MIWI catalytic activity is required for spermatogenesis, indicating that piRNA-guided cleavage is critical for germ cell development. Through identifying meiotic piRNA targets, this study supports the idea that meiotic piRNA populations must be strongly selected to enable successful spermatogenesis, both driving the response away from essential genes and directing the pathway toward mRNA targets that are regulated by small RNAs in meiotic cells.
Keywords: MIWI, pachytene piRNAs, RNA cleavage, ping-pong cycle, exogenous piRNAs, spermatogenesis defect
Abstract
MIWI catalytic activity is required for spermatogenesis, indicating that piRNA-guided cleavage is critical for germ cell development. To identify meiotic piRNA targets, we augmented the mouse piRNA repertoire by introducing a human meiotic piRNA cluster. This triggered a spermatogenesis defect by inappropriately targeting the piRNA machinery to mouse mRNAs essential for germ cell development. Analysis of such de novo targets revealed a signature for pachytene piRNA target recognition. This enabled identification of both transposable elements and meiotically expressed protein-coding genes as targets of native piRNAs. Cleavage of genic targets began at the pachytene stage and resulted in progressive repression through meiosis, driven at least in part via the ping-pong cycle. Our data support the idea that meiotic piRNA populations must be strongly selected to enable successful spermatogenesis, both driving the response away from essential genes and directing the pathway toward mRNA targets that are regulated by small RNAs in meiotic cells.
Beginning at the pachytene stage of meiosis, the Piwi proteins MIWI and MILI bind a highly abundant and extremely diverse class of piRNAs (Aravin et al. 2006; Girard et al. 2006). These are derived from discrete genomic loci, termed pachytene piRNA clusters, from which emanate an extreme diversity of small RNAs. Nearly all of these map perfectly only to the loci from which they are derived. The importance of pachytene piRNAs is inferred from the essential requirement for their binding partner, MIWI, and its catalytic activity for successful sperm development (Deng and Lin 2002; Reuter et al. 2011).
Despite a clear requirement for MIWI in spermatogenesis, multiple conflicting models exist describing the precise molecular function of MIWI and its piRNA partners. Early proposals contemplated an essential role for MIWI in promoting the expression of a subset of microRNAs (miRNAs) in testes (Grivna et al. 2006a,b). The same studies also reported the association of MIWI with polysomes. This was supported by a separate analysis of Mael mutants that showed that reduced levels of meiotic piRNAs corresponded with a reduction in the translation of some spermiogenic mRNAs (Castañeda et al. 2014). Following consideration of the observation that loading of pachytene piRNAs directs MIWI toward the ubiquitin–proteosome pathway for degradation (Zhao et al. 2013), an alternative model emerged, which suggested that in the absence of meiotic piRNAs, MIWI was stabilized so it can inhibit translation. In addition to repressing or activating translation, MIWI has also been implicated in promoting RNA stability (Nishibu et al. 2012; Vourekas et al. 2012). Given that MIWI-associated mRNAs, identified by cross-linking immunoprecipitation (CLIP), were less abundant in Miwi mutants, it was argued that MIWI binding stabilizes bound mRNAs. In this study, MIWI binding did not require meiotic piRNAs but instead must have identified its targets in a way that differs from other Argonaute proteins. Finally, based on the phenotype of Mov10L1 mutants, which lack meiotic piRNAs, MIWI was implicated in promoting DNA damage repair and genome stability, although this could be a direct effect or an indirect consequence of the posited roles of MIWI in gene regulation (Zheng and Wang 2012).
The development of Miwi mutant sperm fails at the round spermatid stage. Mice expressing only a catalytically incompetent MIWI protein have precisely the same phenotype, strongly implicating piRNA-directed cleavage as being critical to germ cell development. Based on 5′RACE, LINE1 elements were strongly implicated as direct targets of MIWI (Reuter et al. 2011). This was true despite the fact that meiotic piRNA clusters were initially reported as being depleted of mobile element sequences as compared with the genome as a whole (Girard et al. 2006). However, recent studies have shown that young, potentially active elements are enriched in meiotic piRNA clusters, and these show a bias in orientation such that antisense piRNAs are preferentially produced (Hirano et al. 2014; KA Wasik, OH Tam, I Falciatori, SR Knott, M Hammell, VV Vagin, and GJ Hannon, in prep.). Despite strong evidence for LINEs as pachytene piRNA targets, it is unclear whether LINE activation is the root cause of the sterility of MIWI mutants. Mice lacking a selected piRNA cluster showed a 15-fold elevation in LINE1 expression but were fully fertile (Xu et al. 2008), suggesting that MIWI must also have other targets whose engagement is important for sperm production. We therefore sought to develop strategies to identify such targets so as to help characterize the role of pachytene piRNAs in spermatogenesis.
Results
Expression of human piRNAs in mouse testes
As a step toward identifying MIWI targets, it was essential to understand the rules that govern piRNA target recognition. To achieve this goal, we sought to produce mice in which we augmented the endogenous piRNA repertoire and then searched for newly acquired cleavage targets. Our hope was that matching these new targets to exogenous piRNAs might reveal rules for piRNA targeting in vivo that would allow us in turn to identify the targets of native piRNAs.
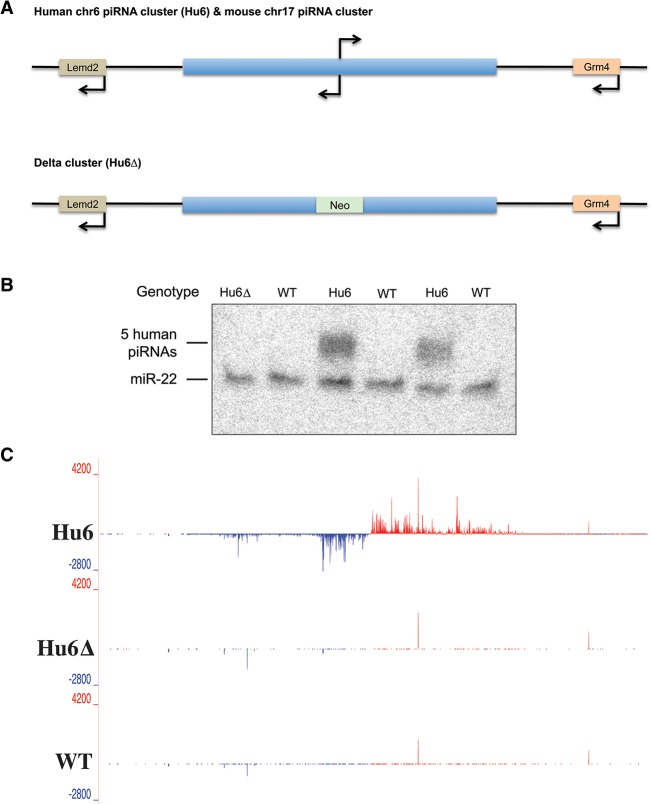
Our strategy was to introduce an entire human piRNA cluster into mice so that its content might contribute to mouse piRNA populations. The sequence of pachytene piRNA clusters is not conserved, yet clusters are found at syntenic locations in mammals (Girard et al. 2006). Given that a major cluster on mouse chromosome 17 (ch17) contributes substantially to mouse piRNA populations, we transferred its ∼80-kb human counterpart, found on human ch6, into mice by bacterial artificial chromosome (BAC) transgenesis (termed Hu6 mice) (Fig. 1A). We also generated mice carrying the human ch6 cluster with an ∼4-kb deletion in its central region (Hu6Δ), presumably removing both transcriptional control elements and the transcription start sites for the divergently synthesized piRNA precursor transcripts (Li et al. 2013). Northern blotting for selected human piRNAs indicated that the cluster was active only in adult mouse testes that contained the intact Hu6 cluster (Fig. 1B). Small RNA sequencing (RNA-seq) of testes total RNA or MIWI immunoprecipitates from transgenic animals also confirmed that piRNAs were produced from the entire human cluster and that these had the characteristic length and first uridine (1U) bias of bona fide primary piRNAs (Fig. 1C; Supplemental Fig. S1).
