Abstract
Cux-1 is a member of a family of homeobox genes structurally related to Drosophila Cut. Mammalian Cut proteins function as transcriptional repressors of genes specifying terminal differentiation in multiple cell lineages. In addition, mammalian Cut proteins serve as cell-cycle-dependent transcriptional factors in proliferating cells, where they function to repress expression of the cyclin kinase inhibitors p21 and p27. Previously we showed that transgenic mice expressing Cux-1 under control of the CMV immediate early gene promoter develop multiorgan hyperplasia. Here we show that mice constitutively expressing Cux-1 exhibit hepatomegaly correlating with an increase in cell proliferation. In addition, the increase in Cux-1 expression in transgenic livers was associated with a decrease in p21, but not p27, expression. Within transgenic livers, Cux-1 was ectopically expressed in a population of small cells, but not in mature hepatocytes, and many of these small cells expressed markers of proliferation. Transgenic livers showed an increase in α-smooth muscle actin, indicating activation of hepatic stellate cells, and an increase in cells expressing chromogranin-A, a marker for hepatocyte precursor cells. Morphological analysis of transgenic livers revealed inflammation, hepatocyte swelling, mixed cell foci, and biliary cell hyperplasia. These results suggest that increased expression of Cux-1 may play a role in the activation of hepatic stem cells, possibly through the repression of the cyclin kinase inhibitor p21.
Keywords: Cux-1, CDP/Cux, p21, p27
INTRODUCTION
Cux-1 is a member of a family of homeobox genes related to the Drosophila cut gene. Mammalian Cut homologues have been identified in human {CCAAT displacement protein (CDP)} [1], mouse (Cux) [2], dog (Clox) [3], and rat (CDP-2) [4]. While these homologues all contain a cut homeodomain and three cut repeats, several truncated Cut proteins have been identified, including testis Cux-1 [5] and CASP [6]. Mammalian cut homologues function as transcriptional repressors of many different genes including γ-globin [7], c-Myc [8], myosin heavy chain [9], NCAM [3], CD8a [10], c-mos [11], gp91-phox [12], p21 [13], and p27 [14]. The binding of Cut proteins to the promoters of these genes appears to be limited to tissues or developmental stages where the target genes are not expressed. Upon terminal differentiation, Cut proteins are down regulated or lose the ability to bind to the promoters, and transcription of the target genes is permitted. Cut proteins function to repress transcription by two different mechanisms: (1) Competition for CCAAT or Sp1 binding site occupancy, preventing activation by the corresponding transcription factors, or (2) active repression via a carboxy terminal repression domain following binding at a distance from the transcription start site [15,16].
Cux-1 is highly and transiently expressed in multiple tissues during embryogenesis [17]. To explore the role of Cux-1 in regulating nephrogenesis, we generated transgenic mice constitutively expressing Cux-1 using the cytomegalovirus immediate early gene promoter. CMV/Cux-1 mice developed hyperplasia in organs in which the transgene was highly expressed [14]. In the kidney, this was associated with down regulation of the cyclin kinase inhibitor p27 [14]. Transient transfection experiments revealed that Cux-1 repressed p27 gene expression [14], supporting its role as a transcriptional regulator of cell cycle progression. Here we report the development of hepatomegaly associated with the chronic expression of Cux-1 in CMV/Cux-1 transgenic mice.
MATERIALS AND METHODS
Generation of Transgenic Mice
The CMV/Cux-1 mice express the full length Cux-1 cDNA under control of the cytomegalovirus (CMV) immediate early gene promoter, and were produced as described earlier using (C57/Bl6 × C3H) F1 mice [14]. Transgene screening was performed by Southern blot analysis of the tail DNA after digestion with appropriate restriction nucleases. Alternatively, transgene screening was performed by PCR analysis using a 5′ primer specific for the CMV promoter and a 3′ primer for the Cux-1 cDNA. Transgenic mice were maintained in accordance with the Institutional Animal Care and Use Committee at the University of Kansas Medical Center.
Anatomical and Histological Analysis
Livers were isolated and weighed from 8-, 10-, and 14-month-old wild type and transgenic mice (three males and three females for each genotype and time point). For histological analysis, livers were fixed in freshly prepared 4% paraformaldehyde in PBS, cryoprotected in 30% sucrose in PBS for 24 h, and frozen in OCT (optimal cutting temperature) compound (Sakura Finetek, Torrance, CA, USA). Alternatively, following fixation, livers were dehydrated with graded ethanols, cleared in xylene, and embedded in paraffin. Slides prepared with 5-µM-thick tissue sections were stained with hematoxylin and eosin. Analysis of liver morphology was performed in a blinded fashion by a board certified veterinary pathologist (D.M.P.). For analysis of fatty change, livers were stained with oil-red-O. Images were captured on a Leica DMR microscope equipped with an Optronics Magnafire digital camera. All images are representative of at least five from each of three wild type or four transgenic livers.
Immunohistochemistry
Endogenous peroxidase was blocked with 3% hydrogen peroxide for 30 min and the samples were then rinsed in PBS. To obtain adequate signal, the slides were treated with antigen unmasking solution (Vector Laboratories, Burlingame, CA, USA) according to manufacturer’s protocol. To reduce background, the sections were blocked for 1 h at RT in 10% normal serum from the species the secondary antibody was made in. Antibody (Ab) dilutions were 1:50 for Cux-1 Ab, 1:100 for Ki-67 Ab, 1:100 for chromogranin-A, 1:100 for α-smooth muscle actin, 1:250 for CD45, 1:250 for CD90.2, and 1:3000 for proliferating cell nuclear antigen (PCNA) in 2% blocking serum in PBS. Slides were incubated at room temperature with 100 µL of antibody in a humid chamber and then washed four times in PBS. Biotin-conjugated secondary antibodies (Vector) were diluted 1:400 in PBS containing 2% serum from the species in which the secondary antibody was generated. After incubation for 1 h at room temperature, slides were washed four times in PBS, then incubated with avidin–biotin–peroxidase complex (ABC-Elite; Vector) and then 3,3′-diaminobenzidine. The tissue sections were then dehydrated with graded ethanols and mounted with Permount (Fisher Scientific, Pittsburgh, PA, USA). Images were captured on a Leica DMR microscope equipped with an Optronics Magnafire digital camera. All images are representative of at least five from each of four wild type or four transgenic kidneys.
Western Blot Analysis
Nuclear extracts from 8 month-old wild type and CMV/Cux-1 transgenic livers (50 µg), prepared as previously described [5], were solubilized in SDS–PAGE sample buffer, electrophoresed on 4–15% gradient polyacrylamide gels, and transferred to nitrocellulose filters, as described previously [14]. The immunoblot was blocked in 5% nonfat dry milk in PBST (PBS containing 0.1% tween 20) for 1 hr at room temperature, before incubation with primary antibodies to Cux-1 (1:100), prepared in blocking solution. After overnight incubation at 4°C, filters were washed three times at room temperature with PBST, and incubated with secondary antibodies (1:10,000 dilution in blocking solution) for 1 h at room temperature. Following three additional washes with PBST, bound antibody was detected by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Pierce, Rockford, IL, USA) according to manufacturer’s instructions, followed by exposure to X-ray film for 1 min. The immunoblot was then incubated sequentially with antibodies to p27 (1:100), p21 (1:100), and β-actin (1:100), to insure equal protein loading. Between Western blotting for each antibody, the immunoblot was stripped by incubation in 100 mM β-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl (pH 6.7) for 1 h at 56°C. Western blotting for each antibody was repeated three times.
Cell Proliferation and White Blood Cell Analysis
OCT embedded sections were analyzed by immunohistochemistry using the rat anti-Ki67 antibody (dilution 1:100) to estimate the degree of cell proliferation. The proliferative index was estimated by counting the number of Ki67-positive cells in five high power fields from four different animals at each time point. For white blood cell analysis paraffin embedded sections were analyzed by immunohistochemistry using a cocktail of the rat anti-CD45 and rat anti-CD90.2 antibodies (dilution 1:250) to estimate the degree of inflammation. The number of white blood cells was estimated by counting the number of CD45/CD90.2 positive cells in five high power fields from four different wild type or transgenic animals.
