alenius M. Combinatorial Activation and Repression by Seven Transcription Factors Specify Drosophila Odorant Receptor Expression. PLoS Biol. 2012;10:e1001280. doi: 10.1371/journal.pbio.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y-H, Yuen RKC, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, He M, et al. Detection of Clinically Relevant Genetic Variants in Autism Spectrum Disorder by Whole-Genome Sequencing. Am. J. Hum. Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Ahn KS, Heo SY, Won JY, Shim H. Gene targeting in mouse embryos mediated by RecA and modified single-stranded oligonucleotides. In Vitro Cell. Dev. Biol. Anim. 2008;44:57–62. doi: 10.1007/s11626-007-9080-y. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do H-H, Weiss G, Enard W, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Research. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Research. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon K-R, Tang Z, Mathe E, Qian J, Sung M-H, Li G, Resch W, Baek S, Pruett N, Grøntved L, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Itou Y, Yanazawa M, Ohsawa M, Suzuki-Migishima R, Umeki Y, Hohjoh H, Yanagawa Y, Shinba T, Itoh M, et al. Three human ARX mutations cause the lissencephaly-like and mental retardation with epilepsy-like pleiotropic phenotypes in mice. Hum. Mol. Genet. 2009;18:3708–3724. doi: 10.1093/hmg/ddp318. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Krystal JH, State MW. Psychiatric Disorders: Diagnosis to Therapy. Cell. 2014;157:201–214. doi: 10.1016/j.cell.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Nizovtseva EV, Polikanov YS, Ulianov SV, Studitsky VM. Distant activation of transcription: mechanisms of enhancer action. Mol. Cell. Biol. 2012;32:4892–4897. doi: 10.1128/MCB.01127-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa D, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development. 2004;131:3319–3331. doi: 10.1242/dev.01220. [DOI] [PubMed] [Google Scholar]
- Kurokawa D, Ohmura T, Sakurai Y, Inoue K, Suda Y, Aizawa S. Otx2 expression in anterior neuroectoderm and forebrain/midbrain is directed by more than six enhancers. Dev. Biol. 2014;387:203–213. doi: 10.1016/j.ydbio.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HC, Meredith DM, Johnson JE. bHLH Factors in Neurogenesis and Neuronal Subtype Specification. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing Cns and Pns. Elsevier; 2013. pp. 333–354. [Google Scholar]
- Lee AT, Gee SM, Vogt D, Patel T, Rubenstein JL, Sohal VS. Pyramidal neurons in prefrontal cortex receive subtype-specific forms of excitation and inhibition. Neuron. 2014;81:61–68. doi: 10.1016/j.neuron.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Daniels S, Sweeney E, Venkataraman S, Devenney PS, Gautier P, Morrison H, Fantes J, Hill RE, Fitzpatrick DR. Enhancer-adoption as a mechanism of human developmental disease. Hum. Mutat. 2011;32:1492–1499. doi: 10.1002/humu.21615. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJH, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905–1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JLR. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cerebral Cortex. 2009a;19(Suppl 1):i96–i106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JLR. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J. Neurosci. 2007;27:3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Garel S, Depew MJ, Tobet S, Rubenstein JLR. DLX5 regulates development of peripheral and central components of the olfactory system. J. Neurosci. 2003;23:568–578. doi: 10.1523/JNEUROSCI.23-02-00568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JLR. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J. Comp. Neurol. 2009b;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JL, Fame RM, Azim E, Shnider SJ, Molyneaux BJ, Arlotta P, Macklis JD. Specification of Cortical Projection Neurons. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing Cns and Pns. Elsevier; 2013. pp. 475–502. [Google Scholar]
- Machon O, van den Bout CJ, Backman M, Røsok Ø, Caubit X, Fromm SH, Geronimo B, Krauss S. Forebrain-specific promoter/enhancer D6 derived from the mouse Dach1 gene controls expression in neural stem cells. Neuroscience. 2002;112:951–966. doi: 10.1016/s0306-4522(02)00053-2. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Page DT, Merzlyak I, Kim C, Tecott LH, Janak PH, Rubenstein JLR, Sur M. Reduced conditioned fear response in mice that lack Dlx1 and show subtype-specific loss of interneurons. J Neurodev Disord. 2009;1:224–236. doi: 10.1007/s11689-009-9025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Favaro R, Lancini C, Vaccari G, Ferri AL, Bertolini J, Tonoli D, Latorre E, Caccia R, Ronchi A, et al. Emx2 is a dose-dependent negative regulator of Sox2 telencephalic enhancers. Nucleic Acids Res. 2012;40:6461–6476. doi: 10.1093/nar/gks295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh E, Fulp C, Gomez E, Nasrallah I, Minarcik J, Sudi J, Christian SL, Mancini G, Labosky P, Dobyns W, et al. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain. 2009;132:1563–1576. doi: 10.1093/brain/awp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Hyman SE. Progress in the genetics of polygenic brain disorders: significant new challenges for neurobiology. Neuron. 2013;80:578–587. doi: 10.1016/j.neuron.2013.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King M-C. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio LA, Huylebroeck D, Higashi Y, Rubenstein JLR. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77:83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. Mouse ENCODE Consortium. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtha M, Tokcaer-Keskin Z, Tang Z, Strino F, Chen X, Wang Y, Xi X, Basilico C, Brown S, Bonneau R, et al. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat. Methods. 2014;11:559–565. doi: 10.1038/nmeth.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and Dynamics of Human Transcription Factor Regulatory Networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for longrange enhancers. Science. 2003;302:413–413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. Rapid and Pervasive Changes in Genome-wide Enhancer Usage during Mammalian Development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc. Natl. Acad. Sci. U.S.A. 2009;106:22358–22363. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega-Pereira S, Kessaris N, Du T, Kimura S, Anderson SA, Marin O. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59:733–745. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DDM, Stocker AM, Zembrzycki A. Area Patterning of the Mammalian Cortex. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing Cns and Pns. Elsevier Inc.; 2013. pp. 61–85. [Google Scholar]
- O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex Targeted Sequencing Identifies Recurrently Mutated Genes in Autism Spectrum Disorders. Science. 2012a;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg N, Stevison L, Wall JD, Ahituv N. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 2013;9:e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetti PR, Noebels JL. Interneuron, interrupted: molecular pathogenesis of ARX mutations and X-linked infantile spasms. Curr. Opin. Neurobiol. 2012;22:859–865. doi: 10.1016/j.conb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, et al. NISC Comparative Sequencing Program. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman K, Golonzhka O, Lindtner S, Nord AS, Taher L, Hoch R, Silberberg SN, Zhang D, Bin Chen, Zeng H, et al. Transcriptional Regulation of Enhancers Active in Protodomains of the Developing Cerebral Cortex. Neuron. 2014;82:989–1003. doi: 10.1016/j.neuron.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee S-I, Cooper GM, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JLR. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletikos M, Sousa AMM, Sedmak G, Meyer KA, Zhu Y, Cheng F, Li M, Kawasawa YI, Sestan N. Temporal Specification and Bilaterality of Human Neocortical Topographic Gene Expression. Neuron. 2013;81:321–332. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitras L, Yu M, Lesage-Pelletier C, MacDonald RB, Gagné J-P, Hatch G, Kelly I, Hamilton SP, Rubenstein JLR, Poirier GG, et al. An SNP in an ultraconserved regulatory element affects Dlx5/Dlx6 regulation in the forebrain. Development. 2010;137:3089–3097. doi: 10.1242/dev.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JLR. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol. Cell. Neurosci. 2009;40:167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006a;314:786–786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Research. 2006b;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JL. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Quint M, Drost H-G, Gabel A, Ullrich KK, Bönn M, Grosse I. A transcriptomic hourglass in plant embryogenesis. Nature. 2012;490:98–101. doi: 10.1038/nature11394. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An Atlas of Combinatorial Transcriptional Regulation in Mouse and Man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo JL, Bessa J, Hidalgo C, Fernández Miñán A, Tena JJ, Roncero Y, Gómez-Skarmeta JL, Casares F. Identification and analysis of conserved cis-regulatory regions of the MEIS1 gene. PLoS ONE. 2012;7:e33617. doi: 10.1371/journal.pone.0033617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JLR, Campbell K. Neurogenesis in the Basal Ganglia. In: Rubenstein JLR, Rakic P, editors. Patterning and Cell Type Specification in the Developing CNS and PNS. Elsevier; 2013. pp. 455–474. [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RJ, Olson EN. Building the heart piece by piece: modularity of cis-elements regulating Nkx2-5 transcription. Development. 1999;126:4187–4192. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker--a web server for aligning two genomic DNA sequences. Genome Research. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold BA, Stanco A, Cho KKA, Potter GB, Kim C, Sohal VS, Rubenstein JLR, Schreiner CE. Chronic reduction in inhibition reduces receptive field size in mouse auditory cortex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13829–13834. doi: 10.1073/pnas.1205909109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genomewide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbereis JC, Nobuta H, Tsai H-H, Heine VM, McKinsey GL, Meijer DH, Howard MA, Petryniak MA, Potter GB, Alberta JA, et al. Olig1 function is required to repress dlx1/2 and interneuron production in Mammalian brain. Neuron. 2014;81:574–587. doi: 10.1016/j.neuron.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat. Genet. 2013;45:1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanco A, Rubenstein JLR. Unpublished. [Google Scholar]
- Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, Brazma A, Adams DJ, Talianidis I, Marioni JC, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–540. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. Developmental Fate and Cellular Maturity Encoded in Human Regulatory DNA Landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y, Kokura K, Kimura J, Kajikawa E, Inoue F, Aizawa S. The same enhancer regulates the earliest Emx2 expression in caudal forebrain primordium, subsequent expression in dorsal telencephalon and later expression in the cortical ventricular zone. Development. 2010;137:2939–2949. doi: 10.1242/dev.048843. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Teng L, He B, Gao P, Gao L, Tan K. Discover context-specific combinatorial transcription factor interactions by integrating diverse ChIP-Seq data sets. Nucleic Acids Res. 2014;42:e24–e24. doi: 10.1093/nar/gkt1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Aydin S, Koch S, Grotewold L, Rüther U. Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development. 2002;129:3045–3054. doi: 10.1242/dev.129.13.3045. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Ng L, Menon V, Martinez S, Lee C-K, Glattfelder K, Sunkin SM, Henry A, Lau C, Dang C, et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014;83:309–323. doi: 10.1016/j.neuron.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou H-H, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berghe V, Stappers E, Vandesande B, Dimidschstein J, Kroes R, Francis A, Conidi A, Lesage F, Dries R, Cazzola S, et al. Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron. 2013;77:70–82. doi: 10.1016/j.neuron.2012.11.009. [DOI] [PubMed] [Google Scholar]
- van Heyningen V, Williamson KA. PAX6 in sensory development. Hum. Mol. Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Villar D, Flicek P, Odom DT. Evolution of transcription factor binding in metazoans -mechanisms and functional implications. Nat. Rev. Genet. 2014;15:221–233. doi: 10.1038/nrg3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA. Functional autonomy of distant-acting human enhancers. Genomics. 2009a;93:509–513. doi: 10.1016/j.ygeno.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009b;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T, Yang Z, Baraban SC, Rubenstein JLR. Lhx6 Directly Regulates Arx and CXCR7 to Determine Cortical Interneuron Fate and Laminar Position. Neuron. 2014;82:350–364. doi: 10.1016/j.neuron.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waclaw RR, Allen ZJ, Bell SM, Erdélyi F, Szabó G, Potter SS, Campbell K. The zinc finger transcription factor Sp8 regulates the generation and diversity of olfactory bulb interneurons. Neuron. 2006;49:503–516. doi: 10.1016/j.neuron.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Waclaw RR, Ehrman LA, Pierani A, Campbell K. Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J. Neurosci. 2010;30:6944–6953. doi: 10.1523/JNEUROSCI.5772-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Long JE, Flandin P, Pla R, Waclaw RR, Campbell K, Rubenstein JLR. Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants. J. Comp. Neurol. 2013;521:1561–1584. doi: 10.1002/cne.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Lufkin T, Rubenstein JLR. Dlx6 regulates molecular properties of the striatum and central nucleus of the amygdala. J. Comp. Neurol. 2011;519:2320–2334. doi: 10.1002/cne.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dye CA, Sohal V, Long JE, Estrada RC, Roztocil T, Lufkin T, Deisseroth K, Baraban SC, Rubenstein JLR. Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J. Neurosci. 2010;30:5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabĕ de Angelis M, Weinmaster G, Rubenstein JLR. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JLR. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J. Comp. Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Vogt D, Blood A, Hermesz E, Westphal H, Rubenstein JLR. Ldb1 is essential for development of Nkx2.1 lineage derived GABAergic and cholinergic neurons in the telencephalon. Dev. Biol. 2014;385:94–106. doi: 10.1016/j.ydbio.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JLR, Westphal H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9005–9010. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q-P, Le TN, Qiu X, Spencer V, de Melo J, Du G, Plews M, Fonseca M, Sun JM, Davie JR, et al. Identification of a direct Dlx homeodomain target in the developing mouse forebrain and retina by optimization of chromatin immunoprecipitation. Nucleic Acids Res. 2004;32:884–892. doi: 10.1093/nar/gkh233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]