Wegener et al. reveal that upregulation of Olig2 in oligodendrocyte progenitor cells enhances their migration and differentiation, leading to precocious myelination in prenatal mice and accelerated remyelination of lesions in adults. The presence of Olig2 in regenerating oligodendrocytes in multiple sclerosis lesions suggests that its upregulation could facilitate myelin repair.
Keywords: multiple sclerosis, remyelination, Olig2, oligodendrocyte, tetracycline system
Abstract
The basic helix-loop-helix transcription factor Olig2 is a key determinant for the specification of neural precursor cells into oligodendrocyte progenitor cells. However, the functional role of Olig2 in oligodendrocyte migration and differentiation remains elusive both during developmental myelination and under demyelinating conditions of the adult central nervous system. To decipher Olig2 functions, we generated transgenic mice (TetOlig2:Sox10rtTA/+) overexpressing Olig2 in Sox10+ oligodendroglial cells in a doxycycline inducible manner. We show that Olig2 overexpression increases the generation of differentiated oligodendrocytes, leading to precocious myelination of the central nervous system. Unexpectedly, we found that gain of Olig2 function in oligodendrocyte progenitor cells enhances their migration rate. To determine whether Olig2 overexpression in adult oligodendrocyte progenitor cells promotes oligodendrocyte regeneration for myelin repair, we induced lysophosphatidylcholine demyelination in the corpus callosum of TetOlig2:Sox10rtTA/+ and control mice. We found that Olig2 overexpression enhanced oligodendrocyte progenitor cell differentiation and remyelination. To assess the relevance of these findings in demyelinating diseases, we also examined OLIG2 expression in multiple sclerosis lesions. We demonstrate that OLIG2 displays a differential expression pattern in multiple sclerosis lesions that correlates with lesion activity. Strikingly, OLIG2 was predominantly detected in NOGO-A+ (now known as RTN4-A) maturing oligodendrocytes, which prevailed in active lesion borders, rather than chronic silent and shadow plaques. Taken together, our data provide proof of principle indicating that OLIG2 overexpression in oligodendrocyte progenitor cells might be a possible therapeutic mechanism for enhancing myelin repair.
Introduction
Developmental studies on oligodendrocyte differentiation have shed light on oligodendrocyte regeneration and myelin repair in response to CNS injury, such as demyelination in multiple sclerosis. As oligodendrocyte regeneration in the adult CNS may be due to the recapitulation of developmental pathways, their study might provide the molecular basis to unravel signalling pathways regulating the migration and differentiation of adult oligodendrocyte progenitor cells (OPCs) (Fancy et al., 2010). Furthermore, it has been shown that despite the presence of endogenous OPCs in chronic lesions of multiple sclerosis (Wolswijk, 1998; Chang et al., 2002), they often fail to remyelinate axons, suggesting a differentiation failure (Arnett et al., 2004; Kuhlmann et al., 2008). To overcome this remyelination deficiency, inducing migration of adult OPCs and/or their differentiation into remyelinating oligodendrocytes represents a rational approach for enhancing repair and neuroprotection in multiple sclerosis. Therefore, the identification of molecular pathways regulating OPC migration and differentiation during development may provide new potential targets for effective remyelinating therapies.
During development, the determination of OPCs is tightly regulated by a complex interplay of intrinsic, extrinsic and epigenetic factors (Rowitch and Kriegstein, 2010). Among intrinsic factors, the basic helix-loop-helix (bHLH) transcription factor Olig2 has critical functions in oligodendrocyte determination (Rowitch, 2004). Olig2 loss- and gain-of-function studies provided compelling support of its requirement in oligodendrocyte specification (Lu et al., 2000, 2002; Zhou et al., 2000; Zhou and Anderson, 2002; Liu et al., 2007; Maire et al., 2010). Furthermore, Olig2 remains expressed in OPCs (Lu et al., 2000; Takebayashi et al., 2000; Zhou et al., 2000), suggesting additional functions during oligodendrocyte differentiation. Indeed, it has been shown that combined Olig2/Nkx2.2 (Zhou et al., 2001) or Olig2/Zfp488 (Wang et al., 2006) overexpression in OPCs promoted their differentiation into myelinating oligodendrocytes. Other reports showed that overexpression of Olig2 alone triggers OPC differentiation (Liu et al., 2007). However, the functional role of Olig2 in OPC differentiation and maturation during development and under pathological conditions has never been investigated in vivo using selective genetic tools.
In this study, we explore the functional role of Olig2 as a potential target regulating oligodendrocyte migration and differentiation during developmental myelination, in lysophosphatidylcholine (LPC)-induced demyelination and in multiple sclerosis. To decipher Olig2 function in OPCs, we generated transgenic mouse lines overexpressing Olig2 in Sox10+ oligodendroglial cells, using inducible tetracycline systems (Gossen and Bujard, 1992; Ludwig et al., 2004). We demonstrate that overexpression of Olig2 alone is sufficient for enhancing OPC migration and differentiation, leading to precocious myelination. Interestingly under demyelinating conditions, inducible Olig2 overexpression increased the remyelination rate of LPC-induced lesions. Similarly, we showed that OLIG2 is upregulated in maturing RTN4-A/NOGO-A+ oligodendrocytes in active and chronic active lesions, suggesting that upregulation of OLIG2 in adult OPCs is required for their proper differentiation and remyelination in multiple sclerosis.
Materials and methods
TetOlig2:Sox10rtTA/+ and TetOlig2:Sox10-Cre/RosaΔtTA/+ transgenic mice
All experiments were performed according to European Community regulations and Inserm ethical committee (authorization A-75-2024; 26/10/2012). TetOlig2 (EGFP<-tetO->Olig2) (Maire et al., 2010) mice were crossed with Sox10rtTA/+ (Tet-On system) (Ludwig et al., 2004) or Sox10-Cre/RosaΔtTA/+ inducer lines (Cre-dependent Tet-Off system) (Matsuoka et al., 2005; Wang et al., 2008). In the RosaΔtTA/+ transgenic line, a neomycin-stop cassette flanked by two LoxP sites, upstream to tTA coding sequence, was inserted by homologous recombination into the Rosa locus, allowing Cre-conditional expression of tTA upon removal of the stop cassette (Wang et al., 2008). Sox10rtTA/+, TetOlig2:Sox10rtTA/+, TetOlig2:Sox10-Cre/RosaΔtTA/+ were genotyped by PCR with different sets of primers: Sox10-R: 5′-ctaggctgtcagagcagacga-3′, Sox10-F: 5′-ctccacctctgataggtcttg-3′, Sox10rtTA-F: 5′-ctcgattggcagggcatcgag-3′, Olig2-R: 5′-tgctggaggaagatgacttgaagcc-3′ and CMV-F: 5′-gcagagctggtttagtgaaccgc-3′; Cre-F: 5′-atgctgtttcactggttatg-3′, Cre-R: 5′-attgcccctgtttcactatc-3′, Rosa-F: 5′-aagggagctgcagtggagta-3′, Rosa-R: 5′-tcatcaaggaaaccctggac-3′ and RosatTA: 5′-ccgaaaatctgtgggaagtc-3′. For developmental studies, doxycycline treatment (6 mg/ml, Sigma-Aldrich) was performed from E10.5 to E15.5/(P)0 or E18.5 to P5/10/15, wild-type and Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ mice were treated similarly for all experiments. TetOlig2:Sox10-Cre/RosaΔtTA/+ mice were not treated with doxycycline.
Tissue preparation, in situ hybridization and immunochemistry
Mouse tissue sections (from E15.5 to P120) or post-mortem multiple sclerosis brain samples were processed for light and transmission electron microscopy (Bachelin et al., 2010; Maire et al., 2010), in situ hybridization with specific riboprobes for Mbp, Plp1 or Myrf (Finzsch et al., 2010; Hornig et al., 2013), or for immunohistochemistry using primary antibodies against APC (clone CC1), GFP, Ki67, MBP, NG2, NOGO-A/RTN4-A, Olig2, PCNA, PDGFRA and Sox10. Sources and working concentrations of primary antibodies are detailed in Supplementary Table 1. Secondary antibodies were used at 1:1000 (Alexa-conjugated) and 1:100 (TRITC or FITC-conjugated). Nuclei were also counterstained with DAPI. In situ hybridization and immunolabelling were visualized using a bright field and fluorescence microscope (Leica). Images were processed using Explora Nova and Photoshop software. Statistical analyses were performed using Student or ANOVA Tukey tests (SigmaStat or GraphPad Prism 5 software).
Reverse transcriptase PCR and quantitative PCR
Total RNA from TetOlig2:Sox10rtTA/+, Sox10rtTA/+ and wild-type were isolated from spinal cords using RNeasy® mini-kit (Qiagen). The concentration of total RNA was assessed by NanoDrop™ (Applied Biosystems) and adjusted to the same concentrations among samples. DNA contamination was eliminated by DNaseI/RNase-free treatment (Promega) and 500 ng aliquots of RNA were used to synthesize first strand cDNA using the SuperScript® Strand II cDNA synthesis (Invitrogen). Three replicate experiments were performed for quantitative PCR analysis using an ABIprism 7000 sequence detection system with TaqMan® gene expression assay probes (Applied Biosystems). Ct numbers were calculated for both reference gene (TBP) and target genes with auto-baseline and auto-threshold. ΔΔCt was used to determine the fold increase.
