To the editor
Most next-generation sequencing (NGS) quality scores are space intensive, redundant and often misleading. In this Correspondence, we recover quality information directly from sequence data using a compression tool named Quartz, rendering such scores redundant and yielding substantially better space and time efficiencies for storage and analysis. Quartz is designed to operate on NGS reads in FASTQ format, but can be trivially modified to discard quality scores in other formats for which scores are paired with sequence information. Discarding 95% of quality scores counterintuitively resulted in improved SNP calling, implying that compression need not come at the expense of accuracy.
Advances in next-generation sequencing (NGS) technologies have produced a deluge of genomic information, outpacing increases in our computational resources1, 2. This avalanche of data enables large-scale population studies (e.g., maps of human genetic variation3, reconstruction of human population history4, and uncovering cell lineage relationships5), but to fully capitalize on these advances, we must develop better technologies to store, transmit, and process genomic data.
The bulk of NGS data typically consists of read sequences, in which each base call is associated with a corresponding quality score, which consumes at least as much storage space as the base calls themselves6. Quality scores are often essential for assessing sequence quality (their main use), filtering low quality reads, assembling genomic sequences, mapping reads to a reference sequence and performing accurate genotyping. Because quality scores require much space to store and transmit, they are a major bottleneck in any sequence analysis pipeline, impacting genomic medicine, environmental genomics, and the ability to find signatures of selection within large sets of closely related sequenced individuals.
At the expense of downstream analysis (e.g. variant calling, genotype phasing, disease gene identification, read mapping, and genome assembly), biomedical researchers have typically discarded quality scores altogether or turned to compression, which has been moderately successful when applied to genomic sequence data7, 8, 9, 10, 11, 12. Quality score compression is usually lossy, meaning that maximum compression is achieved at the expense of the ability to reconstruct the original quality values13, 14. Due to decline in downstream accuracy, such methods are suboptimal for both transmission and indefinite storage of quality scores. To address these limitations, several newly-developed methods exploit sequence data to boost quality score compression using alignments to a reference genome8, 10, 15 or use raw read datasets without reference alignment16; however, reference-based compression requires runtime-costly whole-genome alignments of the NGS dataset, while alignment-free compression applies costly indexing methods directly to the read dataset. On the other hand, quality score recalibration methods, such as found in GATK17, increase variant calling accuracy at the cost of significantly decreasing compressibility of the quality scores (Supplementary Table S1). To our knowledge, no existing approach simultaneously provides a scalable method for terabyte-sized NGS datasets and addresses the degradation of downstream genotyping accuracy that results from lossy compression10.
To achieve scalable analyses, we take advantage of redundancy inherent in NGS read data. Intuitively, the more often we see a read sequence in a dataset, the more confidence we have in its correctness; thus, its quality scores are less informative and useful. However, for longer read sequences (e.g., >100bp), the probability of a read appearing multiple times is extremely low. For such long reads, shorter substrings (k-mers) can instead be used as a proxy to estimate sequence redundancy. By viewing individual read datasets through the lens of k-mer frequencies in a corpus of reads, we are able to ensure that ‘lossiness’ of compression does not deleteriously affect accuracy.
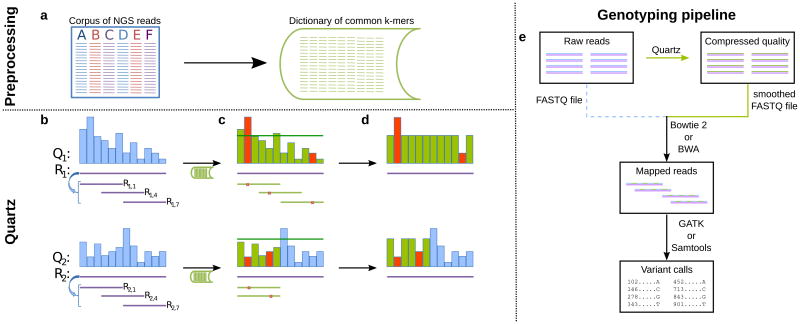
Here we present a highly efficient and scalable compression tool, Quartz (QUAlity score Reduction at Terabyte scale), which compresses quality scores by capitalizing on sequence redundancy. Compression is achieved by smoothing a large fraction of quality score values based on the k-mer neighborhood of their corresponding positions in the read sequences (Fig. 1, left panel). We used the hypothesis that any divergent base in a k-mer likely corresponds to either a SNP or sequencing error; thus, we only preserve quality scores for probable variant locations and compress quality scores of concordant bases by resetting them to a default value. More precisely, frequent k-mers in a large corpus of NGS reads correspond to a theoretical consensus genome with overwhelming probability18; without having to do any explicit mapping, Quartz preserves quality scores at locations that potentially differ from this consensus genome. k-mer frequencies have been used to infer knowledge about the error content of a read sequence—in fact, many sequence-correction and assembly methods directly or indirectly make use of this phenomenon19, 20—but never for quality score compression.
