ABSTRACT
Macrophages regulate tissue immunity, orchestrating the initiation and resolution of antimicrobial immune responses and repair of damaged tissue architecture. Their dysfunction can, thus, manifest in either pro- and anti-inflammatory responses. Indeed, despite the importance of macrophage function in health and disease, the role of tissue-resident macrophages in human immunodeficiency virus (HIV) disease progression remains incompletely defined. Here, we use flow cytometry to assess the phenotypes and functions of macrophages isolated from the spleens, axillary lymph nodes, colons, jejuna, and livers of healthy and chronically simian immunodeficiency virus (SIV)-infected Asian macaques, the prominent nonhuman primate model for HIV infection. Our data demonstrate that macrophages from healthy animals exhibit considerable phenotypic and functional heterogeneity across tissues and across a variety of stimuli. Further, our analysis reveals changes in the lipopolysaccharide (LPS) responsiveness of macrophages isolated from SIV-infected animals. We anticipate that our findings will inform future research into macrophage-directed immunity across a variety of primate diseases.
IMPORTANCE These findings highlight the functional and phenotypic heterogeneity of tissue macrophages in different anatomic sites and as a result of SIV infection. We believe that our data will lead to novel therapeutic interventions aimed at altering the proinflammatory capacity of tissue macrophages in progressively HIV-infected individuals.
INTRODUCTION
Nonhuman primates (NHPs) serve as an ultimate model for studying some human diseases and development. NHPs share upwards of 90% genetic similarity to humans, exhibit a largely parallel developmental program, and mirror human anatomy and anatomic process (1). The zoonotic transmission of pathogens from NHPs to humans remains a significant concern and has been documented to include herpes simian B virus, Marburg virus, and simian immunodeficiency virus (SIV), the ancestral progenitor of human immunodeficiency virus (HIV). Despite the many evolutionarily conserved similarities between human and divergent NHP species, differences in disease outcome are apparent. Importantly, HIV type 1 (HIV-1) infection in humans and SIV infection in Asian macaques result in similar, severe disease progressions, while SIV infection in NHPs of African origin results in a nonpathogenic course of disease (2). Given the limited divergence exhibited between primate species, a complete dissection of immunological processes in NHP species will yield critical information regarding potential mechanisms of disease progression and remission in human patients.
SIV infection in Asian macaques and HIV infection of humans are characterized by persistent immune activation that is attributable, in part, to damage to the gastrointestinal barrier and subsequent microbial translocation (3, 4). The gastrointestinal lumen is estimated to contain 1014 commensal and potentially pathogenic microbes that are kept separate from the underlying tissue and systemic circulation by a physical barrier consisting of epithelial cells and mucus as well as an immunologic barrier, comprised of antimicrobial peptides, antibodies, professional phagocytic cells, and a host of specialized innate and adaptive immune cells (5). During acute, progressive HIV and SIV infections, replication among intestinal CD4+ T cells initiates a pronounced inflammatory state that damages the integrity of the epithelial and immunological barriers and promotes the persistent recruitment and depletion of intestinal CD4+ T cells and in particular, T helper 17 (TH17) cells (3). The translocation of luminal microflora products across the compromised gastrointestinal barrier and into systemic circulation exacerbates inflammation and contributes to continued dysfunction of the host immune system (4, 6). Aberrations in immunological responses are not limited to a loss of CD4+ T cells and extend to inappropriate lymphocyte priming, proliferation, and functional exhaustion and dysfunctional antigen presentation and cytokine secretion by dendritic cells (DCs) (7, 8). Despite the well-documented roles of macrophages in mitigating the translocation of microbial products and in orchestrating appropriate immune responses, a comprehensive understanding of the contributions of tissue-resident macrophages to HIV and SIV disease progression is lacking, with some studies suggesting functional abnormalities within myeloid cells of HIV-infected humans or SIV-infected macaques (9–15).
Accumulating evidence in humans and murine models suggest that macrophages exhibit contextual, functional plasticity. Indeed, intestinal macrophages orchestrate differential T cell immunity, depending on the bacterial antigen, with TH17 cells, TH1 cells, or regulatory T cells (Treg cells) capable of being differentially induced (16–19). Macrophages further exhibit heterogeneity by anatomic location (20). Whereas intestinal macrophages are reported to direct a largely anti-inflammatory program in response to the presence of commensal microflora, the systemic dissemination of microflora promotes an inflammatory response by marginal zone splenic macrophages and an anti-inflammatory response by Kupffer cells (21, 22). Hence, the ability of macrophages to discriminate between pathogenic and commensal species is essential for the development of appropriate immune responses in health and disease.
Given the importance of macrophages in regulating immune processes in health and disease, a systematic assessment of macrophages in NHP health and SIV infection may provide useful insight into their contribution to HIV disease progression in humans. As such, here we identified tissue macrophages by their phagocytic properties directly ex vivo and characterized their phenotype and proinflammatory potential. Our analysis reveals that NHP macrophages exhibit extensive heterogeneity in phenotype and proinflammatory capacity by anatomic location and that SIV infection is associated with alterations of macrophage functionality in response to lipopolysaccharide (LPS) stimulation.
MATERIALS AND METHODS
Animals and tissue processing.
Single-cell suspensions of tissues from 9 uninfected rhesus macaques (RM) (Macaca mulatta) and 14 chronically SIV-infected RM were accessed for this study (Table 1). Single-cell suspensions of cells from the axillary lymph node (ALN), colon, jejunum, liver, and spleen were generated and frozen as previously described (23). SIV-infected animals were infected with SIVmac239, SIVsmE543, or SIVsmE660. Animals were housed and cared for in accordance with standards of the American Association for Accreditation of Laboratory Animal Care (AAALAC) in AAALAC-accredited facilities, and all animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committees of NIH under ASP LMM12. Animals were sacrificed according to the recommendations set forth by the American Veterinary Medical Association Guidelines on Euthanasia.
TABLE 1.
