Abstract
PRH/Hex is a homeodomain protein that plays an important role in early embryonic patterning and hematopoiesis. PRH can act as either a tumor suppressor or an oncogene and its expression is dysregulated in certain types of lymphoid and myeloid leukemias. Aberrant exclusion of PRH from the nuclei has been associated with thyroid and breast cancers and a subset of myeloid leukemias. Accordingly, nuclear localization of PRH was found to be necessary for the inhibition of eIF4E dependent transformation. Since PRH’s nuclear-cytoplasmic localization has been associated with neoplastic transformation we sought to better understand how PRH is transported to the nuclear compartment. Here we report an essential element that controls the mechanism of PRH nucleocytoplasmic trafficking, namely that it is imported into the nuclei by Karyopherin/Importin 7. Kap7 was identified as a binding partner for PRH in a GST pulldown from a HeLa cell protein lysate, followed by mass spectrometry. The Kap7-PRH complex is dissociated in the presence of RanGTP, as expected for a nuclear import complex. Kap7 can bind directly to PRH in a GST-pull down assay with purified proteins, as well as mediates the transport of PRH to the nuclear compartment in a digitonin permeabilized cells assay. Lastly, in vivo depletion of Kap7 dramatically reduces accumulation of PRH in the nucleus. Our data open the way for investigations of the mechanism of perturbed PRH localization in tumors and possible therapeutic interventions.
Keywords: PRH, Hex, importin, karyopherin, nucleocytoplasmic, trafficking
Introduction
The Proline-Rich Homeodomain protein (PRH), also referred to as the Hematopoietically Expressed Homeodomain (Hex) is an orphan homeodomain protein, classified as a member of the tinman family of homeodomain proteins [1]. PRH plays an important role in early embryonic patterning and hematopoiesis. During embryogenesis, its function is essential for the forebrain, liver and thyroid gland development [2]. In hematopoietic cells, PRH is strongly expressed in pluripotent erythromyeloid and B-cell progenitors, and is generally down regulated during differentiation of most of hematopoietic lineages [3, 4]. Besides hematopoietic cells, PRH is expressed in a limited number of adult tissues and cell types including liver, lung, thymus, thyroid and endothelial cells [1, 5–7].
Upregulation of PRH expression has been reported in certain types of lymphoid leukemia, and its overexpression in bone marrow cells of C57BL6 mice induces T cell lymphoma [8, 9]. In contrast, PRH is down regulated in certain types of myeloid leukemia, and when ectopically expressed in NIH 3T3 cells it inhibits eIF4E-mediated oncogenic transformation [10, 11]. Thus, PRH can act as either a tumor suppressor or an oncogene, depending on the cellular context and degree of dysregulated expression.
PRH is distributed in the nuclear and cytoplasmic compartments of cells, and in the nuclei it co-localizes with eIF4E and PML nuclear bodies [11–13]. PRH alters gene expression at multiple levels. At the transcriptional level, PRH acts as a repressor, and at the level of mRNA transport it inhibits eIF4E-dependent nuclear export of specific, “eIF4E-sensitive” transcripts [12, 14, 15]. Both of the aforementioned functions require the presence of PRH in the nuclei.
It has been consistently reported that nuclear exclusion of PRH correlated with its loss of function in certain types of thyroid and breast tumors and myeloid leukemia, [6, 10, 16]. Moreover, inhibition of NFκB activity in leukemia cells, which leads to the restoration of PRH activity, correlates with PRH re-location to the nucleus and co-localization with eIF4E bodies [10]. Conversely, forced expression of PRH leads to the disruption of eIF4E and PML nuclear bodies, dislocation of eIF4E, PML and PRH to the cytoplasm, and inhibition of eIF4E-dependent mRNA transport [12]. Thus, PRH activity depends not only on its level, but also on its subcellular localization, which in turn depends on the rates of nucleocytoplasmic trafficking. Therefore the mechanisms and the cellular factors that govern PRH nuclear import play a pivotal role in the regulation of its activity. Elucidating this mechanism could reveal how the subcellular localization of PRH and its activity are regulated. Furthermore, disruption of these mechanisms could represent a causal factor of oncogenic processes in thyroid and hematopoietic cells.
The vast majority of proteins are imported into the nuclei by forming complexes with carriers that belong to the karyopherin (Kap)/importin family [17–23](for review). The proteins to be imported contain segments known as Nuclear Localization Sequences (NLS), which bind to a Kap α family member, which in turn binds to Kap β1. Alternatively, some NLSs bind directly to a Kap β family member. The cargo-carrier complexes interact with a subset of nuclear pore proteins, and the interaction mediates the passage of the complexes across the nuclear pores [24]. In the nucleus the complexes dissociate due to the interaction of Kap β with the GTPase Ran, which in the nuclei is present in GTP bound form [25, 26]. The cargoes are released and the Kaps are recycled to the cytoplasm.
Here we present information about the PRH nuclear import pathway, namely that karyopherin 7 (Kap 7), mediates PRH nuclear import.
Materials and Methods
cDNA constructs
Human PRH was subcloned via the SpeI and HindIII sites into the bacterial expression vector pET41a (Novagen, Madison, WI), containing N-terminal GST and C-terminal His tags. EGFP was subcloned via PCR from pEGFP-C1 (Clontech, Palo Alto, CA) into the pET41a-PRH construct using the SpeI site. EGFP was subcloned via PCR from pEGFP-C1 (Clontech) into pET41a using the SpeI and KpnI sites for the GST-GFP control. The bacterial expression construct for His-Kap7 was a kind gift from Dirk Görlich [27]. Constructs for bacterial expression encoding GST-GFP-SV40 NLS, GST-Kapβ1 and His-Kapα2 were gifts from Tarick Soliman. The mouse RanBP1 cDNA was subcloned into pGEX6P-1 (Amersham Biosciences, Piscataway, NJ) using the BamH1 and XhoI sites. Human Ran was subloned into pQE30 via the BamHI and HindIII. All PCR amplifications were performed using the Advantage HF2 polymerase (Clontech) except for PRH, which was amplified using GC 2 polymerase (Clontech). All constructs were verified by sequencing. The Xpress-tagged PRH construct was pcDNA3.1AHisXpressPRH as previously described [11]. For expression of GFP-NLS in cells lines, the construct pCMV/myc/nuc/GFP (Invitrogen) was used.
