Abstract
Th1/Th17-type T-cell responses are upregulated in Behcet’s disease (BD). However, signaling pathways associated with this aberrant immune response are not clarified. Whole-genome microarray profiling was performed with human U133 (Plus 2.0) chips using mRNA of isolated CD14+ monocytes and CD4+ T-cells from PBMC in patients with BD (n=9) and healthy controls (HC) (n=9). Flow cytometric analysis of unstimulated (US) and stimulated (PHA) STAT3 and pSTAT3 expressions of PBMCs were also analysed (BD and HC, both n=26). JAK1 was observed to be upregulated in both CD14+ monocytes (1.95 fold) and CD4+ T-lymphocytes (1.40 fold) of BD patients. Using canonical pathway enrichment analysis, JAK/STAT signaling was identified as activated in both CD14+ monocytes (p= 9.55E-03) and in CD4+ lymphocytes (p= 8.13E-04) in BD. Interferon signaling was also prominent among upregulated genes in CD14+ monocytes (p= 5.62E-05). Glucocorticoid receptor signaling and IL-6 signaling were among the most enriched pathways in differentially expressed genes in CD14+ monocytes (p= 2.45E-09, and 1.00E-06, respectively). Basal unstimulated total STAT3 expression was significantly higher in BD (1.2 vs 3.45, p<0.05). The JAK1/STAT3 signaling pathway is activated in BD, possibly through the activation of Th1/Th17-type cytokines such as IL-2, IFNγ, IL-6, IL-17 and IL-23.
Keywords: Behcet’s disease, JAK/STAT signaling, STAT3
INTRODUCTION
Behcet’s disease (BD) is a multi-systemic, chronic inflammatory disorder with a complex genetic background, leading to a pro-inflammatory activation of the innate and adaptive immune systems [1, 2]. Both CD4+ and CD8+ T cells producing Th1 type pro-inflammatory cytokines IL-2, IL-12 and IFN-γ are increased in the peripheral blood (PB) and inflammatory tissues in BD [3, 4]. Th17 cells represent a relatively new subset of T helper cells which mainly produce IL-17A, IL-17F, IL-22 and TNF-α [5]. IL-6, IL-23 and TGF-β, and induce the differentiation and maturation of Th17 cells from naïve T-cells. Th17 cells are suggested to be involved in various disorders such as spondyloarthropathies (SpA), psoriasis and inflammatory bowel disease (IBD), which have strong clinical and genetic similarities with BD [6, 7]. Recent data also implicate the participation of IL-17 and Th17 type responses in BD pathogenesis [8, 9]. Active BD is characterized by high serum levels of IL-6, IL-17, IL-23 and activated Th17 cells [10–12]. Therapies such as interferon-alpha (IFNα) and anti-TNFα agents which are effective in ocular BD, are also shown to suppress Th17 responses [13, 14].
Various cytokines such as IFNs, interleukins and colony-stimulating factors bind to the similar type I/II cytokine superfamily receptors on cell membranes [15], and associate with Janus family of kinases (JAK1-3) membrane proximal domains, leading to their phosphorylation. Recruitment of signal transducer and activator of transcription (STAT) family of transcription factors then modulates gene transcription [16]. IFNγ, GM-CSF, IL-2, IL-6, IL-12, IL-15, IL-17, IL-21, IL-22 and IL-23 activate through various JAK/STAT combinations in the cell surface and are implicated in the pathogenesis of infections, malignancies and autoimmune/inflammatory disorders such as rheumatoid arthritis, IBD and psoriasis [16, 17]. Although proinflammatory and most Th1/Th17-associated cytokines are also implicated in BD pathogenesis, the role of intra-cellular signaling pathways, such as JAK/STAT system, has not been characterized.
In this study, we demonstrated the role of the JAK/STAT pathway in PB CD14+ monocyte and CD4+ T cells with whole genome gene expression analysis and confirmed our results with flow cytometric STAT3 analysis in PB of patients with BD.
RESULTS AND DISCUSSION
When genes with 1.5 fold differences were selected, upregulation of the signals of 1035 probes (738 genes) and downregulation of 1431 probes (1004 genes) were detected in CD14+ monocytes of BD patients, whereas signals from 335 probes (230 genes) upregulated and 68 probes (51 genes) downregulated in CD4+ T cells. JAK1 (Probe ID: 1552611) was upregulated in both CD14+ monocytes (1.95 fold) and CD4+ T- lymphocytes (1.40 fold) of BD patients compared to the controls (supplement 2).
