Abstract
The chromosomal DNA replication in eukaryotic cells begins at replication initation sites, which are marked by the assembly of the pre-replication complexes in early G1. At the G1/S transition, recruitment of additional replication initiation proteins enables origin DNA unwinding and loading of DNA polymerases. We found that depletion of the human DNA helicase B (HDHB) inhibits the initiation of DNA replication, suggesting a role of HDHB in the beginning of the DNA synthesis. To gain insight into the function of HDHB during replication initiation, we examined the physical interactions of purified recombinant HDHB with key initiation proteins. HDHB interacts directly with two initiation factors TopBP1 and Cdc45. In addition we found that both, the N-terminus and helicase domain of HDHB bind to the N-terminus of Cdc45. Furthermore depletion of HDHB from human cells diminishes Cdc45 association with chromatin, suggesting that HDHB may facilitate Cdc45 recruitment at G1/S in human cells.
Keywords: Human DNA Helicase B, Cdc45, DNA replication initiation
Introduction
Eukaryotic chromosome replication is a precisely regulated process that ensures rapid and complete duplication of the genome once per cell cycle. The process consists of two steps separated in time: the assembly of a multi-protein pre-replication complex (pre-RC) on chromosomal origins of replication in late mitosis and early G1 phase, followed by the activation of pre-RCs by S-phase protein kinases [1,2]. From the G1/S transition until mitotic exit at the end of the cell cycle, S-phase and mitotic kinase activities, as well as targeted proteolysis, prevent the assembly of new pre-RCs. Faulty regulation of these processes can lead to genomic instability, apoptosis, and/or tumorigenesis [2,3,4].
The origin recognition complex (ORC) [5], composed of six-subunit Orc1-6, marks potential origins [6]and, together with Cdc6 and Cdt1, recruits and loads the hexameric MCM2-7 (mini-chromosome maintenance) complex to license the origin in G1 [2,7,8,9]. Replication origin unwinding and loading of the DNA polymerases requires additional proteins forming the pre-initiation complex (pre-IC), which includes the GINS complex, Cdc45, Mcm10 and the action of the Dbf4-dependent protein kinase (DDK), and S-phase cyclin-dependent protein kinases (CDK) [10,11,12,13,14,15,16,17,18,19,20,21,22]. Loading of DNA polymerases epsilon, alpha, and delta at the origin is thought to complete the assembly required for initiation.
However, this activation pathway is not yet well understood, particularly in metazoans. In the budding yeast, CDK1 phosphorylates Sld2 and Sld3 proteins, stimulating their binding to two pairs of BRCT-repeats in the conserved protein Dpb11, an essential step for the switch from G1 to S phase [23,24,25]. CDK phosphorylation of yeast Sld3 strengthens its association with Cdc45, enabling Cdc45 loading on chromatin [24,26,27,28]. The Xenopus homolog of Dpb11, Xmus101, is needed to load Cdc45 on chromatin, but unlike Dpb11, it associates with ORC on chromatin prior to S-phase kinase action [19,20,29]. Similarly, TopBP1, the human homolog of Dpb11, interacts with Cdc45 and is required for the G1/S transition [30,31,32]. RecQL4, a conserved vertebrate 3′-5′ DNA helicase of superfamily II, is thought to fulfill the functions of Sld2 in Xenopus extracts because it contains limited Sld2 homology, binds to Xenopus homolog of Dpb11, Xmus101, and is required to initiate replication [33,34,35]. However, unlike Sld2 in budding yeast, RecQL4 is not required for Xmus101 or Cdc45 recruitment to chromatin [33,34]. Recently, treslin was identified as the functional homolog of Sld3 in higher eukaryotes [36,37,38,39,40]. In addition in human cells further protein, such as MTBP were identified, which interact with Treslin/TICRR and regulate DNA replication initiation [36]. Thus, the pathway for assembly and activation of pre-RCs and pre-ICs in vertebrates are more complex and may diverge from that in fungi [24].
DNA helicase B (DHB) was purified as a thermo-labile DNA-dependent ATPase from a temperature-sensitive mouse cell line defective in S phase entry and was later shown to be a robust 5′-3′DNA helicase of superfamily I [41,42,43,44,45]. DHB is conserved among vertebrates, but homologs in lower eukaryotes are not evident by sequence comparison. Characterization of recombinant human DHB (HDHB) confirmed the biochemical properties of the mouse homolog [45]. The protein consists of three functional regions: an uncharacterized N-terminal region, a central helicase domain, and a C-terminal domain containing a nuclear localization signal and a nuclear export signal activated by CDK-dependent phosphorylation of Ser967 at G1/S [46](Figure 1A). Thus HDHB is primarily nuclear during G1 and primarily cytoplasmic during S/G2. Interestingly, HDHB relieves RPA-mediated inhibition of RNA primer synthesis by DNA polymerase alpha-primase on single-stranded DNA, a property typically associated with primosome activity that is needed for replication [44,45,47]. Importantly, microinjection of purified recombinant HDHB protein with a nonfunctional Walker B motif into cells in early G1 inhibited the G1/S transition, whereas the wild type protein had no effect [44]. Taken together, the thermo-labile onset of S phase in mouse cells expressing mutant HDHB and the ability of injected Walker B mutant HDHB protein to block G1/S progression in human cells led us to ask whether endogenous HDHB is required for initiation of chromosomal replication and if so, what role it plays.
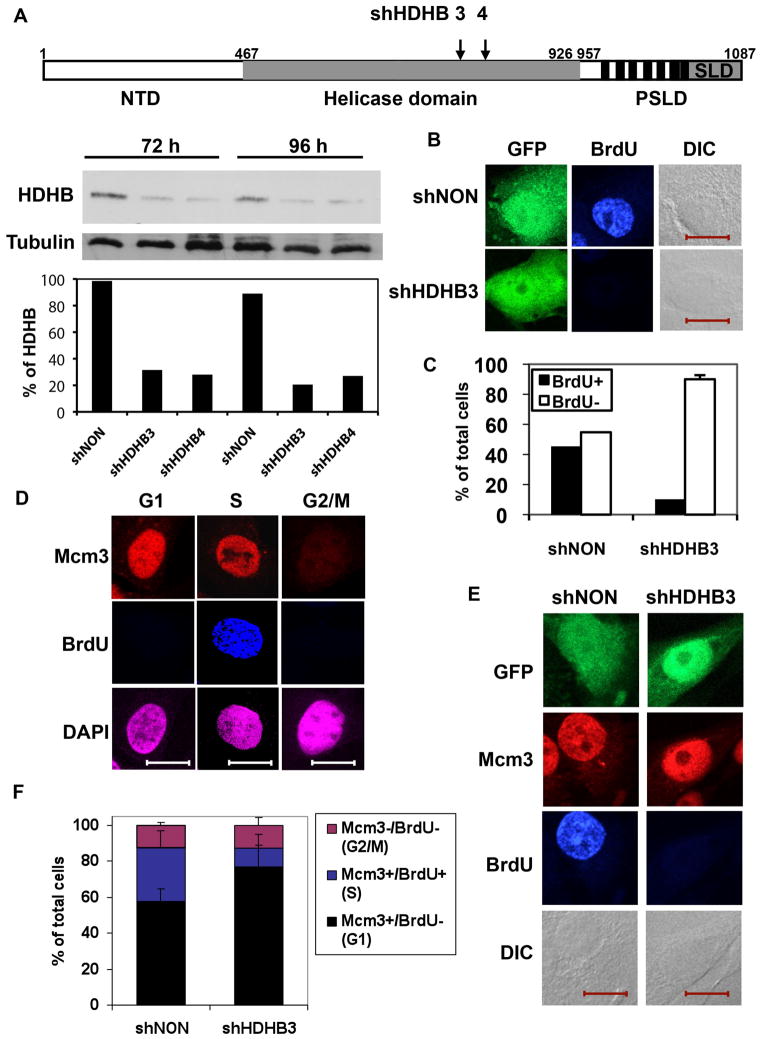
Figure 1. HDHB-depleted cells fail to initiate DNA replication.