Protein expression and purification
Protein expression was carried out as previously described [27, 28]. For bacterial protein expression cultures were grown at 37°C to O.D.600 ~ 1.0 and then shifted to 18°C. After the temperature equilibrated the cultures were induced with 0.1 mM IPTG and grown overnight with shaking at 170 rpm. Recombinant proteins were purified on glutathione-Sepharose 4B (Amersham Biosciences, Piscataway, NJ) or Ni-NTA agarose (Qiagen, Valencia, CA). pQE30-Ran constructs were expressed in the strain M15[pREP4](Qiagen, Valencia, CA). The pET41a constructs were expressed in BL21 (DE3) Codon Plus RIL strain (Stratagene, La Jolla, CA). All other proteins were expressed in the BL21 strain (Novagen, Madison, WI).
HeLa lysate preparation
One ml of packed HeLa cell pellet was rinsed once in ice cold transport buffer (20 mM HEPES–KOH pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM DTT). Cells were hypotonically lysed by adding 1.5 cell pellet volumes of lysis buffer (10 mM HEPES–KOH pH 7.3, 10 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT). Cells were placed on ice in buffer for 10 minutes followed by Dounce homogenization. To rupture the nuclei, the lysate was sonicated 4 times for 10 seconds. Cells were centrifuged at 13,000 g for 15 minutes followed by an additional 1 hour centrifugation at 100,000 g. Supernatants were adjusted to physiological salt levels.
GST pull-downs
GST pull downs were done essentially as described in [28]. GST fusion proteins (2 μg) were bound to glutathione Sepharose 4B. 1 μg of recombinant proteins were mixed with the Glutathione Sepharose-GST fusion proteins in the presence of binding buffer (20 mM HEPES–KOH pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 10% glycerol, 0.1% Tween-20) for 1 hour at 4°C. Beads were pulse spun at 13,000 g in a microcentrifuge. Supernatants were removed and beads were washed 4 x with 0.5 ml of washing buffer (binding buffer supplemented with 150 mM NaCl). Proteins were eluted with SDS-PAGE sample buffer, boiled for 10 min and analyzed by 10% SDS-PAGE following by coomassie blue staining to visualize bound proteins. Where applicable RanGTP was used at a concentration of 4 μM. Ran was loaded with GTP as described [29]. GST pull down assays using HeLa lysate were done essentially as described above. RanBP1 and RanGAP were added to the lysate to a final concentration of 1 μM where applicable. Immobilized GST-PRH and 0.6 ml of HeLa lysate were mixed for 2 hours at room temperature and then overnight at 4°C. Supernatants were removed and beads were washed with 4 × 1 ml of binding buffer. Proteins were eluted with SDS-PAGE sample buffer, boiled for 10 min and analyzed by 10% SDS-PAGE. To visualize the separated protein bands, the gel was stained in Coomassie blue followed by de-staining in 50% methanol/10% acetic acid in water. Protein bands within the range of 90 – 120 kDa were excised by razor blade and submitted to the Rockefeller University Protein Resource Center for mass spectrometry.
Nuclear import assays
Import reactions were performed at room temperature essentially as described [28, 30, 31]. Recombinant Kap 7 (3 μM), Kap β1(2 μM) and Kap α2 (2 μM) and cargoes (1 μM) in transport buffer (20 mM HEPES–KOH pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 250 mM sucrose, 2 mM DTT) were overlaid on digitonin permeabilized HeLa cells and incubated at room temperature for 25 min. A Ran mix (3 μM RanGDP, 0.2 μM NTF2 [32], 0.2 μM RanBP1 and 0.2 μM RNA1p [33] was added along with an energy-regenerating system (5 mM creatine phosphate, 0.25 mM ATP and GTP, and 1 unit of creatine kinase). Cells were fixed using 3% paraformaldehyde for 20 minutes and mounted using Vectashield (Vector Laboratories, Burlingame, CA). Import reactions were visualized by fluorescence microscopy using an Olympus BX60 microscope and a 40x 0.75 N.A. objective. All pictures within an experiment were taken under identical exposure conditions using an Optronics NTSC digital camera and Flashpoint acquisition software (Integrated Technologies, Indianapolis, IN).
Cell culture
The Hep G2 cell line, (cat # HB-8065), and Hek 293 cell line (cat# CRL-1573) (ATCC, Manassas, VA) were grown in Eagle’s Minimum Essential Medium with 10 % fetal bovine serum at 37°C in a 95% air/5% CO2 humid atmosphere.
Silencing of gene expression
The day before silencing RNA (siRNA) transfection, Hep G2 cells or Hek293 cells were seeded in 24 well cell culture plates at a density of ~5×103 per well. When the cells reached ~40–50% confluency (typically 24 hours later), they were transfected with 20 pmol per well with the appropriate Stealth™ siRNA duplex oligoribonucleotides (Invitrogen) complexed with Liptofectamine 2000 (Invitrogen) transfection reagent. Additionally as a transfection control (mock transfection), a no siRNA transfection was performed where cells were treated with Lipofectamine 2000 reagent that was not complexed with siRNA. SiRNA duplex sequences used: Kap 7 (5′ uaagcagauucccucaagcuguugg 3′, 5′ ccaacagcuugagggaaucugcuua 3′), Kap β1(5′ uaaacucuccuccaauagcaagggc 3′, 5′ gcccuugcuauuggaggagaguuua 3′). For gene silencing experiments using Hek 293 cells, cells were co-transfected with plasmids coding for Xpress- tagged PRH, or GFP-NLS along with siRNA. All transfections were performed according to the manufacturer’s instructions for transfecting Stealth™ siRNAs. For experiments depicted in figure 3a and 3b, cells were treated in a similar fashion as above, but were grown on 12 mm poly-L-Lysine coated coverslips (BD Biosciences, San Jose, CA).
