Abstract
The presence of Anopheles (Nyssorhynchus) dunhami Causey in Colombia (Department of Amazonas) is confirmed for the first time through direct comparison of mtDNA cytochrome c oxidase I (COI) barcodes and nuclear rDNA second internal transcribed spacer (ITS2) sequences with topotypic specimens of An. dunhami from Tefé, Brazil. An. dunhami was identified through retrospective correlation of DNA sequences following misidentification as Anopheles nuneztovari s.l. using available morphological keys for Colombian mosquitoes. That An. dunhami occurs in Colombia and also possibly throughout the Amazon Basin, is of importance to vector control programs, as this non-vector species is morphologically similar to known malaria vectors including An. nuneztovari, Anopheles oswaldoi and Anopheles trinkae. Species identification of An. dunhami and differentiation from these closely related species are highly robust using either DNA ITS2 sequences or COI DNA barcode. DNA methods are advocated for future differentiation of these often sympatric taxa in South America.
Keywords: Anopheles dunhami, COI barcodes, ITS2, topotypic, Brazil, Colombia
Anopheles (Nyssorhynchus) dunhami Causey was first described from Tefé, Amazonas (AM), Brazil (Causey 1945), but was placed in synonymy with the malaria vector Anopheles nuneztovari Gabaldón (Lane 1953), where it remained for some decades until taxonomic revisions resulted in its re-elevation (Peyton 1993). In the same study, Peyton also reduced the widespread east-Andean taxon Anopheles trinkae Faran to a junior synonym of An. dunhami, based on similarities in male genitalia (Peyton 1993). Later integrated studies, combining egg morphology, cytogenetics and rDNA sequences of the second internal transcribed spacer (ITS2), confirmed that An. trinkae and An. dunhami were indeed separate taxa and An. trinkae was reinstated to species status (Lounibos et al. 1998). Recently, Calado et al. (2008) reaffirmed the differences between An. dunhami and An. nuneztovari using the morphology of male genitalia and DNA sequences of the ITS2 and a portion of the mitochondrial cytochrome c oxidase I (COI). Overlapping morphological characters between proven malaria vectors An. nuneztovari, An. trinkae and Anopheles oswaldoi (Hayes et al. 1987, Olano et al. 2001, Quiñones et al. 2006, Gutiérrez et al. 2009) and isomorphic non-vector species such as An. dunhami continue to hamper correct species identification in South America and can impact the effectiveness of vector control programs.
As a result of its historically confused taxonomic status, few verified distribution records exist for An. dunhami. Records to date are limited to AM: Tefé (type locality) (Causey 1945, Peyton 1993), Tabatinga (Lounibos et al. 1998) and Parintins (Calado et al. 2008). Although Tabatinga is adjacent to the Colombian border, An. dunhami has never been reported from Colombia and the species is not included in the available morphological keys to Colombian taxa (Suárez et al. 1988, González & Carrejo 2007). Herein, we molecularly characterize topotypic An. dunhami using COI barcodes [after Hebert et al. (2003)] and ITS2 rDNA sequences and, through molecular correlation of sequence data, confirm the presence of An. dunhami in the Colombian Amazon for the first time.
MATERIALS AND METHODS
Mosquito specimens
A total of 4,893 mosquitoes were collected in four departments in Colombia (Antioquia, Amazonas, Caquetá and Norte de Santander) in 2006 as part of the PhD study of Ruiz (2010). Of 574 mosquitoes collected in Amazonas, where An. nuneztovari s.l. is unreported, two specimens were keyed out as An. nuneztovari s.l. using the morphological keys of Suárez et al. (1988) and Faran (1980). DNA was extracted from these two specimens and a further 17 individuals of An. nuneztovari from the departments of Zaragoza (n = 7) and Norte de Santander (n = 10) (Table). Given the morphological similarities between An. nuneztovari and An. dunhami and the reported presence of the latter species in the Brazilian Amazon, DNA was also extracted from abdomens of five archive specimens of An. dunhami collected at the type locality (Tefé, Brazil). Voucher specimens are stored in the mosquito collections of the Smithsonian Institution of National Museum of Natural History (NMNH), Washington DC, USA (Table).