(A) Top; Functional regions of HDHB. An N-terminal domain (NTD) is uncharacterized. The central domain (gray) (residues 467–926) contains the seven superfamily I helicase motifs. The C-terminal domain contains consensus CDK phosphorylation sites (vertical black bars) and a subcellular localization domain (SLD), which together constitute a phosphorylation-dependent subcellular localization domain (PSLD) [46]. The positions of short hairpin (sh) RNAs shHDHB3 and shHDHB4 sequences are indicated. Below; Protein level of HDHB in U2OS cells expressing shHDHB3, shHDHB4, or shNON were analyzed by western blot. Diagram; Western blot was quantified by ImageJ64 program and the percentage of band intensity was calculated for each cell line. (B, C) U2OS cells co-expressing GFP and shHDHB3 or shNON were incubated with BrdU for 45 min, stained with anti-BrdU antibody, and examined by indirect immunofluorescence microscopy and differential interference contrast (DIC). Scale bar, 15 μm. (C) The fraction of BrdU-stained cells expressing shNON (n=82) and shHDHB3 (n=78) in two independent experiments is shown. Brackets indicate standard deviation. (D) U2OS cells synchronized in G1, S and G2/M were incubated with BrdU for 45 min before fixation, stained with antibodies specific for Mcm3 and BrdU, and visualized by indirect immunofluorescence. Scale bar, 15 μm. (E, F) U2OS cells expressing shHDHB3 or shNON were incubated with BrdU for 45 min, and BrdU and Mcm3 were visualized by indirect immunofluorescence. Scale bar, 15 μm. (F) The fraction of BrdU-, Mcm3-, and double-positive cells in (E) was determined in two independent experiments.
Here we studied the interaction between HDHB and replication intiation proteins like TopBP1 and Cdc45. We found that the N-terminus and helicase domain of HDHB binds to the N-teminus of the replication initiation protein Cdc45 in vitro. Furthermore we present evidence that HDHB is required for replication initiation and loading of Cdc45 onto chromatin. Defective loading of Cdc45 on chromatin in HDHB-depleted cells, together with direct physical interactions of HDHB with TopBP1 and Cdc45, suggest that HDHB might have a function in initiation of mammalian chromosomal DNA replication.
Results
HDHB depletion prevents DNA synthesis
Published evidence suggests that HDHB is needed for the G1/S transition, based on a temperature-sensitive arrest of mutant cells at G1/S [43,45] and on inhibition of G1/S progression by a nonfunctional HDHB protein [44]. In mammalian cells depletion of proteins can be approximated by siRNA/shRNA silencing. Thus, if HDHB were required for replication initiation, one would predict that transient depletion of HDHB protein should compromise initiation of DNA replication. To deplete HDHB, we transfected human cells with a pGIPZ vector that co-expresses GFP with an shRNA that targets HDHB (shHDHB3 or 4) (Figure 1A), or with a non-silencing shRNA (shNON) as a negative control. To monitor the effectiveness of shRNA silencing, the levels of HDHB were quantified using Western blot (Figure 1A). We examined DNA synthesis in U2OS cells that had been transfected with pGIPZ plasmids co-expressing GFP and shHDHB3 or shNON for 96 hours and incubated with BrdU for 45 min. BrdU incorporation and GFP expression in transfected cell cultures were analyzed by fluorescence microscopy (Figure 1B). Quantification of these results showed that more than 40% of the cells expressing shNON displayed BrdU incorporation, indicative of robust DNA synthesis. In contrast, only 8% of the cells expressing shHDHB3 incorporated BrdU into DNA (Figure 1C).
If cells partially depleted of HDHB were indeed unable to initiate DNA replication, as suggested by their failure to incorporate BrdU, they should arrest in G1 or G1/S. To test this prediction, we took advantage of the observation that Mcm3 re-localizes in nuclei only in late telophase and thus can be utilized as a marker for cells in G1 [48,49,50]. Indeed, when U2OS cells that had been synchronized and labeled with BrdU were stained for immunofluorescence microscopy, the predicted staining patterns for G1 (Mcm3+/BrdU−), S (Mcm3+/BrdU+), and G2/M (Mcm3−/BrdU−) were observed (Figure 1D). Using this approach, the cell cycle distribution of U2OS cells co-expressing GFP and shNON or shHDHB was determined by their staining patterns (Figure 1E, Suppl. Figure 1). Comparison of the cell cycle distributions revealed that the shHDHB-expressing cells accumulated in G1 with a corresponding reduction in S phase cells relative to the shNON-expressing cells (Figure 1F). Taken together, the evidence suggests that cells depleted of HDHB fail to progress from G1 to S phase.
Many replication initiation proteins like ORC and MCM are expressed only in cycling cells due to transcriptional down-regulation of their genes in quiescent cells [51,52]. To further investigate the role of HDHB during DNA replication initiation, we examined if HDHB is up regulated after entering the cell cycle. Therefore extracts of quiescent primary fibroblast cells were prepared at different times after serum stimulation to promote re-entry into the cell cycle, and HDHB protein levels were monitored by western blotting. The Mcm7 protein, which served as a positive control, was barely detected in serum-deprived cells, but reappeared after serum stimulation (Supple Figure 2C). HDHB protein was still detectable in quiescent cells, but increased substantially after serum stimulation (Suppl. Figure 2C).
Furthermore to investigate whether HDHB is enriched at replication origins, U2OS cells were blocked in G2/M phase and released for 5 hours (G1), or synchronized in G1/S phase, released for 6 (S) or 9 hours (G2/M), and treated with formaldehyde to crosslink chromatin. Chromatin was isolated, sheared, and immunoprecipitated using purified polyclonal HDHB antibody or non-immune control antibody. HDHB enrichment in immunoprecipitated chromatin was measured by quantitative real-time PCR using one primer pair in the replication initiation region of each origin [53,54] (Suppl. Figure 2A; LB48 primers for LaminB2 origin, Suppl. Figure 2B; UPR4 primers for MCM4) and one primer pair in a distal region of each origin (LB2C1, Ex6). In G1 cells, HDHB was significantly enriched on chromatin in the initiation region of each origin relative to the distal region (Suppl. Figure 2A and B). HDHB enrichment in both initiation regions was reduced in S and G2/M phase. The data indicate that HDHB is up regulated after cell cycle entry and is binding at two human replication origins in G1.
