Abstract
Little is known about the molecular mechanisms by which the embryo proper and suspensor of plant embryos activate specific gene sets shortly after fertilization. We analyzed the upstream region of the Scarlet Runner Bean (Phaseolus coccineus) G564 gene in order to understand how genes are activated specifically in the suspensor during early embryo development. Previously, we showed that a 54-bp fragment of the G564 upstream region is sufficient for suspensor transcription and contains at least three required cis-regulatory sequences, including the 10-bp motif (5′-GAAAAGCGAA-3′), the 10 bp-like motif (5′-GAAAAACGAA-3′), and Region 2 motif (partial sequence 5′-TTGGT-3′). Here, we use site-directed mutagenesis experiments in transgenic tobacco globular-stage embryos to identify two additional cis-regulatory elements within the 54-bp cis-regulatory module that are required for G564 suspensor transcription: the Fifth motif (5′-GAGTTA-3′) and a third 10-bp-related sequence (5′-GAAAACCACA-3′). Further deletion of the 54-bp fragment revealed that a 47-bp fragment containing the five motifs (the 10-bp, 10-bp-like, 10-bp-related, Region 2 and Fifth motifs) is sufficient for suspensor transcription, and represents a cis-regulatory module. A consensus sequence for each type of motif was determined by comparing motif sequences shown to activate suspensor transcription. Phylogenetic analyses suggest that the regulation of G564 is evolutionarily conserved. A homologous cis-regulatory module was found upstream of the G564 ortholog in the Common Bean (Phaseolus vulgaris), indicating that the regulation of G564 is evolutionarily conserved in closely related bean species.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-015-0308-z) contains supplementary material, which is available to authorized users.
Keywords: Phaseolus coccineus, Scarlet runner bean, Suspensor, Gene regulatory network, Cis-regulatory elements, Promoter analysis
Introduction
In most higher plants, the zygote divides asymmetrically to form a small apical cell and a large basal cell with distinct developmental fates (Goldberg et al. 1994). The apical cell differentiates into the embryo proper, which will become the next generation plant; whereas, the basal cell generates the hypophysis and suspensor. The hypophysis will be incorporated into the root meristem of the developing embryo proper (Dolan et al. 1993). The suspensor is a terminally differentiated embryonic region that physically connects the embryo proper to the maternal tissues and transports nutrients and growth regulators to the embryo proper before degenerating later during embryogenesis (Yeung and Meinke 1993).
The precise mechanisms that determine the developmental fates of the apical and basal cells remain unknown. A number of studies using individual genes and whole-genome transcriptome profiling have shown that different gene sets are activated in the globular-stage embryo proper and suspensor shortly after fertilization (Weterings et al. 2001; Haecker et al. 2004; Le et al. 2007; Ueda et al. 2011; Belmonte et al. 2013; Slane et al. 2014). Little is known, however, about the cis-regulatory sequences and trans-acting factors that activate genes in different embryonic regions. Nor is it known how these genes are connected into regulatory networks that control their expression in precise spatial and temporal patterns during embryonic development (Peter and Davidson 2015).
Previously, we used the giant embryos of the Scarlet Runner Bean, Phaseolus coccineus, to identify suspensor-specific mRNAs (Weterings et al. 2001; Le et al. 2007; Kawashima and Goldberg 2010). These embryos contain a large, highly specialized suspensor that is particularly suited for region-specific gene expression studies early in embryo development (Le et al. 2007; Kawashima and Goldberg 2010; Henry and Goldberg 2015). One Scarlet Runner Bean suspensor-specific mRNA, designated as G564, which encodes a protein of unknown function, first appears in the basal cells of the four-cell embryo, and then accumulates to a high level in the suspensor at later developmental stages (Weterings et al. 2001). The spatial expression pattern of the G564 gene is controlled primarily at the transcriptional level by sequences in its proximal upstream region (Weterings et al. 2001; Kawashima et al. 2009). The G564 upstream region contains a ~150-bp sequence repeated five times in tandem (Fig. 1a) (Weterings et al. 2001; Kawashima et al. 2009). We dissected the fourth 150-bp repeat and found that it contains a 54-bp positive cis-regulatory module that is sufficient to program suspensor transcription shortly after fertilization, and contains at least three suspensor cis-regulatory elements designated as the (1) 10-bp motif (5′-GAAAAGCGAA-3′), (2) 10-bp-like motif (5′-GAAAAACGAA-3′), and (3) Region 2 motif (5′-TTGGT-3′) (Fig. 1b) (Kawashima et al. 2009). The G564 54-bp suspensor cis-regulatory module is conserved in all five 150-bp repeats, with some sequence differences (Kawashima et al. 2009). We showed that the machinery that activates suspensor transcription using these motifs is conserved among flowering plants (Kawashima et al. 2009).
Fig. 1.