TABLE.
Field codes, collection localities and co-ordinates of specimens of Anopheles dunhami and Anopheles nuneztovari C used in this study, alongside GenBank accessions for second internal transcribed spacer (ITS2) and cytochrome c oxidase I (COI) sequences where appropriate outgroup taxa were chosen based on their close morphological similarity to An. dunhami
| Species | Country, locality (co-ordinates) |
Field code | GenBank ITS2 COI |
|
|---|---|---|---|---|
| An. dunhami | Brazil, Amazonas, Tefé (−03.3207, −64.7235) |
SA26 | HQ020394 | - |
| SA44 | HQ020395 | HQ315878 | ||
| SA79 | HQ020396 | HQ315879 | ||
| SA85 | HQ020397 | HQ315880 | ||
| SA98 | HQ020398 | HQ315881 | ||
| Colombia, Amazonas, Leticia (−04.1158, −69.9522) |
C08921 | HQ020382 | - | |
| C09041 | HQ020383 | HQ315867 | ||
| An. nuneztovari C | Colombia, Antioquia, Zaragoza (07.4847, −74.8679) |
Za-106 | HQ020399 | - |
| Za-110 | HQ020400 | - | ||
| Za-130 | HQ020401 | - | ||
| Za-131 | HQ020402 | - | ||
| Za-158 | HQ020403 | - | ||
| Za-207 | HQ020404 | - | ||
| Za-208 | HQ020405 | - | ||
| Colombia, Norte de Santander, Tibú (08.6403, −72.7372) |
NS-17 | HQ020388 | HQ315872 | |
| NS-62 | HQ020391 | HQ315875 | ||
| NS-71 | HQ020392 | HQ315876 | ||
| NS-72 | HQ020393 | HQ315877 | ||
| NS-130 | HQ020384 | HQ315868 | ||
| NS-135 | HQ020385 | HQ315869 | ||
| NS-139 | HQ020386 | HQ315870 | ||
| NS-144 | HQ020387 | HQ315871 | ||
| NS-243 | HQ020389 | HQ315873 | ||
| NS-319 | HQ020390 | HQ315874 | ||
| Outgroups | ||||
| Anopheles oswaldoi B | Colombia, Antioquia, Nechí (08.1100, −74.7670) |
CO9000 | - | HQ315866 |
| Anopheles rangeli | Brazil, Mato Grosso (−10.2256, −54.9861) |
BR704-40 | - | HQ315865 |
| Anopheles trinkae | Ecuador, Napo, Sardina Yacu (−00.1666, −77.0833) |
SY205 | - | HQ315882 |
| SY227 | - | HQ315883 | ||
Molecular methods
DNA template was acquired using the commercially available DNeasy Blood & Tissue Kit and BioSprint 96 DNA (both QIAgen®, Maryland, USA). Amplification of rDNA ITS2 polymerase chain reaction (PCR) was carried out using the primers of Collins and Paskewitz (1996) and the protocol described by Linton et al. (2001).
Amplification of the 710 bp fragment of the barcoding region of the mitochondrial COI gene was achieved using the primer pair LCO 1490 and HCO 2198 developed by Folmer et al. (1994), using a PCR protocol optimized by the Laboratories of Analytical Biology, Museum Support Center, Smithsonian Institution/NMNH. Each PCR contained 1×NH4 buffer, 0.5 mM each dNTP, 2 mM of MgCl, 0.2 U of Taq polymerase (BioLine, Taunton, MA, USA), 0.3 µM each primer and 1 µL of DNA template and was made up to a total volume of 10 µL using ddH20. The PCR thermocycler parameters included: a single cycle at 95°C for 5 min, followed by 34 cycles of 95°C for 30 s, 48°C for 30 min and 72°C for 45 s, respectively, terminating with a 72°C for 5 min extension step and a 10°C hold. PCR products (both ITS2 and COI) were visualized on 1% agarose gels, containing 0.5 mg/mL of ethidium bromide and purified using ExoSAP-IT® (USB Corporation, Cleveland, Ohio, USA).
