ABSTRACT
The ubiquitous ATP-dependent RNA helicase DDX3X is involved in many cellular functions, including innate immunity, and is a pivotal host factor for hepatitis C virus (HCV) infection. Recently, we showed that DDX3X specifically recognizes the HCV 3′ untranslated region (UTR), leading to the activation of IKK-α and a cascade of lipogenic signaling to facilitate lipid droplet biogenesis and viral assembly (Q. Li, V. Pene, S. Krishnamurthy, H. Cha, and T. J. Liang, Nat Med 19:722–729, 2013, http://dx.doi.org/10.1038/nm.3190). The interaction of DDX3X with HCV core protein seems to be dispensable for its proviral role. In this study, through systematic imaging and biochemical and virologic approaches, we identified a dynamic association between DDX3X and various cellular compartments and viral elements mediating multiple functions of DDX3X in productive HCV infection. Upon HCV infection, the HCV 3′UTR interacts with DDX3X and IKK-α, which redistribute to speckle-like cytoplasmic structures shown to be stress granules (SGs). As viral proteins accumulate in infected cells, DDX3X granules together with SG-associated proteins redistribute and colocalize with HCV core protein around lipid droplets (LDs). IKK-α, however, does not relocate to the LD but translocates to the nucleus. In HCV-infected cells, various HCV nonstructural proteins also interact or colocalize with DDX3X in close proximity to SGs and LDs, consistent with the tight juxtaposition of the replication complex and the assembly site at the surface of LDs. Short interfering RNA (siRNA)-mediated silencing of DDX3X and multiple SG components markedly inhibits HCV infection. Our data suggest that DDX3X initiates a multifaceted cellular program involving dynamic associations with HCV RNA and proteins, IKK-α, SG, and LD surfaces for its crucial role in the HCV life cycle.
IMPORTANCE DDX3X is a proviral host factor for HCV infection. Recently, we showed that DDX3X binds to the HCV 3′UTR, activating IKK-α and cellular lipogenesis to facilitate viral assembly (Q. Li et al., Nat Med 19:722–729, 2013, http://dx.doi.org/10.1038/nm.3190). Here, we report associations of DDX3X with various cellular compartments and viral elements that mediate its multiple functions in the HCV life cycle. Upon infection, the HCV 3′UTR redistributes DDX3X and IKK-α to speckle-like cytoplasmic structures shown to be SGs. Subsequently, interactions between DDX3X, SG, and HCV proteins facilitate the translocation of DDX3X-SG complexes to the LD surface. HCV nonstructural proteins are shown to colocalize with DDX3X in close proximity to SGs and LDs, consistent with the tight juxtaposition of the HCV replication complex and assembly site at the LD surface. Our data demonstrate that DDX3X initiates a multifaceted cellular program involving dynamic associations with HCV elements, IKK-α, SGs, and LDs for its critical role in HCV infection.
INTRODUCTION
Despite recent advances in therapeutics, hepatitis C virus (HCV) infection is still a leading cause of chronic liver disease. Approximately 80% of HCV infections become chronic, with many cases necessitating antiviral treatment. Such a high persistence rate is unusual for a human pathogen and can be attributed to various viral immune evasion strategies (1, 2). Chronic hepatitis C patients have a high risk of developing hepatic steatosis, liver cirrhosis, and hepatocellular carcinoma. While current therapeutic regimens are improving, a protective HCV vaccine still is unavailable (3, 4).
HCV has a single positive-strand RNA genome of about 9.6 kb consisting of two untranslated regions (UTRs) at the 5′ and 3′ termini, respectively, that are needed for translation and replication of viral RNA. Between the 5′ and 3′UTRs, a single open reading frame encoding a large polyprotein is processed further into both structural (core protein, E1, and E2) and nonstructural (P7, NS2, NS3/4A, NS4B, NS5A, and NS5B) proteins. The viral genome replicates at the so-called replication complex (RC), an endoplasmic reticulum (ER) membrane-associated replicase structure engulfing viral nonstructural proteins. The core protein forms the viral nucleocapsid (5), oligomerizes, binds to HCV RNA through its N-terminal domain, and associates with lipid droplets (LDs) and the ER through its C-terminal hydrophobic domain. The association between HCV core protein and the LD is essential for production of infectious viral particles (6, 7). Core protein also recruits viral RCs to LD-associated membranes (6) and induces the accumulation of LDs in hepatocytes to facilitate viral assembly (8, 9).
One strategy for viral escape from the host immune system is to hijack cellular proteins involved in antiviral immunity. DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked (DDX3X), is a ubiquitous, multifunctional ATP-dependent RNA helicase and an RNA-dependent ATPase that is involved in a variety of cellular processes related to RNA processing, such as transcription, mRNA splicing, export and translation, RNA decay, and ribosome biogenesis (10). The precise mechanisms for these functions are not well understood. DDX3X also has been shown to be involved in the cellular stress response and stress granule (SG) assembly independent of its RNA helicase activity (11). SG contains translation-initiation components and specific RNA-binding proteins. The SG is one of the two best-characterized RNA granules, the other being the processing bodies (P bodies) that encompass the mRNA decay machinery (12). DDX3X also has been proposed to act as a viral RNA sensor, signaling intermediate, and transcriptional coactivator (13, 14). Interestingly, several recent studies have reported that despite being involved in the induction of IFN-β mediated by RIG-I-like helicases (15, 16), DDX3X is necessary for the replication of several human-pathogenic viruses that impose major global health threats, including HIV, hepatitis B virus, and poxviruses (17, 18). DDX3X also appears to be a strong proviral host factor for HCV (19, 20). Recently, we showed that the highly structured HCV 3′UTR interacts with DDX3X and then activates I-kappa-B kinase-alpha (IKK-α) to induce cellular lipogenesis and LD biogenesis through a novel innate pathway independent of NF-κB (21). In addition, HCV core protein binds to and redistributes DDX3X to the viral assembly sites around LDs (22, 23); however, this interaction is dispensable not only for HCV replication but also for HCV-triggered activation of the DDX3X–IKK-α pathway (21).
In this study, we aim to dissect the precise role of DDX3X in the HCV life cycle and HCV-triggered cellular stress responses through confocal microscopic, biochemical, and virologic approaches. We demonstrate that DDX3X dynamically interacts with various cellular compartments, including SGs and LDs, IKK-α, and viral elements, conferring multiple functions in productive HCV infection.
MATERIALS AND METHODS
Cell culture.
The human hepatoma cell line Huh-7.5.1 was provided by Francis Chisari (The Scripps Research Institute, La Jolla, CA). Cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (FBS; Omega Scientific). The HCV genotype 2a JFH-1 strain was propagated and viral titer was determined as previously described (24, 25). HCV infection was performed at a multiplicity of infection (MOI) of 0.5. Culture supernatants and cell lysates were collected at various time points postinfection.
Antibodies.
Anti-HCV core protein monoclonal antibody was produced from the α-core 6G7 hybridoma cells provided by Harry Greenberg and Xiaosong He (Stanford University, Palo Alto, CA). Monoclonal antibody 9E10 (α-NS5A) was a gift of Charles Rice (The Rockefeller University, New York, NY). The following antibodies were obtained commercially: mouse (C7-50) α-HCV core protein (MA1-080; Thermo Scientific), mouse (8 G-2) α-HCV NS3 (ab65407; Abcam), rabbit α-HCV NS5B (ab65410; Abcam), rabbit α-DDX3X antibody (A300-474A, Bethyl Laboratories), purified mouse α-human IKK-α (clone B78-1; BD Pharmingen), rabbit α-IKK-α (ab4111; Abcam), mouse α-IKK-α (ab54626; Abcam), mouse α-G3BP (ab59533; Abcam), rabbit α-G3BP (ab56574; Abcam), mouse α-PABP (sc32318; Santa Cruz Biotechnology), rabbit α-PABP (ab21060; Abcam), mouse α-catalase (ab88650; Abcam), rabbit α-catalase (ab1877; Abcam), rabbit α-LC3B (2775; Cell Signaling Technology), rabbit α-DDX6 (ab40684; Abcam), mouse α-EDC4 (sc-8418; Santa Cruz Biotechnology), rabbit α-EDC4 (ab72408; Abcam), α-hemagglutinin (HA) rabbit monoclonal antibody (C29F4; Cell Signaling Technology), mouse α-Flag (F1804; Sigma), monoclonal α-β-tubulin (TUB 2.1; Sigma), mouse IgG2b, κ (clone MPC-11; BD Biosciences), mouse IgG1 monoclonal isotype (ab184443; Abcam), normal rabbit IgG (2729; Cell Signaling Technology), Alexa Fluor 488 goat α-mouse IgG (A11001; Life Technologies), Alexa Fluor 488 goat α-rabbit IgG (A11008; Life Technologies), Alexa Fluor 568 goat α-mouse IgG (A11004; Life Technologies), Alexa Fluor 568 goat α-rabbit IgG (A11011; Life Technologies), Alexa Fluor 647 goat α-mouse IgG (A21235; Life Technologies), Alexa Fluor 647 goat α-rabbit IgG (A21244; Life Technologies), goat α-mouse IgG (A2554; Sigma), and goat α-rabbit IgG (A0545; Sigma).
Plasmid transfection.
HA-IKK-α plasmid was a gift of Ebrahim Zandi (University of Southern California, Los Angeles, CA). pFLAG-CMV-NS3 and pFLAG-CMV-NS5B, both derived from HCV JFH-1 strain, were generated by amplification of the respective viral genes, which then were subcloned into p3xFLAG-CMV-7.1 expression vector (Sigma). DNA transfections were performed using FuGENE 6 transfection reagent (Roche) according to the manufacturer's instructions.
