Abstract
Cereulide and isocereulides A-G are biosynthesized as emetic toxins by Bacillus cereus via a non-ribosomal peptide synthetase (NRPS) called Ces. Although a thiotemplate mechanisms involving cyclo-trimerization of ready-made D-O-Leu-D-Ala-L-O-Val-L-Val via a thioesterase (TE) domain is proposed for cereulide biosynthesis, the exact mechanism is far from being understood. UPLC-TOF MS analysis of B. cereus strains in combination with 13C-labeling experiments now revealed tetra-, octa-, and dodecapeptides of a different sequence, namely (L-O-Val-L-Val-D-O-Leu-D-Ala)1-3, as intermediates of cereulide biosynthesis. Surprisingly, also di-, hexa-, and decadepsipeptides were identified which, together with the structures of the previously reported isocereulides E, F, and G, do not correlate to the currently proposed mechanism for cereulide biosynthesis and violate the canonical NRPS biosynthetic logic. UPLC-TOF MS metabolite analysis and bioinformatic gene cluster analysis highlighted dipeptides rather than single amino or hydroxy acids as the basic modules in tetradepsipeptide assembly and proposed the CesA C-terminal C* domain and the CesB C-terminal TE domain to function as a cooperative esterification and depsipeptide elongation center repeatedly recruiting the action of the C* domain to oligomerize tetradepsipeptides prior to the release of cereulide from the TE domain by macrocyclization.
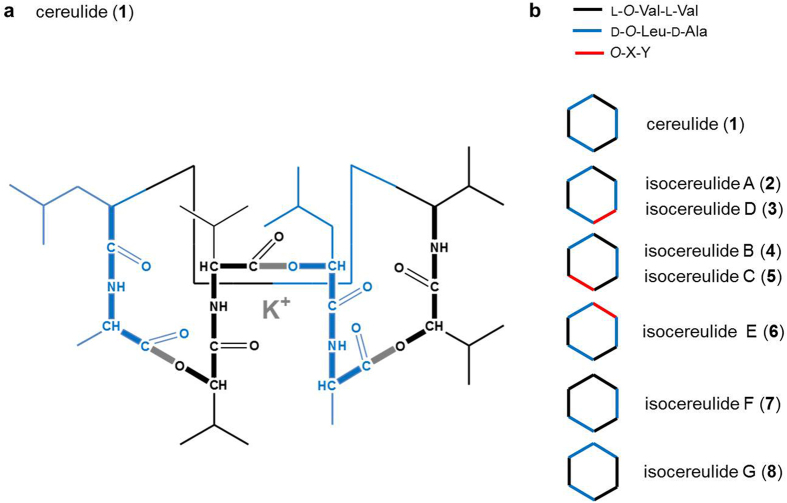
The endospore-forming bacterium Bacillus cereus is a food-borne human pathogen causing diarrhea due to the activity of haemolytic and nonhaemolytic enterotoxins as well as food intoxications induced by the heat-stable, emesis provoking cereulide1,2. Cereulide (1) is a dodecadepsipeptide composed of six α-amino acid and six α-hydroxy acid moieties arranged in three repeating tetradepsipeptide units, namely [D-O-Leu-D-Ala-L-O-Val-L-Val]3 (Fig. 1)3,4.
Figure 1.
(a) Chemical structure of the cyclic dodecadepsipeptide cereulide (1), [D-O-Leu-D-Ala-L-O-Val-L-Val]3, (b) chemical structure of isocereulides A (2) to G (8), O-X-Y indicates the position of the dipeptides L-O-Leu-L-Val (isocereulide A ), D-O-Val-D-Ala (isocereulide B), D-O-Leu-D-Ser (isocereulide C), L-O-Val-L-Ala (isocereulide D), D-O-Ile-D-Ala (isocereulide E).
Biosynthesis of fungal or bacterial cyclooligomer depsipeptides, such as the cyclohexadepsipeptide enniatin from Fusarium species or the cyclooctadepsipeptide bassianolide from Beauveria bassiana, as well as the cyclodecadepsipeptide valinomycin from Streptomyces species or the cyclododecadepsipeptide cereulide from B. cereus is performed by large multi-domain non-ribosomal peptide synthetases (NRPS)5,6,7,8. In B. cereus, the cereulide non-ribosomal peptide synthetase (Ces NRPS) is encoded by the polycistronically transcribed ces gene cluster, which is located on a mega plasmid that is sharing its backbone with the B. anthracis toxin plasmid pX019,10,11. Besides the structural genes for the synthetase (cesA and cesB), the ces gene locus comprises a putative hydrolase (cesH), a phosphopanthetheinyl transferase (cesP), a type II thioesterase (cesT), as well as a putative ABC transporter (cesC/D)9.
