Significance
The lumazine binding protein (LUMP) emits a cyan-colored fluorescence and has the longest average fluorescence lifetime of any genetically encoded fluorescent protein complex. Coupled with a small mass of 20 kDa, LUMP and its fusion with capture sequences are exploited as unique sensors of protein hydrodynamics and are shown to enable quantitative fluorescence anisotropy imaging of specific target proteins in vitro and in vivo. Moreover, the surface location of the lumazine probe is shown to improve the efficiency of Förster resonance energy transfer (FRET) with the Venus acceptor protein compared with CFP, which is used in the development of a new class of FRET-based sensor.
Keywords: fluorescence anisotropy, FRET, fluorescent protein, protein sensor, FLIM
Abstract
The specialized light organ of the ponyfish supports the growth of the bioluminescent symbiont Photobacterium leiognathi. The bioluminescence of P. leiognathi is generated within a heteromeric protein complex composed of the bacterial luciferase and a 20-kDa lumazine binding protein (LUMP), which serves as a Förster resonance energy transfer (FRET) acceptor protein, emitting a cyan-colored fluorescence with an unusually long excited state lifetime of 13.6 ns. The long fluorescence lifetime and small mass of LUMP are exploited for the design of highly optimized encoded sensors for quantitative fluorescence anisotropy (FA) measurements of protein hydrodynamics. In particular, large differences in the FA values of the free and target-bound states of LUMP fusions appended with capture sequences of up to 20 kDa are used in quantitative FA imaging and analysis of target proteins. For example, a fusion protein composed of LUMP and a 5-kDa G protein binding domain is used as an FA sensor to quantify the binding of the GTP-bound cell division control protein 42 homolog (Cdc42) (21 kDa) in solution and within Escherichia coli. Additionally, the long fluorescence lifetime and the surface-bound fluorescent cofactor 6,7-dimethyl-8- (1′-dimethyl-ribityl) lumazine in LUMP are utilized in the design of highly optimized FRET probes that use Venus as an acceptor probe. The efficiency of FRET in a zero-length LUMP-Venus fusion is 62% compared to ∼31% in a related CFP-Venus fusion. The improved FRET efficiency obtained by using LUMP as a donor probe is used in the design of a FRET-optimized genetically encoded LUMP-Venus substrate for thrombin.
The most direct and predictable approach to quantify the binding of a target protein is to measure the change in mass or hydrodynamic volume of a fluorescence biosensor following complexation (1). This measurement is most easily achieved by recording the polarized components of fluorescence emission of the probe in the free and bound states, and by computing the fluorescence anisotropy (FA), where:
| [1] |
Ipara and Iperp are the fluorescence intensity components parallel and perpendicular to the polarization of the excitation, respectively. The FA value increases in the sensor–target complex owing to the larger hydrodynamic volume and slower rate of tumbling (2). In particular, the Perrin–Weber equation expressed in terms of FA values shows that:
| [2] |
where r is the measured FA value and r0 is the limiting FA value that depends predominantly on the angle between the absorption and emission transition dipole moments, and is in praxis the value of the probe in an extremely viscous solvent (3, 4). τf is the excited-state fluorescence lifetime. τc is the rotational correlation time, the time it takes a sphere to rotate through 1 rad, which is related to the hydrodynamic volume V according to:
| [3] |
where η is the solvent viscosity , R is the gas constant, and T is the temperature in Kelvin (3, 4). Eqs. 2 and 3 show that the ideal fluorescent protein probe for FA measurements has a small mass or hydrodynamic volume and has almost equal values of τf and τc (r0 is fixed). In the case of CFP (28 kDa), however, the FA value in buffer at 20 °C is ∼0.340, which is already close to its limiting anisotropy value of 0.390 (5). CFP is therefore an unsuitable probe for FA-based analysis of target biomolecules. Although pyrene (τf > 100 ns) has been used for in vitro analysis of protein hydrodynamics (4), the probe is excited below 350 nm and its delivery and targeting to specific proteins in living cells remain challenging.
The lumazine binding protein (LUMP) from Photobacterium leiognathi is distinct in having the longest average fluorescence lifetime (13.6 ns) of any encoded protein, and it has 67% of the mass of CFP (6, 7). Together, these properties approximate to an ideal genetically encoded FA sensor. LUMP and its fusions with peptide capture sequences are exploited as part of an approach for FA-based detection and imaging of target proteins in solution and in living cells. Moreover, the surface location of the 6,7-dimethyl-8-(1′-dimethyl-ribityl) lumazine (ribityl-lumazine) probe on LUMP is exploited in the design of a new class of genetically encoded sensors for fluorescence lifetime imaging microscopy (FLIM) and Förster resonance energy transfer (FRET). The increase in the measured FRET efficiency of LUMP-Venus fusion proteins compared with CFP-Venus arises, in part, from the close proximity of the donor probe on the surface of LUMP to the acceptor probe within Venus.
Results
FA Properties of LUMP.