HDHB interacts directly with TopBP1 and Cdc45 in vitro
When viewed in combination with earlier data [43,44,45], the new evidence that HDHB might be required for replication initiation suggests that HDHB might interact with proteins that associate with origins prior to G1/S transition. The earliest pre-RC proteins that assemble on chromatin in Xenopus extract include ORC, MCM2-7, and Xmus101 [20], whereas TopPB1 (the human homolog of Xmus101)- and CDK-dependent recruitment of CDC45 is a critical step in the assembly of pre-IC at the origins. We first tested Orc2, Mcm4, and TopBP1 as potential HDHB-interacting candidates. Because we could not co-immunoprecipitate endogenous replication initiation factors with HDHB, we co-infected insect cells with a recombinant baculovirus expressing HDHB and a baculovirus expressing one of the potential interacting proteins. HDHB was then immunoprecipitated from infected cell extracts and co-immunoprecipitation of the pre-RC proteins was monitored by western blots. Neither Orc2 nor Mcm4 was co-immunoprecipitated with HDHB (Figure 2A, lanes 3, 6). However, TopBP1 did co-precipitate with HDHB in the presence of an anti-HDHB monoclonal antibody (Figure 2A, lane 9), and not in the presence of nonimmune IgG (lane 8).
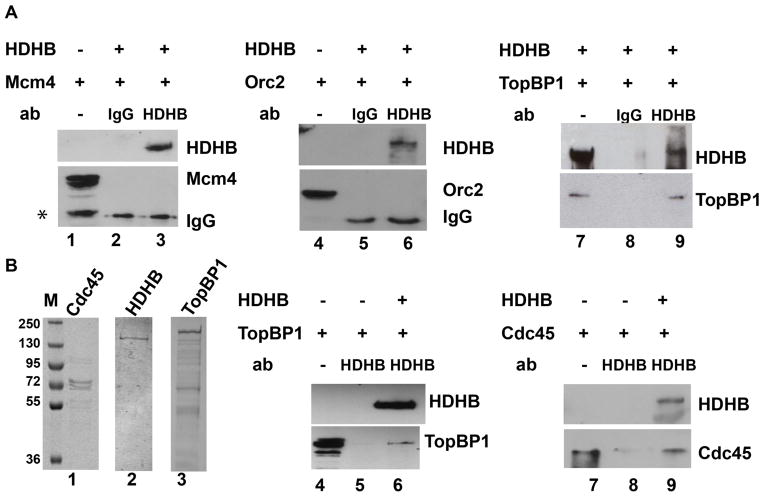
Figure 2. HDHB interacts with TopBP1 and Cdc45 in vitro.
(A) HDHB was immunoprecipitated from extracts of baculovirus-infected insect cells co-expressing HDHB and Mcm4, Orc2, or TopBP1 using non-immune rat IgG (lanes 2, 5, 8) or anti-HDHB 5c9 monoclonal antibody (lanes 3, 6, 9). Co-precipitated proteins were detected by western blot with the antibodies indicated to the right of each panel. Samples of extract from cells infected with baculoviruses expressing only Mcm4 (lane 1), only Orc2 (lane 4), or both HDHB and TopBP1 (lane 7) were electrophoresed in parallel. Asterisk: partially degraded Mcm4 (lane 1). (B) Purified bacterially expressed Cdc45 (lane 1), purified baculovirus-expressed HDHB (lane 2) and TopBP1 (lane 3) were visualized by SDS-PAGE and Coomassie staining. Protein A-agarose beads pre-bound to polyclonal HDHB antibody were incubated with purified TopBP1 or Cdc45 in the absence (lanes 5, 8) or presence of purified HDHB (lanes 6, 9). Samples (20%) of TopBP1 and Cdc45 input are shown in lanes 4 and 7. Precipitated proteins were analyzed by SDS-PAGE and western blot with the indicated antibodies.
To determine whether TopBP1 interacts directly with HDHB, we conducted co-immunoprecipitation experiments using purified recombinant HDHB and TopBP1 proteins (Figure 2B, lanes 2, 3). TopBP1 was immunoprecipitated with purified polyclonal HDHB antibody [55] in the presence, but not absence, of purified HDHB protein (Figure 2B, compare lanes 5, 6). TopBP1 homolog in yeast S. cerev., Dpb11 is involved replication initiation through binding of initiation factors like Cdc45 [23,24,25]. We were wondering whether HDHB might also interact with Cdc45. To address this question, we used purified recombinant Cdc45 and HDHB (Figure 2B, lanes 1, 2) in co-immunoprecipitation experiments. Cdc45 was co-precipitated with the anti-HDHB polyclonal antibody in the presence, but not absence, of purified HDHB protein (Figure 2B, compare lanes 8, 9). The data strongly suggest that HDHB binds directly to both TopBP1 and Cdc45.
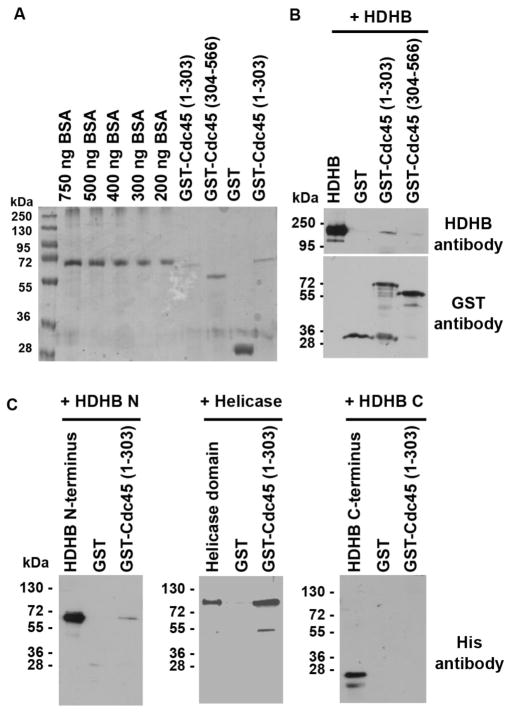
Cdc45 is a key factor in the DNA replication initiation process. To investigate which part of HDHB and which part of Cdc45 directly interacts with HDHB we next performed GST pull-down assays. First we examined which part of Cdc45 binds to purified HDHB protein. Cdc45 does not contain any characteristic protein domains, but a N-terminal region shows sequence similarity to RecJ protein family [56]. To narrow down the HDHB interaction domain of Cdc45, two GST-tagged partial constructs of Cdc45 were used in GST pull-down experiments: an N-terminal construct, containing the RecJ-homology domain (amino acids 1–303) and a C-terminal construct (amino acids 304–566) (Figure 3A). HDHB was immunoprecipitated with GST-Cdc45 N-terminus and C-terminus. However, only a weak interaction was detected between HDHB and the GST-Cdc45 C-terminus (Figure 3B). We next mapped the domains of HDHB that interact with the Cdc45 N-terminus by GST pull-down experiments (Figure 3C). HDHB has a N-terminal domain, helicase domain and C-terminal domain, which contains CDK phosphorylation sites and a subcellular localization domain (SLD) (Figure 1A). With the Cdc45 N-terminus we co-precipitated the N-terminus and helicase domain of HDHB, but not the C-terminus of HDHB. These results show that the N-terminus of Cdc45 physically interacts with both N-terminal and helicase domains of HDHB.
