Abstract
Accurate folding, assembly, localization and maturation of newly synthesized proteins are essential to all cells and requires a high fidelity in the protein biogenesis machineries that mediate these processes. Here, we review our current understanding on how high fidelity is achieved in one of these processes, the co-translational targeting of nascent membrane and secretory proteins by the signal recognition particle (SRP). Recent biochemical, biophysical, and structural studies have elucidated how the correct substrates drive a series of elaborate conformational rearrangements in the SRP and SRP receptor GTPases, which provide effective fidelity checkpoints to reject incorrect substrates and enhance the fidelity of this essential cellular pathway. The mechanisms used by the SRP to ensure fidelity share important conceptual analogies with those used by cellular machineries involved in DNA replication, transcription and translation, and likely represent general principles for other complex cellular pathways.
Keywords: protein biogenesis, signal sequence, GTPases, RNA, ribosome, signal recognition particle
INTRODUCTION
Generation of order and organization is an essential feature of life. Central to the establishment of this organization is the exquisite fidelity of biochemical reactions that mediate every aspect of biology. Efforts to understand mechanisms that give rise to biological fidelity began over half a century ago, in consideration of the accuracy with which genetic information is replicated and decoded. Although the initial hypothesis suggested that base pairing interactions can provide selectivity, subsequent studies indicate that the free energy difference between matched and mismatched base pairs are too small to account for the fidelity required to maintain genome integrity, and far below the observed accuracy of cellular machineries that mediate these processes. For example, the error rates of base pairing interactions in solution are typically 10−3–10−4, whereas DNA and RNA polymerase exhibit error rates of 10−6–10−8 and 10−5, respectively. The error rates of amino acid recognition by tRNA synthetases are on the order of 10−2, whereas the translation process exhibits error rates of <10−3–10−5 (36). These observations led to the recognition that cellular machineries must be able to significantly enhance fidelity, beyond what can be achieved by a simple molecular recognition process.
Investigation of the molecular basis underlying selectivity during gene replication, transcription and translation revealed a variety of mechanisms by which cellular machineries enhance fidelity. For example, beyond preferential binding of the correct substrate, DNA and RNA polymerases use at least three mechanisms to enhance fidelity: (i) the correct substrate preferentially induces conformational changes on the polymerase that lead to high catalytic activity; (ii) incorporation of a mismatched nucleotide introduces ‘pauses’, i.e., the polymerase slows down subsequent rounds of DNA or RNA extension, by introducing off-pathway conformations; and (iii) proof-reading, during which the mis-incorporated nucleotide is removed by exonuclease activities ((77, 135) and references therein). A similar set of strategies was found to enhance fidelity during protein translation by the ribosome: (i) the cognate tRNA preferentially induces conformational changes that trigger GTP hydrolysis by elongation factor (EF)-Tu, a process required for EF-Tu to dissociate from the ribosome; (ii) kinetic proof-reading, in which a near-cognate tRNA is more likely to be irreversibly rejected from the ribosome rather than being accommodated into the active site for peptide bond formation ((97, 117) and references therein); (iii) mis-incorporation of an amino acid causes ribosome slippage that lead to premature release of the nascent polypeptide (156). Analogously, tRNA synthetases can enhance fidelity using a distinct ‘editing site’, which hydrolytically removes the mischarged amino acid (36–38). During pre-mRNA splicing, DExD/H helicases play diverse roles in rejecting or discarding complexes assembled at suboptimal splice sites ((123) and references therein). These studies have contributed significantly to our current conceptual framework for how fidelity arises in biology.
Nevertheless, the requirement to achieve high fidelity is pervasive in all cellular processes, from metabolic reactions, immune responses to signal transduction pathways. Our understanding of the molecular mechanisms that ensure fidelity in these other pathways lags far behind those for the replication and decoding of genetic information. In this review, we discuss recent advances in understanding the fidelity in one of these processes, the biogenesis of nascent proteins after its synthesis by the ribosome.
FIDELITY CONSIDERATIONS DURING PROTEIN BIOGENESIS
Multiplicity of fates during the biogenesis of nascent proteins
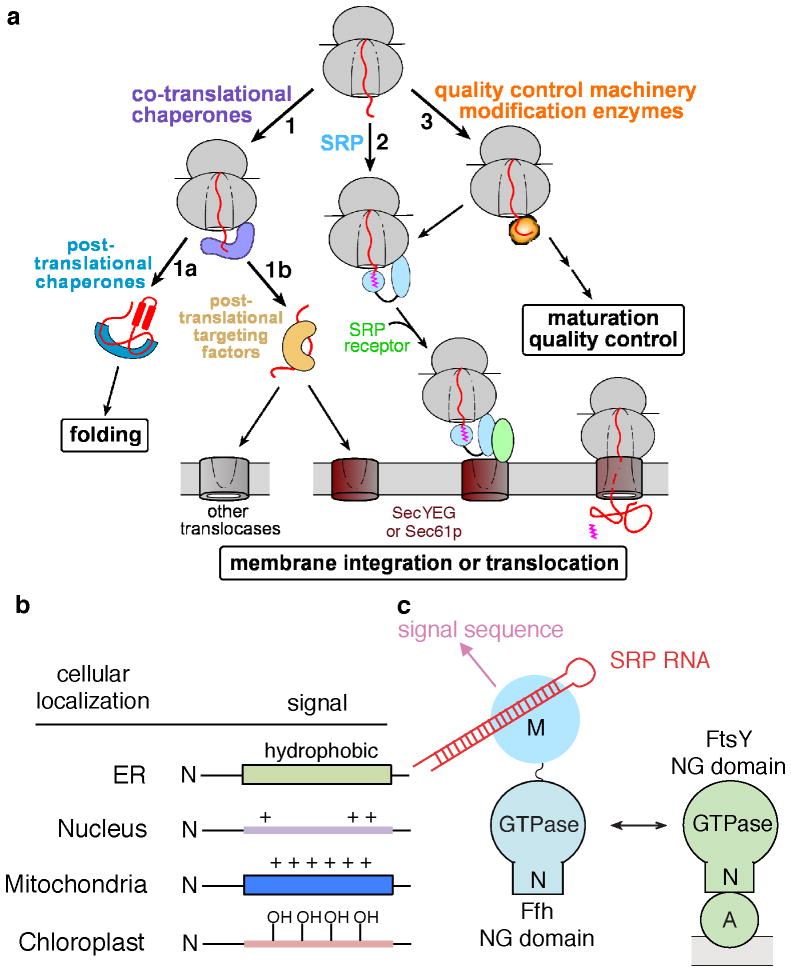
The proper biogenesis and homeostasis of proteins are essential to all cells and requires the correct folding, localization, maturation, and quality control of all the newly synthesized proteins (59, 107). Accumulating data now show that mechanisms to ensure proper protein homeostasis begin at the ribosome exit site, where a variety of protein biogenesis machineries rendezvous and vie for access to the nascent polypeptide (Fig. 1). For example, virtually all newly synthesized proteins interact with ribosome-associated chaperones, including trigger factor and DnaK/DnaJ in bacteria (29, 59, 62, 72, 89, 98), Hsp70 SSB and the NAC (nascent polypeptide associated complex) and RAC (comprised of Zuo and ssz) complexes in eukaryotic cells (3, 4, 64, 147, 151) (Fig. 1, route 1). These early interactions prevent the nascent polypeptide from misfolding and aggregation, thus directing them through more productive folding pathways. Upon the completion of protein synthesis, cytosolic proteins can be passed onto post-translational chaperones, including DnaK/J and GroEL in bacteria (95, 119) or the Hsp70 SSA, Hsp90, and the TriC complex in eukaryotes (59, 87, 88, 136, 155), to attain their correct three-dimensional fold (Fig. 1, 1a). On the other hand, proteins destined for various cellular organelles (or for secretion in bacteria) engage post-translational targeting factors, which maintain the nascent polypeptide in a loosely-folded, translocation-competent state and direct them to translocation sites on the target membrane (Fig. 1, 1b)(26, 28, 30, 46, 52, 60, 93, 122).
Figure 1.
Multiplicity of fates awaiting a newly synthesized protein. (a) A variety of protein biogenesis factors can interact with a nascent protein during and immediately after its synthesis by the ribosome, including co- and post-translational chaperones, co- and post-translational protein targeting machineries, and ribosome-associated modification enzymes and quality control complexes. (b) General features of signal sequences that direct proteins to distinct subcellular organelles. (c) Domain structures of the SRP, which is composed of the SRP54 (Ffh) protein and the SRP RNA (left), and of the bacterial SRP receptor FtsY (right).
In a distinct route, ribosomes translating proteins destined for specific cellular membranes or for the secretory pathway are recognized by and engage co-translational targeting machineries, the most well-established example being the signal recognition particle (SRP) (Fig. 1, route 2)(2, 146). Via interactions with a membrane-localized receptor, the SRP mediates the delivery and unloading of the translating ribosome onto a protein translocation machinery on the target membrane, the SecYEG complex in bacteria and archaea or the Sec61p complex in eukaryotic cells. Through this translocation machinery, the nascent polypeptide is either integrated into the lipid bilayer or translocated across the membrane to enter the secretory pathway (30, 69, 112).
In addition, over 50% of proteins are co-translationally processed by nascent chain-modifying enzymes (76). This includes the peptide deformylase in bacteria, N-acetyl transferase and arginyl transferase in eukaryotic cells, and methionine aminopeptidase in all organisms (Fig. 2, route 3) (76). These covalent modifications are essential to the growth and survival of cells and implicated in the maturation and quality control of the protein (137), although their precise roles and mechanisms remain to be determined. Finally, protein quality control could begin during ongoing protein synthesis. Ribosomes translating damaged mRNAs, such as those lacking a stop codon, are recognized by a ribosome quality control complex (RQC) comprised of the ubiquitin ligase Listerin/Rkr1, the AAA+ ATPase cdc48, Taek2 and Ydr333C (19, 67, 141). RQC mediates the dissociation of the ribosomal subunits and targets the aberrant nascent protein for degradation by the proteasome (19, 67, 141).
Figure 2.
Cargo binding by the SRP is insufficient to ensure a high fidelity of co-translational protein targeting. (a) Incorporation of a fluorescent non-natural amino acid to produce site-specifically labeled RNC. (b) FRET assay to monitor the SRP-RNC interaction. Fluorescence emission spectra are shown for Cm-labeled RNC1A9L (black), BODIPY-Fl labeled SRP (blue), and their complex (red). (c) Design of signal sequence variants for investigation of the fidelity of SRP-dependent protein targeting. Bold highlights the hydrophobic core, and blue highlights the N-terminal signal sequence extension of EspP. (d) Summary of the binding affinities of SRP for RNCs bearing the different signal sequences. Adapted from Figure 1E in reference (160). The green line denotes the cellular SRP concentration of ~400 nM.