In vitro transcription and RNA transfection.
The pUC/JFH-1 plasmid was provided by Takaji Wakita (NIID, Tokyo, Japan). The plasmids pFK-J6/C3 (JC1) and pFK-Con1/C3 were gifts of Ralf Bartenschlager (University of Heidelberg, Germany). The plasmid carrying the HCV 3′UTR was generated as previously described (21). pBlueScript KS(+) enhanced green fluorescent protein (EGFP) plasmid was generated by cloning the EGFP gene into the pKS(+) vector between AvrII and NotI sites. EGFP was amplified from pEGFP-N1 plasmid using primers 5′-CCC CTA GGG ATG GTG AGC AAG GGC GAG-3′ and 3′-GAG GCG GCC GCT TGT ACA GCT CGT CCA TGC-5′. pHCV-CLX-CMV wild-type (WT) plasmid was provided by Michael Niepmann (Justus-Liebig-University-Giessen, Giessen, Germany). R-Luc/JFH-1-replicon was provided by Taka Kato (NIID, Tokyo, Japan). Plasmids were linearized by enzyme digestion and purified by phenol-chloroform-isoamyl alcohol extraction. In vitro transcription was performed by using the MEGAscript T7 kit (Ambion) according to the manufacturer's protocol. RNA was purified by use of TRIzol RNA isolation reagent (Life Technologies), and their quality and quantity were evaluated by electrophoresis and NanoDrop spectrophotometry (Thermo Scientific). RNA transfection was performed using DMRIE-C reagent (Life Technologies) according to the manufacturer's instructions.
siRNA transfection.
SMARTpool short interfering RNA (siRNA) (Dharmacon) was transfected by Oligofectamine (Life Technologies) into Huh-7.5.1 cells at a 50 nM final concentration, using a reverse transfection protocol as previously described (26). Unless otherwise indicated, further treatments or assays typically were performed 72 h after siRNA transfection, when gene-silencing efficiency reached maximal levels.
HCV core protein staining.
Huh-7.5.1 cells were treated with a nontargeting (NT) control or DDX3X SMARTpool siRNA at a concentration of 50 nM for 72 h and then infected with the HCV JFH-1 strain. Cells were fixed 48 h after infection, immunostained, and imaged for HCV core protein expression. Core protein staining was performed as previously described (21).
Quantification of viral RNA.
Total RNA was isolated from whole-cell lysate using the RNeasy minikit (Qiagen) or from culture medium with a QIAamp viral RNA minikit (Qiagen). Intracellular and extracellular copy numbers of HCV RNA were determined by quantitative PCR with the probe, primers, and parameters described previously (27). The relative amount of HCV RNA was normalized to the internal-control human 18S rRNA (Applied Biosystems).
HCV internal ribosome entry site (IRES)-mediated translation assay.
Huh-7.5.1 cells were transfected with the indicated siRNAs for 3 days and then transiently transfected with pHCV-CLX-CMV RNA harboring a firefly luciferase reporter gene. After 24 h, cell lysates were obtained and firefly luciferase activity was measured with the Firefly luciferase assay system (Promega) using a POLARstar Omega multidetection microplate reader (BMG Labtech).
HCV subgenomic replicon assay.
Huh-7.5.1 cells were treated with the indicated siRNAs for 3 days and subsequently were transfected with JFH-1-RLuc subgenomic replicon RNA. Cell lysates were collected at 48 h posttransfection, and Renilla luciferase activity was determined using the Renilla luciferase assay system (Promega).
Immunofluorescence and confocal microscopy.
Cells grown on Lab-Tek II borosilicate 4-well chamber coverslips (Nunc) were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, and incubated with blocking solution in phosphate-buffered saline (PBS) containing 3% bovine serum albumin (BSA) and 10% normal goat serum (Vector Laboratories). Cells then were labeled with appropriate primary antibodies diluted in PBS with 1% BSA and subsequently incubated with Alexa Fluor 488, 568, or 647 secondary antibody (Life Technologies) in PBS with 1% BSA. Nuclei were counterstained with Hoechst 33342 (Life Technologies) at 1:5,000 in PBS. LDs were stained with BODIPY 493/503 (Life Technologies) at 1 μg/ml for 1 h in PBS with 1% BSA. Mitochondria were labeled with MitoTracker red CMXRos (Life Technologies) at 100 nM for 30 min at 37°C. Each step was followed by three washes with PBS. Confocal laser-scanning microscopy analysis was performed with an Axio Observer.Z1 microscope equipped with a Zeiss LSM 5 Live DuoScan system under a 1.4-numeric-aperture oil-immersion 63× objective lens (Carl Zeiss). Images were acquired using ZEN 2009 software (Carl Zeiss). Dual or triple color images were acquired by consecutive scanning with only one laser line active per scan to avoid cross-excitation. Pearson's correlation coefficient (CR), which demonstrates the intensity of colocalization, was determined using ZEN software on selected areas. The CR can range from −1 to 1. A CR of −1 indicates a perfect negative linear relationship between two signals (exclusion), a CR of 0 indicates the absence of any relationship between two signals, and a CR of 1 indicates a perfect colocalization between two signals.
Immunoprecipitation.
Cells were lysed with lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris, pH 8.0) supplemented with complete protease inhibitor cocktail (Roche) for 20 min on ice and then subjected to centrifugation at 14,000 rpm for 20 min. Lysates were immunoprecipitated with the appropriate antibodies at 4°C overnight. The immune complexes were incubated with protein A-agarose beads (KPL) for 1 h, washed five times with wash buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris, pH 8.0), and eluted in Laemmli buffer for SDS-PAGE.
Immunoblotting.
After denaturation at 95°C for 5 min, proteins were separated in a NuPAGE 4 to 12% Bis-Tris gel (Life Technologies) in NuPAGE morpholineethanesulfonic acid (MES) or morpholinepropanesulfonic acid (MOPS) SDS running buffer (Life Technologies). Gels were electroblotted onto a nitrocellulose membrane (Life Technologies) in NuPAGE transfer buffer (Life Technologies). Membranes were probed overnight with appropriate primary antibodies in blocking buffer (2% blocking agent [GE Healthcare] in 0.05% Tris-buffered saline [TBS]-Tween) and washed five times for 5 min with 0.05% TBS-Tween, followed by incubation with horseradish peroxidase-labeled secondary antibody (Sigma) for 1 h. After five washing steps with 0.05% TBS-Tween and an additional washing step in TBS, immunoreactive proteins were detected using ECL Advance (GE Healthcare).
Gene expression assay.
Total cellular RNA was extracted using the RNeasy minikit (Qiagen). cDNA was synthesized using the first-strand cDNA synthesis kit (Roche). The mRNA expression levels of target genes were quantified by quantitative PCR using gene-specific primers and probes (IDT) and TaqMan gene expression master mix (Applied Biosystems) on an ABI 7500 real-time PCR system. Relative transcript levels were calculated using the ΔΔCT method (CT indicates threshold cycle), with 18S rRNA as the normalizing control gene.
Statistical analysis.
The results are presented as the means ± standard deviations (SD). The two-tailed unpaired Student's t test was used for statistical analysis. The level of significance is denoted in each figure (*, P < 0.05; **, P < 0.01).
RESULTS
HCV 3′UTR RNA induces DDX3X and IKK-α redistribution to SGs.
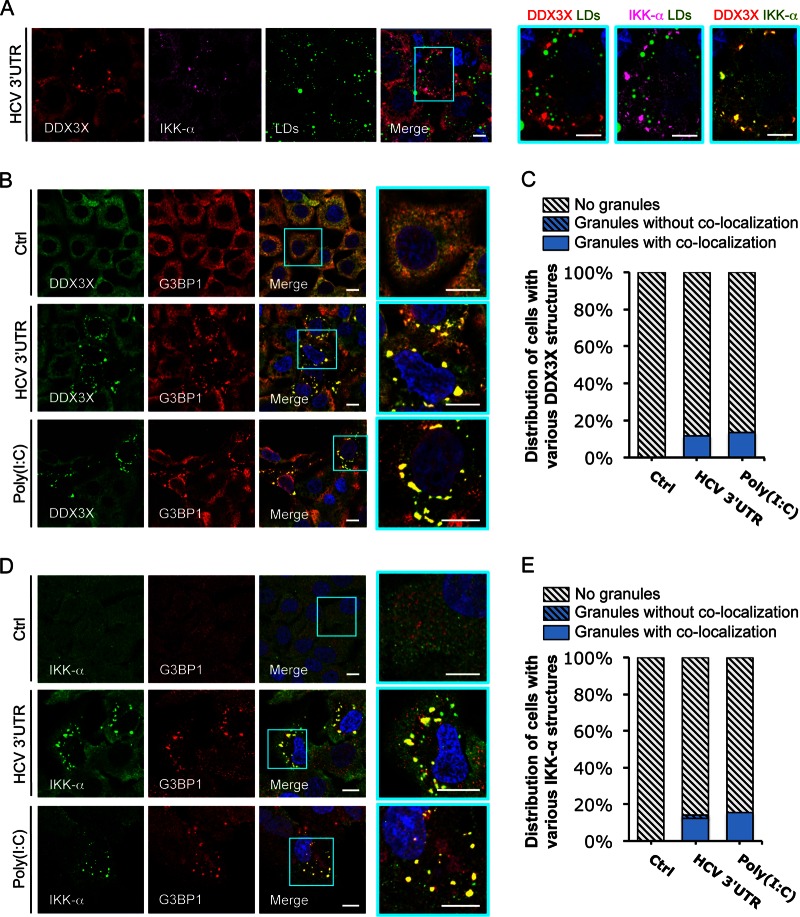
We recently showed that upon HCV infection, HCV RNA, through its 3′UTR complexes with DDX3X, in turn recruits and activates IKK-α to enhance viral assembly (21). DDX3X also has been shown to redistribute to the surface of LDs in HCV-infected cells (23). To further characterize DDX3X-HCV interactions, we first transfected Huh-7.5.1 cells with in vitro-transcribed HCV 3′UTR RNA and performed immunofluorescence and confocal imaging analysis at 24 h posttransfection. DDX3X and IKK-α formed large granular cytoplasmic structures (1 to 2.5 μm in diameter) that did not colocalize with LDs (Fig. 1A). These DDX3X-IKK-α granules stimulated by HCV 3′UTR RNA also did not colocalize with mitochondria (MitoTracker was used as a marker), peroxisomes (catalase as a marker), or autophagosomes (LC3B as a marker) (see Fig. S1A to D in the supplemental material). Intriguingly, they colocalized exclusively with GTPase-activating (SH3 domain) binding protein 1 (G3BP1), a bona fide SG marker (Fig. 1B to E). DDX3X has been shown to specifically bind to RNA containing a complex secondary structure and function as a pattern recognition receptor in innate immunity (14, 21). As such, we tested poly(I·C), a synthetic viral mimetic that serves as a pathogen-associated molecule pattern (PAMP) and potentially can be recognized by DDX3X. In the poly(I·C)-treated cells, we observed that DDX3X and IKK-α also similarly redistributed to the SGs (Fig. 1B to E).