Characteristics of animals used in this study
| Animal | SIV infection status | No. of CD4+ T cells/μl | Plasma viral load (no. of copies of vRNA/ml)a | Presence/absenceb of virus in the following tissue of the animal: |
||||
|---|---|---|---|---|---|---|---|---|
| ALN | Colon | Jejunum | Liver | Spleen | ||||
| Rh4016 | Uninfected | 613 | N/A | N | Y | Y | Y | Y |
| Rh495 | Uninfected | 574 | N/A | Y | Y | Y | N | Y |
| Rh595 | Uninfected | 685 | N/A | Y | Y | Y | N | Y |
| Rh769 | Uninfected | 465 | N/A | Y | Y | Y | N | Y |
| RhDB7H | Uninfected | 526 | N/A | Y | N | N | N | Y |
| RhDB9z | Uninfected | 2,623 | N/A | Y | Y | Y | N | Y |
| RhDBM6 | Uninfected | 692 | N/A | Y | Y | Y | N | Y |
| RhDBv1 | Uninfected | 693 | N/A | Y | Y | Y | Y | Y |
| RhDBxG | Uninfected | 696 | N/A | Y | Y | Y | Y | Y |
| RhDB07 | Chronic SIVmac239 | 317 | 500,000 | Y | Y | Y | N | Y |
| RhDB4e | Chronic SIVmac239 | 565 | 810,000 | Y | Y | Y | Y | Y |
| RhDB92 | Chronic SIVmac239 | 256 | 100,000 | Y | Y | Y | N | Y |
| Rh591 | Chronic SIVsmE543 | 186 | 251,000 | Y | N | Y | N | Y |
| Rh759 | Chronic SIVsmE543 | 526 | 561 | Y | Y | Y | N | Y |
| Rh760 | Chronic SIVsmE543 | 296 | 5,000 | Y | Y | Y | N | Y |
| Rh764 | Chronic SIVsmE543 | 134 | 222,000 | Y | N | Y | N | Y |
| Rh766 | Chronic SIVsmE543 | 927 | 50,000 | Y | Y | Y | N | Y |
| Rh767 | Chronic SIVsmE543 | 163 | 158,000 | Y | Y | Y | N | Y |
| Rh768 | Chronic SIVsmE543 | 265 | 113,000 | Y | Y | Y | N | Y |
| RhCF4j | AIDS SIVmac239 | 241 | 200,000 | Y | N | N | Y | Y |
| RhDB17 | AIDS SIVmac239 | 122 | 92,000 | Y | Y | Y | Y | Y |
| Rh831 | AIDS SIVsmE543-3 | 14 | 752,421 | N | N | N | Y | N |
| Rh809 | AIDS SIVsmE660-FL6 | 20 | 10,348 | N | N | N | Y | N |
Abbreviations: vRNA, viral RNA; N/A, not available.
Use of tissue from each animal is indicated as follows: Y for yes and N for No.
Stimulation and phagocytosis assay.
Single-cell tissue suspensions were thawed into heat-inactivated fetal bovine serum (FBS) (HyClone) containing 10 U/ml DNase I (Roche) and rested for 5 h at 2 × 106 cells/ml in 10% Dulbecco modified Eagle medium (DMEM) (10% fetal calf serum [FCS], 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, and 1.7 mM sodium glutamate) containing 8 U/ml DNase. From liver homogenates, suspensions were spun through a continuous Percoll gradient (53.8% Percoll in 2× phosphate-buffered saline [PBS] allowed to settle overnight), and debris was discarded prior to rest. Liver preparations were spun through a Percoll gradient to remove the significant amount of debris and hepatic cells. Rested cells were resuspended at 2 × 106 cells/ml in 1% DMEM, with or without 107 fluorescent microspheres/ml (Invitrogen FluoSpheres), alone or stimulated with 1 μg/ml LPS, 100 μg/ml Bacillus subtilis peptidoglycan (BS-PGN), or 100 μg/ml Escherichia coli (0111:B4) peptidoglycan (EC-PGN) (Invivogen) at 37°C. After 1 h, all cells were treated with 10 μg/ml brefeldin A and left to incubate for an additional 15 h.
Flow cytometry.
Polychromatic flow cytometry was preformed utilizing a BD LSRFortessa cell analyzer equipped with fluorescence-activated cell sorting (FACS) DiVA software (version 6.1.3; BD). Antibodies against the following targets were used at predetermined concentrations: CD3 (clone SP34-2) APCCy7 (APC stands for allophycocyanin), CD8 (SK1) APCH7, CD11b (ICRF44) Brilliant Violet 605, CD20 (2H7) APCH7, CD45 (D058-1283) phycoerythrin-labeled CF594 (PE-CF594), CD68 (Y1/82A) Alexa Fluor 647, CD206 (19.2) PECy5, and interleukin-1β (IL-1β) (AS10) PE from BD; CD11c (3.9) PerCPe710 (PerCP stands for peridinin chlorophyll protein), IL-6 (MQ2-13A5) Alexa Fluor 700, and tumor necrosis factor alpha (TNF-α) (MAb11) PECy7 from eBioscience; and CD14 (M5E2) Brilliant Violet 650 and HLADR (L243) Brilliant Violet 711 from Biolegend. Cells were stained in the presence of anti-rat serum (eBioscience). Cell viability was assessed using the Live/Dead Aqua Fixable Dead Cell Stain (Invitrogen). Cells were permeabilized with Cytofix/Cytoperm (BD) prior to intracellular staining for cytokines and CD68. The data acquired were analyzed using FlowJo software (v9.6.4; TreeStar). For all analyses, we used a threshold cutoff of 100 defined, gated macrophages.
Statistics.
Statistical analyses for single surface marker or cytokine comparisons were performed using Prism (v6.0; GraphPad Software Inc.). Two-tailed, paired t tests were used to assess differences in frequency of cytokine expression in response between stimuli. Multiple t tests (fewer assumptions) were used to compare the frequency of cytokine expression between tissues and similar stimuli. Mann-Whitney U tests were used to compare cytokine responses across SIV status, with one-tailed tests used to compare macrophage frequency and cytokine expression, and two-tailed tests used to analyze surface marker expression. Statistical differences between the distribution of all surface markers or cytokines (as represented by pie charts) were assessed using the SPICE (v5.35; National Institute of Allergy and Infectious Diseases) permutation test (24). Averaged data are presented as arithmetic means. A P value of less than 0.05 was considered significant.
RESULTS
Macrophage identification.
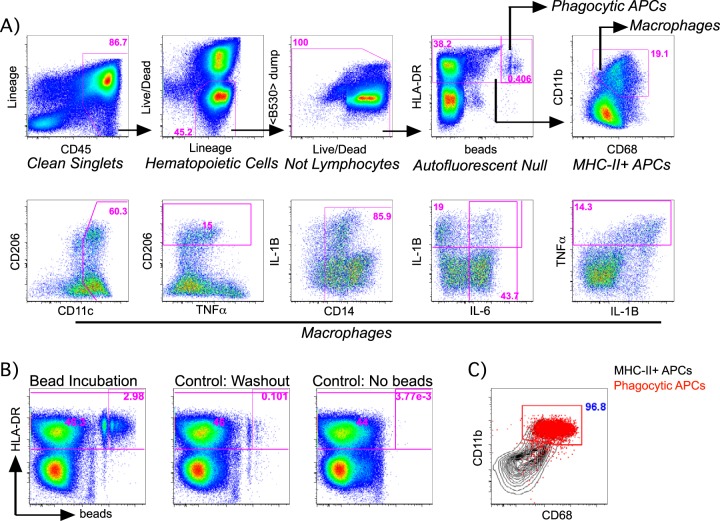
The characterization of NHP macrophages by flow cytometry has been restricted by the availability of cross-reactive reagents and is complicated both by the autofluorescence inherent to macrophages as well as the nonspecific capture of antibodies resulting from their high Fc receptor expression (25, 26). In order to identify macrophages from NHPs, we first incubated single-cell suspensions of RM tissue homogenates with fluorescent microspheres to demarcate specifically phagocytic cells (Fig. 1). We subsequently stained these cells with fluorescently conjugated antibodies against previously described human or murine macrophage markers in the presence of rat serum to limit nonspecific, Fc-mediated antibody capture. By this method, phagocytic cells were found almost exclusively among cells expressing major histocompatibility complex class II (MHC-II) (HLA-DR) (Fig. 1A and B) and were found to coexpress moderate to high levels of CD68 and CD11b (integrin αM) which in combination, have been previously identified as macrophage markers (Fig. 1B and C) (20, 27). Although defining macrophages as HLADR+ CD11b+ CD68+ leukocytes may not encompass all macrophages, the use of these markers captured most phagocytic cells in our cultures and is thus useful in the identification of NHP macrophages. Using this method, we assessed the reactivity of several antibodies and designed a panel and gating strategy for the purpose of characterizing the phenotype and function of NHP tissue macrophages (Fig. 1A).
