Key Points
Regulation of genes required for B-cell progenitor proliferation is exquisitely dependent on Moz gene dosage.
Loss of one Moz allele delays the onset of MYC-driven lymphoma by 3.9-fold.
Abstract
The histone acetyltransferase MOZ (MYST3, KAT6A) is the target of recurrent chromosomal translocations fusing the MOZ gene to CBP, p300, NCOA3, or TIF2 in particularly aggressive cases of acute myeloid leukemia. In this study, we report the role of wild-type MOZ in regulating B-cell progenitor proliferation and hematopoietic malignancy. In the Eμ-Myc model of aggressive pre-B/B-cell lymphoma, the loss of just one allele of Moz increased the median survival of mice by 3.9-fold. MOZ was required to maintain the proliferative capacity of B-cell progenitors, even in the presence of c-MYC overexpression, by directly maintaining the transcriptional activity of genes required for normal B-cell development. Hence, B-cell progenitor numbers were significantly reduced in Moz haploinsufficient animals. Interestingly, we find a significant overlap in genes regulated by MOZ, mixed lineage leukemia 1, and mixed lineage leukemia 1 cofactor menin. This includes Meis1, a TALE class homeobox transcription factor required for B-cell development, characteristically upregulated as a result of MLL1 translocations in leukemia. We demonstrate that MOZ localizes to the Meis1 locus in pre–B-cells and maintains Meis1 expression. Our results suggest that even partial inhibition of MOZ may reduce the proliferative capacity of MEIS1, and HOX-driven lymphoma and leukemia cells.
Introduction
During hematopoiesis, relatively quiescent stem cells differentiate in a step-wise manner through progenitor stages to form mature blood cells. Chromatin modifications, and the nuclear enzymes that produce them, are intimately linked to gene transcription1 and play a central role in regulating hematopoiesis.2-4 Not surprisingly, given the importance of chromatin in regulating hematopoietic stem and progenitor cells, mutations in genes encoding epigenetic regulators are commonly found in leukemia and lymphoma.
The monocytic leukemia zinc finger protein, MOZ (MYST3; KAT6A), regulates chromatin conformation by acetylating histones.5 MOZ was first identified in a recurrent t(8;16)(p11;p13) chromosomal translocation leading to the fusion of MOZ with CBP in cases of acute myeloid leukemia (AML).6 Since its discovery, additional translocation partners of MOZ including p300,7 TIF2,8 and NCOA39 have been described. The MOZ-TIF2 chromosomal translocation has been studied in detail. The MOZ-TIF2 fusion protein confers self-renewing properties upon hematopoietic progenitors leading to transplantable leukemia.10,11 AML arising from MOZ-chromosomal translocations is particularly aggressive.12,13 The median survival of patients with MOZ–translocation-driven leukemia is reported to be between 2 and 10 months post-diagnosis.12,14 This shows that the deregulation of MOZ has potent effects on the progression of hematopoietic malignancies.
Consistent with its role in leukemia, the endogenous Moz gene is essential for the establishment of hematopoietic stem cells (HSCs) during murine development.15 This role of MOZ is dependent on its histone acetyltransferase activity, as mice homozygous for a point mutation that affects its catalytic domain show decreased HSC activity.16 Furthermore, the same mice have decreased numbers of immature B cells, suggesting that MOZ may regulate B-cell development at the chromatin level.
Because the regulation of progenitor proliferation is critical for producing normal numbers of blood cells, and also because MOZ is a chromatin regulator intimately involved in hematopoiesis and leukemia, we have examined the role of MOZ in B-cell progenitors in healthy mice and in a mouse model of MYC-driven lymphoma. We examined Eμ-Myc transgenic mice,17 which model the genetic lesion found in human Burkitt lymphoma, t(8;14)(q24;q32). This translocation brings the Myc gene under the control of an immunoglobulin heavy chain regulatory element. The lymphoid-specific immunoglobulin heavy chain enhancer (Eμ) is first active at the pro–B-cell/pre–B-cell stage and when used to direct the expression of Myc, leads to a marked increase in the proliferation of pre–B-cell progenitors. Eμ-Myc transgenic mice remain free of overt disease until additional cooperating oncogenic mutations arise that prevent apoptotic cell death.18,19
In this study, we show that Moz haploinsufficiency leads to a 3.9-fold increase in the survival of Eμ-Myc lymphoma prone mice. MOZ was required to maintain B-cell progenitor numbers, both in the presence and absence of MYC overexpression. We show that MOZ is essential for maintaining normal transcriptional levels of Meis1, Hoxa7, and Hoxa9, genes commonly overexpressed in leukemia. Our data suggest that even partial inhibition of MOZ could significantly reduce progression of MEIS1- and HOX-gene driven leukemias.
Materials and methods
Mice
Experiments were approved by the Walter and Eliza Hall Animal Ethics Committee and conformed to the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. Mice were C57BL/6 and kept in a 14-hour light and 10-hour dark cycle at 22°C. The Eμ-MycT/+, Moz−, Ink4a-Arf −, and Trp53− alleles have been previously described.17,20-22 Mice suffering from Eμ-Myc–induced lymphoma were euthanized as soon as palpable lymph nodes or spleen, or breathing difficulties were evident. For transplantation assays, 2 million bone marrow (BM) cells from wild-type (WT) and Moz+/− mice were injected into lethally irradiated recipients (2 doses: 5.5 Gy separated by 3 hours). Hematopoietic analyses were carried out on recipients 4 months after transplantation.
Cell culture
Progenitor B-cell cultures were carried out as described by Lee et al.23 Cell viability was determined using propidium iodide and Annexin-V binding.
RNA sequencing
BM pre–B-cells (B220+, CD19+, c-KITneg, and sIgMneg) were isolated by fluorescence-activated cell sorter (FACS) from 3 WT and 3 Moz+/− adult mice. RNA was isolated and sequenced on the Illumina HiSeq 2000 platform. The single-end 50 bp reads were aligned using Subread24 and analyzed using limma and voom.25 Gene set enrichment analyses used Roast,26 which correlates differential expression results from different experiments taking into account the direction and magnitude of expression changes in both experiments. More details are provided in supplemental “Methods” on the Blood Web site.
Statistical analyses
All statistical analyses were carried out using Stata version 12 (Stata Corp., College Station, TX). Data were analyzed using one-factorial analysis of variance, with Moz genotype as the independent factor, followed by Bonferroni’s post hoc test. Mutation frequencies in Moz+/−; Eμ-MycT/+ vs Moz+/+; Eμ-MycT/+ lymphomas (Figure 1) were analyzed using Pearson’s χ2 test.
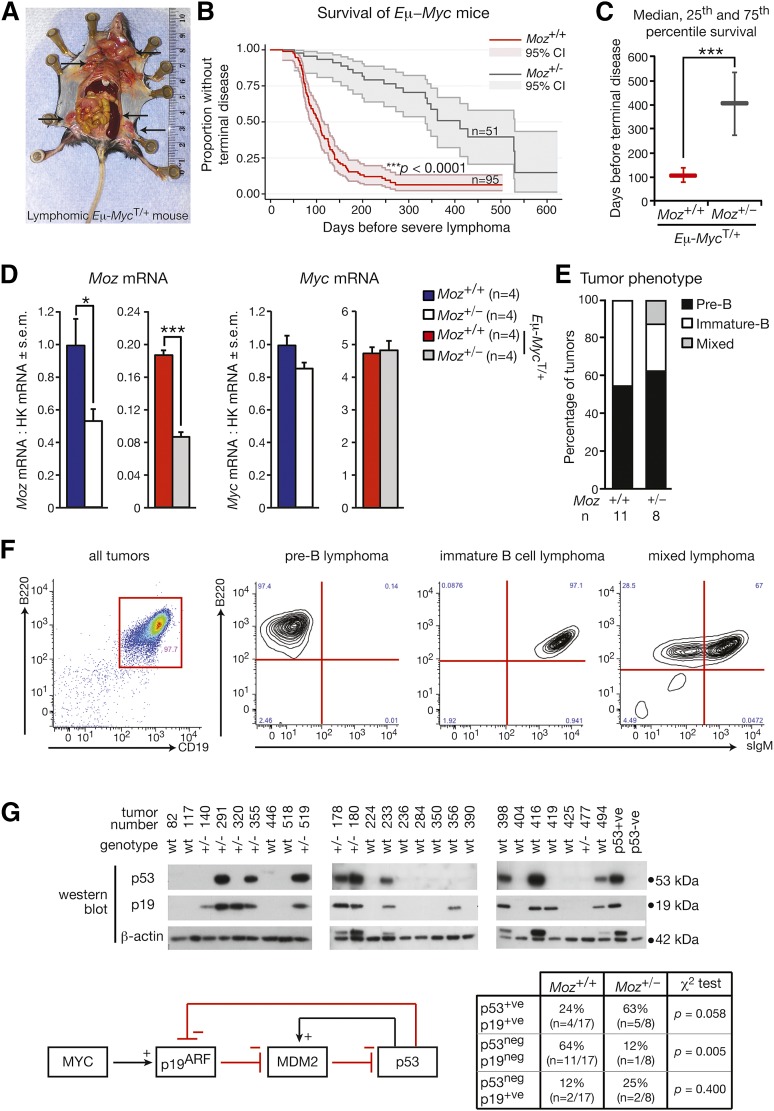
Figure 1.