Given the multiplicity of fates awaiting a nascent polypeptide, highly accurate molecular recognition and regulation at the ribosome exit site is crucial for the proper biogenesis of nascent proteins. Within the first few seconds to minutes after a nascent protein emerges from the ribosome exit tunnel, it must engage with the correct set of protein biogenesis machinery and thus commit to the proper pathway (101). Below, we discuss the issues associated with fidelity in protein localization pathways, which serve as an example for the general challenges faced by cellular machineries in achieving highly accurate protein biogenesis.
Fidelity in protein localization: general principles and challenges
The ‘Signal Hypothesis’, proposed by Blobel and Sabatini in 1971, posits that the cellular destination of a protein is often encoded by cis signals, termed signal sequences, embedded within the nascent polypeptide (15). These signal sequences recruit specific cellular targeting machineries, which in turn mediate the targeted delivery of the nascent protein to the proper cellular destination. In the subsequent three decades, various signal sequences have been identified that encode for localization of nascent proteins to distinct subcellular organelles (Fig. 1b): hydrophobic signal sequences direct the co-translational targeting of proteins to the eukaryotic endoplasmic reticulum or the bacterial periplasmic membrane (142) (see details below); short stretches of amino acids rich in basic residues comprise different types of nuclear localization signals (NLS), which target folded proteins for nuclear import and export by virtue of their interaction with karyopherins (61, 154); mitochondrial targeting peptides are often comprised of positively charged, amphiphilic helices, which can post-translationally direct mitochondria precursor proteins to translocation machineries on the mitochondrial outer and inner membranes (31); N-terminal ‘transit peptides’ rich in hydroxylated residues mediate the post-translational targeting of proteins to the chloroplast translocation machinery for import into the chloroplast stroma (34, 80). The discovery of these distinct classes of signal sequences, and the numerous protein targeting machineries and pathways that utilize them, provide strong evidence for the ‘Signal Hypothesis’ as a general strategy for mediating the localization of proteins to their correct subcellular organelles.
Nevertheless, a quantitative understanding of the molecular mechanisms by which highly accurate substrate selection is achieved during protein localization has been challenging for multiple reasons. First, signal sequences tend to be highly divergent in length, shape, and amino acid composition. For example, signal sequences that engage the SRP are characterized by a core of 8–12 hydrophobic amino acids facilitated by basic residues at the N-terminus (50, 142) and the propensity to adopt α-helical structures (70, 148), but otherwise lack a consensus sequence motif. Thus, targeting machineries like SRP must be sufficiently adaptable to accommodate a variety of degenerate signal sequences (13, 50, 142, 164). Analogous challenges are faced by molecular chaperones, other protein targeting factors, and quality control machinery. The simplistic view that stereospecific complementarity between a substrate and its binding site give rise to high selectivity would be difficult to apply to protein biogenesis pathways. Second, only minor differences distinguish SRP-dependent signal sequences from related ones, for example the signal sequences that engage the post-translational Sec pathway for secretion in bacteria (164). Although a threshold level of hydrophobicity in signal sequences was generally thought to specify the SRP pathway, it has been difficult to define such a ‘threshold’ for SRP-dependent signal sequences (3, 66). Thus despite its flexibility, the SRP must remain highly specific to its correct substrates and be able to effectively discriminate against incorrect substrates based on minor differences. Third, targeting factors are often present in catalytic amounts relative to its cargo proteins and must cycle rapidly between the cytosol and target membrane. For example, translating ribosomes are present at concentrations of 40 – 50 μM in vivo, over 100-fold higher than that of the SRP. Further, the nascent polypeptide loses its competence to be targeted by the SRP when it exceeds a critical length of ~140 amino acids (40, 78, 131); this sets a ‘timer’ during which SRP must complete the targeting reaction. Given the molecular crowding in the cellular environment and the limited time window of action, how the SRP and other protein targeting machineries achieve accurate and timely selection of the correct set of substrates has been a challenging question to address.
In the sections below, we describe recent work that elucidates how high fidelity is achieved during the co-translational targeting of proteins by the SRP pathway (Fig. 1, route 2). The lessons from this pathway draw intriguing analogies to those from the DNA and RNA polymerases, tRNA synthetases, and tRNA selection by the ribosome, suggesting that they could provide generalizable concepts to understand how highly accurate substrate selection is achieved during the biogenesis of nascent proteins in general. Many of these advances in our understanding of the SRP were enabled by the development of high-resolution biophysical assays, which allow quantitative measurements of the efficiency of individual molecular steps in this pathway. These assays are also reviewed here, as they offer generalizable approaches to decipher how efficiency and specificity is achieved during protein biogenesis at both the conceptual and experimental level.
SEQUENTIAL CHECKPOINTS GOVERN FIDELITY OF CO-TRANSLATIONAL PROTEIN TARGETING
Overview of the SRP: composition and interaction partners
The SRP is an ancient and essential ribonucleoprotein particle conserved across all kingdoms of life (for comprehensive reviews, see (2, 73, 146)). In bacterial cells, the SRP is thought to mediate the delivery of integral membrane proteins and a subset of the periplasmic proteome to the cytoplasmic membrane. In higher eukaryotic cells, the SRP becomes the major pathway for delivering membrane and secretory proteins to the endoplasmic reticulum. Although the size and composition of SRP vary significantly across species, the highly simplified bacterial SRP and SRP receptor can replace their mammalian homologues to mediate efficient targeting of mammalian substrates to ER microsomes (14, 110). This demonstrates the remarkable evolutionary conservation of this pathway, and allows the bacterial SRP to serve as a model system for detailed mechanistic dissection of the functional core of this targeting machine.
The bacterial SRP contains the universally conserved SRP54 protein (called Ffh in bacteria) in complex with the 4.5S SRP RNA. Ffh contains two structurally and functionally distinct domains connected by a 30 amino acid linker (Fig. 1c): a methionine-rich M-domain that recognizes signal sequences (55, 68, 74) and binds, with picomolar affinity, to the SRP RNA (7, 68, 74); and an NG-domain comprised of a helical N-domain and a central GTPase, G-domain that together form a structural and functional unit ((43, 44, 91) and references therein). The N-domain mediates interactions with the ribosome (53, 57, 106, 121), and the G-domain mediates GTP binding and hydrolysis by the SRP. Collectively, the NG-domain of Ffh also interacts directly with a highly homologous NG-domain in the SR (32, 41) (Fig. 2c). The bacterial SR, called FtsY, also contains an N-terminal acidic A-domain, which mediates the peripheral association of this receptor with the phospholipid membrane and with the SecYEG translocation machinery (100, 149).
Cargo recognition by the SRP is insufficient to ensure fidelity
Timely and effective capture of cargo is essential for proper initiation of protein targeting. The cargos recognized by the SRP are translating ribosomes bearing signal sequences near the N-terminus of the nascent polypeptide (termed the ribosome-nascent chain complex or RNC). Extensive crosslinking (53, 106, 165), cryoEM (56, 57, 121), and crystallographic (55, 68, 74) analyses showed that SRP54/Ffh recognizes its cargo via a bi-dentate interaction: the signal sequence binds to a groove in the Ffh/SRP54 M-domain comprised almost exclusively of hydrophobic residues, while the protein L23 adjacent to the ribosome exit site interacts with conserved basic residues at the ‘tip’ of the Ffh N-domain. In addition, the Ffh N-domain makes limited contacts with the L29 protein in the vicinity of the ribosome exit site, and its M-domain also contacts ribosomal RNAs and perhaps ribosomal proteins L22 and L24 (56, 57, 121), although these contacts remain to be verified biochemically.
Earlier work, largely based on co-sedimentation and crosslinking data, have led to a simplest view in which the fidelity of co-translational protein targeting could be derived primarily from the weaker binding of SRP to ‘incorrect’ cargos – RNCs bearing no or weak signal sequences. However, a direct experimental test of this model has been challenging due to the lack of quantitative assays that directly report on the energetics of the RNC-SRP interactions. To overcome this problem, several types of biophysical assays have been developed in the last decade mostly based on fluorescence spectroscopy (40, 63, 120, 160). In a recent development (120), amber suppression technology was used to site-specifically incorporate a fluorescent non-natural amino acid (red in Fig. 2a), into the nascent protein during translation. Efficient fluorescence resonance energy transfer (FRET) was observed between the donor dye labeled at the signal sequence on the RNC and an acceptor dye labeled at the Ffh M-domain (Fig. 2b), providing a robust and sensitive assay to measure the equilibrium and kinetics of the SRP-cargo interactions (120).
To test the specificity during the cargo recognition step by the SRP, several groups have compared the binding affinities of SRP for RNCs bearing SRP-dependent and SRP-independent substrates (16, 40, 63, 160). In a systematic study (160), the hydrophobic core of the signal sequence of alkaline phosphatase (phoA), a borderline substrate that primarily uses the post-translational SecB/A pathway (65, 139), was replaced with a combination of leucine and alanine (Fig. 2c). The Leu/Ala ratio was systematically varied to generate signal sequences with varying hydrophobicity (Fig. 2c). Two additional SRP-independent substrates were used as negative controls in this study: (i) the autotransporter EspP, which, despite the presence of a fairly hydrophobic signal sequence, was able to escape the SRP pathway due to an unusual N-terminal extension (Fig. 2c, blue) (104); and (ii) firefly luciferase, a cytosolic protein without a recognizable signal sequence.
A combination of fluorescence anisotropy (160) and FRET (Fig. 2a, b; (120)) measurements showed that, consistent with previous observations, SRP binds tightly to the correct cargos, with equilibrium dissociation constants (Kd) in the subnanomolar to low nanomolar range for RNCs with more than 7 leucines in the signal sequence (Fig. 2d). This binding affinity weakens up to 102-fold as the signal sequence is weakened (Fig. 2d). Similar observations were made in earlier measurements with the mammalian SRP based on an environmentally sensitive fluorescent dye labeled at the signal sequence on the RNC (40), and in a more recent study that uses FRET between donor labeled at L23 and acceptor labeled on the SRP to measure ribosome-SRP binding (63). This latter study further showed that the difference in binding equilibrium derives largely from differences in the kinetic stability of the RNC•SRP complex: SRP can bind rapidly to RNCs bearing both SRP-dependent and SRP-independent substrates, but it dissociates much more slowly from the former.
