Significance
The X chromosome has a different inheritance pattern from the autosomes, direct interaction and potential conflict with the Y chromosome, and fewer copies than the autosomes. Natural selection may, therefore, act differently on the X chromosome. We analyze polymorphism patterns in 10 great ape species using 87 high-coverage whole genomes. We find that the X chromosome contains megabase-sized regions that are almost without variation in most species. No such regions are found on the autosomes. We suggest that independent and very strong selective sweeps are the only plausible explanation for these observations, and we hypothesize that the targets of these sweeps are multicopy testis-expressed genes in a genetic conflict with the Y chromosome for transmission to the next generation.
Keywords: X-chromosome evolution, great apes, selective sweeps, ampliconic genes, meiotic drive
Abstract
The unique inheritance pattern of the X chromosome exposes it to natural selection in a way that is different from that of the autosomes, potentially resulting in accelerated evolution. We perform a comparative analysis of X chromosome polymorphism in 10 great ape species, including humans. In most species, we identify striking megabase-wide regions, where nucleotide diversity is less than 20% of the chromosomal average. Such regions are found exclusively on the X chromosome. The regions overlap partially among species, suggesting that the underlying targets are partly shared among species. The regions have higher proportions of singleton SNPs, higher levels of population differentiation, and a higher nonsynonymous-to-synonymous substitution ratio than the rest of the X chromosome. We show that the extent to which diversity is reduced is incompatible with direct selection or the action of background selection and soft selective sweeps alone, and therefore, we suggest that very strong selective sweeps have independently targeted these specific regions in several species. The only genomic feature that we can identify as strongly associated with loss of diversity is the location of testis-expressed ampliconic genes, which also have reduced diversity around them. We hypothesize that these genes may be responsible for selective sweeps in the form of meiotic drive caused by an intragenomic conflict in male meiosis.
Because beneficial and recessive X-linked mutations are fully exposed to selection in males, the X chromosome may experience increased rates of adaptive evolution (1, 2). For the same reason, a male-beneficial allele may be fixed by selection even when a deleterious effect in females far exceeds the advantageous effect in males (3), making the X chromosome a potential target of sexually antagonistic genes. These unique characteristics as well as the close evolutionary links between the X and Y chromosomes (4, 5) are likely to be responsible for the different gene content of the X chromosome compared with autosomes (6) and the higher nonsynonymous-to-synonymous substitution ratio in X-linked protein coding sequences compared with that of the autosomal ones (7, 8).
Several studies have indicated that the great ape X chromosome experiences more prevalent positive selection and more efficient purifying selection than the autosomes. Using McDonald–Kreitman-type analyses, we have previously reported that, in the Central chimpanzee lineage, 30–40% of amino acid changes on the X chromosomes are estimated to have been fixed by positive selection, whereas the autosomes do not show significant signs of positive selection (9). A similar conclusion was recently reached for the human X chromosome (10). In all great ape species, the X chromosome shows a more prominent decrease in nucleotide diversity around genes than the autosomes (11–14), interpreted as evidence for more efficient selection removing deleterious variants or fixing advantageous variants on the X chromosomes. Recently, Arbiza et al. (14) showed that 14 human populations have similar relationships between the distance from genes and the diversity, suggesting comparable selective efficiency on the X and autosomes across human populations.
Here, we analyze X-chromosome data from the Great Ape Genome Diversity Project (11). Comparisons of X-chromosomes diversity patterns between the different species allow us to search for general properties of X-chromosome evolution. Surprisingly, we find that the diversity of the X chromosome is strongly reduced in megabase-wide regions in great ape species. We examine various possible explanations for this reduction in diversity and conclude that they are compatible only with recurrent hard selective sweeps affecting overlapping regions. We hypothesize that the targets are multicopy testis-expressed genes, which are overrepresented in the affected regions.
Results
We analyzed the polymorphism and divergence data of whole-genome sequences from 3 to 27 individuals from humans, bonobos, four chimpanzee subspecies, two gorilla species, and two orangutan species (Table S1). Filtering (Methods) leaves 1.93 Gb of the autosomes (67.4%) and 105.03 Mb of the X chromosome (67.8%), with 83,232,220 autosomal SNPs and 2,289,265 X-linked SNPs (Table S2). The SNP quality scores on the X chromosomes are comparable with those on the autosomes (Fig. S1).
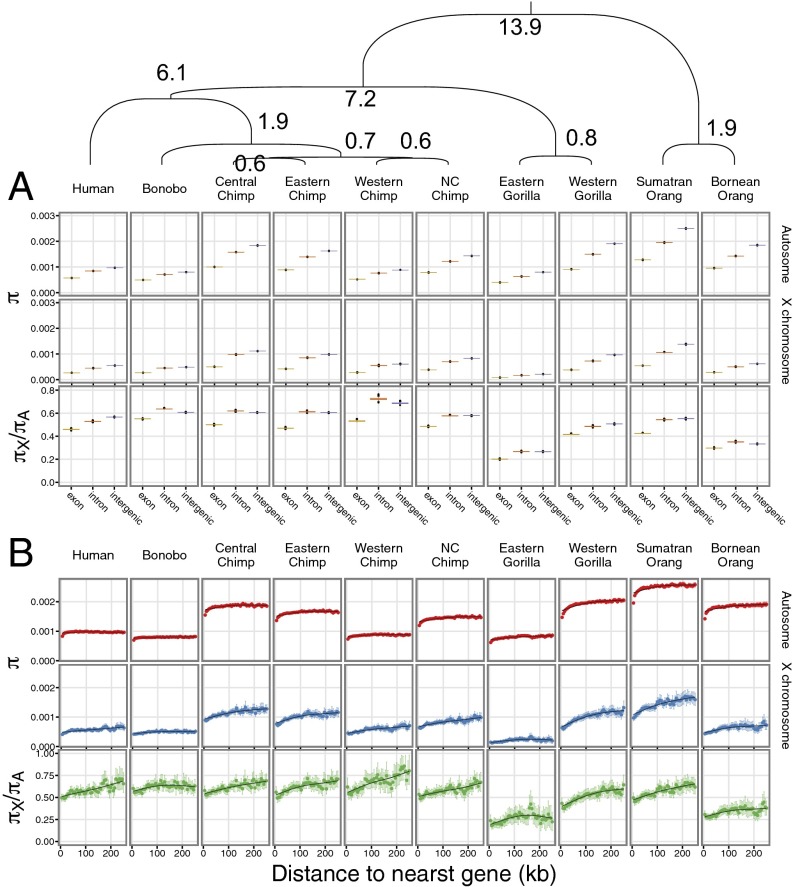
To contrast diversity patterns between autosomes and X chromosomes, we divided the dataset into exonic, intronic, and intergenic regions. We then estimated nucleotide diversity, π, for each functional category in each (sub)species (Fig. 1A). For both autosomes and X chromosomes, π is the lowest for exons followed by introns and then, intergenic regions. Contrasting X-chromosome and autosome diversity shows that the reduction on X, relative to that on autosomes, is largest for exons in all species.
