Abstract
Denitrification in agricultural soils is a major source of N2O. Legume crops enhance N2O emission by providing N-rich residues, thereby stimulating denitrification, both by free-living denitrifying bacteria and by the symbiont (rhizobium) within the nodules. However, there are limited data concerning N2O production and consumption by endosymbiotic bacteria associated with legume crops. It has been reported that the alfalfa endosymbiont Ensifer meliloti strain 1021, despite possessing and expressing the complete set of denitrification enzymes, is unable to grow via nitrate respiration under anoxic conditions. In the present study, we have demonstrated by using a robotized incubation system that this bacterium is able to grow through anaerobic respiration of N2O to N2. N2O reductase (N2OR) activity was not dependent on the presence of nitrogen oxyanions or NO, thus the expression could be induced by oxygen depletion alone. When incubated at pH 6, E. meliloti was unable to reduce N2O, corroborating previous observations found in both, extracted soil bacteria and Paracoccus denitrificans pure cultures, where expression of functional N2O reductase is difficult at low pH. Furthermore, the presence in the medium of highly reduced C-substrates, such as butyrate, negatively affected N2OR activity. The emission of N2O from soils can be lowered if legumes plants are inoculated with rhizobial strains overexpressing N2O reductase. This study demonstrates that strains like E. meliloti 1021, which do not produce N2O but are able to reduce the N2O emitted by other organisms, could act as even better N2O sinks.
Keywords: denitrification, dinitrogen, greenhouse gas, nitric oxide, nitrous oxide reductase
Introduction
The presence of N2O in the atmosphere has been known since 1939 (Adel, 1939). However, its importance to the global environment was not recognized until the early 1970s when scientists hypothesized that N2O released into the atmosphere could activate reactions in the stratosphere that contribute to the depletion of the ozone layer (Crutzen, 1974). The fourth assessment report of the intergovernmental Panel on Climate Change (IPCC, 2007) estimated N2O emissions from both natural and anthropogenic sources to be 8.5–27.7 Tg N2O/year. The terrestrial ecosystems are the main source of N2O, accounting about 65% of total emissions. Agricultural activities are the major sources of N2O emissions, accounting for 60–80% of the anthropogenic N2O sources, mostly as N inputs to agricultural soils (Smith, 2008; Smith et al., 2012). These N2O emissions are likely to increase with the predicted expansion in the use of nitrogenous fertilizers in order to satisfy the escalating demand for food of the growing world population.
A variety of biological pathways are involved in N2O emissions from soils, and it has been estimated that >65% of the atmospheric N2O derives from microbial N transformations, mainly through the processes nitrification and denitrification (Thomson et al., 2012). Of these, denitrification is generally considered to be the largest source of N2O and, depending on the type of microorganisms involved and the environmental conditions, this process can serve not only as source but also as sink for N2O (Thomson et al., 2012). Denitrification is the respiratory reduction of nitrogen oxides (NOx) which enables facultative aerobic bacteria to survive and multiply under oxygen-limiting conditions. During this process nitrate (NO−3) is converted into molecular nitrogen (N2) via nitrite (NO−2) and the gaseous intermediates nitric oxide (NO) and nitrous oxide (N2O) (Zumft, 1997).
In contrast to the variety of N2O sources in soils, removal of N2O is only achieved by the last step of the denitrification process which is catalyzed by the N2O reductase (N2OR) enzyme encoded by the nosZ gene. Recent reports have demonstrated that diverse microbial taxa possess divergent nos clusters with genes that are related yet evolutionarily distinct from the typical nos genes of denitrifiers (Sanford et al., 2012). In fact, phylogenetic analyses of the nosZ gene revealed two distinct clades of nosZ differing in their signal peptides, indicating differences in the translocation pathway to the N2OR across the membrane (Jones et al., 2013). The expression and activity of N2OR is a natural target in the search for options to mitigate N2O emission from agricultural soils (Richardson et al., 2009). A promising mitigation strategy suggested recently is to stimulate N2O reductase by sustaining a high soil pH (Bakken et al., 2012). The latter is motivated by recent demonstrations that reduction of N2O is severely inhibited by suboptimal pH in the model organism Paracoccus denitrificans (Bergaust et al., 2010), in bacterial communities extracted from soils (Liu et al., 2014), and in intact soils (Raut et al., 2012; Qu et al., 2014). Another interesting option would be to alter the composition of the denitrifying community of soils, the objective being to enhance the growth of organisms with high N2O reductase activity. This would be a daunting task if the free-living soil bacteria were the target, but plant-associated bacteria appear more promising.
Rhizobia is a general term that describes bacteria that have the ability to establish N2-fixing symbiosis in legume roots or on the stems of some aquatic leguminous plants. In addition to fixing N2, many rhizobial strains have genes for enzymes of some or all of the four reductase reactions for denitrification. Several studies have reported that legume crops induce N2O emission by providing N-rich residues for decomposition (Baggs et al., 2000). In addition to soil denitrifiers, endosymbiotic bacteria may be partly responsible for this legume-induced N2O emission, since most rhizobia are able to denitrify under free-living and under symbiotic conditions (Bedmar et al., 2005; Delgado et al., 2007; Sanchez et al., 2011). Increased N2O emissions due to degradation of nodules were reported in soybean ecosystems (Inaba et al., 2012). Based on this, Itakura et al. (2013) hypothesized and proved that N2O emission from soil could be reduced by inoculating soybean plants with a nosZ-overexpressing strain of Bradyrhizobium japonicum. This suggests that root nodules of leguminous plants are net sources or sinks for N2O. Thus, the investigation of denitrification among rhizobia may provide novel options for reducing N2O emissions from soils.
Ensifer (formerly Sinorhizobium) meliloti 1021 is a key model organism for studying the symbiotic interaction between rhizobia and plants of the genera Medicago, Melilotus, and Trigonella, that has also been extensively used in previous works to better understand the regulation and symbiotic characterisation of E. meliloti denitrification genes (Bobik et al., 2006; Meilhoc et al., 2010; Horchani et al., 2011). In fact, analysis of the Ensifer meliloti 1021 genome sequence revealed the presence of the napEFDABC, nirK, norECBQD, and nosRZDFYLX denitrification genes encoding a periplasmic nitrate reductase, a copper-containing nitrite reductase, a c-type nitric oxide reductase and a nitrous oxide reductase enzyme, respectively. The involvement of the E. meliloti napA, nirK, norC, and nosZ structural genes in nitrate respiration and in the expression of denitrification enzymes under specific growth conditions (initial oxygen concentrations of 2% and initial cell density of 0.2–0.25) was also demonstrated (Torres et al., 2014). However, this strain has for a long time been considered a partial denitrifier due to its apparent inability to grow under anaerobic conditions with nitrate or nitrite as final electron acceptors (Garcia-Plazaola et al., 1993; Torres et al., 2011a). In order to better understand the truncated denitrification phenotype of E. meliloti 1021, an accurate estimation of the efficiency of the denitrifying process is required. For that purpose, in this work we have used a robotized system which allowed us to simultaneously monitor the O2 consumption, as well as the consumption and production of each NOx during the transition from oxic to anoxic respiration.
The results convincingly demonstrated that this strain (1021) was unable to reduce NO−3 or NO−2 to N2O or N2. In contrast, this bacterium was capable to reduce externally supplied N2O to N2, serving as a terminal electron acceptor during anoxic respiration. Thus, our study expands the current understanding of anaerobic respiration in rhizobia and explores the effect of pH, NOx and type of carbon source on N2O reduction in E. meliloti.
Materials and methods
Bacterial strains, and growth conditions in batch cultures
Ensifer meliloti 1021 (Smr, Meade et al., 1982), and napA (napA::mini-Tn5 Smr, Kmr, Pobigaylo et al., 2006) and nirK (nirK::mini-Tn5 Smr, Kmr, Pobigaylo et al., 2006) mutant strains were used in this study. E. meliloti strains were grown aerobically in 120 mL serum vials containing a triangular magnetic stirring bar and 50 mL of Triptone Yeast (TY) complete medium (Beringer, 1974) at 30°C. All cultures were continuously stirred at 700 rpm to avoid aggregation and ensure complete dispersal of cells. These cultures were then used as inocula into vials containing minimal defined medium (Robertsen et al., 1981) supplemented with or without 10 mM of KNO3 or 5 mM of NaNO2. The influence of carbon susbtrates on N2O uptake capacity was analyzed in minimal medium where the carbon substrate was replaced with either 5 mM of succinate or 5 mM of butyrate as oxidized or reduced carbon sources, respectively. The effect of pH on N2O uptake capacity was also studied in minimal medium strongly buffered (50 mM phosphate buffer) at pHs 6, 7, and 8. In all the treatments the headspace was filled with an initial concentration of O2 of 1 or 2% (12 or 24 μM dissolved O2 at 30°C, respectively). The headspace of experimental vials used to study the N2O reduction capacity was additionally supplied with an initial concentration of N2O of 2% (0.42 mM) or 5% (1.2 mM). To avoid possible external contaminations, antibiotics were added to the cultures at the following concentrations (μg mL−1); streptomycin, 200; kanamycin, 200.