FIG 1.
Representative gating strategy for macrophage identification and characterization. (A) RM macrophages were defined as single cells that did not exhibit evidence of nonspecific binding (clean singlets) or autofluorescence in the B530nm channel (<B530> dump) and were further characterized for the expression of a panel of surface markers and cytokines as shown. Frequencies of the gated populations relative to that of the shown population are in magenta. (B and C) Markers for RM macrophage identification and characterization were identified from among MHC-II+ antigen-presenting cells (APCs) that exhibited evidence of specific fluorescent-bead incorporation compared to controls (B), which revealed that the coexpression of CD11b and CD68 specifically identified these phagocytic cells (red) from among all MHC-II+ APCs (black) (C). (B) Frequencies of phagocytic APCs relative to that of the shown population are in magenta. (C) The frequency of phagocytic APCs characterized as CD11b+ CD68+ is in blue.
Macrophages exhibit extensive phenotypic and functional heterogeneity.
To evaluate the contributions of tissue macrophages to NHP immunological processes, we began by evaluating the distribution of macrophages in RM axillary lymph nodes (ALNs), colons, jejuna, livers, and spleens from SIV-uninfected and SIV-infected animals (Table 1). Irrespective of SIV infection status, the highest frequencies of macrophages (among leukocytes) were found in the colon and liver, while lower frequencies were observed within the ALN, jejunum, and spleen (Fig. 2A). Compared to SIV-uninfected animals, SIV-infected animals exhibited similar or higher macrophage frequencies with significant differences noted in the ALN (P = 0.0224 by one-tailed Mann-Whitney U test) and colon (P = 0.0231).
FIG 2.
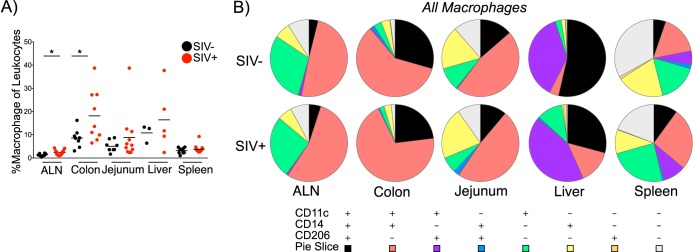
Macrophages exhibit considerable phenotypic heterogeneity by anatomic site. (A) Percent macrophages of leukocytes in the ALN, colon, jejunum, liver, and spleen of SIV-uninfected (SIV−) and SIV-infected (SIV+) RM. Each symbol represents the value for an individual animal. The short line represents the mean for the group of animals. The statistical significance of differences in animals across SIV status was assessed by a one-tailed, Mann-Whitney test. Values that were significantly different (P < 0.05) are indicated by a bar and asterisk. (B) Pie charts depict the relative expression of CD11c, CD14, and/or CD206 by macrophages in the indicated tissues in SIV-uninfected and -infected macaques. The significance of profile differences was assessed by the SPICE software permutation test.
We further evaluated the phenotypes of tissue macrophages in SIV-uninfected and -infected animals by analyzing the expression of CD11c (integrin αX), CD14 (LPS coreceptor), and CD206 (mannose receptor C type I). Although the interpretation of the use of these markers in myeloid cell identification and characterization remains inconclusive in primate and murine studies, the initial use of these markers may provide useful insight into the distribution and function of myeloid cells in NHP health and in progressive SIV infection (20, 27). Interestingly, although among splenic macrophages, CD11c was the only marker to be differentially expressed by SIV status (P = 0.0093 by two-tailed Mann-Whitney U test), extensive heterogeneity was evident across tissues (Fig. 2B; see Fig. S1 in the supplemental material). For example, while the majority of liver macrophages expressed CD206 with or without CD14 expression, the majority of intestinal macrophages expressed CD14 with variable CD206 expression. Of the tissues examined, the spleen was unique in its absence of one or two predominant phenotypes and in having a notable proportion of macrophages that expressed none of the examined markers. The heterogeneity observed across tissues corroborates previous findings in human and murine macrophage studies and likely reflects the unique environments encountered by macrophages in each tissue (20).
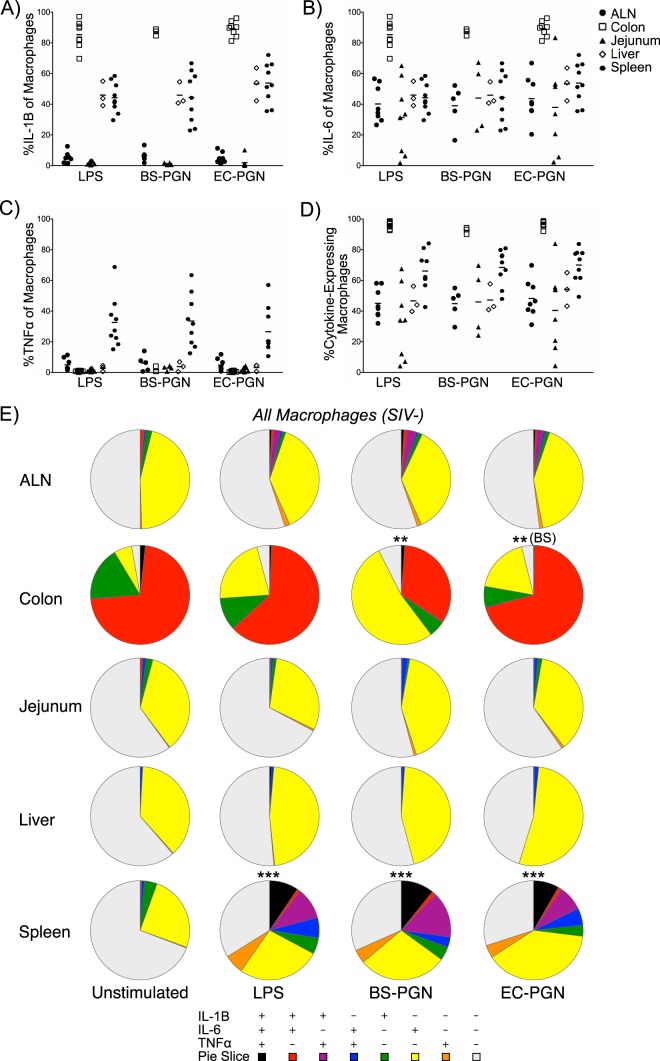
We next sought to determine whether the phenotypic heterogeneity was accompanied by macrophage functional heterogeneity in SIV-uninfected animals. Given the central role of macrophages in shaping adaptive immune responses subsequent to commensal or pathogenic microbe sensing (20), we assessed the ability of tissue macrophages to respond to bacterial stimuli. Specifically, we measured IL-1β, IL-6, and TNF-α expression either directly ex vivo or in response to LPS or peptidoglycan (PGN). While LPS classically induces a proinflammatory Toll-like receptor 4 (TLR4) response, PGN-induced inflammatory TLR2 responses can be confounded by pathogen versus commensal PGN sensing at the level of intracellular NOD2 (28–30). As such, we included PGN from pathogenic Escherichia coli (EC-PGN) and PGN from commensal-like Bacillus subtilis (BS-PGN) (31). At the level of individual cytokine expression (monofunctionality), many differences were noted across distal macrophages and different stimulations (Fig. 3A to D; see tables in the supplemental material). Of interest, mean IL-1β (Fig. 3A; see Table S1A in the supplemental material) and IL-6 (Fig. 3B and Table S2A) expression were significantly higher in colonic macrophages than in all other tissue macrophages with similar simulation conditions, although splenic macrophages were unique in modulating the expression of these cytokines across varied stimuli (Fig. 3A and B and Tables S1B and S2B). Moderate TNF-α expression was observed only among splenic macrophages in response stimulation and at significantly higher levels than in all other tissue macrophages in similar stimulation conditions (Fig. 3C and Table S3A). We additionally compared the ability of tissue macrophages to express any cytokine (IL-6 or IL-1β or TNF-α) to assess potential macrophage anergy. Among similar stimulation conditions, significant differences in function were noted between splenic and colonic macrophages and almost all other tissue macrophages (Fig. 3D and Table S4A), and again, differences between stimulus conditions were observed only among splenic macrophages (Fig. 3D and Table S4B). The high level of spontaneous cytokine expression among colonic macrophages could be attributed to the high degree of bacterial products within the lumen of the colon present while processing the tissue. Collectively, these results suggest that NHP tissue macrophages express both phenotypic and functional heterogeneity.
