ABSTRACT
Influenza A viruses (IAVs) are maintained mainly in wild birds, and despite frequent spillover infections of avian IAVs into mammals, only a small number of viruses have become established in mammalian hosts. A new H3N2 canine influenza virus (CIV) of avian origin emerged in Asia in the mid-2000s and is now circulating in dog populations of China and South Korea, and possibly in Thailand. The emergence of CIV provides new opportunities for zoonotic infections and interspecies transmission. We examined 14,764 complete IAV genomes together with all CIV genomes publicly available since its first isolation until 2013. We show that CIV may have originated as early as 1999 as a result of segment reassortment among Eurasian and North American avian IAV lineages. We also identified amino acid changes that might have played a role in CIV emergence, some of which have not been previously identified in other cross-species jumps. CIV evolves at a lower rate than H3N2 human influenza viruses do, and viral phylogenies exhibit geographical structure compatible with high levels of local transmission. We detected multiple intrasubtypic and heterosubtypic reassortment events, including the acquisition of the NS segment of an H5N1 avian influenza virus that had previously been overlooked. In sum, our results provide insight into the adaptive changes required by avian viruses to establish themselves in mammals and also highlight the potential role of dogs to act as intermediate hosts in which viruses with zoonotic and/or pandemic potential could originate, particularly with an estimated dog population of ∼700 million.
IMPORTANCE Influenza A viruses circulate in humans and animals. This multihost ecology has important implications, as past pandemics were caused by IAVs carrying gene segments of both human and animal origin. Adaptive evolution is central to cross-species jumps, and this is why understanding the evolutionary processes that shape influenza A virus genomes is key to elucidating the mechanisms underpinning viral emergence. An avian-origin canine influenza virus (CIV) has recently emerged in dogs and is spreading in Asia. We reconstructed the evolutionary history of CIV and show that it originated from both Eurasian and North American avian lineages. We also identified the mutations that might have been responsible for the cross-species jump. Finally, we provide evidence of multiple reassortment events between CIV and other influenza viruses (including an H5N1 avian virus). This is a cause for concern, as there is a large global dog population to which humans are highly exposed.
INTRODUCTION
Influenza A viruses (IAVs) constitute a group of segmented, negative-sense RNA viruses that belong to the Orthomyxoviridae and possess a viral genome that encodes up to 14 viral proteins (1). The classification of IAVs is based on the combination of hemagglutinin (HA) and neuraminidase (NA) glycoproteins present on the virus envelope, and to date, 18 HAs and 11 NAs have been reported (2). Wild birds are regarded as the major natural reservoir of IAVs, as most known virus subtypes have been detected in wild birds throughout the world (3). Further, avian IAVs have occasionally jumped the species barrier, causing epizootics and panzootics in domestic birds and various mammalian species, such as horses (4, 5), dogs (6), and pigs (7). Thus, interspecies transmission is one of the most important ecological and epidemiological features of IAVs.
Mutation and reassortment are central to IAV evolution. Like other RNA viruses, IAVs show high mutation rates (8) due to the lack of proofreading activity of the viral polymerase. In addition, segment reassortment (a process by which different IAVs exchange gene segments) allows for rapid incorporation of genetic traits en bloc. Importantly, both processes—mutation and reassortment—have been associated with influenza cross-species transmission and emergence: for example, the former has resulted in the transfer of equine IAV into dogs (9), while the latter led to the emergence of H1N1 swine-origin virus that caused the influenza pandemic of 2009 (10).
IAVs have circulated in humans and other mammals for a long time, and despite evidence of exposure and/or spillover infections (11–13), dogs have historically been considered to be refractory to the establishment of IAVs. However, recently, two canine influenza viruses (CIVs) have emerged, one of equine origin (9) and the other of avian origin (6). H3N8 CIV was first identified in the United States. Phylogenetic analyses showed that this virus originated as a result of a direct interspecies transmission of an equine H3N8 IAV, as all eight segments were closely related to those of an equine virus circulating at the time (9). H3N8 CIV is now enzootic in certain U.S. states such as New York, Pennsylvania, and Colorado (14, 15).
H3N2 CIV was first reported in South Korea in 2007 (6) and shown to be a virus of avian origin. However, a subsequent study showed that H3N2 CIV was already present in China in 2006 (16), and serological surveys using archived sera from dogs in South Korea showed evidence of CIV H3N2 infections as early as 2005 (17). Since then, H3N2 CIVs have often been isolated in dogs in both China and South Korea, indicating that this virus is stably circulating in the Asian canine population (18–22). Moreover, the geographical distribution of this virus is rapidly expanding, as it has also been isolated in dogs in Thailand (23). Notably, H3N2 CIV seems to have a broad host range, as it has been isolated from cats during an outbreak of respiratory disease in a shelter in South Korea (24, 25), and experimental studies have shown that H3N2 CIV can infect ferrets, although transmission in this species seems to be limited (26).
The recent emergence of CIV has important implications. First, the ecological niche of IAVs has increased significantly, as dogs comprise an estimated global population of 700 million (27). Second, as dogs are susceptible to mammalian (equine-origin H3N8 CIV) and avian (avian-origin H3N2 CIV) viruses, they possess all the attributes to become—like pigs—another “mixing vessel” species, particularly if we consider that natural and experimental infections of dogs with human viruses have been reported (12, 28). The fact that dogs are the closest human companions makes reassortment between canine and human viruses more likely to occur. In fact, the isolation of an H3N1 reassortant virus carrying the HA of H3N2 CIV and the seven other genomic segments from the human H1N1 pandemic virus supports this view (29). This is particularly alarming, because the introduction of novel, antigenically distinct glycoproteins within the backbone of human IAVs has previously been associated with pandemics (30).
Detailed studies on the evolution of H3N2 CIV are lacking. To fill this gap, our goal was to infer important aspects of H3N2 CIV evolution such as the source population, the approximate time of origin, the evolutionary rate, and the amino acid sites that might have played a role in the adaptation of CIV to the dog. To this end, we performed a phylogenetic analysis of all publicly available complete CIV genomes using both maximum likelihood (ML) and Bayesian approaches. Further, to determine the source population of H3N2 CIV, we performed phylogenetic analyses of a large genome data set representing 14,764 IAVs from various species.
(Part of this work was submitted by H. Zhu in partial fulfillment of the requirements for the M.Sc. Bioinformatics, Polyomics and Systems Biology Degree at the University of Glasgow, 2014.)
MATERIALS AND METHODS
Sequence data.
Various sequence data sets were sequentially used throughout this study.
(i) Avian-origin H3N2 CIV complete genomes.
Twenty-four canine H3N2 virus complete genomes and one feline H3N2 virus complete genome (alignments provided in Data Set S1 in the supplemental material) were downloaded from the Influenza Virus Resource Database at NCBI (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). A/feline/Korea/01/2010 was included in the data set despite having been isolated in cats, as this virus was essentially an H3N2 CIV that was transmitted to cats during an outbreak initiated in dogs in a shelter (24, 25). A/canine/Guangxi/L2/2013 was excluded from the initial data set, as it was found to be an H3N2 CIV of swine origin (not shown).
(ii) Multiple IAV data set.
A total of 14,764 complete influenza A virus genomes were downloaded from the NCBI Influenza Virus Resource Database. This large data set was reduced in order to make the analysis tractable. To this end, we used CD-HIT (31) to cluster complete genome sequences with identity over 98%, and one sequence within each cluster was selected. After clustering, 878 full genomes remained (alignments provided in Data Set S2 in the supplemental material).