Preparation of incubation vials
120 mL vials containing 50 mL liquid medium were crimp-sealed with rubber septa (Matriks AS, Norway) and aluminum caps to ensure an airtight system. Oxygen from vials was removed by 6 cycles of air evacuation during 360 s and helium (He) filling during 40 s. Constant stirring (400 rpm) was kept to ensure optimal gas exchange between liquid and headspace. Then, vials were injected with the required concentrations of O2 and N2O.
Gas measurements
After inoculation, cultures, blanks, and gas standards were placed in a thermostatic water incubator containing a serial magnetic stirrer at 30°C, with continuous stirring at 700 rpm, and the gas kinetics were monitored in each vial (2 to 3 h intervals). The gas measurements were performed by monitoring the headspace-concentrations of relevant gases (O2, CO2, NO, N2O, and N2) by repeated gas sampling through the rubber septa of the incubation vials as described by Molstad et al. (2007). The gas samples were drawn by a peristaltic pump coupled to an autosampler (Agilent GC Sampler 80), and with each sampling an equal volume of He was pumped back into the vials. This secured that the gas pressure was sustained near 1 atm despite repeated sampling, but diluted the headspace atmosphere (with He). This dilution was taken into account when calculating rates of production/consumption for each time increment (Molstad et al., 2007). The sampling system was coupled to a gas chromatograph (GC) (Agilent GC -7890A) with two 30 m × 0.53 mm id columns: a Porous Layer Open Tubular (PLOT) column for separation of CH4, CO2 and N2O, and a Molsieve column for separation of O2 and N2 (and Ar, Ne). The GC had three detectors: a flame ionization detector (FID), a thermal conductivity detector (TCD), and an electron capture detector (ECD). N2O was detected by both the ECD and TCD, thus securing accurate measurements at near-ambient concentrations (ECD, linear range 0–4 or 0–20 ppmv, depending on detector temperature) and linear response for higher concentrations (TCD). NO concentrations were determined by a Chemoluminiscence NOx analyser (Model 200A, Advanced Pollution Instrumentation, San Diego, USA).
OD600, nitrate and nitrite measurements
Cell densities (OD600), nitrate and nitrite concentrations were measured for each sample. Samples were taken from the liquid phase of the vials throughout the experiment to measure OD600 (0.7 mL sample), NO−3(0.1 mL sample), and NO−2 (0.1 mL sample) using sterile syringes. For determination of NO−3, a 10 μL aliquot was injected into a purge vessel with heating jacket and condenser (ASM 03292) containing 1 M HCl and vanadium (III) chloride. Temperature of vessel was controlled by a circulating water bath at 95°C and cold water for the condenser. In addition, a gas bubble/NaOH trap with Teflon sleeve (ASM 04000) was used to avoid the corrosive effects of HCl. Vanadium (III)/HCl converts nitrite and S-nitrosocompounds to NO, which is transported (by N2) to a chemiluminescence detector Nitric Oxide Analyzer NOA 280i (General Electric). N2 was continuously bubbled through the reducing agent to maintain an anaerobic environment in the system and to transport the NO through the NO analyzer (Walters et al., 1987). The approximate detection limit was 1 pmol NO, equivalent to 0.1 μM (when injecting 10 μL). For determination of NO−2, a 10 μL subsample was injected into a purge vessel (gas bubble/NaOH trap is not needed) containing acetic acid with 1% vol NaI where NO−2 is converted to NO.
Analyses of kinetics of aerobic and anoxic NO−3, NO−2, or N2O respiration
Experimental dataset obtained from the series of incubations were used to determine the kinetics of O2, NO−3 NO−2, or N2O respiration and NO, N2O, and N2 production in order to provide the most accurate information on E. meliloti physiology during the transition from aerobic to NOx anoxic respiration. O2 and NO concentration in the liquid, determined as μM and nM, respectively, was estimated taking into account the partial pressure of these gases at headspace, their solubilities and transport coefficients between headspace and liquid. Additionally, O2 concentration in liquid was estimated respective the O2 respiration rate for each time increment (see Molstad et al., 2007 for details). N2O was analyzed as μmol N2O vial−1, whereas N2 was determined as cumulative net production of N2. All data were corrected for dilution rates and losses by gas sampling, and leaks due to gas diffusion through the rubber septa. The concentrations of NO−3 and NO−2 were determined at different times compared to the gas sampling. However, we needed values for NO−2 concentrations at the same time as the gas sampling in order to estimate electron flow rates. For this reason, polynomial functions [f(t)] were fitted to the measured NO−3 and NO−2 concentrations, and used to estimate NO−2 concentration at the time of gas samplings. Graphical presentations for NO−3 and NO−2 concentrations include both measured data points and the polynomial function.
The apparent growth rates based on O2 consumption (μox), and reduction of any NOx during the anoxic phase (μanox) were estimated by regression [ln (Ve−) against time] for the phases with exponentially increasing rates. Yield (cells pmol−1 e−) calculation was based on the number of cells rendered per pmol electron used by the respiratory terminal oxidases to reduce O2 to H2O during oxic phase (Yieldox) or by the complete set of denitrifying reductases to reduce NO−3 NO−2 or N2O to N2 during anoxic phase (Yieldanox). Vmax is an useful parameter that can tell us the efficiency for O2 and NOx respiration per cell. It estimates the maximal velocities per cell and per hour for the reduction of O2 and NOx. This parameter is based on the fmol of electrons used by the terminal oxidases and denitrifying enzymes to reduce O2 or NOx, respectively, per cell and per hour. For further details regarding these calculations, see Molstad et al. (2007) and Nadeem et al. (2013).
Results
Kinetics of aerobic respiration
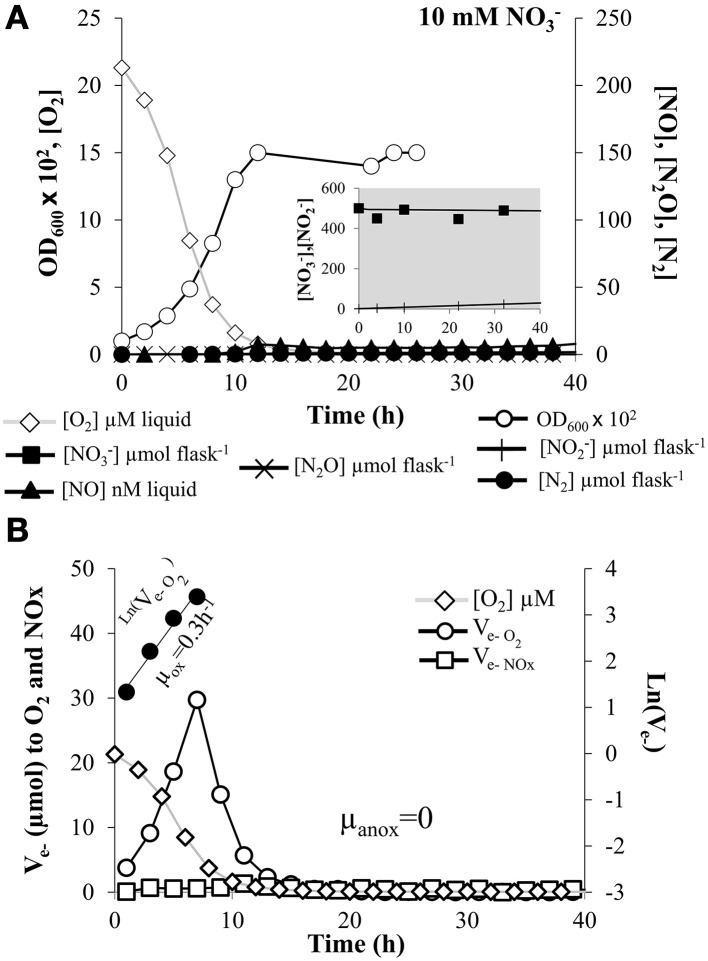
E. meliloti strain 1021 was grown aerobically for 30 h with vigorous stirring (700 rpm) until a maximal optical density at 600 nm (OD600) of ~0.3 to avoid generation of localized anoxic conditions due to cell aggregation. Then, an aliquot was used to inoculate the culture vials to an initial OD600 of 0.01 (8 × 106 cells mL−1). The medium contained either 10 mM of nitrate (Figure 1), 5 mM NO−2 (Figure 2) or 10 mM nitrate plus 5% N2O (1.2 mM N2O concentration in the liquid when in equilibrium with the headspace) (Figure 3). In all the treatments for studying the kinetics of aerobic respiration, the initial O2 concentration in the headspace was 2%. Figure 1A shows the measured OD600, O2, NO, N2O, and N2 concentrations in the medium for a single vial throughout the 40 h incubation in the presence of nitrate. NO−3 depletion and production of NO−2 is also shown (Figure 1A, insert). In nitrate-treated cells, oxygen was consumed within the first 15 h, OD600 increased linearly with the cumulative O2 consumption (r2) = 0.9877, and remained practically constant throughout the anoxic phase. Rates of O2 consumption for each time increment between two samplings were used to calculate electron (e−) flow rates to oxygen (Ve−O2). As shown in Figure 1B, Ve−O2 increased exponentially throughout the first 7 h in proportion with the increase in OD600 (r2 = 0.9105), and declined gradually in response to diminishing O2 concentrations. The initial exponential increase in electron flow during oxic respiration can be taken as an indirect measure of growth rate (μox) (Liu et al., 2013). Thus, the apparent μox estimated by linear regression of ln (Ve−O2) against time was 0.30 (±0.03) h−1 (Figure 1B, Table 1A). The final OD600 was 0.15 (±0.02) (1.60 × 108 cells mL−1, Table 1B) resulting in a yield of 24.6 (±2.8) cells pmol−1e− to O2 (Table 1A). The apparent maximum specific respiration rate, Vmax, which is a useful indicator of the respiration per cell, was 11.6 (±0.5) fmol e− cell−1 h−1 for oxygen respiration in cells grown in the presence of nitrate (Table 1A).