These studies also showed that even the weakest cargos or empty ribosomes bind the SRP with significant affinity, with Kd values in the 80 – 100 nM range (Fig. 2d; (40, 63, 160)). Thus, the interaction of SRP with the ribosome provides a significant driving force for the recruitment of SRP to RNCs. Given the cellular concentrations of SRP (~400 nM in bacteria) and ribosomes (40–50 μM), it is unlikely that the observed differences in cargo binding affinity are sufficient for SRP to discriminate against the incorrect cargos. Most unexpectedly, RNC bearing an SRP-independent substrate, EspP, binds the SRP as tightly as a correct cargo (3A7L) (Fig. 2d). These results strongly suggest that the initial cargo-binding step is insufficient to ensure the accurate selection of substrates by the SRP and hence, subsequent steps in the targeting pathway must provide additional fidelity checkpoints to reject the incorrect cargos. These additional fidelity checkpoints are provided by the SRP and SR GTPases, as described in the next sections.
A novel GTPase cycle for the SRP and SR
The twin GTPases in the SRP and SR represent a novel class of nucleotide hydrolases whose biological activity is regulated by nucleotide-dependent dimerization cycles (6, 24, 47, 82). They are distinguished from the classic signaling GTPases, such as Ras (17, 51), in several aspects: (i) Free Ffh and FtsY exhibit minor structural differences amongst the apo, GDP-, and GTP-bound states (44, 45, 48, 91, 99, 115). Hence, the exchange between nucleotide states per se cannot provide the mechanism to regulate these GTPases. (ii) Even with GTP bound, both Ffh and FtsY by themselves are in an inactive open conformation, exhibiting weak nucleotide affinities and rapid nucleotide dissociation rates (103). Hence, they forego the need for an external Guanine Nucleotide Exchange Factor (GEF) to convert them from the GDP- to GTP-bound state, and the recruitment of a GEF cannot serve as a mechanism to turn these GTPases to the ‘on’-state. (iii) They reciprocally activate the GTPase (or ATPase) activity of one another (103, 108), and thus forego the need to recruit an external GTPase Activating Protein (GAP) to convert them from the GTP- to GDP-bound state. Hence, the recruitment of an external GAP cannot serve as a mechanism to turn these GTPases to the ‘off’-state either.
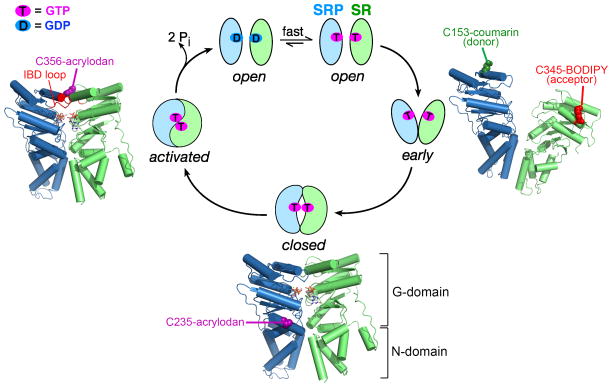
Instead, the GTPase cycles of SRP and SR are driven by a series of discrete conformational changes during their dimerization that culminates in their reciprocal GTPase activation (Fig. 3). Dimerization between the Ffh and FtsY NG domains begins with the assembly of a transient early intermediate, which forms rapidly but is highly unstable (Kd ~ 4–10 μM; Fig. 3, step 1)(158). This intermediate is primarily driven by electrostatic attractions between the N-domains of Ffh and FtsY but lacks stable contacts between their G-domains, and hence can form independently of GTP (Fig. 3, right)(35, 159). Subsequent rearrangements, involving readjustments at the intramolecular N-G domain interface (32, 41, 124, 126) and the removal of an inhibitory N-terminal helix (49, 94, 130), generates a stable closed complex in which the Ffh and FtsY G-domains establish extensive contacts with one another (Kd ~16 – 30 nM; Fig. 3, step 2 and bottom). At the dimer interface, the two GTP molecules directly hydrogen bond via their 3′-OH and γ-phosphoryl oxygens, which further stabilizes the closed complex and confers its specificity for GTP (32, 41). The final GTPase activation step involves readjustments of a conserved catalytic loop, termed the Insertion Box Domain (IBD) loop, in both proteins, during which multiple catalytic residues are positioned to coordinate the GTP molecules, the nucleophilic water and the active site Mg2+ (Fig. 3, step 3). This generates an activated complex conducive to efficient GTP hydrolysis (32, 41, 126). The hydrolysis of GTP then drives the irreversible disassembly of the SRP•FtsY complex (Fig. 3, step 4; (25, 103)), thus completing the cycle.
Figure 3.
A series of conformational rearrangements in the SRP•FtsY dimer drive their GTPase cycle. The SRP and SR NG-domains are in green and blue, respectively. (Right) molecular model of the early intermediate [Protein Data Bank (PDB) 2XKV]. (Bottom and left) co-crystal structure of the Ffh-FtsY NG domain complex in the closed/activated conformation (PDB: 1RJ9). The positions of fluorescence probes that detect each conformational state are depicted in the structures.
A high-resolution understanding of this GTPase cycle was made possible by several complementary sets of tools. First, a set of mutant GTPases and GTP analogues was isolated that specifically lock the SRP•SR GTPase complex at distinct conformational stages (126, 158). This allowed characterization of the structure, energetics, and dynamics of each conformational state during the dimerization cycle of SRP and SR (35, 158, 159, 161). Second, a complete set of fluorescence probes was developed that detect specific conformational states in the GTPase dimer. For example, FRET between donor (coumarin) and acceptor (BODIPY-Fl) probes labeled in the Ffh and FtsY G-domains, respectively, provides a highly sensitive assay that enabled detection of the transient early intermediate (Fig. 3, right panel)(63, 158). Further, this intermediate exhibits a lower FRET value than the closed and activated complexes, so that it can be distinguished from the subsequent conformations (158). An environmentally sensitive probe, acrylodan labeled at residue 235 of Ffh, detects the N-G domain readjustment during the early-to-closed rearrangement and thus specifically detects the closed and activated complexes (Fig. 3, bottom) (161). Finally, acrylodan labeled at residue 356 of FtsY, near its IBD loop, specifically detects the activated complex (Fig. 3, left panel) (161). Importantly, these tools revealed that each of the conformational changes during the SRP/SR GTPase cycle is extensively regulated by the cargo protein and the target membrane (1, 79, 161). These allosteric regulations not only enhance the efficiency of the protein targeting reaction, but also provide additional fidelity checkpoints that help reject the incorrect cargos.
Correct cargos drive rapid assembly of the SRP-SR GTPase complex
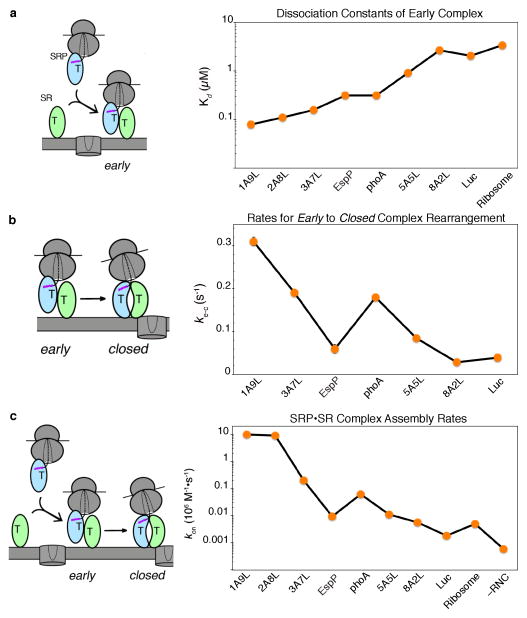
Once a cargo is loaded on the SRP, it must be rapidly delivered to the target membrane via the interaction between the SRP and SR GTPases. However in the absence of any biological cues, the assembly of a stable closed SRP•FtsY GTPase complex is extremely slow (kon ~102–103 M−1s−1; (18, 102, 103)) and insufficient to support protein targeting. These slow rates are primarily due to the labile nature of the early intermediate, >98% of which dissociates before it rearranges into the stable closed complex (158). A resolution to this problem came from the finding that a correct cargo bearing a bona-fide SRP substrate FtsQ (RNCFtsQ) stabilizes the early intermediate over 100-fold (161), such that a RNC•SRP•SR early targeting intermediate can be sufficiently stable to accumulate under physiological conditions. Importantly, stabilization of the early intermediate is observed specifically with the correct cargos, whereas the early targeting complexes formed by incorrect cargos are up to 50-fold less stable (Fig. 4a, step 2) (160). Compared to the correct cargos, the early intermediates formed with the incorrect cargos also appear to be mispositioned and less productive: they exhibit lower FRET efficiencies between donor- and acceptor-labeled Ffh and FtsY (Fig. 3, right), and mediate the early → closed rearrangement 5–10 fold more slowly than the correct cargos (Fig. 4b, step 3)(143, 160). Thus, correct cargos mediate the formation of more stable and productive early targeting intermediates.
Figure 4.
Correct cargos stabilize the early intermediate and mediate faster rearrangement to the closed complex, and thus enabling faster assembly of a stable SRP-FtsY complex. (a) Summary of the equilibrium dissociation constants (Kd) of the early intermediate formed with different cargos (adapted from Fig. 2C in ref. (160)). (b) Summary of the rate constants for the conformational rearrangement from the early to closed complex mediated by the different cargos (adapted from Fig. 2G in ref. (160)). (c) Rate constants for assembly of the stable, GTP-dependent closed complex mediated by the different cargos (adapted from Fig. 3C in ref. (160)).
As would be expected from the combination of these effects, measurements using several independent probes showed that the correct cargos mediate much more rapid assembly of the stable closed SRP•FtsY complex, with up to ~103-fold discrimination between the strongest (RNC1A9L) and weakest (RNCEspP and RNC8A2L) cargos (Fig. 4c). Consistent with these observations, a more recent study showed that RNC bearing the nascent chain of leader peptidase, an SRP-dependent substrate, mediate faster SRP-FtsY complex assembly than non-translating ribosomes (63). In addition, signal peptides or signal sequence fused to SRP54 partially mimic the effect of RNC and stimulate more rapid SRP-FtsY complex assembly (18, 55). Collectively, these results demonstrate that the correct cargos can be rapidly delivered to the target membrane once they are loaded on the SRP, whereas the targeting complexes formed with incorrect cargos are more likely to disassemble and thus exit the SRP pathway prematurely before arrival at the membrane (Fig 6a, steps 2 and 3).