Figure 3. In vivo gene knockdown of Kap7 prevents accumulation of PRH in the nucleus.
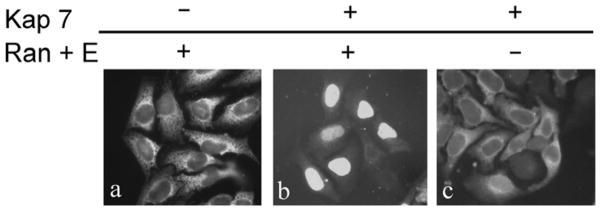
a. Hek 293 cells were co-transfected with a plasmid coding for Xpress-tagged PRH along with either siRNAs targeting Kap7 (panel a, i and ii) or Kapβ1 (panel a, v and vi). Additionally Hek 293 cells were co-transfected with a plasmid coding for GFP-NLS along with either siRNAs targeting Kap7 (panel a, iii and iv) or Kapβ1 (panel a, vii and viii). 96 hours post transfection, cells were washed, fixed, and stained with DAPI (Note that the DAPI stained cells depicted in panels a, i, iii, v, and vii are the same cells as in a, ii, iv, vi, viii, respectively). Immunofluorescence for Xpress-tagged PRH is shown in panels a, ii and vi. Depletion of Kap7 is associated with a striking decrease in the nuclear accumulation of PRH (panel a, ii) as compared to cells transfected with siRNA targeted to deplete Kapβ1 (panel a, vi). Fluorescence for GFP-NLS is shown in panels a, iv and viii. Depletion of Kap7 had no effect on the nuclear accumulation of GFP-NLS (panel a, iv). In contrast, cells transfected with siRNA targeted to deplete Kapβ1 (panel a, viii) exhibited reduced nuclear accumulation of GFP-NLS and a noticeable increase in cytoplasmic accumulation. b. Hep G2 cells were transfected with siRNAs targeted to Kap7 or Kapβ1. 96 hours post transfection, cells were washed, fixed and immunofluorescence for endogenous PRH was performed. Depletion of Kap7 is associated with a striking decrease in the nuclear accumulation of PRH (panel b, ii) as compared to control cells that were not transfected with siRNA (Mock) (panel b, i), or cells transfected with siRNA targeted to deplete Kapβ1 (panel b, iii). c. Quantitative RT-PCR utilizing gene specific DNA primers for Kap7 and Kapβ1 at 72 hours post siRNA transfection. Kap7 mRNA was significantly reduced in cells that were transfected with Kap7 siRNA as compared to a mock transfection, or cells transfected with Kapβ1 siRNA (n = 3, *p<0.005 relative to the mock transfection group). Additionally cells transfected with Kapβ1 siRNA, exhibited a significant reduction of Kapβ1 mRNA as compared to a mock transfection or cells transfected with Kap7 siRNA (n = 3, *p<0.005 relative to the mock transfection group). Error bars represent standard error of the mean (SEM). d. Western blotting using protein specific antibodies confirmed a significant reduction of Kap7 protein in cells that were transfected with Kap7 siRNA, but not cells transfected with Kapβ1 siRNA, or a mock transfection (n = 3, *p<0.005 relative to the mock transfection group). Additionally cells transfected with Kapβ1 siRNA, did not show a reduction of Kap7 protein, but did show a significant reduction of Kapβ1 protein (n = 3, *p<0.005 relative to the mock transfection group). Error bars represent SEM. e. Representative images for western blot data for experiment in c. Rows represent western blot data using respective antibodies against Kapβ1, Kap7 and GAPDH. All samples were normalized against the GAPDH loading control. Columns represent samples treated with siRNAs for Kapβ1 (β1), Kap7 (7), or the mock transfection (M).
Immunofluorescence
For gene silencing experiments, Hep G2 or Hek 293 cells were grown on coverslips and were either fixed at 72 or 96 hours post transfection in 3% paraformaldehyde in 1x phosphate buffered saline (PBS) on ice for 10 minutes followed by three washes in 1x PBS. Cells were permeabilized in 0.1% Triton X-100 for 5 minutes at room temperature followed by three washes in 1xPBS. Cells were incubated with either a rabbit polyclonal anti-PRH antibody [12] or a mouse monoclonal anti-Xpress antibody (Invitrogen) at 1:50 dilution for 2 hours at room temperature, and subsequently with fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) or a donkey anti-mouse antibody at 1:200 dilution for 45 minutes at room temperature. Note that GFP was detected by direct fluorescence. Fluorescence was observed using 100x magnification with a zoom of 2, on a LSM510 confocal microscope (Zeiss). All channels were detected separately, with no cross-talk observed between them. Micrographs represent single sections with a thickness of 300 nm. Experiments were repeated three times with >500 cells in each sample.
Quantitative real-time PCR (qrtPCR)
qRT-PCR was performed using the ΔΔCt method as described previously [34, 35]. 72 hours post transfection, Hep G2 cells were harvested and homogenized in 1ml of TRIzol Reagent (Invitrogen, Carlsbad, CA) with a plastic pestle. RNA isolation was performed according to the manufacturer’s instructions. The isolated total RNA was DNase treated in a 100 μl reaction with 6.8 Krunitz Units of DNase I (Qiagen, Valencia, CA) for 10 min at room temperature followed by total RNA purification using the Rneasy MinElute Kit (Qiagen). RNA purification was performed according to the manufacturer’s instructions. 1 μg for each total RNA sample was converted to cDNA in a 20 μL reaction using reverse transcriptase (Genisphere, Hatfield, PA) with 0.65 μL of 3 μg/μL random hexamers (Invitrogen). qRT-PCR was performed in an ABI 7900 instrument (Applied Biosystems, Foster City, CA) using 16 μL reaction volumes with Quantitect SYBR Green (Qiagen) and gene specific primers from Qiagen: (Kap β1 (cat # QT00096754), Kap 7 (cat # QT01014853), and GAPDH (cat # QT01192646)). Relative gene concentrations were normalized against GAPDH. The cycling parameters utilized for 40 cycles were: 94°C for 15 s, 56°C for 30 s, 76°C for 30 s. All samples were performed in triplicate. Data were analyzed from three independent experiments using Analysis of Variance (ANOVA) and Duncan’s Post-hoc test.