Figure 1. Compressive quality scores.
Quartz algorithm. (a) A dictionary of common k-mers (green lines) in a corpus of NGS reads is generated. The dictionary is generated once for any species. (b) Each read sequence R is broken up into overlapping supporting k-mers (purple). (c) Dictionary k-mers that are within one mismatch from the supporting k-mers are identified (mismatch positions in red). Top: Every position different from a dictionary k-mer is annotated as a possible variant (in red) unless covered by a dictionary k-mer corresponding to a different supporting k-mer, in which case it will nevertheless be marked for correction (in green; e.g. the second mismatch). Other covered positions are also marked for correction as high quality (green). Bottom: When two dictionary k-mers correspond to the same supporting k-mer, all mismatches are preserved, unless the mismatch position is covered by a dictionary k-mer corresponding to a different supporting k-mer as shown top. Uncovered bases are also annotated (blue). (d) Quality scores are smoothed and the scores of all high-quality positions (i.e. bases) are set to a default value. Scores of uncovered and possible variant loci are kept. (e) Quartz can fit into existing genotyping analysis pipelines as an additional processing step between acquisition of raw reads and mapping and genotyping.
Unlike other quality score compression methods, Quartz simultaneously maintains genotyping accuracy while achieving high compression ratios, and is able to do so in orders of magnitude less time. Compression is made possible by Quartz's “coarse” representation of quality scores, which allows it to store quality scores at roughly 0.4 bits per value (from the original size of 8 bits in FASTQ format or 1.4 bits even after standard lossless text compression) (Supplementary Table S2). It is important to note that the compression ratio achieved by Quartz is primarily dependent on the conditional entropy of observing the read sequence, given the local consensus k-mer landscape. This advance is in contrast to lossless compressors, which can reproduce the original quality scores with perfect fidelity but are dependent on the entropy of the quality scores themselves, or to lossy methods that directly reduce the quality score entropy by quality score smoothing procedures13, 6, 21.
