Abstract
In addition to their classical roles as detergents to aid in the process of digestion, bile acids have been identified as important signaling molecules that function through various nuclear and G protein-coupled receptors to regulate a myriad of cellular and molecular functions across both metabolic and nonmetabolic pathways. Signaling via these pathways will vary depending on the tissue and the concentration and chemical structure of the bile acid species. Important determinants of the size and composition of the bile acid pool are their efficient enterohepatic recirculation, their host and microbial metabolism, and the homeostatic feedback mechanisms connecting hepatocytes, enterocytes, and the luminal microbiota. This review focuses on the mammalian intestine, discussing the physiology of bile acid transport, the metabolism of bile acids in the gut, and new developments in our understanding of how intestinal metabolism, particularly by the gut microbiota, affects bile acid signaling.
Keywords: enterohepatic circulation, transporters, nuclear receptors, microbiome
After their synthesis from cholesterol in the hepatocyte, bile acids pass into the small intestine where they promote intestinal absorption of biliary and dietary lipids prior to their return to the liver in the enterohepatic circulation or their excretion in the feces. In addition to their well-studied role in hepatic bile secretion and intestinal absorption of fats and fat-soluble vitamins, bile acids function as signaling molecules with metabolic effects that extend beyond their control of hepatobiliary and intestinal function (1–4). The best studied of these pathways include those regulated via direct ligand-mediated activation of the nuclear receptors farnesoid X receptor (FXR) (NR1H4), pregnane X receptor (PXR) (NR1I2), and vitamin D receptor (VDR) (NR1I1), and via the G protein-coupled receptor (GPCR) TGR5 (GPBAR1) (1, 5). However, bile acids also act through other receptors including muscarinic receptors, the sphingosine-1-phosphate receptor 2, α5,β1-integrin, through signal transduction pathways such as those mediated by Jun N-terminal kinase (JNK), ERK, and Akt (3, 6–8), and through posttranslational interactions with proteins (9). Signaling via these diverse pathways is regulated in part by the local concentration and chemical structure of the bile acid species, which in turn is controlled by hepatic synthesis, host and microbial metabolism, and the enterohepatic circulation. Efficient intestinal absorption and hepatic extraction of bile acids enables a very effective recycling mechanism that conserves and largely restricts bile acids to the intestinal and hepatobiliary compartments. This mechanism facilitates delivery of bile acids in sufficient concentration for efficient micellarization of biliary and dietary lipids, while restricting their signaling potential in other compartments such as the systemic circulation. The major transporters that function to maintain the enterohepatic circulation of bile acids have been identified (10, 11), and the intestinal transporters are shown schematically in Fig. 1. In the liver, bile acids are taken up across the sinusoidal membrane of the hepatocyte by the Na+-taurocholate cotransporting polypeptide (NTCP; SLC10A1) and members of the organic anion transporting polypeptide (OATP) family (OATP1B1, OATP1B3; SLCO1B1, SLCO1B3 in humans; Oatp1b2, Slco1b2 in mice) and exported across the canalicular membrane via the bile salt export pump (BSEP; ABCB11) (10, 12–15). In the intestine, bile acids are taken up into the enterocyte by the apical sodium-dependent bile acid transporter (ASBT; SLC10A2), bind the cytosolic ileal bile acid binding protein [IBABP; fatty acid binding protein 6 (FABP6)], and then are exported across the basolateral membrane by the heteromeric organic solute transporter (OST), OSTα-OSTβ (OSTα, SLC51A; OSTβ, SLC51B) (16–19). Among the “stops along the road” of the enterohepatic circulation that can profoundly affect the function of bile acids, the principal and most complex involves bile acid interactions with luminal microbes and cells of the intestine. This review will explore these aspects by focusing on bile acid metabolism by the mammalian intestine and complements recent reviews of bile acid chemistry and physiology (20, 21), bile acid signaling (22–25), bile acid interactions with the gut microbiota (26), and the regulation of metabolism by bile acids (1, 27).
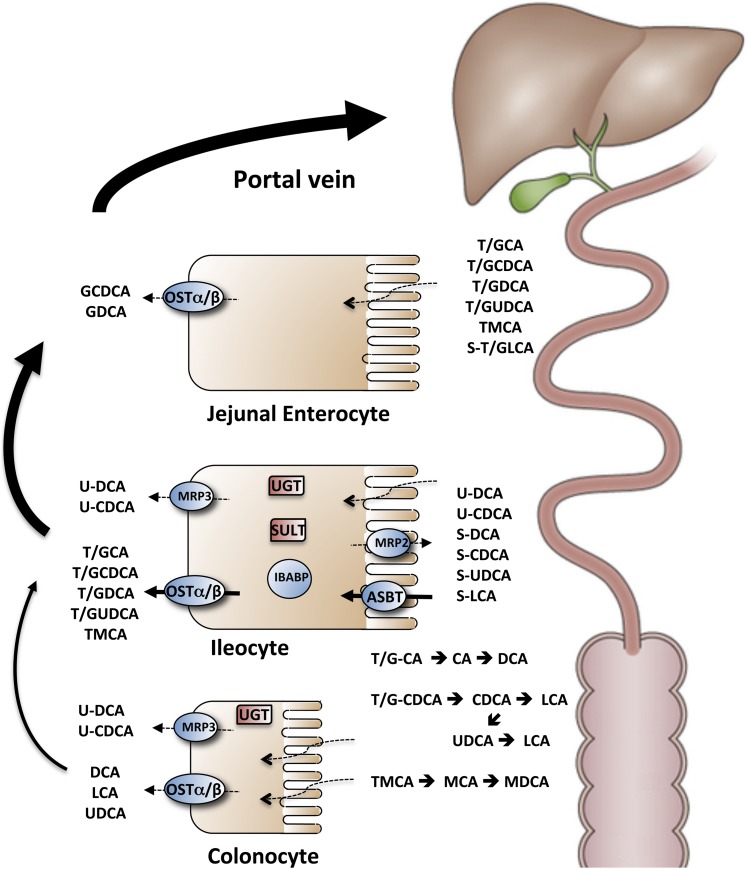
Fig. 1.
Schematic of intestinal transport and metabolism of bile acids. The taurine (T) and glycine (G) conjugated and sulfated (S) primary and secondary bile acids are secreted into bile, stored in the gallbladder, and empty into the small intestine in response to a meal. Bile acids encounter high concentrations of the gut microbiota in the distal ileum and colon and undergo a variety of bacterial transformations including deconjugation, dehydroxylation, and epimerization (black arrows). Passive uptake (dotted lines) occurs down the length of the small intestine for the protonated uncharged fraction of glycine conjugated bile acids and any unconjugated bile acids that are formed. OSTα-OSTβ may participate in the export of those bile acids across the enterocyte or colonocyte basolateral membrane. In the ileum, active uptake of conjugated bile acids across the apical brush border membrane is mediated by the ASBT. The bile acids interact in the cytosol with IBABP, and then are exported across the basolateral membrane by the OSTα-OSTβ. Unconjugated dihydroxy bile acids that are passively absorbed in the distal ileum and colon can be glucuronidated (U) by UGT and exported across the apical membrane by MRP2 or ABCG2 or across the basolateral membrane by MRP3. A fraction of the bile acids taken up in the distal small intestine may also be sulfated by the SULT2A1 and exported back across the apical brush border membrane by MRP2 or ABCG2 for elimination. The bile acids absorbed from the intestine are carried back in the portal circulation to the liver for uptake and may undergo repair by the hepatocyte. Repair includes essentially complete reconjugation with taurine or glycine, reepimerization of iso (β-hydroxy) bile acids to their α-hydroxy form, reduction of oxo groups to hydroxyl groups, and rehydroxylation at the C-7 position to generate the original primary bile acid (in mice but not humans). Adapted with permission from (238).
PHYSIOLOGY AND MOLECULAR MECHANISMS OF INTESTINAL BILE ACID ABSORPTION
Bile acids are synthesized from cholesterol primarily in pericentral hepatocytes through a series of sterol ring hydroxylations and side chain oxidation steps. Bile acids synthesized by the hepatocyte are termed primary bile acids to distinguish them from the secondary bile acids that are formed by a variety of reactions carried out by the gut microbiota, which include dehydroxylation, dehydrogenation (oxidation of a hydroxy group to an oxo group), and epimerization (converting an α-hydroxyl group to a β-hydroxyl group or vice versa). In humans, the primary bile acids are cholic acid (CA) and chenodeoxycholic acid (CDCA), whereas in mice, the primary bile acids include CA, CDCA, and 6-hydroxylated bile acids, mainly muricholic acid (MCA) (28, 29). These are converted to secondary bile acids, which in humans include deoxycholic acid (DCA), ursocholic acid (UCA), ursodeoxycholic acid (UDCA), and lithocholic acid (LCA), and in mice include DCA, UCA, UDCA, murideoxycholic acid (MDCA), and hyodeoxycholic acid (HDCA). Products of bile acid epimerization (iso or epi bile acids) and dehydrogenation (oxo-bile acids) are also present in the gut. The major reactions involved in metabolism of the primary bile acids CA, CDCA, and MCA are summarized in Fig. 2. In the hepatocyte, the newly synthesized bile acids are then N-acyl amidated on the side chain with glycine or taurine at a ratio of approximately 3 to 1 in humans and almost exclusively with taurine (>95%) in mice. After their synthesis, bile acids undergo an enterohepatic circulation where they are secreted by the liver along with other biliary constituents, pass into the small intestine, are reabsorbed from the intestinal lumen, and are carried back in the portal circulation to the liver for efficient extraction and secretion into bile (20). In the lumen of the small intestine, bile acids are present at micellar concentrations and form mixed micelles with dietary lipids and their digestion products such as monoacylglycerols and fatty acids (30–32). Bile acids also solubilize nonpolar lipids such as cholesterol and fat-soluble vitamins, increasing their water-solubility and promoting their diffusion across the unstirred water layer for delivery to the intestinal epithelium (33). Absorption of these other dietary and biliary lipids begins in the proximal and mid-intestine, whereas bile acids are absorbed primarily in the distal small intestine (ileum), with only about 5% of intestinal bile acids escaping reabsorption to undergo fecal elimination. The bile acid molecule is too large to pass paracellularly through the tight junctions of the intestinal epithelium and must be absorbed by transcellular passive or active mechanisms (34). Passive absorption of protonated uncharged bile acid species, unconjugated bile acids, and a small fraction of the glycine conjugates occurs down the length of the intestine (35). However, as most of the bile acid pool is conjugated to taurine or glycine and ionized, their uptake across the apical brush border membrane requires the presence of a transporter, and active carrier-mediated absorption of bile acids is restricted to the ileum (36, 37). Indeed, in all vertebrates examined to date, including primitive vertebrates such as the little skate (Leucoraja erinacea) and sea lamprey (Petromyzon marinus) (38), the ileal epithelium maintains an efficient transport system for the active reclamation of bile acids (39).
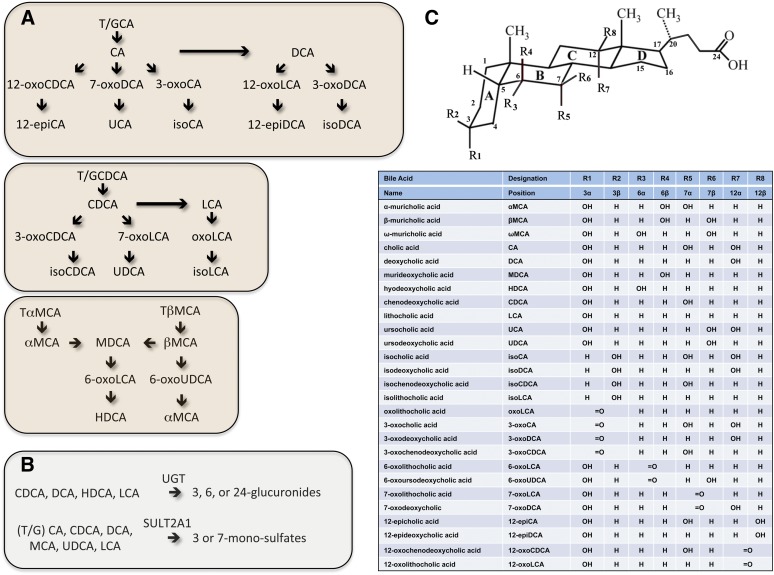
Fig. 2.
Scheme for intestinal phase 2 conjugation and bacterial metabolism of bile acids in humans and mice. A: Bacterial metabolism includes deconjugation, 7-dehydroxylation, oxidation, and epimerization of bile acids. B: The phase 2 conjugation reactions carried out by the gut epithelial cells include glucuronidation and sulfation (sulfonation). C: Structure and abbreviations for the bile acid species. Adapted from (53) using the nomenclature system of Hofmann et al. (239).