Western blotting
Hep G2 protein lysates were prepared by direct lysis of cells in SDS sample buffer 72 hours post transfection. The samples were boiled for 10 min and electrophoresed on 10% Tris-HCl SDS-PAGE and transferred to Immobilon-P membrane (Millipore, Bedford, MA). Membranes were blocked in TTBS buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) with 5% dry milk and then incubated overnight at 4°C in the following primary antibodies at a 1:1000 dilutions: Kap β1 (cat # sc-11367, Santa Cruz Biotech, CA), Kap 7 (cat # ab15840, Abcam, Cambridge, MA), and GAPDH (cat # 9484, Abcam). Blots were then incubated with anti-rabbit conjugated to horseradish peroxidase (Cell Signaling, Danvers, MA) and developed using West Dura chemiluminescent substrate (Pierce Laboratories, Rockford, IL). Western blots were developed in the linear range used for densitometry. Densitometry was conducted using Image J software. To control for inconsistencies in loading, optical densities were normalized to GAPDH protein. Data were analyzed from three independent experiments using ANOVA and Duncan’s Post-hoc test.
Results
Kap7 interacts with PRH
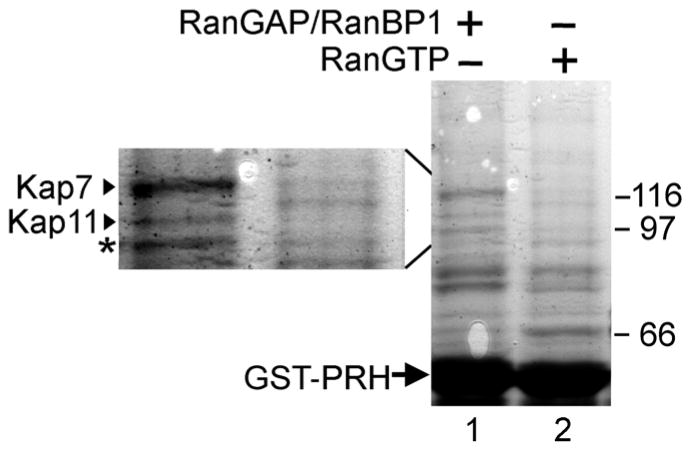
To identify what karyopherin binds to PRH and likely is responsible for its transport to the nuclear compartment, we performed a GST pull down from a HeLa cell protein lysate. HeLa cell protein lysates are routinely used for karyopherin identification studies and have previously been shown to contain most if not all known karyopherins [36]. GST-PRH was expressed in E. coli, purified, immobilized onto glutathione-Sepharose beads and mixed with a total HeLa cell lysate. To facilitate formation of the karyopherin-cargo complexes, recombinant purified RanBP1 and RanGAP were added to the lysate in order to hydrolyze the RanGTP that may be present in the lysate [37] (Fig. 1a, lane 1). As a control, purified recombinant Ran bound to a non-hydrolysable GTP analog was added to a control lysate sample, to promote dissociation of import-karyopherin-cargo complexes (Fig. 1a lane 2). After washing, the proteins retained on the beads were analyzed by SDS-PAGE followed by Coomassie staining. Only one predominant protein band was present within the 95–125 kDa known size range of the Kap β family members (Fig. 1a, lane 1 and lane 1a). Additionally, this predominant protein band was not precipitated in the HeLa lysate treated with RanGTP (Fig. 1a lane 2 and lane 2a), supporting the hypothesis that the band corresponds to a karyopherin that binds to its cargo in an import-karyopherin-cargo complex manner, which is RanGTP sensitive. To identify what protein or proteins were represented by this differentially bound band, the band was cut out of the gel and analyzed by mass spectrometry. Mass spectrometry determined with exceptionally high confidence (expect value = 5.1×10−18), that the band represented Kap7/Importin 7 [27, 31, 38]. The fact that only one major protein was precipitated from the potentially many hundreds of proteins present in the HeLa lysate within the 95–125 KDa range supports the hypothesis that Kap 7 is the Kap with the greatest affinity for PRH and likely the main if not only karyopherin that mediates the transport of PRH to the nuclear compartment.
Figure 1. Kap7 interacts with PRH.
a. Kap7 binds to GST-PRH in a GST-pull down assay from a total protein HeLa cell lysate. GST-PRH was expressed and purified from E. coli, and ~15 μg of GST-PRH was immobilized on glutathione-Sepharose and mixed with total HeLa cell lysate (lanes 1 and 2). Recombinant purified RanBP1 and RanGAP were added to the lysate (lane 1), to hydrolyze endogenous RanGTP. As a control, recombinant purified RanGTP was added to the lysate (lane 2) to promote dissociation of import Kap-cargo complexes. The proteins retained on the beads were analyzed by SDS-PAGE followed by Coomassie staining. A major protein band in the range 95–125 kDa (a characteristic for the Kap β family), was detected and analyzed by mass-spectrometry to be the karyopherin, Kap7. The left side insert (lanes 1a and 2a) is an enlargement of the 95–125 kDa region of the gel. b. Kap7 peptides identified by mass-spectrometry are depicted with highly significant expect values. c. Purified recombinant Kap7 binds directly to GST-PRH in a GST-pull down assay. PRH was expressed as a GST fusion in E. coli, purified and immobilized on glutathione-Sepharose. 1 μg of purified recombinant Kap7 was mixed with 1–2 μg of immobilized GST-PRH. After washing, the proteins retained on the beads were analyzed by SDS-PAGE followed by Coomassie staining. Lane 1 shows 50% of the Kap7 that was used in lanes 2–4. The addition of RanGTP reduced binding of Kap7 to GST-PRH (lane 4) as compared to lane 3, where no RanGTP was added. No binding of Kap7 to the control GST-GFP is detected (lane 2).