OPC cultures and transduction
CG4 cells were cultured in N1/B104 medium (70 V/30 V) composed of Dulbecco’s modified Eagle's medium and N1 supplement with B104 conditioned medium (B104CM, obtained after 3 days of conditioning serum-free N1 medium with the B104 neuroblastoma) (Louis et al., 1992; Avellana-Adalid et al., 1996). Primary mouse OPCs were obtained from P0/P1 newborn c57bl6/J mice (Janvier). Briefly, brains were dissected free of meninges, dissociated with 0.05% trypsin/0.1% DNase. Cells were then plated at the density of 40 000 cells/mm2 in Dulbecco’s modified Eagle’s medium /F12 medium (1:1) supplemented with N2 (1%), B27 (0.5%), insulin (25 μg/ml) glucose (6 mg/ml), HEPES (5 mM), FGF2 (10 ng/ml) and PDGF (10 ng/ml). CG4 cells or mouse OPCs (105 cells/mm2) were transduced with bi-cistronic lentiviral vectors (50 ng of p24): pWPi-Olig2 encoding Olig2 cDNA and EGFP under the control of the ubiquitous EF1-alpha promoter (EF1α->Olig2-IRES-EGFP) or pWPi encoding EGFP alone (EF1α->IRES-EGFP) (Maire et al., 2009). Transduced cells were expanded in their respective proliferation medium and fluorescence-activated cell sorting selected based on GFP (green fluorescent protein) expression.
Migration assays
Spinal cord explants
To assess the role of Olig2 on OPC migration, time-lapse imaging on spinal cord explants were prepared from E13.5 TetOlig2:Sox10rtTA/+ and TetGFP:Sox10rtTA/+ (control) embryos. Briefly, embryos were dissected in 0.1 M phosphate-buffered saline and spinal cords finely dissected. Spinal cords were cut and cultured on polyornithine (PO, 100 µg/ml)/1% matrigel coated dishes in N1 medium, supplemented with 0.5% foetal calf serum and 2 µg/ml of doxycycline (Sigma-Aldrich). After overnight incubation at 37°C with 5% CO2, the migration of GFP+ OPCs in TetOlig2:Sox10rtTA/+ and control TetGFP:Sox10rtTA/+ spinal cord explants was analysed during 24 h using an Axiovert 200 video microscope (Zeiss). Images were taken every 10 min and analysed using MetaMorph® software. Several cells for each explant were followed and n ≥ 10 explants analysed for each genotype.
CG4 and mouse OPC spheres
pWPi- and pWPi-Olig2-transduced CG4 and mouse OPCs oligospheres were plated on polyornithine (100 μg/ml) or polyornithine (100 μg/ml)/laminin (1 μg/ml) -coated glass coverslips in N1B104CM medium (CG4 cells) or N2 supplemented with PDGF (10 ng/ml) and FGF (10 ng/ml, for mouse OPCs). Migration was assessed for 6 h for CG4 (n = 12 spheres) and 24 h for mouse OPCs (n = 10). Ten minute sequential images were taken and analysed using ImageJ software. The distance of migration corresponding to the mean distance achieved between each time interval and the migration velocity (whole distance/time) were quantified in each condition.
Differentiation assays and immunocytochemistry
CG4 oligospheres were dissociated into single cells with 0.05% trypsin, 0.1% glucose and 0.02% EDTA solution and plated onto polyornithine- (100 μg/ml) and laminin- (1 μg/ml; both from Sigma-Aldrich) coated coverslips. For differentiation assays, cells were placed in N1 medium without B104-CM supplemented with 0.5% foetal bovine serum for 3 and 7 days. Live cells were incubated with O4 and GalC primary antibodies (1:10, American Type Culture Collection) for 20 min, washed in 0.1 M phosphate-buffered saline, fixed with 4% paraformaldehyde and then incubated with appropriate secondary antibodies for 30 min. Nuclei were counterstained with DAPI.
Lysophosphatidylcholine-induced demyelination and remyelination analysis
Focal demyelinating lesions were induced by stereotaxic injection of 0.5% LPC in 0.1 M phosphate-buffered saline (Sigma-Aldrich) into the corpus callosum (AP: +0.13 mm; ML: +0.10 mm; DV: −0.17 mm relative to Bregma) as previously described (Nait-Oumesmar et al., 1999). Wild-type and Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ mice were induced 3 weeks before LPC injections and analysed at 7, 14 and 21 days post-injection. Images of transverse ultra-thin sections through the lesion were observed at a magnification of ×6500 and analysed using ImageJ. For remyelination analysis, the percentage of remyelinated axons was quantified over the total number of axons, excluding non-myelinated (axonal diameter ≤1 μm2) and non-demyelinated axons (G-ratio ≤ 0.55).
Multiple sclerosis tissue samples
Post-mortem human multiple sclerosis brain samples and control samples from individuals without neurological diseases were obtained from the UK MS Tissue Bank (Dr R. Reynolds, London). Informed consent had been obtained from each patient and this study was approved by the London Multicenter Research Ethics Committee (MREC 02/2/39). The clinical feature of these post-mortem multiple sclerosis cases and lesions analysed are detailed in Supplementary Table 2. Twelve micrometre frozen sections were performed and processed as previously described (Nait-Oumesmar et al., 2007). Histological assessment of the lesions was performed using Luxol Fast blue/Cresyl violet and Oil-red-O staining. Lesions were classified according to their inflammatory activity (KP1 and MHC-II immunolabelling) and on the basis of histological criteria (Lucchinetti et al., 1996).
Results
Generation of a transgenic model of inducible overexpression of Olig2 in oligodendroglial cells
Although earlier studies indicated that Olig2 cooperates with additional factors to regulate oligodendroglial differentiation (Sun et al., 2001; Zhou et al., 2001; Wang et al., 2006), others suggested that Olig2 overexpression alone is sufficient for inducing OPC differentiation (Liu et al., 2007). However in these studies, overexpression of Olig2 was not selectively targeted to cells of oligodendroglial lineage. To obtain better insights on Olig2 function in vivo, we generated a doxycycline inducible bigenic TetOlig2:Sox10rtTA/+ mouse line (Fig. 1A) using a knock-in strain expressing the reverse tetracycline trans-activator rtTA-M2 under the control of the Sox10 promoter (Sox10rtTA/+) (Ludwig et al., 2004). In the TetOlig2 responder line, the expression of Olig2 and Egfp transgenes are controlled by the tetracycline-responsive element (Gossen and Bujard, 1992; Maire et al., 2010). After doxycycline treatment, we confirmed that only double transgenic TetOlig2:Sox10rtTA/+ mice express GFP, as illustrated in P0 spinal cords (Fig. 1B and C). After doxycycline induction, GFP was specifically targeted in Olig2+ and Sox10+ oligodendroglial cells, including both PDGFRA+ OPCs and CC1+ differentiated oligodendrocytes (Fig. 1D). As expected, the vast majority of Sox10+ cells expressed GFP (93.82 ± 0.53%) and nearly all GFP+ cells were Sox10+ (95.15 ± 0.54%) and Olig2+ (97.08 ± 0.36%). Quantification by real-time PCR revealed a 2-fold increase in Olig2 expression after doxycycline treatment in P0 TetOlig2:Sox10rtTA/+ spinal cord compared to Sox10rtTA/+ (P = 0.009, Tukey test) and wild-type littermates (P = 0.012, Fig. 1E). Expression of Sox10 is also decreased by 2-fold in TetOlig2:Sox10rtTA/+ and Sox10rtTA/+ with respect to wild-type (P = 0.0017 and P = 0.002, respectively; Fig. 1F), indicating that Olig2 gain of function did not rescue Sox10 haplodeficiency. Together, these results indicate that this transgenic model provides a powerful tool to decipher Olig2 function in oligodendroglial cells.
Figure 1.