Antibodies
Commercial reagents used were: rabbit anti-CDP (Cux-1) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA #sc-13024); rat anti-Ki-67 (Dako Cytomation California Inc., Carpinteria, CA, USA #M 7249); mouse anti-PCNA (Sigma-Aldrich, St. Louis, MO, USA #P 8825); rabbit anti-Chromogranin-A (Santa Cruz # sc-13090); rabbit anti-p27 (Santa Cruz # sc-528); mouse anti-p21 (Santa Cruz # sc-6246); mouse anti-β-actin (Sigma # A-5441); rat anti-CD45 (BD Biosciences, Palo Alto, CA, USA # 550539); rat anti-CD90.2 (BD Biosciences # 550543); mouse anti-α-smooth muscle actin (Sigma # A2547); biotin conjugated Goat anti-rabbit IgG (Vector); biotin conjugated rabbit anti-mouse IgG (Vector); biotin conjugated Goat anti-mouse IgG (Vector); and peroxidase conjugated anti-rabbit IgG (Sigma).
Serum Chemistry
Mice were anesthetized with carbon dioxide and then killed by cervical dislocation. Blood (200–500 µL) was collected prior to sacrifice. Serum alanine aminotransferase (ALT) levels were determined using an autoanalyzer (Physicians Reference Laboratory, LLC, Overland Park, KS).
RESULTS
In our previous studies we generated transgenic mice constitutively expressing the homeobox gene Cux-1 using the CMV immediate early gene promoter. Three independent transgenic mouse lines were established, and Southern blotting of mouse genomic DNA showed each lineage had a distinct integration pattern [14]. Two of the three lines, 2189 and 2200, ectopically expressed the transgene at high levels in a number of organs resulting in hyperplasia. While mice from these two lines were indistinguishable from each other, mice from the third line, 2235, did not express the transgene. In the kidney, ectopic expression of Cux-1 was associated with the down regulation of the cyclin kinase inhibitor p27, and transient transfection experiments revealed that Cux-1 represses p27 gene expression. In this study we describe the impact of chronic expression of Cux-1 in the liver.
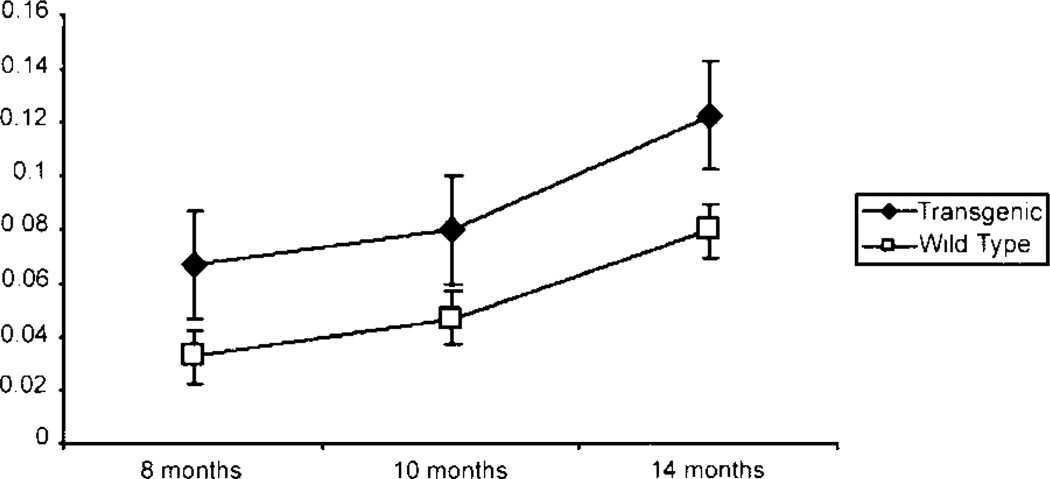
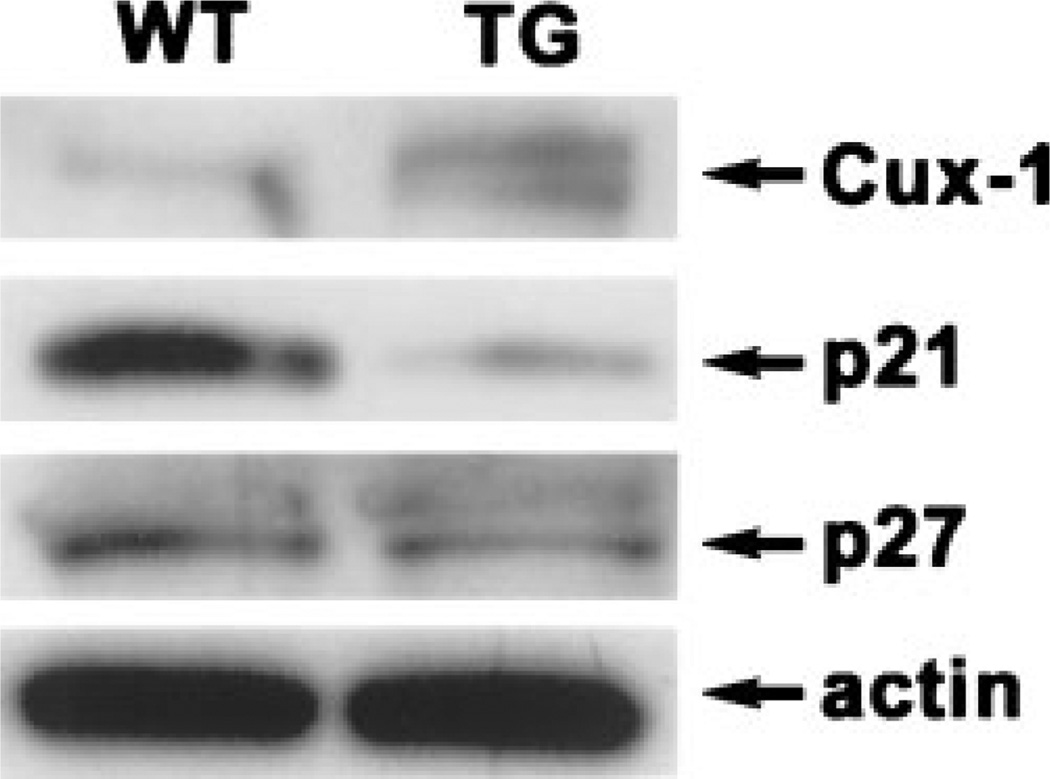
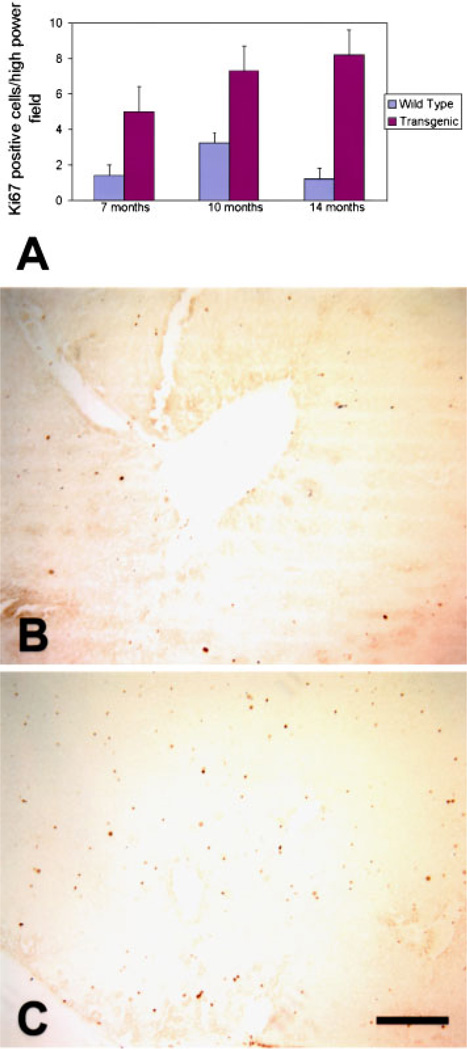
Previously, we showed an increase in liver weight in the transgenic livers, with a two-fold increase at 6 wk of age, an 80% increase at 3 months of age, and a 50% increase at 6 months of age, compared to wild type livers. At these ages, the body weight was not different between transgenic and wild type mice [14]. To determine the effects of chronic Cux-1 expression on liver growth, we compared the liver to body weight ratio of transgenic mice to wild type mice at three different ages ranging from 8 to 14 months. As shown in Figure 1, the liver/body weight ratio was greater in transgenic than wild type mice at all the ages examined. Similar to our previous results, we saw no difference in body weight between wild type and transgenic mice at each of the ages examined. However, the transgenic liver weight was increased at each of the ages examined compared to wild type. Previously we showed that Cux-1 expression was significantly elevated in transgenic livers compared to wild type at 6 wk of age [14]. Figure 2 shows that Cux-1 expression remained elevated in transgenic livers at 8 months of age, compared to wild type. In Cux-1 transgenic kidneys, we showed that ectopic expression correlated with a decrease in p27 gene expression, suggesting a mechanism in which Cux-1 regulates cell proliferation [14]. To determine whether ectopic Cux-1 expression in the liver altered the expression of p21 and/or p27 expression, we evaluated their expression by Western blot analysis. Figure 2 shows that p27 expression was not changed in the transgenic livers. In contrast, p21 expression was reduced in the transgenic liver compared to wild type. To determine whether constitutive expression of Cux-1 in the liver resulted in increased cell proliferation, as described for transgenic kidneys, we counted the number of Ki67 positive cells in sections from 7-, 10-, and 14-month wild type and transgenic livers. At all three ages there were more proliferating cells in transgenic livers than in wild type livers (Figure 3).