Figure 1.
Kinetics of O2 and NO−3 respiration. (A) Absorbance at 600 nm (OD600), O2 consumption, NO−3 depletion (insert), and production of NO−2 (insert), NO, N2O, and N2 by E. meliloti 1021 when incubated in the presence of 10 mM NO−3 in the medium and an initial O2 concentration of 2% in the headspace. (B) The electron flow rate to O2 is shown as log-transformed values for the phases with exponential increase (filled circle symbols). The slopes estimating apparent growth rates were 0.3 (± 0.03) h−1 and 0 for oxic and anoxic phase, respectively. Cultures with an initial OD600 of 0.01 were vigorously stirred at 700 rpm. The result shown is for a single vial. Several replicates were analyzed, with similar results, although the exact timing of events was not the same. However, the consistency of the observations is demonstrated in Table 1 where averages of at least three different cultures are reported.
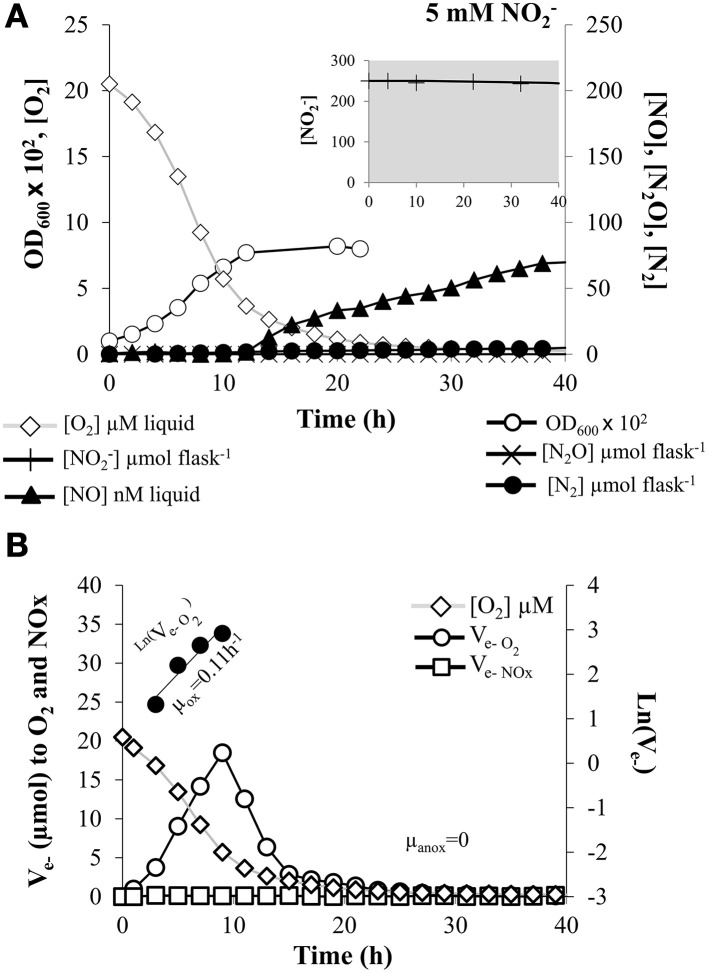
Figure 2.
Kinetics of O2 and NO−2 respiration. (A) Absorbance at 600 nm (OD600), O2 consumption, NO−2 depletion (insert), and production of NO, N2O, and N2 by E. meliloti 1021 when incubated in the presence of 5 mM NO−2 in the medium and an initial O2 concentration of2% in the headspace. (B) The electron flow to O2 shown as log-transformed values for the phases with exponential increase (filled circle symbols). The slopes estimating apparent growth rates were 0.11 (±0.02) h−1 and 0 for oxic and anoxic phase, respectively. Cultures with an initial OD600 of 0.01 were vigorously stirred at 700 rpm. The result shown is for a single vial. Several replicates were analyzed, with similar results, although the exact timing of events was not the same. However, the consistency of the observations is demonstrated in Table 1 where averages of at least three different cultures are reported.
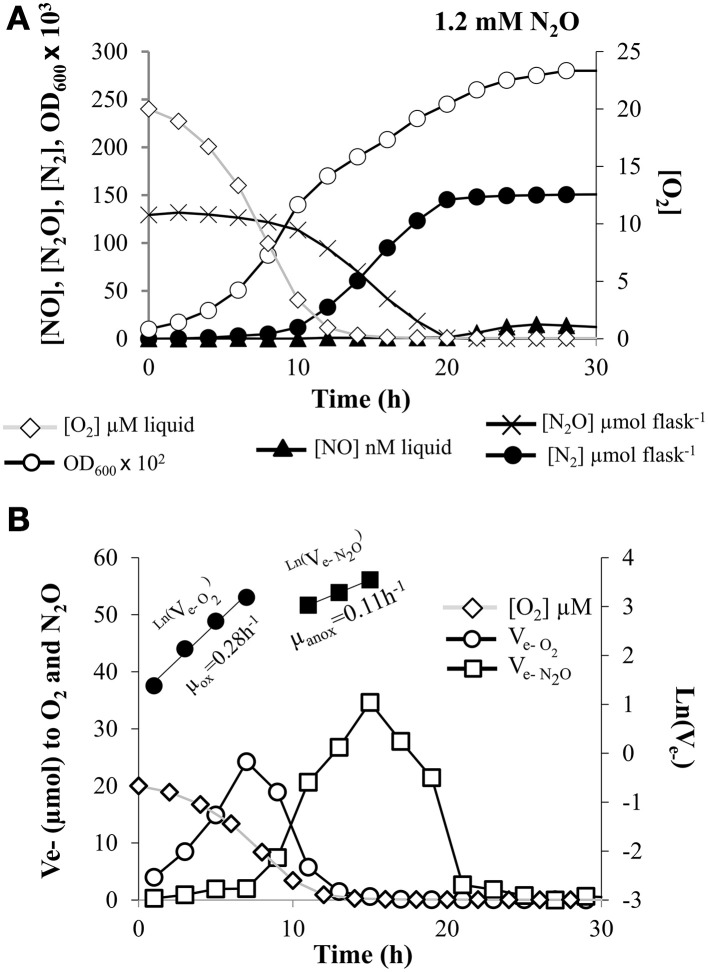
Figure 3.
Kinetics of O2 and N2O respiration. (A) Absorbance at 600 nm (OD600), O2 and N2O depletion, and production of NO, and N2 by E. meliloti 1021 when incubated in the presence of 10 mM NO−3 in the medium and an initial O2 concentration of 2% in the headspace plus 5% N2O. (B) The electron flow to O2 or to N2O is shown as log-transformed values for the oxic (filled circle symbols) and anoxic phase (filled square symbols) with exponential increase. The slopes estimating apparent growth rates were 0.28 (±0.03) h−1 for the oxic phase, and 0.11 (±0.03) h−1 for the anoxic phase, respectively. Cultures with an initial OD600 of 0.01 were vigorously stirred at 700 rpm. The result shown is for a single vial. Three replicates were analyzed in parallel with similar results. Consistency of the observations are demonstrated in Table 1 where averages of at least three different cultures are reported.