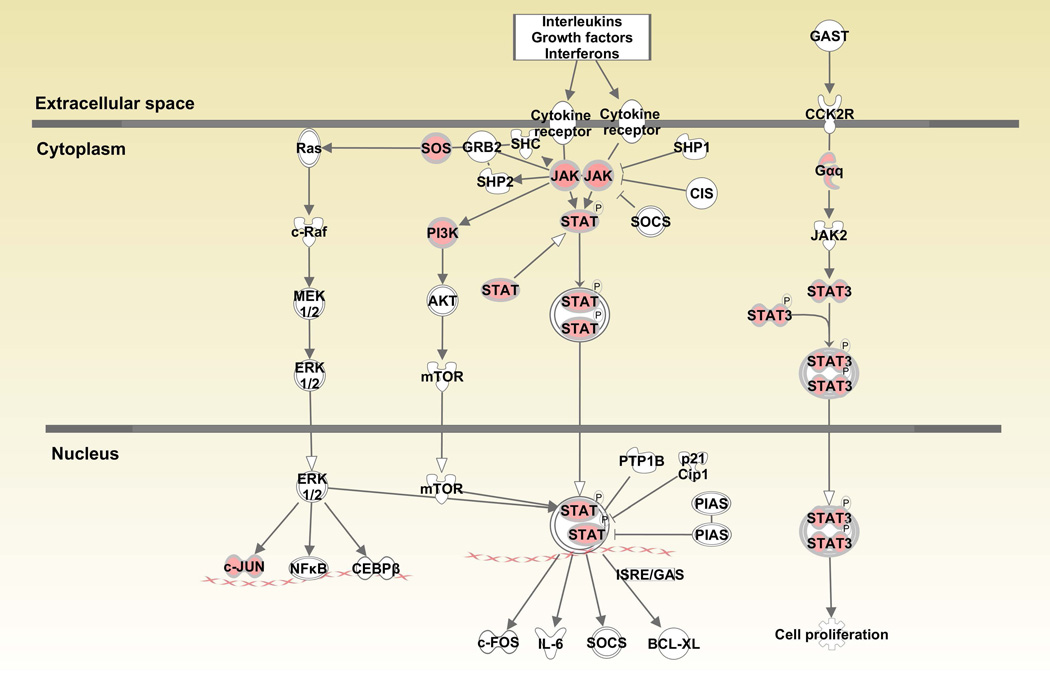
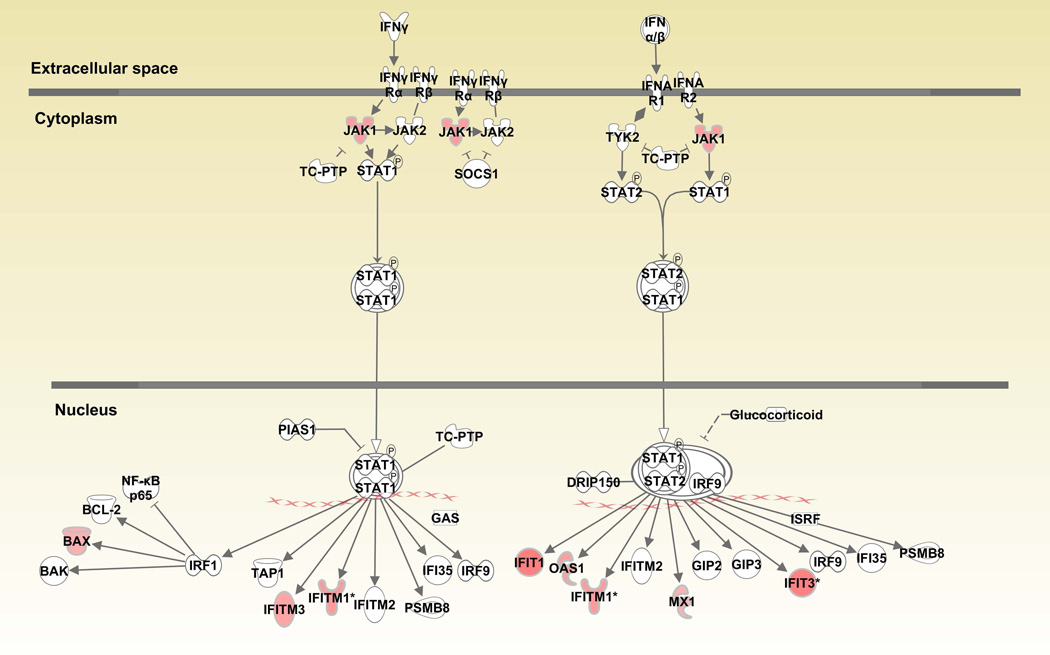
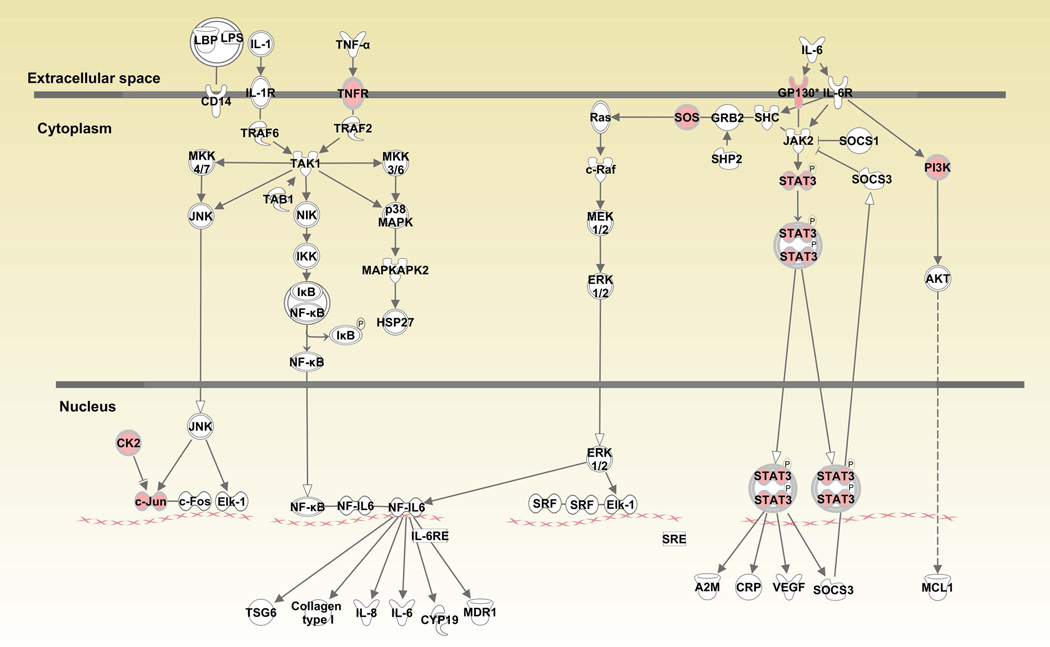
When canonical pathways were analyzed with Ingenuity Pathway Analysis (IPA), various pathways associated with cell growth, differentiation, cytokine and glucocorticoid receptor signaling and anti-bacterial responses were upregulated in monocytes (Table 1 and Supplementary Table 1). “JAK/STAT signaling” (JAK1, JUN, SOS2, SOS1, PIK3R5, GNAQ, PIK3CB, STAT3, p=9.55E-03) was among the upregulated pathways in CD14+ monocytes (Figure 1A). Similarly, “Interferon signaling” (IFIT3, IFIT1, IFITM3, OAS1, JAK1, MX1, BAX, IFITM1, p=5.62E-05) was also prominent among upregulated genes (Figure 1B). Downstream genes in the interferon signaling pathway such as IFIT1/IFIT3 were upregulated over 2 fold. When “IL-6 signaling” was analyzed, gp130 (2.0 fold) in the cell membrane, c-jun (1.71 fold) in the nucleus and STAT3 (1.64 fold) were also upregulated in monocytes from BD patients compared to healthy controls (Figure 1C).
Table 1.
Canonical pathways overrepresented by upregulated genes in CD14+ monocytes in BD patients
| Ingenuity Canonical Pathways |
Molecules | p-values |
|---|---|---|
| Interferon Signaling | IFIT3, IFIT1, IFITM3, OAS1, JAK1, MX1, BAX, IFITM1 | 5.62E-05 |
| Mouse Embryonic Stem Cell Pluripotency | IL6ST, TCF4, JAK1, SOS1, SOS2, PIK3R5, BMPR2, PIK3CB, SMAD5, FZD1, STAT3, TCF3, FZD2 | 1.00E-04 |
| TGF-β Signaling | TGFBR2, IRF7, JUN, CDC42, SOS2, SOS1, SMAD7, BMPR2, SMURF2, VDR, SMAD5, ACVR1B | 2.29E-04 |
| Systemic Lupus Erythematosus Signaling | LSM14B, CD79B, NFATC3, SOS2, PIK3R5, FCGR1A, PTPRC, SNRNP48, CD3G, RNPC3, JUN, CBL, NFAT5, LSM12, SOS1, PRPF6, PIK3CB, FCGR1B | 4.17E-04 |
| EGF Signaling | CSNK2A2, JAK1, JUN, SOS2, MAP3K1, SOS1, PIK3R5, PIK3CB, STAT3 | 4.37E-04 |
| B Cell Receptor Signaling | CD79B, POU2F2, NFATC3, SOS2, MAP3K1, PIK3R5, TCF3, PTPRC, EBF1, CAMK2D, NFAT5, JUN, DAPP1, CDC42, SOS1, PIK3CB, MAP3K3 | 5.50E-04 |
| Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency | IL6ST, RIF1, JAK1, SOS2, SOS1, PIK3R5, BMPR2, PIK3CB, STAT3, FZD1, SMAD5, FZD2 | 1.51E-03 |
| T Cell Receptor Signaling | PTPRC, CD3G, NFAT5, JUN, CBL, NFATC3, SOS2, MAP3K1, SOS1, PIK3R5, PIK3CB | 1.70E-03 |