Sequencing reactions were carried out in both directions using the Big Dye Terminator Kit® (PE Applied BioSystems, Warrington, England) on an ABI 3730 automated sequencer (PE Applied BioSystems). Sequences were edited using Sequencher™ v.4.8 (Genes Codes Corporation, Ann Arbor, MI) and either aligned automatically in ClustalX (Thompson et al. 1997) or manually using MacClade v.4.06 (Maddison & Maddison 2003). Sequence similarities were compared with those available in GenBank using Basic Local Alignment Search Tool (blast.ncbi.nlm.nih.gov/Blast.cgi). Sequence divergence was calculated in MEGA version 4 (Kumar et al. 2008) using the Kimura 2-parameter distance model (K2P) (Kimura 1980). A bootstrap neighbor joining (NJ) tree (Saitou & Nei 1987) was generated in PAUP version 4.0 (Swofford 2003), using 1,000 replicates of the K2P distance matrices.
COI sequences generated formed part of the activities of the Mosquito Barcoding Initiative (project leader YML) and both full specimen and COI sequence data, including electropherograms, are freely available through the Barcode of Life database website at boldsystems.org. GenBank accession for sequences generated in this study are listed in Table.
RESULTS
ITS2 sequences (n = 5) and COI DNA barcodes (n = 4) were generated from five topotypic archive specimens of An. dunhami from Tefé. ITS2 sequences from 19 Colombian Anopheles, morphologically identified as An. nuneztovari using the keys of Faran (1980) and Suárez et al. (1988) revealed two distinct ITS2 sequences, differing at 17 bases [1 transition (G/A), 1 transversion (C/G) and 15 indels] (Fig. 1).
Fig. 1.
alignment showing differing bases of second internal transcribed spacer sequences of Colombian Anopheles dunhami (546 bp; n = 2) and Anopheles nuneztovari C (549 bp; n = 17) generated in this study in comparison to topotypic An. dunhami from Tefé, Brazil (546 bp; n = 5) and other available sequences available in GenBank (360 bp; EU848337-42) (Calado et al. 2008). Bra: Brazil; Col: Colombia; Par: Parintins; N.S.: Norte de Santander; Zar: Zaragoza.
The most frequent ITS2 sequence was shared by seven specimens from Zaragosa (Antioquia) and 10 specimens from Tibú (Norte de Santander), morphologically identified as An. nuneztovari s.l., whereas a different sequence was shared by two specimens collected in Kilómetro 12, Leticia (Amazonas, Colombia). The most frequent sequence was identical to previously published sequences of An. nuneztovari C from Colombia (Sierra et al. 2004) (AY028081-124, AY028128), Brazil (Marrelli et al. 2005) (AF461749) and Venezuela (Fritz et al. 1994) (L22462).
The two specimens from Kilómetro 12, Leticia showed 100% identity with topotypic An. dunhami sequences and three published An. dunhami sequences from Parintins (EU848337, EU848338 and EU848344) (Calado et al. 2008). However, sequence differences were noted between the topotypic and Colombian specimens and the remaining six available An. dunhami sequences (generated from clones) reported from Parintins. Topotypic and Colombian An. dunhami shared 99.7% identity with EU848340 and EU848341 (1/360 variant bases), 99.4% identity with EU848339, EU848343, EU84845, same haplotype (2/360 variant bases) and 98.9% with EU848342 (4/360 variant bases) (Fig. 1).
MtDNA COI DNA barcode sequences (658 bp without primers) were generated for specimens previously identified by ITS2 sequence data as follow: topotypic An. dunhami from Brazil (n = 4) and Colombia (Kilómetro 12, Leticia) (n = 1) and An. nuneztovari C from Norte de Santander (n = 10). COI barcode data were also generated for An. trinkae (n = 2), An. oswaldoi B (n = 1) and An. rangeli (n = 1), as these species can be morphologically mistaken for An. dunhami or An. nuneztovari s.l. in the female adult stage (Faran 1980).