Figure 1.
Increased liver/body weight ratios in transgenic mice. Three male and three female CMV/Cux-1 mice and wild-type littermates were weighed at 8, 10, and 14 months. The livers from these mice were isolated and weighed. The data are plotted as the average liver/body weight ratio for transgenic (filled diamonds) and wild type (open boxes) mice. Standard deviation is indicated.
Figure 2.
Expression of Cux-1, p21, and p27 protein in livers from wild type and transgenic mice. Fifty micrograms of nuclear extract prepared from 8-month-old transgenic and wild type livers was subjected to SDS–PAGE and transferred to nitrocellulose membranes. The presence of Cux-1, p21, p27, and β-actin (as a loading control) was detected by Western blot analysis as described in the ‘‘Materials and Methods.’’ Cux-1 protein was elevated in the transgenic livers compared to wild type. p21 expression was decreased in the transgenic livers, compared to wild type, while p27 expression was unchanged.
Figure 3.
Increased proliferation in transgenic livers. The proportion of proliferating cells is increased in livers from CMV/Cux-1 mice (A). The data are plotted as the number of Ki67 positive cells per high power field. Ki67 positive cells were counted in five high power fields from four different animals at each time point. Cell counts in transgenic animals were obtained from extrafocal regions. Sections are shown from 10-month-old wild type liver (B) and extra-focal region of transgenic liver (C). Standard deviation is indicated by error bars. Original magnification: 100× (Bar, 5 µm). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
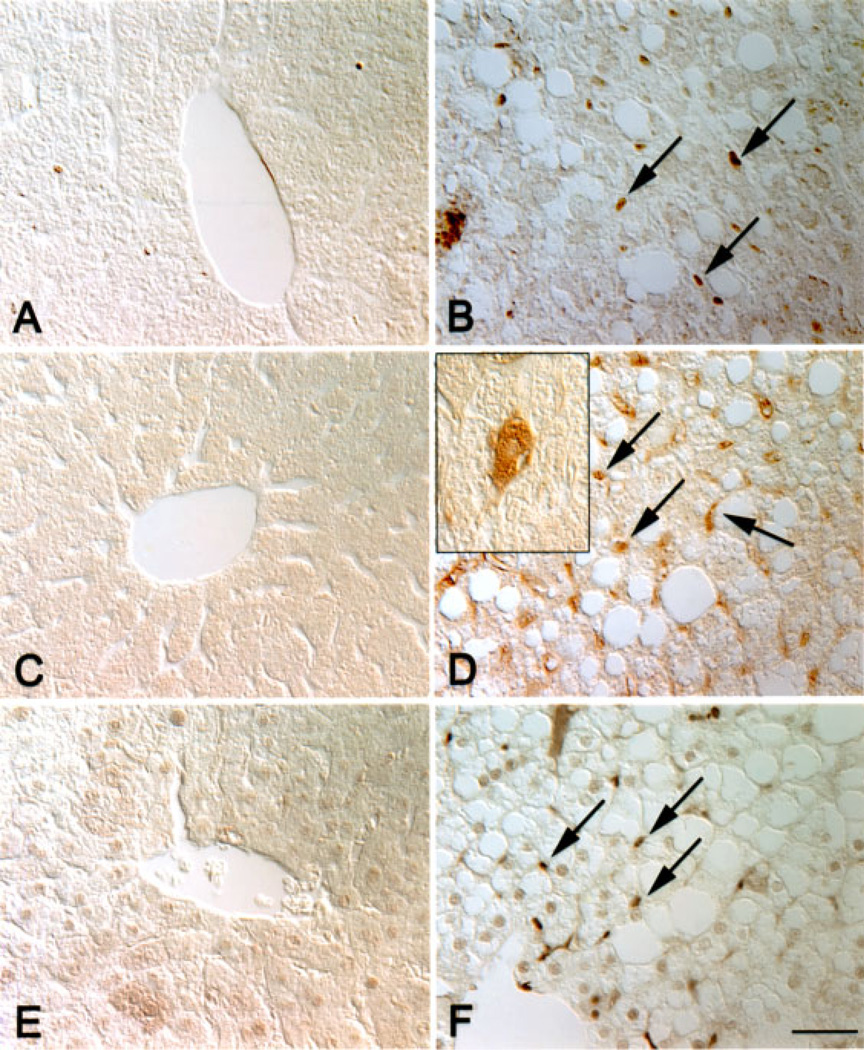
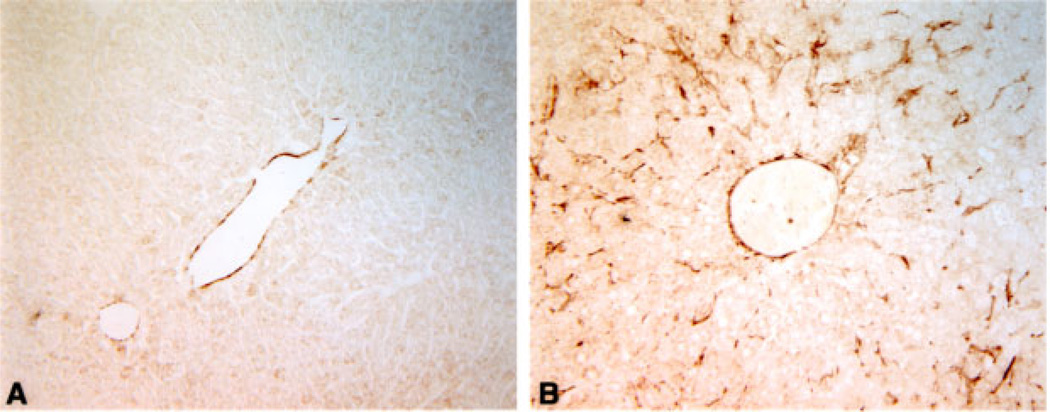
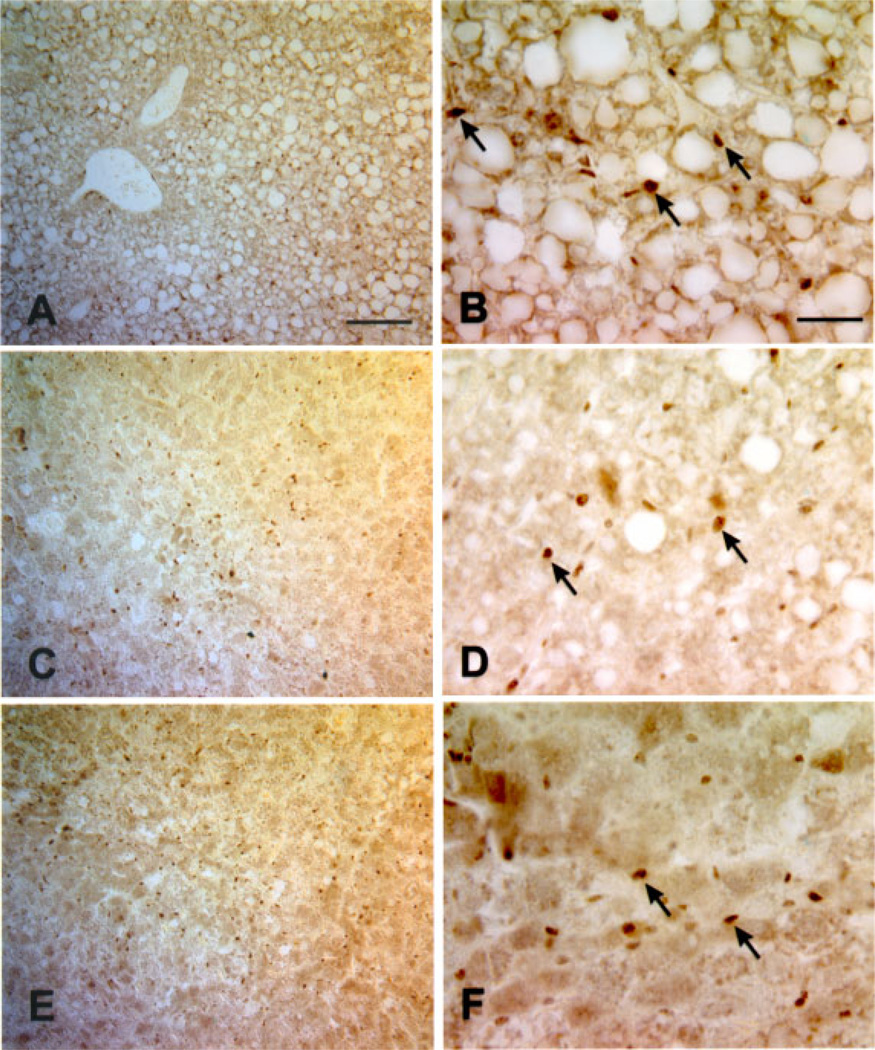
To determine which cells were proliferating, and whether ectopic Cux-1 expression correlated with proliferation, we performed immunohistochemical studies on Cux-1 transgenic and wild type liver sections from mice 8 months of age. Cux-1 protein was not detected in the hepatocytes of either wild type or transgenic livers. However, high levels of Cux-1 protein were observed in a population of small cells in transgenic liver sections (Figure 4B). In contrast, there were relatively few positively staining cells in the wild type liver (Figure 4A). In addition, there was an increase in the number of cells positively stained with antibodies directed against chromogranin-A, a marker for hepatic precursor cells (Figure 4C and D). The chromogranin-A positive cells included both small cells and cells that morphologically resembled hepatocytes (inset in Figure 4D). Moreover, staining with PCNA revealed that most of the proliferating cells in the transgenic liver corresponded to the small cells (Figure 4E and F). To further characterize the transgenic livers, we labeled sections of wild type and transgenic livers from 8-month-old mice with antibodies directed against α-smooth muscle actin. Figure 5A shows that α-smooth muscle actin was found only in the blood vessel wall of the wild type liver. In contrast, there was abundant α-smooth muscle actin staining in cells throughout the parenchyma of the transgenic liver (Figure 5B).