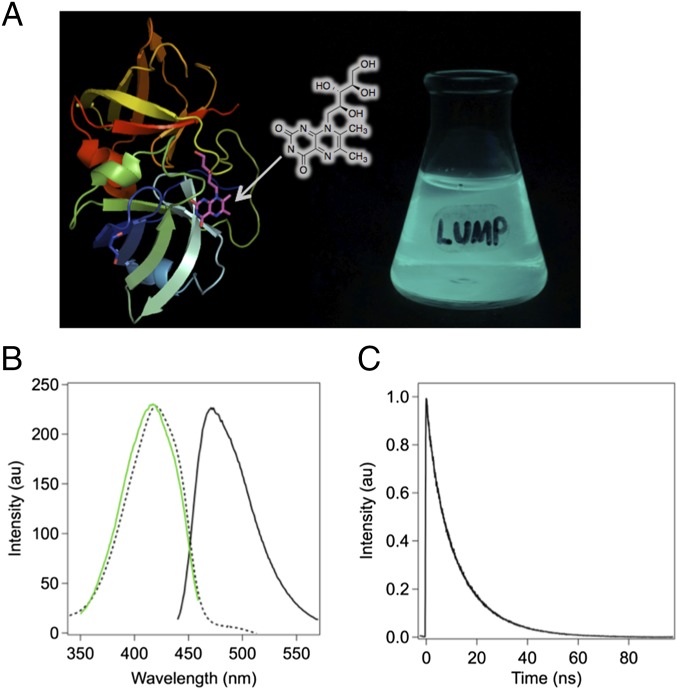
LUMP is a 20-kDa polypeptide composed of two structurally homologous spherical domains (α and β), with the α-subunit binding noncovalently at its surface to ribityl-lumazine (Fig. 1A and Table S1) with a dissociation constant (Kd) of 16 nM at 20 °C (7, 8). Bacteria and most nonmammalian cells biosynthesize ribityl-lumazine, and when transfected with a gene encoding for the α-subunit of LUMP, they produce large quantities of fluorescent protein without the need for exogenous probes (Fig. 1A). The absorption spectrum of the lowest energy (S0-S1) transition in purified LUMP displays a maximum at 420 nm (23,810 cm−1) and a FWHM of 3,468 cm−1. The absorption band is noticeably less complex than the absorption band of CFP, which has overlapping transitions in the lowest energy absorption band (5). LUMP serves as a FRET acceptor in complexes with marine bacterial luciferases, shifting the bioluminescence to a cerulean color (8, 9) (Fig. 1 A and B). Ribityl-lumazine bound to LUMP exhibits an elevated fluorescence quantum yield (Φf = 0.55), and exhibits one of the longest excited state lifetimes (average of 13.6 ns; Fig. 1C) of any natural fluorescent protein (7).
Fig. 1.
Structure and fluorescence properties of LUMP. (A) Crystal structure of LUMP (8) with surface-bound ribityl-lumazine adjoined with a photograph of a flask containing an E. coli culture expressing LUMP. (B) Peak-normalized absorption (dashed line), excitation spectrum (green line) with emission at 470 nm, and emission spectrum (solid line) of LUMP measured in aqueous buffer [20 mM Hepes (pH 7.9), 150 mM NaCl] at 20 °C. (C) Time-resolved fluorescence intensity decay of LUMP. au, arbitrary units.
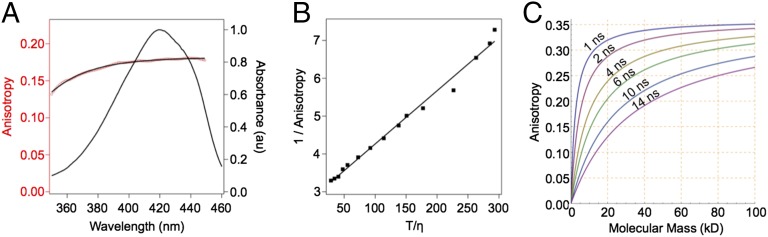
The FA excitation spectrum of purified LUMP (20 kDa) in a viscous medium [75% (mass/vol) sucrose] shows that the S0-S1 transition extends from 380 to 480 nm, reaching a maximum FA value of 0.350 (Fig. 2 A and B). The rotation of polarized emission at high sucrose levels has negligible effects on FA because of the short effective path length (1.67 mm) of the semi-microcuvette. The limiting FA value of LUMP of 0.360 is measured at an even higher viscosity (7). The theoretical maximum FA value of 0.400 is not attained, presumably because the absorption and emission dipoles in the ribityl-lumazine molecule are not colinear (8). The FA values of LUMP measured over a range of viscosities by adding incremental amounts of a concentrated sucrose solution are analyzed using the Perrin–Weber plot (Fig. 2B). A fluorescence lifetime for LUMP of 13.25 ns is calculated from the slope of the line (Fig. 2B), which is similar to the average lifetime of 13.6 ns measured by FLIM (Fig. 1C and Tables S2 and S3).
Fig. 2.
FA properties of LUMP. (A) Excitation anisotropy scan (red) with fluorescence emission at 470 nm superimposed on absorbance scan (black) of LUMP. (B) Perrin–Weber plot of inverse anisotropy vs. T/η, where η is the viscosity (measured in centipoise) in water/sucrose. (C) Simulated plot of anisotropy as a function of molecular mass, assuming 1 nanosecond (ns) τc corresponds to 2.5 kDa.
The expected FA value of a spherical 20-kDa LUMP molecule at 20 °C is computed as 0.133 using Eq. 2 [τf = 13.6 ns, r0 = 0.360 (7)], and assumes τc = 8.0 ns, which is based on a 1-ns increase in τc for every 2.5-kDa increase in mass. The experimentally determined FA value of LUMP is somewhat higher at 0.166 ± 0.002 (Fig. 2A), which suggests that the protein is not strictly spherical. Regardless, the change in FA values between free LUMP at 1 centipoise (cP; 0.166) and its limiting FA value (0.360) are the largest recorded for any genetically encoded fluorescent protein, with the corresponding change for GFP being ∼0.050 (10). The nonlinear relationship between the calculated FA value and protein mass for fluorescently labeled spherical proteins is revealed by fitting Eqs. 2 and 3 for different fluorescent lifetime values (Fig. 2C). The plots clearly reveal the nonlinear relationship between FA and molecular weight. For example, the FA value of a LUMP fusion protein of 25 kDa with a 14-ns excited state lifetime will increase by 41% (from 0.150 to 0.212) on binding to a 25-kDa target protein, and to 0.267 (78% increase) on binding to a 75-kDa protein. Moreover, LUMP would only reach an FA value of 0.324, which is 90% of the limiting FA, on binding to a 325-kDa protein (τc = 140 ns). With a fluorescence lifetime of 13.6 ns (Fig. 1C), one can append capture sequences to LUMP of up to 20 kDa while still achieving a large dynamic range in FA values between the unbound and bound states of the sensor. For example, a sensor composed of a 20-kDa LUMP and a 20-kDa capture protein exhibits a theoretical FA value of 0.195, which increases to 0.281 when in complex with an 80-kDa target protein.
FA-Based Protein Sensors of GTP-Bound Cell Division Control Protein 42 Homolog.