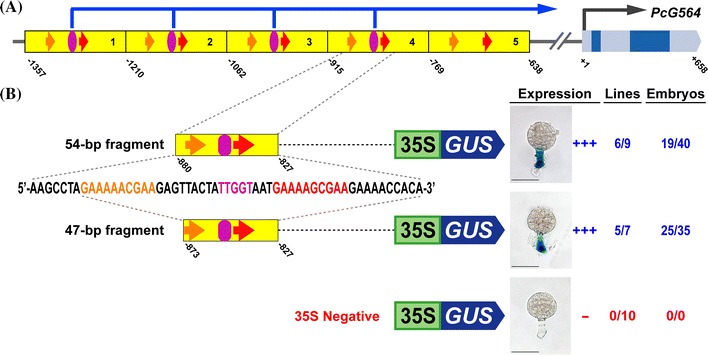
A 47-bp region contains all of the sequences required for suspensor transcription. a Conceptual representation of the G564 gene and upstream region. b Suspensor GUS activity in transgenic globular-stage tobacco embryos containing GOF fragments from the G564 upstream region. Names and conceptual representations of the constructs are to the left of each embryo. Numbers indicate positions relative to the G564 transcription start site (+1) (Weterings et al. 2001). Yellow boxes indicate the 150-bp promoter repeats (Weterings et al. 2001; Kawashima et al. 2009). Red arrows, orange arrows and purple ovals indicate the 10-bp, 10-bp-like, and Region 2 motifs, respectively (Kawashima et al. 2009). 35S/GUS indicates the CaMV 35S minimal promoter/GUS gene. The sequences of the 10-bp, 10-bp-like and Region 2 motifs were taken from Kawashima et al. (2009) and are shown in red, orange, and purple fonts, respectively. +++ in the Expression column indicates that suspensor GUS activity was strong and detected by 2-h incubation for a majority of the GUS-positive lines; − in the Expression column indicates no detectable suspensor GUS activity. Numbers in the Lines column indicate the number of individual transformants displaying suspensor GUS activity over the total number of individual transformants analyzed. Numbers in the Embryos column indicate the number of embryos displaying suspensor GUS activity by 24-h incubation over the total number of analyzed embryos of GUS-positive lines. Photographs were taken after 24-h GUS incubation (Scale bar: 50 μm)
In this paper, we present experiments identifying two additional cis-regulatory elements within the 54-bp cis-regulatory module that are required for G564 suspensor transcription. Mutagenesis of the 54-bp fragment identified a new suspensor cis-regulatory element, the Fifth motif (5′-GAGTTA-3′), and an additional 10-bp sequence, designated the 10-bp-related motif (5′-GAAAACCACA-3′), that are required for suspensor transcription. Further deletion of the 54-bp fragment revealed that a 47-bp fragment containing only the five motifs (the 10-bp, 10-bp-like, 10-bp-related, Region 2 and Fifth motifs) is sufficient for suspensor transcription. We also show that the Scarlet Runner Bean G564 suspensor cis-regulatory module is conserved in the G564 ortholog (PvG564) of the Common Bean, Phaseolus vulgaris, suggesting that these motifs program G564 expression patterns in the giant suspensors of two closely-related bean species. Together, our results have identified five suspensor cis-regulatory elements, three of which are functionally equivalent, that activate G564 transcription specifically in the suspensor shortly after fertilization.
Materials and methods
Generating gain-of-function (GOF) and site-directed mutagenesis constructs
For GOF and mutagenesis analyses of the G564 upstream region, the GOF12 vector described by Kawashima et al. (2009), which contains the G564 54-bp fragment (−880 to −827) upstream of the CaMV 35S minimal promoter (Benfey et al. 1990; Koltunow et al. 1990) and the β-glucuronidase (GUS) reporter gene (Jefferson et al. 1987), was used as a PCR template with primers containing the desired mutation and EcoRI or BamHI restriction sites (Kawashima et al. 2009). Primer sequences used for mutagenesis are listed in Online Resource 1. The amplified DNA fragments were then digested with EcoRI and BamHI and ligated into the EcoRI- and BamHI-digested GOF12 vector. G564 fragment regions in the mutagenesis constructs were sequenced to confirm that they contained the correct mutated bases.
Plant transformation
Tobacco (Nicotiana tabacum cv SR1) plants were transformed and regenerated by using the leaf disk procedure (Horsch et al. 1985). At least six independent transformants were generated for each construct. The promoter/GUS region from each individual transformant was sequenced to ensure that there were no rearrangements. A total of 16 different mutagenesis constructs and 129 individual tobacco transformants were generated in order to carry out this study.
GUS histochemical assay
Transgenic tobacco seeds were harvested 8 days after pollination (DAP). Globular-stage embryos were hand-dissected from seeds and assayed for GUS activity after 2 and 24 h at 37 °C, as described previously (Kawashima et al. 2009). Embryos were photographed under bright-field illumination using a compound microscope (LEICA 5000B). T1 seeds from GUS-negative lines were tested for kanamycin-resistant segregation after selfing to confirm that the T-DNA was not silenced. In total, approximately 800 individual globular-stage embryos were assayed for GUS activity in order to generate the results reported in this study.
Results
A 47-bp fragment contains all of the sequences required for suspensor transcription
We used the Scarlet Runner Bean G564 gene to identify cis-regulatory elements required for suspensor transcription. Previously, we showed that a gain-of-function (GOF) construct containing a 54-bp fragment of the G564 upstream region can program suspensor-specific transcription when fused to a Cauliflower Mosaic Virus (CaMV) 35S minimal promoter/GUS vector and introduced into tobacco plants (Kawashima et al. 2009). This 54-bp fragment contains the previously identified 10-bp motif (5′-GAAAAGCGAA-3′), 10-bp-like motif (5′-GAAAAACGAA-3′), and Region 2 motif that contains the sequence 5′-TTGGT-3′ (Fig. 1b) (Kawashima et al. 2009). To identify the minimal sequence required for G564 suspensor transcription, we deleted the distal 7-bp of the 54-bp fragment, and showed that it produced strong suspensor GUS activity under the direction of the CaMV 35S minimal promoter in transgenic tobacco embryos, similar to that observed for the 54-bp fragment (Fig. 1b). By contrast, a CaMV 35S minimal promoter/GUS construct produced no detectable suspensor GUS activity. Thus, the 47-bp fragment is the minimal cis-regulatory module that can activate G564 suspensor transcription.