Figure 3. N-terminus of Cdc45 physically interacts with the N- and helicase domain of HDHB.
(A) Expression level of GST N- (aa 1–303) and C-terminus (aa 304–566) of Ccd45 was determined by SDS-PAGE and Coomassie staining. (B) GST pull-down experiments; HDHB was added to the GST N- (aa 1–303) and C-terminus (aa 304–566) of Ccd45. Precipitated proteins were analyzed by SDS-PAGE and western blot with the indicated antibodies, either GST or HDHB antibody. (C) GST pull-down experiments; HDHB the N-terminus (aa 1–393), helicase domain (aa 394–958) or C-terminus (aa 957–1088) containing a His-tag was added to the GST N-terminus of Cdc45. Precipitated proteins were analyzed by SDS-PAGE and western blot with the indicated antibodies, either with His-taq or GST antibody.
HDHB promotes recruitment of Cdc45 to chromatin
The physical interactions among HDHB, TopBP1, and Cdc45 suggested the possibility that HDHB might facilitate recruitment of TopBP1 or Cdc45 onto chromatin. If so, depletion of HDHB should result in defective loading of TopBP1 or Cdc45 on chromatin. CDK activity is a phylogenetically conserved requirement for Cdc45 loading on chromatin [14,28,57]. Thus, to develop an assay for Cdc45 loading in single HDHB-depleted cells, U2OS cells were incubated with or without the CDK inhibitor roscovitine [58], which was predicted to arrest cells in late G1 prior to Cdc45 loading on chromatin. The cells were labeled with BrdU, fixed and stained for Cyclin E and BrdU or Cdc45 using two different protocols, and examined by immunofluorescence microscopy. As expected, roscovitine abolished BrdU incorporation, but not Cyclin E staining (Figure 4A), confirming that the drug inhibited initiation of replication. Roscovitine-treated cells fixed with a protocol that included extraction with 0.02% SDS [59] displayed brighter, crisper Cyclin E staining than cells fixed using a standard protocol without SDS (compare Figure 4B with 4A). Using the SDS protocol, bright Cdc45 staining was detected in the absence of roscovitine, but only faint Cdc45 signals were detected in its presence (Figure 4B). Quantification of these results indicated that the fraction of cyclin E-positive cells that were also stained for Cdc45 dropped by 50% in the presence of roscovitine (Figure 4C), validating the approach to monitor Cdc45 loading on chromatin.
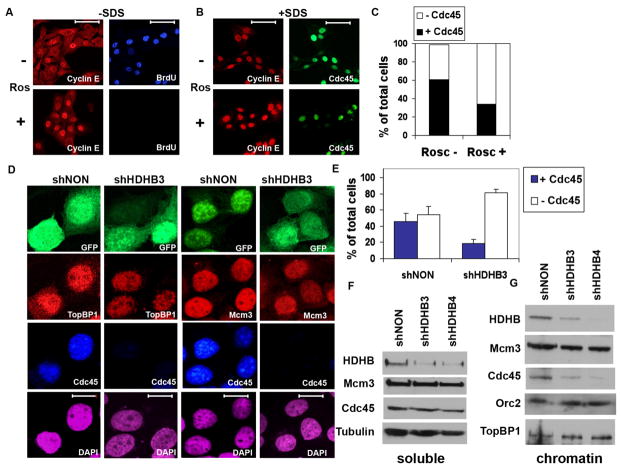
Figure 4. Cdc45 chromatin loading is reduced in HDHB-depleted cells.
(A) U2OS cells cultured with (+) or without (−) 50 μM roscovitine (Ros) as indicated were incubated with BrdU for 30 min, fixed and permeabilized using the standard protocol (−SDS), and stained with anti-BrdU and anti-cyclin E for immunofluorescence microscopy. Scale bar, 70 μm. (B) U2OS cells cultured + or − roscovitine as indicated (+SDS) were fixed with 1 % formaldehyde for 10 min, extracted with 0.1% Triton X-100 and 0.02% SDS [49,59], and stained with anti-cyclin E and anti-Cdc45 for immunofluorescence microscopy. (C) The fraction of Cdc45-positive nuclei in cultures treated + or − roscovitine and stained as in (B) was determined by counting 100 DAPI-stained cells each culture. (D, E) U2OS cells co-expressing GFP and shNON or shHDHB3 were fixed and permeabilized as in (B), and stained with antibodies against Mcm3, TopBP1, and Cdc45 as indicated. Nuclei were stained with DAPI. Scale bar, 15 μm. (E) The fraction of Cdc45-positive cells expressing shNON (n=190) or shHDHB3 (n=183) was determined in two independent experiments. Brackets indicate standard deviation. (F, G) Cells expressing control (shNON) or HDHB shRNA (shHDHB3 and shHDHB4) were fractionated into soluble (F) and chromatin (G) extracts, and analyzed by western blot with the indicated antibodies.
To test whether HDHB plays a role in the recruitment of Cdc45 or TopBP1 to chromatin, we partially depleted HDHB in U2OS cells using shHDHB3 or control shNON plasmids co-expressing GFP. At 72 h after transfection, the cells were analyzed by immunofluorescence microscopy using the SDS fixation protocol (Figure 4D). The fraction of HDHB-depleted cells displaying Mcm3 (52%) and TopBP1 (75%) did not differ significantly from the fraction of control-depleted cells displaying Mcm3 (51%) and TopBP1 (75%) (Figure 4D). These results indicate that HDHB is dispensable for TopBP1 and Mcm3 association with chromatin. In contrast, the fraction of HDHB-depleted cells stained for Cdc45 was strongly reduced compared to that of the control-depleted cells (Figure 4D, E and Suppl. Figure 4). This reduction in HDHB-depleted cells is somewhat greater than the 50% reduction observed in the roscovitine-treated cells (Figure 4C), suggesting that HDHB might function in Cdc45 recruitment or loading on chromatin.