FIG 3.
Macrophage function varies by anatomic site and stimulation condition. (A to D) Absolute percentage of stimulated tissue macrophages from SIV-uninfected macaques expressing IL-1β (A), IL-6 (B), TNF-α (C), or any cytokine (D). Each symbol represents the value for an individual animal. The short line represents the mean for the group of animals. (E) Pie charts depict the relative expression of IL-1β, IL-6, and/or TNF-α by macrophages from the indicated tissues of SIV-uninfected macaques ex vivo (unstimulated) or in response to stimulation with LPS, BS-PGN, or EC-PGN. Asterisks represent statistical significance compared to the value for unstimulated samples for that tissue unless noted otherwise in parentheses. The significance of profile differences was assessed by the SPICE software permutation test.
The individual differences in cytokine secretion described led us to examine the composite functional profile of NHP tissue macrophages. This profiling revealed several interesting findings. ALN, jejunal, and liver macrophages were overwhelmingly nonfunctional (by our measures) or IL-6 monofunctional, with a small percentage of macrophages expressing other or additional cytokines (Fig. 3E; see Fig. S2 in the supplemental material). In these tissues, stimulated macrophages exhibited no significant differences in functionality compared to each other or compared to unstimulated controls, suggesting refractoriness to the particular stimuli applied. In stark contrast to these tissues, few colonic macrophages were nonfunctional even in the absence of stimulation, with a large proportion of cells expressing IL-6 and/or IL-1β. Among stimulated macrophages, only the BS-PGN-stimulated macrophages exhibited a significant difference in profile compared to the unstimulated controls (P = 0.0040 by the SPICE permutation test) and compared to EC-PGN-stimulated controls (P = 0.0052). Among splenic macrophages, unstimulated cells were largely nonfunctional; however, significant differences in functional profiles were noted between unstimulated controls and all stimulated populations (all P < 0.0001). Unlike the responses reported for the other tissues, responding splenic macrophages contained a significant proportion of TNF-α-expressing cells as already described above (Fig. 3, Fig. S2, and Table S3). The profiles described by these analyses are likely reflective of the unique environments and functional requirements of tissue macrophages.
Functional heterogeneity is reflective of anatomic origin.
We examined whether the observed phenotypic differences in tissue macrophages are associated with functional heterogeneity. To assess the relationship between phenotype and function in tissue macrophages, we characterized the functional profile of the most abundant subsets of macrophages in the colon and liver, CD11c+ CD14+ CD206− and CD11c+ CD14− CD206+ macrophages, respectively. Our subset analysis revealed that, irrespective of phenotype, the functional profiles of macrophages from the same anatomic source were more similar to each other than to those of macrophages isolated from a different tissue (Fig. 4; see Fig. S3 in the supplemental material). However, subsets from within the same tissue were not necessarily identical. Although both subsets of liver macrophages behaved similarly—with comparable proportions of nonfunctional and IL-6 monofunctional cells across all stimuli—colonic macrophages exhibited intricacies in their response to stimuli. Compared to unstimulated controls, both examined subsets of BS-PGN-stimulated colonic macrophages responded with a loss of IL-1β cytokine secretion (P = 0.0062 for CD206− macrophages and P = 0.0059 for CD206+ macrophages), resulting in predominantly IL-6 monofunctional subsets. Although the EC-PGN-stimulated CD11c+ CD14+ CD206− colonic macrophage subset similarly responded with IL-1β downregulation compared to unstimulated controls (P = 0.0087), the CD11c+ CD14− CD206+ subset did not exhibit a similar downregulation, instead showing no difference from the unstimulated control. However, both LPS-stimulated (P = 0.0237) and EC-PGN-stimulated (P = 0.0436) CD11c+ CD14− CD206+ macrophages differed significantly from matched-subset BS-PGN-stimulated macrophages. These results indicate that although tissue macrophage function is reflective of anatomic source rather than the phenotypic identity per se, functional intricacies exist between phenotypically distinct macrophages isolated from the same tissue.
FIG 4.
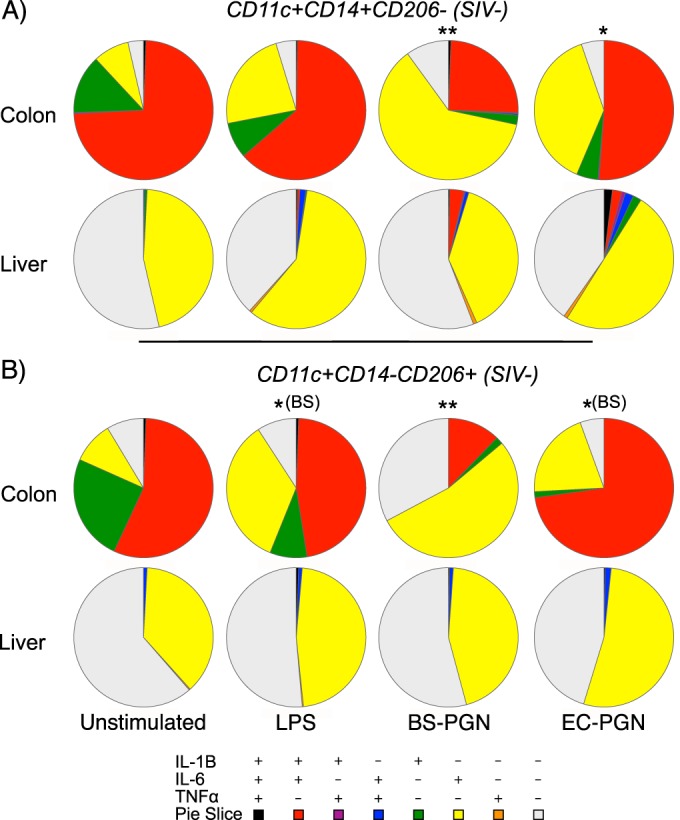
Macrophage phenotype does not dictate functionality. (A and B) Pie charts depicting the relative expression of IL-1β, IL-6, and/or TNF-α in CD11c+ CD14+ CD206− (A) or CD11c+ CD14− CD206− (B) macrophages isolated from the colons or livers of SIV-uninfected RM ex vivo or in response to stimulation with LPS, BS-PGN, or EC-PGN. Asterisks represent statistical significance compared to the value for unstimulated samples for that tissue unless noted otherwise in parentheses. The significance of profile differences was assessed by the SPICE software permutation test.