Figure 1.
Expression of a human piRNA cluster in mouse testes. (A, top) A schematic of the region that contains the bidirectional Hu6 piRNA cluster syntenic to the mouse ch17 cluster is shown. (Bottom) A schematic of the same human region that was engineered to replace the promoter region of the piRNA cluster with a neomycin resistance cassette (Neo) is shown. Arrows indicate transcription start sites for genes and the expected transcription start sites for the piRNA cluster. (B) Northern blotting of human piRNAs. The bands represent the five human piRNAs (30 nucleotides [nt]) and loading control, miR-22 (22 nt), that were blotted in various mouse testis RNA. (C) A genome browser visualization of total small RNAs cloned from Hu6, Hu6▵, and wild-type mouse testes and mapped to human ch6 coordinates 33,820,000–33,903,000. Red and blue signals denote reads that map to the plus and minus strand, respectively, of the genome. Y-axis values denote reads per million mapped reads (RPM).
Human piRNA-guided de novo cleavage patterns reveal a piRNA cleavage signature
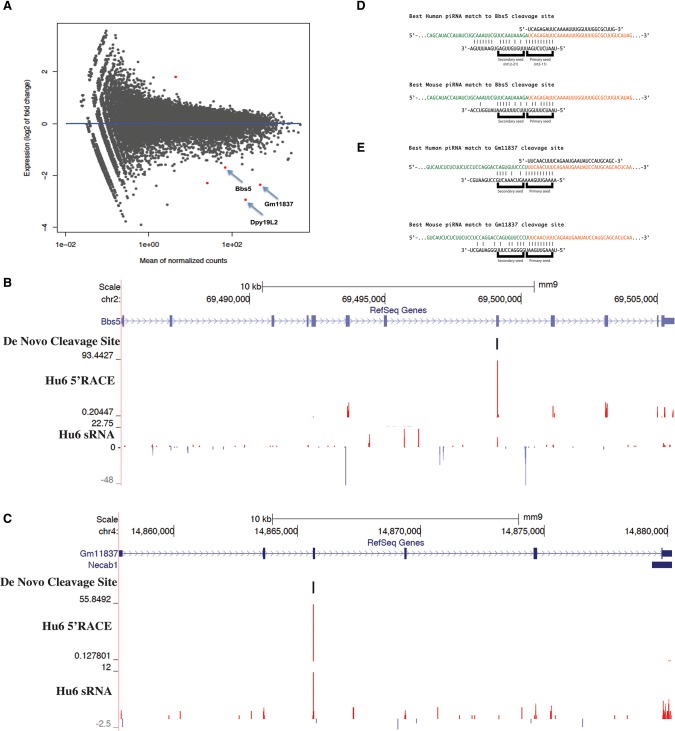
In an effort to identify potential direct cleavage targets of human piRNAs, we performed RNA-seq from sorted Hu6 round spermatids and found that a number of cellular mRNAs were significantly misregulated (P < 0.01) (Fig. 2A). We also used global 5′RACE to detect human piRNA-dependent cleavage sites. We initially noticed 5′RACE tags enriched in the Hu6 mouse that corresponded to genes that were also repressed upon introduction of the human piRNA cluster (Fig. 2B,C). By comparing these cleavage sites with piRNAs present only in Hu6 testes, we noted the presence of sense piRNAs that mapped perfectly to the cleaved mRNAs and began precisely at the cleavage site detected by 5′RACE. This was a strong indication of at least partial operation of the ping-pong cycle in which a cleaved, piRNA-targeted mRNA would be further metabolized to generate a small RNA loaded into a PIWI protein.
Figure 2.
Hu6 human piRNAs target mouse mRNAs for cleavage, thereby revealing a piRNA cleavage signature. (A) A scatter plot depicting the RNA-seq values as sequenced from sorted Hu6 versus wild-type round spermatids. Each dot represents the average RPM of an individual gene. The X-axis denotes the mean of average normalized read counts between both wild-type and Hu6 samples, while the Y-axis denotes the log2 fold change of RNA levels between Hu6 and wild-type samples. Positive and negative Y-axis values denote genes that are up-regulated or down-regulated, respectively, in Hu6 compared with wild-type round spermatids. The red dots denote genes that are significantly misregulated (twofold cutoff). P < 0.01. The arrows identify three genes of interest. The RNA-seq values are the average of two Hu6 or three wild-type biological replicates. (B,C) A genome browser visualization of round spermatid 5′RACE tags and MIWI-bound small RNA reads sequenced from Hu6 testes, located in mouse Bbs5 (B) or Gm11837 (C). The format is the same as in Figure 1C. (D,E) A schematic showing the targeting of mouse Bbs5 (D) and Gm11837 (E) cleavage sites by human piRNAs. The orange sequence denotes the 5′RACE tag enriched in Hu6 round spermatids, and the green sequence denotes the transcript sequence upstream of the cleavage site. The sequence above the 5′RACE tag, if any, depicts a representative Hu6 testis-exclusive sense piRNA that shares a 5′end with the 5′RACE tag. The sequence below the 5′RACE tag is the reverse sequence of a representative intergenic cluster guide piRNA. Each vertical bar denotes a perfect base pair between the cleavage site and the guide piRNA. The Hu6 cluster guide piRNA (top) or native mouse guide piRNA (bottom) that can best complement each cleavage site is shown.
This evidence for piRNA-guided cleavage prompted us to search among human ectopic piRNAs for species capable of targeting these mouse mRNAs. By varying the extent of the required complementarity and the number of tolerated mismatches, we found parameters consistent with targeting of detected cleavage sites by human piRNAs but not by any mouse piRNAs (Fig. 2D,E). These parameters indicated a necessity for perfect matching in nucleotides 2–11 (primary seed), with a maximum of four mismatches being tolerated in nucleotides 12–21 (secondary seed). The aggregate of our observations of exogenous piRNA–target pairs permitted us to derive a set of criteria that could be applied to identify targets of native mouse piRNAs. This involved (1) the comparison of 5′RACE tags in Miwi mutants versus heterozygous littermates, (2) the detection of sense piRNAs at cleavage sites that have the size and nucleotide biases of MIWI-bound piRNAs (Supplemental Fig. S2A,B), and (3) the detection of corresponding pachytene piRNAs that meet the mapping parameters described above (see the Materials and Methods for details).
We hypothesized that the piRNA cleavage signature derived from Hu6 mouse data could be used to identify native piRNA cleavage targets of mouse pachytene piRNAs in wild-type mice. Starting with a list of MIWI-dependent 5′RACE tags that are enriched in Miwi+/− over Miwi−/− spermatogenic cells, we identified and annotated the targets that fulfilled our criteria (Supplemental Fig. S2C,D). Given the vast sequence repertoire of the pachytene piRNA population, it was possible that with the tolerance for four mismatches in the secondary seed, piRNAs could match to identified targets simply by chance. To resolve this issue, we quantified the frequencies of identified antisense piRNAs that match at various secondary seed complementarities to their corresponding targets. We also determined the respective frequencies of randomly selected piRNA secondary seeds, assuming perfect complementarity in the primary seed (see the Materials and Methods). We found that random piRNAs tend to exhibit zero to five matches with identified targets via the secondary seed, which means that antisense piRNAs that complement identified targets with six or more matches do so not simply by chance (Supplemental Fig. S3). Furthermore, we noted that identified antisense piRNAs were also generally better matched to their corresponding targets than the required six secondary seed matches (Supplemental Fig. S3C).
