Abstract
NusA and Rho are essential Escherichia coli proteins that influence transcription elongation and termination. We show that an E. coli derivative unable to express NusA, because its sole nusA gene contains a large deletion/substitution, is viable providing that the bacterium also carries a rho mutation that reduces transcription termination. This Rho-mediated suppression is not allele specific, since either a mutation changing amino acid 134 [rho(E134D)] or a mutation changing amino acid 352 (rho1) allows growth of a nusA-deleted E. coli. However, both rho mutations similarly decrease transcription termination 8- to 9-fold. We propose that the essential role of NusA is to enhance pausing of RNA polymerase at certain sites, permitting tight coupling of transcription and translation. This coupling interferes with Rho access to and/or movement on the nascent RNA and blocks premature termination of transcription. Thus, NusA-dependent coupling should be less important in a mutant with low Rho activity. The fact that E. coli grows without NusA argues that NusA should be considered an accessory factor rather than a subunit of RNA polymerase.
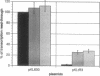
Full text
PDF


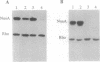
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Aldea M., Maples V. F., Kushner S. R. Generation of a detailed physical and genetic map of the ilv-metE-udp region of the Escherichia coli chromosome. J Mol Biol. 1988 Apr 5;200(3):427–438. doi: 10.1016/0022-2836(88)90533-5. [DOI] [PubMed] [Google Scholar]
- Barik S., Bhattacharya P., Das A. Autogenous regulation of transcription termination factor Rho. J Mol Biol. 1985 Apr 20;182(4):495–508. doi: 10.1016/0022-2836(85)90236-0. [DOI] [PubMed] [Google Scholar]
- Briat J. F., Chamberlin M. J. Identification and characterization of a new transcriptional termination factor from Escherichia coli. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7373–7377. doi: 10.1073/pnas.81.23.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven M. G., Friedman D. I. Analysis of the Escherichia coli nusA10(Cs) allele: relating nucleotide changes to phenotypes. J Bacteriol. 1991 Feb;173(4):1485–1491. doi: 10.1128/jb.173.4.1485-1491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven M. G., Granston A. E., Schauer A. T., Zheng C., Gray T. A., Friedman D. I. Escherichia coli-Salmonella typhimurium hybrid nusA genes: identification of a short motif required for action of the lambda N transcription antitermination protein. J Bacteriol. 1994 Mar;176(5):1394–1404. doi: 10.1128/jb.176.5.1394-1404.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- Das A., Wolska K. Transcription antitermination in vitro by lambda N gene product: requirement for a phage nut site and the products of host nusA, nusB, and nusE genes. Cell. 1984 Aug;38(1):165–173. doi: 10.1016/0092-8674(84)90537-3. [DOI] [PubMed] [Google Scholar]
- Daube S. S., von Hippel P. H. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992 Nov 20;258(5086):1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baron L. S. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974 Mar;58(1):141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Interaction between bacteriophage lambda and its Escherichia coli host. Curr Opin Genet Dev. 1992 Oct;2(5):727–738. doi: 10.1016/s0959-437x(05)80133-9. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R., Johnson L. L., Alessi D., Craven M. G. Transcription-dependent competition for a host factor: the function and optimal sequence of the phage lambda boxA transcription antitermination signal. Genes Dev. 1990 Dec;4(12A):2210–2222. doi: 10.1101/gad.4.12a.2210. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Schauer A. T., Baumann M. R., Baron L. S., Adhya S. L. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. C., Yager T. D., von Hippel P. H. Escherichia coli sigma 70 and NusA proteins. II. Physical properties and self-association states. J Mol Biol. 1991 Jul 20;220(2):325–333. doi: 10.1016/0022-2836(91)90016-y. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981 May;24(2):421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Nodwell J. R., Mason S. W. Transcriptional antitermination. Nature. 1993 Jul 29;364(6436):401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Irani M., Orosz L., Busby S., Taniguchi T., Adhya S. Cyclic AMP-dependent constitutive expression of gal operon: use of repressor titration to isolate operator mutations. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4775–4779. doi: 10.1073/pnas.80.15.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. J., Burgess R. R., Richardson J. P., Gross C. A. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainz M., Roberts J. Structure of transcription elongation complexes in vivo. Science. 1992 Feb 14;255(5046):838–841. doi: 10.1126/science.1536008. [DOI] [PubMed] [Google Scholar]