Table 1.
Summary of oxic and anoxic growth parameters (A)1 Depending on the presence of nitrogen oxides, and the subsequent conversion of the nitrogen oxides present (B)2.
| (A) | ||||||
|---|---|---|---|---|---|---|
| Growth parameters for the oxic phase | Growth parameters for the anoxic phase | |||||
| NOx present (mM) | μox (h-1) | Yieldox (cell pmol-1 e-) | Vmax (fmol e- cell-1 h-1) | μanox (h−1) | Yieldanox (cell pmol-1 e-) | Vmax (fmol e- cell-1 h-1) |
| NO−3(10) | 0.30(±0.03) a | 24.6(±2.8) a | 11.6(±0.5) a | 0 | 0 | 0 |
| NO−2(5) | 0.11(±0.02) b | 14.1(±1.1) b | 8.2(±0.7) b | 0 | 0 | 0 |
| N2O(1.2)3 | 0.28(±0.03)a | 23.1(±6.2) a | 8.9(±0.1) b | 0.11(±0.03) | 18(±0.6) | 5.7(±1.1) |
| (B) | ||||||
| NOx present (mM) | [O2] at onset of NOx-reduction (μMO2) | Max [NO−] in liquid (nM NO) | Final OD (OD600) | |||
| NO−3(10) | 2.7(±1.5) a | 12.4(±2.10) a | 0.90(±0.30) a | 0.15(±0.02) a | ||
| NO−2(5) | 3.0(±0.7) a | 94.2(±16.9) b | 0.18(±0.02) b | 0.08(±0.01) b | ||
| N2O(1.2)3 | 5.9(±2.6) b | 15.0(±1.10) a | 100(±2.50) c | 0.28(±0.05) c | ||
The alternative respiratory substrate (NOx) present in the medium (NO−3 or NO−2) or at headspace (N2O) for each analysis is indicated. All the experimental vials contained an initial O2 concentration of 2% at headspace. Data are means with standard error (in parenthesis) from at least three independent cultures. Values in a column followed by the same lower-case letter are not significantly different according to One-Way ANOVA and the Tukey HSD test at P ≤ 0.05.
1Apparent oxic growth (μox, h−1) and anoxic growth (μanox, h−1) rates based on O2 consumption during the oxic phase or reduction of NO−3, NO−2, or N2O during the anoxic phase. Yield (cells per mole electron) based on increase in OD vs. cumulated consumption of oxygen or reduction of NO−3, NO−2, or N2O, and apparent maximum specific respiration rate (Vmax, fmol electrons cell−1 h−1) during the initial phase (0–5 h) of the experiments (Figures 1, 2).
2The oxygen concentration at the time of the first indications of anoxic respiration (i.e., appearance of NO in the treatments with NO−3 and NO−2, and appearance of significant N2O reduction to N2 in the treatment with N2O).
35 % N2O (150 μmol N2O at 20°C) was injected into each vial, resulting in 1.1 mM N2O in the liquid when in equilibrium with the headspace.
O2 uptake and growth kinetics were also analyzed in cells grown in the presence of 5 mM NO−2 as final electron acceptor (Figure 2). For this treatment, O2 was consumed within the first 30 h of incubation showing a delay in comparison to NO−3 treatment (Figure 2A). As observed in nitrate-treated cells, OD600 also increased during the oxic phase in proportion with O2 consumption, and remained constant during the anoxic phase. The estimated oxic growth rate in the presence of nitrite (linear regression of ln(Ve−O2) against time was μox = 0.11 (±0.02)h−1 (Figure 2B, Table 1A) and the estimated cell yield was only 14.1 (±1.1) cells pmol−1 e− (Table 1A). The estimated Vmax for oxygen respiration in cells grown in the presence of nitrite was 8.2 (±0.7) fmol e− cell−1 h−1 (Table 1A). Thus, the presence of NO−2 in the medium appeared to exert an inhibitory effect on the oxygen respiration by terminal respiratory oxidases, resulting in lower Vmax and cell yield per mol electron compared to cells grown in the presence of nitrate.
Finally, kinetics of O2 respiration were also analyzed when cells were incubated in vials containing minimal medium with 10 mM of NO−3, and an initial concentration of 5% N2O and 2% O2 in the headspace. Figure 3A shows the measured O2, NO, N2O, and N2 for a single vial throughout the 40 h incubation, as well as the OD600. In this case, oxygen was consumed within the first 15 h and the OD600 increased in proportion with the cumulative O2 consumption and continued increasing throughout the anoxic phase. Electron flow rate to O2 increased exponentially with an apparent growth rate (μox) = 0.28 (±0.03) h−1 (Figure 3B, Table 1A). Cell yield resulting from O2 respiration was very similar to that observed in nitrate-treated cells [23.1 (±6.2) cells pmol−1 e− with a Vmax of 8.9 (±0.13) fmol e− cell−1 h−1] (Table 1A).
Kinetics of NO−3 and NO−2 respiration
When cells were cultured with NO−3, there was a very low NO−3 consumption rate as well as very low progressive accumulation of NO−2 throughout the entire anoxic phase (Figure 1A, insert), reaching only ~50 μmol vial−1 (which accounts for 10% of the NO−3-N in the medium). Very low levels of NO were also observed (12.40 ± 2.10 nM) after 40 h incubation (Table 1B, Figure 1A). Production of N2O in the headspace was insignificant and the fraction of NO−3 reduced to N2 at the end of the incubation was also extremely low (0.9 ± 0.3 %) (Table 1B, Figure 1A). When NO−2 was used as final electron acceptor, the first detection of NO occurred as the oxygen concentration in the liquid reached ~3 μM (Figure 2A, Table 1B). During the subsequent anoxic phase, NO continued to accumulate, reaching 94.20 ±16.90 nM levels at the end of the incubation period (Table 1B, Figure 2A). Similarly as for nitrate-treated cells (Figure 1A), production of N2O was undetectable and the total, cumulative production of N2 from the initially provided NO−2-N was also very low (0.18 ± 0.02 %) (Figure 2A, Table 1B). These data show that E. meliloti 1021 was clearly unable to shift effectively to NO−3 or NO−2 based anaerobic respiration. This inability was also confirmed by the lack of increase in measured OD600 throughout the anoxic phase (Figures 1, 2). Thus, the apparent growth rate during either NO−3 or NO−2 anoxic respiration (μanox) was zero (Figures 1B, 2B, Table 1A). Similar growth rates were observed by using 1 mM or 500 μM NO−2 as electron acceptor (data not shown). One possible explanation to the lack of efficient reduction of NO−3 and NO−2 could be that rapid depletion of the oxygen in these cultures may have resulted in entrapment of the bacteria in anoxia, as shown previously for P. denitrificans by Bergaust et al. (2010). To test this hypothesis, we performed a follow-up experiment where the stirring speed was reduced from 700 rpm (used in the experiments reported in Figures 1, 2) to 200 rpm, in order to secure a slow transition from oxic to anoxic conditions in the liquid. These cultures showed the same lack of effective transition to denitrification as cultures with vigorous stirring, despite the fact that the cells with low stirring experienced a progressive O2 limitation during 50 h prior to complete O2 depletion (see Supplementary Figure S1).
Kinetics of N2O respiration
The capacity of E. meliloti 1021 to reduce N2O was examined in vials containing 10 mM NO−3 in the medium plus 5% N2O and 2% O2 initially added to the headspace (Figure 3). As shown in Figure 3A, N2O was consumed rapidly and N2 production followed stoichiometrically the reduction of N2O to its complete depletion (100% of N2O was converted to N2 gas) (Figure 3A, Table 1B). As shown in Figure 3A, N2O reduction was at first detected at an O2 concentration of 5.9 (±2.6) μM (Table 1B). Traces of NO from NO−3 reduction were also detected (15 ±1.1 nM in the liquid; Table 1B). Final OD600 of cells incubated with N2O was clearly higher than that obtained when cells were incubated only with NO−3 or NO−2 as alternative electron acceptors (Table 1B), demonstrating the capacity of E. meliloti to couple N2O reduction with growth.