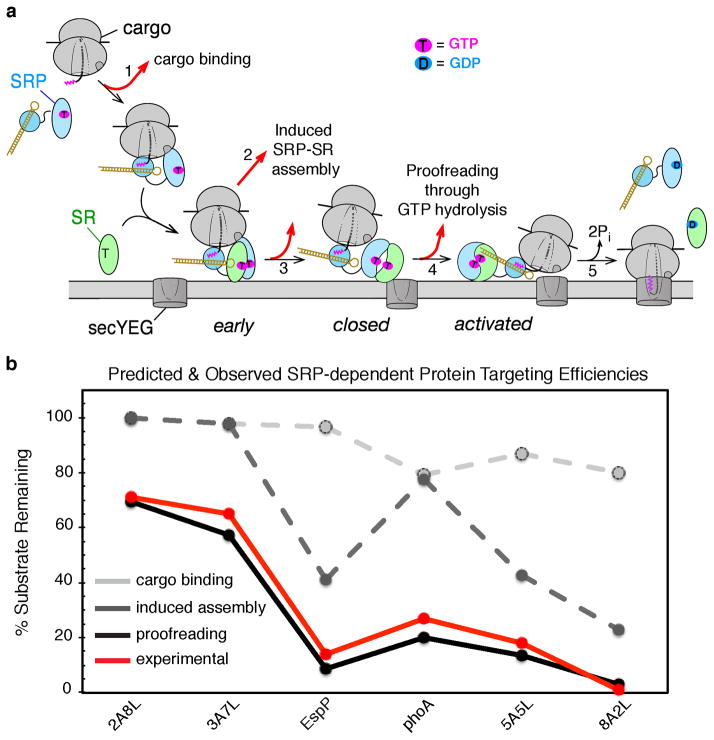
Figure 6.
A sequential series of checkpoints reject incorrect cargos from the SRP pathway. (a) Model for how conformational rearrangements in the SRP/SR GTPases provide the driving force and ensure the fidelity of protein targeting. Step 1, a cargo with a signal sequence (magenta) enters the pathway upon binding SRP. Step 2, the cargo-bound SRP forms a stabilized early intermediate with FtsY. Step 3, association of FtsY with membrane drives the rearrangements from the early intermediate to the closed complex. Step 4, the SecYEG translocon promotes conformational rearrangements that drive GTPase activation and cargo handover. Step 5, the cargo is unloaded from the SRP onto SecYEG, and GTP hydrolysis drives the disassembly and recycling of SRP and FtsY. At each step, the cargo can be either retained in (black arrows) or rejected (red arrows) from the pathway. Color codings are the same as in Figure 3. (b) Predicted fraction of cargos retained in the SRP pathway after cargo binding (light grey), induced SRP-SR assembly (dark grey), and kinetic proofreading through GTP hydrolysis (black). The experimentally determined protein targeting efficiencies are shown in red.
Kinetic proofreading through GTP hydrolysis
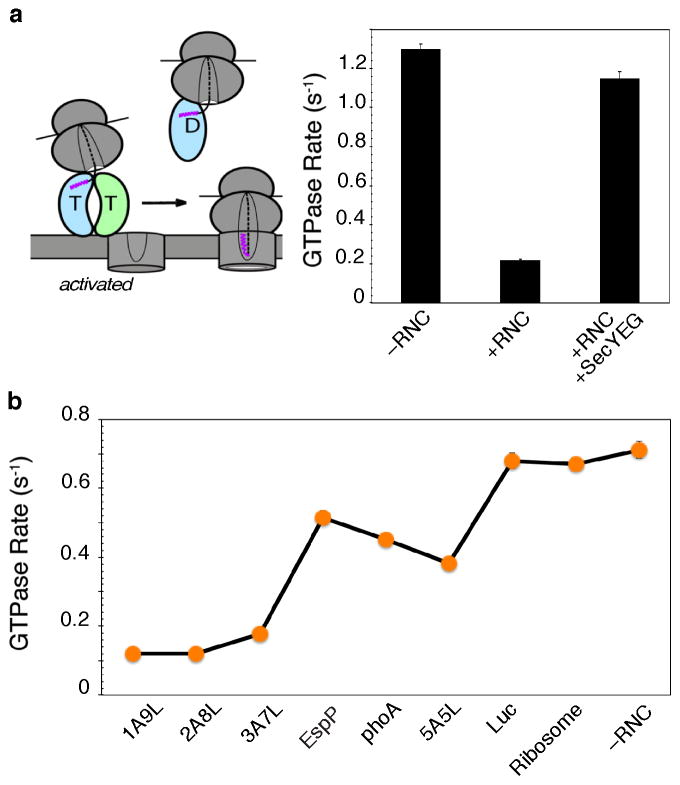
The timing of GTP hydrolysis in the SRP•FtsY complex is critical during the protein targeting reaction. On the one hand, efficient GTP hydrolysis is needed at the end of the reaction to recycle the SRP and SR for additional rounds of targeting (125, 152). On the other hand, premature GTP hydrolysis would lead to abortive targeting reactions before the RNC•SRP•SR targeting complex identifies and successfully transfers the cargo to the SecYEG translocation machinery on the membrane. By itself, a stable SRP•FtsY complex hydrolyzes GTP rapidly (Fig. 5a), with a lifetime of τ ≤ 1 s (103). Importantly, a correct cargo such as RNCFtsQ delays the conformational rearrangement that leads to GTPase activation, such that the observed GTP hydrolysis rate is slowed 6–8 fold (Fig. 5a) (161). This effect, termed ‘pausing’, extends the lifetime of the targeting complex from ≤ 1 s to ~5 s, providing it with an important time window to search for the target membrane and the SecYEG translocon before GTP hydrolysis drives the irreversible dissociation of the targeting complex (Fig. 5a). Consistent with this hypothesis, delayed GTPase activation in the RNC•SRP•SR complex can be reversed by the SecYEG complex (Fig. 5a) (1), indicating that the timing of GTP hydrolysis is exquisitely regulated during the targeting reaction and tightly coupled to the unloading of cargo at the target membrane (Fig. 6a, step 4; more discussions below).
Figure 5.
Correct cargos delay GTP hydrolysis. (a) The correct cargo delays GTP hydrolysis from the RNC-SRP-FtsY complex, which is restored by the SecYEG translocon. (b) Summary of GTPase rate constants from the RNC•SRP•FtsY complex mediated by the different cargos.
Remarkably, the delay of GTPase activation is observed specifically with correct cargos bearing strong signal sequences, but becomes much less pronounced with the incorrect cargos (Fig. 5b)(160). Effectively, GTP hydrolysis is used to set a differential timer for the different cargos: those with SRP-dependent signal sequences have a much longer time window to search for and locate the SecYEG machinery, whereas those with SRP-independent substrates are more likely to be rejected through premature GTP hydrolysis (Fig. 6a, step 4). Such a kinetic partitioning mechanism utilizing the energy of GTP hydrolysis is conceptually analogous to the kinetic proofreading mechanisms observed during tRNA selection by the ribosome described in the INTRODUCTION (116, 117).
Multiple fidelity checkpoints govern substrate selection by the SRP
The quantitative dissection of the bacterial SRP pathway described above supports a novel model in which the fidelity of the protein targeting reaction is achieved through a combination of mechanisms including preferential binding, induced fit, and kinetic proofreading (Fig. 6a). A mathematical simulation, based on the rate and equilibrium constants of the individual steps, the rate of nascent polypeptide elongation, and the cellular concentrations of SRP and FtsY, illustrates how the incorrect cargos are rejected through a sequential series of checkpoints in the pathway including: (i) weaker binding to the SRP (Fig. 6a, step 1 and Fig. 6b, light grey line), (ii) slower delivery to the target membrane (Fig. 6a, steps 2 & 3 and Fig. 6b, dark grey line); and (iii) premature GTP hydrolysis prior to productive unloading of the cargo (Fig. 6a, step 4 and Fig. 6b, black solid line). Remarkably, the results of this simulation faithfully reproduced the experimentally observed pattern of substrate selection in a reconstituted protein targeting assay (Fig. 6b, red line) (160), suggesting that all these fidelity checkpoints are required to effectively reject the incorrect cargos from the pathway.
A critical factor that contributes to the fidelity of the SRP is kinetic competition of the targeting pathway with the elongation of the nascent polypeptide by the ribosome. Multiple lines of evidence have suggested that SRP loses its targeting competence when the nascent proteins exceeds a critical length of ~140 amino acids (39, 133). This gives the SRP pathway a limited time window of 3–5 seconds to complete the protein targeting reaction, such that incorrect cargos that are delivered to the membrane more slowly and hence miss this time window are irreversibly rejected. In vitro and in vivo targeting experiments show that reducing the rate of translation elongation can rescue substrate proteins bearing mutant signal sequences that are otherwise sub-optimal in co-translational protein targeting (78, 96, 157). Similar observations were made either when the SRP subunits were depleted, or when the kinetics of SRP-receptor binding was compromised (96, 157). These data support the earlier proposals by Gierasch (164) and Rapoport (114) in which co-translational protein targeting is in kinetic competition with ongoing translation and contributes to substrate selection by the SRP.
Although a good understanding has been reached for how the SRP achieves accurate substrate selection in a single round of protein targeting, additional mechanisms could contribute to the fidelity of the SRP pathway in vivo. First, competition between the strong and weak cargos may lower the effective concentration of free SRP in vivo; this would allow the differences in SRP’s cargo binding affinities to maker a greater contribution to specificity. Second, during multiple rounds of protein targeting, the faster SRP-FtsY interaction kinetics driven by the correct, than the incorrect, cargos will allow a larger number of the former to be targeted within a given time window. Third, SRP-FtsY interaction kinetics could be slower in vivo than in vitro, as protein diffusion rates tend to be slower within the crowded environment of the cell. This would render the SRP-SR complex assembly step more rate-limiting for the targeting reaction in vivo and thus increase its contribution to the rejection of borderline substrates such as phoA. Finally, the SecYEG translocation machinery, which also provides a binding site for signal sequences (30, 113), could provide an additional fidelity checkpoint to further reject proteins that lack a signal sequence (71). The contribution of these additional factors to the fidelity of the SRP remains to be evaluated.
STRUCTURAL AND MOLECULAR BASIS OF FIDELITY CONTROL
As described above, the fidelity of the SRP pathway is derived in large part by a series of signal sequence-driven conformational rearrangements in the SRP and SR GTPases. Nevertheless, the signal sequence binding site and the GTPase module of SRP are located on two distinct domains, M and NG, that are connected by a 30-amino acid long linker (Fig. 1c). Direct interaction between these two domains has not been demonstrated. This raises the question: how does the SRP sense the information about signal sequence occupancy in its M-domain and transmit this information to its GTPase domains? Recent biochemical, biophysical and structural studies strongly suggest that this role is fulfilled by the other essential and universally conserved component of the SRP, the SRP RNA.