Identification of known piRNA targets validates the piRNA cleavage signature
As existing data point to transposons and particularly LINEs as targets of the meiotic piRNA pathway (Reuter et al. 2011), we first searched mobile element classes for evidence of piRNA-guided cleavage. We found both active LINE1 families and LTR element families harboring putative piRNA cleavage sites (Supplemental Figs. S2D, S4A,B; Goodier et al. 2001). If these transcripts were indeed piRNA targets, we expected the loss of MIWI to increase expression of these elements. While TE expression was similar in wild-type and mutant animals at the earlier pachytene spermatocyte stage, by the later round spermatid stage, transposons identified as targets all displayed greater than threefold increases in expression in Miwi mutants (Supplemental Fig. S4C,D). Thus, the parameters that we developed could effectively identify known meiotic piRNA targets.
Meiotic mRNAs as piRNA targets
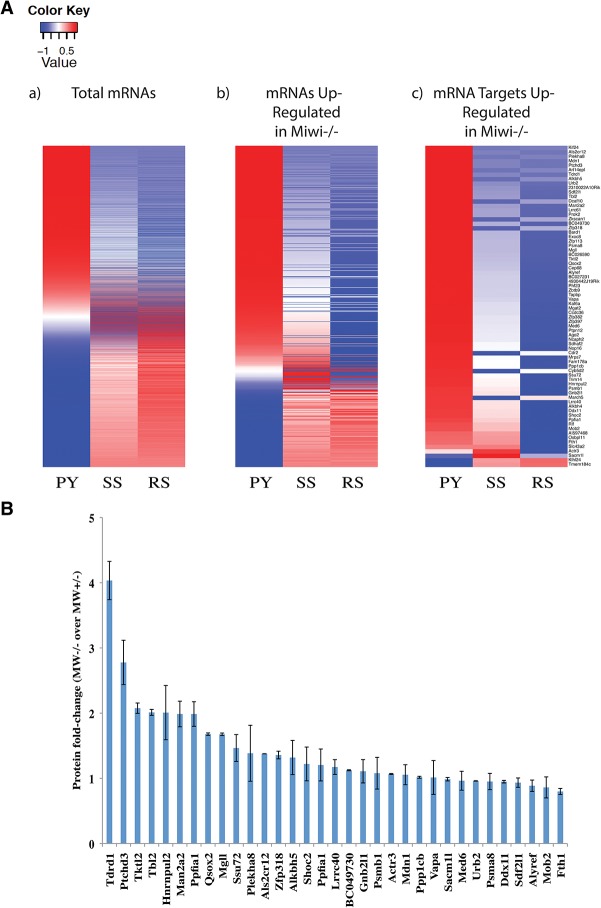
Success in identifying transposons as piRNA targets led us to examine the possibility that meiotic piRNAs might also regulate endogenous mRNAs, as predicted by the fact that increased transposon expression alone seemed insufficient to cause arrest at the round spermatid stage (Xu et al. 2008). This was supported by the fact that a sizeable proportion of our piRNA targets was annotated as protein-coding genes (Supplemental Fig. S2D). Using the criteria established above, we identified mRNAs that showed cleavage patterns and corresponding antisense and sense (mRNA-derived) piRNAs consistent with their being targeted directly by MIWI complexes. By the same logic used for validating transposon targets, we expected these mRNAs to be up-regulated in Miwi−/− round spermatids. Compared with randomly selected mRNAs, our putative targets were more likely to be significantly up-regulated (P < 0.01) (Fig. 3A–C, red dots) or were biased to be more highly expressed in Miwi−/− round spermatids (Fig. 3C, green dots). In total, we identified 330 putative mRNA targets, of which 72 were up-regulated in Miwi−/− round spermatids (Supplemental Tables S1, S2).
Figure 3.
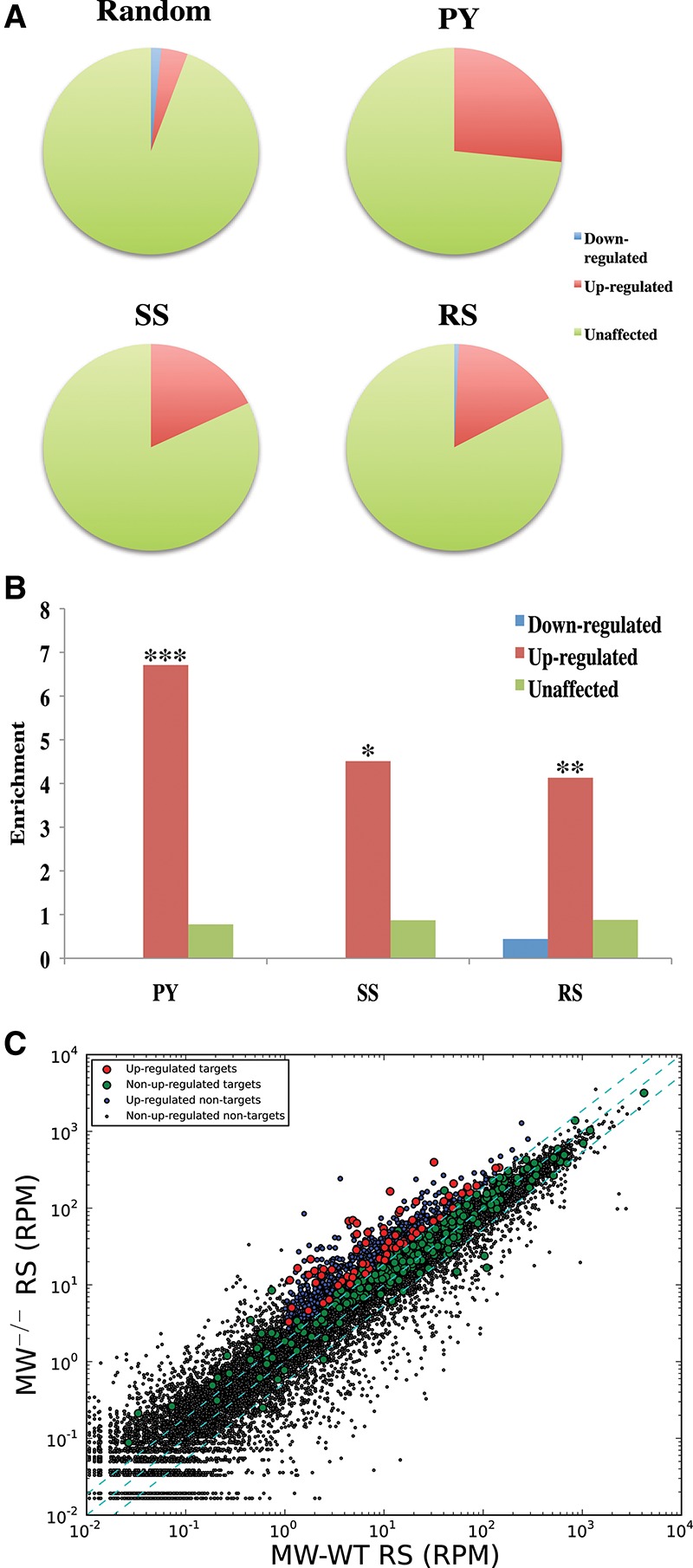
The piRNA cleavage signature identifies meiotic mRNAs as targets of native pachytene piRNAs. (A) The pie charts display proportions of identified mRNA targets, which are significantly misregulated in Miwi knockout versus Miwi wild-type round spermatids (twofold cutoff; P < 0.01) or are not misregulated. “PY,” “SS,” and “RS” denote the proportions for mRNA targets that were identified separately using 5′RACE tags from sorted pachytene spermatocytes, secondary spermatocytes, or round spermatids, respectively. “Random” denotes the same proportions for mRNAs randomly sampled from mRNAs expressed in round spermatids. (B) A bar graph depicting the enrichment of pie chart proportions in A for various meiotic stages over respective proportions in the “Random” pie chart. (*) P < 3 × 10−6; (**) P < 4 × 10−9; (***) P < 7 × 10−26. (C) A scatter plot depicting the RNA-seq values as sequenced from sorted Miwi knockout versus Miwi wild-type round spermatids. Each dot represents the average RPM of an individual gene. The cyan dashed lines (from top left to bottom right) represent a twofold increase cutoff, no fold-change, and a twofold decrease cutoff. The RNA-seq values are the average of five Miwi wild-type or two Miwi knockout biological replicates.