INTESTINAL APICAL BRUSH BORDER MEMBRANE TRANSPORT
Bile acids are reclaimed through a combination of intestinal passive absorption, active transport in the distal ileum, and passive absorption in the colon. The general consensus from numerous studies is that ileal active transport is the major route for conjugated bile acid uptake, whereas the passive or facilitative absorption present along the length of the small intestine may be significant for unconjugated and some glycine-conjugated bile acids (34, 35, 40–42). Although bile acids in most vertebrates are conjugated to taurine, the bile acid pool of humans includes a significant proportion of glycine conjugates (43, 44). A fraction of the glycine conjugates and unconjugated bile acids are protonated under conditions of the acidic surface pH of the enterocyte (45) and can be absorbed by passive diffusion across the apical brush border membrane (35).
The question of whether facilitative carriers such as members of the OATP family participate in intestinal bile acid absorption has not been fully resolved and may differ between species (46, 47). Several OATPs capable of transporting bile acids are expressed in the small intestine (OATP1A2 and OATP2B1 in humans; Oatp1a4, Oatp1a5, and Oatp2b1 in mice) (48, 49), and their role in transporting xenobiotics and endobiotics such as bile acids has been examined recently using OATP-null mouse models (13, 50). In mice engineered to delete the entire Oatp1a/1b locus (encompassing Slco1b2, Slco1a4, Slco1a1, Slco1a6, and Slco1a5), plasma levels of conjugated bile acids were unchanged, whereas unconjugated bile acid levels increased by approximately 13-fold (13). Similar results were obtained for a mouse model where only the Oatp1b2 (Slco1b2) gene was deleted, indicating that this OATP is primarily responsible for mediating hepatic clearance of unconjugated bile acids in mice (50). Because unconjugated bile acids represent only a small fraction of the bile acid pool, the contribution of Oatp1b2 (or other Oatp1a/1b transporters) to intestinal or hepatic uptake of bile acids appears to be small in states where normal physiological pathways are intact. Later studies using mouse models where Oatp1a1 and Oatp1a4 were individually deleted also provided little support for an important direct role of these transporters in liver or intestinal bile acid uptake (51). However, it should be noted that the Oatp1a1- and Oatp1a4-null mice exhibited complex changes in bile acid metabolism and intestinal function, including changes in the gut microbiome (52, 53). OATP2B1 (previously called OATP-B) transports taurocholic acid at acidic but not neutral pH (54, 55), raising the possibility that OATP2B1 may contribute to bile acid transport in the acidic microclimate of the gut. However, an Oatp2b1-null mouse has not yet been reported and its contribution to intestinal bile acid transport has not been specifically examined. To date, the Oatp-null mouse model findings reaffirm the conclusions from an earlier study where ASBT-null mice were fed a diet containing cholestyramine, a bile acid sequestrant that would block passive, facilitative, and active mechanisms for intestinal bile acid absorption. In that study, no further increase in the loss of bile acids in feces was observed in ASBT-null mice fed the cholestyramine as compared with ASBT-null mice fed the control diet, suggesting that alternative non-ASBT mechanisms contribute little to intestinal reclamation of bile acids in the mouse (17).
The ASBT is responsible for active uptake of bile acids in the terminal ileum (10, 56), and properties of the ASBT and other SLC10A transporters have been reviewed recently (57–59). A major role for the ASBT in intestinal bile acid absorption is supported by genetic evidence, where targeted inactivation of the ASBT eliminates enterohepatic cycling of bile acids in mice (17, 60, 61) and ASBT loss-of-function mutations in humans are associated with intestinal bile acid malabsorption (16), and by pharmacological evidence where administration of small molecule inhibitors of the ASBT to animal models reduced intestinal bile acid absorption (62–64). Although this review focuses on the intestine, it should be noted that other epithelia, including renal proximal tubule cells, cholangiocytes lining the biliary tract, and gallbladder epithelial cells, also express the ASBT (39, 65–67). In the ileum and kidney, ASBT functions to absorb the majority of the bile acids in the adjacent lumen and only a small fraction escapes absorption and is excreted in the feces or urine. In contrast, almost all the bile acids that are secreted by the liver into bile will ultimately pass through the biliary tract and gallbladder into the small intestine. In those compartments, the physiological role of the ASBT is still being explored, but most likely involves cholehepatic or cholecystohepatic shunting of bile acids, whereby a fraction of the biliary bile acids is absorbed prematurely and returned directly to hepatocytes, affecting hepatic bile secretion (68, 69) and bile acid enterohepatic cycling (66, 70). ASBT efficiently transports all the major species of bile acids in a sodium-dependent fashion (18, 71), and although two new non-bile acid chemical substrates were recently described (72), no endogenous ASBT substrates other than bile acids have yet been identified.
INTESTINAL INTRACELLULAR BILE ACID TRANSPORT
In contrast to transmembrane transport of bile acids, little is known regarding their intracellular transport in the enterocyte. Because of their detergent properties, it is assumed that bile acids should be bound or sequestered in some fashion within the cell. Just as bile acids are transported in plasma bound to albumin and other proteins (73–75), it is likely that bile acids are bound to cytosolic proteins during their transit through the enterocyte or hepatocyte, and there is no compelling evidence for intracellular vesicular transport (76). Serving this role in the enterocyte is the IBABP (also called the ileal lipid binding protein, ILBP; gene symbol FABP6), an abundant cytosolic protein and member of the fatty acid binding protein family (77). IBABP expression parallels that of the ASBT and OSTα-OSTβ along the longitudinal axis of the small intestine, and its gene expression is strongly induced by bile acids acting through FXR. IBABP binds both fatty acids and bile acids, but with a marked preference for bile acids and a bile acid relative affinity of taurine-conjugated > glycine-conjugated > unconjugated species (78, 79). The solution structure of IBABP from several species has been determined using NMR and X-ray crystallographic techniques, proving additional insight to the mechanism and stoichiometry (two or three bile acid molecules per molecule of IBABP) of ligand binding (80, 81). Based on its abundance, selective high affinity binding of bile acids, coexpression in the same ileal enterocytes as the membrane bile acid transporters, and positive regulation by bile acids, IBABP likely plays an important role in intestinal bile acid transport, enterocyte bile acid sensing, or enterocyte intracellular bile acid signal transduction. However, direct in vivo evidence in support of those roles has only recently begun to emerge. With regard to ileal transcellular transport of bile acids, studies using a whole body FXR-null mouse model, which lacks appreciable IBABP expression, initially suggested that IBABP is not absolutely required for maintenance of ileal bile acid absorption or return to the hepatocyte via the portal circulation (82). Similar results were obtained using an intestine-specific FXR-null mouse where isotope dilution studies found no significant changes in the fractional turnover rate for a cholic acid tracer (83). However, because FXR regulates many different genes and pathways (1), it was difficult to draw specific conclusions regarding the in vivo role of IBABP from those models. New insights to that question come from a recent study using an IBABP-null mouse model (84). In that study, mucosal-to-serosal transport of taurocholate was significantly reduced in ileal gut sacs prepared from male and female IBABP-null mice. But the in vivo phenotype was complex, where loss of IBABP was associated with reduced hepatic cholesterol 7alpha-hydroxylase (Cyp7a1) expression and increased fecal bile acid excretion in male but not female mice. Generally supporting a direct role in trans-ileal transport, the findings also suggested that the loss of IBABP is associated with complex changes in bile acid metabolism. The mechanism could involve enterocyte bile acid sensing, because previous studies in transfected cells found that IBABP affects FXR signaling by direct interaction or by altering delivery of ligand (85, 86).
BASOLATERAL MEMBRANE BILE ACID EXPORT
Whereas several candidate intestinal basolateral membrane transporters were identified over the years (10), the preponderance of evidence indicates that the heteromeric transporter OSTα-OSTβ is responsible for the majority of intestinal basolateral membrane bile acid export (87, 88). Support for this role of OSTα-OSTβ, a heterodimer consisting of a 352-amino acid polytopic membrane protein (OSTα) and a 182-amino acid predicted type I membrane protein (OSTβ), includes: 1) intestinal expression that overlaps the ASBT, 2) plasma membrane localization restricted to the basolateral aspect of the enterocyte, 3) transport specificity that includes the major bile acid species, 4) expression that is strongly induced by bile acids acting through FXR, and 5) impaired intestinal bile acid absorption and altered bile acid metabolism in OSTα-null mice (18, 19). Although expressed at highest levels in the ileum, OSTα-OSTβ is also expressed in the proximal small intestine, cecum, and colon where it may function to export glycine-conjugated or unconjugated bile acids that entered the cell by passive diffusion (35, 36, 89). A fraction of these bile acids would be uncharged when the pH of the intestinal lumen or microclimate overlaying the mucosa is sufficiently low to protonate the bile acid anion (45). However after entry, the bile acids would ionize at the neutral intracellular pH and potentially require a carrier such as OSTα-OSTβ for export. When expressed in transfected cells or Xenopus oocytes, OSTα-OSTβ exhibits bidirectional transport, and studies suggest that it operates by facilitated diffusion, mediating solute uptake or efflux depending on the electrochemical gradient (90). This raises the possibility that OSTα-OSTβ could mediate uptake across the basolateral membrane, delivering bile acids to the interior of the enterocyte under conditions such as cholestasis where plasma bile acid levels rise precipitously. Although a theoretical possibility, several lines of evidence suggest that such intestinal basolateral membrane uptake via OSTα-OSTβ (or another mechanism) does not occur at appreciable levels. This evidence includes: 1) Following bile duct ligation in mouse models, ileal expression of the FXR target genes, FGF15 and OSTα-OSTβ, is reduced despite significant elevations in serum bile acids (91, 92); and 2) In humans, a patient with an isolated NTCP (SLC10A1) deficiency and significantly elevated plasma conjugated bile acid levels had normal plasma levels of FGF19, suggesting that ileal expression of FGF19 was not increased (12). The other membrane transporter that may contribute to basolateral bile acid export is the ABC transporter, multidrug resistance protein (MRP3) (ABCC3) (93). In mice, loss of MRP3 had little effect on intestinal bile acid absorption under normal physiological conditions (94, 95), but under supraphysiological conditions, such as feeding a diet containing cholic acid, MRP3 appeared to be a more important contributor (96). The contribution of human MRP3 to intestinal bile acid transport has not been studied directly. However, in vitro transport studies using isolated membrane vesicles or transfected cells found that human MRP3 avidly transports glucuronide conjugates but has only a weak affinity for native bile acids, suggesting that it does not contribute substantially to transport of nonglucuronidated bile acids under physiological conditions (95, 97, 98).
Unlike ASBT-null mice or patients with ASBT mutations (16, 17, 99), OSTα-null mice exhibit a complex phenotype that includes ileal morphological changes, increased ileal production of FGF15, and reductions in hepatic bile acid synthesis and bile acid pool size (18, 19). This finding of altered gut-liver FXR-fibroblast growth factor 15/19 (FGF15/19) signaling has important physiological and therapeutic implications, as interruption of the enterohepatic circulation of bile acids at the intestinal apical versus basolateral membranes is predicted to have differential and widespread metabolic effects. This point is nicely illustrated in a study comparing the development of hypercholesterolemia and atherosclerosis in apoE-null mice lacking either the ASBT or OSTα (100). Similar to the effects of bile acid sequestrants (101) or ASBT inhibitors (102, 103), levels of plasma and hepatic cholesterol and markers of atherosclerosis development were reduced in apoE-null mice lacking the ASBT. However, these markers were not reduced in apoE-null mice lacking OSTα. The effects of inactivation of OSTα on lipid and glucose metabolism in mice fed a basal diet have recently been reported (104), and include decreased intestinal lipid and cholesterol absorption, improved glucose tolerance and insulin sensitivity, and reduced weight gain. Clearly, additional studies will be required to understand the underlying mechanisms responsible for the complex metabolic effects associated with altering intestinal absorption of bile acids, including the contributions of FGF15/19 and bile acid signaling (105–107).
Additional insight to the in vivo functions of OSTα-OSTβ comes from a study of OSTα-null mice lacking FXR (108). In that study, loss of FXR in OSTα-null mice restored intestinal cholesterol absorption and reversed the decreases in hepatic bile acid synthesis, fecal bile acid excretion, and bile acid pool size. However, the adaptive changes in ileal villus morphology in OSTα-null mice, including the increase in cell number, decrease in villus height, and increase in crypt depth, were not reversed by inactivation of FXR. These findings raise important questions regarding the in vivo functions of OSTα-OSTβ and the role of bile acids in this adaptive response. Whereas the ASBT is expressed almost exclusively in the bile acid-transporting epithelium of ileum, biliary tract, and renal proximal tubules, and appears to have no endogenous substrates other than bile acids (56), OSTα-OSTβ is more widely expressed and is abundant in tissues important for steroid homeostasis (88). Because OSTα-OSTβ also transports endogenous small molecules such as prostaglandins, neurosteroids, and steroid sulfates (109, 110), it is possible that the intestinal adaptation is a response to alterations in transport of a non-bile acid substrate. For example, in vitro studies have shown that OSTα-OSTβ can transport prostaglandin E2, which is important for regeneration of epithelial crypts following intestinal injury (111, 112). Alternatively, it is possible that OSTα-OSTβ has other cellular functions besides solute transport. Although no evidence for such a role has been found in mammalian systems (87, 88), a primitive OSTα-like protein identified in C. elegans was shown to regulate intracellular membrane trafficking and the morphology and protein composition of sensory cilia (113). Finally, no inherited phenotypic defects in the human OSTα or OSTβ genes have been identified, and an OSTβ-null mouse has not yet been reported.