(iii) Expanded IAV data set.
After the maximum likelihood (ML) trees derived from the multiple IAV data set were inferred, we selected a subset of sequences comprising the subtrees containing all H3N2 CIVs and closely related lineages. This data set was expanded to its original number of taxa (before using CD-HIT) as follows: 784 taxa for PB2, 713 taxa for PB1, 829 taxa for PA, 805 taxa for NS1, 452 taxa for NP, 650 taxa for NA, 784 taxa for M1, and 538 taxa for HA. Avian influenza sequences from South Korea were also added to this data set and used in downstream analyses (see Data Set S3 in the supplemental material).
(iv) NS data set.
A total of 31,205 NS sequences were downloaded from the NCBI Influenza Virus Resource Database. We used CD-HIT as described above to reduce this data set and at the same time keep it representative of genetic diversity among IAVs. After clustering was performed, we added all NS sequences derived from canine, equine, and feline IAVs. The final NS data set comprised 775 NS sequences (see Data Set S4 in the supplemental material).
(v) H3N8 CIV data set.
All available H3N8 CIV sequences were downloaded from the NCBI Influenza Virus Resource Database and aligned against their respective H3N2 CIV segments (see Data Set S5 in the supplemental material).
Sequence analysis.
We used MUSCLE (32) to generate individual codon alignments for each segment: PB2 (2,277 nucleotides [nt]), PB1 (2,271 nt), PA (2,148 nt), HA (1698 nt), NP (1,494 nt), NA (1,413 nt), NS (837 nt), and M (981 nt). Alignments were manually edited using MEGA (33). To infer phylogenetic trees, both ML and Bayesian inference approaches were applied by using RAxML (34) and MrBayes (35), respectively. The best-fit nucleotide substitution model was determined by MEGA and applied to all segments for MrBayes and the GTRGAMMA model was used in RAxML. The overall rates of evolutionary change and the time of the most recent common ancestor (TMRCA) were estimated using BEAST (36), assuming a Bayesian skyline coalescent prior with HKY85 plus Gamma nucleotide substitution model and a relaxed (uncorrected exponential lognormal) clock. All chains were run twice independently for 10,000,000 generations with 10% burn-in, and runs were further combined.
We used BaTS (Bayesian Tip-Significance testing) (37) to determine the overall degree of geographical structure among H3N2 CIV sequences. For this analysis, taxon labels were replaced by their geographical location. Given the difference of surface area between China and South Korea, viruses isolated from China were grouped into regions based on proximity (i.e., Zhejiang and Jiangsu were grouped together, as well as Heilongjiang and Liaoning), whereas all viruses from South Korea were considered as derived from a single location. For this analysis, reassortant segments were removed. The association index (AI) and parsimony score (PS) statistic were calculated. Statistical support was obtained by using phylogenetic trees inferred with MrBayes. P values of <0.05 were considered significant.
To visualize intrasubtype reassortment events, whole-genome alignments were generated, and split decomposition networks were inferred using Splitstree 4 (38). The same alignment was used to determine reassortment in RDP4 using a range of methods (39).
To determine the origin of H3N2 CIV, we inferred ML trees using the alignment obtained with the multiple IAV data set described above. After inferring the ML trees, sequences from subtrees containing all H3N2 CIVs and closely related lineages were selected for further analyses with the expanded IAV data set. We also added all the full-length H3N2 avian influenza sequences derived from South Korea to this data set. Maximum likelihood trees were inferred using RAxML and 1,000 rapid bootstraps. Trees were rooted on the oldest sequence.
We used Mesquite (http://mesquiteproject.org) to parsimoniously reconstruct the ancestral amino acid changes along the main trunk branches that gave origin to the H3N2 CIV lineage. Sites that represented amino acid changes of potential adaptive value were compared with those present in H3N8 CIV in order to identify independent (and convergent) mutations.