| Glucocorticoid Receptor Signaling | JAK1, PRKAB2, POU2F2, NFATC3, HSPA1A/HSPA1B, MAP3K1, SOS2, PIK3R5, HSPA6, STAT3, FCGR1A, TGFBR2, CD3G, HSP90B1, JUN, NFAT5, TAF5, SMARCA2, SOS1, PIK3CB, TAF15 | 2.24E-03 |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | IFIH1, IRF7, JUN, DDX58, ZBP1, SIKE1, IFIT2, ISG15 | 3.09E-03 |
| SAPK/JNK Signaling | JUN, NFATC3, CDC42, SOS2, MAP3K1, SOS1, PIK3R5, PIK3CB, MAP4K5, MAP3K3 | 3.31E-03 |
| Wnt/β-catenin Signaling | TCF4, CSNK1G1, FRAT1, GNAQ, BMPR2, FZD1, TCF3, ACVR1B, TGFBR2, CSNK2A2, JUN, NLK, RARA, TLE4, FZD2 | 3.47E-03 |
| PDGF Signaling | CSNK2A2, JAK1, JUN, SOS2, MAP3K1, SOS1, PIK3R5, PIK3CB, STAT3 | 4.37E-03 |
| PKCθ Signaling in T Lymphocytes | CD3G, NFAT5, JUN, CAMK2D, NFATC3, SOS2, MAP3K1, SOS1, PIK3R5, PIK3CB, MAP3K3 | 4.68E-03 |
| IL-2 Signaling | CSNK2A2, JAK1, JUN, SOS2, SOS1, PIK3R5, PIK3CB | 5.89E-03 |
| Molecular Mechanisms of Cancer | TCF4, JAK1, SOS2, PIK3R5, BMPR2, SMAD5, FZD1, CDKN2B, TGFBR2, CAMK2D, NLK, JUN, SOS1, HIPK2, FZD2, SMAD7, GNAQ, BAX, AURKA, TCF3, NBN, CBL, CDC42, RBPJ, PIK3CB | 7.24E-03 |
| ErbB2-ErbB3 Signaling | NRG1, JUN, SOS2, SOS1, PIK3R5, PIK3CB, STAT3 | 7.94E-03 |
| PPARα/RXRα Activation | PRKAB2, CPT1B, CD36, SOS2, GNAQ, BMPR2, NR2C2, ACVR1B, TGFBR2, HSP90B1, JUN, GPD2, SOS1, INSR | 8.51E-03 |
| PTEN Signaling | TGFBR2, CSNK2A2, CBL, CDC42, SOS2, SOS1, PIK3R5, BMPR2, PIK3CB, INSR, IGF2R | 9.12E-03 |
| JAK/Stat Signaling | JAK1, JUN, SOS2, SOS1, PIK3R5, GNAQ, PIK3CB, STAT3 | 9.55E-03 |
| HGF Signaling | JUN, CDC42, SOS2, MAP3K1, SOS1, PIK3R5, PIK3CB, STAT3, MAP3K3, ELF1 | 1.07E-02 |
| Acute Myeloid Leukemia Signaling | TCF4, RARA, SOS2, SOS1, PIK3R5, PIK3CB, STAT3, TCF3 | 1.12E-02 |
| Regulation of IL-2 Expression in Activated and Anergic T Lymphocytes | TGFBR2, CD3G, NFAT5, JUN, NFATC3, SOS2, MAP3K1, SOS1 | 1.29E-02 |
| CD28 Signaling in T Helper Cells | PTPRC, CD3G, NFAT5, JUN, ARPC5L, NFATC3, CDC42, MAP3K1, PIK3R5, PIK3CB | 1.29E-02 |
| CNTF Signaling | IL6ST, JAK1, SOS1, PIK3R5, PIK3CB, STAT3 | 1.35E-02 |
| Epithelial Adherens Junction Signaling | TGFBR2, TCF4, ACTA2, ARPC5L, CDC42, FER, WASL, BMPR2, TCF3, IQGAP1, CLIP1, ACVR1B | 1.38E-02 |