Phagocytic macrophages display unique functional properties.
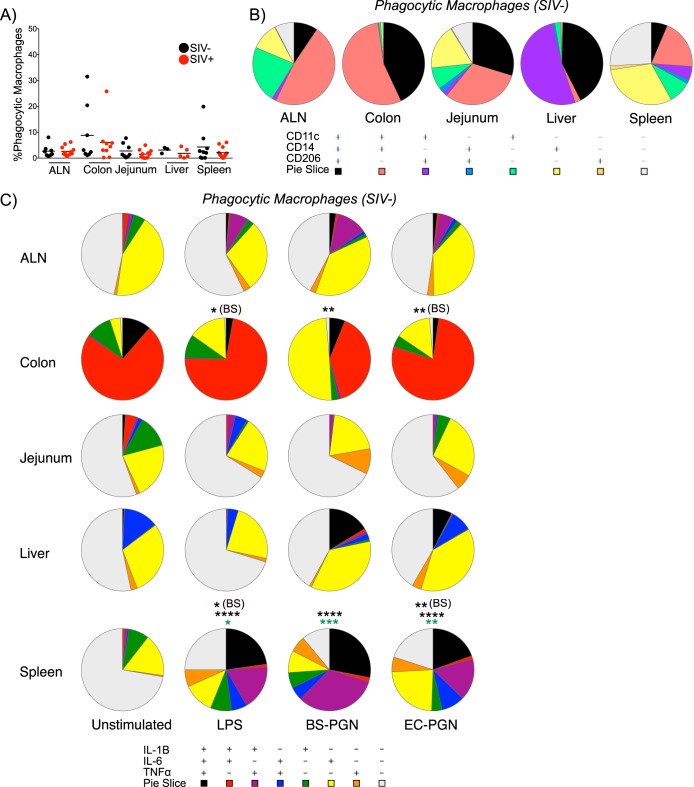
Phagocytosis is essential for the direct ability of macrophages to clear potential pathogens as well as the ability of macrophages to orchestrate antimicrobial responses through cytokine secretion (25, 32, 33). To determine whether differences existed in the frequency, phenotype, and function of phagocytic macrophages by tissue origin, we measured incorporation of fluorescent microspheres as described in the legend to Fig. 1. The mean phagocytic frequency was similar across all tissues as assessed in this manner, and no significant differences were noted between tissues or by SIV infection status, consistent with previous findings (Fig. 5A) (34).
FIG 5.
Phagocytic macrophages are highly responsive to stimuli ex vivo. (A) Percent phagocytic macrophages in the indicated tissues from SIV-uninfected and SIV-infected RM. Each symbol represents the value for an individual animal. The short line represents the mean for the group of animals. The significance of differences across SIV status was assessed by a one-tailed Mann-Whitney test. (B) Pie charts depict the relative expression of CD11c, CD14, and/or CD206 by phagocytic macrophages in the indicated tissues of SIV-uninfected macaques. (C) Pie charts depict the relative expression of IL-1β, IL-6, and/or TNF-α by phagocytic macrophages from the indicated tissues of SIV-uninfected macaques ex vivo or in response to stimulation with LPS, BS-PGN, or EC-PGN. Black asterisks represent significance compared to the value for unstimulated samples for that tissue unless noted otherwise in parentheses. Green asterisks represent significance compared to the value for nonphagocytic macrophages. The significance of profile differences was assessed by the SPICE software permutation test.
We next assessed the phenotypic signature of phagocytic tissue macrophages in uninfected RM. Phagocytic macrophages resembled the bulk population of macrophages within each tissue, with a significant difference noted only in EC-stimulated colonic macrophages compared to paired bulk macrophages (P = 0.0373; Fig. 2B and 5B and Fig. S4A in the supplemental material). Here again, ALN, colonic, and liver macrophages exhibited the presence of one or two dominant phenotype subsets, while the jejunal and splenic macrophages exhibited a greater degree of tissue heterogeneity. These results indicate that phagocytic macrophages do not exhibit a unique phenotypic signature with regard to the markers examined here.
In addition to comparing the phenotypes of phagocytic macrophages to those of the bulk macrophage population for each tissue, we examined whether the functional properties of phagocytic macrophages might differ with anatomic location. Functional profiling revealed that while phagocytic macrophages largely resemble paired bulk macrophages with respect to cytokine secretion, phagocytic macrophages exhibit a greater propensity for polyfunctionality compared to the broader macrophage population, particularly with regard to TNF-α expression (Fig. 3E and 5C; see Fig. S4B in the supplemental material). Among splenic macrophages specifically, a significant difference was noted in the functional profile of phagocytosing versus nonphagocytosing macrophages (P = 0.0251 for LPS, P < 0.0001 for BS-PGN, and P = 0.0125 for EC-PGN). Among phagocytic macrophages, colonic and splenic macrophages were unique in having the functional profile of at least one stimulated group differ significantly from that of the unstimulated controls. Furthermore, LPS- and EC-PGN-stimulated phagocytic macrophages from both the colon (P = 0.0254 and P = 0.0067, respectively) and spleen (P = 0.0368 and P = 0.0017, respectively) differed significantly from BS-PGN-stimulated phagocytic macrophages. These data suggest that phagocytic macrophages, though similar in phenotype and function to bulk macrophages, exhibit unique functional responses in response to stimuli in certain anatomic sites, namely, the colon and spleen.
SIV infection promotes subtle changes in macrophage functionality.