Loss of one allele of Moz increases lymphoma-free survival by 3.9-fold in the Eμ-Myc model. (A) Typical features of an Eμ-Myc lymphoma. Arrows indicate markedly enlarged lymph nodes and spleen, which are characteristic of Eμ-MycT/+ lymphoma. (B) Kaplan-Meier survival curves comparing Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice. Shading represents the 95% confidence interval. (C) Depiction of the median, 25th, and 75th percentile survival periods of Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice. (D) Moz and Myc mRNA levels in pre–B-cells. Gene expression levels were standardized to housekeeping genes Hsp90ab1 and Gapdh, and expression in WT samples was designated as 1. (E) Quantification of tumor immunophenotypes. (F) Representative flow cytometry plots depicting pre-B, immature B, and mixed pre-B/immature B-cell lymphomas. (G) Analysis of p53 and p19ARF status of tumor samples from Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice. High p53 expression is characteristic of most Trp53 mutations, as mutated p53 is no longer able to activate Mdm2, preventing the induction of p53 destruction (see schematic). Normally, high levels of p19ARF are evident in Eμ-Myc transgenic cells; however, upon Arf deletion, p19ARF protein is not detectable. Data above are presented as mean ± SEM. Asterisks indicate a statistically significant difference between Moz+/− and WT, or between Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice at *P < .05 and ***P < .001.
Results
Loss of one allele of Moz increases survival by 3.9-fold in the Eμ-Myc lymphoma model
To determine the function of WT MOZ in hematopoietic malignancy, we crossed Moz+/− mice20 to Eμ-Myc transgenic mice.17 Because Moz homozygous mutants are perinatal lethal,15 we confined our analysis to Moz heterozygous mice (Moz+/−). Similar to previous studies,23,27,28 Eμ-Myc transgenic mice (Eμ-MycT/+) with a WT complement of two Moz alleles developed lymphoma with a median survival age of 105 days (Figure 1A-C). In contrast, Moz+/−; Eμ-MycT/+ mice showed a 3.9-fold increase in survival time, with a median survival age of 411 days (Figure 1B-C) (P < .0001). Moz messenger RNA (mRNA) was decreased 55% in pre-leukemic Moz+/−; Eμ-MycT/+ pre–B-cells (Figure 1D). The expression of the Eμ-Myc transgene in pre-leukemic Moz+/−; Eμ-MycT/+ vs Moz+/+; Eμ-MycT/+ pre–B-cells was similar (Figure 1D).
In Eμ-MycT/+ mice, clonal lymphomas can represent neoplastic counterparts of pre–B-cells, immature B cells, or in some cases, mixed pre-B and immature B cells.27 Similar frequencies of lymphomas originating from pre–B-cells and immature B cells were observed in Moz+/−; Eμ-MycT/+ mice compared with Moz+/+; Eμ-MycT/+ controls (Figure 1E-F). No c-KIT expression was present on any of the lymphomas analyzed, suggesting the absence of pro–B-cells in lymphoma samples (n = 0/25).
In pre-leukemic Eμ-Myc transgenic mice, high levels of apoptosis counteract increased the proliferation in B-lymphoid progenitors.19,29 Lymphoma arises once additional oncogenic mutations, frequently affecting the p53-MDM2-ARF axis, overcome the ability of c-MYC to induce apoptosis.18,28 Mutations in Trp53, as indicated by high p53 and p19ARF expression, as well as deletion of p19ARF (shown by the absence of the p19ARF protein), were both observed in lymphomas isolated from Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice (Figure 1G). Some differences in the frequency of acquisition of Trp53 and Arf mutations in Moz+/−; Eμ-MycT/+ vs Moz+/+; Eμ-MycT/+ mice were observed. Although differences in the frequency of inactivating mutations in p53 did not reach significance, the loss of p19ARF was significantly less common in Moz+/−; Eμ-MycT/+ mice. Nevertheless, mutations in Trp53 and Arf could both lead to terminal lymphoma in Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice.
Moz haploinsufficiency results in reduced numbers of B-cell progenitors
At 4-weeks of age, Moz+/− spleens were 35% smaller than those of WT animals (Figure 2A). Furthermore, the spleens of pre-leukemic Moz+/−; Eμ-MycT/+ mice were half the weight of spleens isolated from Moz+/+; Eμ-MycT/+ mice (Figure 2A). In contrast, Moz haploinsufficiency did not affect overall BM cellularity (Figure 2B).
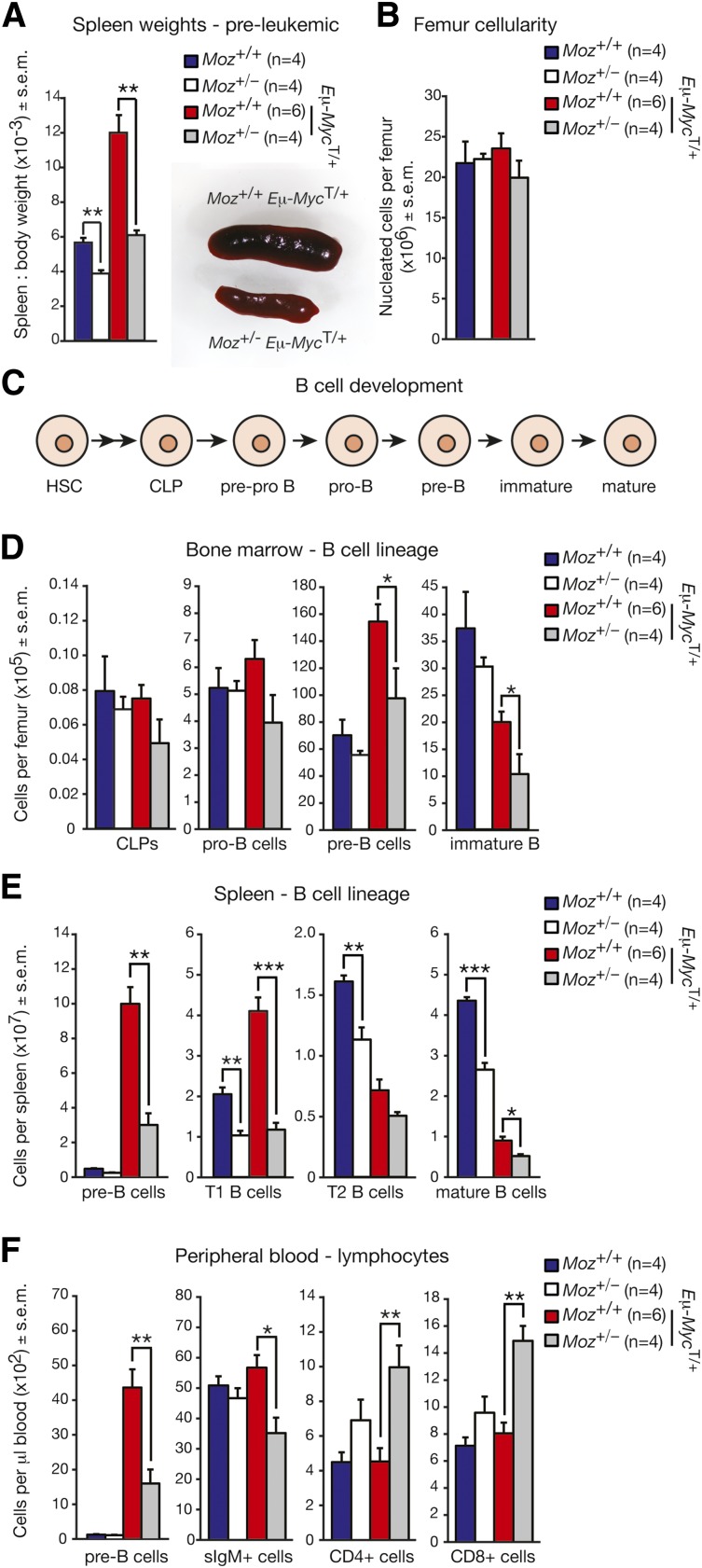
Figure 2.
Pre-leukemic Moz+/−; Eμ-MycT/+ mice have reduced numbers of B-cell progenitors compared with Moz+/+; Eμ-MycT/+ controls. (A) Spleen weights in mice relative to overall body weight. (B) Femoral BM cellularity (nucleated cells only). (C) Outline of B-cell development. HSCs differentiate via multipotent progenitors to form CLPs. CLPs differentiate in a stepwise manner through pre-pro B cells, pro–B-cells, pre–B-cells, immature B cells, and eventually into mature B cells. The different cell types can be detected based on the expression of specific cell surface markers (supplemental Table 8). (D) Numbers of B-cell progenitors in the BM. (E) Numbers of B-cell progenitors and mature B cells in the spleen. (F) Quantification of major peripheral blood lineages. Data above are presented as mean ± SEM. Mice were 3- to 4-weeks old. Asterisks indicate a statistically significant difference between Moz+/− and WT, or between Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice at *P < .05, **P < .01, and ***P < .001. T1/T2, transitory 1/2 B cells.