To corroborate these findings, HDHB-depleted and shNON-depleted cells were fractionated [48] and the soluble (Figure 4F) and chromatin (Figure 4G) extracts were analyzed by western blots. As expected, HDHB protein levels in both the soluble and chromatin extracts declined in shHDHB3- and shHDHB4-transfected cells relative to shNON-transfected cells. Tubulin, Mcm3, and Cdc45 levels in the soluble fraction did not differ significantly in HDHB-depleted relative to shNON-depleted cells (Figure 4F). Moreover, the level of Orc2, Mcm3 and TopBP1 proteins in the chromatin fraction was not affected by HDHB-depletion (Figure 4G). In contrast, the level of Cdc45 in the chromatin fraction was substantially decreased in shHDHB-depleted cells (Figure 4G). Together, these data confirm that HDHB is dispensable for chromatin binding of TopBP1, Orc2, and Mcm3 and suggest that HDHB promotes loading of Cdc45 on chromatin for initiation of replication.
Discussion
The two-phase global regulation of eukaryotic DNA replication - origin licensing during low CDK activity in G1, followed by CDK-dependent initiation at G1/S - is highly conserved [1,2,8,60]. However, the detailed mechanisms that underlie this regulation diverge considerably from budding to fission yeast and among metazoans. At the outset of the studies presented here, two independent lines of evidence suggested that HDHB was needed, directly or indirectly, for the G1/S transition in mammalian cells [43,44,45]. Here we sought to determine whether HDHB interacts physically with DNA replication initiation proteins and is required for replication initiation in human cells.
The results presented here demonstrate that HDHB interacts with replication initiation proteins such as TopBP1 and Cdc45 (Figure 2–3), and promotes DNA replication initiation (Figure 1, 4). These data add significant new evidence that HDHB is needed to initiate chromosomal replication in mammalian cells. Depletion of HDHB in human cells disrupts initiation of replication at a step prior to stable loading of Cdc45 on chromatin (Figure 1, 4). No obvious defect in loading of pre-RC proteins Orc2, Mcm3, or TopBP1 was detected in HDHB-depleted cells (Figure 4D, G). The magnitude of the Cdc45 loading defect in HDHB-depleted cells was comparable to that observed in cells treated with the CDK inhibitor roscovitine (Figure 4A–C), implicating HDHB in a process that loads Cdc45 on chromatin downstream of TopBP1 recruitment. These observations, together with the enrichment of HDHB at replication origins (Suppl. Figure 2A and B) and the ability of HDHB to interact directly with human TopBP1 and Cdc45 suggest that HDHB may have at least some role in initiation or regulation of DNA replication.
HDHB does become phosphorylated at the G1/S transition in human cells [46], raising the question of whether it could be one of the vertebrate CDK targets postulated to be essential for Cdc45 loading on chromatin and activation of origin DNA unwinding [24,25]. The C-terminal domain of HDHB (aa957–1087) contains a nuclear localization signal and a Rev-type nuclear export signal that is dependent on CDK phosphorylation [46]. At G1/S, the bulk of HDHB is phosphorylated at Ser967 and exported to the cytoplasm. It is possible that this HDHB phosphorylation and nuclear export is a crucial step in the G1/S transition. Alternatively, it could contribute to the regulation that prevents re-licensing of chromatin until after mitosis [1,2].
HDHB could be involved in replication initiation and/or in checkpoint control. In G1/S phase it is reported that the binding of Treslin to TopBP1 and the binding of RecQL4 to TopBP1 is critical for initiation of replication [33,37,61]. In addition during activation of the DNA damage checkpoint the interaction between Treslin and TopPB1 is interrupted [36]. Since HDHB binds to TopBP1 and is important for DNA replication initiation, HDHB could together with other replication initiation factors facilitate the recruitment of Cdc45 to replication initiation sites. First, proteins of the pre-RC like Cdc6 and Cdt1 could load HDHB onto the replication initiation sites in G1. Then HDHB could promote together with Treslin, TopBP1 and RecQL4 the loading of Cdc45 in G1/S. At the same time non-chromatin bound HDHB could be exported to the cytoplasm to prevent re-initiation. The copy number of HDHB could be crucial for initiation of replication and partial depletion of HDHB could lead to inability of the cells to enter S phase. Indeed microinjection of purified recombinant HDHB mutant protein into cells in G1 [44] as well as depletion of HDHB by shRNA inhibits cells to progress into S phase.
In addition the interaction of HDHB to TopBP1 could have a function in the DNA damage checkpoint in G1/S or S phase. Given that a small amount of HDHB remains in the nucleus in S phase [62] in stress situations interruption of the binding of HDHB to TopBP1 could trigger suppression of replication initiation from late or unfired origin. These models have to be tested in further studies.
Clues of other potential functions of HDHB in initiation of replication
Potential activity of nuclear HDHB in initiation of chromosomal replication may be the 5′-3′ helicase activity itself [43,44,45]. RecQL4 promotes origin unwinding [15,16] and DNA polymerase loading [33,34]. The N-terminal Sld2-homologous fragment of RecQL4 was unable to substitute for full-length RecQL4 to restore replication activity, whereas a larger fragment substituted at high concentration [33]. Consistent with these observations, a Walker B mutant RecQL4 failed to substitute for wild type protein in Xenopus replication assay [34], suggesting that the 3′-5′ helicase activity of RecQL4 may be a factor to origin DNA unwinding [35]. Since HDHB is localized at replication origin (Suppl. Figure 2A and B) it would be interesting to test whether HDHB contributes to origin unwinding.
Curiously, the most closely related non-vertebrate amino acid sequence homologs of DNA helicase B are bacterial RecD 5′-3′ helicases [44,45]. E. coli RecD operates in complex with the 3′-5′ RecB helicase and RecC as a rapid, highly processive, dual DNA unwinding motor that processes broken duplex DNA for homologous recombination [63]. The relative unwinding rates of the two motors can be regulated, leading to formation of a single-stranded DNA loop ahead of the slower motor. Given the emerging roles of HDHB and RecQL4 in initiation of vertebrate chromosomal replication, it is intriguing to speculate that HDHB and RecQL4 could be helicases that activate or support the MCM complex in unwinding replication origins.
In summary, we have presented evidence that HDHB binds directly to the replication initiation proteins TopBP1 and Cdc45 in vitro. Furthermore depletion of HDHB inhibits initiation of replication and Cdc45 chromatin association at G1/S, but does not affect association of Mcm3 or TopBP1 with chromatin. We suggest that HDHB might have a role in assemble of the replication initiation complex. Future work will be needed to reveal the detailed role(s) of HDHB in initiation of replication.
Materials and methods
Cell culture
U2OS and HCT116 were grown in DMEM with 10% heat-inactivated fetal bovine serum (FBS) and 5% CO2 at 37° C. Cells were blocked in G1/S with 2.2 mM thymidine and in G2/M with 300 ng/ml nocodazole (Sigma) for 16 h. To obtain S-phase cells, cells were blocked in G1/S with 2.2 mM thymidine and released for 6 h. To obtain G1 cells, cells were blocked in G2/M with 300 ng/ml nocodazole (Sigma) and then released for 5 h. High Five insect cells and Sf9 cells were grown in Grace’s insect medium (Invitrogen) supplemented with 10% FBS. U2OS cells were cultured with (+) or without (−) 50 μM roscovitine (Ros) for 24 h.