FIG 1.
HCV 3′UTR RNA induces DDX3X–IKK-α colocalization in stress granules (SGs). Huh-7.5.1 cells were transfected with EGFP control (Ctrl) or HCV 3′UTR RNA for 24 h. Various cellular proteins and compartments were stained and subsequently examined by confocal microscopy. DDX3X (A and B) and G3BP1 (a SG marker) (D) were detected with specific primary rabbit antibodies, and IKK-α (A and D) and G3BP1 (B) were detected with specific primary mouse antibodies, followed by the secondary antibody Alexa Fluor 488 (green), 568 (red), or 647 (magenta). Lipid droplets (LDs) were stained with BODIPY 493-503 (green). Magnifications of selected areas are shown on the right. Scale bars, 10 μm. (A) DDX3X–IKK-α granules and LDs. Colors in the far right picture were digitally changed for better visualization of IKK-α (green instead of magenta) and DDX3X (red). (B) DDX3X and SGs. (C) Quantification of DDX3X structures in cells under various conditions. Confocal microscopic images (6 to 12 randomly selected fields from at least three independent experiments performed for panel B, at least 90 cells under each condition) were counted. Results shown are percentages of cells without DDX3X granules (light gray striped bars) or those with DDX3X granular structures either colocalized with SGs (blue bars) or not colocalized (blue striped bars). (D) IKK-α and SGs. (E) Quantification of IKK-α structures in cells that underwent various treatments. Confocal microscopic images (5 to 10 randomly selected fields from at least three independent experiments performed for panel D, at least 50 cells under each condition) were counted. Results shown are percentages of cells without IKK-α granules (light gray striped bars) or those with IKK-α granules either colocalized with SGs (blue bars) or not colocalized (blue striped bars).
HCV infection triggers DDX3X and IKK-α redistribution to SGs and P bodies.
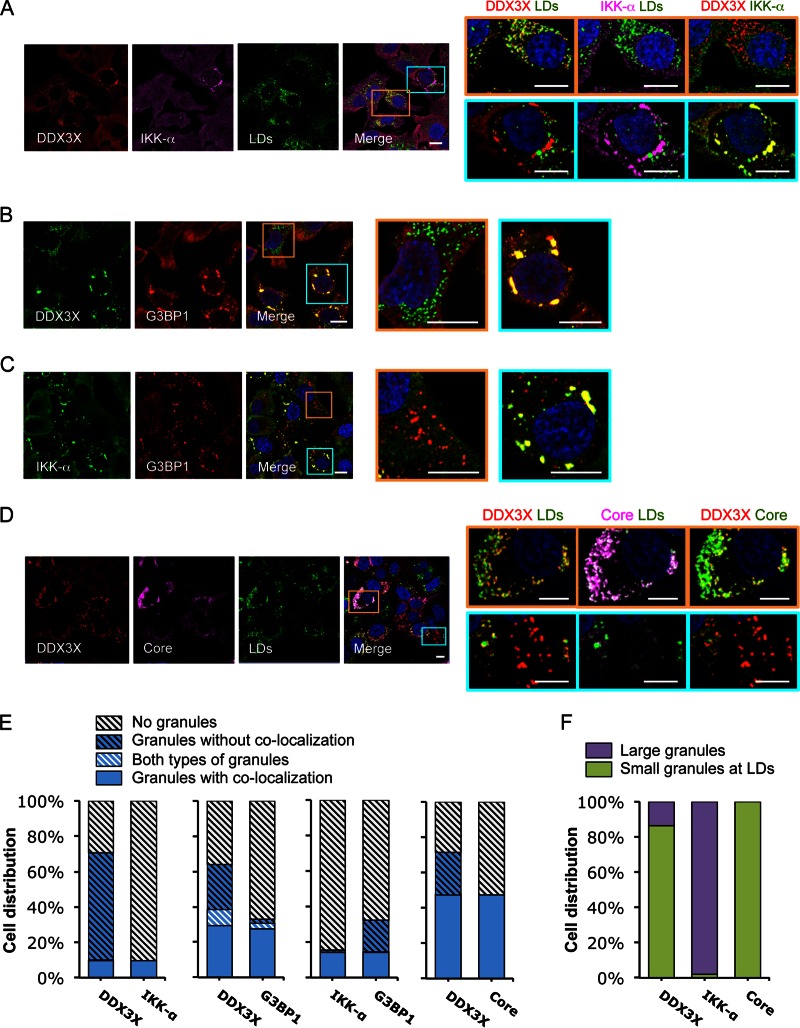
We next examined DDX3X–IKK-α localization in Huh-7.5.1 cells infected with the JFH-1 strain of HCVcc (HCV grown in cell culture). Confocal imaging analyses demonstrated that HCV infection induced large DDX3X–IKK-α granules like the ones described above (1 to 2.5 μm in diameter) that did not colocalize with LDs, mitochondria, peroxisomes, or autophagosomes (Fig. 2A; also see Fig. S1A and C to F, focus on the cells in selected blue areas) but colocalized with SGs (G3BP1 and PABP1 as markers) (Fig. 2B, C, and E; also see Fig. S2A, focus on the cells in selected blue areas). Interestingly, DDX3X, but not IKK-α, also exhibited numerous smaller (<1 μm in diameter) and round structures that surrounded the LDs (Fig. 2A, E, and F; also see Fig. S1E and F, focus on the cells in selected orange areas). These DDX3X structures predominantly colocalized with HCV core protein, whereas the large, irregular-shaped DDX3X granules did not (Fig. 2D to F). Upon HCV infection, DDX3X–IKK-α granules also colocalized with DDX6 and EDC4 (see Fig. S2B and C), markers of P bodies that are closely linked to SGs in stressed cells (28). The physical interaction of DDX3X and G3BP1 also was shown by coimmunoprecipitation assay (see Fig. S2D). Together, these results demonstrate that two forms of DDX3X granular structures are generated by stimulation with HCV infection in hepatocytes: (i) large DDX3X–IKK-α–viral RNA–SG complexes induced by HCV 3′UTR and (ii) smaller DDX3X-core protein complexes around LDs induced by HCV core protein as previously described (22, 23). The differential phenotypes manifested by HCV 3′UTR RNA transfection or HCVcc infection in triggering DDX3X and IKK-α redistribution suggests that a sequential and dynamic relocalization of these proteins from early to late infection exists.
FIG 2.
HCV infection induces formation of two subsets of DDX3X granular structures with distinct subcellular localizations. Huh-7.5.1 cells were infected with JFH-1 HCVcc for 2 days. Various cellular proteins and organelles then were detected by immunofluorescence and confocal microscopy. Scale bars, 10 μm. (A to D) Magnifications of two independent selected areas are shown on the right: small DDX3X-HCV core protein granules at the LD surface (orange area) and large DDX3X–IKK-α granules (blue area). (A) DDX3X with IKK-α or LDs. (B) DDX3X with SGs. (C) IKK-α with SGs. (D) DDX3X, HCV core protein, and LDs. (A and D) Colors in images on the far right were changed for better visualization of IKK-α (A) or HCV core protein (D), shown in green instead of magenta. (E) Quantification of cell distribution of colocalizing granular structures positive for various combinations of proteins. Confocal microscopic images from each comparison group (10 to 25 randomly selected fields from three independent experiments performed for panels A to D, at least 120 cells per group) were counted. The comparison groups are DDX3X with IKK-α (far left), DDX3X with G3BP1 (second from the left), IKK-α with G3BP1 (second from the right), and DDX3X with HCV core protein (far right). Results shown are percentages of cells without any granular structures (light gray striped bars) or those with granular structures either colocalized with each other (blue bars) or not colocalized (blue striped bars). Blue-white striped bars represent percentages of cells with both colocalized and noncolocalized granules. (F) Quantification of cell distribution containing large granules (purple bars) or small granules that colocalize with LDs (green bars) stained positive for DDX3X, IKK-α, or HCV core protein. At least 40 cells for each protein were counted, and percentages of cells with each pattern of granular structure are shown.