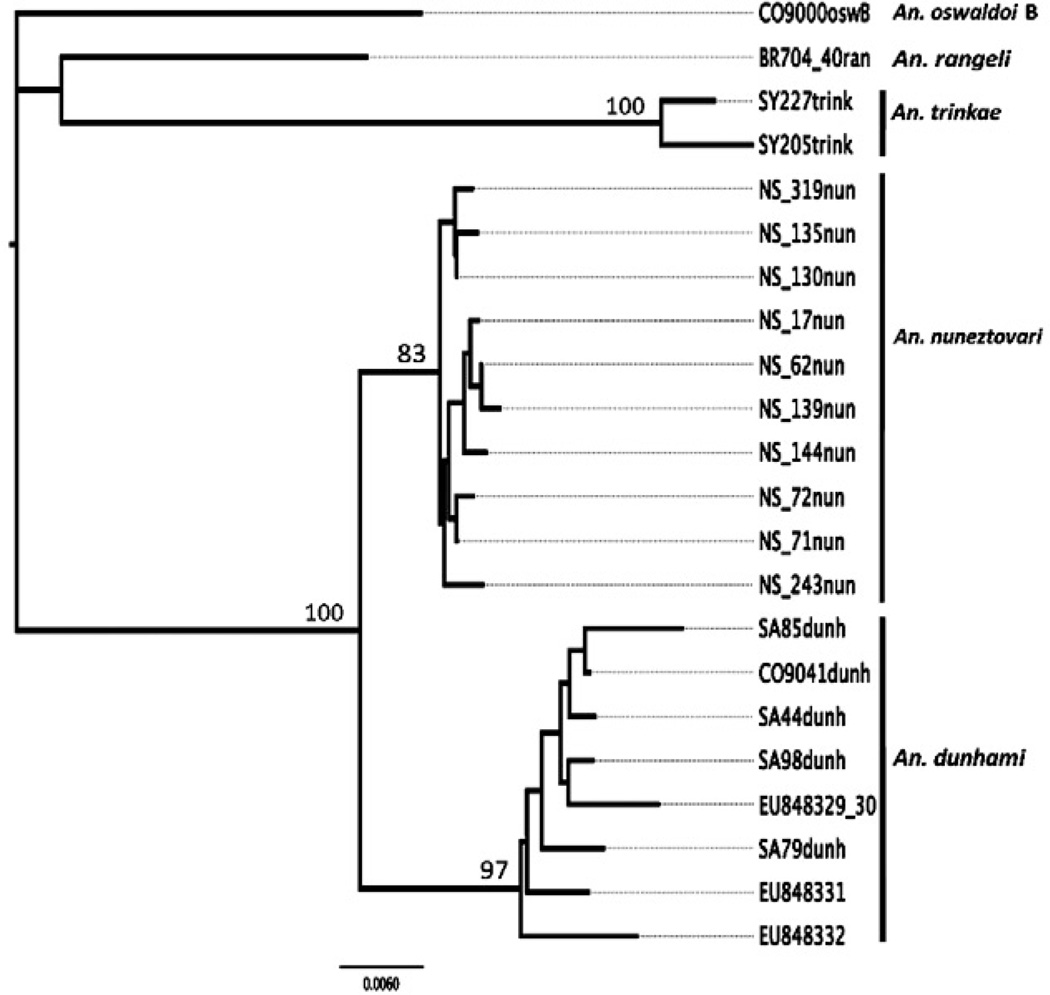
The bootstrap NJ tree (Fig. 2), based on 1,000 replicates of the K2P COI distance matrices (Kimura 1980), reaffirmed the specific identity of topotypic and Colombian An. dunhami and its genetic differentiation from other closely related taxa including An. nuneztovari C, An. trinkae, An. oswaldoi B and An. rangeli obtained by ITS2. Mean intra-specific K2P divergence values were 0.007, 0.004 and 0.010 in An. dunhami (n = 5), An. nuneztovari C (n = 10) and An. trinkae (n = 2), respectively. The mean K2P value between An. dunhami and An. nuneztovari was 0.025, An. dunhami and An. trinkae was 0.093, An. nuneztovari and An. trinkae was 0.085.