A third 10-bp sequence is required for suspensor transcription
Within the 54-bp G564 fragment, there is 10-bp (−836/−827) between the 10-bp motif and the 3′ end of the fragment, in which there is enough space for another suspensor cis-regulatory element (Fig. 1b). To identify additional suspensor cis-regulatory sequences, we generated GOF constructs in which mutated versions of the 54-bp fragment were fused to the CaMV 35S minimal promoter/GUS vector, and introduced these constructs into tobacco plants. A GOF construct in which the 10-bp sequence at −836/−827 (5′-GAAAACCACA-3′) was mutated [m54(45–54)] showed no GUS activity, indicating that there is an additional motif that is required for suspensor transcription (Fig. 2a). Because this 10-bp sequence at −836/−827 is similar to the 10-bp and 10-bp-like motifs, differing by 3-bp, we have designated it as the 10-bp-related motif. Therefore, three 10-bp sequences in the 54-bp fragment are required for suspensor transcription: the 10-bp (−846/−837), 10-bp-like (−873/−864) and 10-bp-related (−836/−827) motifs.
Fig. 2.
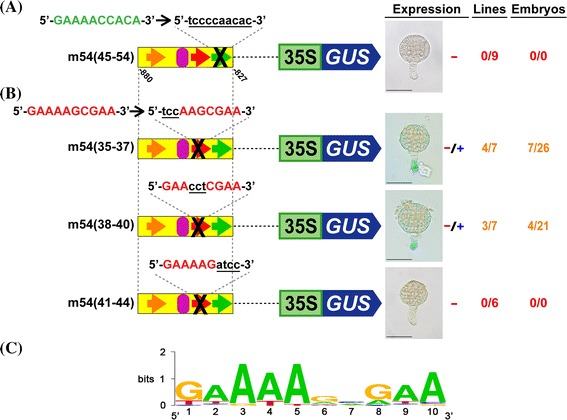
A third 10-bp motif is required for suspensor transcription. a and b Suspensor GUS activity in transgenic tobacco embryos containing 54-bp GOF constructs with mutations within the 10-bp-related motif (a) or 10-bp motif (b). c A consensus 10-bp/10-bp-like/10-bp-related motif sequence was generated from DNA sequences shown to function as the 10-bp motif in transgenic tobacco embryos (Kawashima et al. 2009). Green arrows indicate the 10-bp-related motif. The sequences of the 10-bp and 10-bp-related motifs are shown in red and green fonts, respectively. Black crosses indicate mutations in these motifs. The mutation sequence is written in black, lower case, and underlined font. +++ in the Expression column indicates that suspensor GUS activity was strong and detected by 2-h incubation for a majority of the GUS-positive lines; −/+ in the Expression column indicates that suspensor GUS activity was weak and not detected by 2-h incubation in the majority of embryos from GUS-positive lines; − in the Expression column indicates no detectable suspensor GUS activity. Numbers in the Lines column indicate the number of individual transformants displaying suspensor GUS activity over the total number of individual transformants analyzed. Numbers in the Embryos column indicate the number of embryos displaying suspensor GUS activity by 24-h incubation over the total number of analyzed embryos of GUS-positive lines. Photographs were taken after 24-h GUS incubation (Scale bar: 50 μm)
To determine the core sequence of the 10-bp/10-bp-like/10-bp-related motifs, we took the 10-bp motif as a representative of these three motifs and mutated the first 3-bp, the second 3-bp and the last 4-bp of the 10-bp motif within the 54-bp fragment (Fig. 2b). Although the 10-bp motif is known to tolerate a 3-bp mismatch without affecting the level of suspensor transcription (Kawashima et al. 2009), we hypothesized that the mismatches would not be tolerated if they were all clustered together, rather than being spread throughout the motif. GUS activity in the suspensor was significantly decreased when the first and second 3-bp were mutated [m54(35–37) and m54(38–40)] and completely abolished when the last 4-bp were mutated [m54(41–44)] (Fig. 2b). These results demonstrated that at least one nucleotide in each of these three regions is critical for the function of the 10-bp motif.
Previously, we generated a consensus sequence for the 10-bp motif from divergent 10-bp sequences known to function as the 10-bp motif (Fig. 2c) (Kawashima et al. 2009). Each position of the 10-bp motif can tolerate a mismatch without affecting the level of suspensor transcription. All of the functional 10-bp sequences contain up to three mismatches relative to the 10-bp motif sequence of 5′-GAAAAGCGAA-3′ (Kawashima et al. 2009), and this sequence represents the nucleotide most commonly found in each position of the motif for all of the sequences that function as the 10-bp motif. Figure 2b shows that three mismatches cannot be tolerated if the mismatches are clustered in one part of the 10-bp motif. Taken together, we determined a consensus for the 10-bp/10-bp-like/10-bp-related motifs of 5′-GAAAAGCGAA-3′ with up to three non-adjacent mismatches.
The complete sequence of the Region 2 motif was identified
We previously identified the partial Region 2 motif sequence as 5′-TTGGT-3′ through sequence homology between the 150-bp G564 GUS-positive repeats (first, second and fourth repeats) and the GUS-negative fifth repeat (Fig. 1a) (Kawashima et al. 2009). The Region 2 motif was shown to be functional in the 54-bp fragment by mutating the central guanine at position 29 of the 54-bp fragment to adenine, rendering it inactive (Kawashima et al. 2009). In order to functionally test which nucleotides in Region 2 are required for suspensor transcription, we generated mutations across the Region 2 motif either 2-bp or 3-bp at a time and transferred GOF constructs containing these mutant motifs into transgenic tobacco plants (Fig. 3a). Transversional mutagenesis was used to impose the biggest change on the sequence; however transversional mutations of this GT-rich motif resulted in another GT-rich sequence. Consequently, constructs were also made using adenine substitutions (Fig. 3a).