ENTEROCYTE METABOLISM OF BILE ACIDS
Whereas ileal enterocytes play critical transport and regulatory roles in maintaining bile acid homeostasis, they function in a more limited capacity in mammalian bile acid synthesis and metabolism as compared with hepatocytes or the gut microbiome. Hepatocytes are the major site of synthesis and there is no evidence that the mammalian enterocyte synthesizes bile acids. Interestingly, this is not the case for all vertebrates. Recent studies suggest that bile acid synthesis occurs in both liver and intestine of the sea lamprey, depending upon the developmental stage of this primitive jawless vertebrate (114). In sea lamprey, free-living larvae undergo an apoptotic loss of the hepatic biliary tree and gallbladder during metamorphosis to the juvenile parasitic form. Remarkably, this biliary atresia-like developmental process occurs without apparent liver disease and correlates with an approximately 100-fold reduction in hepatic Cyp7a1 expression and concomitant increase in intestinal Cyp7a1, suggesting that the intestine assumes the major role in bile acid synthesis (114, 115).
As discussed below, a fraction of the conjugated bile acids in the intestinal lumen undergo bacterial deconjugation (cleavage of the amide bond linking the bile acid to glycine or taurine) in the distal small intestine, a process that continues to near completion in the colon. The resultant unconjugated mono- or dihydroxy bile acids are membrane permeable and can undergo passive absorption, whereas unconjugated trihydroxy bile acids, such as cholic acid, are sufficiently hydrophilic as to require a specific transport system (116). After absorption, unconjugated bile acid species do not appear to undergo N-acyl amidation during transport through the enterocyte or colonocyte (117). Instead, reconjugation to taurine or glycine is carried out almost quantitatively by the hepatocyte after the unconjugated bile acid has returned to the liver in the portal circulation, a cycle termed “damage and repair” by Hofmann and Hagey (20). Although N-acyl amidation does not occur, bile acids do undergo limited phase II metabolism in the gut. Unconjugated mono- and dihydroxy bile acids, but not unconjugated trihydroxy, tetrahydroxy, or conjugated bile acids (118, 119), can be glucuronidated on the 3- or 6-hydroxyl or 24-carboxyl positions by microsomal UDP-glucuronosyltransferase (UGT) enzymes expressed in the small intestine and colon (120). In the liver, glucuronidation is not thought to be a major pathway for conjugation of endogenous bile acids in humans under normal physiological conditions (121), but is more important under cholestatic conditions, where glucuronidated bile acids can be exported by MRP3 across the hepatocyte sinusoidal membrane into the blood for urinary elimination (122). Similarly in the small intestine and colon, evidence suggests that glucuronidation of native unconjugated bile acids is limited in normal physiology (123–125). However, under pathophysiological conditions such as colitis, very recent evidence suggests that activation of intestinal UGT gene expression and increased glucuronidation of bile acids plays an important role in altering bile acid signaling and homeostasis and exacerbating bile acid injury in the colon (126). Bile acids that undergo glucuronidation in the enterocyte or colonocyte can be exported by MRP2 (ABCC2) or the breast cancer resistance protein (BCRP) (ABCG2) across the apical membrane back into the gut lumen (127) or by MRP3 across the basolateral membrane into the blood to undergo renal filtration and elimination (128). Another bile acid phase II conjugation reaction is monosulfation (sulfonation) by sulfotransferase (SULT)2A1 (129), with addition of a sulfate group to the 3-hydroxy and 7-hydroxy positions predominating in humans and mice, respectively (129, 130). Sulfation is an important metabolic pathway to detoxify and eliminate bile acids in humans, particularly for the monohydroxy bile acid, LCA (131). After formation in the liver, sulfated LCA N-acyl amidates are secreted into bile and pass into the intestine (131), but are not reabsorbed by the ileal transport system and are eliminated from the body (39, 132). In patients with severe cholestasis, bile acid sulfation and export across the hepatocyte sinusoidal membrane into the blood becomes an important pathway for urinary elimination of bile acids (133, 134). Although liver is the major site of bile acid sulfation, cytosolic bile acid sulfotransferase activity is also detected in the human small intestine, but apparently not the large intestine (135, 136). This raises the prospect that some bile acid species are sulfated by the intestinal epithelium and secreted directly back into the intestinal lumen. Indeed, analysis of human cecal contents found significant amounts of 3-sulfated bile acids, including 3-sulfo-LCA, 3-sulfo-CDCA, 3-sulfo-DCA, and 3-sulfo-UDCA (137). Because biliary excretion is relatively low for sulfate conjugates of CDCA and DCA (138–140), the authors suggested that these sulfates are intestinally derived (137) in a pathway where deconjugated LCA, CDCA, or DCA is absorbed by the enterocyte, sulfated, and exported back into the intestinal lumen by carriers such as MRP2 or BCRP (141, 142). In support of that hypothesis, Caco-2 cells were shown to sulfate LCA and resecreted sulfo-LCA back into the media (143). As compared with humans (129), sulfation is a relatively minor pathway for bile acid metabolism in rats and mice, species that can carry out more extensive phase I hydroxylation of bile acids (44, 144, 145). In those species, the bile acid sulfation was thought to occur mainly in the liver and kidney (146), and only low levels of LCA sulfating activity were detected in mouse small intestine (147). However, more recent analyses using electrospray mass spectrometry showed that mouse fecal samples contain significant proportions of sulfated mono-, di-, and trihydroxy bile acids (148). As these bile acid species are found in only trace amounts in mouse bile (149), it is possible that a similar process of intestinal absorption, sulfation, and resecretion back into the lumen is occurring in mice and may play an underappreciated role in detoxifying intestinal bile acids (150). Finally, in addition to N-acyl amidation, glucuronidation, and sulfation, bile acids are also conjugated with glucose, N-acetylglucosamine, and galactose (151), with glucosidation occurring at the C-3 position, N-acetylglucosaminidation at the C-7β position, and galactosidation at the carboxyl group (152–154). Although the enzyme activity for synthesis of bile acid glucosides is present in human intestinal and colonic mucosa (155) (as well as liver and kidney), the biological significance of this pathway in metabolism of bile acids by the intestine or other tissues is not clear and deserves further investigation.
GUT MICROBIOME METABOLISM OF BILE ACIDS
Metabolism of bile acids by the gut microbiome is complex and has been the subject of numerous recent reviews (26, 148, 156, 157). It is believed that gut bacteria benefit from metabolizing bile acids by acquiring glycine and taurine for subsequent metabolism, and by using bile acids as sinks for the disposal of electrons from fermentation (158). However, it should be noted that bile acids also exert antimicrobial properties, by being directly toxic to bacteria or by stimulating production of antimicrobial factors (159–161), and play an important role in preventing small intestinal bacterial overgrowth (162). Indeed, another prevailing hypothesis is that the gut bacteria metabolize bile acids to thwart this host defense, thereby gaining an advantage over competing microbes (157, 163). Mechanisms responsible for the antimicrobial actions of bile acids have been investigated and include direct interaction with cell membranes (164), with protonated unconjugated bile acid species having more potent effects than conjugated species (161, 165), acidification of the cytoplasm (166), and induction of DNA damage (161). A detergent role for mixed micelles of conjugated bile acids and dietary long chain fatty acids derived from the digestive process has also been proposed to explain the antimicrobial actions of endogenous and exogenously administered conjugated bile acids (159). Additional insight to the anti-microbial mechanisms of action comes from a recent study showing that bile acids cause protein unfolding, protein aggregation, and disulfide stress in bacteria (167). Remarkably, CA and CDCA affected folding of different subsets of bacterial proteins, providing a plausible mechanism for the greater antimicrobial potency of bile acid mixtures. The mechanisms for bacterial resistance to bile acids include expression of efflux pumps and enzymes that biotransform bile acids. The bile acid efflux systems include ATP-dependent (e.g., members of the ABC superfamily) (168–172) and proton motive force-dependent carriers (e.g., members of the resistance-nodulation-division family and major facilitator superfamily) (173, 174) and appear to be a common mechanism for bile acid resistance by gut bacteria. Indeed, expression of many of these efflux systems is bile acid-inducible and their contribution to growth and bile resistance has been demonstrated directly by functional genetics (169, 172, 175). Understandably, much of the research to date has focused on identifying the bacterial genes and their mechanism of action. However, selection for species expressing these efflux systems may be an important part of the mechanism by which bile acids influence the structure of the gut microbiome (26). Moreover, differences in the expression of the bacterial transporters responsible for bile acid uptake (discussed below) and efflux may also play an underappreciated role in controlling the transformation of bile acids, which would in turn affect the size, composition, and hydrophilicity of the host’s bile acid pool.
Primary bile acids undergo a wide array of bacterial transformations including deamidation (deconjugation), oxidation and epimerization of the 3-, 6-, 7-, and 12- hydroxy groups to form oxo or iso epimers, and 7α/β-dehydroxylation (156, 176). These transformations are summarized in Fig. 2. Removal of the N-acyl amidation (glycine or taurine) by conjugated bile salt hydrolases (choloylglycine hydrolase family of enzymes), a reaction called deconjugation (177, 178), usually precedes subsequent microbial biotransformation and is absolutely required for 7α/β-dehydroxylation (179, 180). Because most conjugated bile salt hydrolases are soluble cytosolic proteins, bacteria must also express transporters for uptake of conjugated bile acids. For example, in Lactobacillus johnsonii and Lactobacillus acidophilus, the cbsT1 and cbsT2 genes encoded members of the major facilitator superfamily that facilitate antiport of taurocholate and CA and are part of a polycistronic operon with the bile salt hydrolase (181, 182). Bile salt hydrolase activity is widespread among commensal bacteria, although the levels of enzyme activity may vary widely between individual species (183, 184). The microbiome complement of bile salt hydrolase activity may also differ between host species. For example, a metagenomic analysis revealed bile salt hydrolase enzyme differences between the murine and human microbiota, which likely reflect responses to the bile acid pool composition of the host species (185). Although both glycine and taurine N-acyl amidated bile acid species are extensively deconjugated during transit through the colon prior to their elimination in the feces, evidence from in vitro and in vivo studies suggest that glycine-conjugated bile acids are more susceptible to deconjugation than taurine conjugates (178, 186–188). In contrast, bile acids conjugated to sulfates are relatively resistant to intestinal bacterial metabolism (189, 190), more so for the C-7 sulfated bile acids than the C-3 conjugates (191).
In addition to deconjugation, bile acids undergo additional transformations, the most significant being 7α/β-dehydroxylation, which converts primary bile acids to secondary bile acids (i.e., CA to DCA, CDCA or UDCA to LCA, and MCA to MDCA). These reactions are catalyzed by a group of bacterial genes encoded by a large bile acid inducible (bai) operon, which was identified and extensively characterized by Hylemon and others (156, 192). Among the bai genes is baiG, a member of the major facilitator superfamily and the first bacterial bile acid transporter to be identified and characterized (193). baiG and related genes (194) encode proton-dependent transporters that facilitate the initial uptake of unconjugated CA and CDCA, but not the unconjugated secondary bile acids DCA and LCA (193). After their uptake, 7-dehydroxylation occurs through a complex series of reactions that requires a weak acid to form the CoA derivative (156), and as such is restricted to unconjugated bile acids (179). Whereas many species of commensal bacteria have detectable bile salt hydrolase or hydroxysteroid dehydrogenase activity (which catalyzes bile acid oxidation and epimerization), 7α/β-dehydroxylation activity is present in only a very small fraction of the total gut microbiota, typically a limited number of species in the genus Clostridium (195). Indeed, with the observation that CDCA inhibits Clostridium difficile spore germination, there is increasing interest in developing bile acid analogs that are resistant to bacterial transformations such as 7-dehydroxylation (196). Notably, levels of gut 7α/β-dehydroxylation activity vary many fold between human populations (26) and under different pathophysiological conditions (197, 198).
The metabolism of bile acids by the gut microbiota is not restricted to the described deamidation, dehydroxylation, oxidation, and epimerization reactions and includes additional modifications that are less characterized and merit additional investigation. Indeed, esterified (saponifiable) bile acids have been reported to constitute almost 25% of the total fecal bile acids in humans (199–201). Although bile acid ethyl esters have been reported to form infrequently when samples are heated with ethanolic sodium hydroxide (202), evidence was provided suggesting that the esters were not formed during extraction and processing of the human fecal samples (200). The bile acid esters are likely produced by the gut microbiota, as the saponifiable bile acid fraction decreased from approximately 24 to 9% of total bile acids after antibiotic treatment with the fraction rising back to almost 28% two weeks after the antibiotic course (200). In vitro anaerobic fecal cultures were also shown to produce ethyl LCA esters (203). Among the esterified bile acids identified are ethyl esters (C-3 position) of LCA and isoLCA (203) and C-24 carboxyl esters (200, 204). Another novel metabolite identified in human fecal samples is polydeoxycholate, where chains of deoxycholate were joined at alternating 3α-hydroxy and carboxyl groups (205). The physiological significance of such modifications is unclear. However, the bile acid esters would be more hydrophobic and predicted to be excluded from the fecal water phase, thereby reducing their potential for interaction with the colonocyte and absorption.