Estimation of selection pressures.
Selective pressures on codon sites and individual lineages were estimated for all segments of the H3N2 CIV complete genomes, using Datamonkey (40) (http://www.datamonkey.org/). Four methods, including SLAC, FEL, REL, and MEME were used to estimate selective pressures on individual sites. Branch-site REL was used to test selection along lineages. Only results with P values of <0.1 or a posterior probability of >0.9 detected by more than two methods were considered significant.
RESULTS
Origins of H3N2 CIV.
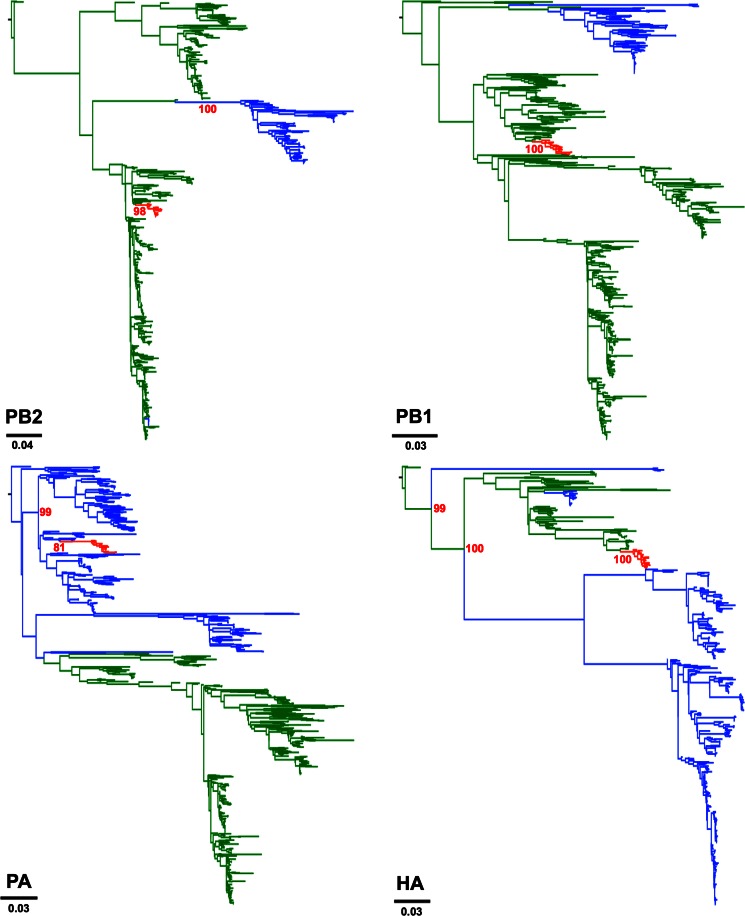
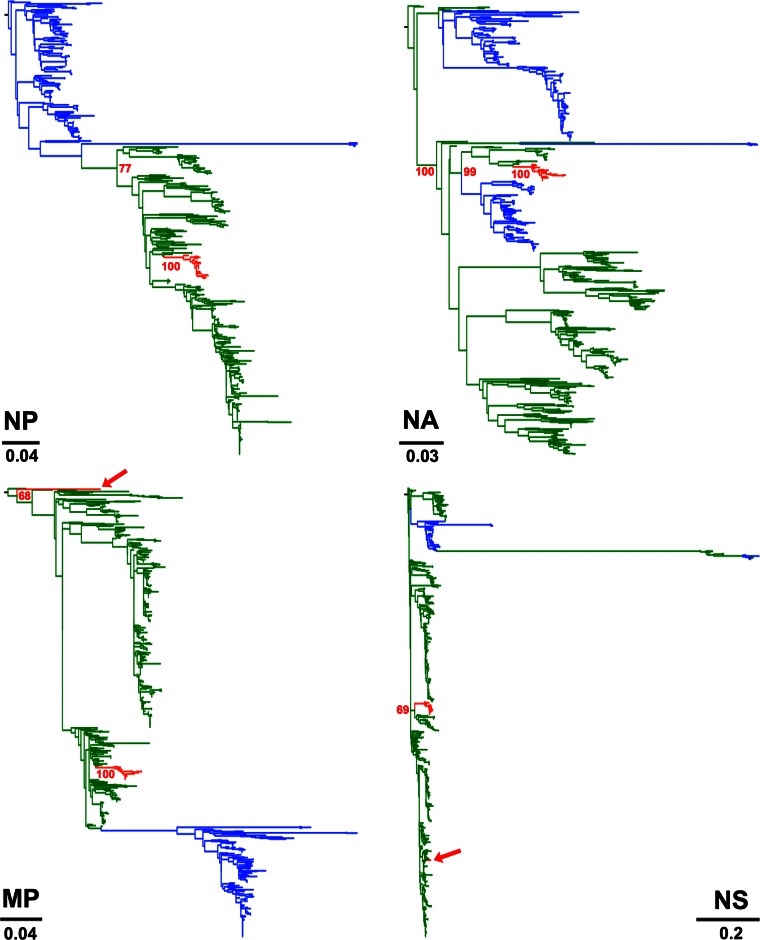
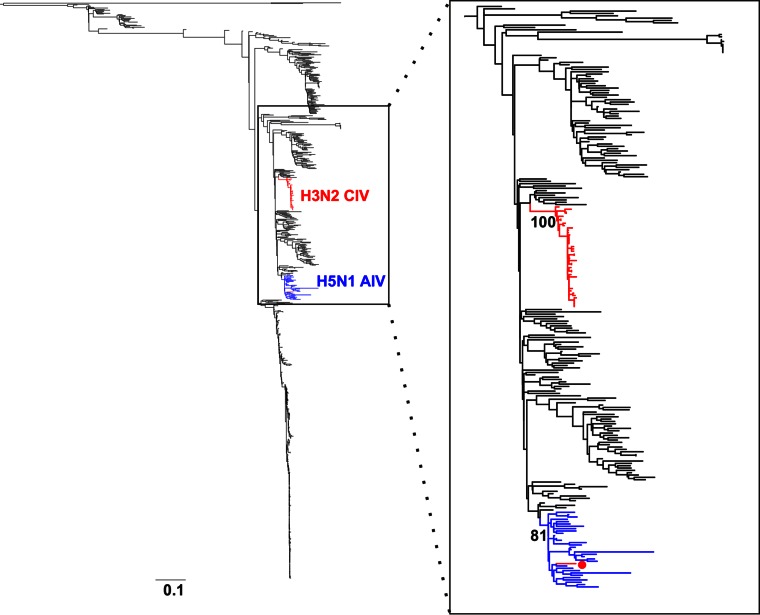
To determine the actual origins of each of the eight genomic segments of H3N2 CIV, we analyzed a large sequence data set that comprised 14,764 publicly available complete genomes of IAVs from all known subtypes. Viral sequences were grouped in 877 clusters based on their identity (sequences that were ≥98% identical were grouped together). We used a single representative from each cluster to infer a ML tree using RAxML. From this first phylogenetic tree, we selected the sequences that were more closely related to the H3N2 CIV branch (usually the branches with closest nodes to those in which CIV was located) and expanded each cluster to their original size. We used the expanded-cluster data set to infer maximum likelihood trees for each segment. As shown in Fig. 1, each segment exhibited a monophyletic origin nested within a well-established Eurasian avian influenza lineage, with the only exception of PA, which grouped with viruses circulating in North America. Except for MP and NS, the phylogenetic relationships between H3N2 CIV and other avian lineages were well supported.
FIG 1.
Origin of H3N2 CIV. Maximum likelihood trees using an expanded subset derived from a phylogeny comprising 877 IAV sequences representing 14,764 IAV genomes. Colored branches represent distinct lineages as follows. The H3N2 CIV lineage is shown in red, Eurasian avian viruses are shown green, and American avian viruses are shown in blue. Bootstrap values of relevant branches are shown. Arrows indicate gene segments of H3N2 CIV that resulted from heterosubtypic reassortment. The bars are drawn to scale.
Consistent with previous reports, the most closely related viruses for each segment were of avian origin. Individual segments displayed different phylogenetic relationships with sequences from IAVs of various subtypes (tree files are available on request). HA and NA were related to H3N2 viruses circulating in South Korea. In turn, PB2 was related to H3N8 viruses from China. PA grouped with high support with H2N2 American avian viruses, PB1 was most closely related to avian influenza viruses (AIVs) from China, Japan, and North America. Notably, the closest ancestor of the CIV lineage was an H5N1 virus from China. The NP segment of H3N2 CIV was most closely related to H3N6 and H5N2 viruses from China and Malaysia, respectively. Overall, our results suggest that complex reassortment events gave rise to H3N2 CIV, including viruses of Eurasian and American lineages.
Amino acid mutations that define the H3N2 CIV lineage.
We determined the amino acid changes that separate the H3N2 CIVs from the ancestral avian viruses in order to identify mutations that could have played a role in H3N2 CIV emergence. To this end, we reconstructed the amino acid changes at the base of the H3N2 CIV lineage. In total, we observed 45 mutations that separate the CIV lineage from the avian lineages (Table 1). Twenty-seven (60%) of the 45 mutations were fixed in the H3N2 CIV genome (i.e., were present in all H3N2 CIV genomes examined), suggesting that they were important for dog adaptation. Importantly, four of the mutations that defined the CIV lineage were also observed in H3N8 CIV (Table 1), suggesting convergent evolution. Table S1 in the supplemental material provides a description of the putative functions of each mutation or of the domains in which mutations were observed.
TABLE 1.