Fig. 3.
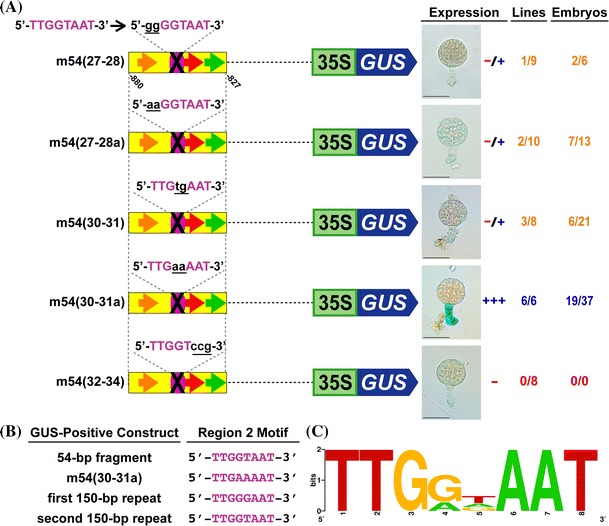
A consensus sequence for the Region 2 motif. a Suspensor GUS activity in transgenic tobacco embryos containing 54-bp GOF constructs with mutations within the Region 2 motif. b Sequences of the Region 2 motif in GOF constructs that activate suspensor transcription. Suspensor GUS activity in transgenic tobacco embryos containing the first and second 150-bp repeats is presented in Kawashima et al. (2009). c A consensus sequence for the Region 2 motif was created by WebLogo (Crooks et al. 2004) using Region 2 motif sequences shown in b that function within the suspensor. Purple ovals indicate the Region 2 motif. Region 2 motif sequences are shown in purple font. Black crosses indicate mutations in this motif. The mutation sequence is written in black, lower case, and underlined font. Other figure details are the same as those outlined in Fig. 1 and 2. Photographs were taken after 24-h GUS incubation (Scale bar: 50 μm)
Mutation of the 5′ TT using either mutagenesis strategy [m54(27–28) and m54(27–28a)] resulted in significantly decreased suspensor GUS activity. Mutation of the central GT to TG [m54(30–31)] also resulted in significantly decreased suspensor GUS activity. By contrast, mutation of GT to AA [m54(30–31a)], creating the sequence 5′-TTGAAAAT-3′, did not alter GUS activity. Finally, mutation of the 3′ AAT [m54(32–34)] abolished GUS activity, demonstrating that the Region 2 motif is longer than previously predicted. Taken together, these results show that the important nucleotides in the Region 2 motif are 5′-TTGGTAAT-3′, and that 5′-TTGAAAAT-3′ can also function as the Region 2 motif (Fig. 3a).
We identified a consensus for the Region 2 motif by comparing Region 2 motif sequences within all 150-bp G564 repeats that we showed previously can drive GUS gene expression in the suspensor of transgenic tobacco embryos (first, second and fourth repeats) (Kawashima et al. 2009). No other sequence element(s) in the 150-bp repeats can compensate for the function of the Region 2 motif, as was shown by a natural point mutation in the fifth repeat that causes a loss of suspensor GUS activity (Fig. 1a) (Kawashima et al. 2009). The sequence of the Region 2 motif in the second repeat and the 54-bp fragment within the fourth repeat is 5′-TTGGTAAT-3′, whereas the Region 2 sequence in the first repeat is 5′-TTGGGAAT-3′ (Fig. 3b). We combined this information with the knowledge that 5′-TTGAAAAT-3′ can also function as the Region 2 motif (Fig. 3a), generating a sequence logo of 5′-TTG(A/G)(A/G/T)AAT-3′ for the Region 2 motif (Fig. 3c). This consensus sequence is representative of the Region 2 motif in each 150-bp G564 upstream repeat, with the exception of the fifth repeat.
A fifth motif is required for suspensor transcription
Within the 54-bp fragment there is 9-bp between the 10-bp-like motif and Region 2 motif—providing enough space for a fifth cis-regulatory element (Fig. 1b). To determine whether there is a fifth suspensor cis-regulatory element, we mutated the sequence 5′-GAGTTAC-3′ in the region between the 10-bp-like and Region 2 motifs in the 54-bp fragment (Fig. 4a). Mutation of 5′-GAGTTAC-3′ [m54(18–24)] abolished suspensor GUS activity completely, indicating the presence of a fifth DNA control element required for suspensor transcription.
Fig. 4.