After their intestinal absorption and return in the enterohepatic circulation, the secondary bile acids generated by the microbiota can undergo “repair” by the hepatocyte. Repair includes essentially complete reconjugation with taurine/glycine, reepimerization of iso (3β-hydroxy) bile acids to their 3α-hydroxy form (206, 207), and reduction of oxo groups to hydroxyl groups (208). Rehydroxylation at the C-7 position to generate the original primary bile acid occurs in many species, including the mouse and rat (44, 209), but does not occur in humans (210, 211). Alternatively, if not absorbed and repaired, the secondary bile acids transit through the colon where they have effects on colonocyte function and oncogenesis (see below).
CONSEQUENCES OF INTESTINAL BILE ACID METABOLISM ON BILE ACID SIGNALING
Metabolism of bile acids influences their fate as well as their signaling properties, and the effect of these bile acid modifications on host signaling via GPCRs, nuclear receptors, and intracellular pathways is being actively explored. The emerging consensus from this work is that these transformations are important contributors to bile acid signaling and gut microbiota-induced changes in whole body physiology (26, 126, 212). For example, microbiota effects upon body weight, adiposity, lipid metabolism, liver disease, intestinal mucosal function, colon carcinogenesis, and cardiovascular function have recently been linked to luminal bile acid metabolism and modulation of bile acid signaling (26, 213–218). Of note, these studies are at the forefront of a new field, and make innovative use of mouse models with altered microbiomes, including models that are germ-free or colonized with specific organisms, including microbiota from humans with select clinical characteristics (214, 216, 219–224). Compared with mice raised conventionally, the pool of bile acids in germ-free mice is less “FXR agonistic,” because the bile and luminal contents include an increased proportion of tauro-β-murocholic acid, an endogenous mouse bile acid species that functions as an FXR antagonist (26, 223). As such, the lack of conventional microbes in the germ-free mouse intestines reduces expression of ileal FXR target genes (e.g., FGF15, IBABP), with consequent increased expression of hepatic genes usually suppressed by FGF15 (Cyp7a1 and others). Intriguingly, reduction of luminal bacteria with antibiotics led to similar but less pronounced effects on bile acid signaling, as those observed in germ-free models. Moreover, administration of a clinically available probiotic mixture, VSL#3, altered the mouse microbiota, leading to increases in bile acid deconjugation, fecal bile acid loss, and the proportion of tauro-β-murocholic acid (220). As in studies with germ-free mice, ileal FGF15 RNA levels were suppressed and hepatic Cyp7a1 levels increased in VSL#3-fed mice. These are but a few examples of the observations from a recently burgeoning field, but they clearly point to significant communication between microbial bile acid metabolism and host physiological responses in gut and liver in ways that are potentially amenable to therapeutic manipulation (225).
Because there are important host bile acid species differences, it is unclear how to extrapolate many of the recent findings from mouse models to humans. For example, humans do not synthesize 6-hydroxylated bile acids such as MCA, so mechanisms whereby gut microbiota alterations produce a bile acid pool with dramatically less FXR activation potential may not be operative in humans. However, alteration of the luminal bile acid composition, hydrophilicity, and concentration have been linked to a variety of human pathophysiological conditions, including: intestinal dysmotility, inflammatory bowel disease phenotypes, nonalcoholic fatty liver disease, and progression of colon cancer (26, 218, 226–228). Increased colonic concentrations of luminal bacteria-derived LCA and DCA are associated with development of colorectal cancer in both mouse models and humans, and the progression of colorectal cancer can be reduced in mice by antibiotic suppression of luminal bacteria (217, 224). The contributors to human colorectal cancer development are complex and certainly extend beyond microbial bile acid metabolism, but recent data link an animal-based diet in humans to shifts in the microbiota favoring species enriched in bile salt hydrolases, with subsequent increased fecal concentrations of the pro-carcinogenic bile acid DCA (229). Altered luminal bile acid composition can also impact mucosal inflammation, in concordance with a response to various dietary signals. In a mouse model of inflammatory colitis, a diet enriched in milk fats led to increased luminal concentrations of taurocholate and growth of Bilophila wadsworthia, which enhanced pro-inflammatory cytokine expression and mucosal inflammation (221), thus linking dietary choices with mucosal damage via altered bile acid homeostasis (230).
Activation of apical membrane TGR5 receptors on enteroendocrine (L) cells in the distal ileum and colon leads to portal release of the incretins GLP-1 and GLP-2. Bile acids are TGR5 agonists, with a potency depending upon their structure (231). When present at sufficient concentration (in states of ASBT suppression, ileal inflammation, bowel resection, colesevelam treatment, or rapid transit), bile acids can increase GLP-1 secretion in both mouse and human models (105, 232, 233). In fact, direct rectal administration of taurocholic acid in healthy male humans induces secretion of GLP-1 and Peptide YY, along with a perceived sense of satiety (234). Clearly, the complexities of bile acid biology extend to membrane receptor biology with a potential to impact upon growth, appetite, and metabolism (235, 236).
Altered composition of bile acids impacts the progression of liver diseases and liver function including signaling related to fibrosis, cholestasis, and obesity-related liver diseases (213, 216). Among the more intriguing overall concepts connecting liver disease to luminal bile acid composition and intestinal function include: bile acid-induced alterations in intestinal permeability that lead to increased transfer of bacterial products to the portal circulation and direct delivery to the liver; modified overall FXR agonist capability of the bile acid pool; and disruptions in the absorption of sterols and fatty acids with delivery of those substances to the liver. In addition, cholestasis leads to less delivery of bile acids into the intestine, reduced ileal FGF15/19 expression, increased bacterial overgrowth and hydrolase activity, and increased gut permeability, contributing to the progression of liver disease (162, 216, 237).
CONCLUSION
New studies, primarily in mice, are expanding our understanding of the role of bile acid signaling in metabolism and the pathogenesis of disease. These studies all point to integrated roles for the utilization and modification of the bile acid pool by resident gut microbiome, with subsequent local and systemic effects on bile acid interactions with surface receptors and transporters, and intracellular targets such as nuclear receptors. Bile acids are thus poised for regional signaling and nutritive functions, and their high concentrations throughout the small intestine offer opportunities for benefit as well as disease pathogenesis. Thus, there are multiple opportunities to potentially exploit the liver-gut-microbiome-bile acid axis for novel therapies to treat gastrointestinal and metabolic diseases.
Acknowledgments
The authors thank Drs. Astrid Kosters and Anuradha Rao for critical reading of the manuscript.
Footnotes
Abbreviations:
- ASBT
- apical sodium-dependent bile acid transporter
- bai
- bile acid inducible
- BCRP
- breast cancer resistance protein
- BSEP
- bile salt export pump
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- Cyp7a1
- cholesterol 7alpha-hydroxylase
- DCA
- deoxycholic acid
- FABP6
- fatty acid binding protein 6
- FGF15/19
- fibroblast growth factor 15/19
- FXR
- farnesoid X receptor
- GPCR
- G protein-coupled receptor
- HDCA
- hyodeoxycholic acid
- IBABP
- ileal bile acid binding protein
- JNK
- Jun N-terminal kinase
- LCA
- lithocholic acid
- MCA
- muricholic acid
- MDCA
- murideoxycholic acid
- MRP
- multidrug resistance protein
- NTCP
- Na+-taurocholate cotransporting polypeptide
- OATP
- organic anion transporting polypeptide
- OST
- organic solute transporter
- SULT
- sulfotransferase
- UCA
- ursocholic acid
- UDCA
- ursodeoxycholic acid
- UGT
- UDP-glucuronosyltransferase
This work was supported by National Institutes of Health Grants DK047987 to P.A.D. and DK056239 to S.J.K.
REFERENCES
- 1.de Aguiar Vallim T. Q., Tarling E. J., Edwards P. A. 2013. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17: 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porez G., Prawitt J., Gross B., Staels B. 2012. Bile acid receptors as targets for the treatment of dyslipidemia and cardiovascular disease. J. Lipid Res. 53: 1723–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., Dent P. 2009. Bile acids as regulatory molecules. J. Lipid Res. 50: 1509–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. 2008. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7: 678–693. [DOI] [PubMed] [Google Scholar]
- 5.Thomas C., Auwerx J., Schoonjans K. 2008. Bile acids and the membrane bile acid receptor TGR5–connecting nutrition and metabolism. Thyroid. 18: 167–174. [DOI] [PubMed] [Google Scholar]
- 6.Gohlke H., Schmitz B., Sommerfeld A., Reinehr R., Haussinger D. 2013. α5 β1-integrins are sensors for tauroursodeoxycholic acid in hepatocytes. Hepatology. 57: 1117–1129. [DOI] [PubMed] [Google Scholar]
- 7.Sheikh Abdul Kadir S. H., Miragoli M., Abu-Hayyeh S., Moshkov A. V., Xie Q., Keitel V., Nikolaev V. O., Williamson C., Gorelik J. 2010. Bile acid-induced arrhythmia is mediated by muscarinic M2 receptors in neonatal rat cardiomyocytes. PLoS ONE. 5: e9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Studer E., Zhou X., Zhao R., Wang Y., Takabe K., Nagahashi M., Pandak W. M., Dent P., Spiegel S., Shi R., et al. 2012. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 55: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anakk S., Bhosale M., Schmidt V. A., Johnson R. L., Finegold M. J., Moore D. D. 2013. Bile acids activate YAP to promote liver carcinogenesis. Cell Reports. 5: 1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson P. A., Lan T., Rao A. 2009. Bile acid transporters. J. Lipid Res. 50: 2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halilbasic E., Claudel T., Trauner M. 2013. Bile acid transporters and regulatory nuclear receptors in the liver and beyond. J. Hepatol. 58: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaz F. M., Paulusma C. C., Huidekoper H., de Ru M., Lim C., Koster J., Ho-Mok K., Bootsma A. H., Groen A. K., Schaap F. G., et al. Sodium taurocholate cotransporting polypeptide (SLC10A1) deficiency: Conjugated hypercholanemia without a clear clinical phenotype. Hepatology.Epub ahead of print. May 28, 2014; 10.1002/hep.27240. [DOI] [PubMed] [Google Scholar]
- 13.van de Steeg E., Wagenaar E., van der Kruijssen C. M., Burggraaff J. E., de Waart D. R., Elferink R. P., Kenworthy K. E., Schinkel A. H. 2010. Organic anion transporting polypeptide 1a/1b-knockout mice provide insights into hepatic handling of bilirubin, bile acids, and drugs. J. Clin. Invest. 120: 2942–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerloff T., Stieger B., Hagenbuch B., Madon J., Landmann L., Roth J., Hofmann A. F., Meier P. J. 1998. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J. Biol. Chem. 273: 10046–10050. [DOI] [PubMed] [Google Scholar]
- 15.Strautnieks S. S., Bull L. N., Knisely A. S., Kocoshis S. A., Dahl N., Arnell H., Sokal E., Dahan K., Childs S., Ling V., et al. 1998. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat. Genet. 20: 233–238. [DOI] [PubMed] [Google Scholar]
- 16.Oelkers P., Kirby L. C., Heubi J. E., Dawson P. A. 1997. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2). J. Clin. Invest. 99: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson P. A., Haywood J., Craddock A. L., Wilson M., Tietjen M., Kluckman K., Maeda N., Parks J. S. 2003. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J. Biol. Chem. 278: 33920–33927. [DOI] [PubMed] [Google Scholar]