Fig. 2.
bootstrap neighbor-joining tree constructed using 1,000 replicates of Kimura two-parameter distance model genetic distance matrices of cytochrome c oxidase unique haplotypes (680 bp) belonging to 23 specimens (only unique haplotype are shown) of Anopheles nuneztovari [Norte de Santander, Colombia (n = 10), GenBank HQ315868-HQ315877], Anopheles dunhami [Kilómetro 12, Amazonas, Colombia (n = 1), HQ315867; Tefé, Amazonas, Brazil (n = 4), HQ315878-HQ315881; EU848330; EU848330 (same haplotype as EU848329), EU848331, EU848332 (Calado et al. (2008)]. Out-groups: Anopheles trinkae (Sardina Yacu, Napo State, Ecuador, HQ315882-HQ315883), Anopheles oswaldoi B (HQ315866) and Anopheles rangeli (HQ315865).
DISCUSSION
This study provides DNA sequence data for topotypic specimens of An. dunhami for the first time, thus establishing the genetic identity of the species. Correlation of the ITS2 rDNA and COI barcodes of Colombian specimens from the Colombian Amazon with topotypic An. dunhami confirms the first record of An. dunhami outside Brazil and comprises a new record for Colombia. That An. dunhami can be morphologically misidentified with vector species, including An. nuneztovari s.l. (as in this study), An. trinkae and An. oswaldoi s.l., is of significance to vector control personnel in Colombia, as it could result in wasted resources, with unnecessary control measures mistakenly being applied against this non-vector species.
Topotypic ITS2 sequences of An. dunhami were less than 1% dissimilar to those reported by Calado et al. (2008). The two remaining GenBank entries labelled as An. dunhami are problematic: U9236 (Tefé) (Lounibos et al. 1998) and AF462378 (Acre, Brazil) (Marrelli et al. 2005). As highlighted by Calado et al. (2008), significant differences exist between the GenBank submission (U9236) and the published sequences in the paper of Lounibos et al. (1998) and the submitted sequence shows 15 base differences (all indels) from our An. dunhami sequences. An author of the present study (JEC) and co-author of Lounibos et al. (1998) confirms that manual sequencing errors are most likely to account for these discrepancies. GenBank entry AF462378 of Marrelli et al. (2005) was more similar (96%) to An. oswaldoi s.s. from Brazil, São Paulo (AF425915), than topotypic An. dunhami 86% (68/496 variant bases).
Although the two An. dunhami specimens detected were field identified by females as An. nuneztovari s.l., these taxa are easily differentiated using either ITS2 or COI sequence data or male genitalia (Calado et al. 2008). An. dunhami and An. nuneztovari C share only 96.9% identity at ITS2 (Fig. 1) and 97.5% at COI (Fig. 2). An. dunhami ITS2 sequences generated in this research share 78–80% identity with An. trinkae sequences in GenBank (U92346, U92355 and Y09075) (Fritz 1998, Lounibos et al. 1998).
Contrary to the low level of intra-specific COI sequence divergence, the mean inter-specific K2P distance between An. dunhami, An. nuneztovari and An. trinkae was 0.067 (range 0.025–0.093). Comparable results were reported in other barcode studies on mosquitoes, i.e. 0.058 between Anopheles fluviatilis and Anopheles minimus (Kumar et al. 2007) and 10.4% (range 0.2–17.2%) for nine genera of Culicidae (Cywinska et al. 2006). In contrast to the ITS2 dataset, using mtDNA markers, An. dunhami and An. nuneztovari appear most closely related with 2.5% sequence divergence, but significantly higher than intra-specific comparisons.