The SRP RNA mediates substrate-induced reorganization of the SRP
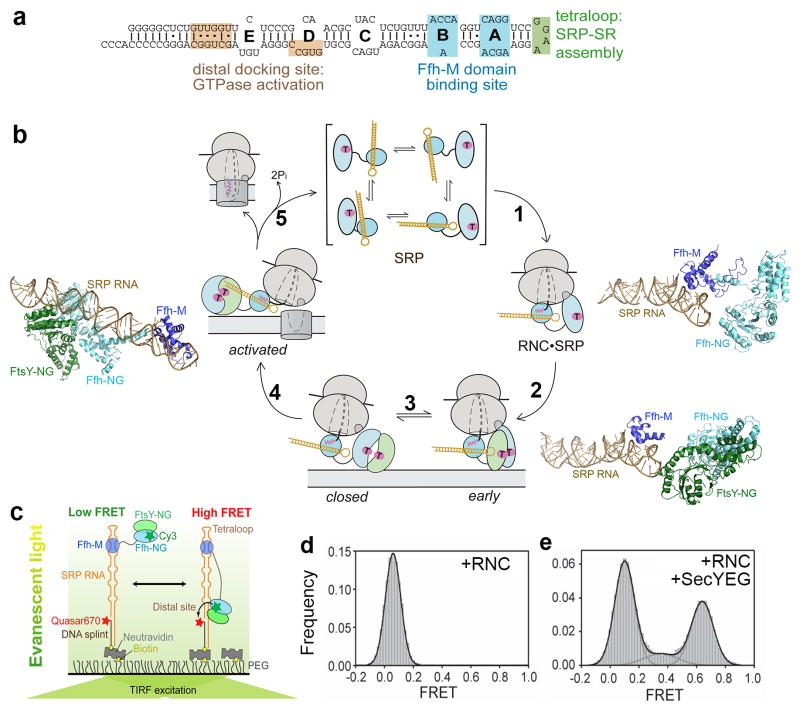
When the mammalian SRP was discovered, the SRP RNA was considered largely as a scaffold that holds multiple SRP protein subunits together in a ribonucleoprotein complex (132, 144, 145). The discovery of the bacterial SRP (13, 14), in which the SRP RNA binds a single protein Ffh, strongly argued against this simple view and suggested a more active role for this RNA. The bacterial 4.5S SRP RNA shares the most evolutionarily conserved domain IV of the SRP RNA, which forms a stable hairpin structure capped by a highly conserved GGAA tetraloop at one end (Fig. 7a). Two internal loops, A and B, adjacent to the tetraloop mediate binding of the SRP RNA to the M-domain of Ffh with picomolar affinity (Fig. 7a) (7, 8). Despite the lack of additional stable interactions between the SRP RNA and the Ffh NG-domain, kinetic analyses demonstrated that the SRP RNA can communicate with the SRP and SR GTPases: it catalyzes the rapid formation of the stable SRP•FtsY GTPase complex, accelerating this otherwise extremely slow process ~102-fold (102, 103). More recent biochemical, structural, and phylogenetic analyses identified a critical interaction between the GGAA tetraloop of the SRP RNA and the conserved basic residues surrounding Lys399 on the lateral surface of FtsY (35, 127, 134). This interaction strongly stabilizes the otherwise highly labile early intermediate, and thus accelerates the formation of the stable closed complex between SRP and FtsY (127, 158).
Figure 7.
SRP RNA-mediated global rearrangement of SRP couples cargo loading and unloading events to the GTPase cycle during protein targeting. (a) Secondary structure of the Escherichia coli 4.5S SRP RNA. The binding sites for the Ffh-M domain (blue) and for FtsY during the GTPase assembly (green) and activation (tan) steps are denoted. (b) Global rearrangement of SRP mediated by the SRP RNA during the protein targeting cycle. Top, free SRP exist in a variety of ‘latent’ conformations not conducive to the recruitment of FtsY. Right, binding of RNC induces a more active conformation of SRP (step 1), in which the SRP RNA tetraloop is properly positioned to interact with the G-domain of SR and hence form a stabilized early targeting complex (step 2). Molecular models derived from cyro-EM reconstructions are shown for the RNC•SRP (right panel) and RNC•SRP•SR early complex (lower right panel); the ribosome was not shown for clarity. Bottom, the GTPases detach from the SRP RNA tetraloop upon formation of the closed complex (step 3). Left, the GTPase complex relocalizes to the distal end of the SRP RNA (step 4), a conformation (left panel; PDB 2XXA) conducive to GTPase activation and cargo unloading (Step 5). All structures are aligned with respect to the SRP RNA. Color codings are the same as in Figure 3. The steps are numbered to be consistent with Figure 6a. (c) The smFRET setup to monitor the dynamic movements of the SRP-FtsY GTPase complex on the SRP RNA. FtsY Cys345 is labeled with Cy3, and the 5′end of the DNA splint is labeled with Quasar 670. (d–e) RNC and SecYEG regulate GTPase movement on the SRP RNA, as shown by the smFRET histograms of the SRP-FtsY complex bound to RNCFtsQ in the absence (d) and presence (e) of the SecYEG translocon. Adapted from reference (128).
Importantly, a series of structural studies strongly suggest that this key interaction is made only when the SRP is bound to ribosomes translating an SRP-dependent substrate. In the free SRP, the orientation of the Ffh M-domain/RNA complex relative to its NG-domain can be highly variable, likely due to the long linker connecting the M- and NG-domains of Ffh. Crystallographic analyses and structural mapping studies have observed at least four different Ffh structures, each showing a distinct inter-domain organization, and the GGAA tetraloop of the SRP RNA points away from the potential Ffh-FtsY interaction surface in most of these structures (Fig. 7b, SRP structural ensemble) (22, 23, 54, 74, 85, 118). Thus, the key interaction between the RNA tetraloop and FtsY-Lys399 would not be favored during the interaction of FtsY with the free SRP. In contrast, cryo-EM analyses of the RNC-SRP complex (21, 57, 121) show that the bidentate interactions of the Ffh M- and NG-domains with the signal sequence and with ribosomal proteins L23/L29 reorganize these domains, allowing the SRP RNA to lie in parallel to the ribosomal surface with its GGAA tetraloop positioned adjacent to the FtsY-interacting surface on the Ffh NG-domain (Fig. 7b, RNC•SRP complex). In this conformation, only modest re-adjustment of SRP is needed for the GGAA tetraloop to establish close contact with Lys399 on the incoming FtsY (Fig. 7b, early intermediate) (35). Thus, binding of a correct cargo optimizes the conformation of SRP, allowing it to establish the key interaction between the RNA tetraloop and FtsY-Lys399 (Fig. 7b, steps 1–2).
Consistent with these structural insights, biochemical analyses show that the ability of the SRP RNA to stabilize the early targeting complex and to accelerate stable SRP-FtsY assembly is only observed when the SRP is bound to correct cargos (127, 129) and, to a lesser extent, to signal peptide or detergents that mimic the effect of signal peptide (18, 55). Incorrect cargos, represented by RNCluciferase and RNCEspP, cannot enable the interaction between FtsY-Lys399 and the GGAA tetraloop to be effectively established and are hence incapable of mediating rapid assembly of the targeting complex (127, 129, 143). A recent cryo-EM analysis of a ‘false’ early targeting complex formed by an incorrect cargo, RNCEsp (143), shows that the Ffh and FtsY NG-domains form a distorted, more flexible complex compared to that observed with a correct cargo, RNCFtsQ. In addition, the FtsY NG-domain is tilted and makes less well-defined contacts with the SRP RNA tetraloop in the ‘false’ early targeting complex. These structural analyses, combined with biochemical evidence, provide a coherent model in which the correct but not the incorrect cargos optimize the conformation of SRP so that the SRP RNA tetraloop is pre-positioned to interact with the incoming FtsY, thus allowing rapid recruitment of the SR to be achieved specifically for the correct cargos.
GTPase movement to the RNA distal end activates GTP hydrolysis
Despite the important role in stabilizing the early intermediate, the SRP RNA tetraloop has no effect on the equilibrium stability of the SRP-FstY complex in the closed/activated states (102, 103), suggesting that this interaction is highly transient and exists only in the early intermediate. Consistent with this notion, the density for the NG-domains of SRP and SR becomes invisible in a cryo-EM structure of the eukaryotic RNC•SRP•SR complex formed with a non-hydrolyzable GTP analogue (58), suggesting their detachment from the tetraloop of the SRP RNA in the vicinity of the ribosome exit site. A recent crystallographic study (5) trapped the SRP•FtsY complex in the closed/activated state and showed that the Ffh•FtsY NG-domain complex is located at the 5′, 3′-distal end of the SRP RNA (Fig. 7a), ~100 Å away from the tetraloop end (Fig. 7b, activated). Biochemical studies identified specific sites at the SRP RNA distal end that stimulates GTP hydrolysis in the activated complex (5), supporting the importance of this alternative RNA-GTPase interaction. Collectively, these results suggest that after the SRP•SR complex initially assembles at the tetraloop end of the SRP RNA, it subsequently docks at the opposite end of the RNA where GTP hydrolysis is fully activated.
These results imply that attainment of the activated state in the Ffh•FtsY GTPase complex is coupled to a global movement of this complex along the two functional ends of the SRP RNA. This large-scale movement is directly demonstrated in a recent study using single-molecule fluorescence-TIRF (total internal reflection) microscopy (128). Individual SRP molecules were immobilized on microscope slides via a DNA splint that hybridizes to a 3′-extension from the SRP RNA (Fig. 7c). The location of the Ffh•FtsY NG-domain complex on the SRP RNA is tracked using FRET between a donor dye on the Ffh or FtsY NG-domain and an acceptor dye near the RNA distal end (Fig. 7c). The analysis showed that occurrence of stable high FRET signals arise specifically under conditions that allow formation of the activated complex, and the frequency of attaining the high FRET state directly correlates with observed GTP hydrolysis rates, providing unequivocal evidence that movement to the SRP RNA distal site is responsible for GTPase activation (128).