Meiotic piRNAs appear roughly at the pachytene stage and build in abundance through the secondary spermatocyte and round spermatid stages. Consistent with this small RNA class regulating endogenous genes, almost all of our proposed mRNA targets of piRNAs showed sharp decreases in wild-type expression as cells progressed from pachytene to round spermatid stages (Fig. 4A, right). Moreover, the degree of differential expression between wild-type and Miwi mutants also increased over this developmental interval (Supplemental Fig. S5A). In contrast, all mRNAs as a class or mRNAs that showed increased expression in Miwi mutants but lacked characteristics of direct targets failed to show this trend (Fig. 4A, left and middle). We note that some of our identified mRNA targets are regulated by the transcription factor A-Myb, which also regulates the transcription of Miwi and pachytene piRNA cluster loci (Li et al. 2013). The resulting regulatory interactions fit into a type 1 incoherent feed-forward loop (Supplemental Fig. S5B), which may further reinforce the expression patterns exhibited by these mRNA targets (Alon 2007).
Figure 4.
Pachytene piRNAs silence target mRNAs throughout meiosis. (A) The heat maps of mRNA abundance levels in pachytene spermatocytes (PY), secondary spermatocytes (SS), or round spermatids (RS) sorted from Miwi wild-type testes. Each horizontal row represents an individual mRNA. Red and blue represent higher and lower abundances, respectively, than the row mean. The color intensities represent the Z-score from each row mean. Each heat map represents mRNAs with at least 1 RPM in Miwi wild-type round spermatids (panel a), mRNAs significantly up-regulated in Miwi−/− round spermatids (panel b), and the 72 identified mRNA targets that are significantly up-regulated in Miwi−/− round spermatids (panel c). The mRNAs in panel c are a subset of those in panel b, which in turn are a subset of those in panel a. mRNA abundances are calculated from the average of five Miwi wild-type biological replicates. (B) The protein fold change (Miwi−/− over Miwi+/−; P < 0.05) of all proteins that are associated with identified mRNA targets and are detectable by iTRAQ. The error bars denote the standard deviations calculated from the average fold change of two biological replicates.
To ask whether targeting by meiotic piRNAs and changes in mRNA levels in Miwi mutants had an effect at the protein level, we performed iTRAQ analyses comparing Miwi+/− and Miwi−/− round spermatids. Out of the subset of proteins encoded by our 72 mRNA targets and detectable by iTRAQ, several showed roughly twofold increases in the absence of MIWI, with TDRD1 standing out as being roughly fourfold increased when piRNA targeting was disrupted (Fig. 4B; Supplemental Fig. S5C,D; Supplemental Table S3).
Pachytene piRNAs silence target mRNAs via the ping-pong cycle
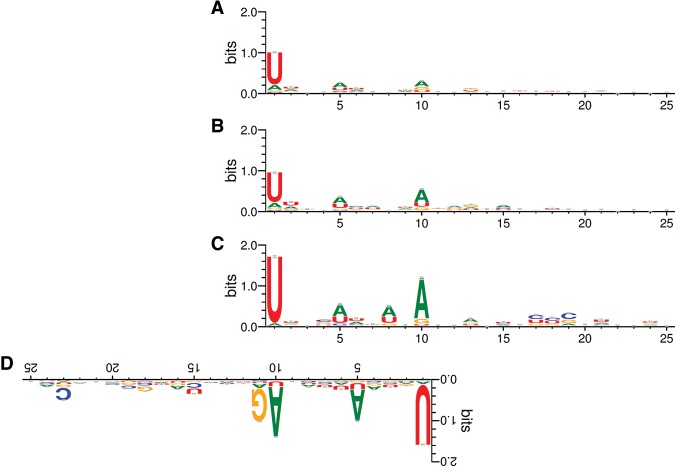
A key feature of the ping-pong cycle is that fragments of target RNAs form new sense piRNAs, with their 5′ ends coinciding with the PIWI-guided cleavage sites. Although this criterion was applied to define pachytene piRNA targets, the ping-pong cycle is known not to operate robustly in meiotic cells (Reuter et al. 2011; Beyret et al. 2012; Watanabe et al. 2015). We therefore asked whether the sense/antisense piRNA pairs generated from mRNA targets exhibited other characteristics of the ping-pong cycle. In order to regenerate the 1U in antisense piRNAs, sense piRNAs that undergo the ping-pong cycle tend to have a 10th adenosine (10A) bias (Brennecke et al. 2007; Gunawardane et al. 2007; Wang et al. 2014b). We noted that before filtering for genes identified as direct piRNA targets, all genic sense piRNAs possessed only a 1U bias (Fig. 5A). If we examined sense piRNAs only from identified protein-coding mRNA targets, we then noticed a slight 10A bias, which was enhanced in the subset of mRNA targets up-regulated in Miwi mutants (Fig. 5B,C). It is important to note that this 10A bias is not generated by the 1U of the antisense piRNA, since the first piRNA nucleotide does not base-pair with its target (Reuter et al. 2011; Nakanishi et al. 2012; Wang et al. 2014b). Furthermore, unlike AUB and MILI (Wang et al. 2014b), MIWI does not seem to have a preference to bind targets that have an adenosine 10 nucleotides (nt) downstream from the cleavage site (Supplemental Fig. S6). Therefore, the enrichment of 10A-biased sense piRNAs within identified mRNAs up-regulated in Miwi mutants indicates that these sense piRNAs use the ping-pong cycle, leading to the post-transcriptional silencing of their host mRNA.
Figure 5.
Genic sense piRNAs exhibit the greatest 10A bias when found in Miwi−/− up-regulated mRNAs that are targeted by antisense guide piRNAs. (A) The nucleotide bias of all genic sense piRNAs that map perfectly to 5′ ends of 5′RACE tags in mRNAs. These piRNAs derive from mRNAs that satisfy all criteria in the piRNA cleavage signature except that they do not necessarily have a complementary antisense guide piRNA. (B) The nucleotide bias of the subset of A exclusive to mRNAs that are identified as pachytene piRNA targets. (C) The nucleotide bias of the subset of B exclusive to mRNAs that are significantly up-regulated in Miwi−/− round spermatids. (D) The nucleotide bias of intergenic cluster guide piRNAs that are complementary to the cleavage sites associated with the sense piRNAs in C, displayed in reverse orientation.