Herein we verified the presence of An. dunhami in Colombia for the first time using both ITS2 sequences and COI barcodes. Although An. dunhami has not been implicated in malaria transmission (Peyton 1993, Lounibos et al. 1998), adult females, the stage most readily collected in epidemiological studies, can be easily misidentified as An. nuneztovari s.l., An. oswaldoi s.l. or An. trinkae, all of which are proven natural vectors of malaria in South America (Faran 1980, Hayes et al. 1987, Herrera et al. 1987, Olano et al. 2001, Quiñones et al. 2006, Gutiérrez et al. 2009). It is possible that An. dunhami may also be a competent vector; however, its role in local malaria transmission is masked by the presence of other morphologically similar species, which are traditionally known as malaria vectors. Knowledge of mosquito species present in vector control regions is important. In Colombia, local vector control strategies vary according to the vector species present and this in turn is driven by the entomological surveys, thus it is now imperative that morphological keys in Colombia are updated to include An. dunhami and to determine novel morphological characters to differentiate this species from An. nuneztovari in the adult female stage if possible. It seems likely that this species will also be present in the Amazonian region of Peru and that its eventual known distribution could include a large portion of the Amazon Basin. Topotypic ITS2 and COI sequences of An. dunhami from Tefé generated in this study will facilitate the identification of this species across its geographical range.
ACKNOWLEDGEMENTS
The authors are especially grateful to La Secretaria de Salud del Amazonas, Colombia. Parts of this research were performed at the Smithsonian Institution under a Memorandum of Understanding between the Walter Reed Army Institute of Research and the Smithsonian Institution, with institutional support provided by both organizations. The material contained within this manuscript has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
Financial support: UNICEF/UNDP/WorldBank/WHO/TDR (A50252 to YML), Canterbury Christ Church University, UK (PhD studentship for FR to DJP and YML), NIH, USA (2R01AI054139 to JEC), Smithsonian Institution, Walter Reed Army Institute of Research
REFERENCES
- Calado DC, Foster PG, Bergo ES, dos Santos CL, Galardo AK, Sallum MA. Resurrection of Anopheles goeldii from synonymy with Anopheles nuneztovari (Diptera, Culicidae) and a new record for Anopheles dunhami in the Brazilian Amazon. Mem Inst Oswaldo Cruz. 2008;103:791–799. doi: 10.1590/s0074-02762008000800009. [DOI] [PubMed] [Google Scholar]
- Causey OR. Description of Anopheles (Nyssorhynchus) dunhami, a new species from the upper Amazon basin. J Natl Malar Soc. 1945;4:231–234. [Google Scholar]
- Collins FH, Paskewitz SM. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol Biol. 1996;5:1–9. doi: 10.1111/j.1365-2583.1996.tb00034.x. [DOI] [PubMed] [Google Scholar]
- Cywinska A, Hunter FF, Hebert PD. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20:413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- Faran ME. Mosquito studies (Diptera: Culicidae) XXXIV. A revision of the Albimanus section of the subgenus Nyssorhynchus of Anopheles . Cont Am Entomol Inst. 1980;15:1–215. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Fritz GN. Sequence analysis of the rDNA internal transcribed spacer 2 of five species of South American human malaria mosquitoes. DNA Seq. 1998;8:215–221. doi: 10.3109/10425179809008454. [DOI] [PubMed] [Google Scholar]
- Fritz GN, Conn J, Cockburn A, Seawright J. Sequence analysis of the ribosomal DNA internal transcribed spacer 2 from populations of Anopheles nuneztovari (Diptera: Culicidae) Mol Biol Evol. 1994;11:406–416. doi: 10.1093/oxfordjournals.molbev.a040122. [DOI] [PubMed] [Google Scholar]