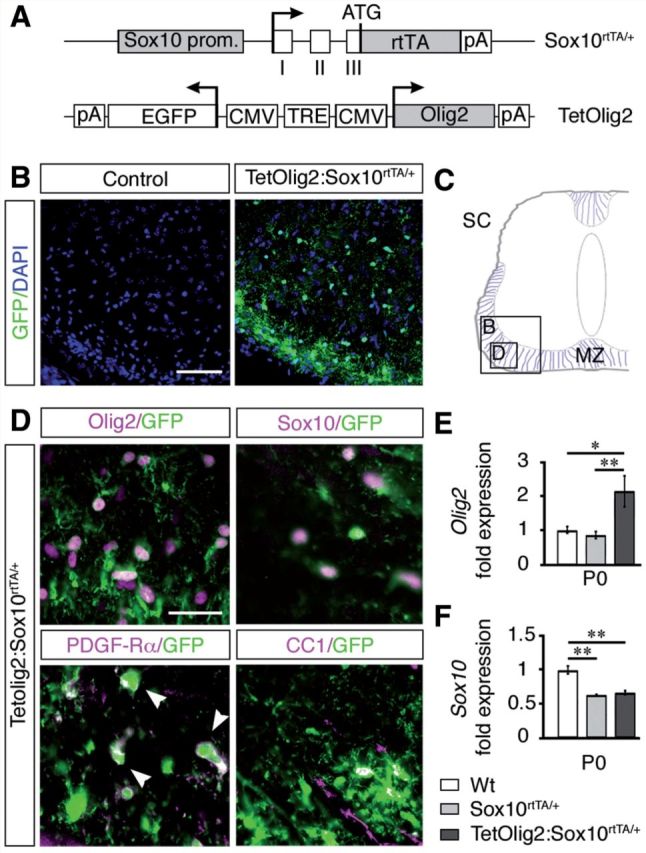
Generation of TetOlig2:Sox10rtTA/+ mouse line. (A) Schematic representation of Sox10rtTA/+ and TetOlig2 alleles that together induce Olig2 overexpression in Sox10+ cells upon doxycycline treatment. Sox10 prom = Sox10 promoter region; open boxes = exons I–III of the Sox10 gene; rtTA = coding sequences for the reverse tetracycline-controlled transactivator; pA = polyadenylation sites; TRE = tetracycline responsive element; CMV = minimal promoter of cytomegalovirus immediate early genes; Olig2 = coding sequence of the Olig2 gene; EGFP = coding sequence of the enhanced green fluorescent protein; ATG = start codon of the Sox10 gene. Arrows indicate transcription starts. (B) GFP immunohistochemistry (green) in control (wild-type or Sox10rtTA/+) and TetOlig2:Sox10rtTA/+ spinal cord at P0. Sections are counterstained with DAPI (blue). Note that GFP was only detected in TetOlig2:Sox10rtTA/+ mice. (C) Schematic representation of the spinal cord (SC) illustrating the marginal zone (MZ) and regions shown in B and D (open boxes). (D) Immunolabellings for GFP (green), Olig2, Sox10, PDGFRA and CC1 (all in red) on spinal cord sections of TetOlig2:Sox10rtTA/+. GFP was detected in Olig2+, Sox10+, PDGFRA+ and CC1+ oligodendroglial cells. (E and F) Quantitative reverse transcriptase PCR for Olig2 and Sox10 (at least n = 3 for each genotype). (E) In P0 TetOlig2:Sox10rtTA/+ mice (dark grey bars), Olig2 expression was significantly increased with respect to wild-type (Wt) (white bars, P = 0.012) and Sox10rtTA/+ littermates (light grey bars, P = 0.009). (F) Sox10 expression level was reduced by half in Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ with respect to wild-type (P = 0.002 and P = 0.0017, respectively). Scale bars: B = 75 µm; D = 20 µm.
Olig2 overexpression increases OPC migration
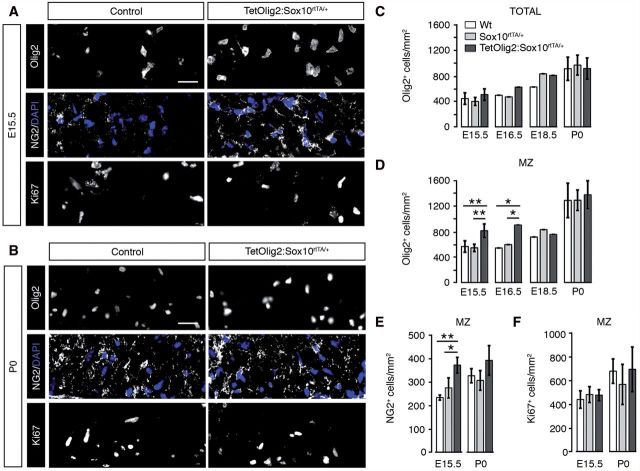
Having validated our Tet-On model, we next compared OPC development in TetOlig2:Sox10rtTA/+, Sox10rtTA/+ and wild-type spinal cords after doxycycline induction from E10.5 to E15.5, E16.5, E18.5 or P0. First, we evaluated the OPC pool using Olig2 and NG2 labelling and found that the overall density of oligodendroglial cells was similar in spinal cords of all three genotypes from E15.5 to P0 (Fig. 2C). However at E15.5 and E16.5, Olig2+ cell density was significantly increased in the marginal zone of the TetOlig2:Sox10rtTA/+ spinal cord compared to Sox10rtTA/+ (PE15.5 = 0.007, PE16.5 = 0.0084) and wild-type (PE15.5 = 0.006, PE16.5 = 0.0084, n ≥ 3; Fig. 2A and D). NG2+ cell density was also increased at E15.5 in the marginal zone of the TetOlig2:Sox10rtTA/+ spinal cord compared to Sox10rtTA/+ (P = 0.031) and wild-type (P = 0.008, n = 3; Fig. 2A and E), confirming that this population represented OPCs. Furthermore, the number of Ki67+ (Fig. 2A, B and F) or terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL)+ cells (Supplementary Fig. 1) was not affected in TetOlig2:Sox10rtTA/+ spinal cord with respect to Sox10rtTA/+ and wild-type littermates, indicating that Olig2 overexpression in OPCs did not alter their proliferation or their survival. Strikingly, increased OPC density in the marginal zone was transient and was no longer detectable from E18.5 to P0 (Fig. 2B, D and E), suggesting that Olig2 gain of function in OPCs promotes their migration during early CNS development.
Figure 2.
Targeted Olig2 overexpression leads to a transient increase of OPCs in the marginal zone. (A and B) Immunohistochemistry for Olig2, NG2 and Ki67 (grey) on spinal cord sections of control (wild-type and Sox10rtTA/+) and TetOlig2:Sox10rtTA/+ embryos at E15.5 (A) and P0 (B). A and B show areas of the marginal zone (MZ). NG2 labelling is shown in combination with DAPI to visualize cells by their nuclei. (C–F) Quantifications of Olig2+ oligodendroglial cells, NG2+ OPCs and Ki67+ cells in the whole spinal cord and in the marginal zone (n ≥ 3 for each genotype). (C) At all time points, the overall density of Olig2+ cells in the spinal cord was not affected in the analysed genotypes. (D) At E15.5 and E16.5, the density of Olig2+ cells was significantly increased in marginal zone of the TetOlig2:Sox10rtTA/+ mice (dark grey bars) with respect to wild-type (white bars, PE15.5 = 0.006, PE16.5 = 0.0084) and Sox10rtTA/+ mice (light grey bars, PE15.5 = 0.007, PE16.5 = 0.0084). (E) The density of NG2+ OPCs was also increased at E15.5 in the marginal zone of TetOlig2:Sox10rtTA/+ compared to wild-type (P = 0.008) and Sox10rtTA/+ (P = 0.031). Differences in Olig2+ or NG2+ OPC density were no longer observed at E18.5 and P0 (D and E). (F) The density of Ki67+ proliferative cells was similar in all genetic strains at E15.5 and P0. Scale bars = 20 µm. Wt = wild-type.
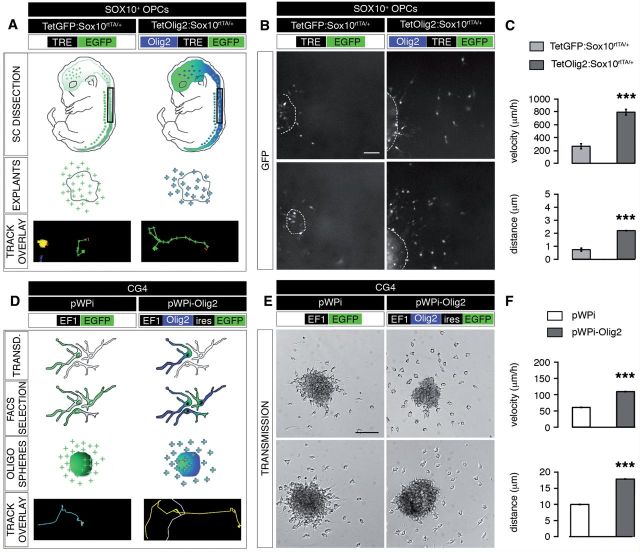
To validate this hypothesis, we performed OPC migration assays using spinal cord explants prepared from E13.5 TetGFP:Sox10rtTA/+ (control) or TetOlig2:Sox10rtTA/+ embryos (Fig. 3A) and analysed the migration of individual GFP+ cells by time-lapse video-microscopy over 24 h (Fig. 3B). Our data demonstrated that the migration velocity of GFP+ OPCs or the covered distance was significantly higher in TetOlig2:Sox10rtTA/+ explants with respect to TetGFP:Sox10rtTA/+ controls (P < 0.001, Fig 3C). To further confirm the effect of Olig2 overexpression on OPC migration, we transduced CG4 cells (Louis et al., 1992) with lentiviral vectors overexpressing Olig2 and GFP (pWPi-Olig2) or GFP alone (pWPi) as a control (Maire et al., 2009) (Fig. 3D). Before analysis, transduced CG4 were FACS-purified based on the expression of GFP (Fig. 3D). Migration of CG4 cells overexpressing Olig2 was then compared to control using time-lapse video-microscopy over 6 h (Fig. 3E). We found that Olig2 overexpression in CG4 cells induced a significant increase of their migration velocity as well as their mean distance of migration (P < 0.001, Fig. 3F). Similar data were obtained in mouse OPCs transduced with pWPi-Olig2 lentivirus (Pvelocity = 0.0067, Pdistance = 0.0249; Supplementary Fig. 2). Taken together, these data demonstrate that Olig2 promotes migration in a cell autonomous manner.