Figure 4.
Increased proliferation of hepatocyte precursor cells in transgenic livers. Wild type (A, C, E) and transgenic (B, D, F) liver sections from 8-month-old mice were labeled with antibodies directed against Cux-1 (A, B), chromogranin-A (C, D), or PCNA (E, F). Cux-1 was ectopically expressed in small cells (arrows in B) between the hepatocytes in transgenic livers, but was not detected in wild type livers (A). Transgenic livers showed an increase in the number of cells labeled positively for chromogranin-A (arrows in D), a marker for hepatocyte precursor cells. Inset in (D) shows that chromogranin-A was detected in cells morphologically resembling hepatocytes, indicating differentiation to the hepatocyte lineage. No chromogranin-A positive cells were observed in the wild type liver (C). The small, fibroblastic shaped cells, but not the hepatocytes, labeled positively for PCNA (arrows in F), indicating that they are proliferating. No PCNA positive cells were detected in the wild type liver (E). Original magnification: 400× (Bar, 50 µm). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 5.
Increased expression of α-smooth muscle actin in transgenic livers. Light micrographs of wild type (A) and transgenic livers (B) from 8-month-old mice labeled with antibodies to α-smooth muscle actin. α-smooth muscle actin was found only in the blood vessel wall of liver isolated from an 8-month-old wild type mouse (A). α-smooth muscle actin staining is observed in cells throughout the parenchyma of liver isolated from an 8-month-old transgenic mouse (B). Original magnification: 200×. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
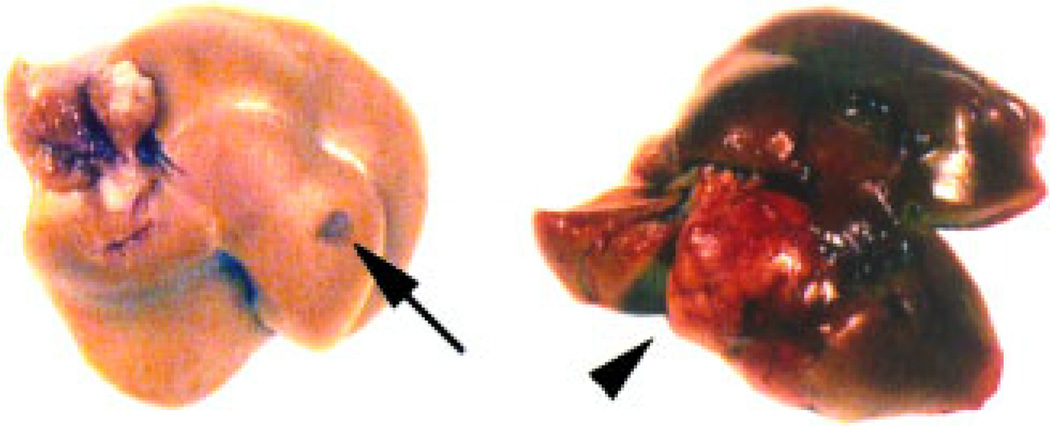
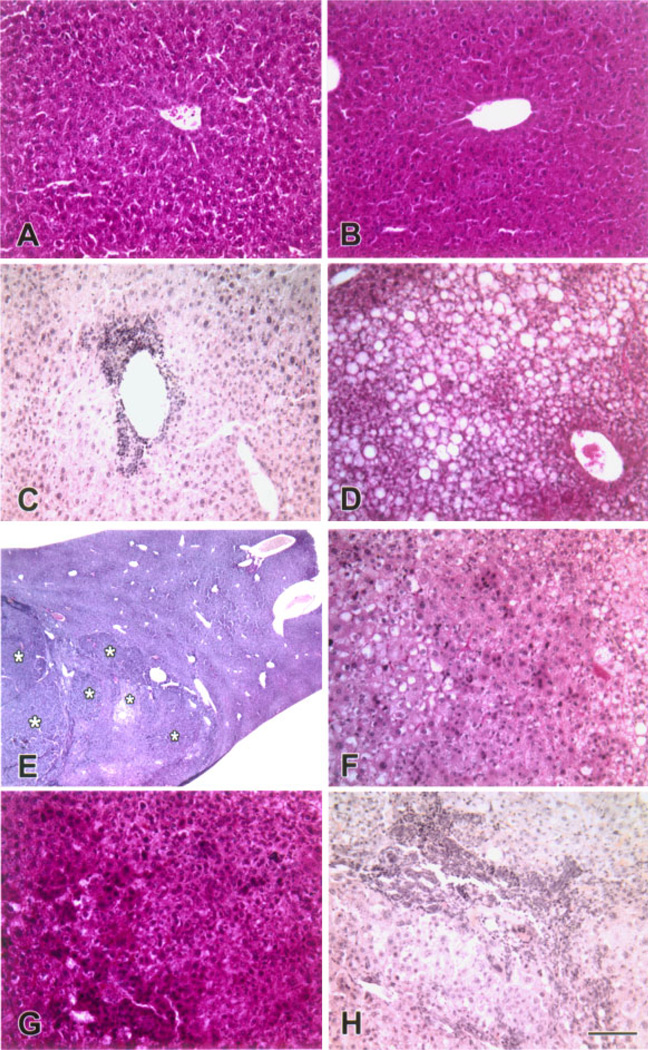
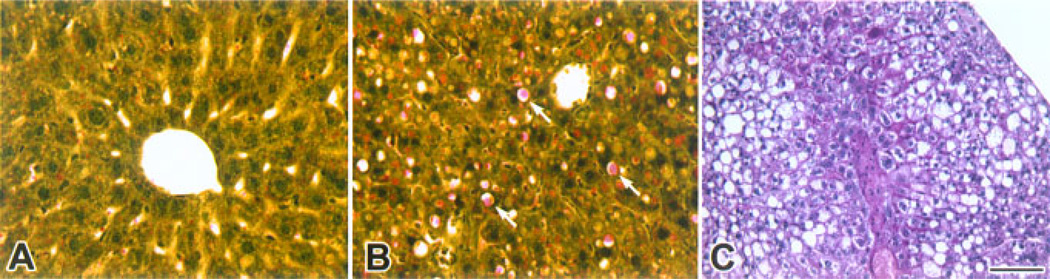
Beginning at 8 months of age, we observed alterations in the appearance of the livers isolated from transgenic mice that ranged from small discolorations to tumors (Figure 6 and summarized in Table 1). To evaluate the morphological changes in transgenic livers, we performed histological analysis of the region containing the nodule on livers isolated from wild type and CMV/Cux-1 mice (results summarized in Table 2). No changes in morphology were observed at 6 months of age (Figure 7A and B). At 8 months of age, most of the transgenic livers showed inflammation (Figure 7C, Table 1), while the wild type livers appeared normal (Table 2). We also observed fatty cell change by 8 months of age. To confirm this, transgenic livers were stained with Oil red-O (Figure 8B). We also observed an increase in PAS positive material in the transgenic livers (Figure 8C). By 10 months of age, mixed cell foci were evident in both male and female transgenic mice (Figure 7F, Table 2), although there was evidence of hepatocellular carcinoma in one out of four wild type male mice. By 14 months of age, inflammation and mixed cell foci were evident in six of six transgenic mice examined, with the appearance of bile duct hyperplasia (Figure 7H, Table 2) and hepatocellular carcinoma in both male and female mice. Again, we observed hepatocellular carcinoma in one out of six wild type male mice (Table 2).