We enumerated B-cell progenitors in 4-week-old WT, Moz+/−, Moz+/+; Eμ-MycT/+ and Moz+/−; Eμ-MycT/+ mice by flow cytometry (Figure 2C; supplemental Table 8). There were no differences in the numbers of BM common lymphoid progenitors (CLPs) or pro–B-cells in WT, Moz+/−, Moz+/+; Eμ-MycT/+ and Moz+/−; Eμ-MycT/+ mice (Figure 2D). However, the numbers of pre–B-cells were reduced by 37%, and the numbers of immature B cells by 50% in the BM of Moz+/−; Eμ-MycT/+ mice compared with Moz+/+; Eμ-MycT/+ controls (Figure 2D). Consistent with the reduced size of Moz+/− spleens, the numbers of mature T1 and T2 B cells were significantly decreased in Moz+/− spleens compared with WT (Figure 2E). Peripheral blood counts revealed a significant reduction in the overall numbers of lymphoid cells in pre-leukemic 4-week-old Moz+/−; Eμ-MycT/+ mice compared with Moz+/+; Eμ-MycT/+ controls (supplemental Table 1). Consistently, a 64% reduction in the numbers of pre–B-cells, and a 38% reduction in the numbers of surface IgM-positive B cells was observed in the peripheral blood of Moz+/−; Eμ-MycT/+ mice compared with Moz+/+; Eμ-MycT/+ littermates (Figure 2F). In addition, reduced numbers of mature B-cell populations were observed in Peyer’s patches and peritoneal lavage of 5-week-old Moz+/− mice compared with WT (supplemental Figure 1). These results revealed a substantial reduction in B lymphopoiesis in Moz+/− mice, with the defect being much more prominent in Eμ-MycT/+ mice compared with those lacking the Eμ-Myc transgene at 4-weeks of age.
The severity of B-cell–deficiency in Moz+/− mice increases with age
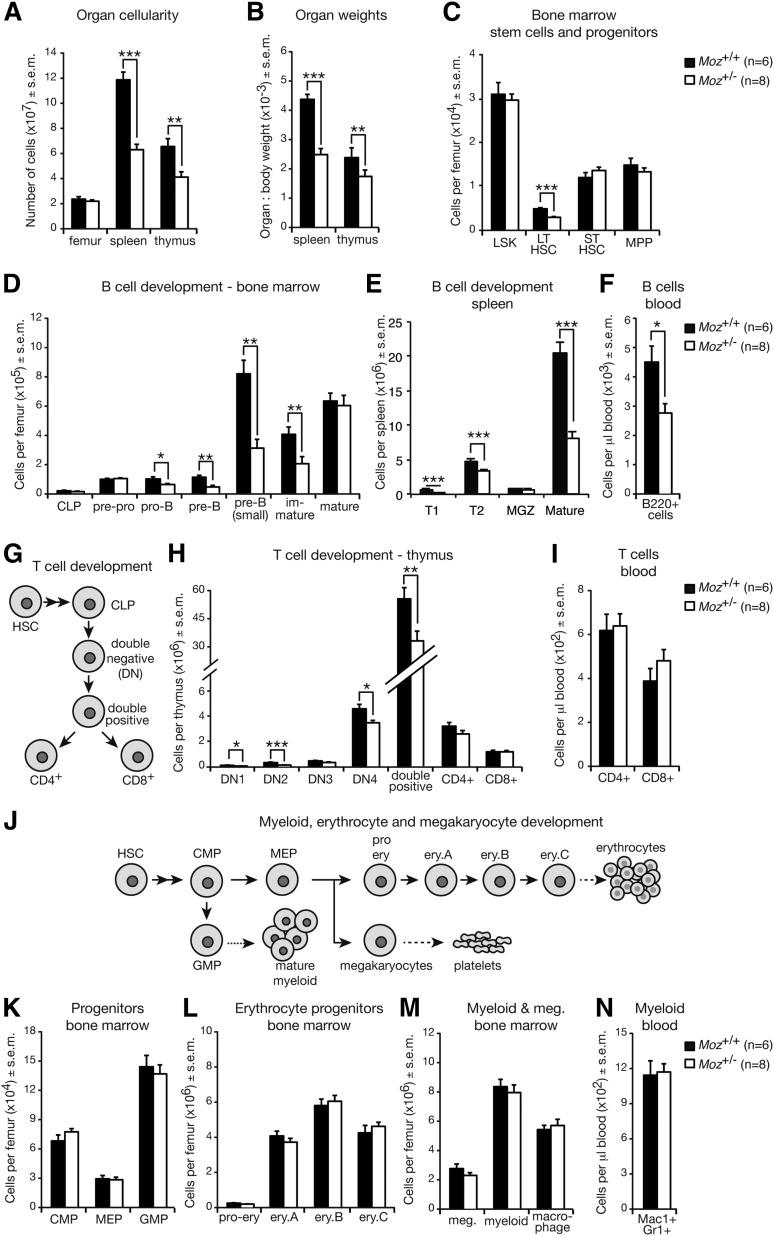
Because the deficiency in B-cell progenitors was more severe in Moz+/− mice possessing the Eμ-Myc transgene than in those without, we investigated whether 18- to 20-week-old adult Moz+/− mice, where B-cell progenitors would have undergone greater rounds of replication, showed more severe defects compared with 4-week-old mice. Moz+/− mice displayed reduced spleen and thymus cellularity and weight compared with WT controls (Figure 3A-B). Consistent with the requirement of MOZ in HSCs,15 long-term HSCs (LT-HSCs) were reduced by around 50% (Figure 3C). In comparison with the mild defects observed in Moz+/− animals vs WT controls at 4-weeks of age, the reduction in B-cell progenitors was much more severe in 18- to 20-week-old Moz+/− mice (Figure 3D-F). Although a reduction in some T-cell progenitors was also evident in Moz+/− mice relative to WT controls (Figure 3G-I), common myeloid progenitors, megakaryocyte erythrocyte progenitors (MEP), granulocyte macrophage progenitors (GMP) (Figure 3J-K), erythrocyte progenitors (Figure 3J,L), and myeloid, macrophage, and megakaryocyte (Figure 3J,M-N) numbers were unaffected.
Figure 3.
Adult 18- to 20-week-old Moz+/− mice display a significant reduction in B-cell progenitors. (A) Femur, spleen, and thymus cellularity of Moz+/− and WT mice. (B) Spleen and thymus weights relative to overall body weight. (C) Quantification of HSC and early hematopoietic progenitors in the BM. (D) Enumeration of B-cell progenitors in the BM. (E) B-cell subset numbers in the spleen. (F) Quantification of B-cell numbers in the peripheral blood. (G) Schematic of T-cell development. CLPs give rise to DN1 cells, which via DN2, DN3, and DN4 intermediaries produce CD4/CD8 double-positive cells. Single CD4+ and CD8+ cells are derived from double-positive cells. (H) Enumeration of T-cell progenitors and CD4+ and CD8+ cells in the thymus. (I) Quantification of T cells in the peripheral blood. (J) Schematic of myeloid, erythrocyte, and megakaryocyte development. (K) Numbers of CLPs, MEPs, and GMPs in the BM. (L) Quantification of erythrocyte progenitors in the BM. (M) Enumeration of myeloid cells and megakaryocytes in the BM. (N) Numbers of myeloid cells in the peripheral blood. Data above are presented as mean ± SEM. Asterisks indicate a statistically significant difference between Moz+/− and WT at *P < .05, **P < .01, and ***P < .001. Cell surface markers used to discriminate between these cell populations are outlined in supplemental Table 9. CMP, common myeloid progenitor; DN, double-negative; Ery, erythrocyte; GMP, granulocyte macrophage progenitor; LSK, lineage negative, Sca-1 positive, c-KIT positive population; LT-HSC, long-term HSC; Meg, megakaryocyte; MEP, megakaryocyte erythrocyte progenitor; MGZ, marginal zone B cells; MPP, multipotent progenitor; ST-HSC, short-term HSC; T1/T2, transitory 1/2 B-cells.