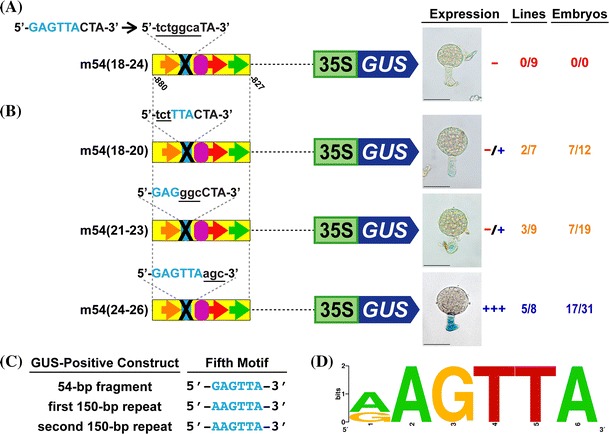
A fifth motif is required for suspensor transcription. a and b Suspensor GUS activity in transgenic tobacco embryos containing 54-bp GOF constructs with mutations within the Fifth motif region. c Sequences of the Fifth motif in GOF constructs that activate suspensor transcription. Suspensor GUS activity in transgenic tobacco embryos containing the first and second 150-bp repeats is presented in Kawashima et al. (2009). d A consensus sequence for the Fifth motif was created by WebLogo (Crooks et al. 2004) using Fifth motif sequences shown in c that function within the suspensor. Blue ovals indicate the Fifth motif. The sequences of the Fifth motif are shown in blue font. Black crosses indicate mutations in this motif. The mutation sequence is written in black, lower case, and underlined font. Other figure details are the same as those outlined in Figs. 1, 2, 3. Photographs were taken after 24-h GUS incubation (Scale bar: 50 μm)
To define the length of the Fifth motif, we generated constructs by mutating the 9-bp between the 10-bp-like motif and Region 2 motif 3-bp at time (Fig. 4b). Mutation of 5′-GAG-3′ [m54(18–20)] and 5′-TTA-3′ [m54(21–23)] caused a significant decrease in GUS activity, indicating that these sequences are part of the Fifth motif. However, mutation of the next 3-bp, 5′-CTA-3′ [m54(24–26)], had no effect on GUS activity. Therefore, the final cytosine mutated in the m54(18–24) construct was not required for suspensor transcription. Taken together, these results showed that the important nucleotides in the Fifth motif are 5′-GAGTTA-3′.
We determined a consensus sequence for the Fifth motif through comparison of the Fifth motif sequence in GUS-positive 150-bp repeats (first, second and fourth) (Fig. 4c) (Kawashima et al. 2009). No other sequence in the 150-bp repeats closely resembles the Fifth motif, so we assume that the sequences in the position of the Fifth motif in the first and second repeats are functional. The sequence of the Fifth motif in the 54-bp fragment within the fourth repeat is 5′-GAGTTA-3′. The sequence in the position of the Fifth motif in the first and second repeats is 5′-AAGTTA-3′. Based on these results, we constructed a sequence logo for the Fifth motif with a consensus sequence of 5′-(A/G)AGTTA-3′ (Fig. 4d).
The 150-bp tandem repeats and suspensor motifs are conserved in the Common Bean G564 upstream region
The recent release of the Common Bean (Phaseolus vulgaris) genome (Schmutz et al. 2014) provides an opportunity to compare G564 expression and promoter sequences in two related bean species with giant suspensors (Fig. 5a) (Henry and Goldberg 2015) that diverged ~2 million years ago (mya) (Delgado-Salinas et al. 2006). We used Illumina sequencing technology to profile the mRNA populations of laser-capture microdissected embryo proper and suspensor regions from Scarlet Runner Bean and Common Bean globular-stage embryos, and mapped RNA-Seq reads to the sequenced Common Bean genome (GEO accession GSE57537). Common Bean G564 mRNA was up-regulated ~140-fold in the globular-stage suspensor relative to the embryo proper, similar to what we observed for Scarlet Runner Bean (Fig. 5b) (Weterings et al. 2001; Henry and Goldberg 2015). We compared the G564 upstream region in Scarlet Runner Bean and Common Bean, and found that the Common Bean G564 promoter also contains the five 150-bp tandem repeats (Fig. 5c–e). These tandem promoter repeats are not found in the upstream region of G564-related genes in soybean or Arabidopsis, which diverged from the Scarlet Runner Bean 19 and 120 mya, respectively (Lavin et al. 2005). Therefore, the 150-bp repeats originated before Scarlet Runner Bean and Common Bean diverged from their common ancestor, but after the divergence of these bean species from their soybean (Glycine max) relative within the Legume family.
Fig. 5.