Figure 6.
Gross appearance of liver lesions in CMV/Cux-1 transgenic mice. Beginning at 8 months of age we observed gross alterations in the appearance of transgenic livers, ranging from small discolorations (arrow) to large tumors (arrowhead). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Table 1.
Animals Exhibiting Liver Nodules
| 8 months | 10 months | 14 months | |
|---|---|---|---|
| CMV/Cux-1 | |||
| Female | 6 (17) | 5 (17) | 3 (12) |
| Male | 4 (10) | 4 (18) | 3 (12) |
| Wild type | |||
| Female | 0 (5) | 1 (6) | 0 (3) |
| Male | 0 (3) | 1 (4) | 0 (3) |
Summarized numbers of animals from CMV/Cux-1 transgenic mice exhibiting gross discolorations and/or nodules on the livers. The total number of animals is listed in parentheses.
Table 2.
Types of Cell Foci in CMV/Cux-1 Transgenic and Non-transgenic Livers
| Type | 8 months | 10 months | 14 months | |
|---|---|---|---|---|
| CMV/Cux-1 transgenic | ||||
| Female | ||||
| Inflammation (non-suppurative) | 4 (6) | 3 (5) | 3 (3) | |
| Mixed cell foci | 0 (6) | 3 (5) | 2 (3) | |
| Oval cell hyperplasia | 0 (6) | 0 (5) | 1 (3) | |
| Hepatocellular carcinoma | 0 (6) | 0 (5) | 1 (3) | |
| Male | ||||
| Inflammation (non-suppurative) | 3 (4) | 4 (4) | 3 (3) | |
| Mixed cell foci | 1 (4) | 4 (4) | 3 (3) | |
| Oval cell hyperplasia | 0 (4) | 0 (4) | 1 (3) | |
| Hepatocellular carcinoma | 0 (4) | 1 (4) | 2 (3) | |
| Non-transgenic | ||||
| Female | ||||
| Inflammation (non-suppurative) | 0 (5) | 1 (6) | 0 (3) | |
| Mixed cell foci | 0 (5) | 1 (6) | 0 (3) | |
| Oval cell hyperplasia | 0 (5) | 1 (6) | 0 (3) | |
| Hepatocellular carcinoma | 0 (5) | 1 (6) | 0 (3) | |
| Male | ||||
| Inflammation (non-suppurative) | 0 (3) | 1 (4) | 0 (3) | |
| Mixed cell foci | 0 (3) | 1 (4) | 0 (3) | |
| Oval cell hyperplasia | 0 (3) | 0 (4) | 0 (3) | |
| Hepatocellular carcinoma | 0 (3) | 1 (4) | 0 (3) | |
Summarized numbers of animals exhibiting preneoplastic lesions and hepatocellular carcinoma in H&E sections of the liver in CMV/Cux-1 transgenic and non-transgenic mice 8, 10, and 14 months after birth. The total number of animals is listed in parentheses.
Figure 7.
Progression of liver lesions in CMV/Cux-1 transgenic mice. Light micrographs of wild type (A) and transgenic livers (B-H). At 6 months of age, transgenic livers (B) are indistinguishable from wild type livers (A). At 8 months of age, non-suppurative inflammation (C) and fatty cell change (D) are observed in transgenic livers. At 10 months of age, multifocal lesions are evident in transgenic livers (asterisks in E). Higher magnification reveals that these are of the mixed cell type (F). At 14 months of age, hepatocellular carcinoma is present in transgenic livers (G). Biliary hyperplasia is also observed (H). Original magnification: (A–D, F–H) 200× (Bar, 100 µm). (E) 13×. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 8.
Fatty cell change in transgenic livers. Wild type (A) and transgenic (B, C) livers were stained with Oil-red-O (A, B) or periodic acid schiff (C) stains. Oil-red-O stain reveals fatty change in transgenic livers (arrows in B) compared to wild type, and an increase in PAS positive material in the transgenic livers. Original magnification: 200× (Bar, 100 µm). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
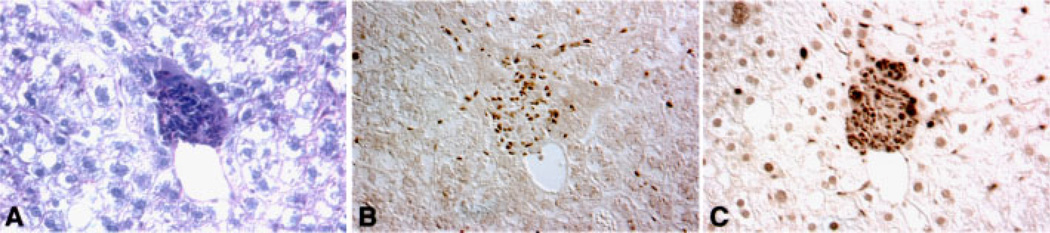
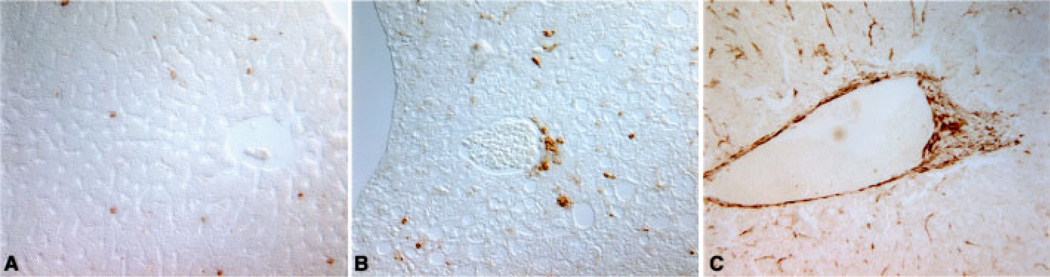
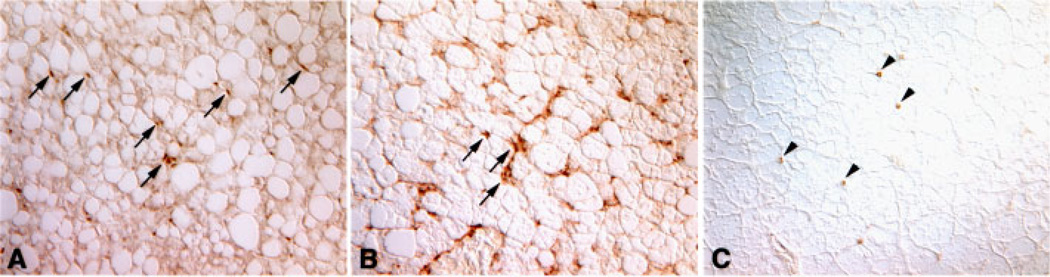
To evaluate expression of Cux-1 protein in the various lesions we performed immunohistochemical studies. Expression of Cux-1 was observed within areas of fatty cell change (Figure 9A and B), within mixed cell foci (Figure 9C and D), and in areas of hepatocellular carcinoma (Figure 9E and F). In all the lesions examined, Cux-1 was expressed in the small cells, but not in hepatocytes. We also observed ectopic Cux-1 expression in small cells in the perivascular area. Adjacent sections of the same liver showed that these cells were highly proliferative and appeared to be morphologically distinct from inflammatory cells (Figure 10). To determine whether these cells could be inflammatory cells and whether the phenotypic changes observed in the transgenic livers were in response to inflammation, we labeled wild type and transgenic liver sections from 8-month-old mice with a cocktail of antibodies directed against leukocytes (CD45 and CD90.2). White blood cells were present both in wild type and transgenic livers, and some inflammatory cells were present in the perivascular region (Figure 11A and B). However, when the number of CD45/CD90.2 positive cells was counted within high power fields, no significant difference between wild type and transgenic was observed (18±2/hpf for TG, 15±2/hpf for WT). Rather, most of the cells found in the perivascular region of the transgenic livers stained positively for α-smooth muscle actin (Figure 11C). To determine whether the tumor formation we observed in the transgenic livers could be in response to inflammation, we compared Cux-1, CD45/CD90.2, and α-smooth muscle actin expression in a section of liver tumor from a 14-month-old transgenic mouse. Figure 12 shows that ectopic expression of Cux-1 was more closely associated with an increase in α-smooth muscle actin positive cells than inflammatory cells within tumors.