Reduction of B-cell progenitors in Moz+/− mice is due to a hematopoietic-intrinsic defect
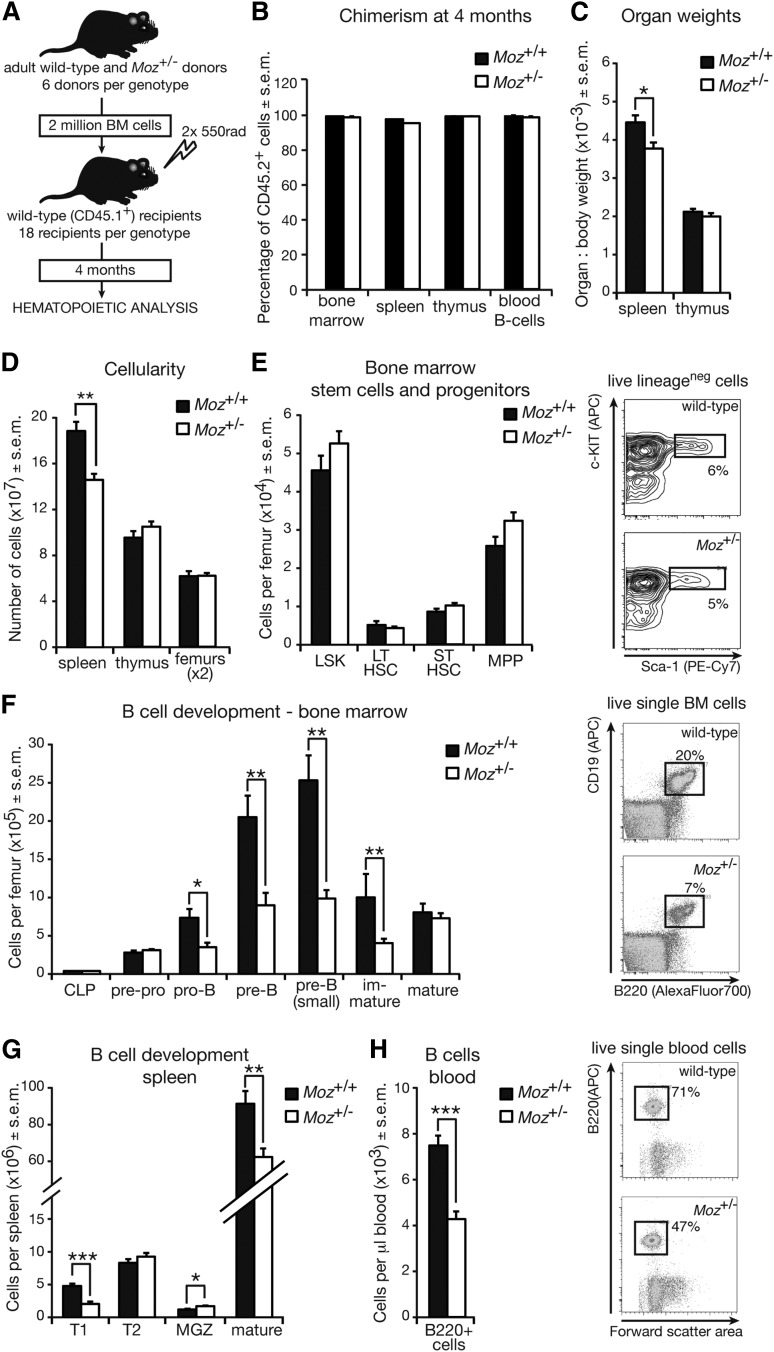
To investigate whether the reduction in B-cell progenitors in Moz+/− mice was intrinsic to the hematopoietic system, we conducted transplantation experiments. We confined our analyses to mice that do not carry the Eμ-Myc transgene, as secondary mutations in the Eμ-Myc model could lead to lymphoma and confound the effects of MOZ on the hematopoietic system. We transplanted 2 million BM cells from 12-week-old WT and Moz+/− donors into lethally irradiated WT recipients (C57BL/6-Ly5 congenic, Figure 4A). The hematopoietic system of recipient mice was analyzed 4 months after transplantation. The contribution of CD45.2+ donor cells was over 95% in the spleen, BM, thymus, and the peripheral blood of recipient mice (Figure 4B). Four months after transplantation, mice receiving Moz+/− cells displayed decreased spleen weight and cellularity compared with recipients of WT BM (Figure 4C). The thymus and BM cellularity and numbers of early progenitors in recipients of Moz+/− BM were similar to controls (Figure 4D-E).
Figure 4.
Defects in B-lymphoid cell development in Moz+/− mice are intrinsic to the hematopoietic system. (A) Experimental design. (B) Chimerism levels in recipient mice of Moz+/− or WT BM cells 4 months after transplantation. (C) Weights of spleen and thymus in recipient mice 4 months after transplantation. (D) Cellularity of spleen, thymus, and femurs (nucleated cells only). (E) Numbers of HSCs and early progenitors in the BM of recipient mice. Right panels show representative flow cytometry plots of the LSK population. (F) Enumeration of B-cell progenitors and mature B cells in the BM of recipient mice. The proportion of BM B lymphocytes in recipient mice is represented in the right panels. (G) Quantification of B-lymphoid cell subsets in the spleens of recipient mice. (H) Quantification of B cells in the peripheral blood of recipient mice. The proportion of B lymphocytes in the peripheral blood of recipient mice is represented in the right panels. Data above are presented as mean ± SEM (n = 6 donors and 18 recipients per genotype). Asterisks indicate a statistically significant difference between recipients of Moz+/− and WT BM at *P < .05, **P < .01, and ***P < .001. Cell surface markers used to discriminate between cell populations are outlined in supplemental Table 9. APC, allophycocyanin.
Consistent with defects observed in adult Moz+/− mice prior to transplantation (Figure 3; supplemental Table 2), there was a > twofold reduction in the numbers of B lymphoid cells from the pro–B-stage onwards in the BM, spleen, and peripheral blood of recipients of Moz+/− BM compared with controls (Figure 4F-H; supplemental Table 3). In contrast, other hematopoietic lineages including the T-lymphoid, erythroid, myeloid, and megakaryocytic lineages were normal in recipients of Moz+/− BM (supplemental Figure 1). These data suggest that hematopoietic defects in Moz+/− and Moz+/−; Eμ-MycT/+ mice are confined primarily to the B-lymphoid lineage and are intrinsic to the hematopoietic system.
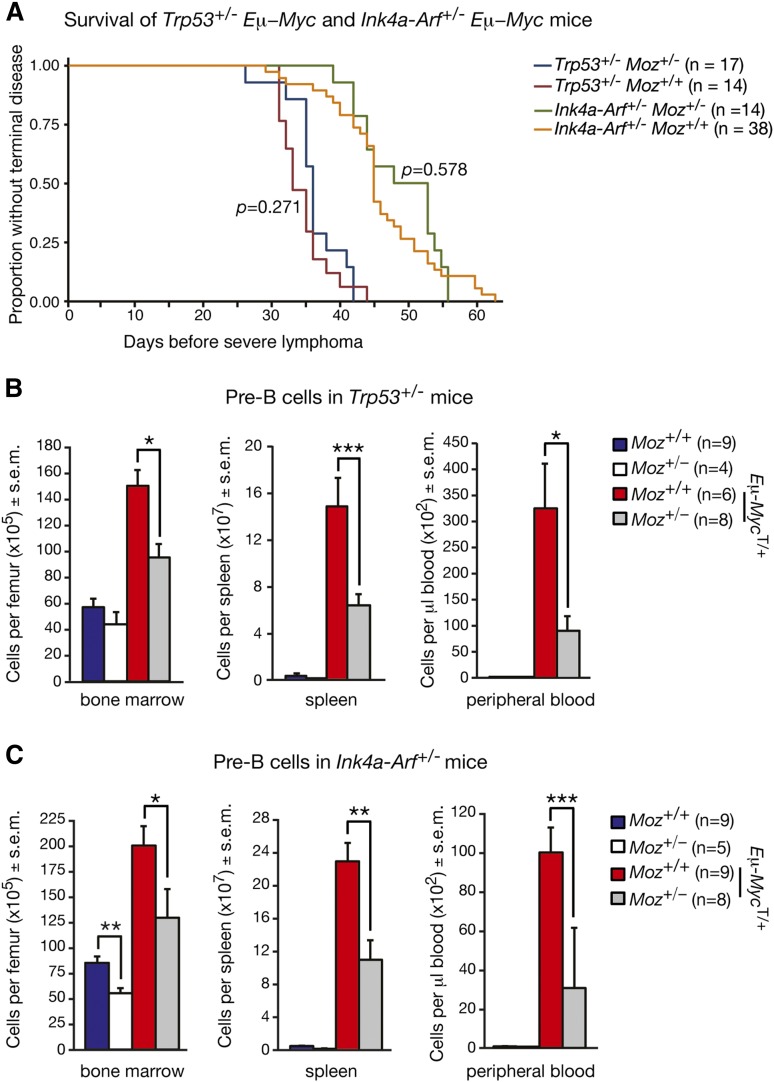
Removal of one allele of Trp53 or Ink4a-Arf accelerates lymphoma in Moz+/−; Eμ-MycT/+ mice to Moz+/+; Eμ-MycT/+ levels
The proteins p53, p16INK4A, and p19ARF are important regulators of cell proliferation, cellular senescence, and apoptosis.30 The deletion of Trp53 or Arf substantially accelerates the development of malignant lymphoma in Eμ-MycT/+ mice.18,31 To ascertain whether defects in B cells caused by Moz haploinsufficiency were related to deregulation of Ink4a-Arf or Trp53, we created compound Moz; Eμ-Myc mice lacking one copy of either Ink4a-Arf or Trp53.