- 18.Rao A., Haywood J., Craddock A. L., Belinsky M. G., Kruh G. D., Dawson P. A. 2008. The organic solute transporter alpha-beta, Ostalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc. Natl. Acad. Sci. USA. 105: 3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballatori N., Fang F., Christian W. V., Li N., Hammond C. L. 2008. Ostalpha-Ostbeta is required for bile acid and conjugated steroid disposition in the intestine, kidney, and liver. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G179–G186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann A. F., Hagey L. R. 2014. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J. Lipid Res. 55: 1553–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwicker B. L., Agellon L. B. 2013. Transport and biological activities of bile acids. Int. J. Biochem. Cell Biol. 45: 1389–1398. [DOI] [PubMed] [Google Scholar]
- 22.Schaap F. G., Trauner M., Jansen P. L. 2014. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 11: 55–67. [DOI] [PubMed] [Google Scholar]
- 23.Chiang J. Y. 2013. Bile acid metabolism and signaling. Compr. Physiol. 3: 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H., Hylemon P. B. 2014. Bile acids are nutrient signaling hormones. Steroids. 86: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T., Chiang J. Y. 2014. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66: 948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridlon J. M., Kang D. J., Hylemon P. B., Bajaj J. S. 2014. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30: 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuipers F., Bloks V. W., Groen A. K. 2014. Beyond intestinal soap–bile acids in metabolic control. Nat. Rev. Endocrinol. 10: 488–498. [DOI] [PubMed] [Google Scholar]
- 28.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 29.Botham K. M., Boyd G. S. 1983. The metabolism of chenodeoxycholic acid to beta-muricholic acid in rat liver. Eur. J. Biochem. 134: 191–196. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann A. F., Borgstroem B. 1964. The intraluminal phase of fat digestion in man: the lipid content of the micellar and oil phases of intestinal content obtained during fat digestion and absorption. J. Clin. Invest. 43: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staggers J. E., Hernell O., Stafford R. J., Carey M. C. 1990. Physical-chemical behavior of dietary and biliary lipids during intestinal digestion and absorption. 1. Phase behavior and aggregation states of model lipid systems patterned after aqueous duodenal contents of healthy adult human beings. Biochemistry. 29: 2028–2040. [DOI] [PubMed] [Google Scholar]
- 32.Hernell O., Staggers J. E., Carey M. C. 1990. Physical-chemical behavior of dietary and biliary lipids during intestinal digestion and absorption. 2. Phase analysis and aggregation states of luminal lipids during duodenal fat digestion in healthy adult human beings. Biochemistry. 29: 2041–2056. [DOI] [PubMed] [Google Scholar]
- 33.Wilson F. A., Sallee V. L., Dietschy J. M. 1971. Unstirred water layers in intestine: rate determinant of fatty acid absorption from micellar solutions. Science. 174: 1031–1033. [DOI] [PubMed] [Google Scholar]
- 34.Dietschy J. M. 1968. Mechanisms for the intestinal absorption of bile acids. J. Lipid Res. 9: 297–309. [PubMed] [Google Scholar]
- 35.Aldini R., Montagnani M., Roda A., Hrelia S., Biagi P. L., Roda E. 1996. Intestinal absorption of bile acids in the rabbit: different transport rates in jejunum and ileum. Gastroenterology. 110: 459–468. [DOI] [PubMed] [Google Scholar]
- 36.Krag E., Phillips S. F. 1974. Active and passive bile acid absorption in man. Perfusion studies of the ileum and jejunum. J. Clin. Invest. 53: 1686–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiff E. R., Small N. C., Dietschy J. M. 1972. Characterization of the kinetics of the passive and active transport mechanisms for bile acid absorption in the small intestine and colon of the rat. J. Clin. Invest. 51: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lionarons D. A., Boyer J. L., Cai S. Y. 2012. Evolution of substrate specificity for the bile salt transporter ASBT (SLC10A2). J. Lipid Res. 53: 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craddock A. L., Love M. W., Daniel R. W., Kirby L. C., Walters H. C., Wong M. H., Dawson P. A. 1998. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am. J. Physiol. 274: G157–G169. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann A. F., Poley J. R. 1972. Role of bile acid malabsorption in pathogenesis of diarrhea and steatorrhea in patients with ileal resection. I. Response to cholestyramine or replacement of dietary long chain triglyceride by medium chain triglyceride. Gastroenterology. 62: 918–934. [PubMed] [Google Scholar]
- 41.Marcus S. N., Schteingart C. D., Marquez M. L., Hofmann A. F., Xia Y., Steinbach J. H., Ton-Nu H. T., Lillienau J., Angellotti M. A., Schmassmann A. 1991. Active absorption of conjugated bile acids in vivo. Kinetic parameters and molecular specificity of the ileal transport system in the rat. Gastroenterology. 100: 212–221. [DOI] [PubMed] [Google Scholar]
- 42.Aldini R., Roda A., Montagnani M., Polimeni C., Lenzi P. L., Cerre C., Galletti G., Roda E. 1994. Hepatic uptake and intestinal absorption of bile acids in the rabbit. Eur. J. Clin. Invest. 24: 691–697. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann A. F., Hagey L. R., Krasowski M. D. 2010. Bile salts of vertebrates: structural variation and possible evolutionary significance. J. Lipid Res. 51: 226–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofmann A. F., Hagey L. R. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65: 2461–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas M. 1983. Determination of acid surface pH in vivo in rat proximal jejunum. Gut. 24: 734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amelsberg A., Jochims C., Richter C. P., Nitsche R., Folsch U. R. 1999. Evidence for an anion exchange mechanism for uptake of conjugated bile acid from the rat jejunum. Am. J. Physiol. 276: G737–G742. [DOI] [PubMed] [Google Scholar]
- 47.Amelsberg A., Schteingart C. D., Ton-Nu H. T., Hofmann A. F. 1996. Carrier-mediated jejunal absorption of conjugated bile acids in the guinea pig. Gastroenterology. 110: 1098–1106. [DOI] [PubMed] [Google Scholar]
- 48.Grandvuinet A. S., Vestergaard H. T., Rapin N., Steffansen B. 2012. Intestinal transporters for endogenic and pharmaceutical organic anions: the challenges of deriving in-vitro kinetic parameters for the prediction of clinically relevant drug-drug interactions. J. Pharm. Pharmacol. 64: 1523–1548. [DOI] [PubMed] [Google Scholar]
- 49.Iusuf D., van de Steeg E., Schinkel A. H. 2012. Functions of OATP1A and 1B transporters in vivo: insights from mouse models. Trends Pharmacol. Sci. 33: 100–108. [DOI] [PubMed] [Google Scholar]
- 50.Csanaky I. L., Lu H., Zhang Y., Ogura K., Choudhuri S., Klaassen C. D. 2011. Organic anion-transporting polypeptide 1b2 (Oatp1b2) is important for the hepatic uptake of unconjugated bile acids: Studies in Oatp1b2-null mice. Hepatology. 53: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong L., Aranibar N., Han Y. H., Zhang Y., Lecureux L., Bhaskaran V., Khandelwal P., Klaassen C. D., Lehman-McKeeman L. D. 2011. Characterization of organic anion-transporting polypeptide (Oatp) 1a1 and 1a4 null mice reveals altered transport function and urinary metabolomic profiles. Toxicol. Sci. 122: 587–597. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Limaye P. B., Lehman-McKeeman L. D., Klaassen C. D. 2012. Dysfunction of organic anion transporting polypeptide 1a1 alters intestinal bacteria and bile acid metabolism in mice. PLoS ONE. 7: e34522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y., Csanaky I. L., Selwyn F. P., Lehman-McKeeman L. D., Klaassen C. D. 2013. Organic anion-transporting polypeptide 1a4 (Oatp1a4) is important for secondary bile acid metabolism. Biochem. Pharmacol. 86: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kullak-Ublick G. A., Ismair M. G., Stieger B., Landmann L., Huber R., Pizzagalli F., Fattinger K., Meier P. J., Hagenbuch B. 2001. Organic anion-transporting polypeptide B (OATP-B) and its functional comparison with three other OATPs of human liver. Gastroenterology. 120: 525–533. [DOI] [PubMed] [Google Scholar]
- 55.Nozawa T., Imai K., Nezu J., Tsuji A., Tamai I. 2004. Functional characterization of pH-sensitive organic anion transporting polypeptide OATP-B in human. J. Pharmacol. Exp. Ther. 308: 438–445. [DOI] [PubMed] [Google Scholar]
- 56.Dawson P. A. 2011. Role of the intestinal bile acid transporters in bile acid and drug disposition. Handb. Exp. Pharmacol. 201: 169–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Claro da Silva T., Polli J. E., Swaan P. W. 2013. The solute carrier family 10 (SLC10): beyond bile acid transport. Mol. Aspects Med. 34: 252–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doring B., Lutteke T., Geyer J., Petzinger E. 2012. The SLC10 carrier family: transport functions and molecular structure. Curr. Top. Membr. 70: 105–168. [DOI] [PubMed] [Google Scholar]
- 59.Anwer M. S., Stieger B. 2014. Sodium-dependent bile salt transporters of the SLC10A transporter family: more than solute transporters. Pflugers Arch. 466: 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jung D., Inagaki T., Gerard R. D., Dawson P. A., Kliewer S. A., Mangelsdorf D. J., Moschetta A. 2007. FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption. J. Lipid Res. 48: 2693–2700. [DOI] [PubMed] [Google Scholar]
- 61.Vivian D., Cheng K., Khurana S., Xu S., Kriel E. H., Dawson P. A., Raufman J. P., Polli J. E. 2014. In vivo performance of a novel fluorinated magnetic resonance imaging agent for functional analysis of bile acid transport. Mol. Pharm. 11: 1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y., Aquino C. J., Cowan D. J., Anderson D. L., Ambroso J. L., Bishop M. J., Boros E. E., Chen L., Cunningham A., Dobbins R. L., et al. 2013. Discovery of a highly potent, nonabsorbable apical sodium-dependent bile acid transporter inhibitor (GSK2330672) for treatment of type 2 diabetes. J. Med. Chem. 56: 5094–5114. [DOI] [PubMed] [Google Scholar]
- 63.Tremont S. J., Lee L. F., Huang H. C., Keller B. T., Banerjee S. C., Both S. R., Carpenter A. J., Wang C. C., Garland D. J., Huang W., et al. 2005. Discovery of potent, nonsystemic apical sodium-codependent bile acid transporter inhibitors (Part 1). J. Med. Chem. 48: 5837–5852. [DOI] [PubMed] [Google Scholar]
- 64.Root C., Smith C. D., Sundseth S. S., Pink H. M., Wilson J. G., Lewis M. C. 2002. Ileal bile acid transporter inhibition, CYP7A1 induction, and antilipemic action of 264W94. J. Lipid Res. 43: 1320–1330. [PubMed] [Google Scholar]
- 65.Lazaridis K. N., Pham L., Tietz P., Marinelli R. A., deGroen P. C., Levine S., Dawson P. A., LaRusso N. F. 1997. Rat cholangiocytes absorb bile acids at their apical domain via the ileal sodium-dependent bile acid transporter. J. Clin. Invest. 100: 2714–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Debray D., Rainteau D., Barbu V., Rouahi M., El Mourabit H., Lerondel S., Rey C., Humbert L., Wendum D., Cottart C. H., et al. 2012. Defects in gallbladder emptying and bile acid homeostasis in mice with cystic fibrosis transmembrane conductance regulator deficiencies. Gastroenterology. 142: 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chignard N., Mergey M., Veissiere D., Parc R., Capeau J., Poupon R., Paul A., Housset C. 2001. Bile acid transport and regulating functions in the human biliary epithelium. Hepatology. 33: 496–503. [DOI] [PubMed] [Google Scholar]
- 68.Yoon Y. B., Hagey L. R., Hofmann A. F., Gurantz D., Michelotti E. L., Steinbach J. H. 1986. Effect of side-chain shortening on the physiologic properties of bile acids: hepatic transport and effect on biliary secretion of 23-nor-ursodeoxycholate in rodents. Gastroenterology. 90: 837–852. [DOI] [PubMed] [Google Scholar]
- 69.Alpini G., Glaser S., Baiocchi L., Francis H., Xia X., Lesage G. 2005. Secretin activation of the apical Na+-dependent bile acid transporter is associated with cholehepatic shunting in rats. Hepatology. 41: 1037–1045. [DOI] [PubMed] [Google Scholar]
- 70.Boyer J. L., Soroka C. J. 2012. A cholecystohepatic shunt pathway: does the gallbladder protect the liver? Gastroenterology. 142: 1416–1419. [DOI] [PubMed] [Google Scholar]
- 71.Kramer W., Stengelin S., Baringhaus K. H., Enhsen A., Heuer H., Becker W., Corsiero D., Girbig F., Noll R., Weyland C. 1999. Substrate specificity of the ileal and the hepatic Na(+)/bile acid cotransporters of the rabbit. I. Transport studies with membrane vesicles and cell lines expressing the cloned transporters. J. Lipid Res. 40: 1604–1617. [PubMed] [Google Scholar]
- 72.Kolhatkar V., Diao L., Acharya C., Mackerell A. D., Jr., Polli J. E. 2012. Identification of novel nonsteroidal compounds as substrates or inhibitors of hASBT. J. Pharm. Sci. 101: 116–126. [DOI] [PubMed] [Google Scholar]