Considering that there are many thousands of proteins in a total HeLa cell lysate, it’s possible that the interaction between PRH and Kap7 is not direct, but rather occurs through an intermediate binding partner. To determine if Kap7 binds directly to PRH, a GST-pull down assay was performed using purified Kap7 protein. GST-PRH was expressed and purified from E. coli, immobilized on glutathione Sepharose and mixed with bacterially purified recombinant Kap7 (Fig. 1c). Kap7 binds to GST-PRH without a need for other factors supporting the hypothesis that the interaction between Kap7 and PRH is direct (Fig. 1c lane 3). As expected for an import-karyopherin-cargo complex, this interaction is disrupted by RanGTP (lane 4).
Kap7 mediates the import of GST-GFP-PRH in digitonin permeabilized cells
To investigate if Kap7 is capable of transporting PRH to the nuclear compartment in cells, a digitonin permeabilized cells assay [39] was performed to test if Kap7 can mediate import of PRH into the nuclei. HeLa cells were grown on glass coverslips and treated with digitonin, which permeabilizes the plasma membrane but leaves the nuclear membrane intact. Import mixes containing purified Kap7 and PRH fused to a GST-GFP tag, together with other factors necessary for nuclear import (purified Ran, RanBP1, RanGAP, NTF2 and a energy regenerating system) [30, 31] were subsequently overlaid on the cells for 25 minutes. The proteins that were not imported into the nuclei were subsequently removed by washing, and the cells were fixed and visualized by fluorescence microscopy. GST-GFP-PRH is imported into the nuclei of the permeabilized cells when Kap7 is present together with Ran, RanBP1, RanGAP, NTF2 and an energy regenerating system (Fig. 2ii). No import of PRH is detectable if Kap7 is absent (Fig. 2i). In the absence of Ran and the energy regenerating system Kap7 does not mediate PRH import (Fig. 2iii). To test for the specificity of Kap7 mediated import, we also examined the import of a GFP tagged SV40 T antigen NLS, which is imported to the nuclear compartment via Kapβ1 and Kapα2 in a Ran and energy dependent manner (Fig. 2viii). As expected, Kap7 cannot mediate the transport of the SV40 NLS to the nuclear compartment (Fig. 2iv, v, vi). Similar experiments were performed using Kapβ1 and Kapα2 to determine if they can mediate transport of PRH to the nuclear compartment. In the presence or absence of Ran and the energy regenerating system, Kapβ1 and Kapα2 did not mediate significant transport of GST-GFP-PRH in digitonin permeabilized cells (Fig. 2x, xi, xii). Lastly we previously showed that another karyopherin, Kap13, could not mediate transport of PRH to the nuclear compartment in a digitonin permeabilized cells assay [28].
Figure 2. Kap7 mediates the import of PRH in digitonin permeabilized cells in a Ran and energy dependent manner.
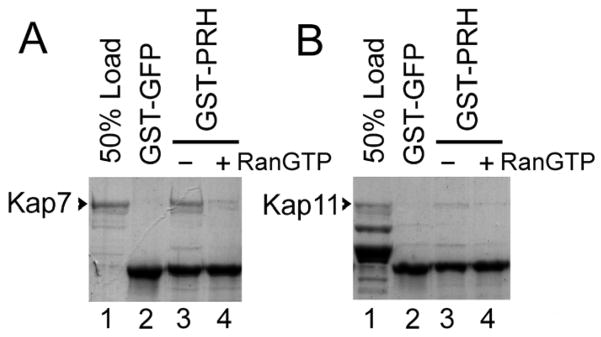
HeLa cells were permeabilized with digitonin, incubated for 25 minutes with the import mixtures, washed and fixed. Only the cargo imported into the nuclei is retained after washing. GST-GFP-PRH is imported into the nuclei in the presence of Kap7, Ran, RanBP1, RanGAP and NTF2 and an energy regenerating system (panel ii), but it is not imported in the absence of Kap7 (i) or in the absence of Ran and the energy regenerating system (iii). As expected, Kap7 cannot mediate the transport of the GFP tagged SV40 NLS to the nuclear compartment (panels iv, v, vi). Similar experiments were performed using Kapβ1 and Kapα2, which can mediate the transport of the SV40 NLS to the nuclear compartment in a Ran and energy dependent manner (panels viii). In the presence or absence of Ran and the energy regenerating system, Kapβ1 and Kapα2 did not mediate significant transport of GST-GFP-PRH to the nuclear compartment (Fig. 2x, xi, xii).