Amino acid changes that differentiate H3N2 CIV from AIVs
| Segment | Positiona | AIV residue (%) | H3N2 CIV residue(s) (%)b | H3N8 CIV residue(s) (%)b |
|---|---|---|---|---|
| PB2 | 76 | T(96.5) | M(93.6)/I(17.4) | T(100) |
| 147 | I(78) | T(100) | V(100) | |
| 365 | M(97) | I(100) | M(100) | |
| 570 | M(94) | V(100) | M(100) | |
| PB1 | 108 | L(96) | I(100) | L(100) |
| 361 | S(94) | N(100) | S(100) | |
| 377 | D(97) | N(78.3)/D(21.7) | D(100) | |
| 517 | I(96) | V(95.64)/I(4.34) | I(100) | |
| 723 | R(97) | Q(93.6)/R(17.4) | R(100) | |
| 744 | M(94) | V(100) | M(100) | |
| PA | 65 | S(93) | Y(100) | S(100) |
| 208 | T(93) | A(100) | T(100) | |
| 234 | D(98) | N(83.3)/D(16.7) | D(100) | |
| 347 | D(99) | N(29.2)/D(70.8) | D(100) | |
| 369 | A(96) | V(100) | A(100) | |
| 441 | M(93) | K(87.5)/R(12.5) | M(95.1)/I(4.9) | |
| 615 | K(94) | R(100) | K(100) | |
| HA | 10 | T(95) | A(100) | T(100) |
| 81 | D(74) | N(100) | Y(98.75)/H(1.25) | |
| 111 | L(96) | I(83.3)/V(16.7)* | I(100) | |
| 172 | D(95) | N(100) | E(8.75)/K(91.25) | |
| 222 | W(90) | L(100)* | L(100) | |
| 435 | H(96) | N(91.7)/H(4.15)/K(4.15) | H(100) | |
| 489 | D(92) | N(100) | Y(100) | |
| NP | 52 | Y(58) | H(95.8)/Y(4.2)* | H(69.8)/Y(30.2) |
| 109 | I(97) | I(62.5)/V(37.5) | I(100) | |
| 125 | N(95) | G(91.7)/N(8.4) | N(100) | |
| 159 | M(95) | L(91.7)/M(8.4) | M(100) | |
| 373 | T(50) | K(95.8)/T(4.2) | T(100) | |
| 428 | A(94) | T(95.8)/A(4.2) | A(100) | |
| 452 | R(79) | K(95.8)/R(4.2)* | K(100) | |
| 473 | N(93) | K(95.8)/N(4.2) | N(100) | |
| NA | 24 | M(95) | L(100) | N/A |
| 48 | N(80) | S(91.7)/N(8.3) | N/A | |
| 54 | E(89) | K(100) | N/A | |
| 81 | P(74) | S(100) | N/A | |
| 143 | D(62) | N(100) | N/A | |
| 156 | P(96) | S(100) | N/A | |
| 372 | S(95) | L(100) | N/A | |
| 432 | R(96) | G(100) | N/A | |
| NS1 | 60 | A(87) | I(100) | A(99.46)/T(0.54) |
| 67 | R(91) | W(100) | Q(100) | |
| 75 | E(85) | K(100) | E(100) | |
| 152 | E(67) | N(100) | E(99.46)/D(0.54) | |
| 172 | E(95) | K(100) | E(100) |
Codon position. H3 numbering is used for codon positions in HA.
Amino acid present at the consensus level. The percentage of each amino acid in our data sets is shown in parentheses. Fixed mutations are shown in bold type. Asterisks indicate convergent mutations between H3N2 and H3N8 CIVs. N/A, not available.
Of the seven mutations in HA that differentiate avian H3N2 viruses from canine viruses, five were fixed among H3N2 CIVs and two were also found in H3N8 CIV. In particular, L222 in the receptor-binding domain of HA has been shown to play an important role in the adaptation of IAVs to the dog both by functional studies using reverse genetics (41) and in structural studies of the HA of H3N8 CIV (42). Mutation D81N could result in altered antigenicity and/or receptor binding (43), as it results in an additional glycosylation site. With regard to NA, seven out of a total of eight amino acid changes displayed 100% prevalence among H3N2 CIVs. In NS, all lineage-defining mutations were observed in all H3N2 CIVs, whereas none of the mutations observed in NP were fixed (Table 1). However, two of the eight amino acid changes in NP were also found in H3N8 viruses: Y52H and R452K. Whether these two mutations play any role in host range needs to be experimentally addressed. With regard to PA, four out of seven lineage-defining mutations were fixed in H3N2 CIV. Of these mutations, amino acid position 615 has been associated with enhanced virulence (44). Interestingly, PB2 lacked all known mutations that have been shown to be important for mammalian adaptation (45), as it displayed E158, T271, G590/Q591, E627, and D701. However, all four PB2 mutations that differentiate H3N2 CIV from its avian ancestor exhibit a high prevalence (>93%). Three of the six mutations that differentiate PB1 of H3N2 CIV from AIVs are fixed (Table 1), and one of these mutations (S361N) is located at a site that has been previously associated with enhanced virulence in humans (46). No CIV lineage-defining amino acid changes were detected for MP, as all observed mutations were synonymous. Overall, our results show that some of the mutations that might have played a role in CIV emergence are consistent with previous influenza virus cross-species jumps and also with influenza virus adaptation to the dog. More importantly, some of the identified amino acid changes have not been previously characterized and might provide novel insight into the mechanisms that allow influenza viruses to adapt to mammals.
Phylogenetic analysis of H3N2 CIVs isolated between 2006 and 2013.
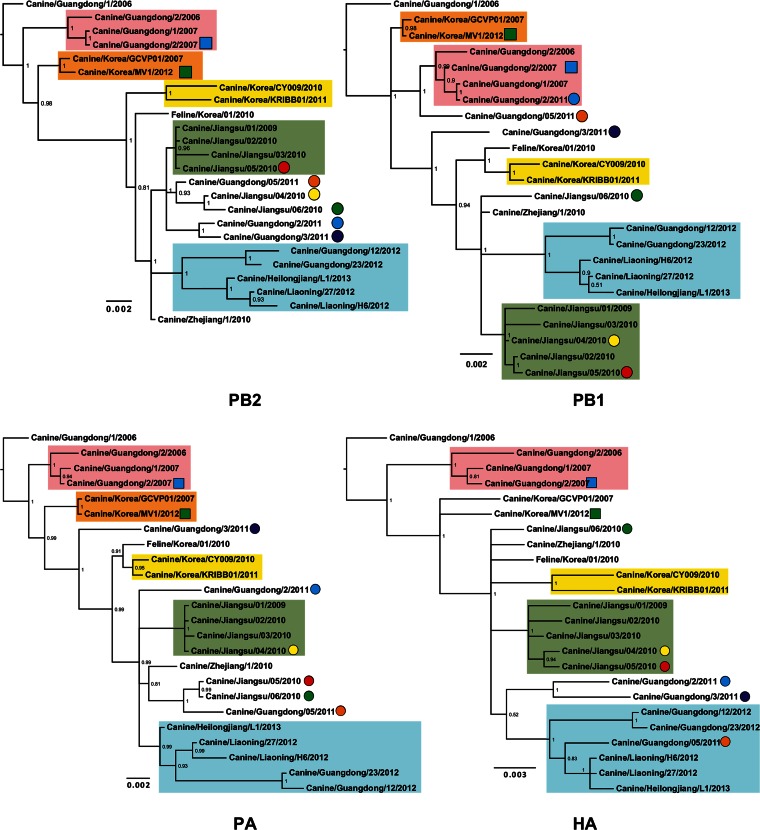
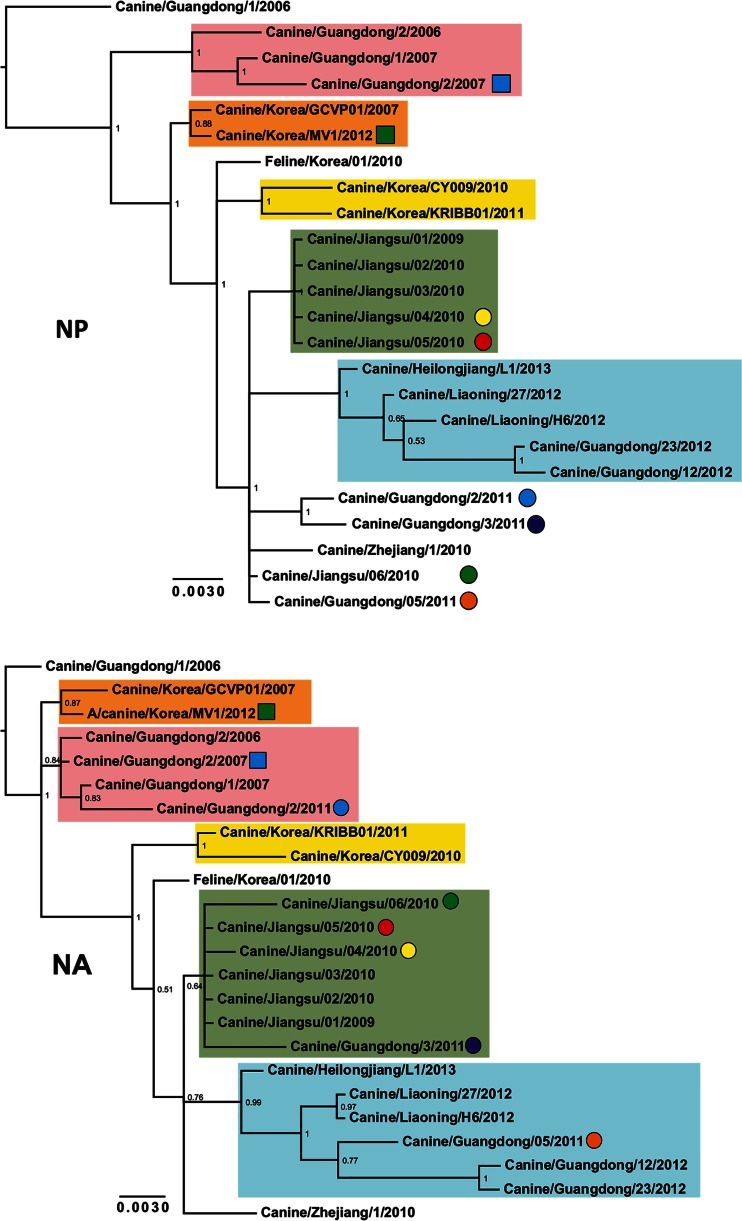
Phylogenetic trees were reconstructed using all CIV H3N2 complete genomes publicly available in GenBank (n = 24 up to 3 October 2014) that had been collected between 2006 and 2013 (see Data Set 1 in the supplemental material). Figure 2 shows consensus, coalescent-approach-based trees for each segment generated using MrBayes. Maximum likelihood trees were also inferred (not shown). In general, both ML and coalescent-model-based trees displayed similar topologies. Trees were rooted with A/canine/Guangdong/1/2006, as this was the oldest H3N2 virus in the data set. As expected, all genomic segments of A/feline/Korea/01/2010 were closely related to CIVs with high posterior probability (Fig. 2), confirming previous reports that this virus originated as a result of a direct interspecies transmission of CIV to cats.