| Role of NFAT in Regulation of the Immune Response | CD3G, JUN, NFAT5, CD79B, CSNK1G1, NFATC3, SOS1, SOS2, GNAQ, PIK3R5, PIK3CB, FCGR1A, FCGR1B | 1.41E-02 |
| Lysine Degradation II | AASDH, AASDHPPT | 1.55E-02 |
| Lysine Degradation V | AASDH, AASDHPPT | 1.55E-02 |
| April Mediated Signaling | NFAT5, JUN, NFATC3, TNFSF13, MAP3K1 | 1.91E-02 |
| Renal Cell Carcinoma Signaling | JUN, CDC42, SOS2, SOS1, PIK3R5, PIK3CB, ARNT | 2.34E-02 |
| IL-4 Signaling | NFAT5, JAK1, NFATC3, SOS2, SOS1, PIK3R5, PIK3CB | 2.51E-02 |
Figure 1.
Canonical pathways activated in CD14+ monocytes in Behcet’s disease. Fig1A: JAK/STAT signaling pathway, Fig. 1B: Interferon signaling pathway, Fig. 1C: Interleukin-6 signaling pathway. Red color indicates genes that are upregulated in patients compared to controls.
In CD4+ T cells, genes related to developmental processes, differentiation and proliferation were activated. Canonical pathways associated with T-cell receptors such as ICOS, CD28 and glucocorticoid receptor signaling, inositol phosphate metabolism and Ca-induced apoptosis are shown to have upregulated molecules in CD4+ T cells of BD patients (Supplementary Tables 2 and 3). JAK/STAT pathway genes in CD4+ T cells were also among the prominent upregulated transcripts (JAK1, MAP2K2, PIK3C2A, SOCS2, STAT2, p=1.78E-03).
A recent study using RNA extracted from total PBMCs demonstrated significant gene expression differences between BD patients and controls [18]. Our ability to directly compare these previously reported results to ours is limited as our study examined specific cell subsets in BD patients and controls. However, our datasets includes 76 and 13 differentially expressed probes in monocytes and CD4+ T cells, respectively, that were also differentially expressed in the previously reported data in total PBMCs. Of particular interest is that two key genes in the neuregulin signaling pathway (EREG and AREG), which were among the top downregulated genes in PBMCs in BD patients [18], were also significantly downregulated in BD monocytes in our study (Supplementary Table 4). Other relevant common downregulated genes in BD monocytes and total PBMCs include protein tyrosine phosphatase receptor type E (PTPRE) and phosphodiesterase 4D cAMP-specific (PDE4D), among others.