- 73.Rudman D., Kendall F. E. 1957. Bile acid content of human serum. II. The binding of cholanic acids by human plasma proteins. J. Clin. Invest. 36: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kramer W. 1995. Identification of the bile acid binding proteins in human serum by photoaffinity labeling. Biochim. Biophys. Acta. 1257: 230–238. [DOI] [PubMed] [Google Scholar]
- 75.Ceryak S., Bouscarel B., Fromm H. 1993. Comparative binding of bile acids to serum lipoproteins and albumin. J. Lipid Res. 34: 1661–1674. [PubMed] [Google Scholar]
- 76.El-Seaidy A. Z., Mills C. O., Elias E., Crawford J. M. 1997. Lack of evidence for vesicle trafficking of fluorescent bile salts in rat hepatocyte couplets. Am. J. Physiol. 272: G298–G309. [DOI] [PubMed] [Google Scholar]
- 77.Smathers R. L., Petersen D. R. 2011. The human fatty acid-binding protein family: evolutionary divergences and functions. Hum. Genomics. 5: 170–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zimmerman A. W., van Moerkerk H. T., Veerkamp J. H. 2001. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int. J. Biochem. Cell Biol. 33: 865–876. [DOI] [PubMed] [Google Scholar]
- 79.Labonté E. D., Li Q., Kay C. M., Agellon L. B. 2003. The relative ligand binding preference of the murine ileal lipid binding protein. Protein Expr. Purif. 28: 25–33. [DOI] [PubMed] [Google Scholar]
- 80.Kramer W., Sauber K., Baringhaus K. H., Kurz M., Stengelin S., Lange G., Corsiero D., Girbig F., Konig W., Weyland C. 2001. Identification of the bile acid-binding site of the ileal lipid-binding protein by photoaffinity labeling, matrix-assisted laser desorption ionization-mass spectrometry, and NMR structure. J. Biol. Chem. 276: 7291–7301. [DOI] [PubMed] [Google Scholar]
- 81.Capaldi S., Saccomani G., Fessas D., Signorelli M., Perduca M., Monaco H. L. 2009. The X-ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule. J. Mol. Biol. 385: 99–116. [DOI] [PubMed] [Google Scholar]
- 82.Kok T., Hulzebos C. V., Wolters H., Havinga R., Agellon L. B., Stellaard F., Shan B., Schwarz M., Kuipers F. 2003. Enterohepatic circulation of bile salts in farnesoid X receptor-deficient mice: efficient intestinal bile salt absorption in the absence of ileal bile acid-binding protein. J. Biol. Chem. 278: 41930–41937. [DOI] [PubMed] [Google Scholar]
- 83.Stroeve J. H., Brufau G., Stellaard F., Gonzalez F. J., Staels B., Kuipers F. 2010. Intestinal FXR-mediated FGF15 production contributes to diurnal control of hepatic bile acid synthesis in mice. Lab. Invest. 90: 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Praslickova D., Torchia E. C., Sugiyama M. G., Magrane E. J., Zwicker B. L., Kolodzieyski L., Agellon L. B. 2012. The ileal lipid binding protein is required for efficient absorption and transport of bile acids in the distal portion of the murine small intestine. PLoS ONE. 7: e50810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakahara M., Furuya N., Takagaki K., Sugaya T., Hirota K., Fukamizu A., Kanda T., Fujii H., Sato R. 2005. Ileal bile acid-binding protein, functionally associated with the farnesoid X receptor or the ileal bile acid transporter, regulates bile acid activity in the small intestine. J. Biol. Chem. 280: 42283–42289. [DOI] [PubMed] [Google Scholar]
- 86.Fang C., Filipp F. V., Smith J. W. 2012. Unusual binding of ursodeoxycholic acid to ileal bile acid binding protein: role in activation of FXRalpha. J. Lipid Res. 53: 664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dawson P. A., Hubbert M. L., Rao A. 2010. Getting the mOST from OST: role of organic solute transporter, OSTalpha-OSTbeta, in bile acid and steroid metabolism. Biochim. Biophys. Acta. 1801: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ballatori N., Christian W. V., Wheeler S. G., Hammond C. L. 2013. The heteromeric organic solute transporter, OSTalpha-OSTbeta/SLC51: a transporter for steroid-derived molecules. Mol. Aspects Med. 34: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mekhjian H. S., Phillips S. F., Hofmann A. F. 1979. Colonic absorption of unconjugated bile acids: perfusion studies in man. Dig. Dis. Sci. 24: 545–550. [DOI] [PubMed] [Google Scholar]
- 90.Ballatori N., Christian W. V., Lee J. Y., Dawson P. A., Soroka C. J., Boyer J. L., Madejczyk M. S., Li N. 2005. OSTalpha-OSTbeta: a major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology. 42: 1270–1279. [DOI] [PubMed] [Google Scholar]
- 91.Wang L., Hartmann P., Haimerl M., Bathena S. P., Sjowall C., Almer S., Alnouti Y., Hofmann A. F., Schnabl B. 2014. Nod2 deficiency protects mice from cholestatic liver disease by increasing renal excretion of bile acids. J. Hepatol. 60: 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mennone A., Soroka C. J., Harry K. M., Boyer J. L. 2010. Role of breast cancer resistance protein in the adaptive response to cholestasis. Drug Metab. Dispos. 38: 1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kruh G. D., Belinsky M. G. 2003. The MRP family of drug efflux pumps. Oncogene. 22: 7537–7552. [DOI] [PubMed] [Google Scholar]
- 94.Belinsky M. G., Dawson P. A., Shchaveleva I., Bain L. J., Wang R., Ling V., Chen Z. S., Grinberg A., Westphal H., Klein-Szanto A., et al. 2005. Analysis of the in vivo functions of Mrp3. Mol. Pharmacol. 68: 160–168. [DOI] [PubMed] [Google Scholar]
- 95.Zelcer N., van de Wetering K., de Waart R., Scheffer G. L., Marschall H. U., Wielinga P. R., Kuil A., Kunne C., Smith A., van der Valk M., et al. 2006. Mice lacking Mrp3 (Abcc3) have normal bile salt transport, but altered hepatic transport of endogenous glucuronides. J. Hepatol. 44: 768–775. [DOI] [PubMed] [Google Scholar]
- 96.Fernández-Barrena M. G., Monte M. J., Latasa M. U., Uriarte I., Vicente E., Chang H. C., Rodriguez-Ortigosa C. M., Elferink R. O., Berasain C., Marin J. J., et al. 2012. Lack of Abcc3 expression impairs bile-acid induced liver growth and delays hepatic regeneration after partial hepatectomy in mice. J. Hepatol. 56: 367–373. [DOI] [PubMed] [Google Scholar]
- 97.Zelcer N., Saeki T., Bot I., Kuil A., Borst P. 2003. Transport of bile acids in multidrug-resistance-protein 3-overexpressing cells co-transfected with the ileal Na+-dependent bile-acid transporter. Biochem. J. 369: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zeng H., Liu G., Rea P. A., Kruh G. D. 2000. Transport of amphipathic anions by human multidrug resistance protein 3. Cancer Res. 60: 4779–4784. [PubMed] [Google Scholar]
- 99.Heubi J. E., Balistreri W. F., Fondacaro J. D., Partin J. C., Schubert W. K. 1982. Primary bile acid malabsorption: defective in vitro ileal active bile acid transport. Gastroenterology. 83: 804–811. [PubMed] [Google Scholar]
- 100.Lan T., Haywood J., Dawson P. A. 2013. Inhibition of ileal apical but not basolateral bile acid transport reduces atherosclerosis in apoE(-)/(-) mice. Atherosclerosis. 229: 374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Terasaka N., Miyazaki A., Kasanuki N., Ito K., Ubukata N., Koieyama T., Kitayama K., Tanimoto T., Maeda N., Inaba T. 2007. ACAT inhibitor pactimibe sulfate (CS-505) reduces and stabilizes atherosclerotic lesions by cholesterol-lowering and direct effects in apolipoprotein E-deficient mice. Atherosclerosis. 190: 239–247. [DOI] [PubMed] [Google Scholar]
- 102.Gälman C., Ostlund-Lindqvist A. M., Bjorquist A., Schreyer S., Svensson L., Angelin B., Rudling M. 2003. Pharmacological interference with intestinal bile acid transport reduces plasma cholesterol in LDL receptor/apoE deficiency. FASEB J. 17: 265–267. [DOI] [PubMed] [Google Scholar]
- 103.Bhat B. G., Rapp S. R., Beaudry J. A., Napawan N., Butteiger D. N., Hall K. A., Null C. L., Luo Y., Keller B. T. 2003. Inhibition of ileal bile acid transport and reduced atherosclerosis in apoE−/− mice by SC-435. J. Lipid Res. 44: 1614–1621. [DOI] [PubMed] [Google Scholar]
- 104.Wheeler S. G., Hammond C. L., Jornayvaz F. R., Samuel V. T., Shulman G. I., Soroka C. J., Boyer J. L., Hinkle P. M., Ballatori N. 2014. Ostα-/- mice exhibit altered expression of intestinal lipid absorption genes, resistance to age-related weight gain, and modestly improved insulin sensitivity. Am. J. Physiol. Gastrointest. Liver Physiol. 306: G425–G438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Potthoff M. J., Potts A., He T., Duarte J. A., Taussig R., Mangelsdorf D. J., Kliewer S. A., Burgess S. C. 2013. Colesevelam suppresses hepatic glycogenolysis by TGR5-mediated induction of GLP-1 action in DIO mice. Am. J. Physiol. Gastrointest. Liver Physiol. 304: G371–G380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Watanabe M., Morimoto K., Houten S. M., Kaneko-Iwasaki N., Sugizaki T., Horai Y., Mataki C., Sato H., Murahashi K., Arita E., et al. 2012. Bile acid binding resin improves metabolic control through the induction of energy expenditure. PLoS ONE. 7: e38286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu X. X., Watts L. M., Manchem V. P., Chakravarty K., Monia B. P., McCaleb M. L., Bhanot S. 2013. Peripheral reduction of FGFR4 with antisense oligonucleotides increases metabolic rate and lowers adiposity in diet-induced obese mice. PLoS ONE. 8: e66923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lan T., Rao A., Haywood J., Kock N. D., Dawson P. A. 2012. Mouse organic solute transporter alpha deficiency alters FGF15 expression and bile acid metabolism. J. Hepatol. 57: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seward D. J., Koh A. S., Boyer J. L., Ballatori N. 2003. Functional complementation between a novel mammalian polygenic transport complex and an evolutionarily ancient organic solute transporter, OSTalpha-OSTbeta. J. Biol. Chem. 278: 27473–27482. [DOI] [PubMed] [Google Scholar]
- 110.Fang F., Christian W. V., Gorman S. G., Cui M., Huang J., Tieu K., Ballatori N. 2010. Neurosteroid transport by the organic solute transporter OSTalpha-OSTbeta. J. Neurochem. 115: 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cohn S. M., Schloemann S., Tessner T., Seibert K., Stenson W. F. 1997. Crypt stem cell survival in the mouse intestinal epithelium is regulated by prostaglandins synthesized through cyclooxygenase-1. J. Clin. Invest. 99: 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tessner T. G., Muhale F., Riehl T. E., Anant S., Stenson W. F. 2004. Prostaglandin E2 reduces radiation-induced epithelial apoptosis through a mechanism involving AKT activation and bax translocation. J. Clin. Invest. 114: 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Olivier-Mason A., Wojtyniak M., Bowie R. V., Nechipurenko I. V., Blacque O. E., Sengupta P. 2013. Transmembrane protein OSTA-1 shapes sensory cilia morphology via regulation of intracellular membrane trafficking in C. elegans. Development. 140: 1560–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yeh C. Y., Chung-Davidson Y. W., Wang H., Li K., Li W. 2012. Intestinal synthesis and secretion of bile salts as an adaptation to developmental biliary atresia in the sea lamprey. Proc. Natl. Acad. Sci. USA. 109: 11419–11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cai S. Y., Lionarons D. A., Hagey L., Soroka C. J., Mennone A., Boyer J. L. 2013. Adult sea lamprey tolerates biliary atresia by altering bile salt composition and renal excretion. Hepatology. 57: 2418–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kamp F., Hamilton J. A., Kamp F., Westerhoff H. V., Hamilton J. A. 1993. Movement of fatty acids, fatty acid analogues, and bile acids across phospholipid bilayers. Biochemistry. 32: 11074–11086. [DOI] [PubMed] [Google Scholar]
- 117.van Berge-Henegouwen G. P., Hofmann A. F. 1977. Pharmacology of chenodeoxycholic acid. II. Absorption and metabolism. Gastroenterology. 73: 300–309. [PubMed] [Google Scholar]
- 118.Trottier J., Verreault M., Grepper S., Monte D., Belanger J., Kaeding J., Caron P., Inaba T. T., Barbier O. 2006. Human UDP-glucuronosyltransferase (UGT)1A3 enzyme conjugates chenodeoxycholic acid in the liver. Hepatology. 44: 1158–1170. [DOI] [PubMed] [Google Scholar]
- 119.Stiehl A., Raedsch R., Rudolph G., Gundert-Remy U., Senn M. 1985. Biliary and urinary excretion of sulfated, glucuronidated and tetrahydroxylated bile acids in cirrhotic patients. Hepatology. 5: 492–495. [DOI] [PubMed] [Google Scholar]
- 120.Matern S., Matern H., Farthmann E. H., Gerok W. 1984. Hepatic and extrahepatic glucuronidation of bile acids in man. Characterization of bile acid uridine 5′-diphosphate-glucuronosyltransferase in hepatic, renal, and intestinal microsomes. J. Clin. Invest. 74: 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hofmann A. F. 2007. Why bile acid glucuronidation is a minor pathway for conjugation of endogenous bile acids in man. Hepatology. 45: 1083–1084, author reply 1084–1085. [DOI] [PubMed] [Google Scholar]