In vivo gene knockdown of Kap7 prevents significant accumulation of PRH in the nucleus
Considering that Kap7 mediates the transport of PRH to the nuclear compartment in a digitonin permeabilized cells assay, we wanted to test the hypothesis that depletion of Kap7 would prevent nuclear accumulation of PRH in vivo, supporting the hypothesis that Kap7 is the main pathway for PRH transport in vivo. Accordingly, we co-transfected the human embryonic kidney cell line, Hek 293 with a plasmid coding for Xpress-tagged PRH and with silencing RNAs (siRNAs) targeted to Kap7, to deplete cells of Kap7 mRNA and protein. 96 hours post transfection, cells were washed, fixed, and immunofluorescence for Xpress-tagged PRH was performed. Depletion of Kap7 is associated with a striking decrease in the nuclear accumulation of PRH in vivo as compared to cells that were transfected with siRNA targeted to deplete Kapβ1 mRNA and protein (Fig. 3a). The fact that depletion of Kapβ1 mRNA and protein did not visibly alter the nuclear accumulation of PRH supports the hypothesis that Kapβ1 does not mediate significant transport of PRH in vivo. To ensure that the deficit in nuclear accumulation of PRH is due to depletion of Kap 7 and not simply due to a generalized nucleocytoplasmic transport deficit, cells were transfected with Kap 7 siRNA and a plasmid coding for the expression of GFP-NLS. The GFP-NLS contains a classical NLS that is recognized and imported into the nuclei via Kapβ1/Kapα and it is not a cargo imported via the Kap7 pathway. We found that siRNA mediated depletion of Kap7 had no effect on nuclear localization of the GFP-NLS protein. In contrast, cells transfected with siRNA to deplete Kapβ1 exhibited reduced nuclear localization of the GFP-NLS protein, where there was a noticeable increase in cytoplasmic localization of GFP-NLS and a significant decrease in nuclear localization of GFP-NLS. These data are consistent with the hypothesis that siRNA mediated depletion of Kap 7 protein does not result in a global deficit in nucleocytoplasmic transport. In addition, depletion of Kap 7 results in a specific deficit for the Kap7 import pathway resulting in decreased PRH nuclear accumulation. To further support the hypothesis that Kap 7 is necessary for the import of PRH, we also examined siRNA mediated depletion of Kap 7 on the nuclear accumulation of endogenous PRH in vivo. Accordingly, we transfected the human hepatoma cell line, Hep G2, which expresses endogenous PRH, with siRNAs targeted to Kap7. 72 and 96 hours post transfection, cells were washed, fixed, and immunofluorescence for endogenous PRH was performed. Depletion of Kap7 is associated with a dramatic decrease in the nuclear accumulation of PRH in vivo as compared to control cells that were not transfected with siRNA (Mock), or cells transfected with siRNA targeted to deplete Kapβ1 mRNA and protein (Fig. 3b). Similar results were obtained for the 72 hours time point (data not shown). To confirm that transfection of siRNAs targeted to Kap7 and Kapβ1 significantly reduced Kap7 and Kapβ1 mRNA and protein in vivo, quantitative RT-PCR and western blotting was were performed from samples harvested 72 hours post siRNA transfection. (Fig. 3. c,d,e). Utilizing gene specific DNA primers, relative levels of mRNA for Kap7 and Kapβ1 were determined via qRT-PCR. The analysis of Variance (ANOVA) revealed a significant effect for group, with the Kap7 siRNA group being significantly different from both the Kapβ1 siRNA and Mock transfection groups (p<.005; Duncan’s test). Additionally cells transfected with Kapβ1 siRNA, did not show a reduction of Kap7 mRNA, but did show a significant reduction of Kapβ1 mRNA (Fig. 3c) where ANOVA revealed a significant effect for group, with the Kapβ1 siRNA group being significantly different from both the Kap7 siRNA and Mock transfection groups (p<.005; Duncan’s test). The analysis of protein levels 72 hrs post transfection also reflected a similar pattern of protein depletion as represented by the mRNA detection experiments. Western blotting using protein specific antibodies confirmed a significant reduction of Kap7 protein in cells that were transfected with Kap7 siRNA, where ANOVA revealed a significant effect for group, with the Kap7 siRNA group being significantly different from both the Kapβ1 siRNA and Mock transfection groups (p<.005; Duncan’s test). Additionally cells transfected with Kapβ1 siRNA, did not show a reduction of Kap7 protein, but did show a significant reduction of Kapβ1 protein (Fig. 3d,e), where ANOVA revealed a significant effect for group, with the Kapβ1 siRNA group being significantly different from both the Kap7 siRNA and Mock transfection groups (p<.005; Duncan’s test).
Discussion
Collectively the data support the hypothesis that Kap7 mediates the transport import of PRH to the nuclear compartment of cells. The binding of endogenous Kap7 from a HeLa cell protein lysate to GST-PRH is robust, as illustrated by the fact that only one major protein band within the size range of Karyopherin β family (95–125kDa) precipitated in the pull down experiment. Additionally the formation of the PRH-Kap7 complex is RanGTP sensitive; consistent with the notion that PRH is an import cargo for Kap7. Kap7 can bind directly to PRH in a GST-pull down assay with bacterially purified proteins and therefore intermediate binding partners are not necessary for this interaction to occur. Kap 7 can also mediate the transport import of PRH to the nuclear compartment in a digitonin permeabilized cells assay. Lastly, in vivo depletion of Kap7 dramatically reduces accumulation of PRH in the nucleus. These data support the conclusion that in the cell types investigated by us Kap7 is the the major, if not the exclusive nuclear import carrier for PRH.
Our data add a new cargo to the small set of proteins known to be imported by Kap7. Other proteins previously identified as Kap7 cargos are the glucocorticoid receptor [40], the four core histones, [41], the linker histone H1, imported by Kap7 in cooperation with Kap β1 [27], the ribosomal proteins L23a, S7 and L5, [31], zinc finger protein EZI [42], Smads [43, 44], p35 [45], and c-jun [46]. Kap 7 was also shown to mediate the import of viral components: human immunodeficiency virus type 1 (HIV-1) Rev protein [47], HIV-1 intracellular reverse transcription complexes and the isolated integrase that is a constituent of these complexes [38, 48] and the adenovirus core protein pVII [49]. In addition, the mammalian and Drosophila homologs of Kap7 mediate the nuclear import of the MAP kinase ERK [50–52].
The Drosophila Kap7 homolog, DIM-7, mediates the transport of the homeodomain protein Caudal [53], which is involved in specifying the anterior-posterior body axis [53, 54]. DIM-7 binds to a segment at the C terminal end of the Caudal homeodomain, which is very similar to the corresponding region of human PRH. In addition, mutations in this same region of PRH strongly inhibit nuclear accumulation of PRH [12]. These observations further substantiate our conclusion that PRH is imported into the nuclei by Kap7.
The information presented here creates the basis for further investigations aimed at understanding the causes of reduced nuclear presence of PRH in certain leukemias and thyroid tumors. For example a straightforward hypothesis to be tested is whether the reduced PRH nuclear concentration in tumors is caused by low levels of Kap7. Detailed knowledge of PRH trafficking mechanism and its regulation may also suggest interventions that could correct PRH localization and inhibit tumorigenesis. Studies of PRH trafficking regulation may also reveal new modes by which PRH controls embryonal development.
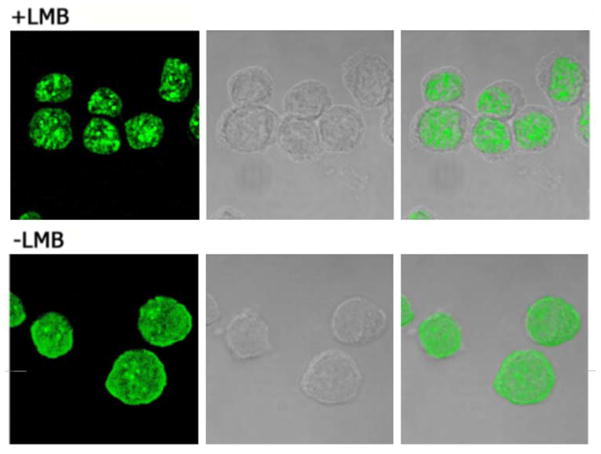
Figure 4. Leptomycin B induces nuclear accumulation of PRH.