Fig. 1.
Diversity levels inside and outside of genes on autosomes and X chromosomes. The phylogenetic relationship among all investigated great apes and the divergence times (in million years ago) are shown along the top. The nucleotide diversity of autosomes and X chromosomes and their ratio in (A) exons, introns, and intergenic regions and (B) 5-kb windows according the distance from the nearest gene (95% confidence intervals from 1,000 bootstrapping iterations) are shown. NC, Nigeria–Cameroon.
We next correlated the intergenic diversity in nonoverlapping bins of 5 kb with physical distance to the nearest gene (Fig. 1B, Top). Similar studies in humans have used genetic mapping distance (11), but because a high-quality recombination map is only available for humans and the autosomes of Western chimpanzees and because recombination rates are known to change rapidly at the fine scale (15), we used physical distance. In all species, π increases with distance from the nearest gene, and this pattern extends farther than 100 kb away from genes in some of the species. The lower diversity around genes is more pronounced for the X chromosomes than the autosomes, which was illustrated by a positive correlation between distance to genes and the ratio of X to autosome diversity (Fig. 1B, Bottom). The latter observation is in line with previous studies reporting a stronger signature of selection on X chromosomes than on autosomes in humans and other great apes (11–14).
For each pair of species, we compared the slope of the diversity with the physical distance to the nearest gene using correlations of the physical distance from genes with the diversity ratio among pairs of species following the approach by Gottipati et al. (13). Demographic differences should not influence the relative slope, but a larger effect of selection on genes will lead to a steeper slope because of hitchhiking and background selection. For the autosomes, gorillas show the steepest relationships followed by orangutans, chimpanzees, bonobos, and humans (Fig. S2). Among the chimpanzees, Nigeria–Cameroon chimpanzees have the steepest slopes. For the X chromosomes, the ranking shifts, because humans have a steeper slope than bonobos, and Western chimpanzees now have the steepest slope among the chimpanzees. These results suggest that selection affects autosomes and X chromosomes differently among species. Among human populations, selection has been reported to affect the X and the autosomes in a similar way (14).
Diversity Patterns Along the X Chromosome.
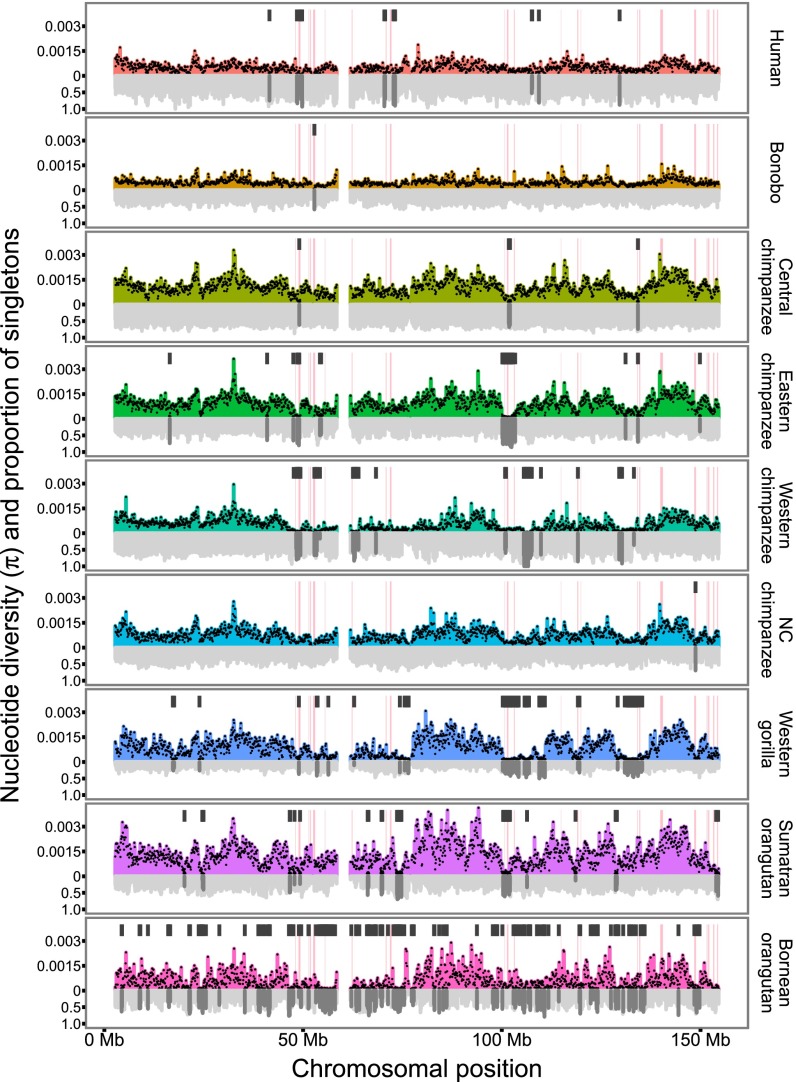
We next calculated π along the X chromosomes using nonoverlapping windows of 100 kb in size. The Eastern lowland gorilla shows an almost complete lack of polymorphism, and because only three X chromosomes were sampled, we chose to omit this species from subsequent analyses. Fig. 2 shows that π is highly variable along the X chromosomes in most species in contrast to similar analysis for autosomes (Fig. S3). We define low-diversity 100-kb windows as windows showing less than 20% of the average X-chromosome diversity, which are represented as black bars in Fig. 2 (the full distribution of π is shown in Fig. S4). We often observe several consecutive windows of low diversity (up to 1.8 Mb); such regions are not found on autosomes (Fig. S3). These regions often overlap among species and even genera, but most regions have normal levels of diversity in at least one species. Fig. 2 also shows the proportion of singletons among SNPs in 100-kb windows below the x axis. In many cases, there is a visible increase in the proportion of singletons in regions of low diversity (see Fig. 4).