Figure 3.
Olig2 overexpression enhances OPC migration in in vitro assays. (A) Schematic representation of the generation and analysis of E13.5 spinal cord (SC) explants for migration studies. Spinal cord explants from doxycycline-induced TetGFP:Sox10rtTA/+ (control) and TetOlig2:Sox10rtTA/+ embryos were dissected and the migration of GFP+ cells was monitored by time-lapse video-microscopy over 24 h. An example of a tracked cell is shown as ‘track overlay’. (B) Representative views of spinal cord explants (dash lines) illustrating the migration of GFP+ OPCs in control and TetOlig2:Sox10rtTA/+ strains after 24 h. (C) The velocity and the migration distance of GFP+ cells were significantly higher in TetOlig2:Sox10rtTA/+ (dark grey bars) with respect to control (light grey bars, P < 0.001). (D) Schematic representation of the generation of oligospheres from transduced CG4 cells for migration assays. Transduction was with pWPi (control) or pWPi-Olig2 lentivirus. (E) Bright field views of pWPi and pWPi-Olig2-transduced oligospheres, 6 h after plating. (F) The velocity and the distance of CG4 cells was significantly increased under Olig2 overexpression conditions (compare dark grey bars to white control bars, P < 0.001). Scale bars: B = 50 µm; E = 100 µm.
Olig2 overexpression enhances OPC differentiation
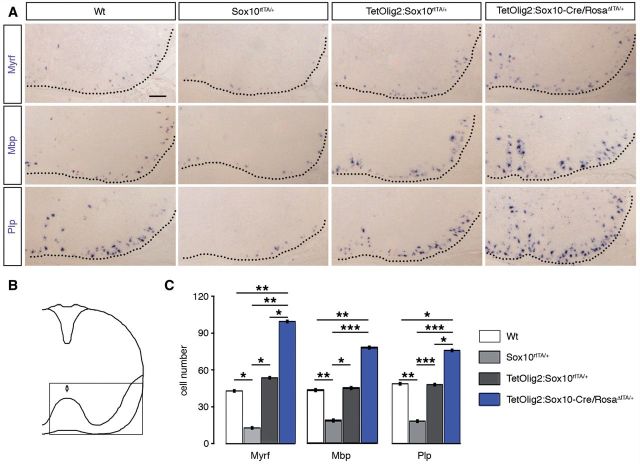
As Olig2 is expressed at all stages of the oligodendroglial lineage, overexpression of this transcription factor might also lead to precocious OPC differentiation. To test this hypothesis, we analysed the expression of Myrf, Mbp and Plp1 by in situ hybridization on wild-type, Sox10rtTA/+, TetOlig2:Sox10rtTA/+ and TetOlig2:Sox10-Cre/RosaΔtTA/+ sections of E18.5 embryos (at least n = 3 for each genotype; Fig. 4). As previously reported, the expression of Myrf, Mbp and Plp1 was significantly decreased in Sox10rtTA/+ deficient embryos with respect to wild-type (PMyrf = 0.031, PMbp = 0.0017, PPlp1 = 0.005; Fig. 4A–C), confirming the slight delay of OPC differentiation in Sox10+/− heterozygous strain (Stolt et al., 2002, 2004). Interestingly, Myrf, Mbp and Plp1 expression were restored to wild-type level in TetOlig2:Sox10rtTA/+ spinal cord, indicating that gain of Olig2 function rescued the delay in OPC differentiation, observed in Sox10rtTA/+ deficient mice (PMyrf = 0.046, PMbp = 0.0202, PPlp1 < 0.001; Fig. 4A–C). As OPC differentiation is slightly delayed in Sox10rtTA/+ deficient mice, one could argue that the effect of Olig2 overexpression might be limited to this genetic background. To exclude this possibility, we generated TetOlig2:Sox10-Cre/Rosa26ΔtTA/+ triple transgenic mice, which had two Sox10 alleles similar to wild-type (Supplementary Fig. 3A–C). Interestingly in this Cre-inducible Tet-Off model, we found that the number of Myrf+, Mbp+ and Plp1+ cells was significantly increased in spinal cords of TetOlig2:Sox10-Cre/Rosa26ΔtTA/+ with respect to both wild-type (PMyrf = 0.0047, PMbp = 0.0076, PPlp1 = 0.0117), Sox10rtTA/+ (PMyrf = 0.002, PMbp < 0.001, PPlp1 < 0.001) and TetOlig2:Sox10rtTA/+ strains. (PMyrf = 0.0382, PPlp1 = 0.0228; Fig. 4A–C). Therefore, our data demonstrated that the pro-differentiation effect of Olig2 gain of function in OPCs is not only restricted to Sox10rtTA/+ heterozygous mice, but also found in mouse strains with physiological expression level of SOX10. It is noteworthy that similar to TetOlig2:Sox10rtTA/+ mice induced since E10.5, TetOlig2:Sox10-Cre/RosaΔtTA/+ transgenic mice are lethal around birth (Supplementary Fig. 3A–C), precluding the use of this transgenic model at postnatal or adult stages.
Figure 4.
Gain of Olig2 function in OPCs leads to their precocious differentiation during embryonic development. (A) In situ hybridization for Myrf, Mbp and Plp1 on E18.5 spinal cord sections from wild-type, Sox10rtTA/+, TetOlig2:Sox10rtTA/+ and TetOlig2:Sox10-Cre/RosaΔtTA/+. Wild-type, Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ were doxycycline treated from E10.5 to E18.5. (B) Schematic representation of the spinal cord indicating the region illustrated in A. (C) The number of Myrf+, Mbp+ and Plp+ cells was significantly decreased in Sox10rtTA/+ mice (light grey bars) with respect to wild-type (white bars, PMyrf = 0.031, PMbp = 0.0017, PPlp1 = 0.005), TetOlig2:Sox10rtTA/+ mice (dark grey bars, PMyrf = 0.046, PMbp = 0.0202, PPlp1 < 0.001) and TetOlig2:Sox10-Cre/RosaΔtTA/+ mice (blue bars, PMyrf = 0.002, PMbp < 0.001, PPlp1 < 0.001). The number of Myrf+, Mbp+ and Plp+ cells was also significantly increased in TetOlig2:Sox10-Cre/RosaΔtTA/+ with respect to wild-type (PMyrf = 0.0047, PMbp = 0.0076, PPlp1 = 0.0117) and TetOlig2:Sox10rtTA/+ for Myrf+ and Plp+ cells (PMyrf = 0.0382, PPlp1 = 0.0228). At least three animals were quantified for each genotype. Scale bar = 100 µm. Wt = wild-type.
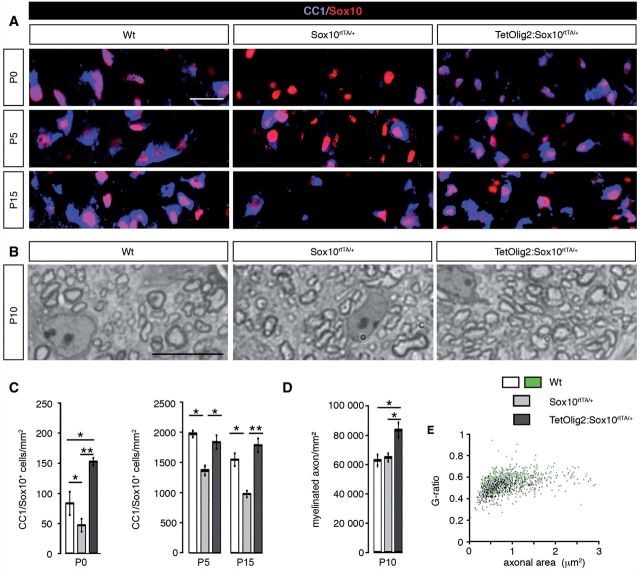
To assess the effect Olig2 gain of function on oligodendrocyte maturation and myelination, we generated TetOlig2:Sox10rtTA/+, Sox10rtTA/+ and wild-type mice induced from E10.5 to P0 or from E18.5 to P5 or P15 (at least n = 3 for each genotype). At these different developmental time points, our data indicated that the density of CC1+/Sox10+ oligodendrocytes was decreased in Sox10rtTA/+ deficient mice with respect to wild-type (PP0 = 0.02, PP5 = 0.03, PP15 = 0.041; Fig. 5A and C). Interestingly, the delay in OPC differentiation was rescued in TetOlig2:Sox10rtTA/+ (PP0 = 0.001, PP5 = 0.036, PP15 = 0.006; Fig. 5A and C). To further validate these data, we performed in vitro differentiation assays using pWi- and pWi-Olig2-transduced CG4 cells. We found that Olig2 overexpression in CG4 cells significantly increased the number of O4+ (P < 0.001; Supplementary Fig. 4A and B) and GalC+ (P = 0.014; Supplementary Fig. 4C and D) oligodendrocytes compared to controls, after 7 days in differentiation medium.