FIG 2.
Phylogenetic trees of 24 complete genomes of H3N2 CIV collected between 2006 and 2013. The phylogenetic trees for each genomic segment were inferred using MrBayes. The colored boxes indicate the stable clades present among all gene segments with the exception of MP and NS. Clade I is shown by pink, clade II is shown by orange, clade III is shown by yellow, clade IV is shown by green, and clade V is shown by light blue. Circles and squares indicate viruses that underwent homosubtypic and heterosubtypic reassortment, respectively. Posterior probability values are shown for each node.
The inferred trees for all genomic segments with the exception of MP and NS exhibited a similar topology, with several viruses consistently clustering together with high posterior probability values. For this reason, we arbitrarily grouped them in five clades (clades I to V [Fig. 2]). Long internal branches separated most clades from the main trunk of the CIV phylogeny. Clade I contains viruses isolated in Guangdong, China, from 2006 to 2007, and clades II (absent in the HA phylogeny) and III contain isolates exclusively from South Korea from 2007 and 2012 and 2010 to 2011, respectively. Clade IV displays viruses isolated in Jiangsu, China, from 2009 and 2010, and clade V contains viruses from 2012 and 2013 collected in Guangdong, Liaoning, and Heilongjiang in China.
In addition to the viruses consistently observed in each clade, individual isolates were occasionally observed for distinct segments (for example Jiangsu/06/2010 in clade IV of the NA tree), suggesting the occurrence of intrasubtypic reassortment (see below).
The phylogenies suggest that viral sequences originating from the same location tend to cluster. We used the parsimony score (PS) and association index (AI) statistics in the BaTS program (37) to test for the presence of geographical structure. To this end, the places of isolation were used as traits and represented by four locations: Guangdong, Jiangsu, and Liaoning in China and South Korea (see Fig. S1 in the supplemental material). We observed that the mean values of PS for each segment were lower than expected, and the average value of AI was low, suggesting a strong association between phylogeny and geographical locations. Table 2 shows the P values for both PS and AI for each segment, indicating that the strength of phylogeny-geography association was significant.
TABLE 2.
Correlation between phylogenies and geographical location for each CIV genomic segment
| Segment | Association index |
Parsimony score |
||
|---|---|---|---|---|
| BaTS estimate (95% HPD interval) | P valuea | BaTS estimate (95% HPD interval) | P valuea | |
| PB2 | 0.05 (0.04–0.07) | 0.00 | 5.02 (5.00–5.00) | 0.00 |
| PB1 | 0.07 (0.03–0.17) | 0.00 | 4.85 (4.00–5.00) | 0.00 |
| PA | 0.12 (0.06–0.15) | 0.00 | 5.66 (5.00–6.00) | 0.00 |
| HA | 0.14 (0.02–0.36) | 0.00 | 5.72 (5.00–7.00) | 0.00 |
| NP | 0.15 (0.04–0.27) | 0.00 | 4.86 (4.00–5.00) | 0.00 |
| NA | 0.27 (0.02–0.54) | 0.00 | 5.93 (5.00–7.00) | 0.00 |
| MP | 0.55 (0.19–0.92) | 0.00 | 6.40 (6.00–7.00) | 0.00 |
| NS | 0.35 (0.08–0.69) | 0.00 | 6.56 (5.00–8.00) | 0.00 |
P value for the BaTS null hypothesis test.
CIV reassorted multiple times with other IAVs.
Two isolates, A/canine/Korea/MV1/2012 and A/canine/Guangdong/2/2007, were located at the tips of unusually long branches in the NS and MP phylogenies, respectively, indicating that those segments were significantly different from other CIVs (see Fig. S2 and Fig. S3 in the supplemental material, respectively). While this was expected for A/canine/Korea/MV1/2012 as it has been shown to be a reassortant virus (47), this result was unexpected for A/canine/Guangdong/2/2007. To identify the origin of the NS segment of A/canine/Guangdong/2/2007, we collected 775 NS sequences from different hosts (see Materials and Methods) and inferred a maximum likelihood phylogenetic tree using RAxML. Our results show that the NS segment of A/canine/Guangdong/2/2007 was more closely related to the NS segments of H5N1 avian viruses than to other CIVs (Fig. 3). These results, together with another report of a human IAV carrying the HA of H3N2 CIV (29) indicate that heterosubtypic reassortment is relatively common for CIV, given the short time since its emergence.
FIG 3.
A/canine/Guangdong/2/2007 reassorted with H5N1 avian viruses. (Left) Maximum likelihood phylogenetic tree for 775 NS sequences derived from IAVs of various subtypes. Branches exhibiting H3N2 CIV and H5N1 avian influenza viruses are shown in red and blue, respectively. (Right) Higher magnification of the clade containing H3N2 CIVs and avian H5N1 sequences. The red circle represents the NS sequence of A/canine/Guangdong/2/2007. Bootstrap values are shown for the relevant nodes. The tree was midrooted.
H3N2 CIVs display extensive intrasubtypic reassortment along their evolutionary history.