Moz+/+; Eμ-MycT/+; Trp53+/− mice developed lymphoma with a median survival of 33 days (Figure 5A). Moz+/−; Eμ-MycT/+; Trp53+/− mice displayed comparable survival kinetics, with a median survival of 36 days. Similarly, the median survival of Moz+/−; Eμ-MycT/+; Ink4a-Arf+/− mice of 48 days was not significantly different from the 45-day median survival of Moz+/+; Eμ-MycT/+; Ink4a-Arf+/− controls (Figure 5A). These data demonstrate that loss of one allele of Trp53 or Arf can overcome the delay in MYC-driven lymphoma development caused by Moz haploinsufficiency.
Figure 5.
Loss of one allele of Trp53 or Ink4a-Arf does not rescue defects in B-cell development imposed by Moz haploinsufficiency. (A) Kaplan-Meier curves depicting lymphoma-free survival of Moz+/−; Eμ-MycT/+; Trp53+/− and Moz+/+; Eμ-MycT/+; Trp53+/− mice, as well as Moz+/−; Eμ-MycT/+; Ink4a-Arf+/− and Moz+/+; Eμ-MycT/+; Ink4a-Arf+/− mice. (B) Quantification of pre–B-cell progenitors in the BM, spleen, and peripheral blood of Moz+/−; Eμ-MycT/+; Trp53+/− and Moz+/+; Eμ-MycT/+; Trp53+/− mice. (C) Enumeration of pre–B-cells in the BM, spleen, and peripheral blood of Moz+/−; Eμ-MycT/+; Ink4a-Arf+/− and Moz+/+; Eμ-MycT/+; Ink4a-Arf+/− mice. Data are presented as mean ± SEM. Asterisks indicate a statistically significant difference between genotypes as indicated at *P < .05, **P < .01, and ***P < .001. Cell surface markers used to discriminate between cell populations are provided in supplemental Table 8.
The reduction in B-lymphoid progenitors in Moz+/−; Eμ-MycT/+ mice is not rescued by the loss of one allele of Trp53 or Ink4a-Arf
We examined whether the deletion of one allele of Trp53 or Ink4a-Arf also resulted in the restoration of B-lymphocyte cell numbers in Moz+/− mice. In the BM, spleen, and peripheral blood, there was approximately a 50% reduction in the numbers of pre–B-cells in Moz+/−; Eμ-MycT/+; Trp53+/− mice compared with Moz+/+; Eμ-MycT/+; Trp53+/− controls (Figure 5B). Similarly, Moz+/−; Eμ-MycT/+; Ink4a-Arf+/− mice harbored significantly fewer pre–B-cells in hematopoietic tissues compared with Moz+/+; Eμ-MycT/+; Ink4a-Arf+/− controls (Figure 5C). Other B-cell progenitors, peripheral blood lymphocytes, and total numbers of white blood cells were also reduced in Moz+/−; Eμ-MycT/+ mice lacking a copy of Trp53 or Ink4a-Arf compared with Moz+/+; Eμ-MycT/+ controls (supplemental Tables 4 and 5; supplemental Figure 3). These data show that the loss of one allele of Trp53 or Ink4a-Arf does not rescue the B-cell development defects observed in Moz+/− mice.
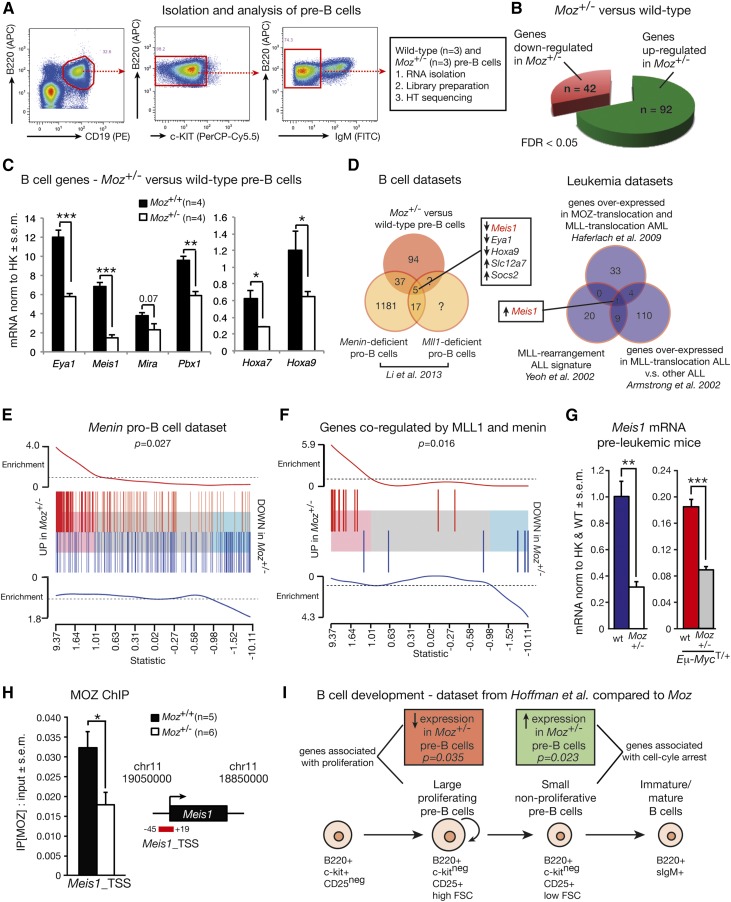
MOZ regulates B-cell development by maintaining expression of key B-cell genes
We performed a transcriptome analysis to identify MOZ target genes in pre–B-cells. Because transgenic Myc potentiates the expression of active genes rather than functioning as an on/off switch,32,33 and because defects in Moz+/− pre–B-cells are evident in both the presence and absence of the Eμ-Myc transgene, we confined our transcriptome analysis to Moz+/− and WT pre–B-cells without the Eμ-Myc transgene. Pre–B-cells were isolated from adult 7- to 10-week-old mice by flow cytometry, and RNA was isolated and submitted for RNA-sequencing (Figure 6A). With a false discovery rate (FDR) cutoff of 5%, there were 42 downregulated genes and 92 upregulated genes in Moz+/− pre–B-cells compared with WT (Figure 6B; supplemental Table 6). Significantly downregulated genes in Moz+/− pre–B-cells included Meis1, Hoxa7 (FDR < 0.05; P < .0001), and Pbx1 (FDR = 0.05; P = .0006). Reduced expression of Eya1, Meis1, Pbx1, and Hoxa7 in Moz+/− pre–B-cells was confirmed in independent samples by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) (Figure 6C). Although Hoxa9 was filtered from the sequencing analysis due to its low expression, we nevertheless tested Hoxa9 expression levels by qRT-PCR because of its importance in regulating B-cell development,34 and because MOZ regulates Hoxa9 expression during embryogenesis and in cord blood cells.20,35 Hoxa9 expression was halved in Moz+/− pre–B-cells compared with WT pre–B-cells (Figure 6C).
Figure 6.
Gene expression analysis comparing Moz+/− and WT pre–B-cells. (A) Experimental design. (B) Number of differentially expressed genes in Moz+/− vs WT pre–B-cells with a FDR cutoff of 5%. A complete list of genes differentially expressed in Moz+/− vs WT pre–B-cells can be found in supplemental Table 6. (C) Confirmation of reduced levels of Eya1, Meis1, Pbx1, Hoxa7, and Hoxa9 in independent samples of Moz+/− pre–B-cells. (D) Comparison of the Moz+/− pre–B-cell expression profile with other published data sets. The “B-cell data sets” schematic Venn diagram shows an overlap between genes differentially expressed in Moz+/− vs WT pre–B-cells, and in Men1- and Mll-deficient pro–B-cells. The “leukemia data sets” compare genes strongly overexpressed in human MLL-translocation ALL, MLL-translocation AML, and MOZ-translocation AML. (E) Enrichment plot showing a positive correlation between the Men1-deficient pro–B-cell signature from Li et al38 and the Moz+/− expression profile. The horizontal axis shows t-statistics for all genes in the Moz+/− data. Red and blue bars mark positions of genes that are up and down, respectively, in the Men1-knockout experiment. Worms show relative enrichment of the Men1-knockout genes relative to uniform ordering. (F) Enrichment plot showing a positive correlation between the Moz+/− expression profile and the overlap genes from Men1 and Mll knockout pro–B-cells from Li et al.38 (G) Meis1 mRNA expression levels in pre–B-cells from pre-leukemic (4-week-old) mice. (H) Binding of MOZ to the Meis1 locus in Moz+/− and WT pre–B-cells. (I) Comparison of the Moz data set with gene expression profiles of B-cell progenitors at different developmental stages.46 Genes strongly expressed in large proliferative pre–B-cells are reduced in Moz+/− pre–B-cells (red box, Roast P = .035), whereas genes enriched in small resting pre–B-cells are upregulated in Moz+/− pre–B-cells (green box, Roast P = .023). Data are presented as mean ± SEM. For qRT-PCR experiments, gene expression levels were normalized to housekeeping genes Hsp90ab1 and Gapdh (C), or housekeeping genes and expression in WT samples (G). Asterisks shown in (C,G,H) indicate a statistically significant difference between Moz+/− and WT, or between Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice at *P < .05, **P < .01, and ***P < .001. ChIP, chromatin immunoprecipitation; FITC, fluorescein isothiocyanate; FSC, forward scatter; HK, housekeeping genes; HT sequencing, high-throughput sequencing; PE, phycoerythrin.