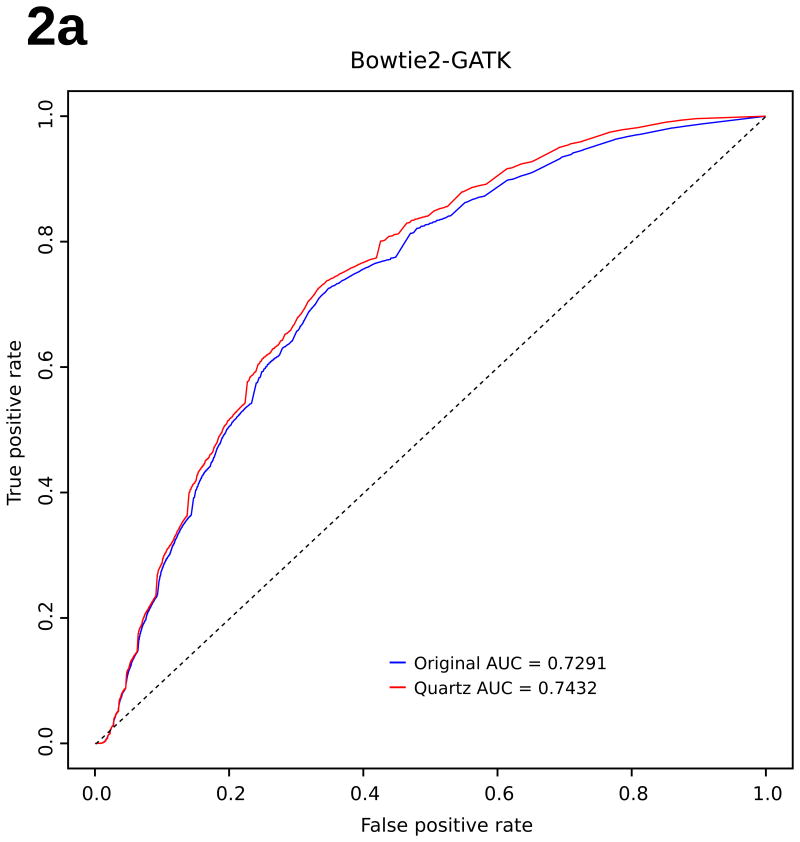
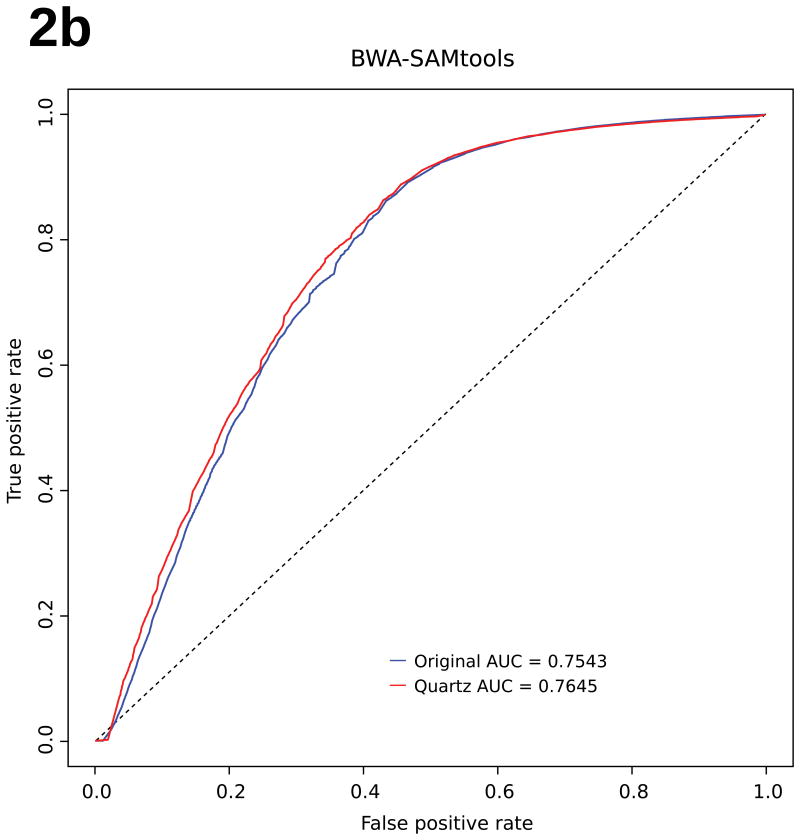
Surprisingly, by taking advantage of the local consensus k-mer landscape, Quartz, while eliminating more than 95% of the quality score information, achieves improved genotyping accuracy compared with using the original, uncompressed quality scores as measured against a trio-validated (i.e. validated against parents' genomes), gold-standard variant dataset for the NA12878 genome from the GATK “best-practices” bundle17 (Fig. 2, Supplementary Information: Methods). We applied both the GATK17 and SAMtools22 pipelines (Fig. 1, right panel) to the compressed quality scores generated by Quartz on a commonly-used NA12878 benchmarking dataset from the 1000 Genomes Project3 (Supplementary Figs. S1-S4). The genotyping accuracy based on Quartz's compressed data consistently outperforms that based on the uncompressed raw quality scores as measured by the area under the receiver operating characteristic (ROC) curve (Supplementary Tables S5-S6); for the experiments in Fig. 2, Quartz compression decreases the number of false positives in the million highest quality variant calls by over 4.5% in several of the pipelines (Supplementary Figs. S1-S2). While this improvement is most pronounced for SNP calls, indel-calling accuracy is also maintained, if not improved, by Quartz compression (Supplementary Figs. S10-S11). This result emerges from the discovery through the application of Quartz that quality score values within an NGS dataset are implicitly encoded in the genomic sequence information with 95% redundancy, so often do not have to be stored separately. This improvement further indicates that compression achieved using Quartz reduces the noise in the raw quality scores, thus leading to better genotyping results. Notably, removing all quality scores (by setting them all to a default value of 50) caused an enormous drop in genotyping accuracy (∼5% decrease in relative ROC AUC) (Supplementary Fig. S5), indicating that retaining quality scores is necessary.