Figure 5.
Gain of Olig2 function in OPCs induces advanced myelination. (A) The marginal zone of wild-type, Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ spinal cord sections was stained with CC1 (blue) and anti-Sox10 (red) antibodies at P0, P5 and P15. (B) Toluidine blue semi-thin sections of wild-type, Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ spinal cord at P10. (C) The number of CC1+/Sox10+ cells was significantly reduced in Sox10rtTA/+ mice (light grey bars) compared to wild-type (white bars, PP0 = 0.02, PP5 = 0.03, PP15 = 0.041) at all postnatal stages analysed. This oligodendroglial differentiation delay was rescued in TetOlig2:Sox10rtTA/+ bigenic mice (dark grey bars, PP0 = 0.001, PP5 = 0.036, PP15 = 0.006) at all stages. Note that the number of CC1+ cells was significantly higher in TetOlig2:Sox10rtTA/+ as in wild-type (Wt) at P0 (P = 0.036). (D) The density of myelinated fibres was significantly increased in TetOlig2:Sox10rtTA/+ compared with Sox10rtTA/+ and wild-type (P = 0.016, P = 0.0056, respectively). (E) G-ratio was comparable to wild-type (green) in all mutant genotypes; n ≥ 3 for each genotype. Scale bars = 20 µm.
Based on these findings, we then compared myelination in TetOlig2:Sox10rtTA/+, Sox10rtTA+/− and wild-type spinal cords, using myelin basic protein (MBP) immunohistochemistry and quantification of myelinated axons by electronic microscopy (Fig. 5B and D and Supplementary Fig. 5). At P0, we found that myelination was precocious in the TetOlig2:Sox10rtTA/+ spinal cord as compared to wild-type and Sox10rtTA/+, which became more obvious at P10 (Supplementary Fig. 5A). Advanced myelination under Olig2 overexpression was also supported by the significant increase of myelinated axons in TetOlig2:Sox10rtTA/+ compared to Sox10rtTA/+ and wild-type at P10 (P = 0.016, P = 0.0056, respectively; Fig. 5D and Supplementary Fig. 5B). Nonetheless, G ratios in ventral spinal cords did not differ significantly between the three genotypes (Fig. 5E), therefore indicating that Olig2 overexpression in OPCs did not alter myelin thickness. Overall, our results demonstrate that gain of Olig2 function in OPCs enhances their differentiation, leading to precocious CNS myelination.
Gain of Olig2 function in adult OPCs accelerates remyelination
In addition to the analysis of the TetOlig2:Sox10rtTA/+ transgenic line during CNS development, we examined whether manipulating expression levels of Olig2 in adult OPCs enhances remyelination of demyelinated CNS lesions. We first verified the functionality of our Tet-On system in the adult brain of TetOlig2:Sox10rtTA/+, Sox10rtTA/+ and wild-type adult mice, treated with doxycycline during 3 weeks. As expected, GFP+ cells were only detected in the adult TetOlig2:Sox10rtTA/+ mouse brain, notably in the corpus callosum, fimbria and striatum (Supplementary Fig. 6A). We also noticed that GFP+ cells were more prominent around subventricular regions, such as the lateral ventricles, whereas only scattered GFP+ cells were detected in white matter tracts like the corpus callosum (Supplementary Fig. 6A). As already described during embryonic and postnatal development, virtually all GFP+ cells expressed Olig2 (100%) and Sox10 (97.16 ± 0.97%; Supplementary Fig. 6B). Furthermore, double immunolabelling for NG2/GFP and CC1/GFP indicated that Olig2 overexpression was selectively targeted in adult OPCs (27.13 ± 4.14%) and mature oligodendrocytes (64.57 ± 2.57%; Supplementary Fig. 6B). GFP labelling was never detected in NeuN+ neurons or GFAP+ astrocytes (data not shown).
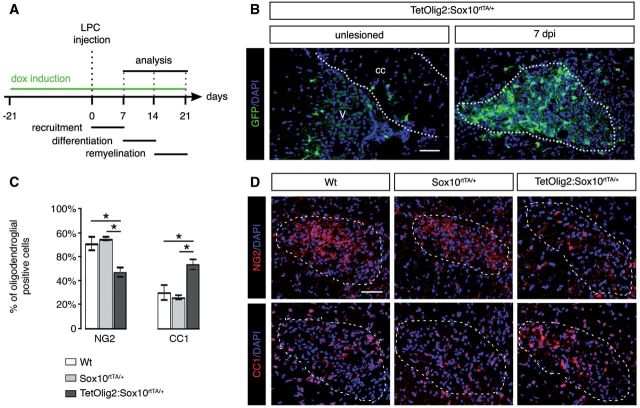
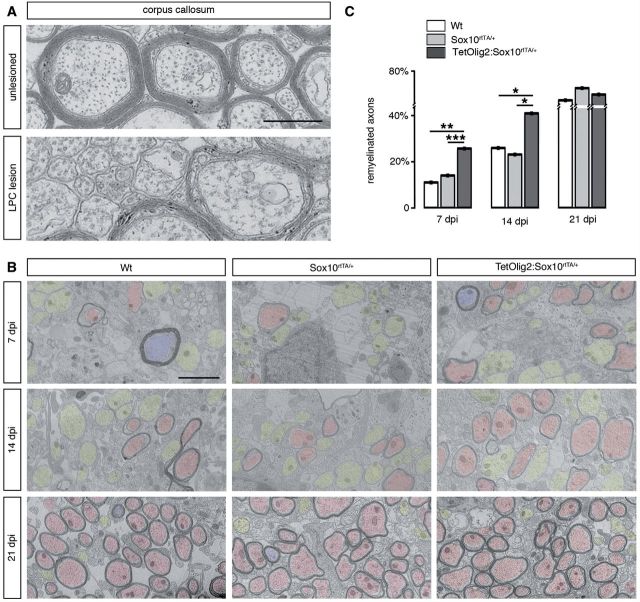
To assess the consequence of Olig2 gain of function in adult OPCs under demyelinating conditions, we performed LPC-induced focal demyelinating lesions in the corpus callosum of adult TetOlig2:Sox10rtTA/+, Sox10rtTA/+ and wild-type mice. Doxycycline treatment was initiated 3 weeks before LPC injections and continued for 7, 14 or 21 days post-injection, corresponding to recruitment, differentiation and remyelination phases of the lesion (Fig. 6A). In doxycycline-treated TetOlig2:Sox10rtTA/+ mice, numerous GFP+ cells were detected in LPC lesions, as illustrated at 7 days post-injection, compared to the non-demyelinated condition, where few GFP+ cells were detected in the corpus callosum (Fig. 6B). To examine the effects of inducible Olig2 on OPC differentiation, we quantified the density of NG2+ OPCs and CC1+ differentiated oligodendrocytes in the lesion at 7 days post-injection (Fig. 6C and D). We found that the percentage of NG2+ OPCs was significantly lower in LPC lesions of TetOlig2:Sox10rtTA/+ mice with respect to Sox10rtTA/+ and wild-type mice (both P < 0.05; Fig 6C). In contrast, the percentage of CC1+ differentiated oligodendrocytes was significantly enhanced in lesions of TetOlig2:Sox10rtTA/+ mice compared to Sox10rtTA/+ and wild-type strains (P < 0.05; Fig. 6C), indicating that Olig2 overexpression promotes OPC differentiation in demyelinated lesions. Although we could not rule out an effect of Olig2 overexpression on OPC migration, the restricted induction of Olig2 gain of function in LPC lesions did not allow us to analyse the consequence of forced Olig2 on OPC migration after demyelination. To determine the effect of Olig2 overexpression on the remyelination rate of LPC lesions, we next quantified the number of remyelinated axons with thin myelin sheaths (Fig. 7A) over the total number of axons on ultra-thin sections through the lesion at 7, 14 and 21 days post-injection (n ≥ 3 for each genotype at each time points). Ultrastructural analysis confirmed the presence of remyelinated and demyelinated axons within the lesions at 7, 14 and 21 days post-injection in all genotypes (Fig. 7B and C). Interestingly, we found that the percentage of remyelinated axons, which increased gradually from 7 to 21 days post-injection in the three genotypes, was significantly higher in LPC lesions of TetOlig2:Sox10rtTA/+ mice at 7 and 14 days post-injection (dpi) with respect to wild-type (P7dpi = 0.0095, P14dpi = 0.0413) and Sox10rtTA+/- mice (P7dpi < 0.001, P14dpi = 0.0343; Fig. 7B and C). Interestingly at 21 days post-injection, the percentage of remyelinated axons within the lesions was nearly similar in the three genotypes (Fig. 7B and C), clearly demonstrating that gain of Olig2 function in adult OPCs accelerates the remyelination rate of demyelinated lesions by inducing OPC differentiation.