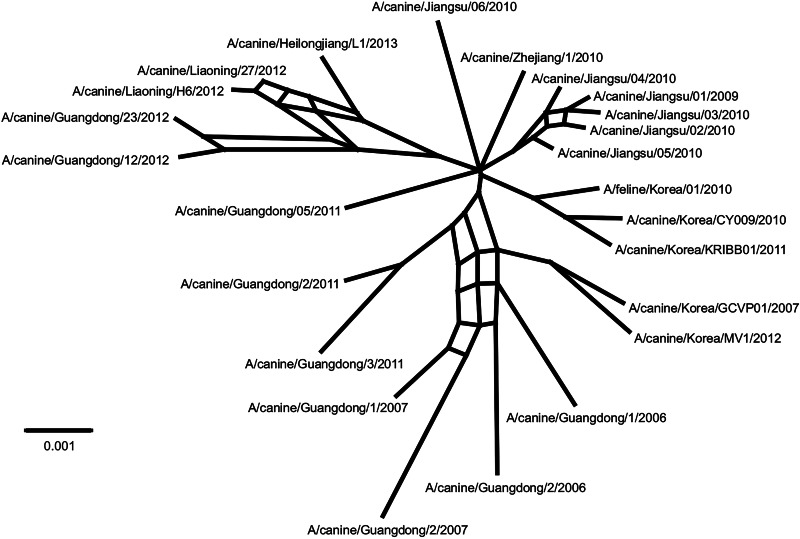
Previous studies have shown that intrasubtypic reassortment is not an uncommon event among IAVs (48, 49). To determine whether this was the case for CIV, we concatenated all the CIV genomes and then generated a split decomposition network using Splitstree. Split networks are aimed at displaying incompatibilities and ambiguous phylogenetic signals within and between data sets (i.e., within a sequence alignment of a given gene from different species or between phylogenetic trees of different genes). If there was no reassortment, no incompatibilities would be observed in the network from the concatenated alignment. The network shown in Fig. 4 displays several parallel edges consistent with reassortment and thus confirms our hypothesis. The results from RDP also support intrasubtypic reassortment based on multiple methods (not shown). Close examination of the phylogenies in Fig. 2 revealed changes in the tree topologies with high support that were consistent with intrasubtypic reassortment. Three sections of the trees showed clear incompatibilities.
FIG 4.
Intrasubtypic reassortment among H3N2 CIVs. The split decomposition network was inferred using Splitstree. Potential reassortment events are shown as reticulation between branches.
For example, A/canine/Guangdong/2/2011 grouped with high support with clade I viruses for PB1 and NA but not for the rest of the genomic segments. A/canine/Guangdong/3/2011 was located in clade IV only in the NA phylogeny, but its PB2, HA, and NP segments grouped with A/canine/Guangdong/2/2011. For A/canine/Guangdong/5/2011, the HA and NA genes were located within clade V, whereas the PB1 gene grouped with clade I viruses, and PB2 and PA genes grouped with viruses isolated in Jiangsu, China, in 2010. Viruses from Jiangsu, China, collected in 2009 and 2010 were also reassortants, as their phylogenetic relationships varied among gene segments: for example, A/canine/Jiangsu/06/2010 was located within clade IV only in the NA tree but formed a separate group with A/canine/Jiangsu/04/2010 for PB2 and another one with A/canine/Jiangsu/05/2010 for PA (Fig. 2).
Evolutionary dynamics and selection analysis of H3N2 CIVs.
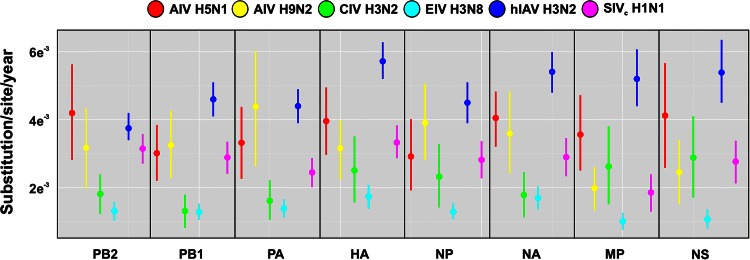
We used BEAST to estimate the overall rates of evolutionary change of H3N2 CIV. The means and 95% highest posterior density (95% HPD) intervals of all genomic segments are shown in Table 2. While rates (measured as the number of substitutions/site/year) were within the expected range for RNA viruses (8), they were lower than the ones published for other IAVs (49–52): for most gene segments (with the exception of MP and NS), H3N2 CIV rates were at the lower end of the estimated HPD intervals (Fig. 5), and significant differences for all the genomic segments were detected only between CIV and H3N2 human influenza virus (the 95% HPD interval did not overlap with the HPD interval of H3N2 human influenza virus). We also used BEAST to estimate the time of the most recent common ancestor in order to date the origins of CIV. Our results showed that CIV originated around 2004 (95% between 1999 and 2006; Table 3), not long before the first isolation in China in 2006.
FIG 5.
Evolutionary rates for avian, canine, human, equine, and swine IAVs. Evolutionary rates for avian (red), canine (green), human (dark blue), equine (light blue) and swine (fuchsia) IAVs are shown. Vertical bars represent 95% HPD of the evolutionary rates estimated by BEAST.
TABLE 3.
Estimates of nucleotide substitution rates and time of the most recent common ancestor for each gene segment of CIV H3N2
| Segment | Mean substitution rate (95% HPD interval) | Mean TMRCA (95% HPD interval) |
|---|---|---|
| HA | 2.44 × 10−3 (1.54 × 10−3, 3.50 × 10−3) | 2004 (2002–2006) |
| MP | 2.57 × 10−3 (1.52 × 10−3, 3.78 × 10−3) | 2005 (2003–2006) |
| NA | 1.93 × 10−3 (1.16 × 10−3, 2.66 × 10−3) | 2005 (2003–2006) |
| NP | 2.30 × 10−3 (1.35 × 10−3, 3.33 × 10−3) | 2003 (1999–2005) |
| NS | 2.83 × 10−3 (1.71 × 10−3, 4.06 × 10−3) | 2004 (2001–2006) |
| PA | 1.60 × 10−3 (1.58 × 10−3, 2.17 × 10−3) | 2004 (2001–2006) |
| PB1 | 1.22 × 10−3 (7.45 × 10−4, 1.74 × 10−3) | 2003 (2000–2005) |
| PB2 | 1.80 × 10−3 (1.22 × 10−3, 2.24 × 10−3) | 2004 (2001–2006) |
Selection analyses showed that only two codons (469 in HA [amino acid position 453 based on H3 numbering] and 723 in the PA) were positively selected. In addition, we used the genetic branch (GA) algorithm to determine whether any individual branch in the CIV phylogenies was under positive selection. No branches under selection were detected.
DISCUSSION
Since the discovery of H3N2 CIV in 2007, most studies have focused on the genetic and virological characterization of individual isolates (6, 16, 18–21, 23, 25, 26) and on clinical and epidemiological investigations (20, 22–24). Here we compiled all the publicly available complete genome sequences of CIV and carried out a comprehensive phylogenetic analysis to elucidate its evolutionary history. Our results confirm previous findings and provide new insight on the origins, epidemiology, and evolution of H3N2 CIV. We determined—with better accuracy than previous reports—the origins of H3N2 CIV and propose an evolutionary pathway that led to the emergence of H3N2 CIV. Clearly, all the gene segments, with the exception of PA, originated from Eurasian avian influenza viruses. The phylogenies for each of those segments suggest that this genetic constellation arose by multiple reassortment events. The American origin of the PA gene can be explained by occasional hemispheric mixing of avian influenza viruses, as this has been previously reported (53). The HA and NA segments were probably derived from H3N2 avian viruses circulating in South Korea and could have been obtained en bloc. Similarly, PB2 and PB1 were likely derived from avian H3N8 and H5N1 viruses from China, respectively, and NP was derived from Asian AIVs of either H5N2 or H3N6 subtypes. We could not infer the evolutionary history of NS and MP, albeit they are clearly of Eurasian origin.