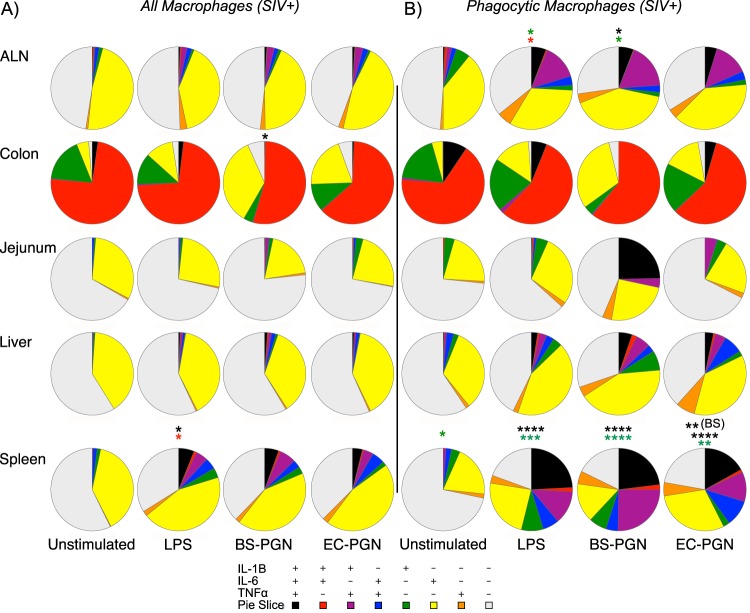
We next wanted to assess potential contributions of abnormalities within macrophages to SIV disease progression. Although previous publications have addressed individual aspects of macrophage function during SIV infection, comprehensive analyses are limited and conflicting (9). To investigate the contributions of macrophages to disease progression during pathogenic SIV infection, we profiled the phenotype and function of macrophages as for macrophages isolated from SIV-uninfected animals. As described above, although SIV infection was characterized by a significantly higher frequency of ALN and colonic macrophages compared to uninfected controls, no significant differences were noted by infection status with regard to phenotype (Fig. 2A). Among tissue macrophages from SIV-infected animals and similar to macrophages from uninfected animals, differences in function compared to unstimulated controls were limited to colonic and splenic macrophages (Fig. 6A; see Fig. S5A in the supplemental material). Among colonic macrophages, BS-PGN-stimulated macrophages differed in functionality compared to unstimulated controls (P = 0.0183), while LPS-stimulated splenic macrophages differed from unstimulated controls (P = 0.0201). Interestingly, of all the tissues and conditions examined, only LPS-stimulated splenic macrophages showed a significant difference in functional profile by SIV infection status (P = 0.0408). This was characterized by an expansion in the relative frequency of IL-6 monofunctional cells, and indeed, a significant difference was noted by infection status in the mean percentage of IL-6 expression (P = 0.0497 by one-tailed Mann-Whitney U test).
FIG 6.
SIV infection is associated with subtle changes in macrophage function. (A and B) Pie charts depict the relative expression of IL-1β, IL-6, and/or TNF-α by bulk macrophages (A) or phagocytic macrophages (B) from the indicated tissues of SIV-infected macaques ex vivo or in response to stimulation with LPS, BS-PGN, or EC-PGN. Black asterisks represent statistical significance compared to the value for unstimulated samples for that tissue unless noted otherwise in parentheses. Green asterisks represent significance compared to the value for matched nonphagocytic macrophages. Red asterisks represent significance compared to the value for the same sample from SIV-uninfected macaques. The significance of profile differences was assessed by the SPICE software permutation test.
In addition to examining the bulk macrophage population, we investigated potential SIV infection-associated differences in the phenotype and function of phagocytic macrophages. No significant differences by SIV infection status were observed in the frequency or phenotype of phagocytic macrophages (Fig. 5 and data not shown). Among phagocytic macrophages from SIV-infected animals, all stimulated phagocytic splenic macrophages differed significantly in function from unstimulated controls (all P < 0.0001) with EC-PGN-stimulated macrophages additionally differing significantly from BS-PGN-stimulated macrophages (P = 0.0103; Fig. 6B and Fig. S5B in the supplemental material). Phagocytic splenic macrophages also differed significantly in profile from matched nonphagocytic macrophages both before stimulation (P = 0.0437) and after stimulation (P = 0.0007 for LPS, P < 0.0001 for BS-PGN, and P = 0.0040 for EC-PGN). Unlike macrophages isolated from uninfected animals, no significant differences were noted between stimulated and unstimulated phagocytic, colonic macrophages from infected animals, and no significant differences were noted in colonic macrophages by SIV infection status. Interestingly, the only differences noted among ALN macrophages in this study were noted in regard to phagocytic macrophages isolated from SIV-infected animals. BS-PGN-stimulated ALN phagocytic macrophages exhibited a significantly different functional profile (P = 0.0229) compared to that of unstimulated controls, characterized predominantly by an expansion in IL-6+ IL-1β+ dual functional responses, and compared to matched nonphagocytic macrophages (P = 0.0151), largely characterized by greater overall function. Furthermore, the LPS-specific responses by phagocytic ALN macrophages differed significantly from nonphagocytic macrophages (P = 0.0142) as driven by greater overall function, and a significant difference was additionally noted in LPS responsiveness between phagocytic ALN macrophages from SIV-infected and -uninfected animals (P = 0.0424), as driven by an expansion of the IL-6+ IL-1β+ response. Collectively, these results indicate that although changes in bulk and phagocytic macrophage phenotype do not differ significantly by SIV status, tissue-specific functional changes in LPS responsiveness are apparent among tissue macrophages from SIV-infected animals.
DISCUSSION
Macrophages are multifaceted regulators of tissue immunity, orchestrating the initiation and resolution of antimicrobial immune responses and the repair of tissue integrity. Given the myriad of responses orchestrated by macrophages, dysfunction can manifest across a spectrum of pro- and anti-inflammatory responses. Despite the known importance of macrophage function to health and disease, the role of macrophages in HIV disease progression remains incompletely defined. Here, we assessed macrophage phenotype and function by flow cytometry in healthy and SIV-infected macaques, a NHP model for HIV infection. Our data demonstrate that macrophages from healthy animals exhibit considerable phenotypic and functional heterogeneity across tissues and across a variety of stimuli. Furthermore, our analysis revealed LPS-specific changes in the function of macrophages isolated from SIV-infected animals. We anticipate that our findings will inform future research into macrophage-directed immunity across a variety of NHP disease models.
Considerable discrepancies exist in the phenotypic characterization of murine and human macrophages, with differences observed across species, anatomic sites, and disease models, which are further confounded by conflicting reports regarding the delineation of myeloid cell subsets (20, 27). For example, although CD14 expression has historically been used to discriminate recently immigrated tissue monocytes from resident tissue macrophages, both resident and inflammatory CD14+ cells otherwise identical to macrophages have been identified in murine and human intestinal tissues (30, 35, 36). To inform the identification of macrophages in NHPs, we identified macrophages as MHC-II+ CD11b+ CD68+ tissue cells and further characterized macrophages based on the expression of CD11c, CD14, and CD206 (Fig. 1). Within our healthy controls, we expected tissue-isolated MHC-II+ CD11b+ CD68+ cells to consist predominantly of resident macrophages, as the accumulation of recent monocyte immigrants is minimal in the absence of proinflammatory signals (30, 36–38). Although our inclusion criteria likely captured some DCs, the expression of CD14 by a majority of our defined macrophages argues against the inclusion of resting DCs in these noninflamed tissues (Fig. 2) (30, 36, 39, 40). Our analysis revealed extensive macrophage heterogeneity across tissues, which is likely reflective of the unique environments encountered at each anatomic site. While the profound diversity of macrophage subsets within the ALN and spleen may reflect the need of these tissues' cellular constituents to prime immune responses to any number of systemic pathogens, the limited and unique diversity of macrophages within the liver and colon may reflect the predominantly immunoregulatory program directed by innate cells at these sites (20). Intriguingly, we observed that colonic and jejunal macrophages exhibit site-specific phenotypic and functional signatures, which may further reflect the unique metabolites and microflora encountered at each intestinal site (Fig. 2 and 3) (41, 42). The evident diversity in NHP tissue macrophage phenotype indicates that the identification of macrophages based on a restrictive subset of surface markers may be insufficient to capture populations responsible for disease progression and remission.