Figure 2. Scaled ROC Curves.
Genotyping accuracy. Scaled ROC curves of genotyping accuracy for NA12878, before (blue) and after Quartz compression (red), using (a) Bowtie 2 and GATK UnifiedGenotyper, and (b) BWA and SAMtools mpileup. Accuracy is improved under both variant-calling pipelines.
Quartz is also scalable for use on large-scale, whole-genome datasets. After a one-time construction of the k-mer dictionary for any given species, quality score compression is orders of magnitude faster than read mapping, genotyping, and other quality score compression methods (Supplementary Table S1 and Supplementary Figs. S6-S7). Additionally, Quartz is especially applicable for large-scale cohort-based sequencing projects, because its improvements in genotyping accuracy are particularly useful when samples have lower depths of sequencing coverage (e.g., 2×-4×) (Supplementary Fig. S8).
Quartz alters quality scores and improves downstream genotyping accuracy, in common with some existing base quality score recalibration tools17 that make use of human genome variation from population-scale sequencing3, for example the GATK BaseRecalibrator. While currently available recalibration tools and Quartz both use distilled information from genome sequences, they differ in several important ways. Most importantly, quality score recalibration tools do not apply compression; in fact, GATK recalibration tends to greatly increase the amount of storage needed, while also requiring much more computing power (Supplementary Table S1); Quartz avoids losing compressibility by using a single default replacement quality value. Furthermore, because recalibration tools such as the GATK BaseRecalibrator employ a list of known SNP locations, reads must first be mapped to the reference. As Quartz uses only k-mer frequencies and Hamming distances, it is possible to apply Quartz compression upstream of mapping, which is crucial for either genome assembly or mapping.
Quartz is the first scalable, sequence-based quality score compression method that can efficiently compress quality scores of terabyte-sized (or larger) sequencing datasets, thereby solving both the problems of indefinite storage and transmission of quality scores. Had our results merely replicated the genotyping accuracy of existing tools such as GATK and SAMTools, we would have still demonstrated order of magnitude improved storage efficiency due to compression, at almost no additional computational cost (Supplementary Materials). However, our results further suggest that a significant proportion of quality score data, despite having been thought entirely essential to downstream analysis, is less informative than the k-mer sequence profiles, and can be discarded without weakening (and sometimes improving), downstream analysis. Even with aggressive lossy data compression, we have shown that it is possible to preserve biologically important data.
A Quartz compression step can be added to almost any pre-existing NGS data processing pipeline. Quartz takes as input a FASTQ file (the standard format for read data) and outputs a smoothed FASTQ file, which can in turn be input into any compression program (e.g., BZIP2 or GZIP) for efficient storage and transmission, or any read mapper (e.g., BWA23, Bowtie 224). Further analysis steps such as variant calling (e.g., using SAMTools, GATK) can be carried out in the usual way. Our optimized and parallelized implementation of Quartz is available, along with a high-quality human genome k-mer dictionary (Supplementary Software). We also provide here preliminary results on how Quartz changes compression levels and variant-calling accuracy on an E. coli genome, indicating Quartz's utility beyond human genomics (Supplementary Fig. S12).