- 122.Perreault M., Bialek A., Trottier J., Verreault M., Caron P., Milkiewicz P., Barbier O. 2013. Role of glucuronidation for hepatic detoxification and urinary elimination of toxic bile acids during biliary obstruction. PLoS ONE. 8: e80994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Radominska-Pandya A., Little J. M., Pandya J. T., Tephly T. R., King C. D., Barone G. W., Raufman J. P. 1998. UDP-glucuronosyltransferases in human intestinal mucosa. Biochim. Biophys. Acta. 1394: 199–208. [DOI] [PubMed] [Google Scholar]
- 124.Marschall H. U., Matern H., Egestad B., Matern S., Sjovall S. 1987. 6 alpha-glucuronidation of hyodeoxycholic acid by human liver, kidney and small bowel microsomes. Biochim. Biophys. Acta. 921: 392–397. [DOI] [PubMed] [Google Scholar]
- 125.Parquet M., Pessah M., Sacquet E., Salvat C., Raizman A., Infante R. 1985. Glucuronidation of bile acids in human liver, intestine and kidney. An in vitro study on hyodeoxycholic acid. FEBS Lett. 189: 183–187. [DOI] [PubMed] [Google Scholar]
- 126.Zhou X., Cao L., Jiang C., Xie Y., Cheng X., Krausz K. W., Qi Y., Sun L., Shah Y. M., Gonzalez F. J., et al. 2014. PPARalpha-UGT axis activation represses intestinal FXR-FGF15 feedback signalling and exacerbates experimental colitis. Nat. Commun. 5: 4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Adachi Y., Suzuki H., Schinkel A. H., Sugiyama Y. 2005. Role of breast cancer resistance protein (Bcrp1/Abcg2) in the extrusion of glucuronide and sulfate conjugates from enterocytes to intestinal lumen. Mol. Pharmacol. 67: 923–928. [DOI] [PubMed] [Google Scholar]
- 128.van de Wetering K., Feddema W., Helms J. B., Brouwers J. F., Borst P. 2009. Targeted metabolomics identifies glucuronides of dietary phytoestrogens as a major class of MRP3 substrates in vivo. Gastroenterology. 137: 1725–1735. [DOI] [PubMed] [Google Scholar]
- 129.Alnouti Y. 2009. Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108: 225–246. [DOI] [PubMed] [Google Scholar]
- 130.Zhang Y., Klaassen C. D. 2010. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51: 3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hofmann A. F. 2004. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab. Rev. 36: 703–722. [DOI] [PubMed] [Google Scholar]
- 132.De Witt E. H., Lack L. 1980. Effects of sulfation patterns on intestinal transport of bile salt sulfate esters. Am. J. Physiol. 238: G34–G39. [DOI] [PubMed] [Google Scholar]
- 133.Raedsch R., Lauterburg B. H., Hofmann A. F. 1981. Altered bile acid metabolism in primary biliary cirrhosis. Dig. Dis. Sci. 26: 394–401. [DOI] [PubMed] [Google Scholar]
- 134.Stiehl A., Earnest D. L., Admirant W. H. 1975. Sulfation and renal excretion of bile salts in patients with cirrhosis of the liver. Gastroenterology. 68: 534–544. [PubMed] [Google Scholar]
- 135.Dew M. J., Hawker P. C., Nutter S., Allan R. N. 1980. Human intestinal sulphation of lithocholate: a new site for bile acid metabolism. Life Sci. 27: 317–323. [DOI] [PubMed] [Google Scholar]
- 136.Lööf L., Wengle B. 1979. Enzymatic sulphation of bile salts in man. Scand. J. Gastroenterol. 14: 513–519. [DOI] [PubMed] [Google Scholar]
- 137.Hamilton J. P., Xie G., Raufman J. P., Hogan S., Griffin T. L., Packard C. A., Chatfield D. A., Hagey L. R., Steinbach J. H., Hofmann A. F. 2007. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G256–G263. [DOI] [PubMed] [Google Scholar]
- 138.Takikawa H., Beppu T., Seyama Y. 1985. Profiles of bile acids and their glucuronide and sulphate conjugates in the serum, urine and bile from patients undergoing bile drainage. Gut. 26: 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Capocaccia L., Attili A. F., Cantafora A., Bracci F., Paciscopi L., Puoti C., Pieche U., Angelico M. 1981. Sulfated bile acids in serum, bile, and urine of cirrhotic patients before and after portacaval anastomosis. Dig. Dis. Sci. 26: 513–517. [DOI] [PubMed] [Google Scholar]
- 140.Niessen K. H., Teufel M., Brugmann G. 1984. Sulphated bile acids in duodenal juice of healthy infants and children compared with sulphated bile acids in paediatric patients with various gastroenterological diseases. Gut. 25: 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Akita H., Suzuki H., Ito K., Kinoshita S., Sato N., Takikawa H., Sugiyama Y. 2001. Characterization of bile acid transport mediated by multidrug resistance associated protein 2 and bile salt export pump. Biochim. Biophys. Acta. 1511: 7–16. [DOI] [PubMed] [Google Scholar]
- 142.Blazquez A. G., Briz O., Romero M. R., Rosales R., Monte M. J., Vaquero J., Macias R. I., Cassio D., Marin J. J. 2012. Characterization of the role of ABCG2 as a bile acid transporter in liver and placenta. Mol. Pharmacol. 81: 273–283. [DOI] [PubMed] [Google Scholar]
- 143.Halvorsen B., Kase B. F., Prydz K., Garagozlian S., Andresen M. S., Kolset S. O. 1999. Sulphation of lithocholic acid in the colon-carcinoma cell line CaCo-2. Biochem. J. 343: 533–539. [PMC free article] [PubMed] [Google Scholar]
- 144.Marschall H. U., Wagner M., Bodin K., Zollner G., Fickert P., Gumhold J., Silbert D., Fuchsbichler A., Sjovall J., Trauner M. 2006. Fxr(-/-) mice adapt to biliary obstruction by enhanced phase I detoxification and renal elimination of bile acids. J. Lipid Res. 47: 582–592. [DOI] [PubMed] [Google Scholar]
- 145.Wang R., Salem M., Yousef I. M., Tuchweber B., Lam P., Childs S. J., Helgason C. D., Ackerley C., Phillips M. J., Ling V. 2001. Targeted inactivation of sister of P-glycoprotein gene (spgp) in mice results in nonprogressive but persistent intrahepatic cholestasis. Proc. Natl. Acad. Sci. USA. 98: 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Alnouti Y., Klaassen C. D. 2006. Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol. Sci. 93: 242–255. [DOI] [PubMed] [Google Scholar]
- 147.Takahashi A., Tanida N., Kang K., Umibe S., Kawaura A., Furukawa K., Hikasa Y., Satomi M., Shimoyama T. 1990. Difference in enzymatic sulfation of bile acids between the mouse and rat. Tokushima J. Exp. Med. 37: 1–8. [PubMed] [Google Scholar]
- 148.Hagey L. R., Krasowski M. D. 2013. Microbial biotransformations of bile acids as detected by electrospray mass spectrometry. Adv. Nutr. 4: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Huang J., Bathena S. P., Csanaky I. L., Alnouti Y. 2011. Simultaneous characterization of bile acids and their sulfate metabolites in mouse liver, plasma, bile, and urine using LC-MS/MS. J. Pharm. Biomed. Anal. 55: 1111–1119. [DOI] [PubMed] [Google Scholar]
- 150.Kuo S. M., Merhige P. M., Hagey L. R. 2013. The effect of dietary prebiotics and probiotics on body weight, large intestine indices, and fecal bile acid profile in wild type and IL10-/- mice. PLoS ONE. 8: e60270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Griffiths W. J., Sjovall J. 2010. Bile acids: analysis in biological fluids and tissues. J. Lipid Res. 51: 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Marschall H. U., Griffiths W. J., Zhang J., Wietholtz H., Matern H., Matern S., Sjovall J. 1994. Positions of conjugation of bile acids with glucose and N-acetylglucosamine in vitro. J. Lipid Res. 35: 1599–1610. [PubMed] [Google Scholar]
- 153.Goto T., Shibata A., Sasaki D., Suzuki N., Hishinuma T., Kakiyama G., Iida T., Mano N., Goto J. 2005. Identification of a novel conjugate in human urine: bile acid acyl galactosides. Steroids. 70: 185–192. [DOI] [PubMed] [Google Scholar]
- 154.Marschall H. U., Matern H., Wietholtz H., Egestad B., Matern S., Sjovall J. 1992. Bile acid N-acetylglucosaminidation. In vivo and in vitro evidence for a selective conjugation reaction of 7 beta-hydroxylated bile acids in humans. J. Clin. Invest. 89: 1981–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Matern H., Matern S. 1987. Formation of bile acid glucosides and dolichyl phosphoglucose by microsomal glucosyltransferases in liver, kidney and intestine of man. Biochim. Biophys. Acta. 921: 1–6. [DOI] [PubMed] [Google Scholar]
- 156.Ridlon J. M., Kang D. J., Hylemon P. B. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 157.Begley M., Gahan C. G., Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29: 625–651. [DOI] [PubMed] [Google Scholar]
- 158.Philipp B. 2011. Bacterial degradation of bile salts. Appl. Microbiol. Biotechnol. 89: 903–915. [DOI] [PubMed] [Google Scholar]
- 159.Hofmann A. F., Eckmann L. 2006. How bile acids confer gut mucosal protection against bacteria. Proc. Natl. Acad. Sci. USA. 103: 4333–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Inagaki T., Moschetta A., Lee Y. K., Peng L., Zhao G., Downes M., Yu R. T., Shelton J. M., Richardson J. A., Repa J. J., et al. 2006. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. USA. 103: 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Merritt M. E., Donaldson J. R. 2009. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 58: 1533–1541. [DOI] [PubMed] [Google Scholar]
- 162.Lorenzo-Zúñiga V., Bartoli R., Planas R., Hofmann A. F., Vinado B., Hagey L. R., Hernandez J. M., Mane J., Alvarez M. A., Ausina V., et al. 2003. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 37: 551–557. [DOI] [PubMed] [Google Scholar]
- 163.De Smet I., Van Hoorde L., Vande Woestyne M., Christiaens H., Verstraete W. 1995. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79: 292–301. [DOI] [PubMed] [Google Scholar]
- 164.Kurdi P., Kawanishi K., Mizutani K., Yokota A. 2006. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J. Bacteriol. 188: 1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sung J. Y., Shaffer E. A., Costerton J. W. 1993. Antibacterial activity of bile salts against common biliary pathogens. Effects of hydrophobicity of the molecule and in the presence of phospholipids. Dig. Dis. Sci. 38: 2104–2112. [DOI] [PubMed] [Google Scholar]
- 166.Bustos A. Y., Saavedra L., de Valdez G. F., Raya R. R., Taranto M. P. 2012. Relationship between bile salt hydrolase activity, changes in the internal pH and tolerance to bile acids in lactic acid bacteria. Biotechnol. Lett. 34: 1511–1518. [DOI] [PubMed] [Google Scholar]
- 167.Cremers C. M., Knoefler D., Vitvitsky V., Banerjee R., Jakob U. 2014. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proc. Natl. Acad. Sci. USA. 111: E1610–E1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yokota A., Veenstra M., Kurdi P., van Veen H. W., Konings W. N. 2000. Cholate resistance in Lactococcus lactis is mediated by an ATP-dependent multispecific organic anion transporter. J. Bacteriol. 182: 5196–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ruiz L., Margolles A., Sánchez B. 2013. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 4: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bustos A. Y., Raya R., de Valdez G. F., Taranto M. P. 2011. Efflux of bile acids in Lactobacillus reuteri is mediated by ATP. Biotechnol. Lett. 33: 2265–2269. [DOI] [PubMed] [Google Scholar]
- 171.Zaidi A. H., Bakkes P. J., Lubelski J., Agustiandari H., Kuipers O. P., Driessen A. J. 2008. The ABC-type multidrug resistance transporter LmrCD is responsible for an extrusion-based mechanism of bile acid resistance in Lactococcus lactis. J. Bacteriol. 190: 7357–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pfeiler E. A., Klaenhammer T. R. 2009. Role of transporter proteins in bile tolerance of Lactobacillus acidophilus. Appl. Environ. Microbiol. 75: 6013–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Thanassi D. G., Cheng L. W., Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179: 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bina X. R., Provenzano D., Nguyen N., Bina J. E. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76: 3595–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ruiz L., Zomer A., O’Connell-Motherway M., van Sinderen D., Margolles A. 2012. Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl. Environ. Microbiol. 78: 1123–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bortolini O., Medici A., Poli S. 1997. Biotransformations on steroid nucleus of bile acids. Steroids. 62: 564–577. [DOI] [PubMed] [Google Scholar]
- 177.Batta A. K., Salen G., Shefer S. 1984. Substrate specificity of cholylglycine hydrolase for the hydrolysis of bile acid conjugates. J. Biol. Chem. 259: 15035–15039. [PubMed] [Google Scholar]
- 178.Huijghebaert S. M., Hofmann A. F. 1986. Influence of the amino acid moiety on deconjugation of bile acid amidates by cholylglycine hydrolase or human fecal cultures. J. Lipid Res. 27: 742–752. [PubMed] [Google Scholar]
- 179.Batta A. K., Salen G., Arora R., Shefer S., Batta M., Person A. 1990. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J. Biol. Chem. 265: 10925–10928. [PubMed] [Google Scholar]