Electron flow to N2O increased with an apparent growth rate (μanox) of 0.11 (±0.03) h−1 estimated by linear regression of ln (Ve−N2O)against time (Figure 3B, Table 1A). Although low rates of electron flow to N2O occurred after 3 h, it increased sharply after 7 h as the electron flow to oxygen decreased due to oxygen depletion. Thus, the cells were evidently able to shift gradually from respiring O2 to N2O, preserving the total electron flow rate essentially unaffected after the depletion of oxygen. As shown in Table 1A, the estimated cell yield from N2O reduction was 18 (±0.6) cells pmole−1 e−. Knowing the yield in cell number per hour and the electron flow rate per hour we could estimate the Vmax for N2O reduction to 5.7 (±1.1) fmol e− cell−1 h−1 (Table 1A).
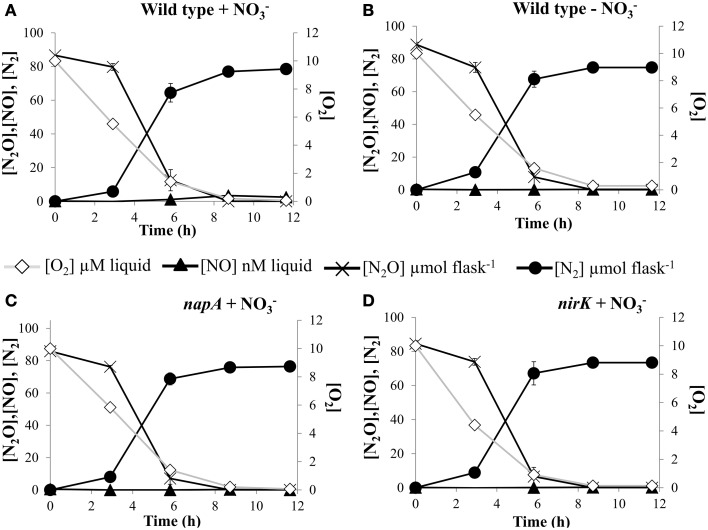
Nox molecules do not trigger N2OR activity in E. meliloti
To evaluate the effect of NOx molecules as inducers of N2OR activity, we measured N2O uptake rates in cultures of E. meliloti 1021 strain that had received 10 mM NO−3 in the medium and compared this with cultures that were not supplemented with NO−3 (Figures 4A,B). The results showed similar N2O consumption as well as N2 production rates for the two treatments. Furthermore, no differences in N2O respiration was found between wild-type cells and strains which were defective in the napA and nirK structural genes when cultured in a medium amended with 10 mM NO−3 (Figures 4A,C,D). The E. meliloti napA or nirK mutants were demonstrated previously to be unable to reduce nitrate and nitrite respectively, to any further NOx intermediary of the denitrification process (Torres et al., 2014). These results suggested that the ability to reduce N2O was not affected by the presence or absence of NO, NO−2, or NO−3.
Figure 4.
NOx effect on kinetics of O2 and N2O depletion. O2 and N2O consumption, and NO and N2 production by E. meliloti 1021 (A,B), and napA (C) and nirK (D) mutant strains when incubated in the presence of 1% O2 plus 2% N2O in the headspace. Cells were incubated in minimal medium with (A,C,D) or without (B) 10 mM NO−3. Cultures with an initial OD600of 0.01 were vigorously stirred at 700 rpm. Plotted values are average of three replicate flasks for each treatment, with standard deviation (SD) as vertical bars (n = 3).
Low pH severely impaires N2O uptake in E. meliloti
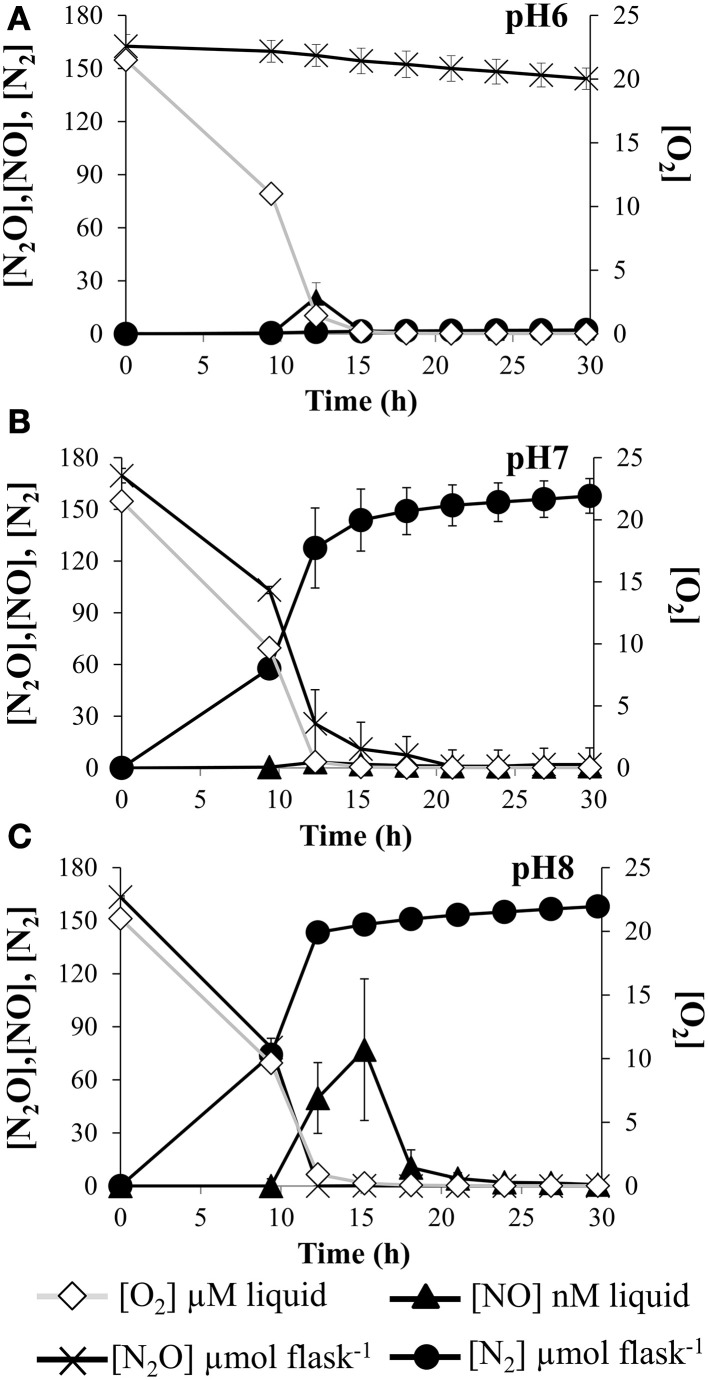
Since pH emerges as a master variable controlling the expression of N2O reductase, in this work we examined the pH effect on the kinetics of N2O reduction. For that purpose, E. meliloti cells were incubated in minimal medium strongly buffered with phosphate buffer, at pH 6, 7, and 8. Firstly, we grew E. meliloti 1021 cells aerobically to exponential (log) phase at pH 7. Then cells were transferred to the experimental vials containing 5% N2O and 2% O2 in the headspace and 10 mM NO−3 in the medium. Rates of O2 consumption were monitored until depletion and no differences were found between treatments. However, N2O reduction to N2 was completely blocked at pH 6 (Figure 5A). Surprisingly, when cells were incubated at pH 8, a significant peak of NO was detected. A negative effect of high pHs on nor expression or Nor activity could explain that transient peak of NO.
Figure 5.
pH effect on kinetics of O2 and N2O depletion. O2 and N2O consumption, and NO and N2production by E. meliloti 1021 when incubated in the presence of 10 mM NO−3 in minimal medium at pH 6 (A), pH 7 (B) and 8 (C), and an initial O2 concentration of 02% in the headspace plus 5% N2O. Cultures with an initial OD600of 0.01 were vigorously stirred at 700 rpm. Plotted values are average of three replicate flasks for each treatment, with standard deviation (SD) as vertical bars (n = 3). The decline in N2O concentration at pH = 6 is due to sampling loss, not biological reduction of N2O to N2.