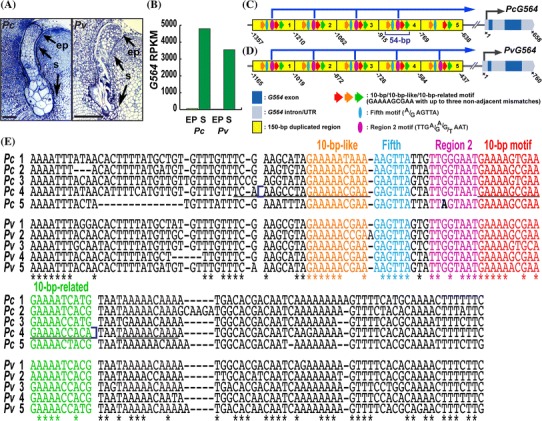
The G564 gene and mRNA abundance in Scarlet Runner Bean and Common Bean. a Plastic section of Scarlet Runner Bean [Phaseolus coccineus (Pc)] globular-stage embryo and paraffin section of Common Bean [Phaseolus vulgaris (Pv)] globular-stage embryo. b Scarlet Runner Bean and Common Bean G564 mRNA prevalences determined by RNA-Seq analysis of laser-captured globular-stage embryo proper and suspensor regions. c and d Conceptual representations of the G564 gene and upstream region in Scarlet Runner Bean (c) and Common Bean (d). Dark blue boxes represent exons. Light blue boxes represent introns and UTRs. Yellow boxes represent 150-bp tandem repeats in the upstream region (Weterings et al. 2001; Kawashima et al. 2009). Red, orange, and green arrows; purple ovals; and blue ovals indicate the 10-bp, 10-bp-like and 10-bp-related motifs; Region 2 motif; and Fifth motif, respectively. Dark blue bracket designates the 54-bp region that was analyzed in this study. Numbers indicate positions relative to the transcription start site (+1). e Nucleotide sequence alignment of the five G564 150-bp tandem repeats. Nucleotides conserved across all five G564 150-bp repeats from Scarlet Runner Bean and Common Bean are indicated by asterisks. The 10-bp, 10-bp-like and 10-bp-related motifs, Region 2 motif and Fifth motif are shown in red, orange, green, purple and blue font, respectively. The bold A in the PcG564 fifth 150-bp repeat is a natural mutation that makes the Region 2 motif non-functional (Kawashima et al. 2009). Sequences shown to function as the 10-bp motif are underlined (Kawashima et al. 2009). The number to the left of the aligned sequences indicates the position of each 150-bp repeat. Gaps were introduced for optimal alignment. ep, embryo proper; Pc, Phaseolus coccineus; Pv, Phaseolus vulgaris; RPKM, reads per kilobase per million; s, suspensor
Sequences nearly identical to the 10-bp, 10-bp-like, 10-bp-related, Region 2 and Fifth motifs were found in each Common Bean 150-bp repeat (Fig. 5e). The one exception was the functional Region 2 motif sequence in the Common Bean fifth repeat, in contrast with the natural mutation in the Scarlet Runner Bean fifth repeat Region 2 sequence (see bold A in Fig. 5e) that makes it non-functional (Kawashima et al. 2009). The suspensor motifs identified in the Common Bean G564 upstream region are most likely functional because the G564 promoter sequences and suspensor-specific expression patterns are nearly indistinguishable in both species (Fig. 5b, e).
Discussion
Five positive cis-regulatory elements are required to activate G564 suspensor transcription
We used the Scarlet Runner Bean G564 gene to identify cis-regulatory elements required for activating suspensor transcription shortly after fertilization. A 54-bp fragment within the fourth 150-bp repeat in the upstream region of G564 is sufficient for suspensor-specific transcription and contains three previously identified suspensor cis-regulatory elements: the 10-bp motif, 10-bp-like motif and the Region 2 motif (Kawashima et al. 2009). Here, we used site-directed mutagenesis of the 54-bp fragment to identify a new cis-regulatory element, the Fifth motif, which is required for suspensor-specific transcription of G564 (Fig. 4a). We also identified a third 10-bp motif, which is required for suspensor transcription (Fig. 2a). Although the five suspensor cis-regulatory elements are densely packed in the G564 suspensor module, these five motifs constitute discrete cis-regulatory elements. The Fifth motif and Region 2 motif are clearly separated by 3 bp (Fig. 5e). Previously, we showed that the Region 2 motif and the 10-bp motif are discrete cis-regulatory elements that can function even when the spacing and orientation are altered (Kawashima et al. 2009).
Positioning of the cis-regulatory elements is flexible
A 47-bp fragment of the G564 upstream region is sufficient for suspensor transcription, and contains the 10-bp motif, 10-bp-like motif, 10-bp-related motif, Region 2 motif and Fifth motif. The 47-bp fragment is the minimal cis-regulatory module that can activate suspensor transcription shortly after fertilization (Fig. 1b), and a change in almost any sequence within this module disrupts suspensor-specific transcription (Fig. 2–4) (Kawashima et al. 2009). The tight arrangement of cis-regulatory elements in the 47-bp suspensor module initially suggested that this module exemplifies the “enhanceosome” model of cis-regulatory module organization, in which the precise arrangement of cis-regulatory elements is essential for promoting cooperative interactions among bound transcription factors (Spitz and Furlong 2012). However, recent cis-regulatory analysis of the Scarlet Runner Bean GA 20-oxidase gene upstream region showed that these same motifs are required for suspensor transcription of GA 20-oxidase even though they have a different order, orientation and spacing (Henry 2014). It is just the presence of the motifs, not their relative positions, that is essential for function, similar to the Drosophila eve stripe two and sog cis-regulatory modules (Peter and Davidson 2015). This suggests that the organization of control elements within the G564 suspensor cis-regulatory module is more representative of the “billboard” enhancer model, in which the relative positioning and spacing of transcription factor binding sites is flexible (Spitz and Furlong 2012).
The organization of the 47-bp module revealed a new property of plant developmental enhancers, whereby multiple copies of a cis-regulatory element are required in close proximity. Within the fourth 150-bp repeat of G564, there are five 10-bp sequences (underlined in Fig. 5e) that have been shown to function as the 10-bp motif when fused to a deletion of the G564 upstream sequence (Kawashima et al. 2009). The 54-bp fragment contains three of these 10-bp motifs, and all three are required to activate suspensor transcription in the context of the 54-bp fragment. Such homotypic clusters of transcription factor binding sites are a common feature of cis-regulatory modules that regulate developmental genes in animals (Lifanov et al. 2003; Cameron and Davidson 2009; Gotea et al. 2010). Our data suggests that homotypic clustering as an organizational principle of developmental cis-regulatory modules is conserved not only in animals, but in the plant kingdom as well.
What transcription factors bind to these suspensor motifs?