Most importantly, this GTPase movement to the RNA distal end is almost completely inhibited by a correct, but not an incorrect, cargo (Fig. 7d) (128). On the other hand, addition of purified SecYEG translocon to the RNC•SRP•FtsY complex effectively restores the occurrence of high FRET states (Fig. 7e)(128), suggesting that the GTPase movement to the RNA distal end is specifically induced at the membrane when the targeting complex encounters the translocation machinery (Fig. 7b, step 4). These results provide a molecular explanation for the effects of these machineries on the observed GTP hydrolysis activity of the SRP•FtsY complex (Fig. 6a). Because both SecYEG and the SRP NG-domain contact the ribosome via the L23 protein and hence must compete for overlapping binding sites (10, 11, 42, 90), the movement of the GTPase complex away from the ribosome exit (while its M-domain is still bound to the signal sequence) also suggests an attractive mechanism for how these two machineries could productively exchange at the ribosome exit site without irreversible loss of cargo (Fig. 6a and 7b, steps 4–5). Most importantly, these results provide an attractive model for how the timing of GTP hydrolysis is spatially and temporally regulated during the targeting reaction and coupled to the eminent unloading of the cargo from the targeting to the translocation machinery.
In summary, the SRP RNA is responsible for mediating the extensive allosteric regulations that the cargo exerts on the SRP and FtsY GTPases, thus providing the molecular linker that couples the loading of a correct cargo to its rapid delivery to and productive unloading at the membrane. As one of the most ancient ribonucleoprotein complexes, studies of the bacterial SRP also demonstrates how a large RNA molecule can provide an active scaffold, not only for holding multiple proteins together but also for coordinating large-scale protein rearrangements, the exchanges of distinct factors, and the precise timing of these events in a complex cellular process. These lessons resonate well with those from the spliceosome (10, 11, 42, 90), and may provide general concepts to understand the role of other large functional RNAs in biology.
PERSPECTIVE
In summary, recent advances in biochemical and biophysical dissections of protein biogenesis pathways make it possible to begin to decipher the molecular basis underlying fidelity of these processes. Although the molecular details differ significantly between protein localization by the SRP and the decoding of genetic information by the polymerases, tRNA synthetases and the ribosome, striking conceptual analogies can be found between the mechanisms that ensure fidelity in all these processes. A recurring theme appears to be the presence of multiple fidelity checkpoints in a pathway, such that incorrect substrates can be discriminated at multiple stages by using a variety of mechanisms including preferential binding, induced fit, editing, and proofreading. This provides a simple and effective principle to enhance specificity, and may be particularly useful in protein biogenesis pathways in which cellular machineries must recognize a diverse set of substrates based on highly degenerate structural and sequence features, while also discriminate between correct and incorrect substrates based on minor differences.
A common strategy used by many pathways is the induced-fit mechanism, in which the correct substrate is much more effective at triggering conformational changes conducive to subsequent steps of the reaction. This can manifest as chemical steps, such as nucleotide incorporation by the polymerases and GTP hydrolysis by EF-Tu during translation, or as noncovalent steps, such as the recruitment of SRP receptor during SRP-dependent protein targeting. A key concept here is that a large fraction of the binding energy with a correct substrate is used to overcome the energetic barrier for otherwise unfavorable conformational changes in the cellular machinery; thus, the more favorable interaction with a correct substrate does not necessarily manifest, in its full extent, in a stronger binding equilibrium. Hence, it will be important experimentally to not only examine the differences in substrate occupancy between correct and incorrect substrates, but also to investigate the functional consequences of these binding interactions.
Another recurring strategy is the introduction of branch points in a pathway, at which the productive forward reaction kinetically competes with irreversible processes that abort the pathway. The conceptual framework for this mechanism has been well established: the correct substrates partition more favorably into the forward reactions than the incorrect substrates, thus amplifying the discrimination against the incorrect substrates (36). This principle was amply demonstrated as editing mechanisms in tRNA synthetases and as proofreading activity during replication, transcription, and tRNA selection by the ribosome (see INTRODUCTION). In the SRP pathway, this principle manifests at two stages: (i) during cargo delivery to the target membrane, at which SRP-FtsY assembly must compete with elongation of the nascent polypeptide; and (ii) at the target membrane, where productive cargo unloading onto the secYEG machinery must compete with GTP hydrolysis that irreversibly disassembles the targeting complex.
A conceptually analogous example has been described for the chaperone SecB that mediates post-translational protein export in bacteria (Fig. 1a, step 1b). Although SecB binds unfolded polypeptides with high affinity, the signal sequence on its substrate proteins does not mediate these interactions and hence could not contribute to selective binding (111). Instead, SecB binding kinetically competes with the irreversible folding/aggregation of the substrate protein; the signal sequence delays these latter processes and thus allows more favorable partitioning of the correct substrates into the export pathway (111). Another example of this principle is recently described for regulated protein quality control of the CD4 receptor by a viral protein Vpu (163). In this system, generation of a mismatched Vpu-CD4 interacting pair causes only minor differences in the rate of CD4 ubiquitylation; however, discrimination against the mismatched Vpu-CD4 pair is increased substantially by kinetic competition from deubiquitylation enzymes that remove the ubiquitin chain on CD4 (163). It remains to be determined whether kinetic partitioning between productive and abortive pathways is a general strategy for ensuring the fidelity of protein biogenesis and quality control pathways.
The ribosome exit site is a crowded environment where numerous protein biogenesis factors interact with the nascent protein (Fig. 1a). Within the cell, the SRP does not act in isolation, but rather must interface with many other factors at the ribosome exit site. Moreover, many of these factors (such as trigger factor, SecA, or the NAC complex) bind the ribosome at or near the same protein, L23 (75), and they preferentially interact with nascent polypeptides via hydrophobic sequences. Given these overlapping preferences, whether and how the different factors compete, collaborate, or interplay with one another via other mechanisms remain to be deciphered. Earlier in vitro analyses in cell extracts have shown that NAC could improve the discrimination of SRP against incorrect SRP substrates (109, 150). More recent studies revealed additional nuances in this interplay: a genome-wide analysis showed that NAC can both enhance or reduce the interaction of SRP with different sets of nascent proteins (3), and crosslinking studies suggest that NAC influences the SRP-cargo interactions in distinct ways depending on the nascent chain length (162). Analogously, trigger factor has been shown to modulate the interaction of SRP with translating ribosomes (9, 20, 33, 81, 138, 140). A coherent model to conceptualize how this molecular interplay contributes to the fidelity of SRP-dependent protein targeting in vivo remains to be established. More broadly, how accurate decisions are made in a timely manner to allow a nascent protein to engage the correct biogenesis factors at the ribosome exit site remain an important and intriguing question to be explored in future investigations.
Accumulating evidence suggest that the ribosome itself may provide the initial platform that examines the nascent protein and facilitate the recruitment of proper biogenesis factor(s) to the exit site. The earliest evidence for this view came from the observation that opening and closing of the Sec61p translocon can be regulated by the presence of transmembrane domains (TMD) of the nascent protein before they emerge from the exit tunnel ribosome (83). A number of more recent biochemical and crosslinking studies found that the presence of a signal sequence inside the ribosome could enhance the binding of SRP to the RNC (12, 16) and help recruit a regulatory protein RAMP4 to the Sec61p translocon (105). In the guided entry of tail-anchored protein (GET) pathway, the Bag6 complex could more favorably associate with the RNC when the C-terminal TMD of the nascent substrate protein emerges inside the ribosome (86). Less direct observations, which also support an active communication between the nascent protein and the ribosome, include the finding of ‘stall’ sequences on the nascent protein that arrest protein synthesis (27, 92) and the observation that nascent polypeptides could assume α-helical folds inside the ribosome tunnel (84, 153). These results raise the intriguing possibility that sequence or structural features of the nascent polypeptide inside the polypeptide exit tunnel could ‘signal’ the ribosome and lead to the recruitment of cellular factors. The nature of ribosome structural changes that underlie these signaling events, whether and how they contribute to the respective protein biogenesis pathways, and the mechanisms ensuring the specificity of these ‘signals’ remain intriguing questions to pursue in future studies.
SUMMARY POINTS.
The ribosome exit site is a crowded environment where the nascent protein must be efficiently and accurately sorted among multiple protein biogenesis factors.
Signal sequences enable nascent proteins to engage the correct protein targeting machinery, and thus be directed to their proper cellular destination. These topogenic sequences are highly degenerate, posing challenges to protein biogenesis machineries in achieving specificity.
SRP recognizes its cargos through bidentate interactions with the signal sequence and the ribosome. However, the SRP-cargo binding affinity is insufficient to ensure the correct selection of substrates by the SRP.
Correct cargos drive conformational rearrangements in the SRP and SRP receptor GTPases necessary for the delivery and unloading of cargo, which provide additional opportunities to reject the incorrect cargo.
The fidelity of substrate selection by the SRP is achieved through the cumulative effects of multiple checkpoints in the pathway.
The SRP RNA orchestrates global rearrangements of the SRP upon cargo binding, thus providing the molecular basis for efficient and accurate co-translational protein targeting.
Induced fit and kinetic partitioning are commonly used strategies to achieve biological specificity.
FUTURE ISSUES.
How do additional factors contribute to the fidelity of SRP in vivo?
Do other protein biogenesis pathways achieve high fidelity using conceptually analogous mechanisms?
How do multiple protein biogenesis machineries interplay at the ribosome exit site to ensure accurate sorting of the nascent protein to the correct pathway?
Does a nascent polypeptide inside the ribosome tunnel ‘signal’ the ribosome to recruit biogenesis factors, and how?