Reduced C-sources attenuates N2O uptake in E. meliloti
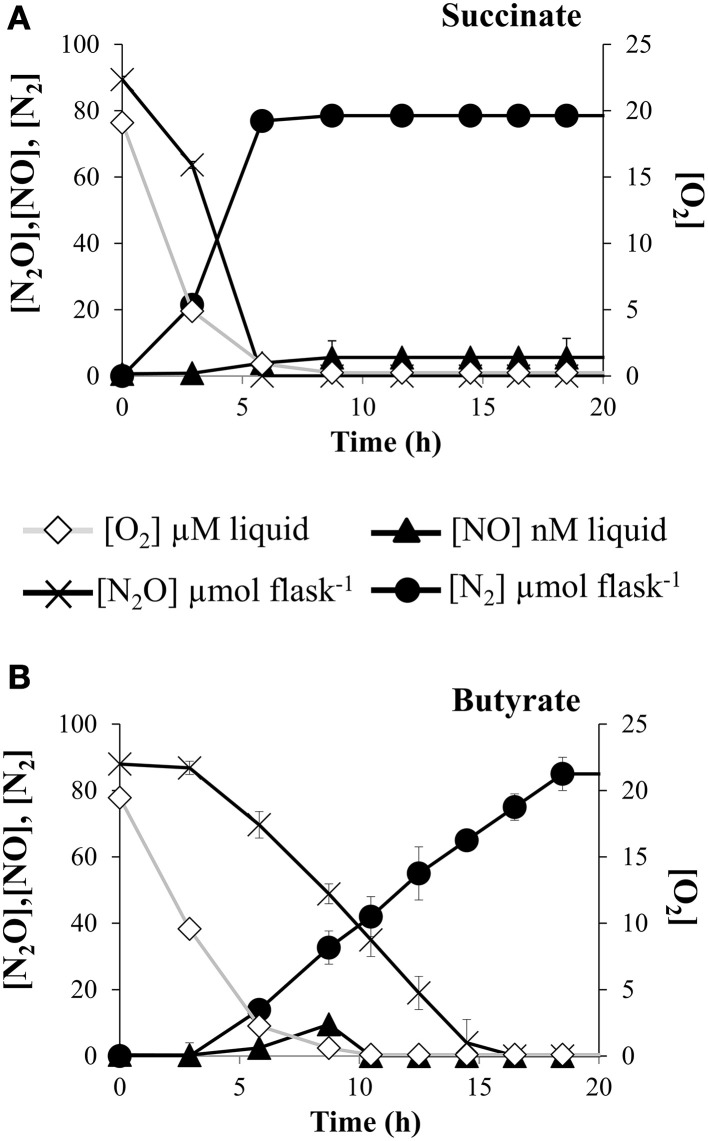
Carbon availability is another key environmental factor affecting N2O production in the field. However, information about the implication of specific forms of reductants in N2O reductase activity is limited. Redox state of the C-sources might influence the amount of electrons available to reduce N2O to N2. For that reason, we tested the capacity of E. meliloti 1021 to reduce N2O in the presence of C-substrates with different redox potential, from highly oxidized as succinate or highly reduced such as butyrate. Aerobically raised cells were collected and inoculated into experimental vials containing minimal medium where glycerol was substituted by either succinate or butyrate. By using the robotized incubation system, rates of O2 respiration occurring previously to N2O consumption were also estimated. We found that O2 respiration from cells incubated in the presence of butyrate was slightly decreased when compared to cells incubated in the presence of succinate (Figures 6A,B). However, rates of N2O consumption were largely dependent on the oxidized or reduced nature of the carbon source. Thus, when butyrate was used as electron donor, the N2O reduction to N2 decreased about 3-fold compared to when succinate was used as the sole carbon substrate (Figures 6A,B).
Figure 6.
C-source effect on kinetics of O2 and N2O depletion. O2 and N2O consumption, and NO and N2 production by E. meliloti 1021 when incubated in the presence of 1% O2 plus 2 % N2O in the headspace, and 10 mM NO−3 in Robertsen's medium which contained succinate (A) or butyrate (B) as carbon sources. Cultures with an initial OD600 of 0.01 were vigorously stirred at 700 rpm. Plotted values are averages of three replicate flasks for each treatment, with standard deviation (SD) as vertical bars (n = 3).
Discussion
In this work, we have used a robotized incubation system designed to simultaneously monitor with high sensitivity real-time changes in concentrations of O2, NO−3, NO−2 NO, N2O, and N2 during the transition from oxic to anoxic respiration. By using this system, we found that E. meliloti 1021 is unable to reduce NO−3 or NO−2 to N2O or N2 during the transition from oxic to anoxic conditions. Consequently, this bacterium was unable to sustain growth during anoxic conditions by using NO−3 or NO−2 as electron acceptors. This is in contrast to recent studies where growth of E. meliloti 1021 was observed during respiration of NO−3 as well as NO−2 (Torres et al., 2011a, 2014). This apparent discrepancy could be due to the different growth conditions and methodological approaches used by Torres et al. (2011a, 2014) and in this work. While they inoculated experimental vials with very high cell density (OD600~ 0.2–0.25), which were shaken at 170 rpm, the initial cell density used in the present work was significantly lower (OD600~ 0.01), and cultures were stirred at 700 rpm. The reason why we used different conditions in this work is to allow an efficient and controlled gas transfer from the headspace to the liquid and prevented cell aggregation and generation of localized micro-oxic spells during the aerobic phase previous to the transition to anaerobic respiration, as well as accumulation of toxic concentration of metabolites resulting from cell respiration. It might be possible that the growth conditions used by Torres et al. (2011a, 2014) provoked generation of anoxic micro-zones preceding total oxygen depletion due to cell aggregation and consequently the induction of E. meliloti 1021 denitrifying machinery would be facilitated. The present work extends the study of denitrification in E. meliloti by performing an estimation of the growth parameters (i.e., μ, yield, Vmax), as well as a precise quantification of NOx gases dynamics during the transition from oxic to anoxic respiration. This approach, never used in rhizobia, allowed us to perform an accurate estimation of the efficiency of the denitrifying process, and is regarded to be more physiologically relevant than previously conducted growth experiments.
When N2O was provided as an alternative electron acceptor, anaerobic respiration, and growth was sustained by reducing N2O to N2. In this context, a recent report showed the ability of B. japonicum USDA110 to grow anaerobically using exogenous N2O as the sole electron acceptor (Sanchez et al., 2013). Growth with N2O as electron acceptor has also been observed in Anaeromyxobacter (Sanford et al., 2012), and in Wolinella, Campylobacter, and Geobacillus (Liu et al., 2008; Kern and Simon, 2009) indicating that the atypical nosZ encodes a functional respiratory terminal N2O reductase in those bacteria. This is unlike Pseudomonas aeruginosa PAO1, which cannot grow on exogenous N2O as the only electron acceptor (Bryan et al., 1985; Zumft and Kroneck, 2007).
It is generally considered that low oxygen concentration is a requirement for expression of the denitrification machinery (van Spanning et al., 2007). Especially the N2OR has been considered as a very O2 labile reductase which is inactivated by the presence of low amounts of O2 (Alefounder and Ferguson, 1982; Coyle et al., 1985; Snyder and Hollocher, 1987). In contrast to these observations, our results suggest that expression of N2OR in E. meliloti might be subjected to a different regulation, in which N2O reduction occurs even in the presence of oxygen concentrations above 8 μM (Figure 3A).
It has been reported that expression and fine-tuning of the denitrification system also requires the presence of key molecules such as NO−3, NO−2, and NO which, through transcriptional factors and their protein-coupled sensory receptors, act as signals that trigger induction of the denitrification pathway (Zumft and Kroneck, 2007; Spiro, 2012). Our results suggested that oxygen limitation was the only prerequisite for maximal expression of N2OR in E. meliloti, although we cannot exclude that N2O is also necessary. The presence of a NOx (NO, NO−2, NO−3) was however not required, since N2OR activity remained at similar levels in the absence or in the presence of NO−3 in wild-type cells. Furthermore, in cells cultured with NO−3, no differences in N2OR activity were observed between wild-type, and the napA or nirK mutant strains where the reduction of NO−3 or NO−2 is blocked, respectively. In fact, previous studies of gene expression proposed that limited oxygen tension alone resulted in induction of the expression of the whole nos operon in E. meliloti (Bobik et al., 2006). In contrast to these findings, transcriptional profile analysis suggested that induction of nosR and nosZ gene expression also requires the presence of nitric oxide (Meilhoc et al., 2010). In line with this, recent studies using qRT-PCR showed that maximal transcription of the E. meliloti nosZ gene occurred when cells were subjected to anoxic conditions in the presence of nitrate (Torres et al., 2014). Similarly to our observations, it was recently reported that P. denitrificans is fully able to reduce N2O in the absence of oxyanions and NO (Bergaust et al., 2012). In contrast, it was proposed that the inability of Pseudomonas aeruginosa PAO1 and Bacillus vireti to grow on exogenous N2O as the only electron acceptor was because these organisms need NO as an inducer of nosZ transcription (Arai et al., 2003).