Dynamic cellular distribution of DDX3X in HCV infection.
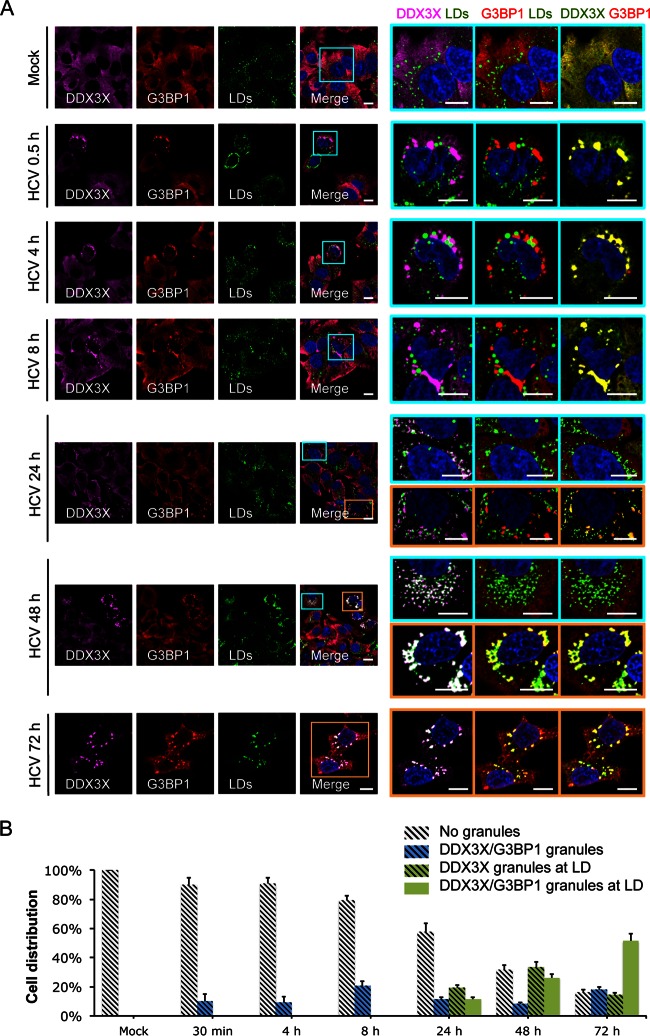
To determine if the various patterns of DDX3X distribution were related to HCV infection kinetics, we performed a time course infection study (Fig. 3; also see Fig. S3 and S4 in the supplemental material). No DDX3X granular structures could be detected at the time of HCV inoculation, but they appeared as early as 30 min postinfection and colocalized exclusively with G3BP1 and PABP1 granules but not with LDs at all early time points (30 min to 8 h) (Fig. 3; also see Fig. S4). These DDX3X granules were not triggered by inoculation of cells with culture supernatant from naive cells or with heat-inactivated JFH-1 HCVcc (see Fig. S5). They may be induced by HCV 3′UTR RNA that is present in newly infected cells either just after inoculation or as the virus spreads and infects naive neighboring cells.
FIG 3.
Dynamic shift of DDX3X subcellular localization during HCV infection. (A) Huh-7.5.1 cells were mock infected or infected with JFH-1 HCVcc for 0.5 to 72 h. DDX3X, G3BP1, and LDs were stained, and their subcellular localizations and associations were examined. Magnifications of selected areas are shown on the right. Colors on the far right were modified for better visualization of DDX3X, shown in green instead of magenta. Scale bars, 10 μm. (B) Quantification of DDX3X colocalization with SGs and/or LDs. Five to 10 confocal microscopic pictures from at least three independent experiments with a total of at least 30 cells were analyzed for colocalization signals. Percentages of cells without granular structures (light gray striped bars) or presenting DDX3X granules that are colocalized with SGs only (blue stripped bars), LDs only (green striped bars), or both SGs and LDs (green bars) are shown.
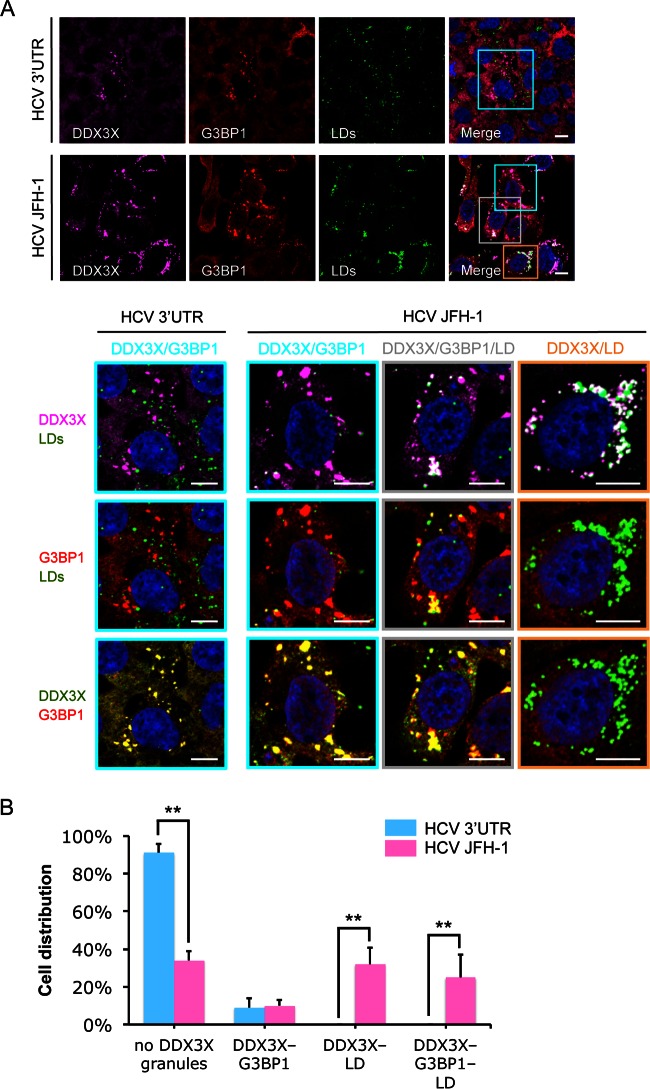
At 24 to 72 h postinfection, when HCV core protein is present in sufficient levels (see Fig. S3A in the supplemental material), the second population of DDX3X structures became evident. These small and numerous DDX3X-positive structures colocalized with core protein around LDs (see Fig. S3). The formation of these structures suggests that DDX3X redistributes with core protein to the LDs when HCV protein expression is robust late during infection. Intriguingly, some DDX3X-core protein-containing structures at the LD surface were stained positive for SG-associated proteins 48 h postinfection (Fig. 3 and 4; also see Fig. S3). All of the observed structures can coexist in the same population of cells but rarely in the same cell, demonstrating the spreading and different stages of infection among cells (Fig. 3 and 4; also see Fig. S3). HCV 3′UTR RNA transfection, in contrast, only stimulated large DDX3X-SG structures (Fig. 4), suggesting that viral protein expression is required for redistribution of DDX3X and G3BP1 granules to LD surfaces.
FIG 4.
Different patterns of DDX3X granular structures coexist in a population of HCV-infected cells. Huh-7.5.1 cells were transfected with HCV 3′UTR RNA for 24 h or infected with JFH-1 HCVcc for 48 h. G3BP1, DDX3X, and LD then were detected by staining, and their associations were evaluated. (A) DDX3X colocalization with SGs or LDs. Magnifications of selected areas are shown in the lower panels. Colors in the lowest panel were modified for better visualization of DDX3X (green instead of magenta) and G3BP1 (red). Scale bars, 10 μm. (B) Quantification of DDX3X granules associated with SGs or LDs in HCV 3′UTR-transfected or HCV-infected cells. Confocal microscopy images were analyzed visually. Cells were classified into four groups according to the presence or absence of DDX3X granules and their association with SGs or LDs. Percentages of cells in the four groups are shown. Five to 10 randomly selected fields with a total of at least 60 cells were counted. Error bars represent ±SD from triplicate experiments. **, P < 0.01.
Functions of DDX3X and SGs in HCV infection.
We previously demonstrated a role of DDX3X in facilitating HCV assembly through triggering an IKK-α-dependent proviral effect upon association of DDX3X with HCV 3′UTR RNA (21). DDX3X also has been shown to be important for HCV replication (19, 20). We silenced DDX3X in Huh-7.5.1 cells using siRNA and performed various HCV life cycle assays (29): quantification of intracellular and extracellular HCV RNA or immunostaining of HCV core protein. Silencing of DDX3X led to a significant inhibition of intracellular as well as extracellular HCV RNA levels and reduced the number of core protein-positive cells after 2 days of infection (see Fig. S6A to C in the supplemental material). The role of DDX3X in the early steps of HCV infection was studied by transfecting DDX3X-silenced Huh-7.5.1 cells with a firefly luciferase-encoded construct driven by the HCV IRES (see Fig. S6D) or a subgenomic JFH-1 replicon encoding Renilla luciferase (see Fig. S6E). One or 2 days after transfection, the firefly or Renilla luciferase activity in cells was determined as a marker of HCV IRES-mediated translation or RNA replication, respectively. Depletion of DDX3X in cells significantly inhibited HCV IRES-mediated translation and viral replication (see Fig. S6D and E). Since HCV RNA triggers the interaction of SGs and DDX3X as shown above, we then knocked down three major SG-nucleating proteins: G3BP1, T-cell-restricted intracellular antigen 1 (TIA1), and its paralog, TIA1 cytotoxic granule-associated RNA binding protein-like 1 (TIAL1). Depletion of these genes by siRNA in hepatocytes resulted in significant reduction of both intracellular and extracellular HCV RNA levels (see Fig. S6F and G). Moreover, G3BP1 silencing also led to a reduction in the number of core protein-positive cells and a 2-fold inhibition of HCV replication (see Fig. S6H and I). These results demonstrate an important role of SGs in productive HCV infection.