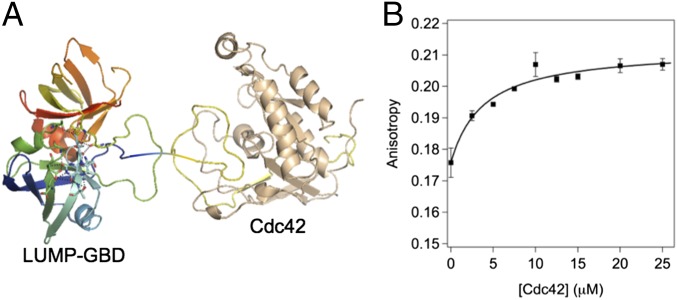
We first developed a LUMP FA sensor for GTP-bound cell division control protein 42 homolog (Cdc42) by appending the 32-aa GTPase binding domain (GBD) from kinase human activated Cdc42 kinase 1 (ACK1; residues 448–489) (11) to the N terminus of LUMP via a flexible six-amino acid linker (GSGSAS; Fig. 3A). LUMP-GBD (25 kDa) binds to Cdc42 specifically with a Kd of 23 nM (11). This fusion protein was purified from Escherichia coli and used for in vitro FA analysis of (GTP)-Cdc42 binding to GBD.
Fig. 3.
FA-based in vitro binding study of LUMP-GBD to Cdc42. (A) Cartoon of crystal structures of LUMP and GBD-Cdc42. (B) FA plot of GBD-LUMP vs. titrated equivalents of Cdc42 in aqueous buffer [20 mM Hepes (pH 7.9), 150 mM NaCl, 2 mM GTP] at 20 °C.
In vitro FA binding study.
The FA value of unbound LUMP-GBD is 0.176 ± 0.004, and it increases to 0.207 ± 0.002 (Δ of ∼18%) when in a stoichiometric complex with GTP-bound Cdc42. These FA values are consistent with those FA values calculated using the Perrin–Weber equation for the free and Cdc42-bound states of LUMP-GBD. Thus, by using a τf of 13.6 ns and a limiting FA of 0.360 for protein spheres of 25 kDa (τc of 10 ns) and 47 kDa (τc of 19 ns), the FA values are calculated as 0.155 and 0.212. These comparisons suggest that LUMP-GBD is not strictly spherical. The binding of a fixed concentration of LUMP-GBD to varying levels of GTP-bound Cdc42 (Fig. 3B) is quantified by FA measurements, with saturation occurring at 1 equivalent of Cdc42 relative to GBD-LUMP (10 μM; Fig. 3B). The increase in FA is shown to be GTP-dependent.
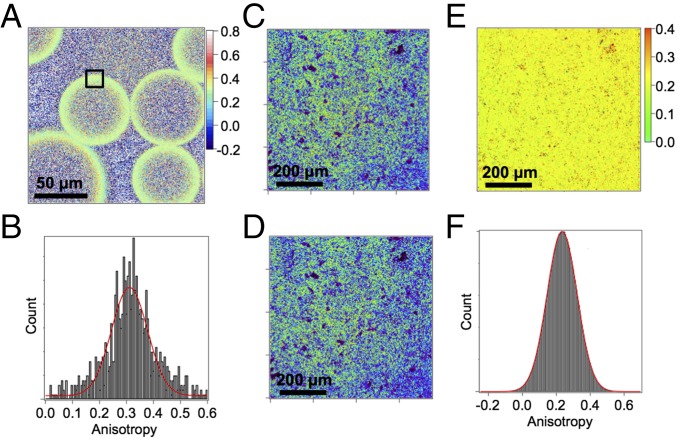
Microscope-based imaging of the polarized fluorescence emission of LUMP is used to compute FA images that quantify the distribution of free and bound populations of the LUMP sensor in a sample (12). In this study, we used a modified confocal fluorescence microscope to record real-time images of the steady-state polarized emission of His-tagged LUMP (23 kDa) bound to nitrilotriacetic acid (NTA)-functionalized agarose beads. The intensity image shows that most of the LUMP is localized to the outer surface of the bead, where, presumably, the bead has the highest level of NTA. FA images and distributions of FA values for LUMP in a sample containing 80 μm of NTA-agarose beads are shown (Fig. 4A). Images were first registered, and the FA was calculated on a pixel-by-pixel basis using Eq. 1. One can see the FA values of His-tagged LUMP at the surface of the bead cluster around 0.310 (Fig. 4B), which is within 14% of the limiting FA value. This result suggests that LUMP molecules are almost completely immobilized when bound to NTA-agarose beads.
Fig. 4.
FA imaging of agarose beads and E. coli. (A) Anisotropy image of His-tagged LUMP on Ni-NTA agarose beads with an average of r (ravg) = 0.310. (B) Anisotropy distribution of the selected region in A with ravg = 0.311 ± 0.002. P-polarized (parallel) emission image (C) and S-polarized (perpendicular) emission images (D) of E. coli cells in PBS buffer expressing GBD-LUMP and Cdc42 (Q61L) in a double-expression vector. (E) Anisotropy image obtained from P- and S-polarized images (C and D) using a G-factor of 0.78. (F) Anisotropy distribution of anisotropy image in E with ravg = 0.233.
A corresponding FA image of His-tagged LUMP in solution with agarose beads lacking the NTA group is composed mostly of free LUMP with an FA value of 0.185. This value is similar to the value measured for His-tagged LUMP in buffer at 20 °C using a SLM-AB2 fluorometer (Aminco) (Fig. S1 A–C). This study highlights an important benefit of using FA images to map the distributions of different molecular forms of the LUMP probe. In particular, because FA values are additive, we can estimate the fraction of LUMP molecules that are free, or that interact transiently with the agarose bead in the FA image. Thus, fractional contributions of each species to the total intensity can be calculated according to the relationship rmeasured = r1f1 + r2f2, where r1 is the FA value of LUMP that is transiently immobilized of fractional intensity f1, r2 is the FA value of unbound LUMP within the bead of fractional intensity f2, and f1 + f2 = 1. This feature of FA imaging is useful because it can be used to quantify the fractions of two populations of the probe in a sample, whereas the intensity image would indicate the presence of a single and uniform population of LUMP molecules. In this particular study, the FA value of His-tagged LUMP outside the bead is 0.185, and inside the bead, the FA value is 0.215 (black and red boxes, respectively, in Fig. S1B). The latter value arises from a mixture of free LUMP (0.185) and LUMP molecules that bind transiently and nonspecifically to the bead (0.310), which represents an FA value that is obtained from a study of NTA-agarose beads (vide infra). Using the relationships above, the percentages of free and transiently bound LUMP within agarose beads are calculated as 76% and 24%, respectively.