Our results clearly showed that E. meliloti 1021 was unable to express N2OR activity at pH 6. This difficulty in expressing N2OR at low pH was observed in P. denitrificans (Bergaust et al., 2010) and in suspensions of extracted soil bacteria (Liu et al., 2014). The phenomenon is ecologically important since there is ample evidence that low soil pH results in high N2O/N2 product ratios of denitrification (Raut et al., 2012; Qu et al., 2014).
Among the environmental factors that influence N2O emissions, and specifically the bacterial N2OR performance, very little is known about the mode in which availability and redox state of C-sources contribute. In this work, the observed attenuated N2OR activity in the presence of highly reduced C-sources could be attributed to a reduced capacity of cells to metabolize more complex C-substrates such as butyrate, causing a lowered electron flow through the respiratory chain, resulting in a reduced electron availability to reduce N2O to N2 by the N2OR (Morley and Baggs, 2011). Alternatively, a reduced efficiency to metabolize butyrate could be due to the fact that its uptake into cell probably requires active transport, and consequently cells may be subjected to periods of reduced N2OR activity (Schalk-otte et al., 2000). Supporting this hypothesis, it was found that N2OR activity was stimulated in the presence of artificial root exudates with easily metabolized C-sources such as glucose, as well as in soils amended with carbohydrates as glucose and starch (Murray et al., 2004; Henry et al., 2008). In addition, a regulatory control on nos transcription could also explain the dependence of the N2OR activity on the redox state of C-sources. In accordance with this, it was recently reported that expression levels of the B. japonicum NorC component of the nitric oxide reductase in wild-type cells, incubated in minimal medium with succinate as the sole C-source, were significantly higher than those observed in cells incubated in the presence of butyrate (Torres et al., 2011b). Similarly, expression of the B. japonicum fixNOQP genes, encoding the high affinity terminal oxidase cbb3, decreased when butyrate was the sole carbon source compared to when malate was used (Bueno et al., 2009).
Taken together, these results showed a novel denitrifying phenotype in E. meliloti 1021, for which the reduction of NO−3, or NO−2 was severely impaired, while N2O was actively reduced. We further demonstrated that the reduction of N2O sustained growth by E. meliloti 1021. To our knowledge this is the first time that it was demonstrated the capacity of E. meliloti to sustain anoxic respiration by using N2O as terminal electron acceptor. Since the effect of pH or C-sources on N2O reductase activity has never been examined in rhizobia, the relevance of this study is to demonstrate that both environmental factors affect N2O reductase activity in the model alfalfa endosymbiont, E. meliloti 1021. Although this strain is a model organism and is not commercially used as inoculant for alfalfa, the results obtained here could be expanded to more competitive and efficient N2-fixers inoculants in order to develop strategies to reduce N2O emissions from alfalfa crops. In fact, despite the large research efforts invested in flux measurement of N2O emissions, progress in developing efficient mitigation options has hitherto been slow. An essential objective should be to understand the underlying mechanisms and factors that affect the regulation of N2O consumption and production, and consequently to improve the product stoichiometry of denitrification (N2O/N2O + N2) in terrestrial ecosystems.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Fondo Europeo de Desarrollo Regional (FEDER)-co-financed grants (AGL2010-18607 and AGL2013-45087-R) from the Ministerio de Economía y Competitividad (Spain). Grant AGR-1968 and support from the Junta de Andalucía to Group BIO-275 are also acknowledged. We thank A. Becker for providing the E. meliloti mutants. EB was supported by a Personal visiting researcher grant – IS-MOBIL (Oslo University, Norway) and from the Consejo Superior de Investigaciones Cientificas JAE-DOC Programme co-financed by European Social Fund (ESF).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00537/abstract
Kinetics of O2 depletion and N2O, NO, and N2 production. E. meliloti 1021 was incubated in the presence of 10 mM NO−3 in minimal medium and an initial O2 concentration of 2% in the headspace. Cultures with an initial OD600 of 0.01 were vigorously stirred at 200 rpm. Plotted values are averages of three replicate flasks for each treatment, with standard deviation (SD) as vertical bars (n = 3).
References
- Adel A. (1939). Note on the atmospheric oxides of nitrogen. Astrophys. J. 90, 627. 10.1086/14412916569421 [DOI] [Google Scholar]
- Alefounder P. R., Ferguson S. J. (1982). Electron transport-linked nitrous oxide synthesis and reduction by Paracoccus dentirificans monitored with an electrode. Biochem. Biophys. Res. Comm. 104, 1149–1155. 10.1016/0006-291X(82)91370-5 [DOI] [PubMed] [Google Scholar]
- Arai H., Mizutani M., Igarashi Y. (2003). Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149, 29–36. 10.1099/mic.0.25936-0 [DOI] [PubMed] [Google Scholar]
- Baggs E. M., Rees R. M., Smith K. A., Vinten A. J. A. (2000). Nitrous oxide emission from soils after incorporation of crop residues. Soil Use Mgmt. 16, 82–87. 10.1111/j.1475-2743.2000.tb00179.x [DOI] [Google Scholar]
- Bakken L. R., Bergaust L., Liu B., Frostegård Å. (2012). Regulation of denitrification at the cellular level a clue to understanding of N2O emissions from soils. Philos. Trans. R Soc. Lond. B Biol. Sci. 367, 1226–1234. 10.1098/rstb.2011.0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedmar E. J., Robles E. F., Delgado M. J. (2005). The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem. Soc. Trans. 33, 141–144. 10.1042/BST0330141 [DOI] [PubMed] [Google Scholar]
- Bergaust L., Mao Y., Bakken L. R., Frosteård Å. (2010). Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrogen oxide reductase in Paracoccus denitrificans. Appl. Environ. Microb. 76, 6387–6396. 10.1128/AEM.00608-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergaust L., van Spanning R., Frostegard Å., Bakken L. R. (2012). Expression of nitrous oxide reductase in Paracoccus denitrificans is regulated by oxygen and nitric oxide through FnrP and NNR. Microbiology 158, 826–834. 10.1099/mic.0.054148-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer J. E. (1974). R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188–198. 10.1099/00221287-84-1-188 [DOI] [PubMed] [Google Scholar]
- Bobik C., Meilhoc E., Batut J. (2006). FixJ: a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J. Bacteriol. 188, 4890–4902. 10.1128/JB.00251-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan B. A., Jeter R. M., Carlson C. A. (1985). Inability of Pseudomonas stutzeri denitrification mutants with the phenotype of Pseudomonas aeruginosa to grow in nitrous oxide. Appl. Environ. Microbiol. 50, 1301–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno E., Richardson D. J., Bedmar E. J., Delgado M. J. (2009). Expression of Bradyrhizobium japonicum cbb3 terminal oxidase under denitrifying conditions is subjected to redox control. FEMS Microbiol. Lett. 1, 20–28. 10.1111/j.1574-6968.2009.01711.x [DOI] [PubMed] [Google Scholar]
- Coyle C. L., Zumft W. G., Kroneck P. M. H., Körner H., Jakob W. (1985). Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina: purification and properties of a novel multicopper enzyme. Eur. J. Biochem. 153, 459–467. 10.1111/j.1432-1033.1985.tb09324.x [DOI] [PubMed] [Google Scholar]
- Crutzen P. J. (1974). Estimates of possible variations in total ozone due to natural causes and human activities. Ambio 3, 201–210. [Google Scholar]
- Delgado M. J., Casella S., Bedmar E. J. (2007). Denitrification in rhizobia-legume symbiosis, in Biology of the Nitrogen Cycle, eds Bothe H., Ferguson S. J., Newton W. E. (Amsterdam: Elservier Science; ), 57–66. [Google Scholar]
- Garcia-Plazaola J. I., Becerril J. M., Arrese-Igor C., Gonzalez-Murua C., Aparicio-Tejo P. M. (1993). The contribution of Rhizobium meliloti to soil denitrification. Plant Soil. 157, 207–213. 10.1007/BF00011049 [DOI] [Google Scholar]
- Henry S., Texier S., Hallet S., Bru D., Dambreville C., Cheneby D., et al. (2008). Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ. Microbiol. 10, 3082–3092. 10.1111/j.1462-2920.2008.01599.x [DOI] [PubMed] [Google Scholar]