Figure 9.
Ectopic expression of Cux-1 increases in lesion progression. Localization of Cux-1 protein in 8-, 10-, and 14-month-old transgenic livers showing fatty cell change (A, B), mixed cell foci (C, D), and hepatocellular carcinoma (E, F). Small Cux-1 positive cells were observed throughout the 8-month-old transgenic livers in areas of fatty cell change (A, and arrows in B). In some areas of 10-month-old transgenic liver, corresponding to mixed cell foci, high concentrations of Cux-1 positive cells were observed (C). Higher magnification revealed that the Cux-1 positive cells corresponded to only small cells (arrows in D), while hepatocytes were not labeled for Cux-1. In a section of 14-month-old transgenic liver corresponding to hepatocellular carcinoma, Cux-1 positive cells were observed (E), and higher magnification revealed that these cells were small cells (arrows in F), but not hepatocytes. Original magnification: (A, C, E) 100× (Bar, 100 µm). (B, D, F) 400× (Bar, 50 µm). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 10.
Co-expression of Cux-1 and PCNA in perivascular cells. Adjacent sections of the same liver isolated from CMV/Cux-1 transgenic were stained with PAS (A), labeled with antibodies directed against Cux-1 (B), and labeled with antibodies directed against PCNA (C), showing that cells ectopically expressing Cux-1 were highly proliferative. Original magnification: 200×. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 11.
Survey of white blood cells in wild type and transgenic liver. Sections of wild type (A) and transgenic (B) livers isolated from 8-month-old mice were labeled with antibodies directed against CD45 (leukocyte common antigen) and CD90.2 to detect all white blood cells. Positive staining cells were detected throughout both livers and cell counting revealed no significant differences in white blood cell number between wild type and transgenic. Labeling with antibodies directed against α-smooth muscle actin (C) showed that the perivascular cells were myofibroblasts. Original magnification: 200×. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 12.
Expression of Cux-1 in liver tumor of CMV/Cux-1 mouse corresponds to α-smooth muscle actin expression. Adjacent sections from the same tumor were labeled with antibodies directed against Cux-1 (A), α-smooth muscle actin (B), and CD45/CD90.2. Cux-1 positive cells show a similar pattern as α-smooth muscle actin positive cells (Arrows in A and B). In contrast, there are relatively few CD45/CD90.2 positive cells in the tumor. Original magnification: 200×. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To evaluate liver function, ALT levels were measured in mice between the ages of 3 and 14 months. In transgenic mice, ALT levels were 86.5±79.7 IU/L (n=86) and in non-transgenic mice ALT levels were 82.4±43.8 IU/L (n=11) (statistically not significant).
DISCUSSION
Mammalian Cut protein expression or activity seems to be limited to proliferating cells in many different tissues [16]. Cux-1 is highly expressed in multiple tissues in the developing embryo, but is down regulated at later stages of development, coinciding with exit from the cell cycle and terminal differentiation [17]. Cell proliferation is positively regulated by cyclins and cyclin-dependent kinases, and negatively regulated by cyclin kinase inhibitors. Cyclins and CDKs are located in the nucleus and together form an active complex [18]. These complexes phosphorylate the protein retinoblastoma (RB), resulting in the release of the transcription factor E2F, which activates genes involved in DNA synthesis [19]. Specific cyclin–CDK complexes are expressed in each phase of the cell cycle. Cyclin kinase inhibitors (CKI) are nuclear proteins that bind to cyclin–CDK complexes preventing phosphorylation of RB and other substrates. This prevents E2F release and subsequent gene activation dependent on E2F, resulting in cell cycle arrest [20,21].
A growing number of studies have demonstrated a role for mammalian cut proteins in cell cycle regulation. The DNA binding activity of Cux-1 was shown to fluctuate during the cell cycle, with highest activity during late G1 and S phase [13,22]. Several of the targets of Cux-1 repression are cell cycle regulated, including the cyclin kinase inhibitors p21 and p27 [13,14]. CDP/cut (Cux-1) was found to be the DNA binding component of the cell cycle regulated histone gene transcription factor HiNF-D [22]. Moreover, this DNA binding activity is developmentally regulated in the liver, with highest activity in the embryonic liver and no activity detected in adult liver [23]. Other growth related targets of Cux-1 include c-myc [8], c-mos [11], and thymidine kinase [24].
Previously, we showed that two independent lines of transgenic mice constitutively expressing the homeobox gene Cux-1 under the direction of the CMV promoter develop multiorgan hyperplasia, including a two-fold increased in liver size, beginning at 6 wk of age [14]. These results provide in vivo evidence for the growing role of mammalian Cut proteins as transcriptional regulators of the cell cycle. In the present study, we describe the effects of chronic expression of Cux-1 in the livers of CMV/Cux-1 mice.
In the CMV/Cux-1 transgenic mice, ectopic Cux-1 expression was restricted to a population of small cells located between the hepatocytes. These cells stained positively for α-smooth muscle actin and PCNA, and were increased in number in the transgenic livers, suggesting that the ectopic expression of Cux-1 in the liver resulted in proliferation of these cells. Hepatic stellate cells (also called Ito cells) are a population of cells that exist in the space between the liver parenchyma and the sinusoidal endothelial cells, and are involved in retanoid storage [25]. In pathological conditions, these cells change morphology and are thought to be the precursor cells to myofibroblast cells found in cirrhosis [26], adenoma [27], and hepatocellular carcinoma [27]. This transformation is accompanied by the increased expression of α-smooth muscle actin [27]. The considerable increase in α-smooth muscle actin observed in the transgenic livers, together with the observed expression of Cux-1 in cells that morphologically resemble fibroblasts, raises the possibility that Cux-1 is ectopically expressed in the hepatic stellate cell lineage resulting in both proliferation and conversion to the myofibroblast phenotype.
We also observed cells that morphologically appeared to be inflammatory cells in the perivascular region of the transgenic livers. And ectopic Cux-1 expression was observed in cells in the perivascular region of the transgenic livers. Taken together, this raised the possibility that the morphologic changes seen in the transgenic livers resulted from an inflammatory response. While we cannot rule out that increased inflammation promotes tumor formation in the livers of the transgenic mice, the expression of Cux-1 within both the perivascular region and the tumors correlated more closely with the number of α-smooth muscle actin positive cells, than the number of CD45/CD90.2 positive cells.