Antibodies against HDHB
Polyclonal rabbit antibodies [46] and monoclonal rat antibodies were described previously [55].
Chromatin fractionation was performed as described [48] with some modifications. Briefly, a total of ~5 ×106 cells were washed with PBS and resuspended in 300 μl solution A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 10 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 10 μg/ml aprotinin, 1 μM leupeptin). Triton X-100 was added to a final concentration of 0.05%, and the cells were incubated on ice for 10 min. Cytoplasmic proteins (S1) were separated from nuclei by centrifugation at 1300× g for 5 min. Isolated nuclei were washed once with solution A and resuspended in 300 μl solution B (3 mM EDTA at pH 8.0, 0.2 mM EGTA, 1 mM DTT). After a 30-min incubation on ice, soluble nuclear proteins (S2) were separated from chromatin (P2) by centrifugation at 1700× g for 5 min. Isolated chromatin was washed once with solution B, resuspended in 300 μl SDS-PAGE sample buffer, and sheared by sonification.
HDHB depletion by shRNA expression
U2OS cells were transfected with GIPZ-HDHB3, HDHB4 or GIPZ-NON (Open Biosystems) (for map and sequence of GIPZ, see http://www.openbiosystems.com/Vector/VectorDetails.aspx?vn=pGIPZ) using FuGENE HD (Roche) according to the manufacturer’s instructions. shHDHB4 (V2LHS_33141): TGCTGTTGACAGTGAGCGCGGCAAGACTGTGATCTAATTATAGTGAAGCCAAGATGTATAATTAGATCACAGTCTTGCCTTGCCTACTGCCTCGGA; shHDHB3 (V2LHS_33143): TGCTGTTGACAGTGAGCGCGCCAGTTCTCAGTCATCTAAATAGTGAAGCCACAGATGTATTTAGATGACTGAGAACTGGCATGCCTACTGCCTCGGA.
Immunofluorescence microscopy
U2OS cells grown on cover slips were washed three times with PBS, fixed with 4% formaldehyde in PBS for 10 min, permeabilized with 0.5 % Triton X-100 for 10 min, incubated with signal enhancer (Image-iT FX; Invitrogen) for 30 min, and probed with primary antibodies: rabbit anti-Mcm3 (1:200) (gift from B. Stillman), rabbit anti-TopBP1 (1:100) (Bethyl), rat anti-BrdU (1:100) (Abcam) or rat anti-Cdc45 C45-3G10 (Bauerschmidt et al, 2007) (1: 50) for 2 h. In Figure 4, cells were fixed and permeabilized as described in the figure legend. For BrdU staining, samples were incubated with primary antibody and 125 U/ml benzonase (Novagen), washed with PBS, and incubated with secondary antibodies (Invitrogen) AlexaFluor 633 anti-rat and AlexaFluor 555 anti-rabbit diluted 1:100 for 1 h. All antibodies were diluted in 10% FBS-PBS. Nuclear DNA in the cells was counterstained with DAPI for 20 min. Cover slips were mounted in ProLong antifade reagent (Molecular Probes, Eugene, OR). Data were collected using an Olympus FV-1000 confocal microscope equipped with three lasers giving excitation lines at 633, 543, 488 nm and UV light at a resolution of 1,024 by 1,024 pixels utilizing a 63x oil immersion objective. The data from the channels were collected sequentially using the appropriate band-pass filters built into the instrument. Data sets were processed using the FV10-ASW 1.6 Viewer software.
Chromatin immunoprecipitation
1 × 108 U2OS cells were washed with PBS and treated with 1% formaldehyde in pre-warmed medium for 5 min at 37°C. Cells were harvested, washed with PBS, and resuspended in 4 ml hypotonic buffer A (10 mM Hepes pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml each of aprotinin, leupeptin, and pepstatin). Cells were lysed by adding Triton X-100 to a final concentration of 0.04% and incubated for 10 min on ice. Samples were centrifuged (4 min, 1300 g, 4°C). Nuclei were washed in ice-cold buffer A. After centrifugation (1300 g, 5 min, 4°C), fixed nuclei were washed with PBS, resuspended in 2.7 ml LSB (10 mM Hepes pH 7.9, 10 mM KCl, 1.5 mM MgCl2) and lysed by adding 300 μl 20% Sarkosyl. The chromatin was transferred onto a 40 ml sucrose cushion (LSB plus 100 mM sucrose) and centrifuged (10 min, 4°C, 2500 g). Supernatant was removed, and the chromatin was resuspended in 4 ml TE and sonicated (Branson sonifier 250-D, 5 output 50 impulses, twice). For partial DNA digests, 3 mM and 10 U micrococcal nuclease (MNase) (Roche) per 1 mg Chromatin were added and CaCl2 incubated for 10 minutes at 37°C. The reaction was stopped by addition of 20 mM EGTA. For immunoprecipitation, 1/10 volume of 11x NET (550 mM Tris-HCl pH 7.4, 1.65 M NaCl, 5.5 mM EDTA, 5.5% NP40) was added to the extract followed by 10 μg affinity-purified polyclonal antibodies against HDHB or as control, 10 μg rabbit IgG. The immunoprecipitation and purification of co-precipitated DNA was performed as described [6].
Real-time PCR analysis was performed according to the manufacturer’s instructions (Roche) using the same parameters and primer pairs as described [53,54]. Enrichment of immunoprecipitated DNA (with HDHB antibody) is defined as the abundance of target sequence detected in the specific HDHB immunoprecipitate minus the abundance of target sequence detected in a non-specific IgG immunoprecipitate, divided by the abundance of target sequence detected in 30 ng of DNA purified from the pre-IP chromatin preparation [64]. When no HDHB enrichment was observed at both the origin and distal region (sum = 0), no value was plotted.
Protein purification
High Five insect cells (Invitrogen) (5 ×108) infected by recombinant HDHB baculovirus for 48 h were lysed in 10 ml of lysis buffer (20 mM Tris-HCl at pH 7.5, 50 mM NaCl, 0.2% (v/v) Nonidet P-40, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 μM leupeptin). Lysate was cleared by centrifugation, and the supernatant was loaded on a 20-ml P11 column (Whatman, Florham Park, NJ) pre-equilibrated in Buffer A (20 mM Tris-HCl at pH 7.5, 50 mM NaCl, 0.02% (v/v) Nonidet P-40, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 μM leupeptin). Proteins were eluted from the column with a 200-ml gradient of NaCl from 50 to 600 mM in Buffer A. The eluate was diluted 10-fold with Buffer A and loaded onto a 1-ml MonoQ column (Amersham Biosciences). Proteins were eluted with a 20-ml gradient of NaCl from 50 to 600 mM in Buffer A and collected in 0.5-ml fractions. Purified baculovirus-expressed TopBP1 was a kind gift from D. Cortez.