In flies, the piRNA initiating the cycle often shows a 1U bias, whereas the piRNA generated in response to cleavage has a 10A bias (Brennecke et al. 2007; Gunawardane et al. 2007). For our targets, both the sense and antisense piRNAs showed a dual 1U/10A bias (Fig. 5C,D). This could reflect a preference for MIWI to bind RNAs with terminal uridines in contrast to flies where AGO3 appears to lack such a preference. Moreover, both sense and antisense piRNAs associate with MIWI, indicating that these meiotic sense/antisense piRNA pairs most likely participate in a homotypic ping-pong cycle.
Augmenting the endogenous pachytene piRNA repertoire causes male sterility
Previous studies had concluded that the pachytene piRNA sequence is irrelevant to its biological function (Vourekas et al. 2012), although studies reported here and those of others strongly support piRNAs acting in a sequence-specific mode. To probe this issue further, we asked whether alterations in the piRNA repertoire might confer a phenotype. In this regard, two independently derived lines of Hu6 transgenic mice were found to be male sterile (Supplemental Table S4). In contrast, Hu6Δ mice that failed to produce human piRNAs were fertile. Thus, human piRNA production was a likely cause for the infertility of the Hu6 mice.
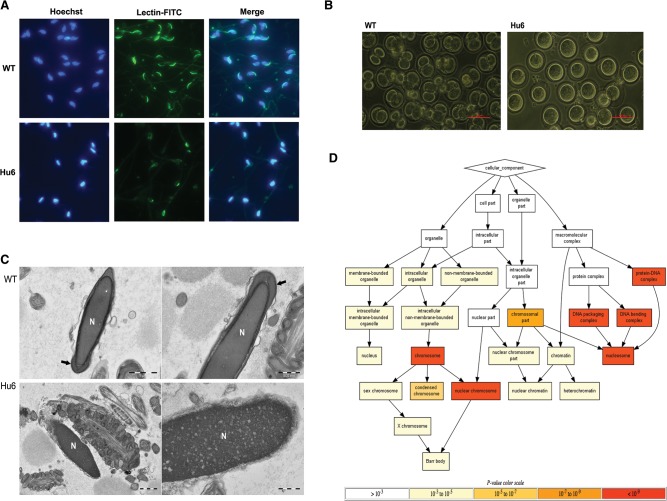
While Hu6 mice produced sperm, these exhibited an unusual morphology. They had rounded sperm heads as opposed to the hook-shaped heads of normal mouse sperm (Supplemental Fig. S7). This sperm head deformity was suggestive of a defect in sperm acrosome development (Yao et al. 2002; Dam et al. 2006). Acrosome staining revealed that, unlike wild-type sperm, which displayed acrosome staining localized to the outer periphery of their hook-shaped heads, Hu6 sperm displayed mislocalized acrosome staining (Fig. 6A). The acrosome is critical in egg penetration during fertilization, but acrosome defects can be compensated for by intracytoplasmic sperm injection (ICSI) (Yao et al. 2002; Wang et al. 2014a). However, Hu6 sperm still could not fertilize eggs via either in vitro fertilization or ICSI, suggesting that these germ cells have defects more pervasive than those that impact the acrosome (Fig. 6B; Supplemental Table S5). This was supported by high-resolution imaging of mutant sperm, which indicated both defects in acrosome development and incomplete compaction of the Hu6 sperm nuclei (Fig. 6C).
Figure 6.
Hu6 males produce defective sperm and are sterile. (A) Representative images of Hoechst and fluorescein isothiocyanate-labeled Pisum sativum agglutinin (FITC-PSA) staining of wild-type (WT) or Hu6 sperm acrosome. (B) Images of eggs 24 h after fertilization via ICSI with either wild-type or Hu6 sperm. The images are representative of two biological replicates for each genotype. Bar, 100 µm. (C) Electron microscope image of wild-type (top) and Hu6 (bottom) sperm. Bars: left, 1 µm; right, 500 nm. (N) Nucleus, (arrow) properly formed acrosome. The images are representative of two biological replicates for each genotype. (D) Gene ontology component analysis of Hu6 round spermatid proteins detected by iTRAQ. The background set is the list of all proteins detected via iTRAQ. The background set was then ranked by protein fold change of Hu6 over wild-type round spermatid, and the top 1% was use as the input set.
During the extensive chromatin remodeling that occurs in spermiogenesis, most core histones are replaced by transition proteins, which in turn are replaced by protamines, resulting in sperm nuclear compaction (Kimmins and Sassone-Corsi 2005). Therefore, we speculated that the incomplete compaction of Hu6 sperm nuclei was caused by incomplete replacement of core histones with protamines. If this was true, protein quantification should reveal increased levels of core histone variants in Hu6 round spermatids as compared with their wild-type counterparts. Indeed, iTRAQ analysis indicated that among proteins with greater abundance in Hu6 round spermatids, core histone variants formed the major class (Fig. 6D; Supplemental Table S6).
It seemed likely that the observed sperm defects were caused by misregulation of germline genes in Hu6 males. Among those genes down-regulated at both the mRNA and protein levels was Dpy19L2 (Fig. 2A; Supplemental Table S6). Dpy19L2 is noteworthy because Dpy19L2−/− mice exhibit male sterility with morphological phenotypes identical to those observed in the Hu6 mice (Pierre et al. 2012; Escoffier et al. 2015; Yassine et al. 2015). Thus, lowered expression of Dpy19L2 was a candidate for a root cause of Hu6 sterility.
We had initial difficulties in identifying a direct cleavage site for human piRNAs within the mouse Dpy19L2 mRNA. We reasoned that this might be due to incomplete coverage by global 5′RACE libraries prepared from small numbers of sorted cells. We therefore applied a relaxed set of criteria that omitted the 5′RACE requirement but relied more on detecting the sense piRNA that is the product of piRNA cleavage. This revealed a cleavage site within the mouse Dpy19L2 mRNA that can be targeted by only a Hu6-specific human piRNA but not any endogenous mouse piRNA (Fig. 7A,B).
Figure 7.
Human piRNA silences mouse Dpy19L2, resulting in male sterility that can be rescued by human Dpy19L2. (A) A schematic showing the targeting of mouse Dpy19L2 mRNA by a Hu6 human piRNA. The format is the same as in Figure 2D, except that the orange sequence denotes the expected cleavage product based on the detected sense piRNA rather than on a 5′RACE tag. (B) A schematic showing the targeting of the mouse Dpy19L2 sequence in A by a mouse piRNA that best matches it. Note that there are eight mismatches within the secondary seed (antisense piRNA nucleotides 12–21). (C) A schematic showing the targeting of the human Dpy19L2 mRNA sequence orthologous to the targeted mouse Dpy19L2 sequence in A by the same Hu6 human piRNA in A, with two mismatches in the primary seed. (D) Genetic cross between a double-transgenic male containing both the Hu6 cluster and human Dpy19L2 gene and a wild-type female. The genotypic frequency of F1 offspring is displayed. Rescue of Hu6 male sterility by HgDpy19L2 was observed in at least three double-transgenic males.
In contrast to the mouse gene, the human Dpy19L2 gene (HgDpy19L2) did not contain sequences that were targets of the human piRNA that directed cleavage of the orthologous mouse gene (Fig. 7C). We therefore produced doubly transgenic animals containing the Hu6 cluster and the human Dpy19L2 gene. Not only did the presence of HgDpy19L2 rescue the male sterility defect caused by human piRNAs, the doubly transgenic male parent was also able to transmit its Hu6 cluster and HgDpy19L2 transgenes to its progeny (Fig. 7D).