K562 cells in culture were treated with leptomycin B (upper panels) or vehicle only (lower panels). After treatment the cells were fixed and PRH was detected using anti PRH antibodies followed by FITC-labeled secondary antibodies; left-side panels: confocal fluorescence images; middle panels: phase-contrast; right-side: overlay of the phase-contrast and PRH signal. The cytoplasmic areas, visible as rings in the phase-contrast images, are devoid of PRH in the leptomycin-treated cells (upper right panel), but not in the control cells (lower right).
Figure 5. The NLS recognized in PRH by Kap7 most likely includes the basic residues at the C-terminal of the homeodomain.
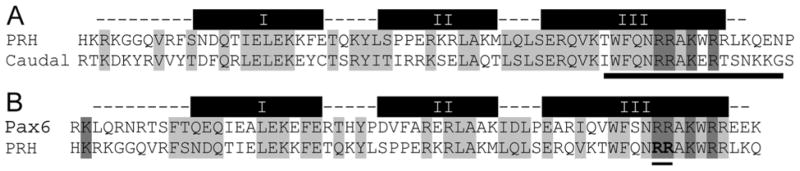
A. PRH and Caudal, which share similar import pathways via Kap7, share high similarity within the domain responsible for binding to Kap7. The homeodomain regions of PRH and Caudal (dashed lines) are aligned. The thick bars represent α helical regions. Residues highlighted in gray are similar between PRH and Caudal. The underlined region of Caudal confers binding to Kap7 (48). Conserved basic residues within this region are marked by dark gray.
Acknowledgments
We thank Dr. Elias Coutavas for valuable suggestions and Drs. Dirk Görlich, and Tarick Soliman for DNA constructs. This work was supported by grants from the National Institutes of Health (R01 GM057569 to AR and RO1 98571 to KLBB). KLBB holds a Canada Research Chair in Molecular Biology of the Cell Nucleus. IT is a Special Fellow of the Leukemia and Lymphoma Society, USA.
References
- 1.Crompton MR, Bartlett TJ, MacGregor AD, et al. Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res. 1992;20:5661–5667. doi: 10.1093/nar/20.21.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez Barbera JP, Clements M, Thomas P, et al. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development. 2000;127:2433–2445. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- 3.Manfioletti G, Gattei V, Buratti E, et al. Differential expression of a novel proline-rich homeobox gene (Prh) in human hematolymphopoietic cells. Blood. 1995;85:1237–1245. [PubMed] [Google Scholar]
- 4.Jayaraman PS, Frampton J, Goodwin G. The homeodomain protein PRH influences the differentiation of haematopoietic cells. Leuk Res. 2000;24:1023–1031. doi: 10.1016/s0145-2126(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 5.Hromas R, Radich J, Collins S. PCR cloning of an orphan homeobox gene (PRH) preferentially expressed in myeloid and liver cells. Biochem Biophys Res Commun. 1993;195:976–983. doi: 10.1006/bbrc.1993.2140. [DOI] [PubMed] [Google Scholar]
- 6.D’Elia AV, Tell G, Russo D, et al. Expression and localization of the homeodomain-containing protein HEX in human thyroid tumors. J Clin Endocrinol Metab. 2002;87:1376–1383. doi: 10.1210/jcem.87.3.8344. [DOI] [PubMed] [Google Scholar]
- 7.Bedford FK, Ashworth A, Enver T, et al. HEX: a novel homeobox gene expressed during haematopoiesis and conserved between mouse and human. Nucleic Acids Res. 1993;21:1245–1249. doi: 10.1093/nar/21.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen GM, Justice MJ. Activation of Hex and mEg5 by retroviral insertion may contribute to mouse B-cell leukemia. Oncogene. 1999;18:6531–6539. doi: 10.1038/sj.onc.1203023. [DOI] [PubMed] [Google Scholar]
- 9.George A, Morse HC, 3rd, Justice MJ. The homeobox gene Hex induces T-cell-derived lymphomas when overexpressed in hematopoietic precursor cells. Oncogene. 2003;22:6764–6773. doi: 10.1038/sj.onc.1206822. [DOI] [PubMed] [Google Scholar]
- 10.Topisirovic I, Guzman ML, McConnell MJ, et al. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol. 2003;23:8992–9002. doi: 10.1128/MCB.23.24.8992-9002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topcu Z, Mack DL, Hromas RA, et al. The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: a RING may link hematopoiesis and growth control. Oncogene. 1999;18:7091–7100. doi: 10.1038/sj.onc.1203201. [DOI] [PubMed] [Google Scholar]
- 12.Topisirovic I, Culjkovic B, Cohen N, et al. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 2003;22:689–703. doi: 10.1093/emboj/cdg069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topisirovic I, Kentsis A, Perez JM, et al. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol Cell Biol. 2005;25:1100–1112. doi: 10.1128/MCB.25.3.1100-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiral M, Bess K, Goodwin G, et al. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J Biol Chem. 2001;276:2961–2970. doi: 10.1074/jbc.M004948200. [DOI] [PubMed] [Google Scholar]
- 15.Swingler TE, Bess KL, Yao J, et al. The proline-rich homeodomain protein recruits members of the Groucho/Transducin-like enhancer of split protein family to co-repress transcription in hematopoietic cells. J Biol Chem. 2004;279:34938–34947. doi: 10.1074/jbc.M404488200. [DOI] [PubMed] [Google Scholar]
- 16.Puppin C, Puglisi F, Pellizzari L, et al. HEX expression and localization in normal mammary gland and breast carcinoma. BMC Cancer. 2006;6:192. doi: 10.1186/1471-2407-6-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weis K. Nucleocytoplasmic transport: cargo trafficking across the border. Curr Opin Cell Biol. 2002;14:328–335. doi: 10.1016/s0955-0674(02)00337-x. [DOI] [PubMed] [Google Scholar]
- 19.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 20.Quimby BB, Corbett AH. Nuclear transport mechanisms. Cell Mol Life Sci. 2001;58:1766–1773. doi: 10.1007/PL00000816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komeili A, O’Shea EK. New perspectives on nuclear transport. Annu Rev Genet. 2001;35:341–364. doi: 10.1146/annurev.genet.35.102401.090720. [DOI] [PubMed] [Google Scholar]