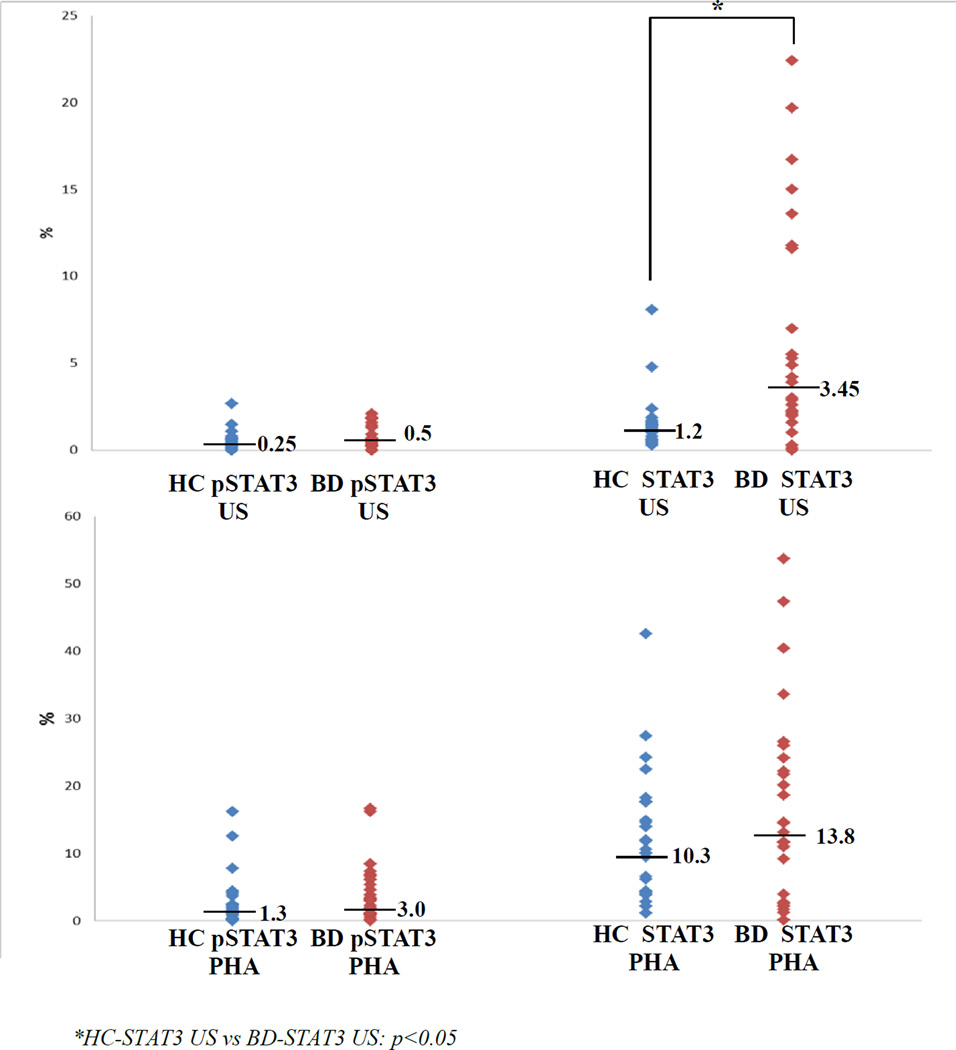
When patients with BD were compared in basal, unstimulated (US) and stimulated conditions (with PHA) for pSTAT3 and total STAT3 expressions, basal US total STAT3 expression was significantly higher in BD (1.2 (0.3–8.1) vs 3.45 (0–22.4), p<0.05)(Figure 2). No correlations were observed between total STAT3 levels in BD patients and any disease manifestation, disease duration, age, gender and treatments.
Figure 2.
STAT3 and pSTAT3 expressions in PBMCs of BD patients and controls.
After stimulations, both pSTAT3 and STAT3 expressions significantly increased compared to baseline, however no differences were observed between BD (pSTAT3: US: 0.5 (0–2.1) vs PHA: 3.0 (0–16.6); STAT3: US: 3.45 (0–22.4) vs PHA: 13.8 (0.1–53.7)) and healthy controls (pSTAT3: US: 0.25 (0–2.7) vs PHA: 1.3 (0–16.2); STAT3: US: 1.2 (0.3–8.1) vs PHA: 10.3 (1.1–42.6)) (Figure 2).
JAK/STAT signaling pathways are crucial for the activation of innate and adaptive immune systems. IFN-γR, IL-2R and IL-6R signal through JAK1, pairing with JAK2 or JAK3, whereas IL-12 and IL-23 activate through JAK2/Tyk2 pathway [19]. Downstream, STAT1 is required for IL-2, IFN-γ and IL-6, whereas STAT3 is associated with IL-2, IL-6, IL-12 and IL-23. The anti-inflammatory cytokine IL-10 also activates the JAK1/STAT3 pathway, regulating SOCS3 [15]. STAT3 was critical in modulating the balance of Th17 and regulatory T cells, as well as in promoting CD4+ T cell proliferation. STAT3 bound to multiple genes, especially IL-6, is involved in Th17 cell differentiation, cell activation, proliferation and survival, regulating both expression and epigenetic modifications. STAT3 also plays an important role in the IFN-γ signaling pathway, which is highly involved in most autoimmune processes. Thus, STAT3 orchestrates multiple critical aspects of T cell function in inflammation and homeostasis [20].