Fig. 2.
Diversity and proportion of singletons along X chromosomes. The nucleotide diversity (colored bars with black dots) and the proportion of singleton polymorphisms among SNPs (gray bar) in nonoverlapping 100-kb windows of called sequence are shown for each species. Eastern lowland gorilla is omitted because of insufficient data. At the top of each panel, black bars indicate windows where the nucleotide diversity is less than 20% of the mean diversity for the X chromosome of the same species. The locations of ampliconic regions of human are shown as pink rectangles. NC, Nigeria–Cameroon.
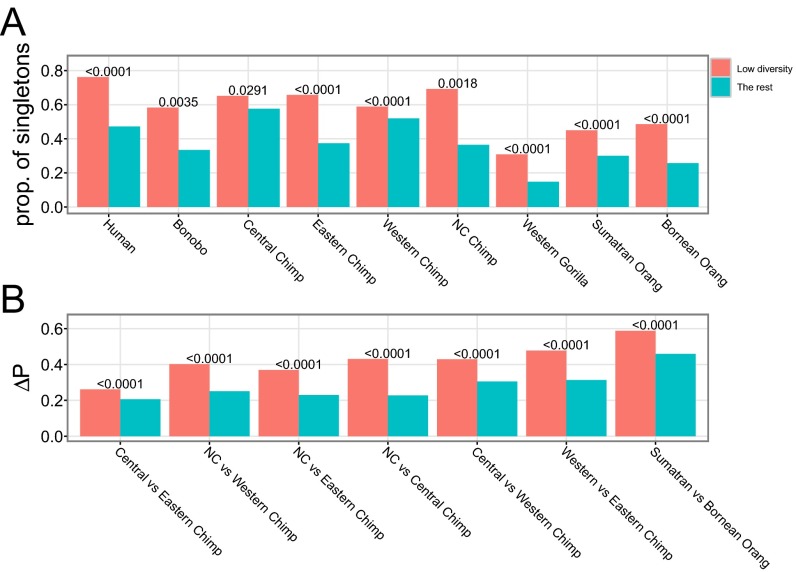
Fig. 4.
Evidence for hard sweeps. (A) The proportion of singleton polymorphisms in SNPs and (B) population differentiation (ΔP) between six pairs of four chimpanzee subspecies and two orangutan species. The differences in the two statistics between regions of reduced diversity (red) and the remaining X chromosome (green) were tested using one-sided bootstrapping tests with 10,000 replicates, and P values are shown above each pair of bar plots. NC, Nigeria–Cameroon.
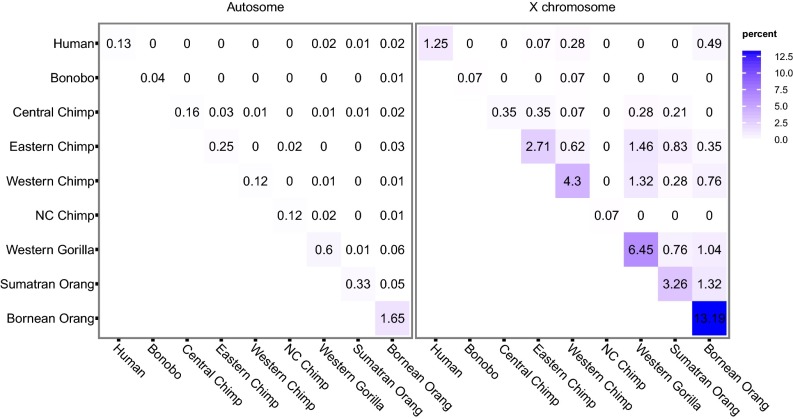
Along the diagonals in Fig. 3, we show the proportion of low-diversity windows for autosomes and X chromosomes. The fraction of low-diversity windows is highest in the Bornean orangutan (13.2%) and lowest in the bonobo (0.07%) and the Nigeria–Cameroon chimpanzee (0.07%). Within the chimpanzees, the Western chimpanzee has the highest faction of low-diversity windows (4.30%). The low-diversity windows are more abundant on the X chromosomes than on the autosomes by 1.95- to 36.30-fold, except for in the Nigeria–Cameroon chimpanzees (0.59-fold). Off-diagonal cells in Fig. 3 show the proportion of low-diversity windows in both members of a pair of species. The proportion of these is much larger for the X than the autosomes (overall 13-fold, P < 10−16; one-tailed Fisher’s exact test), and when we compare species from different genera, sharing is even larger (23-fold, P < 10−16).