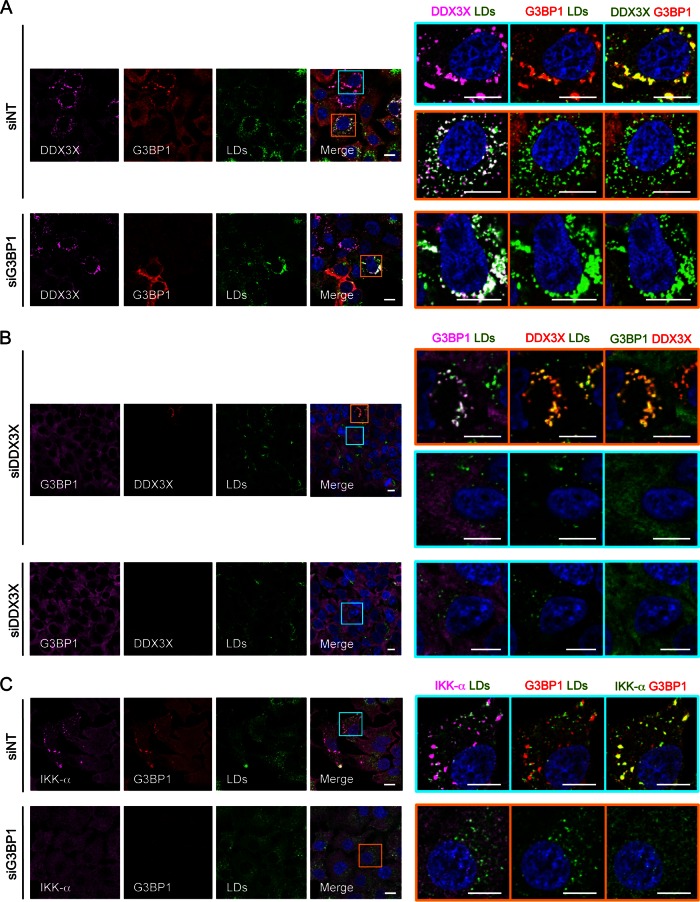
To further address the interactions of SGs with DDX3X and IKK-α in HCV infection, we depleted various SG-associated proteins by siRNAs. Silencing of G3BP1 or PKR (protein kinase R), but not IKK-α or PABP1 expression, abrogated the formation of HCV-induced SGs detected with a specific anti-G3BP1 antibody (Fig. 5; also see Fig. S7 in the supplemental material). The eIF2α kinase PKR activation is a prerequisite of SG formation upon cellular stress involving the presence of double-stranded RNA (such as the HCV replication intermediate) (30). Intriguingly, in cells deprived of DDX3X, no SG was induced either (Fig. 5B), indicating that DDX3X expression is necessary for HCV-triggered SG formation. Moreover, the association of DDX3X with the large SG was observed in infected cells despite the silencing of IKK-α expression (see Fig. S7A), consistent with our previous finding that DDX3X functions upstream of IKK-α in this signaling cascade (21). Cells treated with PKR siRNA were unable to aggregate IKK-α upon HCV infection (see Fig. S7B), and in siG3BP1-transfected cells, neither IKK-α nor PAPB1 granules were detected (Fig. 5C; also see Fig. S7C). However, DDX3X staining in complex with HCV core protein on the surface of LDs still was detected in these cells (Fig. 5A; also see Fig. S7A). These observations are consistent with the notion that colocalization of DDX3X and HCV core protein on LDs is independent of the SG–DDX3X–IKK-α-mediated signaling pathway (21, 22). These results also suggest the importance of SGs in the recruitment of IKK-α by DDX3X upon HCV infection.
FIG 5.
Effect of G3BP1 or DDX3X silencing on SG formation and DDX3X–IKK-α localization. Huh-7.5.1 cells were treated with nontargeting control (siNT) or G3BP1 or DDX3X siRNA for 3 days and then infected with JFH-1 HCVcc for 2 days. LDs and various indicated proteins then were stained and analyzed for their subcellular localizations and associations. Magnifications of the selected area are shown on the right. In the majority of siG3BP1- or siDDX3X-treated cells, G3BP1 or DDX3X was efficiently depleted. A few cells still showed G3BP1 or DDX3X expression; therefore, SG-DDX3X-LD colocalization still exists in these cells (A, lower, and B, upper). (A) Small DDX3X granules are associated with LDs in G3BP1-silenced cells. (B) DDX3X depletion abrogated SG formation. (C) Lack of IKK-α granules in G3BP1-silenced cells. Colors of pictures on the far right were modified for better visualization of DDX3X (A), G3BP1 (B), or IKK-α (C), shown in green instead of magenta. Scale bars, 10 μm.
Interaction of DDX3X and IKK-α with HCV nonstructural proteins.
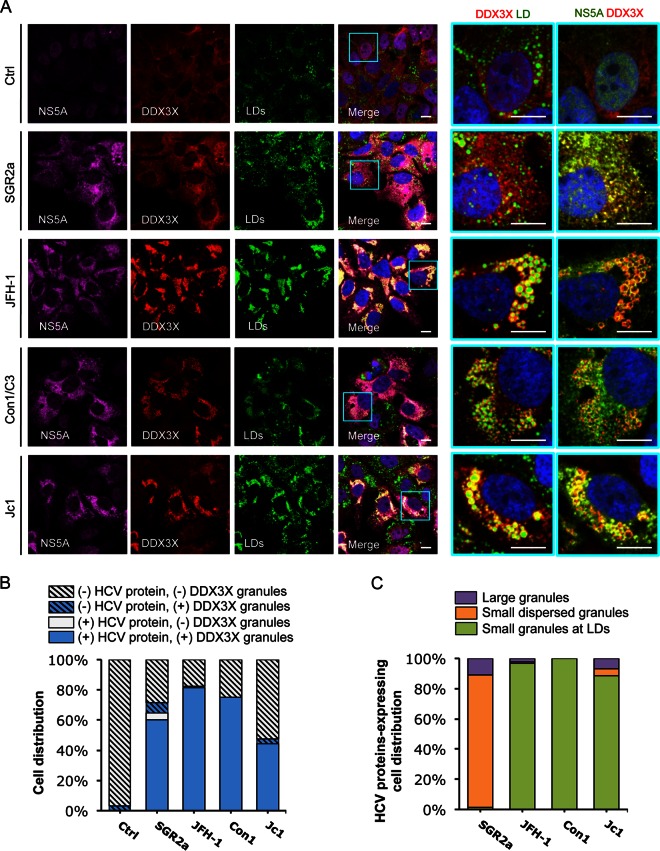
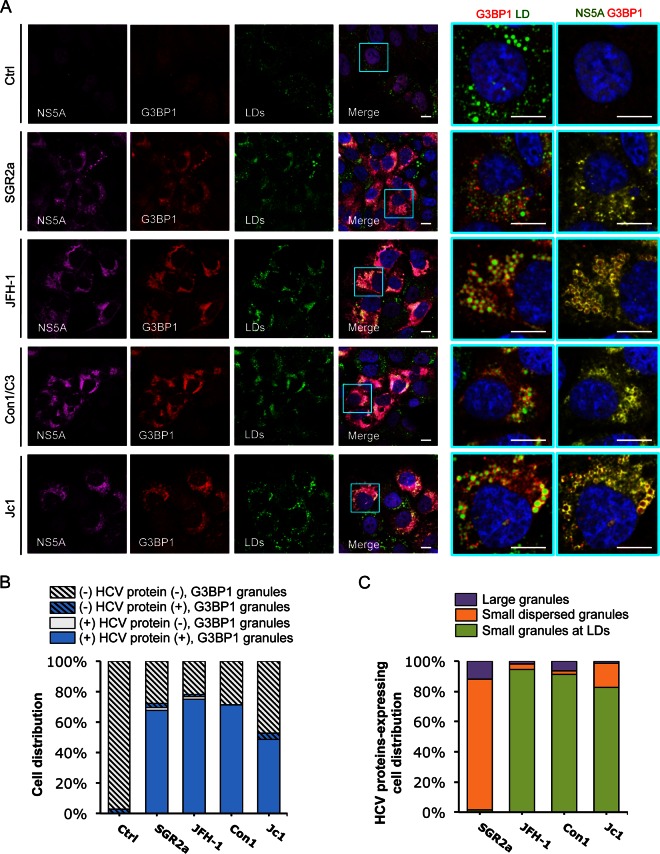
To explore the potential role of various HCV proteins in the dynamics of DDX3X-SG redistribution, we studied the effects of various HCV expression constructs in Huh-7.5.1 cells: a subgenomic replicon of JFH-1 encoding only the nonstructural proteins, the complete genome of JFH-1, and two chimeras of JFH-1, Con1/C3 and Jc1, which expressed the structural proteins of either Con1 (genotype 1b) or J6 (genotype 2a), respectively (Fig. 6 and 7). At 2 days posttransfection, cells were examined by immunostaining and confocal microscopy. When transfected with a control RNA, most cells displayed weak and diffuse DDX3X staining (Fig. 6A). About 3% of cells acquired DDX3X granules (Fig. 6B), which could be attributed to spontaneous activation of DDX3X during the electroporation process for RNA transfection. Upon transfection of any of the four abovementioned HCV constructs, the vast majority of the cells produced HCV proteins (NS5A or NS3) at detectable levels, and most of these viral protein-expressing cells displayed DDX3X granules (Fig. 6A and B). In cells harboring the subgenomic replicon of JFH-1, DDX3X-containing granules colocalized extensively with NS5A in the ER-derived membranous web, presumably at the RC, but colocalized only a little with LDs (Fig. 6A and C). In contrast, upon transfection of full-length HCV genome (JFH-1, Con1/C3, or Jc1), DDX3X colocalized predominantly with NS5A at the surface of LDs (Fig. 6A and C). Similar patterns of NS5A or LD association were observed for SGs with G3BP1 staining (Fig. 7). These data suggest that HCV nonstructural proteins mediate the localization of DDX3X-SG complex to the membranous web on the ER. The data also confirm the role of HCV core protein in mediating DDX3X-SG redistribution to LDs.