JAK/STAT pathway-associated cytokines and Th subsets are shown to be activated in BD [1]. Both IL-12 activated, IFN-γ secreting Th1 and IL-23 activated Th17 cell subsets are observed to be elevated in PB and tissues in BD [3, 4, 21, 22]. Levels of IL-17, IL-23, IL-12/23p40, and IFN-γ in serum and supernatants are significantly elevated [10, 23]. The IL-6 signaling pathway, which is upregulated in our study, is implicated especially in the pathogenesis of neuro-BD and IL-6 has been suggested as a biomarker in CSF analysis [24].
Unstimulated and PHA-stimulated pSTAT3 expressions, although higher in BD, were not significantly different between the study groups in our study. However, pSTAT3 expression is found to be upregulated in BD, in a different setting with anti-CD3/28 antibody stimulation and suggested to be related to Notch pathway activation [25]. Most of total STAT3, observed to be elevated in our samples, seems to be unphosphorylated (U-STAT3). Recently, interest has increased in the functional roles of U-STATs. Ligand-dependent increases in the concentrations of U-STATs are shown to drive the expression of genes that are distinct from those activated by pSTATs. U-STAT3 binds to unphosphorylated NFκB (U-NFκB), in competition with IκB, and the resulting U-STAT3/U-NFκB complex is demonstrated to accumulate in the nucleus [26]. Following long term IL-6 exposure, concentrations of endogenous U-STAT3 is increased and it competes effectively with IκB for U-NFκB, to form a novel transcription factor that induces RANTES expression [27]. This function of U-STAT3 seems clearly different from the absolute requirement for tyrosine phosphorylation that enables STAT3 dimers to bind to GAS motifs (IFN-activating sequences). STAT3 can also enter the nucleus independently of its phosphorylation, shuffling between cytoplasm and nucleus [26]. STAT3 has been shown to bind to some transcription factors such as CRE-binding protein on the JunB promoter, and has effects on CRE-like sites in the C/EBPβ promoter and on the glucocorticoid response element. Recently, it was also demonstrated that U-STAT3 binds to AT-rich DNA sequence sites and recognizes specific DNA structures, such as 4-way junctions and DNA nodes, within negatively supercoiled plasmid DNA [28]. These structures are important for chromatin organization and suggest a role for U-STAT3 as a chromatin/genome organizer. Our data might, therefore, suggest that upregulated U-STAT3 in BD patients can have intracellular effects other than phosphorylation in BD pathogenesis.
The JAK1/STAT3 pathway is also studied in some immune, inflammatory disorders (now termed autoinflammatory) with similar clinical and genetic features to BD, such as IBD, SpAs and psoriasis [16, 17]. A common genetic background associated with JAK/STAT activating cytokines such as IL-23R and IL-12R is present in BD, IBD and psoriasis [7]. Recently, three genetic variants in JAK1 gene is reported to be associated with BD in Chinese patients with ocular disease [29]. However, a functional role of these single nucleotide polymorphisms (SNP) could not be shown. Similarly, another study in the same population also showed that a STAT3 SNP seems to be associated with susceptibility to BD [30], but not confirmed in a Spanish population [31]. STAT3 is also shown to be prominent in some animal models of chronic experimental uveitis and vasculitis, which have similarities to severe BD manifestations [32, 33].
In conclusion, we have demonstrated that JAK1/STAT3 signaling pathway is activated in BD, possibly through elevated serum and tissue expressions of Th1/Th17 type cytokines. In addition to current treatments such as IFNα and anti-TNFα agents that suppress Th17 cells, more direct therapies aiming JAK/STAT associated cytokines such as ustekinumab (anti-IL-12/23) and recently approved tofacitinib that specifically inhibit JAK1/3 may be new therapeutic options for BD [13, 14, 16, 34].
PATIENTS AND METHODS
Patients
Nine patients with Behcet’s Disease followed in the multi-disciplinary Behcet’s Clinic of Marmara University Hospital and 9 healthy controls were studied in the first phase of the study for the microarray analysis of CD14+ monocyte and CD4+ T lymphocytes. In the second phase, when flow cytometric analysis of STAT3 and pSTAT3 levels were investigated, a larger group of patients with BD (n=26, 15 F/11 M, mean age: 35.3±9.1 years) and healthy controls (n=26, 19 F/7 M, mean age: 33.9 ±10.5 years) were recruited. All patients fulfilled the ISG Criteria for the classification of BD and had a mean disease duration of 6.9±4.6 years [35]. Oral ulcers were present in all patients, 96% (n=25) had cutaneous lesions, 81% (n=21) had genital ulcers, 31% (n=8) had musculoskeletal involvement and 50% (n=13) had pathergy positivity. Among major manifestations, 19% (n=5) of the patients had vascular, 12% (n=3) had ocular and 4% (n=1) had central-nervous system disease. Ten patients (38%) were under immunosuppressive treatment (prednisolone >10 mg/d and/or azathioprine). Patients studied for microarray analysis had similar characteristics to the whole group (9/9 patients with mucocutaneous, 3/9 with musculoskeletal, 2/9 with vascular and 1/9 with ocular manifestations). All patients had at least one active BD-associated disease manifestation at the time of blood sampling. The study was approved by the local Ethical Committee of Marmara University and informed consent was taken.