Glossary
- Protein targeting
the process of delivering newly synthesized protein to specific organelles in the cell
- Cotranslational
molecular interactions and pathways that occur while the nascent protein is still being synthesized and attached to the translating ribosome
- Post-translational
molecular interactions and pathways that occur after the nascent protein is released from the ribosome
- Translocon
protein complexes that mediate the vectorial translocation across or lateral integration of proteins into the membrane bilayer. Used interchangeably with ‘translocation machinery’ and ‘translocase’
- Signal sequence
transferable element on the nascent polypeptide that enables the protein to engage with protein targeting machineries and mediates proper localization of the protein
- Amber suppression
a technology that allows site-specific incorporation of a non-natural amino acid into a protein in response to an amber codon during translation. Most often, this utilizes an engineered tRNA that can compete with the release factor in recognizing the amber codon
- AAA+ ATPases
a family of ATPases Associated with diverse cellular Activities that mediate ATP-dependent remodeling of protein complexes or aggregates
Acronyms
- SRP
signal recognition particle
- SR
signal recognition particle receptor
- RNC
ribosome-nascent chain complex
- GTPase
guanosine 5′-triphosphate (GTP) hydrolase
- ATPase
adenosine 5′-triphosphate (ATP) hydrolase
- Hsp70
the 70-kDa heat shock proteins, a family of molecular chaperones that facilitate protein folding and biogenesis. Homologs include DnaK in bacterial and SSA and SSB in yeast cells
- Hsp90
the 90-kDa heat shock proteins
- TriC complex
the eukaryotic chaperonin that provides favorable conditions for a correct protein folding process
- NAC
nascent polypeptide associated complex, a heterodimeric complex that binds eukaryotic ribosomes in close proximity to the emerging nascent protein
- RAC
ribosome-associated complex, a yeast heterodimer complex formed by Zuotin (zuo) and Ssz, which are members of the conserved Hsp40 and Hsp70 chaperones families
- SecYEG
a trimeric protein complex consisting of the subunits SecY, SecE, and SecG, which together form the minimal component of the bacterial translocon. The homologue in mammalian cells is termed the Sec61p complex, comprised of the α, β, and γ subunits
- RQC
ribosome quality control complex, which mediates ubiquitin-dependent degradation of polypeptides on stalled ribosomes and signals stress to Hsf1
- GET
guided entry of tail-anchored proteins, a pathway that mediates the post-translational targeting of tail-anchored membrane proteins to the endoplasmic reticulum
Literature Cited
- 1.Akopian D, Dalai K, Shen K, Duong F, Shan S. SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J Biol Chem. 2013 doi: 10.1083/jcb.201208045. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopian D, Shen K, Zhang X, Shan S. Signal recognition particle: an essential protein-targeting machine. Ann Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. Plos Biology. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albanese V, Yam AY-W, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 5.Ataide SF, Schmitz N, Shenk K, Ke A, Shan S, Doudna A, Ban N. The crystal structure of the Signal Recognition Particle in complex with its receptor. Science. 2011;381:881–86. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bange G, Kummerer N, Grudnik P, Lindner R, Petzold G, Kressler D, Hurt E, Wild K, Sinning I. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nature Struct Molec Biol. 2011;18:1376–80. doi: 10.1038/nsmb.2141. [DOI] [PubMed] [Google Scholar]
- 7.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–39. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 8.Batey RT, Sagar MB, Doudna JA. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J Mol Biol. 2001;307:229–46. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- 9.Beck K, Wu L-F, Brunner J, Muller M. Discrimination between SRP- ad SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 2000;19:134–43. doi: 10.1093/emboj/19.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker T, Bhushan S, Jarasch A, Armache J-P, Funes S, Jossinet F, Gumbart J, Mielke T, Berninhausen O, Schulten K, Westhof E, Gilmore R, Mandon E, Beckmann R. Structure of Monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1367–73. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckmann R, Spahn CMT, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–72. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 12.Berndt U, Oellerer S, Zhang Y, Johnson A, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci. 2009;106:1398–403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–86. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein HD, Zopf D, Freymann DM, Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escheirchia coli homolog. Proc Natl Acad Sci U S A. 1993;90:5229–34. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blobel G, Sabatini DD. Ribosome-membrane interaction in eukaryotic cells. In: Manson LA, editor. Biomembranes. New York: Plenum; 1971. pp. 193–95. [Google Scholar]
- 16.Bornemann T, Jockel J, Rodnina MV, Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol. 2008;15:494–99. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- 17.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–28. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 18.Bradshaw N, Neher SB, Booth DS, Walter P. Signal sequences activate the catalytic switch of SRP RNA. Science. 2009;323:127–30. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandman O, Stewart-Omstein J, Wong D, Larson A, Williams CC, Li G-W, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–54. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buskiewicz I, Deuerling E, Gu SQ, Jockel J, Rodnina MV, Bukau B, Wintermeyer W. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc Natl Acad Sci U S A. 2004;101:7902–06. doi: 10.1073/pnas.0402231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buskiewicz I, Jockel J, Rodnina MV, Wintermeyer W. Conformation of the signal recognition particle in ribosomal targeting complexes. RNA. 2009;15:44–54. doi: 10.1261/rna.1285609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buskiewicz I, Kubarenko A, Peske F, Rodnina MV, Wintermeyer W. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA. 2005;11:947–57. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J Mol Biol. 2005;351:417–30. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 24.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465:435–40. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connolly T, Rapiejko PJ, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–73. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 26.Cross BCS, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nature Rev Mol Cell Biol. 2009;10:255–64. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 27.Cruz-Vera LR, Gong M, Yanofsky C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc Natl Acad Sci U S A. 2006;103:3598–603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins faciliates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–05. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- 29.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–96. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 30.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–67. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 31.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: Common principles and physiological networks. Biochim Biophys Acta. 2013;1833:274–85. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 32.Egea PF, Shan S, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–21. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 33.Eisner G, Moser M, Schafer U, Beck K, Muller M. Alternative recruitment of signal recognition particle and trigger factor to the signal sequence of a growing nascent polypeptide. J Biol Chem. 2006;281:7172–79. doi: 10.1074/jbc.M511388200. [DOI] [PubMed] [Google Scholar]
- 34.Emanuelsson O, Nielson H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–84. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estrozi LF, Boehringer D, Shan S, Ban N, Schaffitzel C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat Struct Mol Biol. 2011;18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fersht A. Enzyme Structure and Mechanism. New York: Freeman and Company; 1984. pp. 384–99. [Google Scholar]
- 37.Fersht AR, Dingwall C. Evidence for the double-sieve editing mechanism in protein synthesis. Steric exclusion of isoleucine by valyl-tRNA synthetases. Biochemistry. 1979;18:2627–31. doi: 10.1021/bi00579a030. [DOI] [PubMed] [Google Scholar]
- 38.Fersht AR, Kaethner MM. Enzyme hyperspecificity. Rejection of threonine by the valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry. 1976;15:3342–46. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 39.Flanagan JJ, Chen JC, Miao YW, Shao YL, Lin JL, et al. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. Journal of Biological Chemistry. 2003;278:18628–37. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 40.Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, Johnson AE. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem. 2003;278:18628–37. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 41.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase Core of the SRP Targeting Complex. Science. 2004;303:373–77. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraunfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nature Struct Molec Biol. 2011;18:614–21. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freymann DM, Walter P. GTPases in protein translocation and protein elongation. In: Hall A, editor. Frontiers in Molecular Biology: GTPases. London: Oxford University Press; 2000. pp. 222–43. [Google Scholar]
- 44.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–64. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 45.Freymann DM, Keenan RJ, Stroud RM, Walter P. Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP. Nat Struct Biol. 1999;6:793–801. doi: 10.1038/11572. [DOI] [PubMed] [Google Scholar]
- 46.Frobel J, Rose P, Muller M. Twin-arginine-dependent translocation of folded proteins. Philos Trans R Soc Lond B Biol Sci. 2012;367:1029–46. doi: 10.1098/rstb.2011.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nature Rev Mol Cell Biol. 2009;10:423–29. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 48.Gawronski-Salerno J, Coon YJS, Focia PJ, Freymann DM. X-ray structure of the T. Aquaticus Ftsy:GDP complex suggests functional roles for the C-terminal helix of the SRP GTPases. Proteins. 2006;66:984–95. doi: 10.1002/prot.21200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gawronski-Salerno J, Freymann DM. Structure of the GMPPNP-stabilized NG domain complex of the SRP GTPases Ffh and FtsY. J Struct Biol. 2007;158:122–28. doi: 10.1016/j.jsb.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gierasch LM. Signal sequences. Biochemistry. 1989;28:923–30. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- 51.Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–49. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 52.Goldshmidt H, Sheiner L, Butikofer P, Roditi I, Uliel S, Gunzel M, Engstler M, Michaeli S. Role of protein translocation pathways across the endoplasmic reticulum in Trypanosoma brucei. J Biol Chem. 2008;283:32085–98. doi: 10.1074/jbc.M801499200. [DOI] [PubMed] [Google Scholar]
- 53.Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–73. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hainzl T, Huang S, Sauer-Eriksson AE. Interaction of signal-recognition particle 54 GTPase domain and signal recognition particle RNA in the free signal-recognition particle. Proc Natl Acad Sci. 2007;104:14911–16. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hainzl T, Huang S, Merilainen G, Brannstrom K, Sauer-Eriksson AE. Structural basis of signal sequence recognition by the signal recognition particle. Nature Struct Molec Biol. 2011;18:389–91. doi: 10.1038/nsmb.1994. [DOI] [PubMed] [Google Scholar]
- 56.Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–14. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 57.Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–11. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 58.Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006;312:745–47. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- 59.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 60.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12:787–98. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–25. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 62.Hoffmann A, Becker AH, Zachmann-Brand B, Deuerling E, Bukau B, Kramer G. Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol Cell. 2012;48:63–74. doi: 10.1016/j.molcel.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 63.Holtkamp W, Lee S, Bornemann T, Senyushkina T, Rodnina MV, Wintermeyer W. Dynamic switch of the signal recognition particle from scanning to targeting. Nature Struct Molec Biol. 2012;19:1332–37. doi: 10.1038/nsmb.2421. [DOI] [PubMed] [Google Scholar]
- 64.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nature Struct Molec Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 65.Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J Bacteriol. 2005;187:2983–91. doi: 10.1128/JB.187.9.2983-2991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J Bacteriol. 2005;187:2983–91. doi: 10.1128/JB.187.9.2983-2991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–24. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–10. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson AE, van Waes MA. The translocon: A dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;18:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 70.Jones JD, McKnight CJ, Gierasch LM. Biophysical studies of signal peptides: implications for signal sequence functions and the involvement of lipid in protein export. J Bioenerg Biomembr. 1990;22:213–32. doi: 10.1007/BF00763166. [DOI] [PubMed] [Google Scholar]
- 71.Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–70. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 72.Kaiser CM, Chang H-C, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–60. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- 73.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–75. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 74.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–91. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 75.Knoops K, Schoehn G, Schaffitzel C. Cryo-electron microscopy of ribosomal complexes in cotranslational folding, targeting, and translocation. Wiley Inbterdiscip Rev RNA. 2011 doi: 10.1002/wrna.119. [DOI] [PubMed] [Google Scholar]