Functional heterogeneity between leukocytes from distinct anatomic sites has been well described and contributes to the magnitude and balance of immunoregulation within each compartment (20). Thus, while intestinal tissues persistently encountering microflora maintain and balance a high frequency of effector and regulatory lymphocytes, such as CD4+ TH17 and Treg cells, the liver maintains only a small cadre of resident lymphocytes, which are largely directed toward a regulatory phenotype (5, 22). Our assessment of NHP tissue macrophages is in line with this heterogeneity and reveals that while the functional profiles of ALN, jejunal, and hepatic macrophages are similar with regard to the cytokines measured, colonic and splenic macrophages exhibit unique and more-varied responses (Fig. 3). Splenic macrophages alone exhibited a canonical proinflammatory response to TLR stimuli—with approximately two-thirds of cells expressing no cytokine prior to stimulation and a similar frequency expressing a host of traditionally inflammatory cytokines after stimulation. Interestingly, in all tissues and irrespective of stimulation, we observed a notable frequency of IL-6-expressing macrophages, with or without additional cytokine secretion. Although largely considered proinflammatory, IL-6 exerts several essential anti-inflammatory and regulatory functions, such as promoting epithelial cell regeneration, inhibiting epithelial cell and T cell apoptosis, and promoting the recruitment and maturation of mononuclear cells and TH17 cells at mucosal barrier sites (43). These and the traditional proinflammatory activities of IL-6 are modulated by the balance of IL-6 receptor (IL-6R) and immunosuppressive IL-10, IL-10R (43–45). In this regard, the apparent refractoriness of nonsplenic macrophages in our cultures to most stimuli may reflect programmed tolerance or induced cis or trans immunosuppression and is in line with the conventional hypothesis that proinflammatory macrophages arise largely from the differentiation of newly immigrated monocytes under inflammatory conditions (30, 37, 38).
The gastrointestinal tract is composed of several distinct segments that are unique in histology, function, and commensal microflora. Supporting recent data describing varied leukocyte frequency and function along the murine intestinal tract (46), our analysis of NHP intestinal macrophages revealed phenotypic and functional differences between cells isolated from the small and large intestines (Fig. 2 and 3). The greater phenotypic diversity observed within jejunal macrophages lies in contrast to the greater functional diversity exhibited by colonic macrophages. In addition to maintaining a larger frequency of macrophages, colonic macrophages were significantly more functional than jejunal macrophages, with higher frequencies secreting IL-1β and IL-6. Although these results contrast with previous findings describing a largely anti-inflammatory function for intestinal macrophages (30, 36, 47), our findings do not preclude the presence of balanced immunoregulatory responses by macrophages or by other regulatory cells in culture and in vivo. Of interest, we observed a significant difference in the functional response of colonic macrophages to stimulation with BS-PGN compared to the same stimulation of macrophages in other anatomic sites. This difference manifested largely in a decrease in IL-1β+ IL-6+ macrophages and a reciprocal increase in IL-6 monofunctional cells and was absent from phagocytic cells from SIV-infected animals. This particular response to BS-PGN stimulation was not observed in macrophages isolated from other examined tissues. The only other difference observed in response to BS-PGN stimulation was a significant upregulation of IL-1β expression by phagocytic splenic macrophages, irrespective of infection status. The three distinct outcomes observed in response to BS-PGN stimulation—anergy, decreased IL-1β, and increased IL-1β—are likely a reflection of tissue-specific priming (or a lack thereof) with intestinal macrophages reflecting nuances due to site-specific microflora (36, 41). Further study into the disparities exhibited by anatomically diverse intestinal NHP macrophages may yield critical insight into mechanisms of human disease progression.
Microbial translocation and dysbiosis have been identified as prominent contributors to disease progression in SIV-infected macaques and HIV-infected individuals (4, 6). Given the well-described role of macrophages in bacterial clearance and the initiation of antimicrobial adaptive immune responses, we hypothesized that aberrant macrophage function might contribute to SIV, and hence, HIV disease progression. SIV infection was associated with a significant increase in the frequency of macrophages within the ALN and colon compared to uninfected control animals (Fig. 2). Compared to macrophages from uninfected controls, significant functional differences among macrophages from SIV-infected animals were limited to LPS responsiveness, with bulk splenic macrophages exhibiting an expansion of IL-6 monofunctional cells and phagocytic ALN macrophages exhibiting greater overall functionality (Fig. 6). LPS is found at significantly elevated levels in HIV-infected humans and SIV-infected macaques during chronic infection (4), and thus, while LPS-associated changes were not surprising, the limited scope of changes suggests that macrophages may not directly promote a proinflammatory milieu. Rather, suboptimal immune responses by macrophages might contribute to microbial persistence and translocation and thereby, sustained inflammation as mediated by other leukocytes. Alternatively, the limited changes in proinflammatory cytokine secretion described here may mask changes in anti-inflammatory cytokine regulation, such as those observed for follicular TH cells from HIV-infected humans (48).
Previous investigations into the contributions of macrophages to NHP disease progression have predominantly measured individual markers or functions in parallel from individual tissues (9, 14, 15). While these studies have certainly informed our understanding of macrophages in human health and disease, a comprehensive and systemic understanding of macrophage phenotype and function is essential. Our cross-sectional analysis of macrophages in macaques reveals that great phenotypic and functional heterogeneity are evident among tissue macrophages, with chronic SIV infection resulting in very specific changes in LPS responsiveness. A full assessment of these and potential counterregulatory functions will be essential in weighing the contributions of macrophages to SIV disease progression and determining the appropriate functional balances required for the maintenance and restoration of immune competence in NHP and human health.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Heather Cronise, JoAnne Swerczek, Richard Herbert, and all the veterinary staff at the NIH animal center.
Funding for this study was provided by the Division of Intramural Research/NIAID/NIH.
The content of this publication does not necessarily reflect the views or policies of DHHS, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00005-15.