Fig. 3.
Proportion of shared regions with reduced diversity. Heat maps for the proportion of shared 100-kb windows with reduced diversity among species in the autosomes and the X chromosomes. The diagonal shows the percentage of windows with less than 20% of the chromosomal average π. The off-diagonal cells show the percentage of windows satisfying this condition in both of two species. NC, Nigeria–Cameroon.
Evidence for Hard Selective Sweeps.
To test whether the low-diversity windows can be partly explained by large genomic regions with lower mutation rates, we compared the divergence from the human reference genome in the low-diversity windows and the rest of the X chromosome. In most species, low-diversity windows also have a lower divergence compared with the rest of the X chromosome (Fig. S5A). However, this lower divergence does not necessarily imply a lower mutation rate. If the low diversity that we observe in the extant species is caused by selection, a similar effect could have reduced diversity in the ancestral species, which would then result in a lower divergence between the descendent species because of shorter coalescent time in the ancestor. The observed difference in divergence between low-diversity windows and the rest of X chromosomes is largest in chimpanzees (20.0%), intermediate in gorillas (9.7%), and lowest in orangutans (4.7%). This inverse relationship of divergence with phylogenetic distance from the human is expected if selection in the ancestral species explains the differences in divergence, because the proportion of time that lineages spend in the ancestral species is larger when the divergence is small. Therefore, selection is expected to contribute most to chimpanzees divergence and least to orangutan divergence, because the chimpanzees diverged most recently, and the orangutans diverged most anciently (11). To address this further, we estimated the diversity in the human–chimpanzee ancestor from the amount of incomplete lineage sorting (16) (Methods). When this ancestral diversity is taken into account, the divergence for low-diversity regions is not significantly different from the rest of the X chromosome (Fig. S5B), arguing that the reductions in divergence are caused by selection in ancestral species rather than reductions in mutation rates.
Close inbreeding may also reduce the diversity locally in the genome (17). However, because we have a relatively large sample size and because the autosomes do not show similar patterns (Fig. S3), inbreeding is unlikely to contribute.
These results leave natural selection on the X chromosome as the sole explanation. Because at most, 10% of the genome seems to be under direct evolutionary constraint (18), reduction in diversity to less than 20% of the mean in large regions must mainly result from indirect selection in the form of background selection (negative selection) or hitchhiking (positive selection). We performed calculations based on existing theory (19) to estimate the expected maximal impact of background selection on diversity (details in SI Text). Within realistic ranges of selection coefficients and proportions of sequence under constraint, background selection is not expected to reduce diversity to less than 75% of the diversity (Fig. S6). To explain the observed levels of diversity, we need to assume a highly unrealistic fraction of the genome under direct selection (such as >80%). Furthermore, if background selection was solely responsible for reduced diversity, we would expect the regions to be almost entirely shared among species.
Computer simulations of soft and hard sweeps, summarized in Table 1 (details in SI Text), show that, even with large selection coefficients (up to s = 0.5) and a low allele frequency at the onset of positive selection (p0 = 0.01), soft sweeps would not reduce diversity to 25% of the chromosomal mean in regions larger than 200 kb (Fig. S7). Thus, a very large number of soft selective sweeps is required to explain the low diversity, and several independent soft sweeps would be needed to reduce diversity to 20% in a region larger than 1 Mb. With hard selective sweeps and effective population sizes in the range of 10,000–100,000, a selection coefficient of 0.1 is expected to reduce diversity to 75% of the original diversity in 1.7- to 2.2-Mb regions and 25% in regions of 400 kb only. For a single sweep to explain depressions below 25% in regions larger than 1 Mb, a selection coefficient larger than 0.5 is required. It is, therefore, more likely that the low-diversity regions on the X chromosome are the result of several sweeps.
Table 1.
Diversity reduction by soft and hard sweeps
| Ne and s | Hard sweep | Soft sweep | ||||
| To 75% | To 25% | p0 = 0.01 | p0 = 0.1 | |||
| To 75% | To 25% | To 75% | To 25% | |||
| 10,000 | ||||||
| 0.01 | 0.3 | 0.1 | 0.4 | <0.1 | 0.1 | <0.1 |
| 0.05 | 1.4 | 0.2 | 1.0 | 0.2 | 0.3 | <0.1 |
| 0.1 | 2.2 | 0.4 | 1.4 | 0.2 | 0.3 | <0.1 |
| 0.2 | 4.1 | 0.8 | 1.9 | 0.3 | 0.3 | <0.1 |
| 0.5 | 10.2 | 2.4 | 2.5 | 0.2 | 0.2 | <0.1 |
| 50,000 | ||||||
| 0.01 | 0.3 | 0.1 | 0.2 | <0.1 | 0.1 | <0.1 |
| 0.05 | 1.0 | 0.2 | 0.4 | 0.1 | 0.1 | <0.1 |
| 0.1 | 1.7 | 0.4 | 0.5 | 0.1 | <0.1 | <0.1 |
| 0.2 | 3.5 | 0.6 | 0.5 | 0.1 | 0.1 | <0.1 |
| 0.5 | 9.0 | 1.8 | 0.5 | 0.1 | 0.1 | <0.1 |
| 100,000 | ||||||
| 0.01 | 0.2 | <0.1 | 0.1 | <0.1 | <0.1 | <0.1 |
| 0.05 | 0.9 | 0.2 | 0.2 | <0.1 | <0.1 | <0.1 |
| 0.1 | 1.7 | 0.4 | 0.2 | <0.1 | <0.1 | <0.1 |
| 0.2 | 3.0 | 0.7 | 0.3 | <0.1 | <0.1 | <0.1 |
| 0.5 | 8.3 | 1.7 | 0.3 | <0.1 | <0.1 | <0.1 |
Summary of expected length of reduced diversity (in megabases) caused by soft and hard sweeps as a function of effective population size (N), selection coefficient (s), and the proportion of beneficial allele onset of selection (p0). Details are in SI Text.