FIG 6.
HCV NS5A protein colocalizes with DDX3X and LDs. Huh-7.5.1 cells were transfected with in vitro-transcribed EGFP control RNA (Ctrl), subgenomic replicon RNA of HCV genotype 2a (SGR2a), genomic-length JFH-1, Con1/C3, or Jc1 chimeric RNA for 48 h. DDX3X, HCV NS5A, and LDs were stained and characterized for their localizations and associations. (A) Confocal microscopic analyses of DDX3X, NS5A, and LD staining in cells treated with various HCV RNAs. Magnifications of selected areas are shown on the right. Colors of pictures on the far right were digitally changed for better visualization of NS5A (green instead of magenta) and DDX3X (red). Scale bars, 10 μm. (B) Quantification of cell distribution for DDX3X granules in HCV protein-negative or -positive cells. Confocal microscopic images (6 to 12 per condition from at least three independent experiments with at least 80 cells in total) were analyzed for the presence or absence of DDX3X granules in HCV protein-positive or -negative cells, as determined by detection of NS5A (A), NS3 (not shown), or core protein (not shown). Results shown are percentages of cells that are HCV protein negative and either with (blue striped bars) or without (light gray striped bars) DDX3X granules and cells that are HCV protein positive and either with (blue bars) or without (light gray bars) DDX3X granules. (C) Characterization and quantification of DDX3X-positive structures in HCV protein-positive cells. HCV protein-positive cells with DDX3X granules (blue bars from panel B) were further classified into three groups: cells with a few large DDX3X granules not associated with LDs (purple bars), cells with DDX3X granules dispersed throughout the ER (orange bars, probably representing HCV replicative intermediates), and cells with small and numerous DDX3X granules localized to LDs (green bars). Percentages of cells in each group are shown (a total of at least 40 cells).
FIG 7.
Colocalization of HCV NS5A protein with SGs. In vitro-transcribed control RNA (Ctrl), SGR2a, JFH-1, Con1/C3, or Jc1 chimera RNA-transfected Huh-7.5.1 cells (as described for Fig. 6) were stained for HCV NS5A, SGs, and LDs. (A) Confocal microscopic imaging of G3BP1 (for SGs), NS5A, and LDs. Magnifications of selected areas are shown on the right. Colors in pictures on the far right were digitally changed for better visualization of NS5A (green instead of magenta) and G3BP1 (red). Scale bars, 10 μm. (B) Quantification of SGs in HCV protein-negative or -positive cells. Six to 12 randomly selected microscopic images from at least three independent experiments with a total of at least 80 cells were analyzed for the presence or absence of SGs in HCV protein-positive or -negative cells. Results shown are percentages of cells that are HCV protein negative and either with (blue striped bars) or without (light gray striped bars) SGs and cells that are HCV protein positive and either with (blue bars) or without (light gray bars) SGs. (C) Characterization of SGs in HCV protein-positive cells. This population of cells, which also presents SGs (blue bars in panel B), were further classified into three groups: cells with a few large SGs not associated with LDs (purple bars), cells with G3BP1 granules dispersed throughout the ER (orange bars), and cells with small and numerous SGs localized to LDs (green bars). Percentages of HCV-positive cells (in a total of at least 40 cells) in each group are shown.
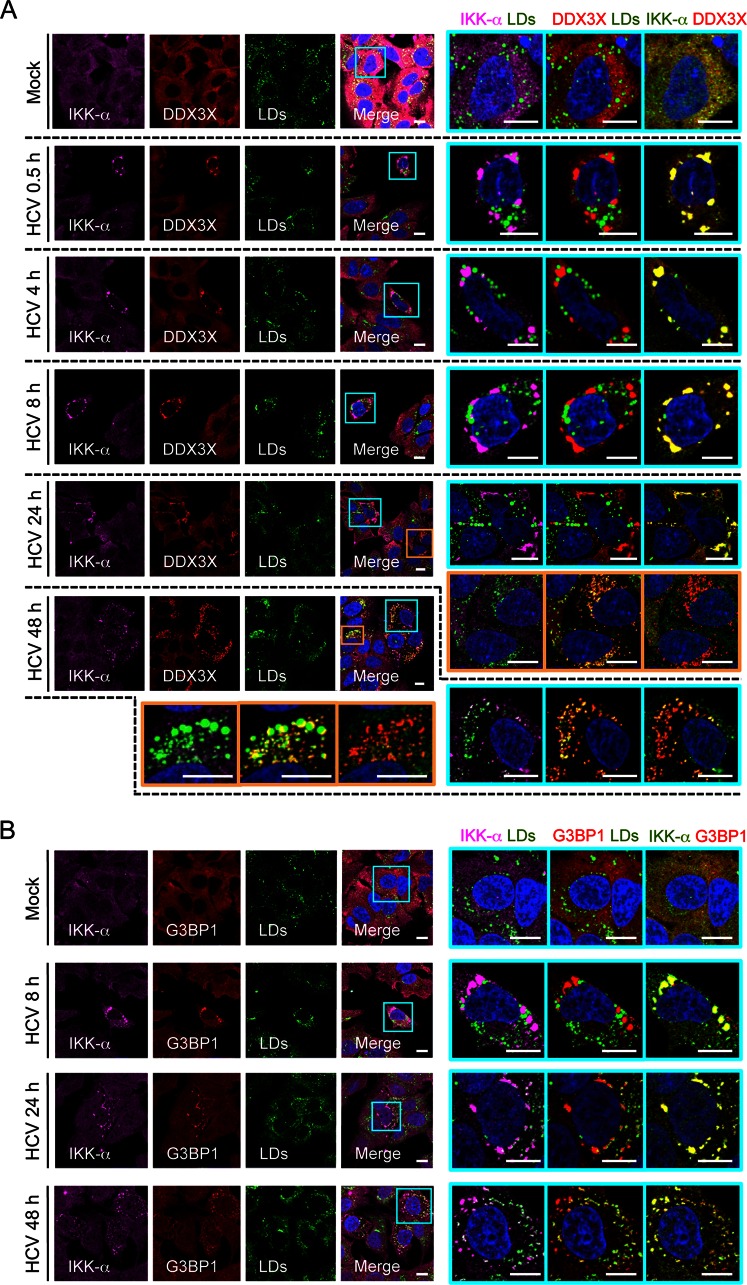
We previously demonstrated that the HCV 3′UTR mediates the formation of a DDX3X–IKK-α complex, leading to IKK-α activation and nuclear translocation for downstream transcriptional functions (21). To further investigate the dynamic process of IKK-α localization and association with DDX3X-SGs during HCV infection, we performed an HCV infection time course experiment (Fig. 8). As early as 30 min postinfection, IKK-α granules started to appear and colocalized with DDX3X and SGs but not with LDs at any time point (Fig. 8). At 24 h postinfection, a smaller population of DDX3X colocalizing with LDs was observed in a subset of cells as described in the legend to Fig. 3; however, IKK-α was not detected in these structures (Fig. 8A). Moreover, in cells that exhibited DDX3X and LD colocalization, the granular cytoplasmic structures of IKK-α no longer existed (Fig. 8A).
FIG 8.
Associations between IKK-α, DDX3X, SGs, and LDs upon HCV infection. Huh-7.5.1 cells were mock infected or infected with JFH-1 HCVcc and then stained for IKK-α, DDX3X, SGs, and LDs at various time points. (A) Association of IKK-α with DDX3X. (B) Association of IKK-α with SGs. Magnifications of selected areas are shown on the right. Colors in pictures on the far right were digitally changed for better visualization of IKK-α, shown in green instead of magenta. Scale bars, 10 μm.