In vivo binding study.
We subsequently carried out FA imaging of LUMP (20 kDa) in live bacteria. Confocal images of the polarized emission components (parallel and perpendicular; Fig. 4 C and D) of LUMP fluorescence are used to calculate the FA value for every pixel in the image field. The images are obtained using a Zeiss LSM710 laser scanning confocal microscope adapted for real-time imaging of the polarized components of the emission (Methods). The images are acquired using a 20× (N.A. = 0.8) objective so as to eliminate the high NA depolarization effect (12). The cerulean emission from untransfected bacteria is negligible compared with the cerulean emission from LUMP-transfected cells. Microscopy-derived values of FA are calibrated and validated by carrying out comparative measurements on the same solution of LUMP (20 kDa) with an SLM-AB2 fluorometer and the FA microscope. The FA value recorded for pure LUMP at 20 °C in a cuvette is 0.166 ± 0.005, and in bacteria the FA is measured at 0.180 ± 0.002. Previous FA imaging and translational diffusion studies have shown that the microviscosity of the cytoplasm in living cells is approximately twofold to fivefold the microviscosity of water (13, 14). The τc of LUMP will therefore increase by a small amount, resulting in a small increase in FA in cells as observed by E. coli FA imaging.
We next extended FA imaging to quantify the hydrodynamic properties of LUMP-GBD (25 kDa) in living bacteria that coexpress constitutively active Cdc42 (Q61L; 20 kDa). Coexpression of GBD-LUMP and Cdc42 using the petDuet (EMD Millipore) double-expression vector results in equal amounts of the respective proteins in the cell, with concomitant formation of a 1:1 GBD-LUMP/Cdc42 complex. The FA value of LUMP-GBD in E. coli is 0.233 ± 0.004 (Fig. 4 E and F), which is consistent with the formation of the 45-kDa heterodimeric (LUMP-GBD/Cdc42) complex. The FA value for the complex recorded in live bacteria is again slightly higher than in solution, indicating that the probe experiences a modestly higher viscosity compared with the viscosity measured at 1 cP in buffer at 20 °C (0.207 ± 0.002).
LUMP as an Improved Donor Probe in FRET with Venus.
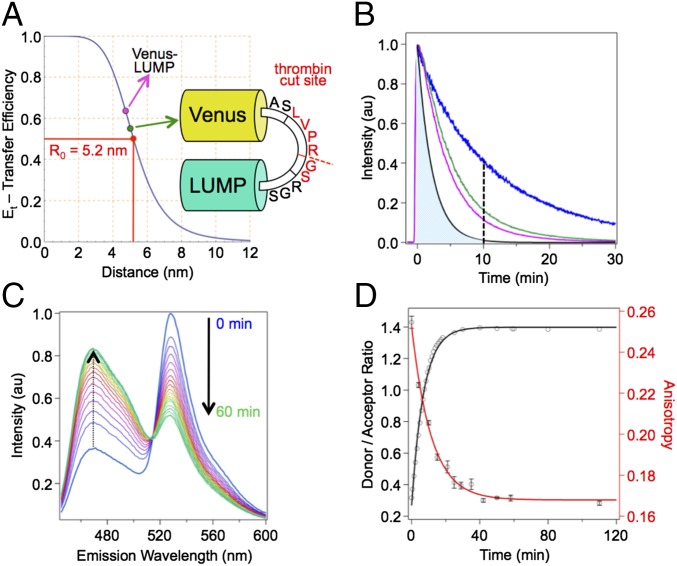
Having developed LUMP as a probe for quantitative FA-based detection and imaging of target proteins, we evaluated LUMP as a donor probe in FRET with the Venus acceptor protein. We have found that LUMP fusions with Venus exhibit robust FRET, which we argue results primarily from the surface location of the ribityl-lumazine on LUMP (Fig. 1A), which reduces the distance between the LUMP donor and Venus acceptor probe in the FRET sensor compared with the distance in a corresponding CFP-Venus fusion protein. The fluorescence lifetime of LUMP linked to Venus by a zero-length (Ala-Ser) linker is measured as 5.2 ns, whereas LUMP alone has an average lifetime of 13.6 ns (Figs. 1C and 5B), yielding a FRET efficiency of 0.62. Using a Förster radius (R0) for LUMP-Venus of 5.2 nm (Fig. 5A), which is calculated from the Förster equation [R0 = 0.0211 (n−4⋅ΦD⋅κ2⋅J)1/6], and measured values of the quantum yield of LUMP (ΦD = 0.55) (7), extinction coefficient for Venus of 92,200 M−1⋅cm−1 at 528 nm (15), spectral overlap integral (J) of 1.964 × 1015 M−1⋅cm−1⋅nm (4), randomized orientations of the donor and acceptor dipoles (κ2 = 2/3) (16), and a refractive index of 1.3346 (17), we calculate an average distance between the donor and acceptor dipoles in the LUMP-Venus fusion protein of 4.8 nm. The same FRET efficiency value of 0.62 was calculated from measuring the integrated emission intensities of pure LUMP and the zero-length Venus-LUMP fusion protein at equal concentrations (Fig. 5A and Fig. S2). The FRET efficiency calculated in the LUMP-Venus fusion protein is twofold higher than the FRET efficiency measured in an optimized zero-length CFP-Venus fusion protein (0.31) (18). Given that the acceptor probe in Venus is buried within the protein matrix and is, on average, ∼2 nm from the surface of Venus, our study suggests that the ribityl-lumazine dipole is within 3 nm of the Venus protein surface.
Fig. 5.