Hard selective sweeps should cause distortions to the site–frequency spectrum. In particular, the fraction of singleton SNPs should be higher in the low-diversity regions than the regions with normal diversity in the presence of hard sweeps (20). In each 100-kb window, we computed the proportion of singletons and found that the low-diversity windows have a significantly higher proportion of singleton polymorphism (Fig. 4A). Tajima’s D, which uses information from the complete site frequency spectrum, was found to be significantly lower in low-diversity regions than in the rest of the X chromosome, which is in agreement with the pattern of proportion of singletons (Fig. S5C).
We further estimated population divergence as the average allele frequency differences, which should be higher in regions affected by population-specific hard sweeps (21), between six pairs of four chimpanzee subspecies and two orangutan species (Methods). Absolute population divergence is significantly higher in low-diversity regions than in the rest of the X chromosome in all comparisons (Fig. 4B). A similar pattern is seen for population divergence measured by FST (Fig. S5D).
Finally, we contrasted the ratio of nonsynonymous to synonymous substitutions between low-diversity windows shared by multiple species and the remaining X chromosome. Nucleotide substitutions were identified by comparing a nucleotide of each species with an ancestral allele of the great apes, and whether a substitution was nonsynonymous or synonymous was determined by the comparison with the human annotation. Then, we counted the number of nonsynonymous and synonymous substitutions observed in the phylogenetic tree of the great apes. The windows with low diversity have a higher nonsynonymous-to-synonymous substitution ratio than the rest of X chromosome by 23.3% (P = 0.0002; one-tailed Fisher’s exact test). The overrepresented nonsynonymous substitutions in the low-diversity windows are compatible with both increased positive selection on protein changes and relaxed purifying selection caused by indirect selection and different gene content of these regions.
Effect of Recombination Rate.
The physical extent of a selective sweep is inversely correlated with recombination rate, and the power to observe them is, therefore, higher in regions of low recombination (22). We, therefore, contrasted the recombination rate from the human recombination map from pedigree data (23) in low-diversity regions with the rest of the X chromosome. The recombination rate of low-diversity regions is between 22.6% and 74.1% of the recombination rate in the remainder of the X chromosome in the different species, except Nigeria–Cameroon chimpanzees, in which the recombination rate is higher by 58.4% in the low-diversity regions (Fig. S5E). Regions in which at least one species has low diversity have an average recombination rate of 65.8% for the rest of the X chromosome. We conclude that reduced recombination of low-diversity regions contributes to their physical extent approximately by a factor of two.
Low-Diversity Regions Associated with X-Linked Ampliconic Regions.
If the low-diversity regions are the result of hard selective sweeps, it is puzzling why sweeps of such magnitude would be private to the X chromosome. We searched for associations between the low-diversity windows and X chromosome-specific properties. We first performed a gene ontology analysis to test whether genes in specific functional categories are overrepresented in the overlaps of low-diversity windows but found no gene ontology term to be significantly overrepresented.
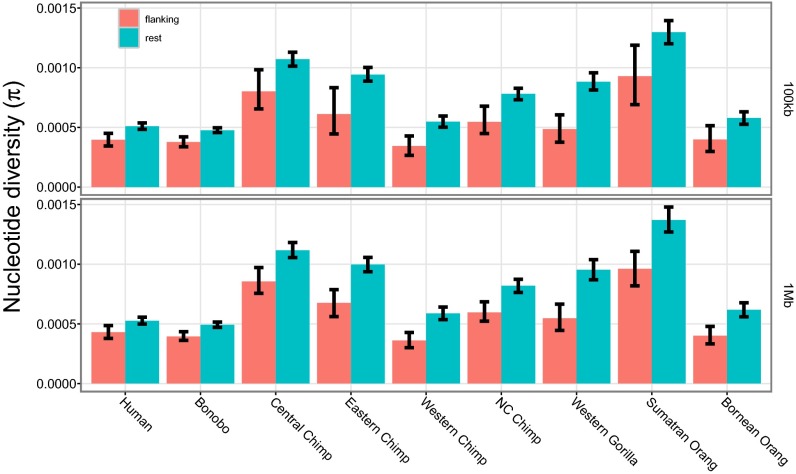
We next considered X-linked ampliconic genes. Mueller et al. (24) recently characterized this X-specific phenomenon in detail. In humans, the X chromosome has 27 independent regions of 100–700 kb in size that contain multiple >99% identical copies of protein coding genes, which are predominantly or exclusively expressed in testis. Only 31% of ampliconic genes are shared with mouse compared with 95% for single-copy genes (24), showing a rapid turnover of these regions. In the mouse, these genes are expressed postmeiotically (25). The ampliconic regions show a striking concordance with low-diversity regions that are most strongly affected by selection in the species (Fig. 2, pink rectangles). To test whether ampliconic genes are associated with lower diversity in regions around them, we contrasted π in the flanking regions of the amplicons (100 kb and 1 Mb) with the rest of the X chromosome. In all species, we observe that π is significantly lower in the flanking regions than in the rest of the X chromosome (Fig. 5). The diversity is difficult to estimate reliably in the ampliconic regions (Fig. S8A), because estimates may be biased by mapping artifacts caused by the repetitive nature of the ampliconic regions; therefore, we put more emphasis on the signatures in the flanking regions presented in Fig. 5. We do not detect a difference in the proportion of singleton polymorphisms between the flanking regions of the amplicons and the rest of the X chromosome (Fig. S8B) but find a higher level of population differentiation in all available (sub)species comparisons (Fig. S8C). Finally, we tested using Fisher’s exact tests whether the flanking regions contain a higher proportion of low-diversity 100-kb windows than the rest of the X chromosomes. We found that low-diversity windows are overrepresented in the flanking regions in all species (Fig. S8D).