We compared our Moz+/− pre–B-cell transcriptional profile with previously published data sets. Like MOZ, the trithorax group protein mixed lineage leukemia (MLL) is also critical for maintaining HSCs, body segment identity, and proper expression of Hox genes during embryonic development.36,37 Interestingly, MLL and its complex member menin 1 (MEN1) also maintain B-cell progenitors.38 Li et al38 analyzed genes misexpressed by the deletion of Mll or Men1 in pro–B-cells. We compared this study to our Moz pre–B-cell data set and identified 3 genes downregulated in the absence of MOZ, MLL, and MEN1, being Meis1, Hoxa9, and Eya1 (Figure 6D). Overall, we found a strong and significant correlation between the Moz data set and genes regulated by MLL and menin (Figure 6E-F).
Interestingly, MEIS1, along with HOXA7 or HOXA9, was also found to be strongly upregulated in human MOZ and MLL translocation-driven leukemia39-42 (Figure 6D). Meis1 mRNA was halved in Moz+/−; Eμ-MycT/+ pre–B-cells isolated from pre-leukemic 4-week-old mice compared with their Moz+/+; Eμ-MycT/+ counterparts (Figure 6G). To determine whether Meis1 was indeed a direct target of MOZ, we carried out chromatin immunoprecipitation (ChIP) analysis using an antibody to MOZ. We found strong binding of MOZ to the transcription start site of Meis1 (Figure 6H). The binding of MOZ to the transcription start site of Meis1 was halved in Moz+/− pre–B-cells, further confirming that the Meis1 locus is a direct target of MOZ (P = .023). Consistent with a hierarchy of MOZ maintaining Meis1, and MEIS1 maintaining the expression of its target genes, genes identified as direct targets of MEIS1 by ChIP43 were significantly downregulated in Moz+/− pre–B-cells compared with controls (Roast P < .001). In contrast, PBX1 and PREP target genes identified in the same study were not significantly decreased in Moz+/− pre–B-cells compared with their WT counterparts (Roast P = .08). Together, these data identify MOZ as a direct regulator of Meis1.
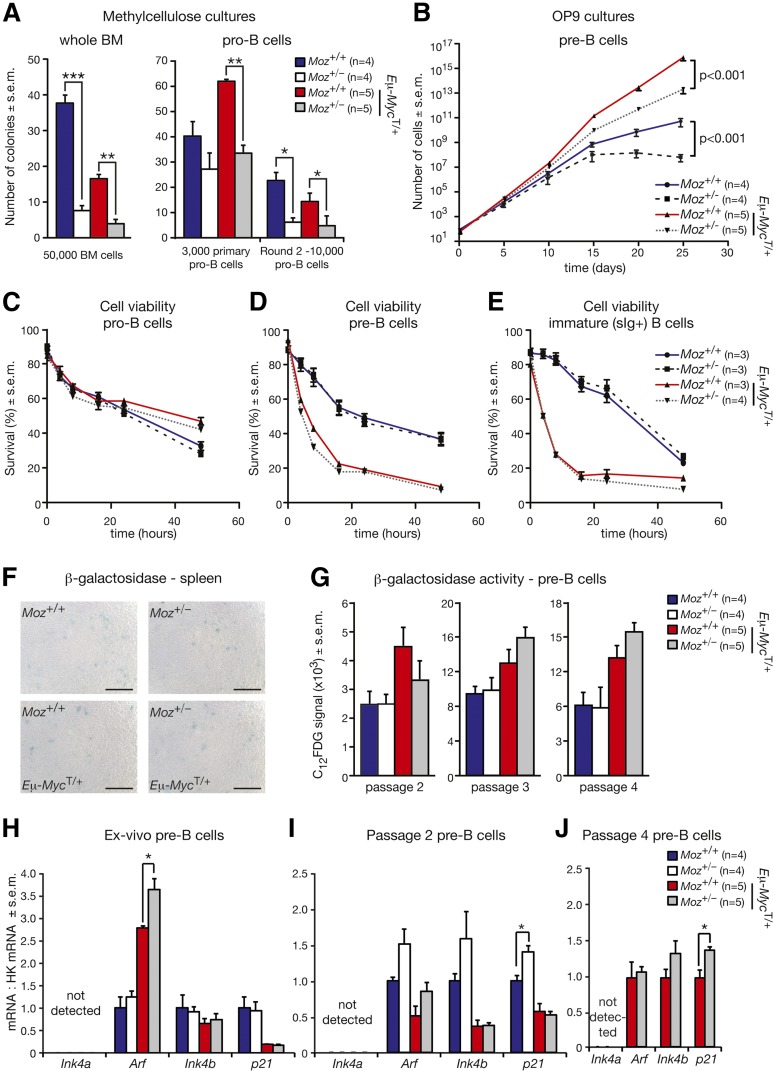
Moz+/− pro–B-cells and pre–B-cells display abnormally reduced proliferative capacity
Overexpression of Meis1 and Hoxa9 in myeloid progenitors has previously been shown to confer extensive self-renewal ability and inhibition of cellular differentiation in myeloid progenitors.44,45 We therefore investigated whether Moz haploinsufficiency resulted in decreased proliferative capacity of B-cell progenitors. We compared the gene expression profile of Moz+/− vs WT pre–B-cells with previously published data sets of proliferating large pre–B-cells and resting small pre–B-cells.46 Interestingly, genes strongly expressed in proliferating large pre–B-cells were downregulated in Moz+/− pre–B-cells (Roast P = .035), whereas genes that become activated when pre–B-cells transition to resting small pre–B-cells were upregulated in Moz+/− pre–B-cells (Figure 6I, Roast P = .023). The expression of immunoglobulin chain genes was upregulated in Moz+/− cells (supplemental Table 7).
To formally test the proliferative capacity of Moz+/− B-cell progenitors, we conducted colony formation and proliferation assays in vitro. Plating of 50 000 WT BM cells in methylcellulose cultures in the presence of the cytokine interleukin 7 (IL-7) resulted in 36 colonies (Figure 7A). In contrast, only 8 colonies were present in cultures of Moz+/− BM cells, which also tended to be smaller in size. Similarly, the numbers of colonies in Moz+/−; Eμ-MycT/+ cultures were more than halved compared with Moz+/+; Eμ-MycT/+ controls (Figure 7A). Thus, the loss of a single Moz allele reduces the clonogenic potential of B-lymphocyte progenitors.
Figure 7.
Moz+/− B-cell progenitors display reduced proliferative capacity. (A) Enumeration of colony formation in methylcellulose culture after plating of unfractionated BM cells and FACS-purified pro–B-cells. (B) Growth curves of pre–B-cells cultured on OP9 cells in the presence of IL-7. After collection by FACS, 500 pre–B-cells were plated and passaged every 5 days. (C-E) Assessment of the survival of (C) pro-B, (D) pre-B, and (E) surface IgM/IgD-positive (sIg+) B cells. At each time point, the proportions of viable cells were determined by staining with propidium iodide and Annexin-V. (F) β-galactosidase activity in spleens from Moz+/− and control Moz+/+ mice (n = 6 to 8 per genotype). Scale bars = 100 μm. (G) Quantification of β-galactosidase activity in pre–B-cells of the indicated genotypes cultured over 25 days. At passages 2, 3, and 4, cells were stained for B-cell markers (B220 and CD19), and incubated with a fluorescent substrate for the β-galactosidase enzyme (C12FDG). β-galactosidase activity in pre–B-cells was quantified using flow cytometry. (H-J) Levels of Ink4a, Arf, Ink4b, and p21 mRNA levels in pre–B-cells, (H) ex vivo, (I) at passage 2, and (J) at passage 4. Insufficient Moz+/− and WT pre–B-cells were available at passage 4 for analysis. Data are presented as mean ± SEM. Asterisks shown in (A,H-J) indicate a statistically significant difference between Moz+/− and WT mice, or between Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice at *P < .05, **P < .01, and ***P < .001. Gene expression levels of Ink4a, Arf, Ink4b, and p21 in (H-J) were standardized to housekeeping genes Hsp90ab1 and Gapdh, and expression in WT samples was designated 1.
To specifically examine whether the reduction of colony formation in the presence of IL-7 was related to pro–B-cells, 3000 FACS-sorted pro–B-cells were plated in a methylcellulose medium with IL-7. Moz+/+; Eμ-MycT/+ pro–B-cells generated 64 colonies, whereas Moz+/−; Eμ-MycT/+ pro–B-cells produced only 34 colonies (Figure 7A). In the second round of plating, the numbers of colonies in Moz+/− cultures were more than halved compared with control Moz+/+ cultures both in the presence and absence of the Eμ-Myc transgene (Figure 7A). These data demonstrate that Moz haploinsufficiency causes a decrease in the colony formation capacity of pro–B-cells in a cell intrinsic manner.