With improvements in sequencing technologies increasing the pace at which genomic data is generated, quality scores will require ever greater amounts of storage space; compressive quality scores will become crucial to fully realizing the potential of large-scale genomics. We show here that unlike previous results21, the twin goals of compression and accuracy do not have to be at odds with each other. Although total compression comes at the cost of accuracy (Supplementary Fig. S6) and quality score recalibration generally decreases compressibility (Supplementary Table S1), there is a happy medium at which we can get good compression and improved accuracy. The Quartz software will greatly benefit any researchers who are generating, storing, mapping, or analyzing large amounts of DNA, RNA, Chip-seq, or exome sequencing data.
Supplementary Material
Acknowledgments
We thank L. Cowen and N. Daniels for helpful discussions and comments. Y.W.Y. gratefully acknowledges support from the Fannie and John Hertz Foundation. D.Y., J.P., and B.B. are partially supported by NIH GM108348.
Footnotes
Software: available at http://quartz.csail.mit.edu and in Supplementary Software.
Competing Financial Interests. The authors declare no competing financial interests.
Contributor Information
Y. William Yu, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA; Department of Mathematics at MIT.
Deniz Yorukoglu, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Jian Peng, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA; Department of Mathematics at MIT.
Bonnie Berger, Email: bab@mit.edu, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA; Department of Mathematics at MIT.
References
- 1.Berger B, Peng J, Singh M. Nature Reviews Genetics. 2013;14:333–346. doi: 10.1038/nrg3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn SD. Science. 2011;331:728–729. doi: 10.1126/science.1197891. [DOI] [PubMed] [Google Scholar]
- 3.The 1000 Genomes Project Consortium. Nature. 2012;491:56–65. [Google Scholar]
- 4.Veeramah KR, Hammer MF. Nature Reviews Genetics. 2014;15:149–162. doi: 10.1038/nrg3625. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro E, Biezuner T, Linnarsson L. Nature Reviews Genetics. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 6.Bonfield JK, Mahoney MV. PloS one. 2013;8(3):e59190. doi: 10.1371/journal.pone.0059190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apostolico A, Lonardi S. Data Compression Conference, 2000. Proceedings. 2000;2000:143–152. [Google Scholar]
- 8.Kozanitis C, Saunders C, Kruglyak S, Bafna V, Varghese G. Journal of Computational Biology. 2011;18(3):401–413. doi: 10.1089/cmb.2010.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones DC, Ruzzo WL, Peng X, Katze MG. Nucleic Acids Research. 2012;40(22):e171–e171. doi: 10.1093/nar/gks754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritz MHY, Leinonen R, Cochrane G, Birney E. Genome Research. 2011;21:734–740. doi: 10.1101/gr.114819.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deorowicz S, Grabowski S. Bioinformatics. 2011;27(6):860–862. doi: 10.1093/bioinformatics/btr014. [DOI] [PubMed] [Google Scholar]
- 12.Loh PR, Baym M, Berger B. Nature Biotechnology. 2012;30:627–630. doi: 10.1038/nbt.2241. [DOI] [PubMed] [Google Scholar]
- 13.Ochoa I, et al. BMC Bioinformatics. 2013;14:187. doi: 10.1186/1471-2105-14-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hach F, Numanagic I, Alkan C, Sahinalp SC. Bioinformatics. 2012;28(23):3051–3057. doi: 10.1093/bioinformatics/bts593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christley S, Lu Y, Li C, Xie X. Bioinformatics. 2009;25(2):274–275. doi: 10.1093/bioinformatics/btn582. [DOI] [PubMed] [Google Scholar]
- 16.Janin L, Rosone G, Cox AJ. Bioinformatics. 2013;29(19):2490–2493. [Google Scholar]
- 17.DePristo MA, et al. Nature Genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu YW, Yorukoglu D, Berger B. Research in Computational Molecular Biology, Springer International Publishing. 2014;8394:385–399. doi: 10.1007/978-3-319-05269-4_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley DR, Schatz MC, Salzberg SL. Genome Biology. 2010;11(11):R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabherr MG, et al. Nature Biotechnology. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canovas R, Moffat A, Turpin A. Bioinformatics. 2014;30(15):2130–2136. doi: 10.1093/bioinformatics/btu183. [DOI] [PubMed] [Google Scholar]
- 22.Li H, et al. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B, Salzberg SL. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.