Kinetic study of 10 μM Venus-thrombin-LUMP in aqueous buffer [20 mM Hepes (pH 7.9), 150 mM NaCl] at 20 °C with addition of 2.5 ng/μL thrombin protease. (A) FRET efficiency summary of minimum linker Venus-LUMP (cyan dot) and Venus-thrombin-LUMP (green dot) FRET probes relative to the R0 of 5.2 nm. (B) Fluorescence decays of LUMP and FRET probes. The simulated fluorescence decay of CFP with lifetime τf = 2.2 ns (black line), LUMP (blue line), Venus-thrombin-LUMP (green line), and Venus-LUMP (cyan line) are shown. The dashed line illustrates a potential 10-ns time gate resulting in almost complete suppression of CFP emission (illustrated by the shaded blue area to the left of the dashed line) relative to LUMP emission. (C) Fluorescence emission spectra of Venus-thrombin-LUMP vs. time after addition of thrombin. The time course is visualized with a color shift from blue to magenta to yellow to green. (D) FA and donor/acceptor ratio of fluorescence emission at 470 nm of Venus-thrombin-LUMP vs. time.
This improvement in FRET efficiency in protein fusions of LUMP with Venus is exploited in the design of LUMP-derived FRET-based sensors that exhibit a large change in FRET between their intact and dissociated forms. In particular, a genetically encoded FRET-based thrombin sensor is introduced that is composed of LUMP, which is fused to Venus via an 11-residue linker that harbors a thrombin cleavage site (ASLVPR//GSRGS, where the proteolysis site is denoted by the symbol //, and the underlined residues represent the thrombin recognition sequence) (19) (Fig. 5A). The efficiency of FRET in this substrate of 51% is calculated from the ratio of the integrated fluorescence intensities of LUMP in the intact and cleaved substrates. The corresponding average distance between the donor and acceptor dipoles in the substrate is 5.2 nm. The change in the emission spectrum of the FRET substrate during proteolysis by thrombin is marked by a large increase in LUMP emission and a dramatic decrease in the sensitized emission of Venus, with the donor/acceptor ratio increasing ∼4.7-fold (Fig. 5C). The activity of thrombin activity may also be measured using the large change in FA values of LUMP in the intact and proteolyzed forms of the substrate (Fig. 5D). Using a fluorescence lifetime of 13.6 ns for purified LUMP derived from the intensity decay (Fig. 5B), and a ratio of LUMP emission in the no-FRET and FRET states of the thrombin substrate of 2.6, we arrive at a calculated fluorescence lifetime for the intact substrate of 6.6 ns, which is close to the 5.5-ns lifetime computed from the decay of the donor emission (Fig. 5B). These lifetime data are used in the Perrin–Weber equation to calculate FA values for the 51-kDa LUMP-Venus substrate of 0.255 ± 0.003, which assumes that the molecule is spherical with τc of 20 ns and decreases to 0.166 ± 0.001 in the proteolyzed sensor (Fig. 5D). Thus, as we highlighted in an earlier study (20), the FA value of the donor probe can also be used to quantify the reaction.
Discussion
FA is a powerful technique that allows the quantification of target molecules in a sample with high specificity and selectivity with a single fluorescent sensor and without the need for wash steps, a quality bearing significant potential to quantify target proteins within living cells (21). We have emphasized in this study that the ideal FA sensor has a fluorescence lifetime (τf) that is similar to, or shorter than, the τc of the free sensor. Unfortunately, CFP and related genetically encoded fluorescent proteins have short fluorescence lifetimes (<2.25 ns) (22) and large molecular masses (∼28 kDa) that translate into large values of τc (∼11 ns) that far exceed τf (τf << τc). Although small molecule probes, including pyrene, have much longer fluorescence lifetimes (>30 ns), they require high-energy excitation (<350 nm) (3) and are difficult to use in targeting specific proteins in living cells. In contrast, genetically encoded LUMP has a far longer fluorescence lifetime (13.6 ns) than CFP and, combined with its smaller mass (20 kDa; τc of ∼8 ns), results in the unique condition where τf > τc, which establishes LUMP as an exemplary FA-based sensor to quantify protein interactions. Protein fusions of LUMP can be optimized to bind to a variety of high-value protein targets in vitro as well as in E. coli. The FA value of a LUMP sensor is computed from recordings of the polarized emission of a sample at a single color and is related to the time constant for protein tumbling, with the smaller hydrodynamic volume of the unbound FA sensor tumbling more quickly than the target-bound sensor. Unlike FRET sensors, which must be designed from scratch for each target to optimize the change in FRET efficiency on binding to a target molecule, the time constants for tumbling of a protein fusion of LUMP are easily predicted for the free and bound states of the sensor by using the Perrin–Weber equation. This feature greatly simplifies the interpretation of FA data, as well as the design of LUMP-derived FA sensors, because one simply needs to ensure that the calculated FA values for a particular sensor differ in its free and complexed states by a value that significantly exceeds the error in the measurement of 0.001 FA units. We also note that FA is easily and rapidly imaged with a fluorescence microscope, which we have shown also allows for the dynamic and spatially resolved observation of FA (12, 23, 24) of LUMP-derived sensors in living cells. We have further shown that the long fluorescence lifetime and small mass of LUMP allows one to develop FA sensors appended with capture proteins as large as 20 kDa, while generating a large dynamic range of FA values (from ∼0.20 to ∼0.360) to quantify the amount and distribution of a target protein in a sample or cell. The large dynamic range of FA values between the free and bound states of LUMP sensors should also permit FA-based imaging of stepwise precomplexation reactions (25). Moreover, our studies have shown that LUMP sensors can be used to quantify specific targets in living bacteria and, by extension, to investigate proteome-wide, high-throughput analyses of protein interactions. We note that it would not be possible to develop a related high-throughput approach using FRET, because each sensor would require separate optimization to generate an adequate change in FRET efficiency on binding to a specific target protein.