We then cultured FACS-purified pre–B-cells on OP9 stromal cells in the presence of IL-7 and carried out serial passaging. Remarkably, there was a 239-fold reduction in the accumulation of Moz+/−; Eμ-MycT/+ pre–B-cells compared with Moz+/+; Eμ-MycT/+ controls over 5 passages (Figure 7B). A similar effect was observed in pre–B-cells in the absence of the Eμ-Myc transgene. Compared with WT pre–B-cells, there was a 400-fold reduction in the accumulation of Moz+/− pre–B-cells over the 25-day culture (Figure 7B).
To determine the effects of Moz haploinsufficiency on the cell cycle during B-cell development, we stained BM and splenic B cells with Ki-67 and DAPI, as well as for markers identifying pro–B-cells, pre–B-cells, and immature B cells. Cycling cells stain positive for Ki-67 and DNA content distinguishes G1, S, and G2/M phases of the cell cycle.47 Cell-cycle phases were similar in Moz+/−; Eμ-MycT/+ and Moz+/+; and Eμ-MycT/+ in the BM. In contrast, in the spleen, significantly more Moz+/−; Eμ-MycT/+ pre–B-cells were found in G2/M phases of the cell cycle compared with Moz+/+; Eμ-MycT/+ pre–B-cells (supplemental Figure 4). This suggests that Moz+/−; Eμ-MycT/+ pre–B-cells may progress more slowly through the G2/M phases of the cell cycle. Moz+/−; Eμ-MycT/+ mice also display a reduced number of Ki-67 low splenic pro–B-cells and pre–B-cells, consistent with a smaller number of small pre–B-cells.
Moz haploinsufficiency does not affect cell viability
We then investigated whether Moz haploinsufficiency affected cell viability. Pro-B, pre-B, and surface IgM/IgD-positive (designated sIg+) B cells were collected by FACS and cultured in the absence of cytokines. In pro–B-cell cultures, there was no significant difference in the proportion of viable cells between Moz+/− and WT cultures, or between Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ pro–B-cells (Figure 7C). Deregulated MYC expression does not only promote cell growth and proliferation, but also increases the propensity of cells to undergo apoptosis under conditions of stress.29,48 Thus, there were fewer viable pre-B and sIg+ cells in cultures of Eμ-MycT/+ cells (Figure 7D-E). However, in both pre-B and sIg+ cultures, there was no difference in the viability of cells isolated from Moz+/− and WT mice, or between cells from Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ mice (Figure 7D-E). Furthermore, we did not detect increased caspase 3 activity in splenic extracts from Moz+/− mice compared with controls both in the presence and absence of the Eμ-Myc transgene (supplemental Figure 5). These results rule out the possibility that the large decrease in cell numbers observed in Moz+/− pro-B and pre-B cultures was related to differences in cell survival.
Moz haploinsufficiency does not affect senescence levels in B-cell progenitors
A recent study suggested that MOZ inhibits cellular senescence by repressing the Ink4a-Arf locus.49 Therefore, we stained spleen samples for the senescence marker β-galactosidase. Both in the presence and absence of the Eμ-Myc transgene, only a very small number of β-galactosidase positive cells were observed (Figure 7F). There was no evidence of increased senescence in Moz+/− spleens compared with Moz+/+ controls. We examined the levels of β-galactosidase activity in cultured pre–B-cells by flow cytometry using the β-galactosidase substrate C12FDG. At passages 2, 3, and 4 of pre–B-cell cultures, there were no significant differences in β-galactosidase activity between Moz+/− and WT pre–B-cells, or between Moz+/−; Eμ-MycT/+ and Moz+/+; Eμ-MycT/+ pre–B-cells (Figure 7G). Moreover, Moz haploinsufficiency had no consistent effects on the expression of senescence markers and mediators Ink4a, Arf, Ink4b, and p21 at passages 0, 2, and 4 in pre–B-cell cultures (Figure 7H-J).
Discussion
MOZ is a critical transcriptional regulator of genes required for proper embryonic development, and particularly for the development of HSC populations.15,16,20,50 Although much effort has focused on understanding the biology of MOZ–translocation-driven leukemia, the role of normal MOZ in supporting proliferation during oncogenesis has not been examined. In the current study, we observed that the loss of just one allele of Moz caused a 3.9-fold increase in lymphoma-free survival in the Eμ-Myc model of MYC-driven lymphoma. This delay in lymphoma development was related to a reduction in the number of B-cell progenitors, which is the cell population from which lymphomas arise in Eμ-Myc mice after cooperating oncogenic lesions have been acquired. Thus, reduced B-cell numbers in Moz+/− mice presented reduced targets for secondary cooperating mutations that are required for lymphoma. The reduction in B-cell progenitors resulting from the loss of one allele of Moz could not be restored by the loss of Trp53 or Ink4a-Arf. Rather, using gene expression profiling, we identified Pbx1, Meis1, Hoxa7, and Hoxa9 as critical genes that require MOZ for normal levels of expression. We found that MOZ binds directly to the Meis1 locus and is critical for maintaining Meis1 expression. This study establishes normal levels of MOZ as a key requirement for B-cell progenitor population expansion in both healthy and MYC-driven pre-leukemic states.
PBX1, MEIS1, and HOX proteins are important regulators of B-cell development. PBX1, originally identified in translocations leading to pre–B-cell leukemia, is a key regulator of HSCs and the B-lymphoid cell lineage, although its importance diminishes after the CLP stage.51 Mice lacking Hoxa9 were reported to have abnormally reduced numbers of B-cell progenitors from the pro–B-cell stage.52 Similarly, specific loss of Meis1 in the hematopoietic compartment results in a marked reduction in all B-progenitor cells.53 Interestingly, HOXA9 and MEIS1 can form heterodimers that bind DNA independently of other proteins.52 Together with our results, these observations suggest that the dependence of Meis1 and Hoxa9 gene expression on MOZ is important for the maintenance of B-cell progenitors.
Previous work has shown that MOZ is required for normal levels of Hox gene expression during mouse embryogenesis in human cord blood cells and in differentiating embryonic stem cells.20,35,54 Similarly, the trithorax group protein MLL maintains Hox gene expression.36 Interestingly, MOZ complexes with MLL in 293T cells and the K562 leukemia-derived cell line, and reduction of MOZ or MLL expression leads to reduced occupancy of the other protein at HOX loci in mouse embryos and human cord blood cells.20,35 In this study, we identified a strong correlation between genes co-regulated by MOZ, MLL, and its complex member menin in B-cell progenitors. In particular, expression levels of Meis1, Hoxa9, and Eya1 were reduced in all 3 data sets. Our results argue for a model where MOZ and MLL collaborate to directly maintain the expression of Meis1, Hoxa9, and Eya1.
MLL chromosomal translocations underlie ∼10% of all B-cell lineage acute lymphoblastic leukemia (ALL) cases55 and 80% of infant ALL.56 One common feature of these types of leukemia is the strong expression of MEIS1 and HOX-A cluster genes.40-42 In this context, it is interesting that genes that drive leukemia development downstream of MLL-translocations, in particular MEIS1 and HOXA9, are also important regulators of B-cell development. Both MOZ and MLL translocations are associated with poor prognosis.12,14,42 Survival rates for ALL patients, particularly those presenting with chromosomal translocations involving MLL, are poor in both adults and infants.57,58 Since upregulation of MEIS1 and HOX gene expression is characteristic of aggressive forms of leukemia and MOZ is required to maintain Meis1 and Hox gene expression, we have performed a high throughput small molecule screen to identify novel inhibitors of MOZ with the aim of ameliorating deleterious gene expression in hematologic malignancies.59
In conclusion, we have shown that the loss of one allele of Moz leads to a significant increase in disease-free survival in the Eμ-Myc lymphoma model. Our study identifies MOZ as a central regulator of gene networks that is required to maintain the proliferative capacity of B-cell progenitors in healthy and the MYC-driven pre-leukemic state. Based on our work, we postulate that even partial inhibition of MOZ activity might be able to inhibit progression of HOX- and MEIS1-driven hematopoietic malignancies.
Acknowledgments
The authors thank R. Cobb, N.L. Downer, F. Dabrowski, and the Walter and Eliza Hall Institute’s FACS laboratory for their excellent technical support.