Figure 6.
Olig2 overexpression enhances OPC differentiation in LPC lesions. (A) Time schedule of doxycycline induction and histological analysis in the LPC lesion paradigm. Histological analyses were performed after demyelination of the corpus callosum at 7, 14 and 21 days post-injection. (B) Sagittal sections through corpus callosum (cc) and lateral ventricle (v) were labelled for GFP and counterstained with DAPI in unlesioned and lesioned TetOlig2:Sox10rtTA/+ mice at 7 days post-injection (dpi). (C) Quantification of NG2+ OPCs and CC1+ oligodendrocytes at 7 days post-injection in lesions. The density of NG2+ OPCs was significantly reduced in TetOlig2:Sox10rtTA/+ mice (dark grey bars) compared to wild-type (white bars) and Sox10rtTA/+ mice (light grey bars, P < 0.05), while the density of CC1+ oligodendrocytes was increased (P < 0.05, D). (D) LPC lesions of TetOlig2:Sox10rtTA/+ were stained for NG2 and CC1 at 7 days post-injection, and counterstained with DAPI. Dashed lines in panels B and D delineate corpus callosum or lesion areas. Scale bars = 100 µm. dpi = days post-injection.
Figure 7.
Gain of Olig2 function in adult OPCs accelerates remyelination. (A) Electronic micrographs of unlesioned and LPC demyelinated corpus callosum. In unlesioned corpus callosum, axons of small calibres are unmyelinated. In demyelinated lesions, remyelinated axons are characterized by a thin myelin sheath. (B) Quantification of remyelinated axons in lesions of doxycycline treated wild-type (white bars), Sox10rtTA/+ (light grey bars) and TetOlig2:Sox10rtTA/+ (dark grey bars) mice at 7, 14 and 21 days post-injection (dpi). The percentage of remyelinated axons was significantly increased in TetOlig2:Sox10rtTA/+ at 7 and 14 days post-injection, compared to wild-type (P7dpi = 0.0095, P14dpi = 0.0413) or Sox10rtTA/+ mice (P7dpi < 0.001, P14dpi = 0.0343). However at 21 days post-injection, the percentage of remyelinated axons was similar in the three groups. (C) Electron micrographs illustrating remyelination of the lesions in wild-type, Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ mice at 7, 14 and 21 days post-injection. Remyelinated, demyelinated and non-demyelinated axons were colour-coded in red, yellow and blue, respectively. Scale bars: A = 500 nm; C = 2 μm.
OLIG2 expression is upregulated in differentiating OPCs in multiple sclerosis lesions
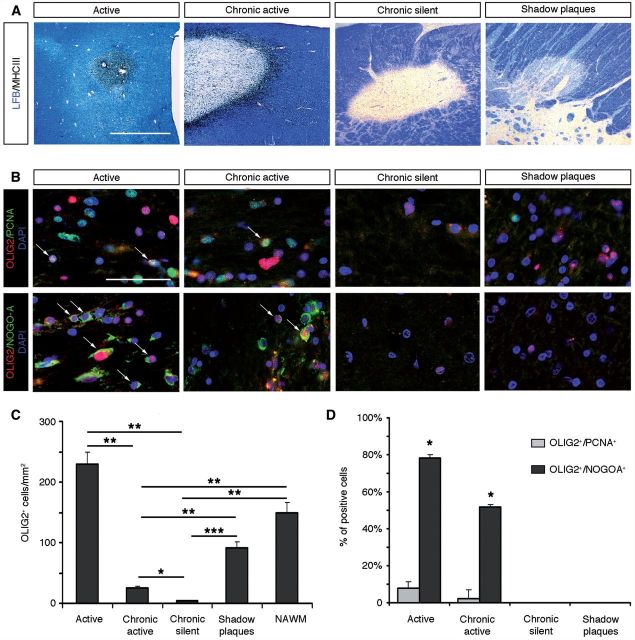
To extend our analysis of OLIG2 function in demyelinating diseases, we analysed its expression pattern in multiple sclerosis lesions. Following experimental demyelination, several transcription factors, which are key determinants of cell differentiation, are upregulated in OPCs. In these models, changes in expression levels of NKX2-2, OLIG1/2 and SOX10 transcription factors are associated with the activation of endogenous OPCs in response to demyelination. However, the existence of such events in multiple sclerosis has so far only rarely been analysed. We analysed OLIG2 expression by immunohistochemical analysis in eight multiple sclerosis brain samples, containing chronic active (n = 10), chronic silent (n = 4) and remyelinated lesions (n = 2) as well as normal appearing white matter (Supplementary Table 2 and Fig. 8A). Immunolabelling for OLIG2 and PCNA or NOGO-A confirmed its specific expression in immature and differentiated oligodendrocytes, respectively (Fig. 8B). Interestingly, our data showed differential expression levels of OLIG2, according to the activity of the lesion. Indeed, the density of OLIG2+ cells was significantly higher in active lesions than in chronic active or chronic lesions (P = 0.0025 and P = 0.0047, respectively; Fig. 8C). These data suggest that increased levels of OLIG2 in active and chronic active multiple sclerosis plaques were mainly associated with the activation of endogenous OPCs. Consistent with this hypothesis, intermediate density of OLIG2+ oligodendrocytes was detected in shadow plaques (Fig. 8C). Importantly, quantification of OLIG2+ cells expressing PCNA (OPCs) or NOGO-A, a marker of maturing oligodendrocytes, indicated that OLIG2 expression occurred predominantly in differentiating oligodendrocytes in active lesion borders of multiple sclerosis (Fig. 8D). Altogether, our data suggest that upregulation of OLIG2 in adult OPCs might play a critical function in oligodendrocyte regeneration and remyelination in multiple sclerosis.
Figure 8.
OLIG2 is mainly expressed in maturing NOGO-A+ oligodendrocytes in multiple sclerosis lesions. (A) Luxol Fast blue/MHC II staining of post-mortem multiple sclerosis brain sections, illustrating typical active, chronic active, chronic silent lesions, and remyelinated shadow plaques. (B) Immunolabellings for OLIG2 (red), PCNA and NOGO-A (both in green) were performed on the different lesion subtypes. PCNA was used to identify OPCs and NOGO-A to label differentiating oligodendrocytes. Nuclei were counterstained with DAPI. (C) Quantification of OLIG2+ cell density in the different subtypes of multiple sclerosis lesions and normal-appearing white matter (NAWM). The density of OLIG2+ cells is significantly increased in active lesions compared to chronic active and chronic lesions (P = 0.0025 and P = 0.0047, respectively). The density in chronic active lesions is also significantly reduced compared to chronic silent lesions, shadow plaques and normal-appearing white matter (P = 0.0029, P = 0.0024 and P = 0.0016, respectively) and in chronic silent lesions versus shadow plaques and normal-appearing white matter (P < 0.001 and P = 0.0019, respectively). (D) Quantifications of OLIG2+ cells expressing PCNA or NOGO-A, according to lesion activity. OLIG2+/NOGO-A+ maturing oligodendrocytes were mainly detected in active lesions with respect to shadow plaques and chronic silent lesions (P ≤ 0.05). Scale bars: A = 1 mm; B = 50 µm.
Discussion
Olig2 plays a critical role in oligodendrocyte specification during development. However, its functional role in oligodendrocyte differentiation during development and in demyelinating diseases, such as multiple sclerosis, remains poorly understood. The main objectives of the present study aimed to test whether forced expression of Olig2 in OPCs affects cell migration and differentiation in a cell-autonomous manner, both during developmental myelination and under pathological conditions in adulthood. Here, we show that specific Olig2 overexpression in oligodendroglial lineage cells in vivo and in vitro promotes (i) OPC migration; and (ii) OPC differentiation into mature oligodendrocytes; and subsequently leads to (iii) precocious CNS myelination. Importantly, we provide compelling evidence indicating that gain of Olig2 function in adult OPCs promotes remyelination. We also demonstrate that OLIG2 expression is preferentially upregulated in NOGO-A+ maturing OPCs in active multiple sclerosis lesions. Altogether, these data provide proof of concept that targeting OLIG2 expression in oligodendrocytes could be a suitable mechanism to promote myelin regeneration in CNS demyelinating disorders.