BL21 bacteria cells were transformed with His-Cdc45-pRSET [65] (gift of A. Dutta), Cdc45-pGEX4T-1, CDC45N-pGEX4T-1, CDC45C-pGEX4T-1 (gift from F. Haenel) or TopBP1-pGEX-6p-1 plasmid [66] (gift of W.C. Lin). HDHB mutants (N-, C- terminus and helicase domain) were cloned in pET28 vector. HDHB mutants and Cdc45 were expressed after IPTG (1 mM) induction for 3 h at 37°C. Bacterial cells were centrifuged at 4000 × g for 10 min and resuspended in Lysis buffer (10 mM Na2S2O5, 10% glycerol, 0.5% Triton X-100, 0.4 mg/ml lysozyme). Cell extracts were sonicated on ice (10 times, 10 s bursts at 300 W) and centrifuged at 10,000 × g for 30 min at 4 °C. His-tagged HDHB mutants and Cdc45 was purified over Ni-NTA agarose (Qiagen) according the manufacturer’s instructions. GST-tagged TopBP1 was expressed for 19 h at room temperature (Liu et al, 2003). For GST-Cdc45, GST-Cdc45N, GST-Ccd45C and GST-TopBP1 purification, bacteria were lysed with PBS containing 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 μM leupeptin, 1 mM EDTA, 1 mM EGTA, and 1 mg/ml lysozyme. After 10 min on ice, 1 % NP-40 and 7 mM MgCl2 were added and the cells were sonicated (6 times, 20 s bursts at 500 W). After centrifugation at 10,000 × g for 20 min (4°C), cell lysate was mixed with 0.2 % polyethyleneimine and centrifuged at 10,000 × g for 20 min at 4°C. GST-tagged TopBP1 was purified over glutathione-agarose (Sigma) according the manufacturer’s instructions. HDHB, GST-TopBP1, and His-Cdc45 proteins were dialyzed overnight in 30 mM Hepes pH 7.9, 7 mM Mg(Ac)2, 1 mM DTT, 10 μM ZnCl2. 200 ng of HDHB, GST-TopBP1, and His-Cdc45 proteins were analyzed by SDS-PAGE.
Co-immunoprecipitation from crude extract
1 × 107 High Five Insect cells were infected by recombinant baculoviruses expressing HDHB, Orc2 (gift from A. Dutta), Mcm4 (gift from D. Schaarschmidt), TopBP1 (gift from D. Cortez), or a combination of viruses as indicated in Figure 2A for 48 h. Insect cells were lysed in 1 ml of Buffer A (20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.2% (v/v) Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml each of aprotinin, leupeptin, and pepstatin) for 5 min on ice. After sonification (10 s impulse, output 5 for 3 times), lysate was centrifuged for 30 min, 13000 × g, at 4°C. This supernatant was rotated with antibodies overnight at 4°C and then with Protein A-Sepharose beads for 2 h. Beads were washed with 10 ml Buffer A plus NP-40 and resuspended in 20 μl SDS-PAGE sample buffer for SDS-PAGE. Western blots were analyzed by chemiluminescence using rat monoclonal anti-HDHB 5c9 (1: 10000), rabbit anti-TopBP1 (1:5000), rabbit anti-Orc2 (1:5000), and rabbit anti-Mcm4 (1: 5000) antibodies (provided by Aloys Schepers) [67].
Co-immunoprecipitation of purified proteins
Protein A beads (20 μl) pre-bound to polyclonal anti-HDHB antibody were incubated with or without 1 or 1.5 μg of purified recombinant HDHB for 1 h at 4°C in 2% milk in binding buffer (30 mM K-Hepes pH 7.8, 10 mM KCl, 7 mM MgCl2). Purified TopBP1 or GST-TopBP1 (5 μg) or His-Cdc45 (4 μg) was added and incubated for an hour. Beads were washed twice with binding buffer and three times with wash buffer (30 mM K-Hepes pH 7.8, 75 mM KCl, 7 mM MgCl2, 0.25% inositol, 0.01% NP-40, 10 μM ZnCl2). The bound proteins were separated on 8.5% or 10% SDS-PAGE and detected by western blotting.
GST pull down experiments
Glutathione–Sepharose beads pre-bound with 5 pmol of either GST-tagged Cdc45N (1–303) or Cdc45C (304–566) protein fragments or GST alone were incubated with 5 pmol of HDHB or HDHB mutants (N- or C-terminus or helicase domain) for 2 h at 4°C in 2% milk in binding buffer. Beads were washed three times with wash buffer. Proteins were eluted from the beads using SDS sample buffer and separated by SDS/10 % PAGE. We loaded 30% of input and 100% of the co-immunoprecipitated proteins. Proteins were detected by western blotting using His- or GST- or HDHB-antibody.
Supplementary Material
Acknowledgments
We thank D. Cortez, A. Dutta, F. Haenel, X. Jiang, W. C. Lin, A. Rokas, D. Schaarschmidt, B. Stillman and E. Warren for reagents and discussion. JG, GG and EF designed experiments, JG and GG performed experiments, JG and GG analyzed data and wrote the manuscript. The financial support of NIH (GM52948 to EF and P30 CA068485 to the Vanderbilt-Ingram Cancer Center), Helmholtz Center Munich, and Vanderbilt University are gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes & Development. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 2.Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement? Nature reviews Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Current opinion in cell biology. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau E, Tsuji T, Guo L, Lu SH, Jiang W. The role of pre-replicative complex (pre-RC) components in oncogenesis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:3786–3794. doi: 10.1096/fj.07-8900rev. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Current opinion in cell biology. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Gerhardt J, Jafar S, Spindler MP, Ott E, Schepers A. Identification of new human origins of DNA replication by an origin-trapping assay. Molecular and cellular biology. 2006;26:7731–7746. doi: 10.1128/MCB.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borlado LR, Mendez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis. 2008;29:237–243. doi: 10.1093/carcin/bgm268. [DOI] [PubMed] [Google Scholar]
- 8.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Molecular cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 10.Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic acids research. 2009;37:2087–2095. doi: 10.1093/nar/gkp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes & development. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, et al. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes & development. 2003;17:1141–1152. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. The Journal of biological chemistry. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- 14.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Molecular cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 15.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nature cell biology. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 16.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Molecular cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 18.Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, et al. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes & development. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Molecular cell. 2002;9:233–240. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- 20.Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, et al. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. The Journal of cell biology. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Molecular cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. The EMBO journal. 2009;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Botchan M. Cell biology: a switch for S phase. Nature. 2007;445:272–274. doi: 10.1038/445272a. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 25.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 26.Kamimura Y, Tak YS, Sugino A, Araki H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. The EMBO journal. 2001;20:2097–2107. doi: 10.1093/emboj/20.8.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima R, Masukata H. SpSld3 Is Required for Loading and Maintenance of SpCdc45 on Chromatin in DNA Replication in Fission Yeast. Molecular Biology of the Cell. 2002;13:1462–1472. doi: 10.1091/mbc.02-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. The EMBO journal. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon Y, Lee KY, Ko MJ, Lee YS, Kang S, et al. Human TopBP1 participates in cyclin E/CDK2 activation and preinitiation complex assembly during G1/S transition. The Journal of biological chemistry. 2007;282:14882–14890. doi: 10.1074/jbc.M609116200. [DOI] [PubMed] [Google Scholar]