In contrast to the hepatic stellate cells, which do not differentiate into hepatocytes, a class of hepatocyte precursor cells, called oval cells, is believed to exist [28]. These cells are thought to exist in the canals of Hering and bile ductules and differentiate into hepatocytes or biliary epithelial cells [29]. Chromogranin-A is expressed in hepatic progenitor cells and in intermediate hepatocyte-like cells [30,31]. After loss of hepatocytes and/or cholangiocytes, hepatocyte precursor cells are believed to proliferate and differentiate towards the biliary and/or hepatocyte lineage (reviewed in [31–33]). Activation of hepatocyte precursor cells has been described in a number of liver diseases, including acute liver necrosis [34], hemochromatosis [35], alcoholic liver disease [36,37], and chronic viral hepatitis [35,38]. Although we observed high levels of Cux-1 expression in the fibroblast shaped cells, we did not detect Cux-1 expression in intermediate hepatocyte like cells, or in mature hepatocytes, suggesting that Cux-1 was not expressed in this lineage. However, we did detect chromogranin A expression both in small cells and in larger cells that morphologically resembled hepatocytes, suggesting the differentiation of hepatocyte precursor cells toward the hepatocyte lineage.
A number of non-hepatocytic epithelial cell lines have been isolated from liver and shown to differentiate into hepatocytes or biliary epithelial cells, suggesting they are hepatic progenitor cells [39,40]. Interestingly, co-culturing these cells with hepatic stellate cells resulted in the differentiation of the progenitor cells into hepatocytes [41]. Accordingly, one possible explanation for the hepatomegaly observed in the Cux-1 transgenic mice, is that an increased number of hepatic stellate cells influences the differentiation of hepatic progenitor cells into hepatocytes. Thus, the increased liver growth we observed in the Cux-1 transgenic mice, prior to any pathological condition, could result from the differentiation of hepatic precursor cells into hepatocytes as a reaction to an increased number of hepatic stellate cells, and not from a direct effect of Cux-1 expression in hepatocytes. Since the hepatic stellate cells are considered to be a mesenchymal cell type this would be consistent with the predicted expression of the transgene [25,42]. However, expression of lacZ under the control of the CMV promoter was detected in the livers of embryonic transgenic mice [43], so we cannot rule out the possibility that Cux-1 is expressed in hepatic precursor cells before differentiating into hepatocytes in the Cux-1 transgenic livers.
We also observed a progression of liver lesions beginning with inflammation and leading to the development of mixed cell foci and hyperplasia. While we observed hepatocellular carcinoma in some transgenic animals, we also observed HCC in wild type animals. This may be explained by the fact that the transgenic mice were generated on a C57Bl/6 × C3H mixed background, and C3H male mice show an increased incidence of HCC [44]. While the primary phenotype we observed in the Cux-1 transgenic livers was hyperplasia of small cells that are likely hepatic stellate cells, one possibility is that activation of these cells in C57Bl/6 × C3H mice contributed to the incidence of tumors and hepatocellular carcinoma. Of interest, rearrangements or deletions of the chromosome region 7q22, the site of the human Cux-1 gene (CUTL1), frequently occur in uterine leiomyomas [45], acute myeloid leukemia [46], and hepatocellular carcinoma [47].
In the livers from transgenic mice, the ectopic expression of Cux-1 correlated with a down regulation of p21. In contrast, the expression of p27 was unchanged between wild type and transgenic livers. In our previous studies, we showed that ectopic expression of Cux-1 in the kidney resulted in the down regulation of p27, leading to hyperplasia [14]. One possible explanation of our results is that Cux-1 regulates cell proliferation by different mechanisms in the transgenic kidney and liver. The ability of the liver to readily undergo regeneration following partial hepatectomy suggests that cell proliferation is regulated by a different mechanism in the liver than in the kidney. Interestingly, targeted expression of p21 in the livers of transgenic mice prevents cell cycle progression and regeneration after partial hepatectomy [48]. Thus, a down regulation of p21 is required for cell cycle entry. Moreover, the activation of hepatic stellate cells is regulated by p21 [49]. Treatment of hepatic stellate cells with the antioxidant (NAC), used in the treatment of some liver diseases, resulted in the upregulation of p21 through the Sp1 transcription activator-dependent mechanism, leading to cell cycle arrest [49]. Since Cux-1 represses p21 by competing with Sp1 for binding site occupancy [13], one possible mechanism for the activation and proliferation of hepatic stellate cells is the upregulation of Cux-1. A number of studies have demonstrated a decrease in hepatoma cell proliferation associated with the expression of p21 [50,51]. And, a decrease in p21 expression is observed in human patients with HCC [52]. Thus, the down regulation of p21 expression may be an important factor in the progression of liver lesions in these mice.
ACKNOWLEDGMENTS
We thank Rosetta Barkley and Eileen Roach for expert technical assistance. We thank Dr. John Bradfield for many helpful discussions. This work was supported by the American Heart Association (G.B.V.H.), by NIH COBRE award 1 P20 RR15563 (G.B.V.H.), and by NIH grant DK-58377 (G.B.V.H.).
REFERENCES
- 1.Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 2.Valarche I, Tissier-Seta JP, Hirsch MR, Martinez S, Goridis C, Brunet JF. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 3.Andres V, Nadal-Ginard B, Mahdavi V. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 1992;116:321–334. doi: 10.1242/dev.116.2.321. [DOI] [PubMed] [Google Scholar]
- 4.Yoon SO, Chikaraishi DM. Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J Biol Chem. 1994;269:18453–18462. [PubMed] [Google Scholar]
- 5.Vanden Heuvel GB, Quaggin SE, Igarashi P. A unique variant of a homeobox gene related to Drosophila cut is expressed in mouse testis. Biol Reprod. 1996;55:731–739. doi: 10.1095/biolreprod55.4.731. [DOI] [PubMed] [Google Scholar]
- 6.Lievens PM, Tufarelli C, Donady JJ, Stagg A, Neufeld EJ. CASP, a novel, highly conserved alternative-splicing product of the CDP/cut/cux gene, lacks cut-repeat and homeo DNA-binding domains, and interacts with full-length CDP in vitro. Gene. 1997;197:73–81. doi: 10.1016/s0378-1119(97)00243-6. [DOI] [PubMed] [Google Scholar]
- 7.Superti-Furga G, Barberis A, Schreiber E, Busslinger M. The protein CDP, but not CP1, footprints on the CCAAT region of the gamma-globin gene in unfractionated B-cell extracts. Biochim Biophys Acta. 1989;1007:237–242. doi: 10.1016/0167-4781(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 8.Dufort D, Nepveu A. The human cut homeodomain protein represses transcription from the c-myc promoter. Mol Cell Biol. 1994;14:4251–4257. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banan M, Rojas IC, Lee WH, et al. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- 10.Higgy NA, Tarnasky HA, Valarche I, Nepveu A, van der Hoorn FA. Cux/CDP homeodomain protein binds to an enhancer in the rat c-mos locus and represses its activity. Biochim Biophys Acta. 1997;1351:313–324. doi: 10.1016/s0167-4781(96)00221-7. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Bramblett D, Zhu Q, et al. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skalnik DG, Strauss EC, Orkin SH. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 13.Coqueret O, Berube G, Nepveu A. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which down modulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledford AW, Brantley JG, Kemeny G, et al. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in down regulation of p27kip1 expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev Biol. 2002;245:157–171. doi: 10.1006/dbio.2002.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailly F, Berube G, Harada R, Mao PL, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms. Active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346–5357. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- 17.Vanden Heuvel GB, Bodmer R, McConnell KR, Nagami GT, Igarashi P. Expression of a cut-related homeobox gene in developing and polycystic mouse kidney. Kidney Int. 1996;50:453–461. doi: 10.1038/ki.1996.336. [DOI] [PubMed] [Google Scholar]