Fig. 5.
Diversity around ampliconic regions. Upper shows π in the 100-kb flanking regions of the amplicons and the rest of the X chromosome in each species. Lower shows the same but for 1-Mb flanking regions. The error bars indicate 95% confidence intervals from 1,000 bootstrapping iterations resampled from 1-Mb windows. NC, Nigeria–Cameroon.
Discussion
We have presented the observation that large regions of the X chromosome and not the autosomes have strongly reduced diversity in the great apes. Because unique characteristics of the X chromosome (such as GC content or repeat density) might cause lower sequence quality, we did several tests to investigate whether biases in mapping and SNP calling could potentially contribute to this result. We find that low-diversity windows have a higher SNP quality score than the rest of the regions of the X chromosome (8,276 for low diversity and 7,801 for the rest, P < 0.0001; one-tailed bootstrapping test). The proportion of variants removed by filtering is slightly lower in low-diversity windows but not significantly so (22.2% for low diversity and 23.0% for the rest of the X chromosome, P = 0.0643). We also found that the low-diversity windows have a comparable proportion of included sequence to the rest of windows (73.0% for low diversity and 72.6% for the rest, P = 0.6689). Thus, sequencing and mapping artifacts do not seem to differ in a way that can explain our observations.
Our analysis suggests that the X chromosomes of the great apes are targets of exceptionally strong positive selection recurrently affecting orthologous targets over many megabases. The strongest selective sweep observed in human populations is associated with the lactase gene and has an estimated selection coefficient of 9–19% in the Scandinavian population (26). Even this level of selection affects diversity only in an 800-kb region.
In a search for X chromosome-specific phenomena that could cause recurrent sweeps, we found that, in all species, low-diversity regions strongly associate with the positions of X-linked ampliconic regions, assuming that all species have ampliconic regions in the same places as in the human genome. The function of genes in ampliconic regions is not well-understood, but their evolution seems to be very dynamic, and it is likely that copy number changes often occur, because only 31% of ampliconic genes are shared between human and mouse compared with 95% for single-copy genes (24). The ampliconic genes are expressed predominantly in testis, and therefore, it is tempting to hypothesize that X-linked ampliconic regions are associated with selection for gene expression at some stage during or after male meiosis. We hypothesize that such selection could mediate an intragenomic conflict with Y-linked elements. In this scenario, duplication in an X-linked ampliconic region would be strongly selected if this results in the preferential transmission of the X chromosome. We believe that recurrent segregation distortions of several percentages are more easily envisioned than similar selection coefficients acting on organismal fitness alone. In response to the fixation of a segregation distorter, any compensatory mutation on the Y chromosomes will be under very strong selection to balance the sex ratio as shown from theoretical prediction (27–29) and in empirical observations from Drosophila (30–32).
In line with this conjecture, correlated gene amplification in mouse between sexually antagonistic X-linked Slx genes and Y-linked Sly genes in rodents has been suggested to be a consequence of intragenomic conflict between X and Y chromosomes to balance the sex ratio (33–35). A recent report in mouse also suggests that amplification of Y-linked ampliconic genes is driven by the gene amplification of the gametologous X-linked gene pairs to restore an optimal sex ratio (36). However, in great apes, ampliconic regions on the Y and X are of different origin (37, 38), suggesting that a mechanism to adjust sex ratio may not involve correlated amplification between gametologous gene pairs.
The great ape species differ greatly in their demographic history, long-term effective population sizes, and breeding systems. Each of these factors potentially plays a role in the extent of selective sweeps depending on the underlying mechanism. The widest regions of diversity loss on the X chromosomes are found in Western lowland gorillas and orangutans followed by chimpanzees and humans and finally, bonobos. Among the chimpanzees, the Western chimpanzee has the widest regions, with reduced diversity on the X chromosomes. Thus, the width and combined size of the regions with reduced diversity do not generally reflect population size estimates, where gorillas and orangutans have similar effective population sizes as Central chimpanzees and the Western chimpanzee has a smaller effective population size than the other chimpanzees (11). The expected width of a specific selective sweep is smaller in a large population than in a small population, because the time to fixation is shorter in a small population (Table 1). However, if the occurrence of new mutations limits the number of selective sweeps, the total effect of sweeps is dependent on census population size rather than effective population size. In addition, for sufficiently strong selective sweeps, the probability of fixation is almost completely determined by the selection coefficient and only weakly dependent on effective population size.
The correlation between regions of reduced diversity and testis-expressed ampliconic regions suggests two possible mechanisms creating repeated signatures of selective sweeps: meiotic drive and sperm competition. Under sperm competition, we expect most sweeps in species with more sperm competition, whereas under meiotic drive, the prediction is reversed, because sex chromosome meiotic drive has been reported to lead to reduction in production of either X or Y carrying sperms (39, 40); strong sperm competition may, thus, reduce the effect of meiotic drive, because sperm containing a driving variant will be disadvantageous in the competition among males. Among the great ape species, using the ratio of testicle to body weight as a proxy for sperm competition, gorillas have the least sperm competition followed by orangutans, humans, chimpanzees, and then, bonobos (41, 42). This ordering of species is negatively correlated with the proportion of reduced diversity windows (Spearman’s ρ = −0.72, P = 0.028) (Fig. S9), which fits the prediction of meiotic drive.