- 76.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–97. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 77.Kunkel TaBK. DNA replication fidelity. Ann Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 78.Lakkaraju AKK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–51. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lam VQ, Akopian D, Rome M, Shen Y, Henningsen D, Shan S. Lipid activation of the SRP receptor provides spatial coordination of protein targeting. J Cell Biol. 2010;190:623–35. doi: 10.1083/jcb.201004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee DW, Jung C, Hwang I. Cytosolic events involved in chloroplast protein targeting. Biochim Biophys Acta. 2013;1833:245–52. doi: 10.1016/j.bbamcr.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 81.Lee HC, Bernstein HD. Trigger factor retards protein export in Escherichia coli. J Biol Chem. 2002;277:43527–35. doi: 10.1074/jbc.M205950200. [DOI] [PubMed] [Google Scholar]
- 82.Leipe DD, Wolf YI, Koonin EV, Aravid L. Classificaiton and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 83.Liao S, Lin J, Do H, Johnson AE. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 84.Lu J, Deutsch C. Folding Zones inside the ribosomal exit tunnel. Nature Struct Molec Biol. 2005;12:1123–29. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 85.Mainprize IL, BEniac DR, Falkovskala E, Cleverley RM, Gierasch LM, Ottensmeyer FP, Andrews DW. The structure of Escherichia coli signal recognition particle revealed by scanning transmission electron microscopy. Mol Biol Cell. 2006;17:5063–74. doi: 10.1091/mbc.E06-05-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan R, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–24. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McClellan AJ, Xia Y. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–35. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 88.Melville MW, McClellan AJ, Meyer AS, Darvaeu A, Frydman J. The Hsp70 and TriC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumer suppressor complex. Mol Cell Biol. 2003;23:3141–51. doi: 10.1128/MCB.23.9.3141-3151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, Deuerling E. Molecular mechanism and structure of trigger factor bound to the translating ribosome. EMBO J. 2008;27:1622–32. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.MItra K, Schaffitzel C, Shaikh T, Tanna F, Jenni S, Brooks CL, Ban N, Frank J. Structure of the E. coli protein conducting channel bound to a translating ribosome. Nature. 2005;438:318–24. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Montoya G, Svensson C, Luirink J, Sinning I. Crystal structure of the NG domain from the signal recognition particle receptor FtsY. Nature. 1997;385:365–68. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 92.Nakatogawa HaIK. Secretion motor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell. 2001;7:185–92. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 93.Natale P, Bruser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–56. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 94.Neher SB, Bradshaw N, Floor SN, Gross JD, Walter P. SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction. Nat Struct Mol Biol. 2008;15:916–23. doi: 10.1038/nsmb.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Netzer WJ, Hartl FU. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- 96.Ogg SC, Walter P. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell. 1995;81:1075–84. doi: 10.1016/s0092-8674(05)80012-1. [DOI] [PubMed] [Google Scholar]
- 97.Ogle JMaRV. Structural insights into translational fidelity. Ann Rev Biochem. 2005;74:129–77. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 98.Oh E, Beck AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichls RJ, Typas A, Gross CA, Kramer G, Weissman JS, Bukau B. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Padmanabhan W, Freymann DM. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase site. Structure. 2001;9:859–63. doi: 10.1016/s0969-2126(01)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parlitz R, Eitan A, Stjepanovic G, Bahari L, Bange G, Bibi E, Sinning I. Escherichia coli signal recognition particle receptor FtsY contains an essential and autonomous membrane-binding amphipathic helix. J Biol Chem. 2007;282:32176–84. doi: 10.1074/jbc.M705430200. [DOI] [PubMed] [Google Scholar]
- 101.Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol Cell. 2013;49:411–21. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–43. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- 103.Peluso P, Shan S, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–33. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 104.Peterson JH, Szabady RL, Bernstein HD. An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the secYEG complex. J Biol Chem. 2006;281:9038–48. doi: 10.1074/jbc.M508681200. [DOI] [PubMed] [Google Scholar]
- 105.Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–48. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 107.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Ann Rev Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 108.Powers T, Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–24. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 109.Powers T, Walter P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr Biol. 1996;6:331–38. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- 110.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–86. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Randall LL, Hardy SJS. High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. TIBS. 1995;20:65–69. doi: 10.1016/s0968-0004(00)88959-8. [DOI] [PubMed] [Google Scholar]
- 112.Rapaport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 113.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–69. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 114.Rapoport TA, Heinrich R, Walter P, Schulmeister T. Mathematical modeling of the effects of the signal recognition particle on translation and translocation of proteins across the endoplasmic reticulum membrane. J Mol Biol. 1987;195:621–36. doi: 10.1016/0022-2836(87)90186-0. [DOI] [PubMed] [Google Scholar]
- 115.Reyes CL, Rutenber E, Walter P, Stroud RM. X-ray structures of the signal recognition particle receptor reveal targeting cycle intermediates. PloS ONE. 2007;2:e607. doi: 10.1371/journal.pone.0000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rodnina MV, Wintermeyer W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. TIBS. 2001;26:124–30. doi: 10.1016/s0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- 117.Rodnina MVaWW. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Ann Rev Biochem. 2001;70:415–35. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 118.Rosendal KR, Wild K, Montoya G, Sinning I. Crystal structure of the complete core of archaeal signal recognition paricle and implications for interdomain communication. Proc Natl Acad Sci. 2003;100:14701–06. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and allostery of the chaperonin GroEL. J Mol Biol. 2013;425:1476–87. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 120.Saraogi I, Zhang D, Chandrasekaran S, Shan S. Site-specific fluorescent labeling of nascent proteins on the translating ribosome. J Am Chem Soc. 2011;133:14936–39. doi: 10.1021/ja206626g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–06. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 122.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–45. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. TIBS. 2012;37:263–73. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shan S, Walter P. Induced Nucleotide Specificity in a GTPase. Proc Natl Acad Sci USA. 2003;100:4480–85. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shan S, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of SRP and its receptor drive initiation of protein translocation. J Cell Biol. 2007;178:611–20. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shan S, Stroud R, Walter P. Mechanism of association and reciprocal activation of two GTPases. Plos Biology. 2004;2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shen K, Shan S. A transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc Natl Acad Sci U S A. 2010;107:7698–703. doi: 10.1073/pnas.1002968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shen K, Arslan S, Akopian D, Ha T, Shan S. Activated GTPase movement on an RNA scaffold drives cotranslational protein targeting. Nature. 2012;492:271–75. doi: 10.1038/nature11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shen K, Zhang X, Shan S. Synergiestic action between the SRP RNA and translating ribosome allows efficient delivery of correct cargos during co-translational protein targeting. RNA. 2011;17:892–902. doi: 10.1261/rna.2610411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shepotinovskaya IV, Freymann DM. Conformational change of the N-domain on formation of the complex between the GTPase domains of Thermus aquaticus Ffh and FtsY. Biochemica et Biophysica Acta. 2001;1597:107–14. doi: 10.1016/s0167-4838(02)00287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Siegel V, Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 1988;7:1769–75. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Siegel V, Walter P. Each of the activities of Signal Recognition Particle (SRP) is contained within a distinct domain: Analysis of biochemical mutants of SRP. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- 133.Siegel V, Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain-length. Embo Journal. 1988;7:1769–75. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spanggord RJ, Siu F, Ke A, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nature Struct Molec Biol. 2005;12:1116–22. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 135.Sydow JFaCP. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19:732–39. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 136.Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Linquist S. Quantitative analysis of Hsp90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pahtway. Ann Rev Biochem. 2012;81:261–89. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ullers RS, Houben ENG, Raine A, Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J Cell Biol. 2003;161:679–84. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife. 2013;3:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.von Heijne G. Signal sequences: The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 143.von Loeffeilholz O, Knoops K, Ariosa A, Zhang X, Karuppasamy M, Huard K, Schoehn G, Berger I, Shan S, Schaffitzel C. Structural basis of signal sequence surveillance and selection by the SRP-FtsY complex. Nature Struct Molec Biol. 2013;20:604–10. doi: 10.1038/nsmb.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–98. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 145.Walter P, Blobel G. Disassembly and reconstitution of signal recognition particle. Cell. 1983;34:525–33. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- 146.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Ann Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 147.Wang S, Sakai H, Wiedmann M. NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J Cell Biol. 1995;130:519–28. doi: 10.1083/jcb.130.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Z, Jones J, Rizo J, Gierasch LM. Membrane-bound conformation of a signal peptide: A transferred nuclear overhauser effect analysis. Biochemistry. 1993;32:13991–99. doi: 10.1021/bi00213a032. [DOI] [PubMed] [Google Scholar]
- 149.Weiche B, Burk J, Angelini S, Schiltz E, Thumfart JO, Koch H-G. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. J Mol Biol. 2008;377:761–73. doi: 10.1016/j.jmb.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 150.Wickner W. The nascent-polypeptide-associated complex: Having a “NAC” for fidelity in translocation. Proc Natl Acad Sci U S A. 1995;92:9433–34. doi: 10.1073/pnas.92.21.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Willmund F, Del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wilson C, Connolly T, Morrison T, Gilmore R. Integration of membrane proteins into the endoplasmic reticulum requires GTP. J Cell Biol. 1988;107:69–77. doi: 10.1083/jcb.107.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differe in FRET-detected foldng far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–36. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 154.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-b proteins. Curr Opin Struct Biol. 2010;20:782–90. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yam AY, Xia Y, Lin H-TJ, Burlingame A, Gerstein M, Frydman J. Defining the TriC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nature Struct Molec Biol. 2008;15:1255–62. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zaher HaGR. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–66. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang D, Shan SO. Translation elongation regulates substrate selection by the signal recognition particle. J Biol Chem. 2012;287:7652–60. doi: 10.1074/jbc.M111.325001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang X, Kung S, Shan S. Demonstration of a two-step mechanism for assembly of the SRP-SRP receptor complex: implications for the catalytic role of SRP RNA. J Mol Biol. 2008;381:581–93. doi: 10.1016/j.jmb.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang X, Lam VQ, Mou Y, Kimura T, Chung J, Chandrasekar S, Winkler J, Mayo S, Shan S. Direct visualization reveals dynamics of a transient intermediate during proten assembly. Proc Natl Acad Sci U S A. 2011;108:6450–55. doi: 10.1073/pnas.1019051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang X, Rashid R, Wang K, Shan S. Sequential checkpoints govern fidelity during co-translational protein targeting. Science. 2010;328:757–60. doi: 10.1126/science.1186743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang X, Schaffitzel C, Ban N, Shan S. Multiple conformational changes in a GTPase complex regulate protein targeting. Proc Natl Acad Sci. 2009;106:1754–59. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang Y, Berndt U, Golz H, Tais A, Oellerer S, Wolfle T, Fitzke E, Rospert S. NAC functions as a modulator of SRP during the early steps of protein targeting to the ER. Mol Biol Cell. 2012 doi: 10.1091/mbc.E12-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang Z-R, Bonifacino JS, Hegde RS. Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell. 2013;154:609–22. doi: 10.1016/j.cell.2013.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zheng N, Gierasch LM. Signal sequences: the same yet different. Cell. 1996;86:849–52. doi: 10.1016/s0092-8674(00)80159-2. [DOI] [PubMed] [Google Scholar]
- 165.Zopf D, Bernstein HD, Johnson AE, Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 1990;9:4511–17. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]