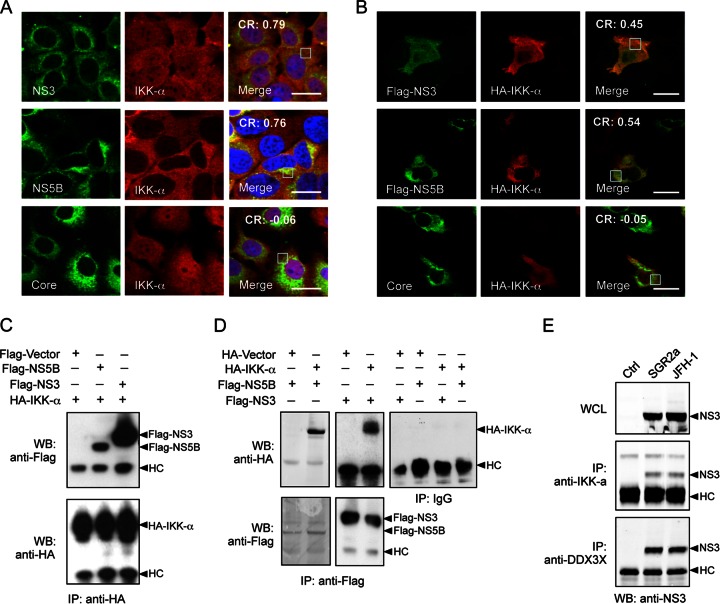
Lastly, HCV infection or transfection of plasmid constructs expressing individual HCV proteins showed that both endogenously and exogenously expressed IKK-α colocalized with NS3 and NS5B but not with core protein, as demonstrated by immunofluorescence and coimmunoprecipitation assays (Fig. 9A to D). NS3 protein also was coimmunoprecipitated with DDX3X and IKK-α in cells transfected with subgenomic or full-length HCV RNA (Fig. 9E). Therefore, HCV nonstructural proteins seem to be important for the association among IKK-α, DDX3X, and SGs. Unlike DDX3X, IKK-α does not interact with HCV core protein for subsequent trafficking to the LD surface. Instead, IKK-α from the initial SG-associated complex is phosphorylated and subsequently translocated to the nucleus, as suggested by our previous study (21).
FIG 9.
Interaction of IKK-α with HCV nonstructural proteins. (A and B) Colocalization of IKK-α with HCV NS3 and NS5B proteins. Huh-7.5.1 cells were infected with JFH-1 HCVcc for 48 h (A) or were transfected with various plasmids encoding HCV proteins for 24 h (B). The subcellular localization of IKK-α and its association with HCV proteins were detected and analyzed by immunofluorescence and confocal microscopy. Scale bars, 20 μm. Pearson's correlation coefficient (CR) was determined on the selected areas (in white boxes). (C and D) Cells were transfected with plasmids encoding various Flag-tagged HCV proteins and/or HA-IKK-α for 24 h. Immunoprecipitation (IP) with anti-HA, anti-Flag, or IgG control antibody was performed, followed by Western blotting (WB) using anti-Flag or anti-HA antibody. (E) Cells were transfected with in vitro-transcribed EGFP control RNA (Ctrl), SGR2a, or JFH-1 RNA for 48 h. Immunoprecipitation on whole-cell lysate (WCL) with anti-IKK-α or anti-DDX3X antibody was conducted, followed by Western blotting using anti-NS3 antibody. HC, IgG heavy chain.
DISCUSSION
We and other groups have previously demonstrated that DDX3X interplays with multiple cellular compartments and HCV elements and exerts an important role in HCV replication and assembly (19–21, 31). The mechanisms underlying these DDX3X-mediated effects remain elusive; in particular, how DDX3X is involved in multiple concurrent cellular machineries that are exploited by HCV for efficient infection is largely unknown. In this study, applying systematic imaging and virologic approaches, we examined the dynamic subcellular localization and association of DDX3X with various cellular components and viral elements and explored the intrinsic functions of DDX3X in the HCV life cycle and HCV-mediated cellular responses.
Once entering host cells, HCV releases its genomic RNA, the 3′UTR of which interacts with DDX3X and initiates a cascade of signaling processes, including PKR-mediated stress granule formation, DDX3X and IKK-α association, and recruitment of DDX3X–IKK-α complex to the SGs (Fig. 10). We recently demonstrated that IKK-α, a major I-κB kinase for activation of the NF-κB pathway, exerts a proviral role in HCV assembly through the induction of host cell lipogenic gene expression and lipid droplet formation. This transcriptional role of IKK-α is independent of NF-κB but is subsequent to HCV 3′UTR-DDX3X interaction (21). DDX3X, bound to and activated by the HCV 3′UTR, interacts with and recruits IKK-α to the SGs, leading to its phosphorylation and nuclear translocation through an unidentified mechanism. Here, we show that DDX3X–IKK-α association and activation take place at the stress granules but not in other subcellular organelles, such as the autophagosome, mitochondria, peroxisome, or lipid droplets, that have been suggested as the sites for HCV replication and production or implicated in the antiviral cellular responses that restrain HCV infection.
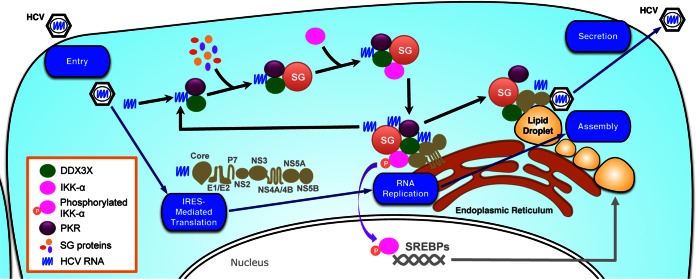
FIG 10.
Proposed model for dynamic localization and association of DDX3X and IKK-α with various cellular compartments during HCV life cycle. Using an HCV life cycle map as the background, the dynamic interactions of DDX3X, IKK-α, SG, LD, and HCV elements are shown. HCV infection, through the 3′UTR RNA, stimulates PKR- and DDX3X-dependent SG formation, which then recruits IKK-α, leading to IKK-α activation at the SGs. The SG-associated DDX3X–IKK-α complex also interacts with HCV nonstructural proteins, which, together with viral genome RNA, constitute the HCV replicase machinery on the ER-derived membranous web. The formation of functional complexes harboring the HCV 3′UTR, DDX3X, IKK-α, and SGs is further amplified by viral replication by providing more HCV genomic RNA. Late in HCV infection, DDX3X and SGs, via interaction with the core protein, redistribute from the ER to the surface of LDs where assembly occurs. In contrast, the transcription factor IKK-α does not relocate to the LDs but translocates to the nucleus for a role in activating cellular lipogenesis and LD biogenesis.
Stress granules are cytoplasmic macromolecular structures of stalled translation initiation complexes that are induced in response to various stress conditions, including viral infection (32). The formation of SGs is driven by aggregation of several key RNA binding proteins, such as TIA-1, TIA-1-related protein (TIAR), and G3BP1 (33). Cells depleted of either G3BP1, TIA-1, or TIAR subsequently are deficient in SG formation (34–36). The primary role of SGs in translation suppression and RNA decay suggests this cellular event impacts the viral life cycle and can be manipulated by viruses for their own survival. Indeed, SG formation is a hallmark of infection for many RNA viruses, albeit with a range of phenotypes. Generally, it appears to be inhibited during infection by most viruses, including influenza virus, HIV-1, rotavirus, and two important members of the Flaviviridae family, West Nile virus (WNV) and dengue virus (37). Since SGs potentially are antiviral by sequestering and altering cell components that are pivotal for viral replication, the antagonization of SG formation by these viruses may be a viral strategy to subvert the cellular stress responses that restrict infections (37). Several other viruses, such as reoviruses (38), respiratory syncytial virus (RSV) (39), and coronaviruses (40, 41), in contrast, induce and coopt SG responses as part of their replication cycle.
HCV, a hepatotropic RNA virus, also has been shown to manipulate SG formation and interact with SG components for survival and efficient propagation. During HCV infection, G3BP1 is associated with the viral NS5B protein and the negative-strand viral RNA; thus, it constitutes part of the HCV replication complexes (RCs) (42). HCV also recruits other components of SGs to the viral RCs, and these SG markers play an important role in HCV genome amplification (43). Using live-cell imaging technology, two recent studies showed that the assembly and disassembly of SGs is a highly dynamic (oscillating) process in HCV-infected cells (30, 44). In addition, HCV-triggered SG formation is associated with delayed cell division, stalled translation, and prolonged cell survival, suggesting that HCV exploits this important stress response to establish persistent infection in hepatocytes (30). Apart from the effect on HCV RNA replication, the SG proteins have been shown to be required for either HCV assembly (G3BP1 and TIA-1) or secretion (TIAR) (31).
Here, we show that SGs, stimulated by HCV 3′UTR RNA in infected cells, provide a platform for DDX3X and IKK-α association and activation (Fig. 10). The SG-associated DDX3X–IKK-α complex binds to multiple HCV nonstructural proteins, which, together with viral genomic RNA, establish the HCV replicase machinery on the membranous web derived from the ER membrane. Late in HCV infection, DDX3X and SGs redistribute from the ER to LDs (Fig. 10). In the absence of core protein expression (for example, in cells with subgenomic replicon RNA transfection), the DDX3X-SG complex does not translocate to the LDs but is retained in the yet-to-be fully established HCV RNA machinery within the ER-derived membranous web. IKK-α does not relocate to the LDs either but instead translocates to the nucleus upon phosphorylation for a role in transcriptionally regulating cellular lipogenesis (Fig. 10) (21).
The relocation of SGs to LDs at the late step of viral infection also has been reported by Ariumi et al. They have shown that HCV hijacks the SGs as well as P-body contents around LDs for enhancement of viral production (43). However, how the SG components are redistributed to the LDs and their contingent roles in modulating HCV assembly still are unclear. DDX3X has been shown to interact with HCV core protein, but this interaction is dispensable for productive HCV infection (22); hence, it is not likely to mediate SG relocalization, as the relocated, LD-associated SG proteins ultimately are required for HCV assembly (31). In addition, the major SG component G3BP1 does not colocalize with an ectopically expressed core protein, which mainly localizes to LDs, suggesting that other cellular or viral factors (such as nonstructural proteins) contribute to the redistribution of SGs to the LDs in association with HCV core protein (45).