Sample collection and microarray analysis
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh blood samples using density gradient centrifugation (Amersham Biosciences, Uppsala, Sweden). Monocytes and T-helper cells were purified using magnetic bead separation from PBMCs (Miltenyi Biotec, Cologne, Germany). The purity of isolated cell populations was confirmed by flow cytometry analysis using fluorochrome-conjugated antibodies against CD4 for T-helper lymphocytes and CD14 for monocytes and was over 90% for both cell populations. Total RNA of purified cell populations was extracted using RNeasy Mini kit (Qiagen, Hilden, Germany) according to the protocol recommended by the manufacturer. Total RNA (>50 ng/ul (80–156)) from monocytes was available from 8 patients and 9 controls, and from CD4+ T cells from all 9 patients and 3 controls. Total RNA was then hybridized to GeneChip Human Genome U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA, USA) following manufacturer's protocols.
Gene expression data normalization and statistical analysis
Raw microarray data were processed using Affymetrix Expression Console version 1.1 (http://www.affymetrix.com/browse/level_sevensoftware_products_only.jsp?productId=131414&categoryId=35623#) to extract gene level intensities using RMA method. The subsequent analysis was performed using the R statistical environment (http://www.R-project.org) and BioConductor packages [36]. Quality control of the data included a comparison of boxplots of gene expression level among the conditions, and hierarchical clustering of condition-specific gene expression profiles to identify potential outliers. The data were quantile normalized and deposited in GEO along with raw .CEL files (GEO accession number GSE61399). (All data conform with the Minimal Information About a Microarray Experiment (MIAME) guidelines.
Differentially expressed probes were identified by Significance Analysis of Microarrays (SAM) [37]. A fold change cutoff of 1.5 was imposed to filter out less differentially expressed probes, and the delta was adjusted to keep false discovery rate <10%. Owing to the fact that some probes may detect expression of different exons, probe lists were converted to gene lists keeping all gene names. Gene lists were analyzed by Ingenuity Pathway Analysis (IPA; Ingenuity® Systems, Redwood City, CA, http://www.ingenuity.com), a web-based bioinformatics tool. Each gene list was tested against the full Affymetrix Human Gene 1.0 ST Array dataset to identify significantly overrepresented functions and canonical pathways as compared to the same number of randomly selected genes. The most significant functions and canonical pathways were selected and genes overrepresented in the functions and pathways were identified.
Flow Cytometric Analysis of STAT3 and pSTAT3 levels
For intracellular STAT3 and phosphorylated STAT3 (pSTAT3) level analysis, freshly isolated PBMCs were suspended in complete RPMI 1640 medium supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin and streptomycin (all from Sigma-Aldrich Inc., St Louis, MO, USA). 1x106 PBMCs in 1 ml were either left unstimulated or were stimulated with 10 µg/ml phytohaemagglutinin (PHA, Sigma-Aldrich Inc.) for 72 hours. PMBCs were then washed and fixed with BD Cytofix™ buffer (BD Biosciences, Franklin Lakes, NJ, USA) for 20 minutes. Following the washing step, fixed cells were permeabilized by using BD Phosflow™ Perm Buffer III (BD Biosciences) for 10 minutes. Unstimulated and PHA stimulated cells were then stained with allophycocyanin (APC) conjugated anti-STAT3 (M59-50) monoclonal antibody which recognizes STAT3 regardless of phosphorylation status. For the detection of phosphorylated STAT3, phycoerythrin (PE) conjugated anti-STAT3 (pS727) monoclonal antibody which recognizes the S727-phosphorylated form of STAT3 isoform 1 was used. All samples were also stained with isotypic controls. STAT3 and pSTAT3 levels of PBMCs were analysed with FACSCanto flow cytometry using Diva software (BD Biosciences). Results were expressed as median (range) of pSTAT3/STAT3-expressing mononuclear cells in PB and non-parametric Mann-Whitney-U test was used for comparisons.
Supplementary Material
ACKNOWLEDGEMENTS
This study is supported through grants of Turkish Scientific and Technical Council (TUBITAK, No: 106S320) and Marmara University (BAPKO, SAG-BGS-290107-0022 and SAG-A-090512-0132). JDW and MGD were supported by NIH 5P20GM103636-02.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
REFERENCES
- 1.Direskeneli H. Autoimmunity vs autoinflammation in Behcet's disease: do we oversimplify a complex disorder? Rheumatology (Oxford) 2006;45(12):1461–1465. doi: 10.1093/rheumatology/kel329. [DOI] [PubMed] [Google Scholar]
- 2.Hughes T, et al. Identification of multiple independent susceptibility loci in the HLA region in Behcet's disease. Nat Genet. 2013;45(3):319–324. doi: 10.1038/ng.2551. [DOI] [PubMed] [Google Scholar]
- 3.Frassanito M, et al. Th1 polarization of the immune response in Behcet's disease. Arthritis Rheum. 1999;42:1967–1974. doi: 10.1002/1529-0131(199909)42:9<1967::AID-ANR24>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Imamura Y, et al. Involvement of Th1 cells and heat shock protein 60 in the pathogenesis of intestinal Behcet's disease. Clin Exp Immunol. 2005;139(2):371–378. doi: 10.1111/j.1365-2249.2005.02695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annunziato F, et al. Type 17 T helper cells-origins, features and possible roles in rheumatic disease. Nat Rev Rheumatol. 2009;5(6):325–331. doi: 10.1038/nrrheum.2009.80. [DOI] [PubMed] [Google Scholar]