The TetOlig2 mouse provides a powerful genetic tool to unravel Olig2 function in myelination and myelin repair
So far, the role of Olig2 in oligodendrocyte differentiation has been hampered by the lack of specific loss- and gain of function studies in oligodendroglial cells. Previous Olig2 gain of function in neural precursor cells leads to controversial interpretations of its function in cell differentiation (Sun et al., 2001; Zhou et al., 2001; Wang et al., 2006; Liu et al., 2007). These studies mainly used electroporation in the developing chick neural tube or viral transduction of neuroepithelial cells but did not target specifically oligodendroglial cells. To clarify the effect of Olig2 on OPC differentiation, we generated innovative transgenic mouse models. For this purpose, TetOlig2 mice were crossed with inducer lines expressing rtTA (Ludwig et al., 2004) or Cre-dependent tTA under the control of the Sox10 promoter (Matsuoka et al., 2005; Wang et al., 2008). These models provided selective genetic tools for controlled and inducible expression of Olig2 in oligodendroglial cells, at embryonic and postnatal stages. However, it is noteworthy that TetOlig2:Sox10-Cre/RosaΔtTA/+ transgenic mice were not viable and died around birth due to severe developmental malformations, including abnormal facial skull and open eyes (Supplementary Fig. 3). These defects were likely related to ectopic expression of Olig2 in neural crest derivatives and their resulting altered development.
An unexpected role for Olig2 in OPC migration
Our developmental analysis of TetOlig2:Sox10rtTA/+ mice revealed that overexpression of Olig2 during early embryogenesis increased the migration of OPCs in the developing spinal cord. We showed that this effect was not related to increased cell proliferation or cell survival, as the number of Ki67+ or TUNEL+ apoptotic cells was not affected. These data were also confirmed by time-lapse video-microscopy both in mouse OPCs and CG4 cell cultures, as well as in spinal cord explants derived from controls and TetOlig2:Sox10rtTA/+ transgenic mouse embryos. Similarly, one could speculate that overexpression of Olig2 in adult OPCs following demyelination might also promote their migration towards the lesions. Nonetheless, in TetOlig2:Sox10rtTA/+ mice, the restricted induction of Olig2 gain of function in OPCs within LPC-induced demyelinated lesions did not allow us to assess the impact of Olig2 overexpression on OPC migration under pathological conditions. Olig2 has been previously implicated in the migration of the glioblastoma cell line U12-1 (Tabu et al., 2007), in which it inhibited cell migration. In contrast, our study indicates that Olig2 promotes OPC migration. This discrepancy may be explained by the fact that Olig2 function in cell migration may differ according to the cell type analysed. One could speculate that Olig2 may promote or inhibit cell migration according to its binding partners. Indeed the physiological effects of bHLH transcription factors are critically dependent to the association with their binding partners (Ross et al., 2003). Interestingly, the involvement of Olig2 in cell migration is also supported by its ability to regulate the expression of RhoGTPases and LIM kinases, two major regulators of cytoskeletal reorganization (Tabu et al., 2007). A comparative gene expression profiling analysis between Olig2-overexpressing and control OPCs will shed light on critical effectors that regulate Olig2-dependent migration. Intriguingly, although bHLH transcription factors, including Olig2, have been extensively studied for their properties in cell fate specification in various tissues, recent reports also link these factors to cell migration. For instance, Hand2 enhances the expression of metalloproteinases (Yin et al., 2010) and Twist1 regulates several genes involved in cytoskeleton reorganization (Mikheeva et al., 2010). Furthermore Ngn1 (now known as Neurog1), NGN2 (now known as Neurog2), Mash1 (now known as Ascl1) and Neurod1, which are key determinants in neuronal determination, also modulate cell migration during cortical embryogenesis (Ge et al., 2006). Taken together, these studies suggest that bHLH transcription factors, closely related to Olig2, direct both neural stem cell fate and migration of committed cells.
Olig2 gain of function in OPCs promotes differentiation into myelinating oligodendrocytes
Our Olig2 overexpressing transgenic mouse models allowed us to precisely decipher the function of this factor in OPC differentiation. Our current analyses of TetOlig2:Sox10rtTA/+ and TetOlig2:Sox10-Cre/RosaΔtTA/+ provide clear evidence that forced expression of Olig2 in OPCs enhances the density of differentiated oligodendrocytes in presumptive white matter of spinal cord, at different developmental time points (Richardson et al., 1997). Interestingly, these data were also confirmed by increased expression of Myrf, Mbp and Plp1 transcripts in TetOlig2:Sox10rtTA/+ and TetOlig2:Sox10-Cre/RosaΔtTA/+ spinal cords at E18.5. Furthermore, PCR analysis showed an increase of Plp1/Dm20 ratio in TetOlig2:Sox10rtTA/+ mice compared with Sox10rtTA/+ and wild-type littermates at E15.5 (data not shown). Lastly, lentiviral transduction of Olig2 in OPCs enhanced their differentiation into O4+ and GalC+ oligodendrocytes. Strikingly, we found that Olig2 overexpression in OPCs is able to rescue the differentiation delay, previously reported in Sox10rtTA/+ heterozygous mice (Stolt et al., 2002, 2004). Still, this pro-differentiation effect of gain of Olig2 function was not restricted to Sox10rtTA/+ heterozygous mice but also observed in TetOlig2:Sox10-Cre/RosaΔtTA/+ mice that have physiological levels of Sox10. The differentiating effect of Olig2 overexpression in OPCs is also reflected in the precocious myelination in TetOlig2:Sox10rtTA/+ transgenic spinal cord relative to Sox10rtTA/+ and wild-type animals, as assessed by in situ hybridization for Mbp, Plp1 and Myrf and quantification of myelinated axons. Our data are in agreement with the recent analysis of the specific deletion of Olig2 in NG2+ and CNP+ OPCs, indicating that Olig2 function is required for differentiation and myelination (Zhu et al., 2012; Mei et al., 2013).
To obtain additional insight into the molecular mechanisms underlying the pro-differentiation effect of Olig2, we performed gene expression profiling by real-time PCR, focusing on critical regulators of the myelination process. We checked whether Olig2 overexpression may control Olig1 and Sox10 gene expression, as Olig2 binds to the U2 enhancer element of the Sox10 promoter (Kuspert et al., 2011). Our data did not reveal any significant modification of Olig1 or Sox10 expression level (data not shown), suggesting that other transcriptional pathways are likely involved in the pro-differentiation effect of Olig2. In this regard, Nkx2.2 is an interesting candidate as it interacts genetically with both Olig2 and Sox10 (Liu et al., 2007). Moreover, cooperation of overexpressed Olig2 with other regulatory factors such as Zfp488 (Wang et al., 2006) or Brg1 may also promote the transcription of major myelin genes (Yu et al., 2013). Overall our findings provide compelling evidence that Olig2 overexpression enhances OPC differentiation that subsequently leads to an acceleration of the myelination process.
Olig2, a putative target for oligodendrocyte regeneration in demyelinating diseases?
To determine whether Olig2 gain of function may promote oligodendrocyte regeneration and remyelination under pathological conditions, adult TetOlig2:Sox10rtTA/+ mice and control littermates were subjected to focal demyelination induced by LPC injections. Interestingly, targeted Olig2 overexpression in adult OPCs potentiates the remyelination rate of demyelinated lesions by inducing OPC differentiation. These findings are particularly relevant for multiple sclerosis, in which a differentiation block of adult OPCs might be a major cause of the remyelination failure (Arnett et al., 2004; Kuhlmann et al., 2008). In general, the repair capacity of adult OPCs is still poorly understood (Fancy et al., 2010). We quantified OLIG2-expressing cells in active, chronic active and chronic silent lesions, as well as in remyelinated shadow plaques and normal-appearing white matter of multiple sclerosis brain samples. Interestingly, we detected differential expression levels of OLIG2 according to lesion activity, which are higher in active than in chronic silent lesions and normal appearing white matter. These data suggest that upregulation of OLIG2 in endogenous OPCs within active lesions may be linked to their ability to regenerate remyelinating oligodendrocytes. This hypothesis is also supported by our immunohistological analysis demonstrating that OLIG2 was mainly detected in NOGO-A+ maturing oligodendrocytes. However, the upregulation of OLIG2 expression alone might be insufficient to promote oligodendrocyte regeneration in active multiple sclerosis lesions and may require additional co-factors, such as NKX2-2 or MASH1 (Nakatani et al., 2013).
Our data support the notion that targeting OLIG2 expression may promote oligodendrocyte regeneration for myelin repair in CNS demyelinating diseases. For a translation towards therapeutics, a pharmacological and selective modulation of OLIG2 expression in adult OPCs is required. Although transcription factors are not easily targeted by drugs, it has been recently demonstrated that stapled peptides, a new class of synthetic factors that are cell-permeable and stabilized alpha-helical peptides, modulate critical protein–protein interactions and thus are able to regulate selectively intracellular biological targets (Kim et al., 2011). For instance, selective stapled peptides modulating Notch (Moellering et al., 2009) or Wnt/β-catenin (Cui et al., 2013) signalling pathways have been developed in cancer research. The development of specific stapled peptides modulating OLIG2 biological activity could be of therapeutic value for enhancing remyelination. Still, this strategy requires a better understanding of the structural and functional domains of OLIG2 that participate in interactions with other factors (Sun et al., 2003; Samanta and Kessler, 2004). E-box proteins have been shown to be required for the transcriptional activity of OLIG2, whereas NKX2-2 and ID proteins have been implicated in oligodendrocyte and astrocyte determination. Furthermore, studies indicate that OLIG2 functions are critically regulated by phosphorylation mechanisms (Li et al., 2011; Sun et al., 2011). The use of small agonists of the sonic hedgehog signalling pathway could be also envisioned to modulate OLIG2 expression and to promote CNS remyelination (Ferent et al., 2013). Although targeted OLIG2 gain of function in adult OPCs seems a rational way to promote myelin regeneration in demyelinating diseases, the development of such strategies will undoubtedly require better insights into the molecular mechanisms of this master transcription factor.