Among the early initiation factors present in SGs is the poly(A)-binding protein 1 (PABP1). PABP1 stimulates initiation factor recruitment to the mRNA, leading to mRNA circularization and efficient translation (14). In a previous study, PABP1 was shown to directly bind to DDX3X, and downregulation of DDX3X in cells interfered with SG assembly (11). This is consistent with our finding that DDX3X knockdown significantly impaired HCV-induced SG formation (Fig. 5B). We also showed that silencing of G3BP1 completely abolished the formation of SGs as well as the formation of DDX3X–IKK-α granules (Fig. 5A and C). These data further confirmed the important role of G3BP1 in SG formation as previously described (35) and implicated an intrinsic role of SG in DDX3X–IKK-α activation for the downstream role in facilitating HCV assembly. In addition, depletion of either DDX3X or the SG component G3BP1 or TIA paralogs significantly reduced viral RNA levels in cells, demonstrating the involvement of DDX3X-SG complexes in HCV replication. This effect was further supported by the presence of HCV nonstructural proteins in the DDX3X-SG complex in the context of either full-length or subgenomic HCV replicon transfection. Moreover, G3BP1 directly binds to HCV NS5B protein and associates with viral RNA (42), implying that SGs participate in the formation of the HCV RC that controls viral RNA replication.
DDX3X previously has been shown to (i) function as an eIF4E-inhibitory protein to specifically repress cap-dependent translation by trapping eIF4E in a translationally inactive complex (46); (ii) preferentially promote the translation initiation of structured 5′UTR regions (47, 48); (iii) associate with the cytoplasmic multisubunit translation initiation factor eIF3 (49), which is involved in HCV translation, by specifically binding to the IRES in the HCV 5′UTR (50, 51); and (iv) interact with HCV core protein to inhibit translation of capped but not uncapped RNA (52). As all of these effects of DDX3X can be attributed to SG functions, we extrapolate that HCV may hijack DDX3X-SG complexes to regulate the expression of viral and cellular proteins for its own advantage.
It is known that LDs are essential for the assembly of infectious HCV particles, and that HCV core protein plays a key role in this process. The core protein promotes the accumulation of LDs to facilitate virus assembly. It has been proposed that HCV core protein recruits nonstructural proteins, mainly NS5A, viral RNA, and the RC- to LD-associated membranes (6), and that NS5A plays a dual role in HCV replication and assembly and meanwhile acts as a switch between these two processes (53, 54). In addition to NS5A, other nonstructural proteins, such as NS2, NS3, NS4A, and NS4B, were shown to be critical for HCV assembly (44, 55–57). Moreover, HCV core protein can interact with NS5B and modulate its RNA-dependent RNA polymerase activity (58). The complex and dynamic interactions of HCV components with DDX3X and SG proteins in LD-associated membranes would allow the tight control of each step of the HCV life cycle as well as potential inhibition of innate antiviral responses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Helen Cha, Keng-Hsin Lan, and Zongyi Hu for technical assistance. We also thank all members of the Liver Diseases Branch, NIDDK, for discussion and support.
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), U.S. National Institutes of Health (NIH).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03197-14.
REFERENCES
- 1.Heim MH. 2013. Innate immunity and HCV. J Hepatol 58:564–574. doi: 10.1016/j.jhep.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Thimme R, Binder M, Bartenschlager R. 2012. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS Microbiol Rev 36:663–683. doi: 10.1111/j.1574-6976.2011.00319.x. [DOI] [PubMed] [Google Scholar]
- 3.Liang TJ. 2013. Current progress in development of hepatitis C virus vaccines. Nat Med 19:869–878. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang TJ, Ghany MG. 2013. Current and future therapies for hepatitis C virus infection. N Engl J Med 368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLauchlan J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J Viral Hepatitis 7:2–14. doi: 10.1046/j.1365-2893.2000.00201.x. [DOI] [PubMed] [Google Scholar]
- 6.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 7.Rouillé Y, Helle F, Delgrange D, Roingeard P, Voisset C, Blanchard E, Belouzard S, McKeating J, Patel AH, Maertens G, Wakita T, Wychowski C, Dubuisson J. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol 80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abid K, Pazienza V, de Gottardi A, Rubbia-Brandt L, Conne B, Pugnale P, Rossi C, Mangia A, Negro F. 2005. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol 42:744–751. doi: 10.1016/j.jhep.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 10.Rocak S, Linder P. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 11.Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. 2012. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J 441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 12.Adjibade P, Mazroui R. 2014. Control of mRNA turnover: Implication of cytoplasmic RNA granules. Semin Cell Dev Biol 34:15–23. doi: 10.1016/j.semcdb.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Schroder M. 2010. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochemical Pharmacol 79:297–306. doi: 10.1016/j.bcp.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 14.Shih JW, Lee YH. 2014. Human DExD/H RNA helicases: Emerging roles in stress survival regulation. Clin Chim Acta 436C:45–58. doi: 10.1016/j.cca.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Oshiumi H, Sakai K, Matsumoto M, Seya T. 2010. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol 40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 16.Schroder M, Baran M, Bowie AG. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J 27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Ge LL, Li PP, Wang Y, Dai JJ, Sun MX, Huang L, Shen ZQ, Hu XC, Ishag H, Mao X. 2014. Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions. Virology 449:70–81. doi: 10.1016/j.virol.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder M. 2011. Viruses and the human DEAD-box helicase DDX3: inhibition or exploitation? Biochem Soc Trans 39:679–683. doi: 10.1042/BST0390679. [DOI] [PubMed] [Google Scholar]
- 19.Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol 81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A 104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. 2013. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med 19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angus AG, Dalrymple D, Boulant S, McGivern DR, Clayton RF, Scott MJ, Adair R, Graham S, Owsianka AM, Targett-Adams P, Li K, Wakita T, McLauchlan J, Lemon SM, Patel AH. 2010. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol 91:122–132. doi: 10.1099/vir.0.015909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owsianka AM, Patel AH. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257:330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- 24.Kato T, Matsumura T, Heller T, Saito S, Sapp RK, Murthy K, Wakita T, Liang TJ. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol 81:4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A 106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YY, Zhang BH, Ishii K, Liang TJ. 2010. Novel function of CD81 in controlling hepatitis C virus replication. J Virol 84:3396–3407. doi: 10.1128/JVI.02391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonas S, Izaurralde E. 2013. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev 27:2628–2641. doi: 10.1101/gad.227843.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Zhang YY, Chiu S, Hu Z, Lan KH, Cha H, Sodroski C, Zhang F, Hsu CS, Thomas E, Liang TJ. 2014. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog 10:e1004163. doi: 10.1371/journal.ppat.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, Mazur J, Bankhead P, Hiet MS, Kallis S, Alvisi G, Samuel CE, Lohmann V, Kaderali L, Rohr K, Frese M, Stoecklin G, Bartenschlager R. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garaigorta U, Heim MH, Boyd B, Wieland S, Chisari FV. 2012. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J Virol 86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 37.White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol 20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. 2006. Reovirus induces and benefits from an integrated cellular stress response. J Virol 80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE Jr. 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol 84:12274–12284. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol 9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sola I, Galan C, Mateos-Gomez PA, Palacio L, Zuniga S, Cruz JL, Almazan F, Enjuanes L. 2011. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J Virol 85:5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi Z, Pan T, Wu X, Song W, Wang S, Xu Y, Rice CM, Macdonald MR, Yuan Z. 2011. Hepatitis C virus coopts Ras-GTPase-activating protein-binding protein 1 for its genome replication. J Virol 85:6996–7004. doi: 10.1128/JVI.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones DM, Patel AH, Targett-Adams P, McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J Virol 83:2163–2177. doi: 10.1128/JVI.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pager CT, Schutz S, Abraham TM, Luo G, Sarnow P. 2013. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology 435:472–484. doi: 10.1016/j.virol.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shih JW, Tsai TY, Chao CH, Wu Lee YH. 2008. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- 47.Geissler R, Golbik RP, Behrens SE. 2012. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res 40:4998–5011. doi: 10.1093/nar/gks070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai MC, Lee YH, Tarn WY. 2008. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell 19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. 2008. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res 36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hellen CU, Pestova TV. 1999. Translation of hepatitis C virus RNA. J Viral Hepatitis 6:79–87. doi: 10.1046/j.1365-2893.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 51.Sun C, Querol-Audi J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JH. 2013. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res 41:7512–7521. doi: 10.1093/nar/gkt510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mamiya N, Worman HJ. 1999. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem 274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- 53.Gawlik K, Baugh J, Chatterji U, Lim PJ, Bobardt MD, Gallay PA. 2014. HCV core residues critical for infectivity also are involved in core-NS5A complex formation. PLoS One 9:e88866. doi: 10.1371/journal.pone.0088866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes M, Griffin S, Harris M. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J Gen Virol 90:1329–1334. doi: 10.1099/vir.0.009332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mousseau G, Kota S, Takahashi V, Frick DN, Strosberg AD. 2011. Dimerization-driven interaction of hepatitis C virus core protein with NS3 helicase. J Gen Virol 92:101–111. doi: 10.1099/vir.0.023325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phan T, Kohlway A, Dimberu P, Pyle AM, Lindenbach BD. 2011. The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly. J Virol 85:1193–1204. doi: 10.1128/JVI.01889-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Heliot L, Rouille Y, Dubuisson J. 2011. NS2 protein of hepatitis C virus interacts with structural and nonstructural proteins towards virus assembly. PLoS Pathog 7:e1001278. doi: 10.1371/journal.ppat.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen Y, Cheng Kao C. 2014. The hepatitis C virus core protein can modulate RNA-dependent RNA synthesis by the 2a polymerase. Virus Res 189C:165–176. doi: 10.1016/j.virusres.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.