The maximum FRET efficiency between CFP and Venus in zero-length fusion proteins is ∼31% (18), and it is usually much lower in specific CFP-Venus FRET sensors because the intervening capture sequence leads to a further separation of the CFP and Venus dipoles (12, 14). The surface location of the ribityl-lumazine donor probe in LUMP provides an opportunity to increase FRET efficiency in fusions with a Venus acceptor probe compared with FRET efficiency in fusions using a CFP donor, where the fluorophore is buried in the protein matrix. We also note that although the longer fluorescence lifetime of ribityl-lumazine in LUMP-Venus fusion proteins does not affect the Förster distance, R0, directly, it does affect FRET efficiency, because the rate of energy transfer is inversely related to the lifetime of the donor. Moreover, one might expect LUMP and Venus to undergo more extensive conformational dynamics during the 13.6-ns excited state lifetime [compared with CFP’s 2.25-ns lifetime (20)] that could favorably affect the orientation and proximity of their dipoles, and thereby increase FRET efficiency. These features were demonstrated in the LUMP-Venus fusion protein linked by a two-amino acid (AS) bridge. The protein exhibits a FRET efficiency of 62%. In comparison, an optimized zero-length CFP-YFP probe exhibits a FRET efficiency of 31% (18). We exploited the improved FRET properties imparted by LUMP in the development of a FRET sensor for thrombin activity that displays a dramatic change in the intensities of the donor and acceptor emissions with a concomitant decrease in the FA value of LUMP on proteolysis owing to a decrease in mass and the increase in the fluorescence lifetime of the LUMP product.
The twofold difference in the fluorescence lifetime of LUMP in the FRET and no-FRET states of the thrombin sensor also highlights the potential of using LUMP as a genetically encoded probe for FLIM and for FLIM-based imaging of FRET. We note that CFP and related proteins have short fluorescence lifetimes [<3 ns (20)] that limit the utility of this class of probe for FLIM. The transfection of cells with specific LUMP fusion proteins would therefore provide a means to extend the dynamic range of FLIM and FLIM-FRET (22), as can be seen from a comparison of the intensity decay curves of LUMP and CFP (Fig. 5B). Finally, we note that the longer lifetime of LUMP could also be exploited as a novel probe for ground state-stimulated emission depletion microscopy (26).
Methods
Plasmid Preparation.
The LUMP gene (P. leiognathi, gene: lumP, accession no. Q06877 of the Universal Protein Resource UniProt) was synthesized by Genewiz, Inc., with Nhe1 and Not1 restriction sites at the 5′ and 3′ ends and was subcloned into our pSKB3 vector that is based on Novagen’s pET-28a vector but where the thrombin site is replaced by a tobacco etch virus (TEV) cleavage site. The gene insert GBD(ACK) was synthesized by Genewiz, Inc., with Nde1 and Nhe1 restriction sites at the 5′ and 3′ ends and was subcloned into a pSKB3 vector with a downstream Not1 restriction site. LUMP with Nhe1 and Not1 restriction sites at the 5′ and 3′ ends was PCR-amplified using forward and reverse primers (forward: 5′-AGC GCA GCT AGC TTT AGA GGT ATT GTT CAA GGT-3′ and reverse: 5′-GAG TGC GGC CGC CTA CCA TTC ATT TAA-3′) and cloned into the GBD(ACK)-containing vector to make the GBD-LUMP construct. All sequences were verified by primer-guided sequencing. GBD-LUMP was subcloned into the multiple cloning site 1 (MCS1) of the petDuet-1 vector (Novagen) with restriction sites Nco1 and HindIII using forward and reverse primers (forward: 5′-ATA TAT CC ATG GGC CTG AGC GCA CAG GAC-3′ and reverse: 5′-TAT TAT AAG CTT GAG TGC GGC CGC CTA CCA TTC-3′). The gene of WT Cdc42 (Homo sapiens) was obtained from Addgene (plasmid 12201: pGEX-Cdc42) and PCR-amplified with NdeI and XhoI restriction sites using forward and reverse primers (forward: 5′-GCG CAT ATG CAG ACA ATT AAG TGT GTT GTT GTG GGC-3′ and reverse: 5′-TAT TAT CTC GAG TCA TAG CAG CAC ACA CCT-3′) and subcloned into the petDuet-1 vector at MCS2. Finally, Cdc42 was mutated to its constitutively active form (Q61L) by site-directed mutagenesis using the KAPA HiFi HotStart ReadyMix PCR Kit (KAPA BioSystems) (forward primer: 5′-GAC TTT TTG GTA CTG CAG GGC TAG AGG ATT ATG ATA GAT TAC-3′ and reverse primer: 5′-GTA ATC TAT CAT AAT CCT CTA GCC CTG CAG TAC CAA AAA GTC-3′).
Protein Expression and Purification.
Plasmids were transformed into E. coli BL21 (DE3). Starter cultures (lysogeny broth, 50 mg/L kanamycin) were inoculated from single colonies, grown at 37 °C and used for 1:50 inoculation of 1-L cultures (terrific broth, 50 mg/L kanamycin). Cultures were grown to an OD of around 0.5, cooled for 20 min at 16 °C, induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside, and grown overnight at 16 °C. Cells were harvested by centrifugation for 15 min at 4,000 × g at 4 °C and either washed with PBS and stored as a pellet at −80 °C or directly resuspended in 20 mL of lysis buffer [20 mM Hepes (pH 7.9); 300 mM NaCl; 10 mM imidazole; half of a tablet of Pierce Protease Inhibitor Tablet, EDTA-free (ThermoScientific); 1 mM PMSF; 2 mg of lysozyme]. After incubation for 20 min at 4 °C, the cells were lysed with an Avestin C3 homogenizer, followed by 20 min of centrifugation at 24,000 × g. The supernatant was filtered through a 40-μm Steriflip filter (Millipore), loaded onto a 5-mL Ni-NTA column (Protino; Machery Nagel), and washed with buffer A [20 mM Hepes (pH 7.9), 300 mM NaCl]. The column was washed with 30 mL of washing buffer [20 mM Hepes (pH 7.9), 300 mM NaCl, 25 mM imidazole] and eluted with elution buffer [20 mM Hepes (pH 7.9), 300 mM NaCl, 250 mM imidazole]. Imidazole was removed by exchanging against buffer A with a 10DG desalting column (BioRad), followed by overnight incubation at 4 °C with 1:50 molar equivalents of TEV protease to remove the N-terminal His-tag. The sample was incubated with 0.5 mL of Ni-NTA agarose for 2 h, and the cleaved protein was eluted with 10 mM imidazole containing buffer A. After concentrating the protein with a centrifugal spin concentrator (Millipore), a final size exclusion chromatography run (Superdex 75 10/300 GL; GE Healthcare) with an Akta purifier against buffer B [20 mM Hepes, 150 mM NaCl, (pH 7.9)] yielded proteins of typically >95% purity, as characterized by SDS/PAGE and electrospray ionization (ESI) MS to validate the mass of all proteins, as well as the mass of the corresponding cofactor (ribityl-lumazine). MS measurements of proteins and cofactors were performed using a Thermo LTQ-Orbitrap-XL mass spectrometer equipped with an ESI source. This instrument is located in the QB3/Chemistry Mass Spectrometry Facility at the University of California, Berkeley. Purified proteins were stored on ice for up to 1 d or frozen in liquid nitrogen in small aliquots and stored at −80 °C. Spectroscopic studies showed no change in the absorption or fluorescence properties of thawed proteins.