This study was funded by grants from the National Health and Medical Research Council (NHMRC) project (1030704, 1008699, and 1010851), the NHMRC program (1054618, 1016647, and 1016701), NHMRC research fellowships (1058892) (G.K.S.), (1058344) (W.S.A.), (1003435) (T.T.), (575512) (A.K.V.), and (1020363) (A.S.), Lady Tata Research Award (S.G.), Cancer Council Victoria postdoctoral fellowship (S.G.), Leukemia and Lymphoma Society (SCOR grant 7001-13) (A.S.), Australian Research Council future fellowship (S.L.N.), Independent Research Institutes Infrastructure Support Scheme from the Australian Government’s NHMRC, the Australia Cancer Research Fund, and the Victorian State Government Operational Infrastructure Support.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.N.S., S.C.W.L., F.E.-S., H.K.V., S.H.M.P., S.G., and T.T. carried out experiments; B.N.S., Y.H., G.K.S., T.T., and A.K.V. analyzed data; T.T. conceived and initiated the project; A.K.V. and T.T. supervised the project; B.N.S., A.S., G.K.S., A.K.V., and T.T. wrote the manuscript; and A.S., W.S.A., and S.L.N. contributed ideas, planned experiments, and interpreted results.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tim Thomas, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Melbourne 3052, Victoria, Australia; e-mail: tthomas@wehi.edu.au; and Anne K. Voss, 1G Royal Parade, Melbourne 3052, Victoria, Australia; e-mail: avoss@wehi.edu.au.
References
- 1.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 2.Cui K, Zang C, Roh TY, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4(1):80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler JS, Dent SY. The role of chromatin modifiers in normal and malignant hematopoiesis. Blood. 2013;121(16):3076–3084. doi: 10.1182/blood-2012-10-451237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26(47):6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 5.Doyon Y, Cayrou C, Ullah M, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21(1):51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Borrow J, Stanton VP, Jr, Andresen JM, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14(1):33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 7.Chaffanet M, Gressin L, Preudhomme C, Soenen-Cornu V, Birnbaum D, Pébusque MJ. MOZ is fused to p300 in an acute monocytic leukemia with t(8;22). Genes Chromosomes Cancer. 2000;28(2):138–144. doi: 10.1002/(sici)1098-2264(200006)28:2<138::aid-gcc2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91(9):3127–3133. [PubMed] [Google Scholar]
- 9.Esteyries S, Perot C, Adelaide J, et al. NCOA3, a new fusion partner for MOZ/MYST3 in M5 acute myeloid leukemia. Leukemia. 2008;22(3):663–665. doi: 10.1038/sj.leu.2404930. [DOI] [PubMed] [Google Scholar]
- 10.Deguchi K, Ayton PM, Carapeti M, et al. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3(3):259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 11.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6(6):587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Sun T, Wu E. Acute monoblastic leukemia with t(8;16): a distinct clinicopathologic entity; report of a case and review of the literature. Am J Hematol. 2001;66(3):207–212. doi: 10.1002/1096-8652(200103)66:3<207::aid-ajh1046>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 13.Stark B, Resnitzky P, Jeison M, et al. A distinct subtype of M4/M5 acute myeloblastic leukemia (AML) associated with t(8:16)(p11:p13), in a patient with the variant t(8:19)(p11:q13)—case report and review of the literature. Leuk Res. 1995;19(6):367–379. doi: 10.1016/0145-2126(94)00150-9. [DOI] [PubMed] [Google Scholar]
- 14.Gervais C, Murati A, Helias C, et al. Groupe Francophone de Cytogénétique Hématologique. Acute myeloid leukaemia with 8p11 (MYST3) rearrangement: an integrated cytologic, cytogenetic and molecular study by the groupe francophone de cytogénétique hématologique. Leukemia. 2008;22(8):1567–1575. doi: 10.1038/leu.2008.128. [DOI] [PubMed] [Google Scholar]
- 15.Thomas T, Corcoran LM, Gugasyan R, et al. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20(9):1175–1186. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Campo FM, Borrow J, Kouskoff V, Lacaud G. The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood. 2009;113(20):4866–4874. doi: 10.1182/blood-2008-04-152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 18.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348(6299):331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- 20.Voss AK, Collin C, Dixon MP, Thomas T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell. 2009;17(5):674–686. doi: 10.1016/j.devcel.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 22.Jacks T, Remington L, Williams BO, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4(1):1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 23.Lee SC, Phipson B, Hyland CD, et al. Polycomb repressive complex 2 (PRC2) suppresses Eμ-myc lymphoma. Blood. 2013;122(15):2654–2663. doi: 10.1182/blood-2013-02-484055. [DOI] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law CW, Chen Y, Shi W, Smyth GK. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26(17):2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167(2):353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalak EM, Jansen ES, Happo L, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16(5):684–696. doi: 10.1038/cdd.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strasser A, Elefanty AG, Harris AW, Cory S. Progenitor tumours from Emu-bcl-2-myc transgenic mice have lymphomyeloid differentiation potential and reveal developmental differences in cell survival. EMBO J. 1996;15(15):3823–3834. [PMC free article] [PubMed] [Google Scholar]
- 30.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2(10):731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13(20):2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin CY, Lovén J, Rahl PB, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie Z, Hu G, Wei G, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence HJ, Christensen J, Fong S, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106(12):3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paggetti J, Largeot A, Aucagne R, et al. Crosstalk between leukemia-associated proteins MOZ and MLL regulates HOX gene expression in human cord blood CD34+ cells. Oncogene. 2010;29(36):5019–5031. doi: 10.1038/onc.2010.254. [DOI] [PubMed] [Google Scholar]
- 36.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378(6556):505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 37.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li BE, Gan T, Meyerson M, Rabbitts TH, Ernst P. Distinct pathways regulated by menin and by MLL1 in hematopoietic stem cells and developing B cells. Blood. 2013;122(12):2039–2046. doi: 10.1182/blood-2013-03-486647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30(1):41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 40.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 41.Camós M, Esteve J, Jares P, et al. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66(14):6947–6954. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- 42.Haferlach T, Kohlmann A, Klein HU, et al. AML with translocation t(8;16)(p11;p13) demonstrates unique cytomorphological, cytogenetic, molecular and prognostic features. Leukemia. 2009;23(5):934–943. doi: 10.1038/leu.2008.388. [DOI] [PubMed] [Google Scholar]
- 43.Penkov D, Mateos San Martín D, Fernandez-Díaz LC, et al. Analysis of the DNA-binding profile and function of TALE homeoproteins reveals their specialization and specific interactions with Hox genes/proteins. Cell Reports. 2013;3(4):1321–1333. doi: 10.1016/j.celrep.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 44.Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci USA. 2001;98(23):13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calvo KR, Sykes DB, Pasillas M, Kamps MP. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol Cell Biol. 2000;20(9):3274–3285. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann R, Seidl T, Neeb M, Rolink A, Melchers F. Changes in gene expression profiles in developing B cells of murine bone marrow. Genome Res. 2002;12(1):98–111. doi: 10.1101/gr.201501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao Z, Ke Z, Gorbunova V, Seluanov A. Replicatively senescent cells are arrested in G1 and G2 phases. Aging (Albany NY) 2012;4(6):431–435. doi: 10.18632/aging.100467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evan GI, Wyllie AH, Gilbert CS, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Campo FM, Costa G, Lie ALM, Stifani S, Kouskoff V, Lacaud G. MOZ-mediated repression of p16(INK) (4) (a) is critical for the self-renewal of neural and hematopoietic stem cells. Stem Cells. 2014;32(6):1591–1601. doi: 10.1002/stem.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voss AK, Vanyai HK, Collin C, et al. MOZ regulates the Tbx1 locus, and Moz mutation partially phenocopies DiGeorge syndrome. Dev Cell. 2012;23(3):652–663. doi: 10.1016/j.devcel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanyal M, Tung JW, Karsunky H, et al. B-cell development fails in the absence of the Pbx1 proto-oncogene. Blood. 2007;109(10):4191–4199. doi: 10.1182/blood-2006-10-054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen WF, Montgomery JC, Rozenfeld S, et al. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17(11):6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ariki R, Morikawa S, Mabuchi Y, et al. Homeodomain transcription factor Meis1 is a critical regulator of adult bone marrow hematopoiesis. PLoS ONE. 2014;9(2):e87646. doi: 10.1371/journal.pone.0087646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheikh BN, Downer NL, Kueh AJ, Thomas T, Voss AK. Excessive versus physiologically relevant levels of retinoic acid in embryonic stem cell differentiation. Stem Cells. 2014;32(6):1451–1458. doi: 10.1002/stem.1604. [DOI] [PubMed] [Google Scholar]
- 55.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 56.Pieters R, Schrappe M, De Lorenzo P, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370(9583):240–250. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 57.Hilden JM, Dinndorf PA, Meerbaum SO, et al. Children’s Oncology Group. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood. 2006;108(2):441–451. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moorman AV, Harrison CJ, Buck GA, et al. Adult Leukaemia Working Party, Medical Research Council/National Cancer Research Institute. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109(8):3189–3197. doi: 10.1182/blood-2006-10-051912. [DOI] [PubMed] [Google Scholar]
- 59.Falk H, Connor T, Yang H, et al. An efficient high-throughput screening method for MYST family acetyltransferases, a new class of epigenetic drug targets. J Biomol Screen. 2011;16(10):1196–1205. doi: 10.1177/1087057111421631. [DOI] [PubMed] [Google Scholar]