- 31.Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. The Journal of biological chemistry. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt U, Wollmann Y, Franke C, Grosse F, Saluz HP, et al. Characterization of the interaction between the human DNA topoisomerase IIbeta-binding protein 1 (TopBP1) and the cell division cycle 45 (Cdc45) protein. The Biochemical journal. 2008;409:169–177. doi: 10.1042/BJ20070872. [DOI] [PubMed] [Google Scholar]
- 33.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Molecular and cellular biology. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, et al. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Ward T, Yao X, Wu J. Chk1 prevents abnormal mitosis of S-phase HeLa cells containing DNA damage. Chinese Science Bulletin. 2009;54:4205–4213. [Google Scholar]
- 36.Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, et al. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Current biology: CB. 2011;21:1152–1157. doi: 10.1016/j.cub.2011.05.057. [DOI] [PubMed] [Google Scholar]
- 37.Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller AC, Keaton MA, Dutta A. DNA replication: mammalian Treslin-TopBP1 interaction mirrors yeast Sld3-Dpb11. Current biology: CB. 2011;21:R638–640. doi: 10.1016/j.cub.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumagai A, Shevchenko A, Dunphy WG. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. The Journal of cell biology. 2011;193:995–1007. doi: 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itou H, Muramatsu S, Shirakihara Y, Araki H. Crystal structure of the homology domain of the eukaryotic DNA replication proteins Sld3/Treslin. Structure. 2014;22:1341–1347. doi: 10.1016/j.str.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Seki M, Enomoto T, Hanaoka F, Yamada M. DNA-dependent adenosinetriphosphatase B from mouse FM3A cells has DNA helicase activity. Biochemistry. 1987;26:2924–2928. doi: 10.1021/bi00384a038. [DOI] [PubMed] [Google Scholar]
- 42.Seki M, Enomoto T, Yanagisawa J, Hanaoka F, Ui M. Further characterization of DNA helicase activity of mouse DNA-dependent adenosinetriphosphatase B (DNA helicase B) Biochemistry. 1988;27:1766–1771. doi: 10.1021/bi00405a057. [DOI] [PubMed] [Google Scholar]
- 43.Eki T, Hanaoka F, Kohda T, Seki M, Ui M, et al. Efficient replication of polyomavirus DNA in a cell-free system supplemented with Escherichia coli single-stranded DNA binding protein, which exhibits species-specificity in the requirement for DNA polymerase alpha-primase. Journal of biochemistry. 1995;118:435–441. doi: 10.1093/oxfordjournals.jbchem.a124926. [DOI] [PubMed] [Google Scholar]
- 44.Taneja P, Gu J, Peng R, Carrick R, Uchiumi F, et al. A dominant-negative mutant of human DNA helicase B blocks the onset of chromosomal DNA replication. The Journal of biological chemistry. 2002;277:40853–40861. doi: 10.1074/jbc.M208067200. [DOI] [PubMed] [Google Scholar]
- 45.Tada S, Kobayashi T, Omori A, Kusa Y, Okumura N, et al. Molecular cloning of a cDNA encoding mouse DNA helicase B, which has homology to Escherichia coli RecD protein, and identification of a mutation in the DNA helicase B from tsFT848 temperature-sensitive DNA replication mutant cells. Nucleic acids research. 2001;29:3835–3840. doi: 10.1093/nar/29.18.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitoh A, Tada S, Katada T, Enomoto T. Stimulation of mouse DNA primase-catalyzed oligoribonucleotide synthesis by mouse DNA helicase B. Nucleic acids research. 1995;23:2014–2018. doi: 10.1093/nar/23.11.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Méndez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Molecular and cellular biology. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krude T, Musahl C, Laskey RA, Knippers R. Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. Journal of cell science. 1996;109 (Pt 2):309–318. doi: 10.1242/jcs.109.2.309. [DOI] [PubMed] [Google Scholar]
- 50.Prasanth SG, Mendez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohtani K, DeGregori J, Leone G, Herendeen DR, Kelly TJ, et al. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Molecular and cellular biology. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, et al. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ladenburger EM, Keller C, Knippers R. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol Cell Biol. 2002;22:1036–1048. doi: 10.1128/MCB.22.4.1036-1048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaarschmidt D, Ladenburger EM, Keller C, Knippers R. Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 2002;30:4176–4185. doi: 10.1093/nar/gkf532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guler GD, Liu H, Vaithiyalingam S, Arnett DR, Kremmer E, et al. Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress. The Journal of biological chemistry. 2012;287:6469–6481. doi: 10.1074/jbc.M111.324582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanchez-Pulido L, Ponting CP. Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics. 2011;27:1885–1888. doi: 10.1093/bioinformatics/btr332. [DOI] [PubMed] [Google Scholar]
- 57.Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Molecular and cellular biology. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Azevedo WF, Leclerc S, Meijer L, Havlicek L, Strnad M, et al. Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. European journal of biochemistry/FEBS. 1997;243:518–526. doi: 10.1111/j.1432-1033.1997.0518a.x. [DOI] [PubMed] [Google Scholar]
- 59.Bauerschmidt C, Pollok S, Kremmer E, Nasheuer HP, Grosse F. Interactions of human Cdc45 with the Mcm2-7 complex, the GINS complex, and DNA polymerases delta and epsilon during S phase. Genes to cells: devoted to molecular & cellular mechanisms. 2007;12:745–758. doi: 10.1111/j.1365-2443.2007.01090.x. [DOI] [PubMed] [Google Scholar]
- 60.Diffley JFX. Regulation of early events in chromosome replication. Current biology: CB. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 61.Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, et al. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes & development. 2010;24:183–194. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu J, Xia X, Yan P, Liu H, Podust VN, et al. Cell Cycle-dependent Regulation of a Human DNA Helicase That Localizes in DNA Damage Foci. Molecular Biology of the Cell. 2004;15:3320–3332. doi: 10.1091/mbc.E04-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiology and molecular biology reviews: MMBR. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gray SJ, Gerhardt J, Doerfler W, Small LE, Fanning E. An origin of DNA replication in the promoter region of the human fragile X mental retardation (FMR1) gene. Molecular and cellular biology. 2007;27:426–437. doi: 10.1128/MCB.01382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saha P, Thome KC, Yamaguchi R, Hou Z, Weremowicz S, et al. The human homolog of Saccharomyces cerevisiae CDC45. The Journal of biological chemistry. 1998;273:18205–18209. doi: 10.1074/jbc.273.29.18205. [DOI] [PubMed] [Google Scholar]
- 66.Liu K, Lin F-t, Ruppert JM, Lin W-c. Regulation of E2F1 by BRCT Domain-Containing Protein TopBP1. Society. 2003;23:3287–3304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, et al. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. Journal of cell science. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.