- González R, Carrejo NS. Claves y notas de distribución. Universidad del Valle, Cali: 2007. Introducción al estudio taxonómico de Anopheles de Colombia ; p. 237. [Google Scholar]
- Gutiérrez LA, González JJ, Gómez GF, Castro MI, Rosero DA, Luckhart S, Conn JE, Correa MM. Species composition and natural infectivity of anthropophilic Anopheles (Diptera: Culicidae) in the states of Córdoba and Antioquia, Northwestern Colombia. Mem Inst Oswaldo Cruz. 2009;104:1117–1124. doi: 10.1590/s0074-02762009000800008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J, Calderon G, Falcon R, Zambrano V. Newly incriminated anopheline vectors of human malaria parasites in Junin Department, Peru. J Am Mosq Control Assoc. 1987;3:418–422. [PubMed] [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera S, Suárez MF, Sanchez GI, Quiñones ML, de Herrera M. Uso de la técnica radioinmunoensayo IRMA en Anopheles de Colombia para la detección de esporozoitos de Plasmodium . Colomb Med. 1987;18:2–6. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) J Med Entomol. 2007;44:1–7. doi: 10.1603/0022-2585(2007)44[1:dbcdso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J. Neotropical Culicidae. Vol. 1. São Paulo: University of São Paulo; 1953. p. 548. [Google Scholar]
- Linton Y-M, Harbach RE, Anthony TG, Chang MS, Asmad M. Morphological and molecular identity of Anopheles (Cellia) sundaicus (Diptera: Culicidae), the nominotypical member of a malaria vector species complex in Southeast Asia. Syst Entomol. 2001;26:357–366. [Google Scholar]
- Lounibos LP, Wilkerson RC, Conn JE, Hribar LJ, Fritz GN, Danoff-Burg JA. Morphological, molecular and chromosomal discrimination of cryptic Anopheles (Nyssorhynchus) (Diptera: Culicidae) from South America. J Med Entomol. 1998;35:830–838. doi: 10.1093/jmedent/35.5.830. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. MacClade: analysis of phylogeny and character evolution, version 4.06. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Marrelli MT, Floeter-Winter LM, Malafronte RS, Tadei WP, Lourenço-de-Oliveira R, Flores-Mendoza C, Marinotti O. Amazonian malaria vector anopheline relationships interpreted from ITS2 rDNA sequences. Med Vet Entomol. 2005;19:208–218. doi: 10.1111/j.0269-283X.2005.00558.x. [DOI] [PubMed] [Google Scholar]
- Olano VA, Brochero HL, Sáenz R, Quiñones ML, Molina JA. Mapas preliminares de la distribución de especies de Anopheles vectores de malaria en Colombia. Biomedica (Bogotá) 2001;21:402–408. [Google Scholar]
- Peyton EL. Anopheles (Nyssorhynchus) dunhami, resurrected from synonymy with Anopheles nuneztovari and validated as a senior synonym of Anopheles trinkae (Diptera: Culicidae) Mosq Syst. 1993;25:151–156. [Google Scholar]
- Quiñones ML, Ruiz F, Calle DA, Harbach RE, Erazo HF, Linton Y-M. Incrimination of Anopheles (Nyssorhynchus) rangeli and An. (Nys.) oswaldoi as natural vectors of Plasmodium vivax in Southern Colombia. Mem Inst Oswaldo Cruz. 2006;101:617–623. doi: 10.1590/s0074-02762006000600007. [DOI] [PubMed] [Google Scholar]
- Ruiz F. PhD Thesis. Canterbury: Canterbury Christ Church University; 2010. Systematics of the Anopheles (Nyssorhynchus) oswaldoi group in Latin America ; p. 189. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sierra DM, Velez ID, Linton Y-M. Malaria vector Anopheles (Nyssorhynchus) nuneztovari comprises one genetic species in Colombia based on homogeneity of nuclear ITS2 rDNA. J Med Entomol. 2004;41:302–307. doi: 10.1603/0022-2585-41.3.302. [DOI] [PubMed] [Google Scholar]
- Suárez MF, Quiñones M, Fleming GA, Robayo M. Guía introductoria a la morfología de Anopheles y clave para determinación de las principales especies de Colombia. Bogotá: DCD, Ministerio de Salud; 1988. p. 120. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony, version 4.0b 10. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal-X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]