Our analysis also reveals important epidemiological features of CIV. The geographical association observed in the phylogenies suggests that CIV might be transmitted at high levels within local dog populations and that once it is introduced in a given area, it tends to establish local lineages that evolve in situ. This is consistent with the high level of intrasubtypic reassortment observed within single locations. Geographic clustering has also been observed for H3N8 CIV (54). While contact heterogeneity seems to be the main cause of the phylodynamics patterns observed for H3N8 CIV (54), the reasons underlying H3N2 CIV evolutionary and epidemiological dynamics are unclear and will require further investigation.
More importantly from a public health perspective, avian influenza viruses of different subtypes such as H5N1, H9N2, and H5N2 have been isolated in dogs (11, 55, 56). Further, H3N2 CIV has reassorted with other IAVs on at least two independent occasions. While the reassortment event between CIV and human/swine H1N1 IAV has been previously reported (47), we showed phylogenetic evidence that the NS of A/canine/Guangdong/2/2007 was indeed closely related to H5N1 avian influenza viruses. This important finding was not described by the authors of the original report because they focused their analysis only on the HA and NA genes (16). It is alarming that despite being a relatively “young” virus (we estimated the time of emergence to ∼2004), CIV has already reassorted with human (29, 47) and avian (this report) influenza viruses. Such reassortment events included various gene segments (HA, MP, and NS) within the genetic background of both human and canine IAVs, clearly showing that the gene pool of avian, human, and canine viruses is indeed compatible. As the geographical range of CIV is rapidly expanding and both human and avian IAVs are prevalent in Asia, it is likely that further reassortants will appear.
It is unknown why influenza A virus lineages apparently did not establish in dogs before the late 1990s and early 2000s. The respiratory tract of the dog possesses sialic acids with both α2,6 and α2,3 linkages that can be used as receptors by IAVs (57), and recent studies have shown that H3N2 CIV preferentially binds α2,3-linked sialic acids (41). Moreover, dogs have clearly been exposed to horses and poultry for a long time before equine and avian influenza viruses jumped the species barrier.
The ecology of influenza viruses has significantly changed now that CIVs are stably circulating in dogs. Although census information is patchy, the global canine population has been estimated to be ∼700 million (27). If we consider that dogs are used as companion and working animals as well as a source of food, it is feasible to think that they can become—like pigs—another “mixing vessel” species in which avian, human, and canine viruses can reassort and originate new viruses with pandemic potential. Our data and previous work (29, 47) support this view. However, differences in life span and population structure might play a role in the generation of pandemic viruses. CIV is rapidly spreading in Asia (it has been detected in China, South Korea, and Thailand), where there is a high prevalence of avian and human influenza viruses, and our data suggest that once H3N2 CIV enters a susceptible dog population, it is likely to persist. The fact that H3N2 viruses are established in humans highlights the zoonotic potential of H3N2 CIV, although it is not known whether there is cross protection between human and canine IAVs.
In sum, we have performed a detailed characterization of the phylogenetic history of H3N2 CIV. Surveillance studies in dogs are needed to monitor the geographical spread and seasonal patterns of H3N2 CIV infections as well as the detection of reassortant viruses. Experimental studies are needed in order to determine the zoonotic—and pandemic—potential of CIV and whether prior infection with seasonal H3N2 protects humans against CIV. Finally, CIV vaccines should also be developed to control the spread of this virus.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Medical Research Council grant G0801822. P.R.M. and J.H. are funded by the Medical Research Council of the United Kingdom.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03395-14.
REFERENCES
- 1.Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog 8:e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waddell GH, Teigland MB, Sigel MM. 1963. A new influenza virus associated with equine respiratory disease. J Am Vet Med Assoc 143:587–590. [PubMed] [Google Scholar]
- 5.Guo Y, Wang M, Kawaoka Y, Gorman O, Ito T, Saito T, Webster RG. 1992. Characterization of a new avian-like influenza A virus from horses in China. Virology 188:245–255. doi: 10.1016/0042-6822(92)90754-D. [DOI] [PubMed] [Google Scholar]
- 6.Song D, Kang B, Lee C, Jung K, Ha G, Kang D, Park S, Park B, Oh J. 2008. Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis 14:741–746. doi: 10.3201/eid1405.071471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. 1981. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull World Health Organ 59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, Chen L, Smith C, Hill RC, Ferro P, Pompey J, Bright RA, Medina MJ, Johnson CM, Olsen CW, Cox NJ, Klimov AI, Katz JM, Donis RO. 2005. Transmission of equine influenza virus to dogs. Science 310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 10.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 11.Songserm T, Amonsin A, Jam-on R, Sae-Heng N, Pariyothorn N, Payungporn S, Theamboonlers A, Chutinimitkul S, Thanawongnuwech R, Poovorawan Y. 2006. Fatal avian influenza A H5N1 in a dog. Emerg Infect Dis 12:1744–1747. doi: 10.3201/eid1211.060542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundon WG, De Benedictis P, Viale E, Capua I. 2010. Serologic evidence of pandemic (H1N1) 2009 infection in dogs, Italy. Emerg Infect Dis 16:2019–2021. doi: 10.3201/eid1612.100514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin D, Sun S, Du L, Ma J, Fan L, Pu J, Sun Y, Zhao J, Sun H, Liu J. 2012. Natural and experimental infection of dogs with pandemic H1N1/2009 influenza virus. J Gen Virol 93:119–123. [DOI] [PubMed] [Google Scholar]
- 14.Pecoraro HL, Bennett S, Huyvaert KP, Spindel ME, Landolt GA. 2014. Epidemiology and ecology of H3N8 canine influenza viruses in US shelter dogs. J Vet Intern Med 28:311–318. doi: 10.1111/jvim.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayward JJ, Dubovi EJ, Scarlett JM, Janeczko S, Holmes EC, Parrish CR. 2010. Microevolution of canine influenza virus in shelters and its molecular epidemiology in the United States. J Virol 84:12636–12645. doi: 10.1128/JVI.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Shi Z, Jiao P, Zhang G, Zhong Z, Tian W, Long LP, Cai Z, Zhu X, Liao M, Wan XF. 2010. Avian-origin H3N2 canine influenza A viruses in Southern China. Infect Genet Evol 10:1286–1288. doi: 10.1016/j.meegid.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YN, Lee DH, Lee HJ, Park JK, Yuk SS, Sung HJ, Park HM, Lee JB, Park SY, Choi IS, Song CS. 2012. Evidence of H3N2 canine influenza virus infection before 2007. Vet Rec 171:477. doi: 10.1136/vr.100718. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Jia K, Qi W, Zhang M, Sun L, Liang H, Du G, Tan L, Shao Z, Ye J, Sun L, Cao Z, Chen Y, Zhou P, Su S, Li S. 2013. Genetic characterization of avian-origin H3N2 canine influenza viruses isolated from Guangdong during 2006-2012. Virus Genes 46:558–562. doi: 10.1007/s11262-013-0893-3. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Zhao Y, Zeng X, Lu C, Liu Y. 2012. Genetic and pathobiologic characterization of H3N2 canine influenza viruses isolated in the Jiangsu Province of China in 2009-2010. Vet Microbiol 158:247–258. doi: 10.1016/j.vetmic.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Liu C, Liu F, Liu D, Chen Y, Zhang H, Qu L, Li Y, Xia D, Liu M. 2014. Identification and genetic characterization of avian-origin H3N2 canine influenza viruses isolated from the Liaoning province of China in 2012. Virus Genes 49:342–347. doi: 10.1007/s11262-014-1092-6. [DOI] [PubMed] [Google Scholar]