- Horchani F., Prevot M., Boscari A., Evangelisti E., Meilhoc E., Bruand C., et al. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155, 1023–1036. 10.1104/pp.110.166140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba S., Ikenishi F., Itakura M., Kikuchi M., Eda S., Chiba N., et al. (2012). N2O emission from degraded soybean nodules depends on denitrification by Bradyrhizobium japonicum and other microbes in the rhizosphere. Microbes Environ. 27, 470–476. 10.1264/jsme2.ME12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC. (2007). Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, 2007. Cambridge: Cambridge University Press. [Google Scholar]
- Itakura M., Uchida Y., Akiyama H., Takada Y., Shimomura Y., Morimoto S., et al. (2013). Mitigation of nitrous oxide emissions from soils by Bradyrhizobium japonicum inoculation. Nat. Clim. Change. 3, 208–212. 10.1038/nclimate1734 [DOI] [Google Scholar]
- Jones C. M., Graf D. R., Bru D., Philippot L., Hallin S. (2013). The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 7, 417–426. 10.1038/ismej.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern M., Simon J. (2009). Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochim. Biophys. Acta 1787, 646–656. 10.1016/j.bbabio.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Liu B., Frostegård Å., Bakken L. R. (2014). Impaired reduction of N2O to N2 in acid soil is due to a post transcriptional interference with the expression of nosZ. MBio 5, e01383–e01314. 10.1128/mBio.01383-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Mao Y., Bergaust L., Bakken L. R., Frostegård Å. (2013). Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ. Microbiol. 15, 2816–2828. 10.1111/1462-2920.12142 [DOI] [PubMed] [Google Scholar]
- Liu X., Gao C., Zhang A., Jin P., Wang L., Feng L. (2008). The nos gene cluster from gram-positive bacterium Geobacillus thermodenitrificans NG80-2 and functional characterization of the recombinant NosZ. FEMS Microbiol. Lett. 289, 46–52. 10.1111/j.1574-6968.2008.01362.x [DOI] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. (1982). Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhoc E., Cam Y., Skapski A., Bruand C. (2010). The response to nitric oxide of the nitrogen-fixing symbiont Sinorhizobium meliloti. Mol. Plant Microbe Interact. 23, 748–759. 10.1094/MPMI-23-6-0748 [DOI] [PubMed] [Google Scholar]
- Molstad L., Dörsch P., Bakken L. R. (2007). Robotized incubation system for monitoring gases (O2, NO, N2O N2) in denitrifying cultures. J. Microbiol. Methods 71, 202–211. 10.1016/j.mimet.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Morley N., Baggs E. M. (2011). Carbon and oxygen controls on N2O and N2 production during nitrate reduction. Soil Biol. Biochem. 42, 1864–1871. 10.1016/j.soilbio.2010.07.008 [DOI] [Google Scholar]
- Murray P. J., Hatcha D. J., Dixona E. R., Stevensb R. J., Laughlinc R. J., Jarvisa S. C. (2004). Denitrification potential in a grassland subsoil: effect of carbon substrates. Soil Biol. Biochem. 36, 545–547. 10.1016/j.soilbio.2003.10.020 [DOI] [Google Scholar]
- Nadeem S., Dörsch P., Bakken L. R. (2013). The significance of early accumulation of nanomolar concentrations of NO as an inducer of denitrification. FEMS Microbiol. Ecol. 83, 672–684. 10.1111/1574-6941.12024 [DOI] [PubMed] [Google Scholar]
- Pobigaylo N., Wetter D., Szymczak S., Schiller U., Kurtz S., Meyer F., et al. (2006). Construction of a large signature-tagged mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl. Environ. Microbiol. 72, 4329–4337. 10.1128/AEM.03072-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., Wang J., Almøy T., Bakken L. R. (2014). Excessive use of nitrogen in Chinese agriculture results in high N2O/(N2O+N2) product ratio of denitrification, primarily due to acidification of the soils. Glob. Change Biol. 20, 1685–1698. 10.1111/gcb.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raut N., Dörsch P., Sitaula B., Bakken L. R. (2012). Soil acidification by intensified crop production in South East Asia results in higher N2O/(N2+N2O) product ratio of denitrification. Soil Biol. Biochem. 55, 104–112. 10.1016/j.soilbio.2012.06.011 [DOI] [Google Scholar]
- Richardson D., Felgate H., Watmought N., Thomson A., Baggs E. (2009). Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle–could enzymatic regulation hold the key? Trends Biotechnol. 27, 388–397. 10.1016/j.tibtech.2009.03.009 [DOI] [PubMed] [Google Scholar]
- Robertsen B. K., Aman P., Darvill A. G., McNeil M., Albersheim P. (1981). Host-symbiont interactions. V. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant. Physiol. 67, 389–400. 10.1104/pp.67.3.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C., Bedmar E. J., Delgado M. J. (2011). Denitrification in Legume-associated endosymbiotic Bacteria, in Nitrogen Cycling in Bacteria, ed Moir J. W. B. (Norfolk, VA: Caister Academic Press; ), 197–210. [Google Scholar]
- Sanchez C., Itakura M., Mitsui H., Minamisawa K. (2013). Linked expressions of nap and nos genes in a Bradyrhizobium japonicum mutant with increased N2O reductase activity. Appl. Environ. Microbiol. 79, 4178–4180. 10.1128/AEM.00703-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford R. A., Wagner D. D., Wu Q., Chee-Sanford J. C., Thomas S. H., Cruz-García C., et al. (2012). Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. U.S.A. 109, 19709–19714. 10.1073/pnas.1211238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk-otte S., Seviour R. J., Kuenen J. G., Jetten M. S. M. (2000). Nitrous oxide (N2O) production by Alcaligenes faecalis during feast and famine regimes. Water Res. 7, 2080–2088. 10.1016/S0043-1354(99)00374-7 [DOI] [Google Scholar]
- Smith K. A., Mosier A. R., Crutzen P. J., Winiwarter W. (2012). The role of N2O derived from biofuels, and from agriculture in general, in Earth's climate. Philos. Trans. R Soc. Lond. B Biol. Sci. 367, 1169–1174. 10.1098/rstb.2011.0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. (2008). Greenhouse gas mitigation in agriculture. Philos. Trans. R Soc. Lond. B Biol. Sci. 363, 789–813. 10.1098/rstb.2007.2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder S. W., Hollocher T. C. (1987). Purification and some characteristics of nitrous oxide reductase from Paracoccus denitrifcans. J. Biol. Chem. 262, 6515–6525. [PubMed] [Google Scholar]
- Spiro S. (2012). Nitrous oxide production and consumption: regulation of gene expression by gas-sensitive transcription factors. Philos. Trans. R Soc. Lond. B Biol. Sci. 367, 1213–1225. 10.1098/rstb.2011.0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A. J., Giannopoulos G., Pretty J., Baggs E. M., Richardson D. J. (2012). Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos. Trans. R Soc. Lond. B Biol. Sci. 367, 1157–1168. 10.1098/rstb.2011.0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. J., Bueno E., Mesa S., Bedmar E. J., Delgado M. J. (2011b). Emerging complexity in the denitrification regulatory network of Bradyrhizobium japonicum. Biochem. Soc. Trans. 39, 284–288. 10.1042/BST0390284 [DOI] [PubMed] [Google Scholar]
- Torres M. J., Rubia M. I., Bedmar E. J., Delgado M. J. (2011a). Denitrification in Sinorhizobium meliloti. Biochem. Soc. Trans. 39, 1886–1889. 10.1042/BST20110733 [DOI] [PubMed] [Google Scholar]
- Torres M. J., Rubia M. I., de la Peña T. C., Pueyo J. J., Bedmar E. J., Delgado M. J. (2014). Genetic basis for denitrification in Ensifer meliloti. BMC Microbiol. 14, 142–151. 10.1186/1471-2180-14-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spanning R. J., Richardson D. J., Ferguson S. J. (2007). Introduction to the biochemistry and molecular biology of denitrification, in Biology of the Nitrogen Cycle, eds Bothe H., Ferguson S. J., Newton W. E. (Amsterdam: Elservier Science; ), 3–20. 10.1016/B978-044452857-5.50002-3 [DOI] [Google Scholar]
- Walters C. L., Gillatt P. N., Palmer R. C., Smith P. L. (1987). A rapid method for the determination of nitrate and nitrite by chemiluminescence. Food Addit. Contam. 4, 133–140. 10.1080/02652038709373624 [DOI] [PubMed] [Google Scholar]
- Zumft W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G., Kroneck P. M. H. (2007). Respiratory transformation of nitrous oxide (N2O) to dinitrogen by bacteria and archaea. Ad. in Microb. Physiol. 52, 107–227. 10.1016/S0065-2911(06)52003-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kinetics of O2 depletion and N2O, NO, and N2 production. E. meliloti 1021 was incubated in the presence of 10 mM NO−3 in minimal medium and an initial O2 concentration of 2% in the headspace. Cultures with an initial OD600 of 0.01 were vigorously stirred at 200 rpm. Plotted values are averages of three replicate flasks for each treatment, with standard deviation (SD) as vertical bars (n = 3).