We also note that orangutans have many reduced diversity regions that do not coincide with the positions of human ampliconic regions. However, the orangutan is phylogenetically most distant from humans, from whom the positions of ampliconic regions are taken. Dynamic evolution of these regions would predict an inverse relationship between phylogenetic distance and extent of overlap to human ampliconic regions. Our hypothesis that ampliconic regions are responsible for extreme loss of diversity can be tested by determining the species-specific positioning of ampliconic regions rather than using the positions in the human genome. We predict that the association should increase if the hypothesis is true. Direct expression analyses at different stages during spermatogenesis and individual differences may also reveal whether escape from meiotic sex chromosome inactivation (25) or interference with XY body formation is a source of meiotic drive.
Methods
Preparation of the Dataset.
We used the pipelines and the variants from the Great Ape Genome Diversity Consortium (biologiaevolutiva.org/greatape/data.html) (11). The sequences were mapped against hg18. We excluded all positions that were not called in all of the great ape species. Then, we excluded pseudoautosomal regions of X chromosomes. All heterozygous positions on X chromosomes in at least one male were also excluded.
The sequences were annotated with refSeqGenes downloaded from the table browser at the UCSC Genome Bioinformatics Site (genome.ucsc.edu/index.html). If any position can be included as multiple functional categories, we annotated the position with the following priority: exons, introns, and intergenic sequences. The nonsynonymous and synonymous variants were determined using ANNOVAR software (43).
Diversity Estimation.
The nucleotide diversity, π, was calculated using the formula by Nei and Li (44) on the variants called in the Great Ape Genome Diversity Consortium that passed the variant filtering used there (11). For exons, introns, and intergenic sequences, we calculated π separately. The diversity according to the physical distance from the nearest gene was calculated as follows.: (i) for each position, we calculated the distance from the nearest transcript, (ii) the positions were sorted by the distance, (iii) the positions were grouped according to the distance by 5 kb, and (iv) π was calculated for each group. The 95% confidence intervals were obtained by the nonparametric bootstrapping resampled among 1-Mb nonoverlapping windows with 1,000 replicates.
Divergence Normalization in Pan.
The average of density of sites with X-chromosomal incomplete lineage sorting between human and chimpanzee is 10.26% (16), which corresponds to the ancestral effective population size (NeAns) of humans, bonobos, and chimpanzees that is equal to 54,103. The NeAns of reduced diversity windows was estimated with the ratio of the diversity in these windows to diversity of the rest of windows. The NeAns of the rest of windows on the X chromosomes was estimated in the same way. The number of generations between extant humans and each Pan species (NgenH−BC) was calculated by
where ngenBC is the number of generations in bonobo–chimpanzee lineage after complete split from human lineage. We inferred ngenBC to be 244,000 based on assumptions that the speciation time is 6.1 Mya and the generation time is 25 y. The divergence levels were normalized by dividing the DXY values (45) with the relative differences of NgenH−BC between low-diversity windows and the rest of windows.
The Putative Sweep Analysis.
We calculated π in nonoverlapping 100-kb windows across the genome and excluded windows where less than 20% of the sites were called.
The index of allelic differentiation (ΔP) between populations A and B was calculated using the following formula:
where AFA,i and AFB,i are the allele frequencies of the ith SNP in populations A and B, respectively, and no. SNP is the total number of SNPs in the window. The FST values are calculated using the methods by Weir and Cockerham (46) with the Genepop software (47).
The calculations on the possible effects of background selection and the simulations to explore selective sweeps are described in SI Text. The list of ampliconic regions was obtained from the literature (24). The gene ontology test was performed using the GOrilla software (48).
Supplementary Material
Acknowledgments
We thank Brian Charlesworth, Andy Clark, David Reich, and Freddy B. Christiansen for comments on the manuscript. The work was supported by the European Commission-funded 7th Framework Programme for Research and Technological Development NEXTGENE Project (to T.M. and M.H.S.) and a grant from the Danish Council for Independent Research (to M.H.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.C.P. is a guest editor invited by the Editorial Board.
2A complete list of contributors to the Great Ape Genome Diversity Project can be found in SI Text.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419306112/-/DCSupplemental.