Given that relaxed target selection criteria allowed us to identify a functionally relevant cleavage site in Dpy19L2, there seems a high likelihood that the 5′ RACE libraries prepared from sorted cells were far from saturation. We therefore searched among mouse mRNAs for additional potential cleavage targets that met all of our criteria except for a detectable RACE product. This enabled us to identify a list of 628 additional potential targets of native mouse meiotic piRNAs, although each of these must be viewed with less confidence than those having a detectable RACE product (Supplemental Table S7).
Discussion
An in vivo pachytene piRNA cleavage signature
Unlike embryonic piRNAs in mammals, meiotic piRNAs have stubbornly resisted efforts to fully understand their functions. These small RNA populations are remarkably diverse and generally match perfectly only to those sites from which they are produced. This negates a simple strategy of searching for targets by matching piRNAs to complementary sequences to which they might direct PIWI proteins to act in trans. Here, we derived an in vivo piRNA cleavage signature for identifying cleavage targets of meiotic piRNAs, which relied on the addition of human piRNAs to the mouse repertoire and the identification of newly acquired cleavage targets among mouse transcripts. The surprising yet most diagnostic feature of this signature is the appearance of target-derived sense piRNAs that map to the 5′ ends of cleaved RNA products. We were able to validate the predictive power of our signature through the identification of known targets of meiotic piRNAs among transposable elements.
Identification of meiotic mRNAs as cleavage targets of pachytene piRNAs
Through the application of our predictive criteria, we found that a substantial number of mouse mRNAs are targeted by the meiotic piRNA pathway (Supplemental Figs. S8, S9). Consistent with their regulation by piRNAs, these mRNAs fell in abundance as wild-type cells progressed from the pachytene spermatocyte to round spermatid stages, and their abundance increased in Miwi mutants. It is tempting to speculate that misexpression of these protein-coding mRNA targets in Miwi mutants might contribute to the arrest at the round spermatid stage. Among the mRNAs regulated by meiotic piRNAs is Tdrd1. Although TDRD1 is essential for cells to successfully complete meiosis, increased expression of TDRD1 and its mislocalization in Miwi mutants could potentially contribute to an arrest at the round spermatid stage (Kojima et al. 2009).
Pachytene piRNA-guided RNA cleavage was first reported through the use of global 5′RACE to identify a cleavage site within a transposon target (Reuter et al. 2011). Our data confirmed this finding and expanded it to include protein-coding mRNAs as piRNA targets. We note that other groups have recently reported observing mRNA silencing by pachytene piRNAs (Gou et al. 2014; Watanabe et al. 2015; Zhang et al. 2015). However, direct cleavage regulation was not demonstrated with 5′RACE in most cases. Also, one report indicates that MIWI and pachytene piRNAs degrade mRNAs in elongated spermatids despite the fact that Miwi−/− germ cells arrest at the earlier round spermatid stage. In this case, the mRNA degradation that they reported appears to be carried out by a MIWI cofactor, CAF1, rather than MIWI itself.
Significance of pachytene piRNAs engaging in the ping-pong cycle
In the process of identifying native targets of pachytene piRNAs, we noted that MIWI cleavage produced genic sense piRNAs that paired with intergenic antisense piRNAs to undergo a homotypic ping-pong cycle. This was unforeseen given that pachytene piRNAs have long been assumed to avoid such an amplification loop (Reuter et al. 2011; Beyret et al. 2012). This assumption was recently reiterated by a group that reported observing mRNA silencing by pachytene piRNAs (Watanabe et al. 2015). This discrepancy is likely related to their identification of pachytene piRNA targets primarily using a RNA-seq comparison between Miwi+/− and Miwi−/− without filtering for genes that do not possess a piRNA-directed cleavage site. Thus, this strategy has the potential to identify both direct (Fig. 3C, red dots) and indirect (Fig. 3C, blue dots) targets. A recent study had also reported the observation of intergenic antisense piRNAs that target mRNAs and have both a 1U and 10A bias, similar to what we report here (Fig. 5D; Zhang et al. 2015). Given that we see a strong 10A signal only in genic sense piRNAs derived from identified mRNA targets that are up-regulated in Miwi mutants, the ping-pong cycle is likely a functional driving force in the silencing of pachytene piRNA targets.
Sterility caused by the introduction of human piRNAs argues for the sequence-specific function of this small RNA class
While the observation of piRNA–mRNA ping-pong cycling strongly implies the importance of mRNA silencing to spermatogenesis, it was critical to ask whether altering this silencing process impacted spermatogenesis. Other groups have investigated this question by augmenting the pachytene piRNA repertoire in the presence of wild-type MIWI but failed to see any phenotypic defects (Xu et al. 2008; Watanabe et al. 2015). Here, we found that simply adding a single human piRNA cluster to the mouse piRNA repertoire created profound consequences, causing the production of abnormal and completely nonfunctional sperm.
Expression of human piRNAs led to the silencing of a germline gene, Dpy19L2, that is essential for spermiogenesis. The resultant reduction in Dpy19L2 likely caused defects in both acrosome development and nuclear compaction that occur during sperm formation, leading to male sterility. In accord with this hypothesis, introduction of the human ortholog of Dpy19L2 was able to fully rescue the male sterility of Hu6 mice. Thus, despite the seeming randomness of the highly diverse mouse meiotic piRNAs, there must be stringent evolutionary selection to prevent targeting of mouse genes essential for sperm development.
Just as their abnormal function can negate fertility, sperm development relies on the proper function of native mouse piRNAs. Considered together, our data indicate a closely linked coevolution of meiotic piRNA sequences and protein-coding genes involved in sperm development. Certainly, these sequences must be strongly selected for target avoidance. However, our results indicate that they also evolved to play a role in positively regulating progression into and through spermiogenesis by repressing mRNAs whose inappropriate expression might impair sperm development. Our data support the idea that meiotic piRNA populations must be evolutionarily constrained to enable successful spermatogenesis, both driving the response away from essential genes and directing the pathway toward mRNA targets that are regulated by piRNAs in meiotic cells.
Materials and methods
piRNA targeting criteria
An intergenic pachytene piRNA cluster MIWI-immunoprecipitated small RNA (>28 nt) is a guide piRNA that targets a cleavage sequence (−11 nt to +10 nt of cleavage site) if it first overlaps the 5′ end of the corresponding 5′RACE tag in an antisense manner by exactly 10 nt, allowing for a mismatch in piRNA nucleotide 1. In addition, it must have perfect complementarity in the primary seed (piRNA nucleotides 2–11) and tolerate at most four mismatches in the secondary seed (piRNA nucleotides 12–21). The complementarity cutoff and seed definitions are based on criteria that allow mouse Bbs5 and Gm11837 to be targeted by only Hu6 human piRNAs but not a single native mouse piRNA and also take into consideration previous in vitro small RNA-based cleavage assays (Reuter et al. 2011; Nakanishi et al. 2012).
piRNA cleavage signature
A mRNA or transposon was identified as a piRNA-guided cleavage site if it fulfilled these four criteria. First, the mRNA or transposon was not located in previously annotated intergenic pachytene piRNA clusters (Li et al. 2013). Second, the mRNA or transposon contained a 5′RACE tag where the average 5′RACE RPM of Miwi+/− biological replicates was at least 10 RPM and at least 10-fold enriched compared with Miwi−/− biological replicates. Third, there was a MIWI-bound sense piRNA (>28 nt) that mapped perfectly to the 5′ end of the 5′RACE tag. Fourth, there was a guide piRNA that can target the corresponding cleavage sequence (−11 nt to +10 nt of cleavage site). Target identifications were carried out using at least two biological replicates of 5′RACE data per cell stage per genotype and two biological replicates of MIWI-immunoprecipitated small RNA data. The relaxed cleavage signature was similar to the regular piRNA cleavage signature except that it did not require detection of a 5′RACE tag, and the cleavage site was demarcated by the MIWI-bound sense piRNA.