We used PLACE (Higo et al. 1999), TRANSFAC (Matys et al. 2003), JASPAR (Mathelier et al. 2014) and the RegSite Databases of Plant Regulatory Elements (http://softberry.com) to check whether any known plant cis-elements are present in the 47-bp G564 suspensor cis-regulatory module. No known plant cis-elements closely match the 10-bp/10-bp-like/10-bp-related motif or the Region 2 motif sequences. By contrast, the Fifth motif consensus sequence [5′-(A/G)AGTTA-3′] resembles the MYB transcription factor binding site 5′-TAACTG-3′ or, in the reverse strand orientation, 5′-CAGTTA-3′ (Urao et al. 1993; Martin and Paz-Ares 1997). The Scarlet Runner Bean G564 upstream region can program transcription in the suspensors of transgenic tobacco embryos (Figs. 1–4) (Kawashima et al. 2009) and Arabidopsis embryos (Kawashima et al. 2009), indicating that the factors binding to these five cis-regulatory elements must be spatially localized within suspensors across the plant kingdom. We used a yeast one-hybrid system to show that several Arabidopsis MYB transcription factors can bind to the G564 54-bp module, supporting the notion that the Fifth motif utilizes a suspensor MYB transcription factor (Henry 2014). The identity of the specific MYB transcription factor binding the Fifth motif, and the transcription factors that interact with the other G564 suspensor control elements remain to be determined.
A model for G564 suspensor transcription
How do these five cis-elements activate transcription in the suspensor? Figure 6 illustrates a heuristic model for G564 suspensor transcription by the 47-bp module. This model assumes that the 10-bp, 10-bp-like and 10-bp-related motifs each bind to a transcription factor X because these motifs have been shown to be functionally the same (Kawashima et al. 2009). The Region 2 motif binds a different transcription factor Y. The Fifth motif binds a MYB transcription factor. A notable feature of the 47-bp suspensor control module is the close proximity of all five control elements. This suggests that three copies of transcription factor X, one transcription factor Y and one MYB transcription factor interact to form a compact complex that interacts with the basal transcriptional machinery to activate transcription in the suspensor shortly after fertilization (Fig. 6). In order to produce the suspensor-specific expression pattern of G564, at least one of these three types of transcription factors must be spatially localized, or active specifically, within the suspensor, and all three types of transcription factors must be present at least as early as the 4-cell embryo, because G564 is expressed in the two basal cells of the 4-cell embryo (Weterings et al. 2001). The presence of an intact suspensor control module in most of the 150-bp repeats upstream of the G564 gene might serve to enhance G564 transcription to a high level in the Scarlet Runner Bean giant suspensor, allowing G564 mRNA to accumulate to high levels (Fig. 5b-e) (Weterings et al. 2001).
Fig. 6.
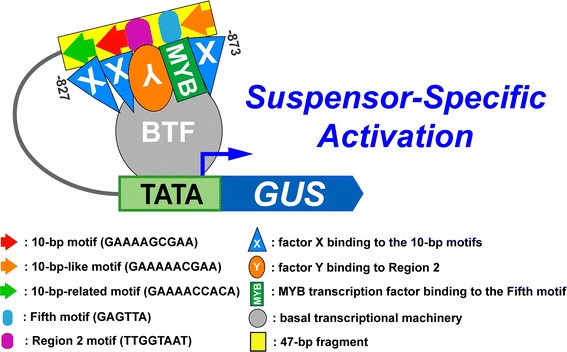
A G564 suspensor-specific cis-control module. Yellow box indicates the 47-bp suspensor cis-regulatory module (−873 to −827) from the fourth G564 150-bp repeat. Red, orange, and green arrows; purple ovals; and blue ovals indicate the 10-bp, 10-bp-like, and 10-bp-related motifs; Region 2 motif; and Fifth motif, respectively. An unknown transcription factor X (blue triangle) binds to the 10-bp, 10-bp-like and 10-bp-related motifs. An unknown transcription factor Y (orange oval) binds to the Region 2 motif. An unknown MYB-family transcription factor (green rectangle) binds to the Fifth motif. The X, Y and MYB transcription factors form a complex to recruit the basal transcriptional machinery that activates G564 transcription within the suspensor. BTF, basal transcriptional machinery; TATA, RNA polymerase II binding site
Conservation of the suspensor cis-regulatory elements in the upstream regions of other suspensor-active genes
Whether the G564 suspensor cis-regulatory elements form part of an extended suspensor-specific gene regulatory network (Peter and Davidson 2015) remains to be determined. A consensus sequence for each of the three types of motif was found in the upstream region of Scarlet Runner Bean genes, such as GA 20-oxidase and Wox9-like, which are highly expressed in the suspensor (Le et al. 2007; Le 2013; Henry 2014; Henry and Goldberg 2015). Recently, we showed that the G564 suspensor control motifs are also required for the region-specific transcription of the GA 20-oxidase gene (Henry 2014). Thus, the 10-bp/10-bp-like/10-bp-related, Region 2, and Fifth suspensor regulatory sequences are required for the spatially-restricted transcription of at least two Scarlet Runner Bean genes.
What about the functionality of these motifs in the promoters of suspensor-specific genes in other plants, such as Arabidopsis? Because the Scarlet Runner Bean G564 upstream region can program suspensor transcription in several plants, including Arabidopsis (Kawashima et al. 2009), we speculate that the G564 suspensor cis-regulatory elements program transcription of genes expressed in the suspensor across the plant kingdom. Although each of these suspensor motifs occur throughout the genome by chance, a cluster of all three types of motif within a few hundred base pairs of sequence is unlikely to occur randomly (Peter and Davidson 2015) and suggests a functional suspensor cis-regulatory module. As an example, a specific region of the Arabidopsis WOX8 promoter was shown to activate transcription shortly after fertilization in the zygote and the suspensor of a globular-stage embryo (Ueda et al. 2011), and we found the G564 suspensor control motifs in this promoter region. Further studies will be required to determine whether the suspensor cis-regulatory elements identified in G564 are also required to program transcription of WOX8 and other genes in plant suspensor regions.