REFERENCES
- 1.Rogers J, Gibbs RA. 2014. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat Rev Genet 15:347–359. doi: 10.1038/nrg3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. 2012. Natural SIV hosts: showing AIDS the door. Science 335:1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchetti G, Tincati C, Silvestri G. 2013. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev 26:2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 5.Littman DR, Pamer EG. 2011. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, Deeks SG, Hunt PW, Lynch SV, McCune JM. 2013. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, Tabb B, Canary LA, Dang Q, Hirsch VM, Alter G, Belkaid Y, Lifson JD, Silvestri G, Milner JD, Paiardini M, Haddad EK, Brenchley JM. 2012. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol 5:646–657. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sodora DL, Silvestri G. 2008. Immune activation and AIDS pathogenesis. AIDS 22:439–446. doi: 10.1097/QAD.0b013e3282f2dbe7. [DOI] [PubMed] [Google Scholar]
- 9.Collini P, Noursadeghi M, Sabroe I, Miller RF, Dockrell DH. 2010. Monocyte and macrophage dysfunction as a cause of HIV-1 induced dysfunction of innate immunity. Curr Mol Med 10:727–740. doi: 10.2174/156652410793384141. [DOI] [PubMed] [Google Scholar]
- 10.Allers K, Fehr M, Conrad K, Epple HJ, Schurmann D, Geelhaar-Karsch A, Schinnerling K, Moos V, Schneider T. 2014. Macrophages accumulate in the gut mucosa of untreated HIV-infected patients. J Infect Dis 209:739–748. doi: 10.1093/infdis/jit547. [DOI] [PubMed] [Google Scholar]
- 11.Koziel H, Eichbaum Q, Kruskal BA, Pinkston P, Rogers RA, Armstrong MY, Richards FF, Rose RM, Ezekowitz RA. 1998. Reduced binding and phagocytosis of Pneumocystis carinii by alveolar macrophages from persons infected with HIV-1 correlates with mannose receptor downregulation. J Clin Invest 102:1332–1344. doi: 10.1172/JCI560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon MA, Gordon SB, Musaya L, Zijlstra EE, Molyneux ME, Read RC. 2007. Primary macrophages from HIV-infected adults show dysregulated cytokine responses to Salmonella, but normal internalization and killing. AIDS 21:2399–2408. doi: 10.1097/QAD.0b013e3282f25107. [DOI] [PubMed] [Google Scholar]
- 13.Gordon SB, Molyneux ME, Boeree MJ, Kanyanda S, Chaponda M, Squire SB, Read RC. 2001. Opsonic phagocytosis of Streptococcus pneumoniae by alveolar macrophages is not impaired in human immunodeficiency virus-infected Malawian adults. J Infect Dis 184:1345–1349. doi: 10.1086/324080. [DOI] [PubMed] [Google Scholar]
- 14.Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, Hirsch VM, Silvestri G, Douek DC, Miller CJ, Haase AT, Lifson J, Brenchley JM. 2010. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog 6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sopper S, Demuth M, Stahl-Hennig C, Hunsmann G, Plesker R, Coulibaly C, Czub S, Ceska M, Koutsilieri E, Riederer P, Brinkmann R, Katz M, ter Meulen V. 1996. The effect of simian immunodeficiency virus infection in vitro and in vivo on the cytokine production of isolated microglia and peripheral macrophages from rhesus monkey. Virology 220:320–329. doi: 10.1006/viro.1996.0320. [DOI] [PubMed] [Google Scholar]
- 16.Schreiber HA, Loschko J, Karssemeijer RA, Escolano A, Meredith MM, Mucida D, Guermonprez P, Nussenzweig MC. 2013. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J Exp Med 210:2025–2039. doi: 10.1084/jem.20130903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi A, Sato T, Kamada N, Mikami Y, Matsuoka K, Hisamatsu T, Hibi T, Roers A, Yagita H, Ohteki T, Yoshimura A, Kanai T. 2013. A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe 13:711–722. doi: 10.1016/j.chom.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw MH, Kamada N, Kim YG, Núñez G. 2012. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med 209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray PJ, Wynn TA. 2011. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Haan JM, Kraal G. 2012. Innate immune functions of macrophage subpopulations in the spleen. J Innate Immun 4:437–445. doi: 10.1159/000335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiegs G, Lohse AW. 2010. Immune tolerance: what is unique about the liver. J Autoimmun 34:1–6. doi: 10.1016/j.jaut.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Klatt NR, Harris LD, Vinton CL, Sung H, Briant JA, Tabb B, Morcock D, McGinty JW, Lifson JD, Lafont BA, Martin MA, Levine AD, Estes JD, Brenchley JM. 2010. Compromised gastrointestinal integrity in pigtail macaques is associated with increased microbial translocation, immune activation, and IL-17 production in the absence of SIV infection. Mucosal Immunol 3:387–398. doi: 10.1038/mi.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roederer M, Nozzi JL, Nason MC. 2011. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. 2014. The function of Fcgamma receptors in dendritic cells and macrophages. Nat Rev Immunol 14:94–108. doi: 10.1038/nri3582. [DOI] [PubMed] [Google Scholar]
- 26.Li F, Yang M, Wang L, Williamson I, Tian F, Qin M, Shah PK, Sharifi BG. 2012. Autofluorescence contributes to false-positive intracellular Foxp3 staining in macrophages: a lesson learned from flow cytometry. J Immunol Methods 386:101–107. doi: 10.1016/j.jim.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, Yona S. 2014. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol 14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe T, Kitani A, Murray PJ, Wakatsuki Y, Fuss IJ, Strober W. 2006. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity 25:473–485. doi: 10.1016/j.immuni.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W. 2008. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest 118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, Shakhar G, Halpern Z, Jung S. 2012. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 31.Mazza P. 1994. The use of Bacillus subtilis as an antidiarrhoeal microorganism. Boll Chim Farm 133:3–18. [PubMed] [Google Scholar]
- 32.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 33.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. 2014. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol 14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 34.Calantone N, Wu F, Klase Z, Deleage C, Perkins M, Matsuda K, Thompson EA, Ortiz AM, Vinton CL, Ourmanov I, Lore K, Douek DC, Estes JD, Hirsch VM, Brenchley JM. 2014. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity 41:493–502. doi: 10.1016/j.immuni.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, Sakuraba A, Kitazume MT, Sugita A, Koganei K, Akagawa KS, Hibi T. 2008. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest 118:2269–2280. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. 2012. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med 209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, Guilliams M, Malissen B, Agace WW, Mowat AM. 2013. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol 6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume DA, Perlman H, Malissen B, Zelzer E, Jung S. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahnke K, Becher E, Ricciardi-Castagnoli P, Luger TA, Schwarz T, Grabbe S. 1997. CD14 is expressed by subsets of murine dendritic cells and upregulated by lipopolysaccharide. Adv Exp Med Biol 417:145–159. doi: 10.1007/978-1-4757-9966-8_25. [DOI] [PubMed] [Google Scholar]
- 40.Pickl WF, Majdic O, Kohl P, Stöckl J, Riedl E, Scheinecker C, Bello-Fernandez C, Knapp W. 1996. Molecular and functional characteristics of dendritic cells generated from highly purified CD14+ peripheral blood monocytes. J Immunol 157:3850–3859. [PubMed] [Google Scholar]
- 41.Sartor RB. 2008. Microbial influences in inflammatory bowel diseases. Gastroenterology 134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 42.Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, Klatt NR, Canary L, Estes JD, Brenchley JM. 14 January 2015. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. 2011. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta 1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 44.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. 2001. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 45.Niemand C, Nimmesgern A, Haan S, Fischer P, Schaper F, Rossaint R, Heinrich PC, Müller-Newen G. 2003. Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially modulated by suppressor of cytokine signaling 3. J Immunol 170:3263–3272. doi: 10.4049/jimmunol.170.6.3263. [DOI] [PubMed] [Google Scholar]
- 46.Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. 2011. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol 187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, Smith PD. 2005. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest 115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Koup RA. 2012. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest 122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.