Transgenic mouse generation
The RP11-1024C2 human BAC was used to expressed the human Dpy19L2 gene in HgDpy19L2 mice. The RP11-119C22 human BAC was used to express a human ch6 piRNA cluster in Hu6 mice. The BAC used to generate Hu6▵ mice was recombineered from RP11-119C22 by replacing a predicted 4007-base-pair (bp) promoter (as defined by forward, ACCCAACCAATTGTCAGTCAGAAAATGTTTAAATTTACCTATAGCCTGGA; and reverse, GAGGGGAATTTCCTGCCGGCTCTTGCATTCCTCAAAGGTATATACAATTA primer sequences) of the bidirectional human piRNA cluster with a neomycin resistance cassette by Red/ET recombination (Gene Bridges, K002). BACs were extracted from 250-mL bacterial cultures with Nucleobond Xtra Maxi EF (Thermo Fisher Scientific, NC0325254), linearized with PI-SceI (New England Biolabs, R0696), and dialyzed on a 0.025-μm membrane filter (Millipore, VSWP02500) with dialysis buffer (10 mM Tris at pH 7.5, 0.1 mM EDTA, 100 mM NaCl, 10.4 µg/mL spermine tetrahydrochloride, 17.8 µg/mL spermidine trihydrochloride). Transgenic animals were generated by pronuclear microinjection of BACs into B6SJL F2 (C57BL/6 × SJL, Jax Laboratory) mouse eggs at the Rodent Genetic Engineering Core at New York University or at the Gene Targeting Shared Resource at Cold Spring Harbor Laboratory. Positive mice were genotyped by PCR. All mice used in our experiments were maintained according to the guidelines of the Cold Spring Harbor Laboratory Institutional Animal Care and Use Committee.
Spermatogenic cell isolation
Total germ cells were first isolated from adult (2- to 6-mo-old) mouse testes. Pachytene spermatocytes, secondary spermatocytes, and round spermatids were subsequently sorted as described (Bastos et al. 2005), with minor modifications. Briefly, testes were isolated from one adult mouse for each sorting experiment, and the tunica was removed. Tissue was digested for 45 min at 37°C in dissociation buffer (25 mg of collagenase A, 25 mg of Dispase II, 2.5 mg of DNase I in HBSS). Cells were resuspended at a concentration of 1 × 106 cells per milliliter in HBSS+ (HBSS, 25 mM Hepes, 1.5 mM EDTA, 5% FBS) and stained with 5 mg/mL Hoechst (Sigma, 33342) for 30 min at 37°C. Immediately before analysis, 1 mg/mL propidium iodide was added for dead cell exclusion. Analysis was performed in a five-laser AriaII cell sorter (Becton Dickinson). Hoechst was excited with a UV laser at 350 nm, and fluorescence was recorded with a 450/50 filter (Hoechst blue) and 670LP filter (Hoechst red). A 505LP filter was used to separate the emission wavelengths. The three different cell types were gated based on their Hoechst blue–red profile in scatter plots as well as Hoechst blue fluorescence and forward and side scatter properties. After applying gates, cells were collected directly into Trizol-LS for RNA isolation.
Total RNA preparation
Adult mouse testis tissue or sorted spermatogenic cells were lysed in Trizol or Trizol-LS reagent, and total RNA was isolated according to the manufacturer's recommendations.
Antibodies
The rabbit polyclonal antibody against MIWI (MIWI-N3) was produced by injecting the synthesized peptide PTEGDLVGRGRQRGM into two rabbits (Covance, Inc.). The antibody was purified from immunized sera using peptide affinity purification (Thermo Fisher Scientific, 44999). Antibody quality was assessed via Western blotting of testis lysate and immunoprecipitation of ∼30-nt testis small RNAs.
Immunoprecipitation of MIWI-bound small RNAs
Both mouse testes were homogenized in 1 mL of lysis buffer (10 mM Hepes at pH 7, 100 mM KCl, 1.5 mM MgCl2, 0.5% NP-40, 1% Triton X-100, 10% glycerol). The lysate was then clarified on a tabletop centrifuge for 30 min, and the supernatant was passed through a 45-µm filter. The lysate was incubated with 6 µg of rabbit anti-MIWI-N3 polyclonal antibody and 4 mL of NT2 buffer (25 mM Tris at pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 0.05% NP-40) while rotating for 3 h at 4°C. One-hundred microliters of protein A-agarose beads (Roche, 11134515001) was prewashed in NT2 buffer and preblocked with 5% BSA before being incubated with the lysate antibody mixture while rotating for 2 h at 4°C. Beads were then washed twice in 1.4 mL of NT2 buffer, twice in 1.4 mL of NT-2 (with additional 150 mM NaCl), and twice with 1.4 mL of NT-2 (with additional 350 mM NaCl). MIWI-immunoprecipitated small RNAs were eluted by treating beads with 400 µg of proteinase K (New England Biolabs, P8107S) and 1× proteinase K buffer (100 mM Tris at pH 7.5, 12.5 mM EDTA, 150 mM NaCl, 1% [w/v] SDS) for 1 h at 60°C with light agitation. Finally, eluted RNA was extracted with phenol–chloroform. A quarter of the eluted RNA was visualized with 5′ end labeling to verify the precipitation of ∼30-nt RNA species, and the remainder was used for small RNA cloning.
Small RNA cloning
Total RNA or MIWI-immunoprecipitated RNA (19–33 nt) was first size-selected on a 15% denaturing polyacrylamide gel. Size-selected small RNAs were then cloned as described (Goh et al. 2014). Libraries were sequenced on an Illumina HiSeq SE36 or SE76 platform.
5′RACE and stranded RNA-seq
Total RNA from sorted pachytene spermatocytes, secondary spermatocytes, or round spermatids was treated with RQ1 DNase (Promega, M6101) for 30 min at 37°C, depleted of ribosomal RNA using the Ribo-Zero kit (Epicentre, MRZH11124), and used to generate global 5′RACE libraries as described (Reuter et al. 2011). Post-amplification libraries were size-selected on a BluePippin 2% agarose chip (Sage Science, BDF2010) using a cutoff of 180–350 bp. Ribo-depleted RNA was also used for strand-specific RNA libraries, which were constructed using the ScriptSeq version 2 RNA-seq library preparation kit (Epicentre, SSV21124). Both global 5′RACE and ScriptSeq libraries were sequenced on an Illumina HiSeq SE76 platform.
Northern blotting
Total RNA was separated on a 15% denaturing polyacrylamide gel. RNA was then transferred from the gel onto a Hybond+ membrane in 0.5× TBE at 20 V for 90 min. The membrane was UV cross-linked at 1200 µJ/cm2 and then hybridized in Ultrahyb hybridization buffer for 1 h. We used five different DNA oligonucleotides for Northern blotting: GGCCTCACGCTCCTGATCTGTAAAA, TATCATGCAGTTGTATTCCATTGTA, AAAGACGTTTCCCTTTCAACTCTA, TACTAATCTATCTTCCCAACATCA, and CATCCATCTAAAGTCTCTCCAAATA. These oligonucleotides are complementary to five different human piRNAs in the Hu6 piRNA cluster, as determined from previous small RNA-seq data of human testes (Girard et al. 2006). The Northern blotting oligonucleotides were 5′-labeled, pooled, and hybridized to the membrane overnight. The membrane was then washed twice with 2× SSC and 0.1% SDS and twice with 1× SSC and 0.1% SDS. The washed membrane was exposed onto a phosphoplate overnight before being viewed on a Fuji FLA-5000 PhosphorImager. All hybridization and wash steps were carried out at 37°C.