- 21.Teng Q, Zhang X, Xu D, Zhou J, Dai X, Chen Z, Li Z. 2013. Characterization of an H3N2 canine influenza virus isolated from Tibetan mastiffs in China. Vet Microbiol 162:345–352. doi: 10.1016/j.vetmic.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Su S, Li HT, Zhao FR, Chen JD, Xie JX, Chen ZM, Huang Z, Hu YM, Zhang MZ, Tan LK, Zhang GH, Li SJ. 2013. Avian-origin H3N2 canine influenza virus circulating in farmed dogs in Guangdong, China. Infect Genet Evol 14:444–449. doi: 10.1016/j.meegid.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 23.Bunpapong N, Nonthabenjawan N, Chaiwong S, Tangwangvivat R, Boonyapisitsopa S, Jairak W, Tuanudom R, Prakairungnamthip D, Suradhat S, Thanawongnuwech R, Amonsin A. 2014. Genetic characterization of canine influenza A virus (H3N2) in Thailand. Virus Genes 48:56–63. doi: 10.1007/s11262-013-0978-z. [DOI] [PubMed] [Google Scholar]
- 24.Jeoung HY, Lim SI, Shin BH, Lim JA, Song JY, Song DS, Kang BK, Moon HJ, An DJ. 2013. A novel canine influenza H3N2 virus isolated from cats in an animal shelter. Vet Microbiol 165:281–286. doi: 10.1016/j.vetmic.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Song DS, An DJ, Moon HJ, Yeom MJ, Jeong HY, Jeong WS, Park SJ, Kim HK, Han SY, Oh JS, Park BK, Kim JK, Poo H, Webster RG, Jung K, Kang BK. 2011. Interspecies transmission of the canine influenza H3N2 virus to domestic cats in South Korea, 2010. J Gen Virol 92:2350–2355. doi: 10.1099/vir.0.033522-0. [DOI] [PubMed] [Google Scholar]
- 26.Lee YN, Lee DH, Park JK, Yuk SS, Kwon JH, Nahm SS, Lee JB, Park SY, Choi IS, Song CS. 2013. Experimental infection and natural contact exposure of ferrets with canine influenza virus (H3N2). J Gen Virol 94:293–297. doi: 10.1099/vir.0.042473-0. [DOI] [PubMed] [Google Scholar]
- 27.Hughes J, Macdonald DW. 2013. A review of the interactions between free-roaming domestic dogs and wildlife. Biol Conserv 157:341–351. doi: 10.1016/j.biocon.2012.07.005. [DOI] [Google Scholar]
- 28.Song D, Kim H, Na W, Hong M, Park SJ, Moon H, Kang B, Lyoo KS, Yeom M, Jeong DG, An DJ, Kim JK. 2015. Canine susceptibility to human influenza viruses (A/pdm09 H1N1, A/H3N2, and B). J Gen Virol 96:254–258. doi: 10.1099/vir.0.070821-0. [DOI] [PubMed] [Google Scholar]
- 29.Song D, Moon HJ, An DJ, Jeoung HY, Kim H, Yeom MJ, Hong M, Nam JH, Park SJ, Park BK, Oh JS, Song M, Webster RG, Kim JK, Kang BK. 2012. A novel reassortant canine H3N1 influenza virus between pandemic H1N1 and canine H3N2 influenza viruses in Korea. J Gen Virol 93:551–554. doi: 10.1099/vir.0.037739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webby RJ, Webster RG. 2001. Emergence of influenza A viruses. Philos Trans R Soc London B Biol Sci 356:1817–1828. doi: 10.1098/rstb.2001.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 36.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker J, Rambaut A, Pybus OG. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol 8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 39.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang G, Li S, Blackmon S, Ye J, Bradley KC, Cooley J, Smith D, Hanson L, Cardona C, Steinhauer DA, Webby R, Liao M, Wan XF. 2013. Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol 94:2599–2608. doi: 10.1099/vir.0.054692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins PJ, Vachieri SG, Haire LF, Ogrodowicz RW, Martin SR, Walker PA, Xiong X, Gamblin SJ, Skehel JJ. 2014. Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A 111:11175–11180. doi: 10.1073/pnas.1406606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. 2014. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6:1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A 102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cauldwell AV, Long JS, Moncorge O, Barclay WS. 2014. Viral determinants of influenza A virus host range. J Gen Virol 95:1193–1210. doi: 10.1099/vir.0.062836-0. [DOI] [PubMed] [Google Scholar]
- 46.Allen JE, Gardner SN, Vitalis EA, Slezak TR. 2009. Conserved amino acid markers from past influenza pandemic strains. BMC Microbiol 9:77. doi: 10.1186/1471-2180-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon H, Hong M, Kim JK, Seon B, Na W, Park SJ, An DJ, Jeoung HY, Kim DJ, Kim JM, Kim SH, Webby RJ, Webster RG, Kang BK, Song D. 2014. H3N2 canine influenza virus with the matrix gene from the pandemic A/H1N1 virus: infection dynamics in dogs and ferrets. Epidemiol Infect 143:772–780. doi: 10.1017/S0950268814001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, St George K, Taylor J, Spiro DJ, Sengamalay NA, Ghedin E, Taubenberger JK, Holmes EC. 2008. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog 4:e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murcia PR, Wood JL, Holmes EC. 2011. Genome-scale evolution and phylodynamics of equine H3N8 influenza A virus. J Virol 85:5312–5322. doi: 10.1128/JVI.02619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunham EJ, Dugan VG, Kaser EK, Perkins SE, Brown IH, Holmes EC, Taubenberger JK. 2009. Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J Virol 83:5485–5494. doi: 10.1128/JVI.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen R, Holmes EC. 2006. Avian influenza virus exhibits rapid evolutionary dynamics. Mol Biol Evol 23:2336–2341. doi: 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- 52.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog 4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalziel BD, Huang K, Geoghegan JL, Arinaminpathy N, Dubovi EJ, Grenfell BT, Ellner SP, Holmes EC, Parrish CR. 2014. Contact heterogeneity, rather than transmission efficiency, limits the emergence and spread of canine influenza virus. PLoS Pathog 10:e1004455. doi: 10.1371/journal.ppat.1004455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun X, Xu X, Liu Q, Liang D, Li C, He Q, Jiang J, Cui Y, Li J, Zheng L, Guo J, Xiong Y, Yan J. 2013. Evidence of avian-like H9N2 influenza A virus among dogs in Guangxi, China. Infect Genet Evol 20:471–475. doi: 10.1016/j.meegid.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 56.Zhan GJ, Ling ZS, Zhu YL, Jiang SJ, Xie ZJ. 2012. Genetic characterization of a novel influenza A virus H5N2 isolated from a dog in China. Vet Microbiol 155:409–416. doi: 10.1016/j.vetmic.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 57.Daly JM, Blunden AS, Macrae S, Miller J, Bowman SJ, Kolodziejek J, Nowotny N, Smith KC. 2008. Transmission of equine influenza virus to English foxhounds. Emerg Infect Dis 14:461–464. doi: 10.3201/eid1403.070643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.