Fluorescence Studies.
The fluorescence properties of purified LUMP and its fusion proteins were characterized by using an SLM-AB2 fluorometer (Aminco). The excitation anisotropy spectrum was recorded in buffer at 20 °C. The emission wavelength was set at 470 nm, and the excitation was scanned from 380 to 450 nm. FA measurements were recorded on dilute and clarified protein solutions at 20 °C in buffer at the indicated excitation and emission wavelengths. The Perrin–Weber plot was carried out by recording anisotropies at varying sucrose concentrations with viscosity values taken from the CRC Handbook of Chemistry and Physics (94th Ed) (27). The fluorescence lifetime of LUMP was calculated from the slope of the Perrin–Weber plot assuming a spherically shaped protein. The limiting FA value for LUMP is calculated from the reciprocal of the y-intercept.
FA Imaging.
Zeiss LSM710.
Images were recorded with a laser scanning confocal microscope (Zeiss LSM 710, inverse AxioObserver) with a plan-apochromat M27 with a magnification of 20× (N.A. = 0.8) at room temperature. An anisotropy map was created on a pixel-by-pixel basis according to:
where S1 and S2 are the P- and S-polarized images, respectively, and G is the G-factor that is determined by referencing the image anisotropy of LUMP to its known anisotropy (r = 0.166) as measured on the SLM-AB2 fluorometer. The LSM710 confocal microscope is equipped with polarization accessories (Carl Zeiss MicroImaging GmbH).
Image analysis.
Anisotropy images were calculated in Igor Pro (version 6.35, Wavemetrics) using a customized procedure that computes the anisotropy image (*_ani) from the S-image (*_S1) and P-image (*_S2) using a user-defined G-factor according to:
The procedure converts the bit image into a floating point image and does not set negative intensity values to zero.
The following code can be copied into an Igor procedure file, compiled, and run from the command line with aniscalc=(name=“image”) while omitting “_S1” and “_S2” of the original file names, in which the output anisotropy image is appended with the ending “_ani”:
#pragma rtGlobals = 3
#include <Waves Average>
Function aniscalc([name])
string name
wave import_S1 = $(name+“_S1”)
wave import_S2 = $(name+“_S2”)
wavestats/Q import_S1
variable row = dimsize(import_S1, 0)
variable column = dimsize(import_S1, 1)
make/O/N=(row, column) $(name+“_ani”)
wave ani = $(name+“_ani”)
ani = (import_S1 - 0.78*import_S2)/(import_S1 + 0.78*2*import_S2)
End
Anisotropy Simulation.
We have chosen to plot anisotropy as a function of molecular size with (i) τf, (ii) limiting anisotropy (r0), and (iii) a τc-to-molecular weight (MW) conversion factor as parameters that can be varied interactively in the simulation. We have chosen to perform this calculation in Mathematica (28) with the “Manipulate” function (Mathematica version 10.1, Wolfram Research). “Factor MW-to-τc” is the conversion factor that converts τc to MW based on a spherical tumbler by the relation: τc = factor × MW. The Mathematica code is shown below:
Manipulate[
Plot[(1/r0*(1 + tf/(f*MW)))^ −1, {MW, 0, 100}, Axes → {True, True},
AxesLabel → {MW - (kDa), r − Anisotropy},
LabelStyle → Directive[Black, Bold], GridLines → Automatic,
PlotStyle → Thickness[0.005], GridLinesStyle → Directive[Orange, Dashed],
PlotRange → {{0, 100}, {0, 0.36}}], {{r0, 0.36, “limiting anisotropy”},
0.3, 0.66}, {{tf, 4.5, “fluorescence lifetime”}, 0.1, 40},
{{f, 0.4, “factor MW→ tc”}, 0, 1}]
Time-Resolved Fluorescence Measurements.
All time-resolved measurements were made on a Leica TC SP2 inverted confocal microscope. The samples were excited by a Delta Diode 375-nm picosecond-pulsed laser (Horiba) operated at 8 MHz, and fluorescence was collected by an HPM 100-40 hybrid detector (Becker & Hickl). Fluorescence from the donor was selected by using a 455/70 bandpass filter (Chroma). Time-correlated single-photon counting was performed with an SPC-150 card (Becker & Hickl). Solutions of fluorescent proteins were imaged in a 385-well plate. The laser was scanned through an objective with a magnification of 20× (N.A. = 0.5) onto the samples for 60 s to reconstitute single decays. The time-resolved fluorescence decays were imported into Origin software for background subtraction, normalization, and fitting. Duplicate curves for lifetime analysis were identical and were averaged to constitute single curves for each sample. A small irregularity in the decays was visible before the peak. This irregularity may be caused by filter fluorescence or reflection in the setup. This small predecay was disregarded in analysis and assumed to have negligible effect because its intensity was less than 1% of the fluorescence emission peak. Analysis of the residuals for monoexponential and biexponential decay models led to the choice of a biexponential fit:
Average fluorescence lifetimes (τavg) are calculated according to:
Supplementary Material
Acknowledgments
We thank Dr. Anthony Iavarone for his assistance with MS measurements. The work was supported by NIH Grant 1R21CA191067-01 (to G.M.) and a Laboratory Directed Research and Development award from the Division of Physical Biosciences at the Lawrence Berkeley Laboratory.