Conclusion
To conclude, the data presented here, and elsewhere (Kawashima et al. 2009) identify a suspensor cis-regulatory module responsible for the developmental regulation of G564 expression. The suspensor module contains three types of cis-regulatory elements, one of which is required in three copies. Future studies are needed to identify other genes regulated by this module, and the identity of the transcription factors that bind to these cis-regulatory elements. With this knowledge, we can begin to build a gene regulatory network specific to the basal cell and suspensor lineage, which is activated shortly after fertilization.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1 Oligonucleotide sequences for generating G564 promoter constructs (PDF 63 kb)
Acknowledgments
We are grateful to the current and former members of our laboratory who have helped with different aspects of this project, as well as Professors Eric Davidson and John Harada for invaluable insights. This work was funded by grants from the National Science Foundation Plant Genome Program, United States Department of Energy, and United States Department of Agriculture. Kelli Henry was supported, in part, by a National Institutes of Health Pre-doctoral Traineeship. Tomokazu Kawashima was supported by a Nakajima Foundation Predoctoral Fellowship.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, Le BH, Drews GN, Brady SM, Goldberg RB, Harada JJ. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA. 2013;110(5):E435–E444. doi: 10.1073/pnas.1222061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua NH. Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 1990;9(6):1677–1684. doi: 10.1002/j.1460-2075.1990.tb08291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RA, Davidson EH. Flexibility of transcription factor target site position in conserved cis-regulatory modules. Dev Biol. 2009;336(1):122–135. doi: 10.1016/j.ydbio.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Salinas A, Bibler R, Lavin M. Phylogeny of the genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Syst Bot. 2006;31(4):779–791. doi: 10.1600/036364406779695960. [DOI] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119(1):71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266(5185):605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20(5):565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131(3):657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Henry KF. Identifying cis-regulatory elements and trans-acting factors that activate transcription in the suspensor of plant embryos. Los Angeles: University of California; 2014. [Google Scholar]
- Henry KF, Goldberg RB. Using giant scarlet runner bean embryos to uncover regulatory networks controlling suspensor gene activity. Front Plant Sci. 2015;6:44. doi: 10.3389/fpls.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry J, Hoffman N, Eichholtz D, Rogers S, Fraley R. A simple and general method for transferring genes into plants. Science. 1985;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Goldberg RB. The suspensor: not just suspending the embryo. Trends Plant Sci. 2010;15(1):23–30. doi: 10.1016/j.tplants.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Wang X, Henry KF, Bi Y, Weterings K, Goldberg RB. Identification of cis-regulatory sequences that activate transcription in the suspensor of plant embryos. Proc Natl Acad Sci USA. 2009;106(9):3627–3632. doi: 10.1073/pnas.0813276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2(12):1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol. 2005;54(4):575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Le BH. Using genome-wide approaches to dissect seed development. Los Angeles: University of California; 2013. [Google Scholar]
- Le BH, Wagmaister JA, Kawashima T, Bui AQ, Harada JJ, Goldberg RB. Using genomics to study legume seed development. Plant Physiol. 2007;144(2):562–574. doi: 10.1104/pp.107.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifanov AP, Makeev VJ, Nazina AG, Papatsenko DA. Homotypic regulatory clusters in Drosophila. Genome Res. 2003;13(4):579–588. doi: 10.1101/gr.668403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet: TIG. 1997;13(2):67–73. doi: 10.1016/S0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, Lim J, Shyr C, Tan G, Zhou M, Lenhard B, Sandelin A, Wasserman WW. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter I, Davidson EH. Genomic control process: development and evolution. London: Academic Press; 2015. [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, Torres-Torres M, Geffroy V, Moghaddam SM, Gao D, Abernathy B, Barry K, Blair M, Brick MA, Chovatia M, Gepts P, Goodstein DM, Gonzales M, Hellsten U, Hyten DL, Jia G, Kelly JD, Kudrna D, Lee R, Richard MM, Miklas PN, Osorno JM, Rodrigues J, Thareau V, Urrea CA, Wang M, Yu Y, Zhang M, Wing RA, Cregan PB, Rokhsar DS, Jackson SA. A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet. 2014;46(7):707–713. doi: 10.1038/ng.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slane D, Kong J, Berendzen KW, Kilian J, Henschen A, Kolb M, Schmid M, Harter K, Mayer U, De Smet I, Bayer M, Jurgens G. Cell type-specific transcriptome analysis in the early Arabidopsis thaliana embryo. Development. 2014;141(24):4831–4840. doi: 10.1242/dev.116459. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Ueda M, Zhang Z, Laux T (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20(2):264–270. doi:10.1016/j.devcel.2011.01.009 [DOI] [PubMed]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5(11):1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings K, Apuya NR, Bi Y, Fischer RL, Harada JJ, Goldberg RB. Regional localization of suspensor mRNAs during early embryo development. Plant Cell. 2001;13(11):2409–2425. doi: 10.1105/tpc.13.11.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung EC, Meinke DW. Embryogenesis in Angiosperms: development of the Suspensor. Plant Cell. 1993;5(10):1371–1381. doi: 10.1105/tpc.5.10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource 1 Oligonucleotide sequences for generating G564 promoter constructs (PDF 63 kb)