- 180.Stellwag E. J., Hylemon P. B. 1979. 7alpha-Dehydroxylation of cholic acid and chenodeoxycholic acid by Clostridium leptum. J. Lipid Res. 20: 325–333. [PubMed] [Google Scholar]
- 181.Elkins C. A., Moser S. A., Savage D. C. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology. 147: 3403–3412. [DOI] [PubMed] [Google Scholar]
- 182.Elkins C. A., Savage D. C. 2003. CbsT2 from Lactobacillus johnsonii 100–100 is a transport protein of the major facilitator superfamily that facilitates bile acid antiport. J. Mol. Microbiol. Biotechnol. 6: 76–87. [DOI] [PubMed] [Google Scholar]
- 183.Tanaka H., Doesburg K., Iwasaki T., Mierau I. 1999. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 82: 2530–2535. [DOI] [PubMed] [Google Scholar]
- 184.Choi S. B., Lew L. C., Yeo S. K., Nair Parvathy S., Liong M. T. Probiotics and the BSH-related cholesterol lowering mechanism: a Jekyll and Hyde scenario. Crit. Rev. Biotechnol. Epub ahead of print. February 27, 2014; 10.3109/07388551.2014.889077. [DOI] [PubMed] [Google Scholar]
- 185.Jones B. V., Begley M., Hill C., Gahan C. G., Marchesi J. R. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA. 105: 13580–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hepner G. W., Hofmann A. F., Thomas P. J. 1972. Metabolism of steroid and amino acid moieties of conjugated bile acids in man. II. Glycine-conjugated dihydroxy bile acids. J. Clin. Invest. 51: 1898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hepner G. W., Sturman J. A., Hofmann A. F., Thomas P. J. 1973. Metabolism of steroid and amino acid moieties of conjugated bile acids in man. 3. Cholyltaurine (taurocholic acid). J. Clin. Invest. 52: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang R., Barnes S., Diasio R. B. 1992. Differential intestinal deconjugation of taurine and glycine bile acid N-acyl amidates in rats. Am. J. Physiol. 262: G351–G358. [DOI] [PubMed] [Google Scholar]
- 189.Cowen A. E., Korman M. G., Hofmann A. F., Cass O. W., Coffin S. B. 1975. Metabolism of lithocholate in healthy man. II. Enterohepatic circulation. Gastroenterology. 69: 67–76. [PubMed] [Google Scholar]
- 190.Eyssen H., Van Eldere J., Parmentier G., Huijghebaert S., Mertens J. 1985. Influence of microbial bile salt desulfation upon the fecal excretion of bile salts in gnotobiotic rats. J. Steroid Biochem. 22: 547–554. [DOI] [PubMed] [Google Scholar]
- 191.Huijghebaert S., Parmentier G., Eyssen H. 1984. Specificity of bile salt sulfatase activity in man, mouse and rat intestinal microflora. J. Steroid Biochem. 20: 907–912. [DOI] [PubMed] [Google Scholar]
- 192.Hylemon P. B., Harder J. 1998. Biotransformation of monoterpenes, bile acids, and other isoprenoids in anaerobic ecosystems. FEMS Microbiol. Rev. 22: 475–488. [DOI] [PubMed] [Google Scholar]
- 193.Mallonee D. H., Hylemon P. B. 1996. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J. Bacteriol. 178: 7053–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wells J. E., Hylemon P. B. 2000. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66: 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wells J. E., Berr F., Thomas L. A., Dowling R. H., Hylemon P. B. 2000. Isolation and characterization of cholic acid 7alpha-dehydroxylating fecal bacteria from cholesterol gallstone patients. J. Hepatol. 32: 4–10. [DOI] [PubMed] [Google Scholar]
- 196.Sorg J. A., Sonenshein A. L. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 192: 4983–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kakiyama G., Hylemon P. B., Zhou H., Pandak W. M., Heuman D. M., Kang D. J., Takei H., Nittono H., Ridlon J. M., Fuchs M., et al. 2014. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am. J. Physiol. Gastrointest. Liver Physiol. 306: G929–G937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kakiyama G., Pandak W. M., Gillevet P. M., Hylemon P. B., Heuman D. M., Daita K., Takei H., Muto A., Nittono H., Ridlon J. M., et al. 2013. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 58: 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Norman A. 1964. Faecal excretion products of cholic acid in man. Br. J. Nutr. 18: 173–186. [DOI] [PubMed] [Google Scholar]
- 200.Korpela J. T., Fotsis T., Adlercreutz H. 1986. Multicomponent analysis of bile acids in faeces by anion exchange and capillary column gas-liquid chromatography: application in oxytetracycline treated subjects. J. Steroid Biochem. 25: 277–284. [DOI] [PubMed] [Google Scholar]
- 201.Korpela J. T., Korpela R., Adlercreutz H. 1992. Fecal bile acid metabolic pattern after administration of different types of bread. Gastroenterology. 103: 1246–1253. [DOI] [PubMed] [Google Scholar]
- 202.Harris W. S., Marai L., Myher J. J., Subbiah M. T. 1977. Bile acid ethyl esters. Their infrequent formation during routine bile acid analysis and identification by gas chromatography and mass spectrometry. J. Chromatogr. 131: 437–441. [DOI] [PubMed] [Google Scholar]
- 203.Kelsey M. I., Sexton S. A. 1976. The biosynthesis of ethyl esters of lithocholic acid and isolithocholic acid by rat intestinal microflora. J. Steroid Biochem. 7: 641–647. [DOI] [PubMed] [Google Scholar]
- 204.Norman A., Palmer R. H. 1964. Metabolites of lithocholic acid-24-C-14 in human bile and feces. J. Lab. Clin. Med. 63: 986–1001. [PubMed] [Google Scholar]
- 205.Benson G. M., Haskins N. J., Eckers C., Moore P. J., Reid D. G., Mitchell R. C., Waghmare S., Suckling K. E. 1993. Polydeoxycholate in human and hamster feces: a major product of cholate metabolism. J. Lipid Res. 34: 2121–2134. [PubMed] [Google Scholar]
- 206.Shefer S., Salen G., Hauser S., Dayal B., Batta A. K. 1982. Metabolism of iso-bile acids in the rat. J. Biol. Chem. 257: 1401–1406. [PubMed] [Google Scholar]
- 207.Marschall H. U., Broome U., Einarsson C., Alvelius G., Thomas H. G., Matern S. 2001. Isoursodeoxycholic acid: metabolism and therapeutic effects in primary biliary cirrhosis. J. Lipid Res. 42: 735–742. [PubMed] [Google Scholar]
- 208.Odermatt A., Da Cunha T., Penno C. A., Chandsawangbhuwana C., Reichert C., Wolf A., Dong M., Baker M. E. 2011. Hepatic reduction of the secondary bile acid 7-oxolithocholic acid is mediated by 11beta-hydroxysteroid dehydrogenase 1. Biochem. J. 436: 621–629. [DOI] [PubMed] [Google Scholar]
- 209.Kunne C., Acco A., Hohenester S., Duijst S., de Waart D. R., Zamanbin A., Oude Elferink R. P. 2013. Defective bile salt biosynthesis and hydroxylation in mice with reduced cytochrome P450 activity. Hepatology. 57: 1509–1517. [DOI] [PubMed] [Google Scholar]
- 210.Hofmann A. F., Cravetto C., Molino G., Belforte G., Bona B. 1987. Simulation of the metabolism and enterohepatic circulation of endogenous deoxycholic acid in humans using a physiologic pharmacokinetic model for bile acid metabolism. Gastroenterology. 93: 693–709. [DOI] [PubMed] [Google Scholar]
- 211.Hanson R. F., Williams G. 1971. Metabolism of deoxycholic acid in bile fistula patients. J. Lipid Res. 12: 688–691. [PubMed] [Google Scholar]
- 212.Li F., Jiang C., Krausz K. W., Li Y., Albert I., Hao H., Fabre K. M., Mitchell J. B., Patterson A. D., Gonzalez F. J. 2013. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4: 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Abu-Shanab A., Quigley E. M. 2010. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 7: 691–701. [DOI] [PubMed] [Google Scholar]
- 214.Nieuwdorp M., Gilijamse P. W., Pai N., Kaplan L. M. 2014. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 146: 1525–1533. [DOI] [PubMed] [Google Scholar]
- 215.Ridlon J. M., Alves J. M., Hylemon P. B., Bajaj J. S. 2013. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 4: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schnabl B., Brenner D. A. 2014. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 146: 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Schwabe R. F., Jobin C. 2013. The microbiome and cancer. Nat. Rev. Cancer. 13: 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Swann J. R., Want E. J., Geier F. M., Spagou K., Wilson I. D., Sidaway J. E., Nicholson J. K., Holmes E. 2011. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. USA. 108(Suppl 1): 4523–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.De Minicis S., Rychlicki C., Agostinelli L., Saccomanno S., Candelaresi C., Trozzi L., Mingarelli E., Facinelli B., Magi G., Palmieri C., et al. 2014. Dysbiosis contributes to fibrogenesis in the course of chronic liver injury in mice. Hepatology. 59: 1738–1749. [DOI] [PubMed] [Google Scholar]
- 220.Degirolamo C., Rainaldi S., Bovenga F., Murzilli S., Moschetta A. 2014. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Reports. 7: 12–18. [DOI] [PubMed] [Google Scholar]
- 221.Devkota S., Wang Y., Musch M. W., Leone V., Fehlner-Peach H., Nadimpalli A., Antonopoulos D. A., Jabri B., Chang E. B. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 487: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Jones M. L., Tomaro-Duchesneau C., Prakash S. 2014. The gut microbiome, probiotics, bile acids axis, and human health. Trends Microbiol. 22: 306–308. [DOI] [PubMed] [Google Scholar]
- 223.Sayin S. I., Wahlstrom A., Felin J., Jantti S., Marschall H. U., Bamberg K., Angelin B., Hyotylainen T., Oresic M., Backhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 224.Sears C. L., Garrett W. S. 2014. Microbes, microbiota, and colon cancer. Cell Host Microbe. 15: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Holmes E., Kinross J., Gibson G. R., Burcelin R., Jia W., Pettersson S., Nicholson J. K. 2012. Therapeutic modulation of microbiota-host metabolic interactions. Sci. Transl. Med. 4: 137rv6. [DOI] [PubMed] [Google Scholar]
- 226.Camilleri M. 2014. Advances in understanding of bile acid diarrhea. Expert Rev. Gastroenterol. Hepatol. 8: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jones M. L., Martoni C. J., Ganopolsky J. G., Labbe A., Prakash S. 2014. The human microbiome and bile acid metabolism: dysbiosis, dysmetabolism, disease and intervention. Expert Opin. Biol. Ther. 14: 467–482. [DOI] [PubMed] [Google Scholar]
- 228.Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science. 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- 229.David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., Ling A. V., Devlin A. S., Varma Y., Fischbach M. A., et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Duboc H., Rajca S., Rainteau D., Benarous D., Maubert M. A., Quervain E., Thomas G., Barbu V., Humbert L., Despras G., et al. 2013. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 62: 531–539. [DOI] [PubMed] [Google Scholar]
- 231.Sato H., Macchiarulo A., Thomas C., Gioiello A., Une M., Hofmann A. F., Saladin R., Schoonjans K., Pellicciari R., Auwerx J. 2008. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J. Med. Chem. 51: 1831–1841. [DOI] [PubMed] [Google Scholar]
- 232.Harach T., Pols T. W., Nomura M., Maida A., Watanabe M., Auwerx J., Schoonjans K. 2012. TGR5 potentiates GLP-1 secretion in response to anionic exchange resins. Sci. Rep. 2: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Parker H. E., Wallis K., le Roux C. W., Wong K. Y., Reimann F., Gribble F. M. 2012. Molecular mechanisms underlying bile acid-stimulated glucagon-like peptide-1 secretion. Br. J. Pharmacol. 165: 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wu T., Bound M. J., Standfield S. D., Gedulin B., Jones K. L., Horowitz M., Rayner C. K. 2013. Effects of rectal administration of taurocholic acid on glucagon-like peptide-1 and peptide YY secretion in healthy humans. Diabetes Obes. Metab. 15: 474–477. [DOI] [PubMed] [Google Scholar]
- 235.Ridaura V. K., Faith J. J., Rey F. E., Cheng J., Duncan A. E., Kau A. L., Griffin N. W., Lombard V., Henrissat B., Bain J. R., et al. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 341: 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yokota A., Fukiya S., Islam K. B., Ooka T., Ogura Y., Hayashi T., Hagio M., Ishizuka S. 2012. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes. 3: 455–459. [DOI] [PubMed] [Google Scholar]
- 237.Fouts D. E., Torralba M., Nelson K. E., Brenner D. A., Schnabl B. 2012. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J. Hepatol. 56: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Walters J. R. 2014. Bile acid diarrhoea and FGF19: new views on diagnosis, pathogenesis and therapy. Nat. Rev. Gastroenterol. Hepatol. 11: 426–434. [DOI] [PubMed] [Google Scholar]
- 239.Hofmann A. F., Sjovall J., Kurz G., Radominska A., Schteingart C. D., Tint G. S., Vlahcevic Z. R., Setchell K. D. 1992. A proposed nomenclature for bile acids. J. Lipid Res. 33: 599–604. [PubMed] [Google Scholar]