- 18.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 19.Nevins JR. E2F: A link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 20.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 21.Peter M, Herskowitz I. Joining the complex: Cyclin-dependent kinase inhibitory proteins and the cell cycle. Cell. 1994;79:181–184. doi: 10.1016/0092-8674(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 22.van Wijnen AJ, van Gurp MF, de Ridder MC, et al. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: A mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Wijnen AJ, Choi TK, Owen TA, et al. Involvement of the cell cycle-regulated nuclear factor HiNF-D in cell growth control of a human H4 histone gene during hepatic development in transgenic mice. Proc Natl Acad Sci USA. 1991;88:2573–2577. doi: 10.1073/pnas.88.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim EC, Lau JS, Rawlings S, Lee AS. Positive and negative regulation of the human thymidine kinase promoter mediated by CCAAT binding transcription factors NF-Y/CBF, dbpA, and CDP/cut. Cell Growth Differ. 1997;12:1329–1338. [PubMed] [Google Scholar]
- 25.Senoo H. Structure and function of hepatic stellate cells. Med Electron Microsc. 2004;37:3–15. doi: 10.1007/s00795-003-0230-3. [DOI] [PubMed] [Google Scholar]
- 26.Cassiman D, Libbrecht L, Desmet V, Denef C, Roskams T. Hepatic stellate cell/myofibroblast subpopulations in fibrotic human and rat livers. J Hepatol. 2002;36:200–209. doi: 10.1016/s0168-8278(01)00260-4. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt-Graff A, Kruger S, Bochard F, Gabbiani G, Denk H. Modulation of alpha smooth muscle actin and desmin expression in perisinusoidal cells of normal and diseased human livers. Am J Pathol. 1991;138:1233–1242. [PMC free article] [PubMed] [Google Scholar]
- 28.Thorgeirsson SS. Hepatic stem cells in liver regeneration. FASEB J. 1996;10:1249–1256. [PubMed] [Google Scholar]
- 29.Lazaro CA, Rhim JA, Yamada Y, Fausto N. Generation of hepatocytes from oval cell precursors in culture. Cancer Res. 1998;58:5514–5522. [PubMed] [Google Scholar]
- 30.Roskams T, De Vos R, Van Eyken P, Myazaki H, Van Damme B, Desmet V. Hepatic OV-6 expression in human liver disease and rat experiments: Evidence for hepatic progenitor cells in man. J Hepatol. 1998;29:455–463. doi: 10.1016/s0168-8278(98)80065-2. [DOI] [PubMed] [Google Scholar]
- 31.Libbrecht L, Roskams T. Hepatic progenitor cells in human liver diseases. Semin Cell Dev Biol. 2002;13:389–396. doi: 10.1016/s1084952102001258. [DOI] [PubMed] [Google Scholar]
- 32.Dabeva MD, Alpini G, Hurston E, Shafritz DA. Models for hepatic progenitor cell activation. Proc Soc Exp Biol Med. 1993;204:242–252. doi: 10.3181/00379727-204-43660. [DOI] [PubMed] [Google Scholar]
- 33.Bustos M, Sangro B, Alzuguren P, et al. Liver damage using suicide genes. A model for oval cell activation. Am J Pathol. 2000;157:549–559. doi: 10.1016/S0002-9440(10)64565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujita M, Furukawa H, Hattori M, Todo S, Ishida Y, Nagashima K. Sequential observation of liver cell regeneration after massive hepatic necrosis in auxiliary partial orthotopic liver transplantation. Mod Pathol. 2000;13:152–157. doi: 10.1038/modpathol.3880029. [DOI] [PubMed] [Google Scholar]
- 35.Lowes KN, Brennan BA, Yeoh GC, Olynyk JK. Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol. 1999;154:537–541. doi: 10.1016/S0002-9440(10)65299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray MB, Mendenhall CL, French SW, Gartside PS. Bile duct changes in alcoholic liver disease. The Veterans Administration Cooperative Study Group. Liver. 1993;13:36–45. doi: 10.1111/j.1600-0676.1993.tb00603.x. [DOI] [PubMed] [Google Scholar]
- 37.Sell S. Comparison of liver progenitor cells in human atypical ductular reactions with those seen in experimental models of liver injury. Hepatology. 1998;27:317–331. doi: 10.1002/hep.510270202. [DOI] [PubMed] [Google Scholar]
- 38.Libbrecht L, Desmet V, Van Damme B, Roskams T. Deep intralobular extension of human hepatic ‘progenitor cells’ correlates with parenchymal inflammation in chronic viral hepatitis: Can ‘progenitor cells’ migrate? J Pathol. 2000;192:373–378. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH700>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Coleman WB, McCullough KD, Esch GL, et al. Evaluation of the differentiation potential of WB-F344 rat liver epithelial stem-like cells in vivo. Differentiation to hepatocytes after transplantation into dipeptidylpeptidase-IV-deficient rat liver. Am J Pathol. 1997;151:353–359. [PMC free article] [PubMed] [Google Scholar]
- 40.Tsao MS, Smith JD, Nelson KG, Grisham JW. A diploid epithelial cell line from normal adult rat liver with phenotypic properties of ‘oval’ cells. Exp Cell Res. 1984;154:38–52. doi: 10.1016/0014-4827(84)90666-9. [DOI] [PubMed] [Google Scholar]
- 41.Nagai H, Terada K, Watanabe G, et al. Differentiation of liver epithelial (stem-like) cells into hepatocytes induced by coculture with hepatic stellate cells. Biochem Biophys Res Commun. 2002;293:1420–1425. doi: 10.1016/S0006-291X(02)00406-0. [DOI] [PubMed] [Google Scholar]
- 42.Bhunchet E, Wake K. Role of mesenchymal cell populations in porcine serum-induced rat liver fibrosis. Hepatology. 1992;16:1452–1473. doi: 10.1002/hep.1840160623. [DOI] [PubMed] [Google Scholar]
- 43.Baskar JF, Smith PP, Ciment GS, et al. Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J Virol. 1996;70:3215–3226. doi: 10.1128/jvi.70.5.3215-3226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drinkwater N, Hanigan M, Kemp C. Genetic and epigenetic promotion of murine hepatocarcinogenesis. In: Stevenson D, McClain RM, Popp JA, Slaga TJ, Ward JM, Pitot HC, editors. Mouse Liver Carcinogenesis, Mechanisms and Species Comparisons. New York: Alan R. Liss, Inc.; 1990. pp. 163–176. [PubMed] [Google Scholar]
- 45.Zeng WR, Scherer SW, Koutsilieris M, et al. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene. 1997;14:2355–2365. doi: 10.1038/sj.onc.1201076. [DOI] [PubMed] [Google Scholar]
- 46.Fenaux P, Preudhomme C, Lai JL, Morel P, Beuscart R, Bauters F. Cytogenetics and their prognostic value in de novo acute myeloid leukaemia: A report on 283 cases. Br J Haematol. 1989;73:61–67. doi: 10.1111/j.1365-2141.1989.tb00221.x. [DOI] [PubMed] [Google Scholar]
- 47.Parada LA, Hallen M, Tranberg KG, et al. Frequent rearrangements of chromosomes 1, 7, and 8 in primary liver cancer. Genes Chromosomes Cancer. 1998;23:26–35. doi: 10.1002/(sici)1098-2264(199809)23:1<26::aid-gcc5>3.3.co;2-g. [DOI] [PubMed] [Google Scholar]
- 48.Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 49.Kim KY, Rhim T, Choi I, Kim SS. N-acetylcysteine induces cell cycle arrest in hepatic stellate cells through its reducing activity. J Biol Chem. 2001;276:40591–40598. doi: 10.1074/jbc.M100975200. [DOI] [PubMed] [Google Scholar]
- 50.Qin LF, Ng IO. Exogenous expression of p21(WAF1/CIP1) exerts cell growth inhibition and enhances sensitivity to cisplatin in hepatoma cells. Cancer Lett. 2001;172:7–15. doi: 10.1016/s0304-3835(01)00701-7. [DOI] [PubMed] [Google Scholar]
- 51.Suzui M, Masuda M, Lim JT, Albanese C, Pestell RG, Weinstein IB. Growth inhibition of human hepatoma cells by acyclic retinoid is associated with induction of p21(CIP1) and inhibition of expression of cyclin D1. Cancer Res. 2002;62:3997–4006. [PubMed] [Google Scholar]
- 52.Shi YZ, Hui AM, Takayama T, Li X, Cui X, Makuuchi M. Reduced p21(WAF1/CIP1) protein expression is predominantly related to altered p53 in hepatocellular carcinomas. Br J Cancer. 2000;83:50–55. doi: 10.1054/bjoc.2000.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]