Contributor Information
Collaborators: Javier Prado-Martinez, Peter H. Sudmant, Jeffrey M. Kidd, Heng Li, Joanna L. Kelley, Belen Lorente-Galdos, Krishna R. Veeramah, August E. Woerner, Timothy D. O’Connor, Gabriel Santpere, Alexander Cagan, Christoph Theunert, Ferran Casals, Hafid Laayouni, Kasper Munch, Asger Hobolth, Anders E. Halager, Maika Malig, Jessica Hernandez-Rodriguez, Irene Hernando-Herraez, Kay Prüfer, Marc Pybus, Laurel Johnstone, Michael Lachmann, Can Alkan, Dorina Twigg, Natalia Petit, Carl Baker, Fereydoun Hormozdiari, Marcos Fernandez-Callejo, Marc Dabad, Michael L. Wilson, Laurie Stevison, Cristina Camprubí, Tiago Carvalho, Aurora Ruiz-Herrera, Laura Vives, Marta Mele, Teresa Abello, Ivanela Kondova, Ronald E. Bontrop, Anne Pusey, Felix Lankester, John A. Kiyang, Richard A. Bergl, Elizabeth Lonsdorf, Simon Myers, Mario Ventura, Pascal Gagneux, David Comas, Hans Siegismund, Julie Blanc, Lidia Agueda-Calpena, Marta Gut, Lucinda Fulton, Sarah A. Tishkoff, James C. Mullikin, Richard K. Wilson, Ivo G. Gut, Mary Katherine Gonder, Oliver A. Ryder, Beatrice H. Hahn, Arcadi Navarro, Joshua M. Akey, Jaume Bertranpetit, David Reich, Thomas Mailund, Mikkel H. Schierup, Christina Hvilsom, Aida M. Andrés, Jeffrey D. Wall, Carlos D. Bustamante, Michael F. Hammer, Evan E. Eichler, and Tomas Marques-Bonet
References
- 1.Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130(1):113–146. [Google Scholar]
- 2.Vicoso B, Charlesworth B. Evolution on the X chromosome: Unusual patterns and processes. Nat Rev Genet. 2006;7(8):645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 3.Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38(4):735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 4.Cortez D, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508(7497):488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 5.Bellott DW, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508(7497):494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vallender EJ, Lahn BT. How mammalian sex chromosomes acquired their peculiar gene content. BioEssays. 2004;26(2):159–169. doi: 10.1002/bies.10393. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3(6):e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meisel RP, Connallon T. The faster-X effect: Integrating theory and data. Trends Genet. 2013;29(9):537–544. doi: 10.1016/j.tig.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hvilsom C, et al. Extensive X-linked adaptive evolution in central chimpanzees. Proc Natl Acad Sci USA. 2012;109(6):2054–2059. doi: 10.1073/pnas.1106877109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veeramah KR, Gutenkunst RN, Woerner AE, Watkins JC, Hammer MF. Evidence for increased levels of positive and negative selection on the X chromosome versus autosomes in humans. Mol Biol Evol. 2014;31(9):2267–2282. doi: 10.1093/molbev/msu166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499(7459):471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer MF, et al. The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nat Genet. 2010;42(10):830–831. doi: 10.1038/ng.651. [DOI] [PubMed] [Google Scholar]
- 13.Gottipati S, Arbiza L, Siepel A, Clark AG, Keinan A. Analyses of X-linked and autosomal genetic variation in population-scale whole genome sequencing. Nat Genet. 2011;43(8):741–743. doi: 10.1038/ng.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arbiza L, Gottipati S, Siepel A, Keinan A. Contrasting X-linked and autosomal diversity across 14 human populations. Am J Hum Genet. 2014;94(6):827–844. doi: 10.1016/j.ajhg.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auton A, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336(6078):193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charlesworth D. Effects of inbreeding on the genetic diversity of populations. Philos Trans R Soc Lond B Biol Sci. 2003;358(1434):1051–1070. doi: 10.1098/rstb.2003.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLOS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durrett R. Natural Selection. Probability Models for DNA Sequence Evolution. Springer; Berlin: 2008. pp. 191–248. [Google Scholar]
- 20.Kim Y. Allele frequency distribution under recurrent selective sweeps. Genetics. 2006;172(3):1967–1978. doi: 10.1534/genetics.105.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephan W, Mitchell SJ. Reduced levels of DNA polymorphism and fixed between-population differences in the centromeric region of Drosophila ananassae. Genetics. 1992;132(4):1039–1045. doi: 10.1093/genetics/132.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23(1):23–35. [PubMed] [Google Scholar]
- 23.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31(3):241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 24.Mueller JL, et al. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet. 2013;45(9):1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller JL, et al. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet. 2008;40(6):794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74(6):1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank SA. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45(2):262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 28.Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol Evol. 2010;25(4):215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: A new explanation of Haldane’s rule and related phenomena. Genetics. 1991;128(4):841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyer KA, Charlesworth B, Jaenike J. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc Natl Acad Sci USA. 2007;104(5):1587–1592. doi: 10.1073/pnas.0605578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Presgraves DC, Gérard PR, Cherukuri A, Lyttle TW. Large-scale selective sweep among Segregation Distorter chromosomes in African populations of Drosophila melanogaster. PLoS Genet. 2009;5(5):e1000463. doi: 10.1371/journal.pgen.1000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unckless RL, Larracuente AM, Clark AG. Sex-ratio meiotic drive and Y-linked resistance in Drosophila affinis. Genetics. 2015;199(3):831–840. doi: 10.1534/genetics.114.173948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis PJI, Bacon J, Affara NA. Association of Sly with sex-linked gene amplification during mouse evolution: A side effect of genomic conflict in spermatids? Hum Mol Genet. 2011;20(15):3010–3021. doi: 10.1093/hmg/ddr204. [DOI] [PubMed] [Google Scholar]
- 34.Cocquet J, et al. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8(9):e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cocquet J, et al. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 2009;7(11):e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soh YQS, et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159(4):800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes JF, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463(7280):536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhowmick BK, Satta Y, Takahata N. The origin and evolution of human ampliconic gene families and ampliconic structure. Genome Res. 2007;17(4):441–450. doi: 10.1101/gr.5734907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaenike J. Sex chromosome meiotic drive. Annu Rev Ecol Syst. 2001;32:25–49. [Google Scholar]
- 40.Rutkowska J, Badyaev AV. Review. Meiotic drive and sex determination: Molecular and cytological mechanisms of sex ratio adjustment in birds. Philos Trans R Soc Lond B Biol Sci. 2008;363(1497):1675–1686. doi: 10.1098/rstb.2007.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harcourt AH, Harvey PH, Larson SG, Short RV. Testis weight, body weight and breeding system in primates. Nature. 1981;293(5827):55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- 42.Dixson AF. Sperm Competition. Primate Sexuality: Comparative Studies of the Prosimians, Monkeys, Apes, and Humans. 2nd Ed. Oxford Univ Press; New York: 2012. pp. 298–333. [Google Scholar]
- 43.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nei M, Jin L. Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol. 1989;6(3):290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- 46.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 47.Raymond M, Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86(3):248–249. [Google Scholar]
- 48.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.