- 22.Marelli M, Dilworth DJ, Wozniak RW, et al. The dynamics of karyopherin-mediated nuclear transport. Biochem Cell Biol. 2001;79:603–612. [PubMed] [Google Scholar]
- 23.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bednenko J, Cingolani G, Gerace L. Nucleocytoplasmic transport: navigating the channel. Traffic. 2003;4:127–135. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 25.Steggerda SM, Paschal BM. Regulation of nuclear import and export by the GTPase Ran. Int Rev Cytol. 2002;217:41–91. doi: 10.1016/s0074-7696(02)17012-4. [DOI] [PubMed] [Google Scholar]
- 26.Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol. 2000;12:302–307. doi: 10.1016/s0955-0674(00)00093-4. [DOI] [PubMed] [Google Scholar]
- 27.Jakel S, Albig W, Kutay U, et al. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ploski JE, Shamsher MK, Radu A. Paired-type homeodomain transcription factors are imported into the nucleus by karyopherin 13. Mol Cell Biol. 2004;24:4824–4834. doi: 10.1128/MCB.24.11.4824-4834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lounsbury KM, Beddow AL, Macara IG. A family of proteins that stabilize the Ran/TC4 GTPase in its GTP-bound conformation. J Biol Chem. 1994;269:11285–11290. [PubMed] [Google Scholar]
- 30.Moore MS, Blobel G. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- 31.Jakel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moroianu J, Hijikata M, Blobel G, et al. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci U S A. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Floer M, Blobel G. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- 34.Ploski JE, Newton SS, Duman RS. Electroconvulsive seizure-induced gene expression profile of the hippocampus dentate gyrus granule cell layer. J Neurochem. 2006;99:1122–1132. doi: 10.1111/j.1471-4159.2006.04156.x. [DOI] [PubMed] [Google Scholar]
- 35.Ploski JE, Pierre VJ, Smucny J, et al. The activity-regulated cytoskeletal-associated protein (Arc/Arg3.1) is required for memory consolidation of pavlovian fear conditioning in the lateral amygdala. J Neurosci. 2008;28:12383–12395. doi: 10.1523/JNEUROSCI.1662-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorlich D, Dabrowski M, Bischoff FR, et al. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bischoff FR, Krebber H, Smirnova E, et al. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fassati A, Gorlich D, Harrison I, et al. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 2003;22:3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedman ND, Yamamoto KR. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol Biol Cell. 2004;15:2276–2286. doi: 10.1091/mbc.E03-11-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baake M, Bauerle M, Doenecke D, et al. Core histones and linker histones are imported into the nucleus by different pathways. Eur J Cell Biol. 2001;80:669–677. doi: 10.1078/0171-9335-00208. [DOI] [PubMed] [Google Scholar]
- 42.Saijou E, Itoh T, Kim KW, et al. Nucleocytoplasmic shuttling of the zinc finger protein EZI Is mediated by importin-7-dependent nuclear import and CRM1-independent export mechanisms. J Biol Chem. 2007;282:32327–32337. doi: 10.1074/jbc.M706793200. [DOI] [PubMed] [Google Scholar]
- 43.Xu L, Yao X, Chen X, et al. Msk is required for nuclear import of TGF-{beta}/BMP-activated Smads. J Cell Biol. 2007;178:981–994. doi: 10.1083/jcb.200703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao X, Chen X, Cottonham C, et al. Preferential utilization of Imp7/8 in nuclear import of Smads. J Biol Chem. 2008;283:22867–22874. doi: 10.1074/jbc.M801320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu X, Choi YK, Qu D, et al. Identification of nuclear import mechanisms for the neuronal Cdk5 activator. J Biol Chem. 2006;281:39014–39021. doi: 10.1074/jbc.M512663200. [DOI] [PubMed] [Google Scholar]
- 46.Waldmann I, Walde S, Kehlenbach RH. Nuclear import of c-Jun is mediated by multiple transport receptors. J Biol Chem. 2007;282:27685–27692. doi: 10.1074/jbc.M703301200. [DOI] [PubMed] [Google Scholar]
- 47.Arnold M, Nath A, Hauber J, et al. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J Biol Chem. 2006;281:20883–20890. doi: 10.1074/jbc.M602189200. [DOI] [PubMed] [Google Scholar]
- 48.Ao Z, Huang G, Yao H, et al. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J Biol Chem. 2007;282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- 49.Wodrich H, Cassany A, D’Angelo MA, et al. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J Virol. 2006;80:9608–9618. doi: 10.1128/JVI.00850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenzen JA, Baker SE, Denhez F, et al. Nuclear import of activated D-ERK by DIM-7, an importin family member encoded by the gene moleskin. Development. 2001;128:1403–1414. doi: 10.1242/dev.128.8.1403. [DOI] [PubMed] [Google Scholar]
- 51.James BP, Bunch TA, Krishnamoorthy S, et al. Nuclear localization of the ERK MAP kinase mediated by Drosophila alphaPS2betaPS integrin and importin-7. Mol Biol Cell. 2007;18:4190–4199. doi: 10.1091/mbc.E06-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol Cell. 2008;31:850–861. doi: 10.1016/j.molcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Mlodzik M, Gehring WJ. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987;48:465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- 54.Moreno E, Morata G. Caudal is the Hox gene that specifies the most posterior Drosophile segment. Nature. 1999;400:873–877. doi: 10.1038/23709. [DOI] [PubMed] [Google Scholar]