Acknowledgements
We are grateful to the UK MS Tissue Bank (Pr Richards Reynolds, Imperial College, London, UK) for providing post-mortem MS brain samples and the Ecole des Neurosciences de Paris for lentiviral productions. We thank Drs Aurelia Sahel and Violetta Zujovic for critical reading of the manuscript, Simone Hillgärtner, Christian Schmitt for their fruitful advice for the analysis of the Tet:Sox10-Cre/RosaΔtTA/+ mice, Dr Karelle Benardais and Baptiste Wilmet for their technical help in mouse demyelinated lesions and the Plateforme d’Imagerie Pitié-Salpêtrière (Dominique Languy) for their assistance in electronic microscopy.
Glossary
Abbreviations
- bHLH
basic helix-loop-helix
- E
embryonic Day
- GFP
green fluorescent protein
- LPC
lysophosphatidylcholine
- NOGO
neurite outgrowth inhibitor (now known as RTN4)
- OPC
oligodendrocyte progenitor cell
- P
postnatal Day
Funding
This study was supported by grants from the French MS Foundation (ARSEP), the National Multiple Sclerosis Society, USA (Award TR 3762-A-1) and has received funding from the program ‘Investissements d’avenir’ ANR-10-IAIHU-06. A.W. was a recipient of fellowships from the French Ministry of Education and Research (MNRT) and La Ligue Française Contre la Sclérose en Plaques (LFSEP).
Supplementary material
Supplementary material is available at Brain online.
References
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306:2111–15. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Avellana-Adalid V, Nait-Oumesmar B, Lachapelle F, Baron-Van Evercooren A. Expansion of rat oligodendrocyte progenitors into proliferative “oligospheres” that retain differentiation potential. J Neurosci Res. 1996;45:558–70. doi: 10.1002/(SICI)1097-4547(19960901)45:5<558::AID-JNR6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Bachelin C, Zujovic V, Buchet D, Mallet J, Baron-Van Evercooren A. Ectopic expression of polysialylated neural cell adhesion molecule in adult macaque Schwann cells promotes their migration and remyelination potential in the central nervous system. Brain. 2010;133:406–20. doi: 10.1093/brain/awp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–73. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- Cui HK, Zhao B, Li Y, Guo Y, Hu H, Liu L, et al. Design of stapled alpha-helical peptides to specifically activate Wnt/beta-catenin signaling. Cell Res. 2013;23:581–4. doi: 10.1038/cr.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Kotter MR, Harrington EP, Huang JK, Zhao C, Rowitch DH, et al. Overcoming remyelination failure in multiple sclerosis and other myelin disorders. Exp Neurol. 2010;225:18–23. doi: 10.1016/j.expneurol.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Ferent J, Zimmer C, Durbec P, Ruat M, Traiffort E. Sonic Hedgehog signaling is a positive oligodendrocyte regulator during demyelination. J Neurosci. 2013;33:1759–72. doi: 10.1523/JNEUROSCI.3334-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm ER, Bosl MR, et al. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J Cell Biol. 2010;189:701–12. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci USA. 2006;103:1319–24. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig J, Frob F, Vogl MR, Hermans-Borgmeyer I, Tamm ER, Wegner M. The transcription factors Sox10 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genet. 2013;9:e1003907. doi: 10.1371/journal.pgen.1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Grossmann TN, Verdine GL. Synthesis of all-hydrocarbon stapled alpha-helical peptides by ring-closing olefin metathesis. Nat Protoc. 2011;6:761–71. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- Kuhlmann T, Miron V, Cui Q, Wegner C, Antel J, Bruck W. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain. 2008;131:1749–58. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- Kuspert M, Hammer A, Bosl MR, Wegner M. Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 2011;39:1280–93. doi: 10.1093/nar/gkq951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, de Faria JP, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69:918–29. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–93. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Louis JC, Magal E, Muir D, Manthorpe M, Varon S. CG-4, a new bipotential glial cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J Neurosci Res. 1992;31:193–204. doi: 10.1002/jnr.490310125. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, et al. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–29. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lucchinetti CF, Bruck W, Rodriguez M, Lassmann H. Distinct patterns of multiple sclerosis pathology indicates heterogeneity on pathogenesis. Brain Pathol. 1996;6:259–74. doi: 10.1111/j.1750-3639.1996.tb00854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Schlierf B, Schardt A, Nave KA, Wegner M. Sox10-rtTA mouse line for tetracycline-inducible expression of transgenes in neural crest cells and oligodendrocytes. Genesis. 2004;40:171–5. doi: 10.1002/gene.20083. [DOI] [PubMed] [Google Scholar]
- Maire CL, Buchet D, Kerninon C, Deboux C, Baron-Van Evercooren A, Nait-Oumesmar B. Directing human neural stem/precursor cells into oligodendrocytes by overexpression of Olig2 transcription factor. J Neurosci Res. 2009;87:3438–46. doi: 10.1002/jnr.22194. [DOI] [PubMed] [Google Scholar]
- Maire CL, Wegener A, Kerninon C, Nait Oumesmar B. Gain-of-function of Olig transcription factors enhances oligodendrogenesis and myelination. Stem Cells. 2010;28:1611–22. doi: 10.1002/stem.480. [DOI] [PubMed] [Google Scholar]
- Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, et al. Neural crest origins of the neck and shoulder. Nature. 2005;436:347–55. doi: 10.1038/nature03837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Wang H, Liu S, Niu J, Wang L, He Y, et al. Stage-specific deletion of Olig2 conveys opposing functions on differentiation and maturation of oligodendrocytes. J Neurosci. 2013;33:8454–62. doi: 10.1523/JNEUROSCI.2453-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheeva SA, Mikheev AM, Petit A, Beyer R, Oxford RG, Khorasani L, et al. TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer. 2010;9:194. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–8. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nait-Oumesmar B, Decker L, Lachapelle F, Avellana-Adalid V, Bachelin C, Baron-Van Evercooren A. Progenitor cells of the adult mouse subventricular zone proliferate, migrate and differentiate into oligodendrocytes after demyelination. Eur J Neurosci. 1999;11:4357–66. doi: 10.1046/j.1460-9568.1999.00873.x. [DOI] [PubMed] [Google Scholar]
- Nait-Oumesmar B, Picard-Riera N, Kerninon C, Decker L, Seilhean D, Hoglinger GU, et al. Activation of the subventricular zone in multiple sclerosis: evidence for early glial progenitors. Proc Natl Acad Sci USA. 2007;104:4694–9. doi: 10.1073/pnas.0606835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani H, Martin E, Hassani H, Clavairoly A, Maire CL, Viadieu A, et al. Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination. J Neurosci. 2013;33:9752–68. doi: 10.1523/JNEUROSCI.0805-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Pringle NP, Yu WP, Hall AC. Origins of spinal cord oligodendrocytes: possible developmental and evolutionary relationships with motor neurons. Dev Neurosci. 1997;19:58–68. doi: 10.1159/000111186. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–19. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–22. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–42. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–58. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–70. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci. 2003;23:9547–56. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Echelard Y, Lu R, Yuk DI, Kaing S, Stiles CD, et al. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol. 2001;11:1413–20. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- Sun Y, Meijer DH, Alberta JA, Mehta S, Kane MF, Tien AC, et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906–17. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabu K, Ohba Y, Suzuki T, Makino Y, Kimura T, Ohnishi A, et al. Oligodendrocyte lineage transcription factor 2 inhibits the motility of a human glial tumor cell line by activating RhoA. Mol Cancer Res. 2007;5:1099–109. doi: 10.1158/1541-7786.MCR-07-0096. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, et al. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–8. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Wang L, Sharma K, Deng HX, Siddique T, Grisotti G, Liu E, et al. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol Dis. 2008;29:400–8. doi: 10.1016/j.nbd.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Dulin J, Wu H, Hurlock E, Lee SE, Jansson K, et al. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–98. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- Wolswijk G. Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J Neurosci. 1998;18:601–9. doi: 10.1523/JNEUROSCI.18-02-00601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Kikuchi K, Hochgreb T, Poss KD, Stainier DY. Hand2 regulates extracellular matrix remodeling essential for gut-looping morphogenesis in zebrafish. Dev Cell. 2010;18:973–84. doi: 10.1016/j.devcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–61. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–43. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zuo H, Maher BJ, Serwanski DR, LoTurco JJ, Lu QR, et al. Olig2-dependent developmental fate switch of NG2 cells. Development. 2012;139:2299–307. doi: 10.1242/dev.078873. [DOI] [PMC free article] [PubMed] [Google Scholar]