Footnotes
Conflict of interest statement: G.M., A.C.H., and Y.Y. have filed a patent on the probes detailed in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424021112/-/DCSupplemental.
References
- 1.Weber G. Polarization of the fluorescence of macromolecules. I. Theory and experimental method. Biochem J. 1952;51(2):145–155. doi: 10.1042/bj0510145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jablonski A. On the notion of emission anisotropy. Bull Acad Pol Sci Biol. 1960;8:259–264. [Google Scholar]
- 3.Jameson DM. Introduction to Fluorescence. CRC; Boca Raton, FL: 2014. pp. 75–99. [Google Scholar]
- 4.Valeur B. Molecular Fluorescence Principles and Applications, 5.6. Wiley; Hoboken, NJ: 2001. Effect of rotational Brownian motion. [Google Scholar]
- 5.Sarkar P, Koushik SV, Vogel SS, Gryczynski I, Gryczynski Z. Photophysical properties of Cerulean and Venus fluorescent proteins. J Biomed Opt. 2009;14(3):034047. doi: 10.1117/1.3156842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visser AJWG, Lee J. Lumazine protein from the bioluminescent bacterium Photobacterium phosphoreum. A fluorescence study of the protein-ligand equilibrium. Biochemistry. 1980;19(18):4366–4372. doi: 10.1021/bi00559a033. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, O’Kane DJ, Visser AJWG. Spectral properties and function of two lumazine proteins from Photobacterium. Biochemistry. 1985;24(6):1476–1483. doi: 10.1021/bi00327a028. [DOI] [PubMed] [Google Scholar]
- 8.Chatwell L, et al. Structure of lumazine protein, an optical transponder of luminescent bacteria. J Mol Biol. 2008;382(1):44–55. doi: 10.1016/j.jmb.2008.06.052. [DOI] [PubMed] [Google Scholar]
- 9.Sato Y, et al. Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl- 8-(1′-D-ribityl) lumazine, and its analogues, riboflavin and flavin mononucleotide, at high resolution. J Bacteriol. 2010;192(1):127–133. doi: 10.1128/JB.01015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volkmer A, Subramaniam V, Birch DJS, Jovin TM. One- and two-photon excited fluorescence lifetimes and anisotropy decays of green fluorescent proteins. Biophys J. 2000;78(3):1589–1598. doi: 10.1016/S0006-3495(00)76711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mott HR, et al. Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature. 1999;399(6734):384–388. doi: 10.1038/20732. [DOI] [PubMed] [Google Scholar]
- 12.Yan Y, Marriott G. Fluorescence resonance energy transfer imaging microscopy and fluorescence polarization imaging microscopy. Methods Enzymol. 2003;360:561–580. doi: 10.1016/s0076-6879(03)60128-x. [DOI] [PubMed] [Google Scholar]
- 13.Mastro AM, Babich MA, Taylor WD, Keith AD. Diffusion of a small molecule in the cytoplasm of mammalian cells. Proc Natl Acad Sci USA. 1984;81(11):3414–3418. doi: 10.1073/pnas.81.11.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao S, et al. Optical lock-in detection of FRET using synthetic and genetically encoded optical switches. Biophys J. 2008;94(11):4515–4524. doi: 10.1529/biophysj.107.124859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 16.Dale RE, Eisinger J, Blumberg WE. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys J. 1979;26(2):161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. FluorTools (2014) a|e - UV-Vis-IR Spectral Software, Version 1.2. Available at www.fluortools.com. Accessed October 10, 2015.
- 18.van der Krogt GNM, Ogink J, Ponsioen B, Jalink K. A comparison of donor-acceptor pairs for genetically encoded FRET sensors: Application to the Epac cAMP sensor as an example. PLoS One. 2008;3(4):e1916. doi: 10.1371/journal.pone.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi T, Doolittle RF. Amino acid sequence studies on factor XIII and the peptide released during its activation by thrombin. Biochemistry. 1974;13(4):750–756. doi: 10.1021/bi00701a018. [DOI] [PubMed] [Google Scholar]
- 20.Heidecker M, Yan-Marriott Y, Marriott G. Proximity relationships and structural dynamics of the phalloidin binding site of actin filaments in solution and on single actin filaments on heavy meromyosin. Biochemistry. 1995;34(35):11017–11025. doi: 10.1021/bi00035a007. [DOI] [PubMed] [Google Scholar]
- 21.Yan Y, Marriott G. Analysis of protein interactions using fluorescence technologies. Curr Opin Chem Biol. 2003;7(5):635–640. doi: 10.1016/j.cbpa.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Laine R, et al. Fluorescence lifetime readouts of Troponin-C-based calcium FRET sensors: A quantitative comparison of CFP and mTFP1 as donor fluorophores. PLoS One. 2012;7(11):e49200. doi: 10.1371/journal.pone.0049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dix JA, Verkman AS. Mapping of fluorescence anisotropy in living cells by ratio imaging. Application to cytoplasmic viscosity. Biophys J. 1990;57(2):231–240. doi: 10.1016/S0006-3495(90)82526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattheyses AL, Hoppe AD, Axelrod D. Polarized fluorescence resonance energy transfer microscopy. Biophys J. 2004;87(4):2787–2797. doi: 10.1529/biophysj.103.036194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Z, Huang CC, Tan W. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal Chem. 2006;78(5):1478–1484. doi: 10.1021/ac0517601. [DOI] [PubMed] [Google Scholar]
- 26.Vicidomini G, et al. STED nanoscopy with time-gated detection: theoretical and experimental aspects. PLoS One. 2013;8(1):e54421. doi: 10.1371/journal.pone.0054421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes WM, editor. 2013. CRC Handbook of Chemistry and Physics (CRC Press, Boca Raton, FL), 94th Ed. [Google Scholar]
- 28. Wolfram Research (2015) Mathematica (Wolfram Research, Inc., Champaign, IL), Version 10.1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.