The biosynthesis of cereulide is proposed to follow the general principle of NRPS multi-enzyme complexes that show a modular organization. Every single module, delivering a specific monomer to the growing peptide, can be subdivided into conserved domains that carry all essential information for recognition, activation, and modification of the substrate. The adenylation (A) domain is responsible for specific recognition and activation of the precursors by adenylation using ATP as co-substrate. A peptidyl carrier protein (PCP) captures the precursor adenylates as covalent thioesters via a phosphopantetheine linker12. A characteristic feature of CesA and CesB is the existence of two A domains which recognize, select, and reduce α-ketocarboxylic acids to α-hydroxyacyl-S-pantetheinyl intermediates. The chiral reduction step is catalyzed by ketoreductase (KR) domains embedded in the corresponding A domains CesA1 and CesB1, respectively. Chain elongation of the intermediate is mediated by a condensation (C) domain catalyzing the nucleophilic attack of the downstream PCP-bound acceptor monomer on the activated thioester of the upstream PCP-bound donor monomer13, thus giving rise to the dipeptide intermediate D-O-Leu-D-Ala-S-PCP. Due to their enantioselectivity, they are also thought to act as selective filters, facilitating the production of NRPS products in a directed manner14. The CesA’s terminal C domain is proposed to work in trans with the D-O-Leu-D-Ala peptide as donor and the hydroxy group of the CesB1 PCP-thioester bound L-O-Val, followed by CesB2 PCP-bound L-Val as nucleophilic acceptors8. Elongation to the D-O-Leu-D-Ala-L-O-Val-L-Val chain is presumably followed by transfer to the hydroxy group of a serine of the thioesterase (TE) domain, while a second tetradepsipeptide builds up on the adjacent PCP of CesB. The thioesterase (TE) domain is able to release the target peptide by either hydrolysis or cyclization12,15,16 and is supposed to condense tetradepsipeptides, first to produce an octadepsipeptidyl-O-TE and then a decadepsipeptidyl-O-TE, followed by macrolactonization to form the cyclic twelve residue product cereulide8,9.
Interestingly, recent UPLC-TOF MS profiling of 78 B. cereus strains showed a high variability in cereulide production and indicated a unique pattern of 18 previously unknown cereulide variants, among which isocereulide A-G (2-8; Fig. 1) could be unequivocally identified by means of ion-trap MSn sequencing, 13C-labeling experiments, and post-hydrolytic dipeptide and enantioselective amino acid analysis17,18. The structures of some isocereulides differed from that of cereulide by substitution of L-O-Val by L-O-Leu (isocereulide A), L-Val by L-Ala (isocereulide D), D-Ala by D-Ser (isocereulide C), and D-O-Leu by D-O-Val (isocereulide B), thus indicating a relaxed substrate specificity of the A domains and an inefficient proof reading by CesT18. However, the structures of isocereulides E (6) and F (7), as well as isocereulide G (8), which shows another constitutional arrangement of the depsidipeptides D-O-Leu-D-Ala and L-O-Val-L-Val when compared to the isobaric cereulide, violate the canonical NRPS biosynthesis logic as suggested for cereulide production.
To gain novel insights into the non-ribosomal peptide assembly leading to cereulide and isocereulides, the presence of depsipeptide intermediates in a variety of B. cereus strains was screened by UPLC-TOF MS and their abundance was correlated with the cereulide productivity of different B. cereus strains. In addition, the ces gene locus was reinvestigated using bioinformatic approaches.