Determination of sterility
Sexually mature males (2–6 mo old) were crossed with CD-1 females. Males are considered sterile if no pups were born after at least 2 mo of crossing.
Preparation of paraffin-embedded testis sections for microscopy
Mouse testes were extracted from adult mice and fixed in Bouin's solution (Sigma, HT10132) overnight at 4°C and then washed for 30 min twice in 50% ethanol and twice in 70% ethanol. After fixation, testis samples were embedded in OCT blocks and sectioned to produce 6-µm sections that were separated by 30 µm. Slides were stained with hematoxylin and eosin, and images were acquired with a Nikon Eclipse Ti microscope.
Staining of sperm acrosome by fluorescein isothiocyanate-labeled Pisum sativum agglutinin (FITC-PSA)
Mouse cauda epididymis was isolated and dissected in 1× DPBS. Sperm were counted, and 15,000 sperm were spotted onto a slide, permeabilized with methanol, and then stained with FITC-PSA as described (Mendoza et al. 1992) and subsequently with Hoechst (Sigma, H33342). Images were acquired with the Nikon Eclipse Ti microscope.
iTRAQ
Sorted round spermatids were pelleted, frozen in liquid nitrogen, and then sent to the Proteomics Shared Resources at Cold Spring Harbor Laboratory for protein extraction and quantification via iTRAQ, which was performed as described (Scuoppo et al. 2012).
Supplementary Material
Acknowledgments
We thank P. Zamore, C. Tipping, and G. Farley for the gift of Miwi−/− mice; S.Y. Kim for assisting in generating the HgDpy19L2, Hu6, and Hu6▵ transgenic mice and assisting in IVF and ICSI; R. Pillai and M. Reuter for advice on global 5′RACE library preparation; E. Lee, E. Hodges, and E. Ghiban for handling library sequencing; S. Boettcher for managing laboratory equipment and reagents; M. Mosquera for mouse genotyping; M. Cahn, J. Coblentz, G. Munoz, and L. Bianco for mouse husbandry; D. Pappin for assisting in iTRAQ; and C. Carlston, J. Ipsaro, V. Vagin, C. Hammell, M. Timmermans, and V. Ambros for helpful discussions. This work was supported by the National Institutes of Health R37 grant GM062534-14 to G.J.H. iTRAQ was performed with assistance from the Cold Spring Harbor Laboratory Proteomics Shared Resource, which is supported by Cancer Center support grant 5P30CA045508. W.S.S.G. is a McClintock Fellow of the Watson School of Biological Sciences and is supported by the NSS Scholarship from the Agency for Science, Technology and Research, Singapore. O.H.T. is supported by a fellowship of the Human Frontier Science Program. R.B. is supported by the Starr Centennial Scholarship from the Watson School of Biological Sciences. G.J.H. is a Howard Hughes Medical Institute Investigator.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.260455.115.
References
- Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8: 450–461. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. [DOI] [PubMed] [Google Scholar]
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. 2005. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry 65A: 40–49. [DOI] [PubMed] [Google Scholar]
- Beyret E, Liu N, Lin H. 2012. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res 22: 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Castañeda J, Genzor P, van der Heijden GW, Sarkeshik A, Yates JR, Ingolia NT, Bortvin A. 2014. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J 33: 1999–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam AHDM, Feenstra I, Westphal JR, Ramos L, van Golde RJT, Kremer JAM. 2006. Globozoospermia revisited. Hum Reprod Update 13: 63–75. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. [DOI] [PubMed] [Google Scholar]
- Escoffier J, Yassine S, Lee HC, Martinez G, Delaroche J, Coutton C, Karaouzène T, Zouari R, Metzler-Guillemain C, Pernet-Gallay K, et al. 2015. Subcellular localization of phospholipase Cζ in human sperm and its absence in DPY19L2-deficient sperm are consistent with its role in oocyte activation. Mol Hum Reprod 21: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. [DOI] [PubMed] [Google Scholar]
- Goh WSS, Seah JWE, Harrison EJ, Chen C, Hammell CM, Hannon GJ. 2014. A genome-wide RNAi screen identifies factors required for distinct stages of C. elegans piRNA biogenesis. Genes Dev 28: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH. 2001. A novel active L1 retrotransposon subfamily in the mouse. Genome Res 11: 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou L-T, Dai P, Yang J-H, Xue Y, Hu Y-P, Zhou Y, Kang J-Y, Wang X, Li H, Hua M-M, et al. 2014. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res 24: 680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. 2006a. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. 2006b. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci 103: 13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Hirano T, Iwasaki YW, Lin ZYC, Imamura M, Seki NM, Sasaki E, Saito K, Okano H, Siomi MC, Siomi H. 2014. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 20: 1223–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589. [DOI] [PubMed] [Google Scholar]
- Kojima K, Kuramochi-Miyagawa S, Chuma S, Tanaka T, Nakatsuji N, Kimura T, Nakano T. 2009. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells 14: 1155–1165. [DOI] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza C, Carreras A, Moos J, Tesarik J. 1992. Distinction between true acrosome reaction and degenerative acrosome loss by a one-step staining method using pisum sativum agglutinin. J Reprod Fertil 95: 755–763. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. 2012. Structure of yeast Argonaute with guide RNA. Nature 486: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibu T, Hayashida Y, Tani S, Kurono S, Kojima-Kita K, Ukekawa R, Kurokawa T, Kuramochi-Miyagawa S, Nakano T, Inoue K, et al. 2012. Identification of MIWI-associated poly(A) RNAs by immunoprecipitation with an anti-MIWI monoclonal antibody. Biosci Trends 6: 248–261. [DOI] [PubMed] [Google Scholar]
- Pierre V, Martinez G, Coutton C, Delaroche J, Yassine S, Novella C, Pernet-Gallay K, Hennebicq S, Ray PF, Arnoult C. 2012. Absence of Dpy19l2, a new inner nuclear membrane protein, causes globozoospermia in mice by preventing the anchoring of the acrosome to the nucleus. Development 139: 2955–2965. [DOI] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. [DOI] [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, et al. 2012. A tumour suppressor network relying on the polyamine–hypusine axis. Nature 487: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wan H, Li X, Liu W, Chen Q, Wang Y, Yang L, Tang H, Zhang X, Duan E, et al. 2014a. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res 24: 852–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yoshikawa M, Han BW, Izumi N, Tomari Y, Weng Z, Zamore PD. 2014b. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol Cell 56: 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Cheng E-C, Zhong M, Lin H. 2015. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res 25: 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, Hecht NB. 2008. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol Reprod 79: 51–57. [DOI] [PubMed] [Google Scholar]
- Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, Yanagida K, Sato A, Toshimori K, Noda T. 2002. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci 99: 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine S, Escoffier J, Martinez G, Coutton C, Karaouzène T, Zouari R, Ravanat J-L, Metzler-Guillemain C, Lee HC, Fissore R, et al. 2015. Dpy19l2-deficient globozoospermic sperm display altered genome packaging and DNA damage that compromises the initiation of embryo development. Mol Hum Reprod 21: 169–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Kang J-Y, Gou L-T, Wang J, Xue Y, Skogerboe G, Dai P, Huang D-W, Chen R, Fu X-D, et al. 2015. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res 25: 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Gou L-T, Zhang M, Zu L-D, Hua M-M, Hua Y, Shi H-J, Li Y, Li J, Li D, et al. 2013. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell 24: 13–25. [DOI] [PubMed] [Google Scholar]
- Zheng K, Wang PJ. 2012. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 8: e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.