- 6.Hreggvidsdottir HS, Noordenbos T, Baeten DL. Inflammatory pathways in spondyloarthritis. Mol Immunol. 2014;57(1):28–37. doi: 10.1016/j.molimm.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Remmers EF, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet's disease. Nat Genet. 2010;42(8):698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamzaoui K. Th17 cells in Behcet's disease: a new immunoregulatory axis. Clin Exp Rheumatol. 2011;29(4 Suppl 67):S71–S76. [PubMed] [Google Scholar]
- 9.Direskeneli H, Fujita H, Akdis CA. Regulation of TH17 and regulatory T cells in patients with Behcet disease. J Allergy Clin Immunol. 2011;128(3):665–666. doi: 10.1016/j.jaci.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Chi W, et al. Upregulated IL-23 and IL-17 in Behcet patients with active uveitis. Invest Ophthalmol Vis Sci. 2008;49(7):3058–3064. doi: 10.1167/iovs.07-1390. [DOI] [PubMed] [Google Scholar]
- 11.Geri G, et al. Critical role of IL-21 in modulating TH17 and regulatory T cells in Behcet disease. J Allergy Clin Immunol. 2011;128(3):655–664. doi: 10.1016/j.jaci.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 12.Hamzaoui K, et al. Expression of Th-17 and RORgammat mRNA in Behcet's Disease. Med Sci Monit. 2011;17(4):CR227–CR234. doi: 10.12659/MSM.881720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, et al. IFN-alpha blocks IL-17 production by peripheral blood mononuclear cells in Behcet's disease. Rheumatology (Oxford) 2011;50(2):293–298. doi: 10.1093/rheumatology/keq330. [DOI] [PubMed] [Google Scholar]
- 14.Sugita S, et al. Inhibition of Th17 differentiation by anti-TNF-alpha therapy in uveitis patients with Behcet's disease. Arthritis Res Ther. 2012;14(3):R99. doi: 10.1186/ar3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178(5):2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 16.O'Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368(2):161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coskun M, et al. Involvement of JAK/STAT signaling in the pathogenesis of inflammatory bowel disease. Pharmacol Res. 2013;76:1–8. doi: 10.1016/j.phrs.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Xavier JM, et al. Gene expression profiling and association studies implicate the neuregulin signaling pathway in Behcet's disease susceptibility. J Mol Med (Berl) 2013;91(8):1013–1023. doi: 10.1007/s00109-013-1022-4. [DOI] [PubMed] [Google Scholar]
- 19.O'Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36(4):542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32(5):605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, et al. Imbalance of Th17 to Th1 cells in Behcet's disease. Clin Exp Rheumatol. 2010;28(4 Suppl 60):S16–S19. [PubMed] [Google Scholar]
- 22.Shimizu J, et al. Excessive CD4+ T cells co-expressing interleukin-17 and interferon-gamma in patients with Behcet's disease. Clin Exp Immunol. 2012;168(1):68–74. doi: 10.1111/j.1365-2249.2011.04543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na SY, et al. Up-regulation of Th17 and related cytokines in Behcet's disease corresponding to disease activity. Clin Exp Rheumatol. 2013;31(3 Suppl 77):32–40. [PubMed] [Google Scholar]
- 24.Akman-Demir G, et al. Interleukin-6 in neuro-Behcet's disease: association with disease subsets and long-term outcome. Cytokine. 2008;44(3):373–376. doi: 10.1016/j.cyto.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Qi J, et al. Increased Notch pathway activation in Behcet's disease. Rheumatology (Oxford) 2014 doi: 10.1093/rheumatology/ket438. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18(4):443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21(11):1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timofeeva OA, et al. Mechanisms of unphosphorylated STAT3 transcription factor binding to DNA. J Biol Chem. 2012;287(17):14192–14200. doi: 10.1074/jbc.M111.323899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou S, et al. Genetic variants in the JAK1 gene confer higher risk of Behcet's disease with ocular involvement in Han Chinese. Hum Genet. 2013;132(9):1049–1058. doi: 10.1007/s00439-013-1312-5. [DOI] [PubMed] [Google Scholar]
- 30.Hu K, et al. JAK2 and STAT3 polymorphisms in a Han Chinese population with Behcet's disease. Invest Ophthalmol Vis Sci. 2012;53(1):538–541. doi: 10.1167/iovs.11-8440. [DOI] [PubMed] [Google Scholar]
- 31.Cenit MC, et al. Influence of the STAT3 genetic variants in the susceptibility to psoriatic arthritis and Behcet's disease. Hum Immunol. 2013;74(2):230–233. doi: 10.1016/j.humimm.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 32.Oh HM, et al. Autoreactive memory CD4+ T lymphocytes that mediate chronic uveitis reside in the bone marrow through STAT3-dependent mechanisms. J Immunol. 2011;187(6):3338–3346. doi: 10.4049/jimmunol.1004019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovacic JC, et al. Stat3-dependent acute Rantes production in vascular smooth muscle cells modulates inflammation following arterial injury in mice. J Clin Invest. 2010;120(1):303–314. doi: 10.1172/JCI40364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeilding N, et al. Development of the IL-12/23 antagonist ustekinumab in psoriasis: past, present, and future perspectives--an update. Ann N Y Acad Sci. 2012;1263:1–12. doi: 10.1111/j.1749-6632.2012.06670.x. [DOI] [PubMed] [Google Scholar]
- 35.Criteria for diagnosis of Behçet's Disease. Lancet. 1990;335:1078–1080. International, et al. [PubMed] [Google Scholar]
- 36.Team RDC. R: A Language and Environment for Statistical Computing. 2013 [Google Scholar]
- 37.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.