Results and discussion
UPLC-TOF MS metabolite analysis of B. cereus strains
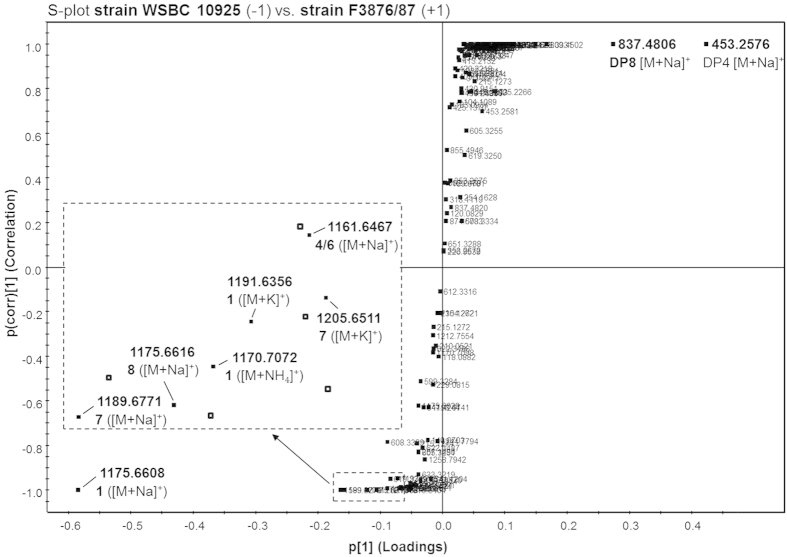
In order to investigate differences in the metabolite profile of cereulide low-medium and high producers, ethanol extracts were prepared from cultures of strains WSBC 10925 and F3876/87, respectively, and analyzed by means of UPLC-ESI TOF MS. To visualize similarities and differences between the samples, S-plots of data pairs of accurate mass and retention time of each metabolite were calculated based on orthogonal partial least squares discriminant analysis (OPLS-DA). As the y-axis of the S-plot denotes confidence of a metabolite’s contribution to the group difference and the x-axis denotes the contribution of a particular metabolite to the group difference, the S-plot indicated the ion m/z 1175.6608 to show by far the highest difference in abundance in high producer strain WSBC 10925, whereas the ions m/z 453.2576 and m/z 837.4806 showed significantly higher abundance in the cereulide low-medium producer strain F3876/87 (Fig. 2). The pseudo molecular ion m/z 1175.6608 ([M+Na]+) was unequivocally identified as sodium adduct (C57H96N6O18Na) of cereulide (1) and, together with the corresponding [M+K]+ (m/z 1191.6356) and [M+NH4]+ ion (m/z 1170.7072), was confirmed by co-chromatography with a purified cereulide reference17,18. Moreover, the pseudomolecular ions m/z 1189.6771 ([M+Na]+) and m/z 1205.6511 ([M+K]+) were assigned as isocereulide F (7), m/z 1175.6616 ([M+Na]+) as isocereulide G (8), and m/z 1161.6467 ([M+Na]+) as a mixture of the isobaric isocereulides B (4) and E (6) by means of co-chromatography with purified reference materials.
Figure 2.
S-Plot of UPLC-TOF MS data of cereulide high producing strain WSBC 10925 vs. low-medium producer F3876/87 with metabolite assignment for cereulide (1) with m/z 1175.6608 ([M+Na]+), m/z 1191.6356 ([M+K]+) and m/z 1170.7072 ([M+NH4]+), isocereulide F (7) with m/z 1189.6771 ([M+Na]+) and m/z 1205.6511 ([M+K]+), isocereulide G (8) with m/z 1175.6616 ([M+Na]+), isocereulide B and E (4, 6) with m/z 1161.6467 ([M+Na]+), and the open-chain tetradepsipeptide (DP4; m/z 453.2576; C21H38N2O7Na, -1.5 ppm) and octadepsipeptide (DP8; m/z 837.4806; C40H70N4O13Na, -4.7 ppm).
On the basis of accurate mass data, the sum formula of the ions m/z 453.2576 and 837.4806, detected with high abundance in strain F3876/87, were calculated to be C21H38N2O7Na (–1.5 ppm) and C40H70N4O13Na (–4.7 ppm), respectively. Interestingly, these sum formula were in good agreement with the ethyl esters of the open-chain tetradepsipeptide D-O-Leu-D-Ala-L-O-Val-L-Val and the octadepsipeptide (D-O-Leu-D-Ala-L-O-Val-L-Val)2 proposed to be key intermediates in cereulide biosynthesis (Fig. 2)8.
Structure determination of candidate depsipeptide esters
In order to determine the chemical structures of the candidate depsipeptide esters detected, the fraction containing the target peptide esters with m/z 453.2576 and 837.4806, respectively, were isolated from the ethanolic extract of strain F4810/72 by means of RP-HPLC. As an example the target compound with m/z 453.2576 is shown in Fig. 3a. Alkaline hydrolysis of the isolated depsipeptides produced dipeptides as reported recently for cereulide and isocereulides18. UPLC-TOF MS analysis of the alkaline hydrolysate of the candidate tetradepsipeptide ethyl ester (Fig. 3b) and comparison with synthetic reference peptides led to the identification of D-O-Leu-D-Ala (m/z 202.1073, [M-H]–) and L-O-Val-L-Val (m/z 216.1238, [M-H]–), thus matching the cleavage peptides of cereulide’s D-O-Leu-D-Ala-L-O-Val-L-Val tetradepsipeptide motif18. Similarly, alkaline hydrolysis of the candidate octadepsipeptide (m/z 837.4806), followed by UPLC-TOF MS analysis, also revealed D-O-Leu-D-Ala (m/z 202.1073, [M-H]–) and L-O-Val-L-Val (m/z 216.1238, [M-H]–) as cleavage peptides as expected to be released from (D-O-Leu-D-Ala-L-O-Val-L-Val)2-OEt (data not shown).