- Kalman M., Murphy H., Cashel M. rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biol. 1991 Sep;3(9):886–895. [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J Mol Biol. 1992 May 20;225(2):221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- Kung H., Spears C., Weissbach H. Purification and properties of a soluble factor required for the deoxyribonucleic acid-directed in vitro synthesis of beta-galactosidase. J Biol Chem. 1975 Feb 25;250(4):1556–1562. [PubMed] [Google Scholar]
- Lau L. F., Roberts J. W., Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J Biol Chem. 1983 Aug 10;258(15):9391–9397. [PubMed] [Google Scholar]
- Lee D. N., Landick R. Structure of RNA and DNA chains in paused transcription complexes containing Escherichia coli RNA polymerase. J Mol Biol. 1992 Dec 5;228(3):759–777. doi: 10.1016/0022-2836(92)90862-e. [DOI] [PubMed] [Google Scholar]
- Li J., Mason S. W., Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993 Jan;7(1):161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- Linn T., Greenblatt J. The NusA and NusG proteins of Escherichia coli increase the in vitro readthrough frequency of a transcriptional attenuator preceding the gene for the beta subunit of RNA polymerase. J Biol Chem. 1992 Jan 25;267(3):1449–1454. [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Nakamura Y., Mizusawa S., Court D. L., Tsugawa A. Regulatory defects of a conditionally lethal nusAts mutant of Escherichia coli. Positive and negative modulator roles of NusA protein in vivo. J Mol Biol. 1986 May 5;189(1):103–111. doi: 10.1016/0022-2836(86)90384-0. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Uchida H. Isolation of conditionally lethal amber mutations affecting synthesis of the nusA protein of Escherichia coli. Mol Gen Genet. 1983;190(2):196–203. doi: 10.1007/BF00330640. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Carey J. L., 3rd rho Factors from polarity suppressor mutants with defects in their RNA interactions. J Biol Chem. 1982 May 25;257(10):5767–5771. [PubMed] [Google Scholar]
- Roberts J. W. RNA and protein elements of E. coli and lambda transcription antitermination complexes. Cell. 1993 Mar 12;72(5):653–655. doi: 10.1016/0092-8674(93)90394-6. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Ruteshouser E. C., Richardson J. P. Identification and characterization of transcription termination sites in the Escherichia coli lacZ gene. J Mol Biol. 1989 Jul 5;208(1):23–43. doi: 10.1016/0022-2836(89)90085-5. [DOI] [PubMed] [Google Scholar]
- Saito M., Tsugawa A., Egawa K., Nakamura Y. Revised sequence of the nusA gene of Escherichia coli and identification of nusA11 (ts) and nusA1 mutations which cause changes in a hydrophobic amino acid cluster. Mol Gen Genet. 1986 Nov;205(2):380–382. doi: 10.1007/BF00430455. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. Binding of rho factor to Escherichia coli RNA polymerase mediated by nusA protein. J Biol Chem. 1984 Dec 25;259(24):15000–15002. [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987 Jun 20;195(4):809–818. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Morgan E. A. Nus A protein affects transcriptional pausing and termination in vitro by binding to different sites on the transcription complex. Biochemistry. 1988 Jul 26;27(15):5622–5627. doi: 10.1021/bi00415a034. [DOI] [PubMed] [Google Scholar]
- Sternberg N. L., Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- Sullivan S. L., Gottesman M. E. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992 Mar 6;68(5):989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- Ward D. F., Gottesman M. E. The nus mutations affect transcription termination in Escherichia coli. Nature. 1981 Jul 16;292(5820):212–215. doi: 10.1038/292212a0. [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Lindahl L. Ribosomal protein L4 and transcription factor NusA have separable roles in mediating terminating of transcription within the leader of the S10 operon of Escherichia coli. Genes Dev. 1992 Dec;6(12B):2655–2662. doi: 10.1101/gad.6.12b.2655. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]