Figure 3.
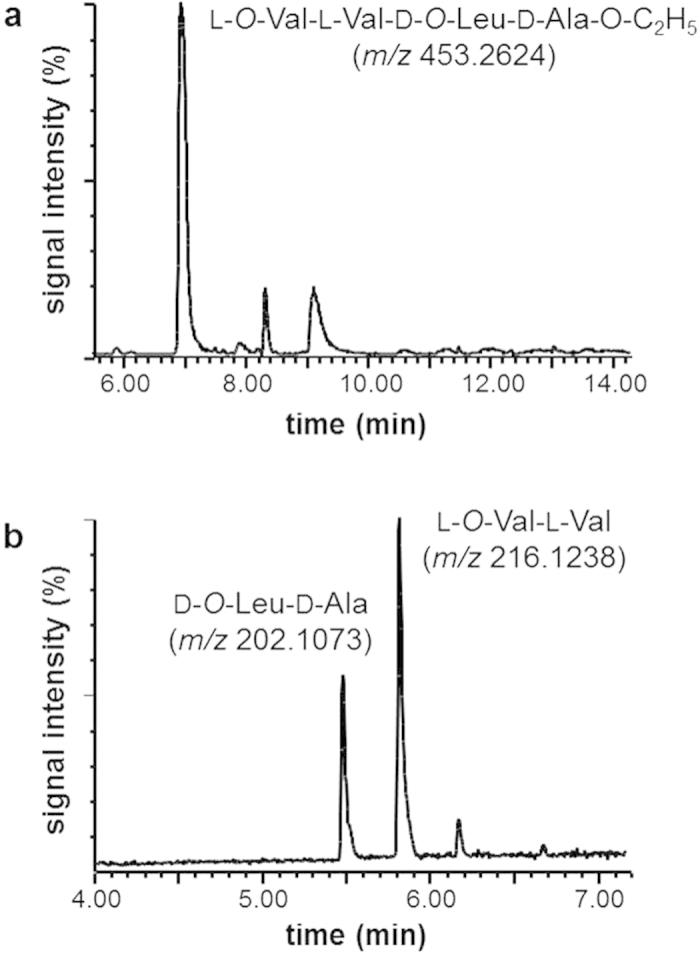
UPLC-TOF MS analysis of (a) isolated tetradepsipeptide (DP4; m/z 453.2576; ESI+) and (b) dipeptides D-O-Leu-D-Ala (m/z 202.1073, [M-H]–) and L-O-Val-L-Val (m/z 216.1238, [M-H]–) after alkaline hydrolysis.
To unequivocally confirm the proposed structure of the ethyl ester of the tetradepsipeptide (m/z 453.2576) and the octadepsipeptide (m/z 837.4806), respectively, in a first set of experiments cell pellets of strain F3876/87 were extracted with MeOH or MeCN/H2O (70/30, v/v) instead of EtOH and the extract was analyzed for depsipeptide methyl esters as well as the free depsipeptides by UPLC-TOF MS. UPLC-TOF MS analysis of the MeOH extract clearly revealed the methyl esters of the expected tetra- (DP4) and octadepsipeptide (DP8) and, in addition, the methyl ester of the dodecadepsipeptide (DP12) with m/z 1207.6941 (C58H100N6O19Na, +1.0 ppm) as the predominant compounds (Fig. 4). Sursprisingly, also two didepsipeptides with m/z 240.1212 (DP2a; C10H19NO4Na, +0.1 ppm) and m/z 254.1368 (DP2b; C11H21NO4Na, +2.0 ppm), two hexadepsipeptides with m/z 624.3472 (DP6a; C29H51N3O10Na, +0.8 ppm) and m/z 638.3629 (DP6b; C30H53N3O10Na, +0.9 ppm), and two decadepsipeptide methyl esters with m/z 1008.5733 (DP10a; C48H83N5O16Na, +0.7 ppm) and m/z 1022.5889 (DP10b; C49H85N5O16Na, +0.8 ppm) were identified at somewhat lower abundance (Fig. 4, Table 1). Also, extraction of the cell pellets with MeCN/H2O (70/30, v/v) released the corresponding free oligodepsipeptides DP2 to DP12 (Table 1).
Figure 4.
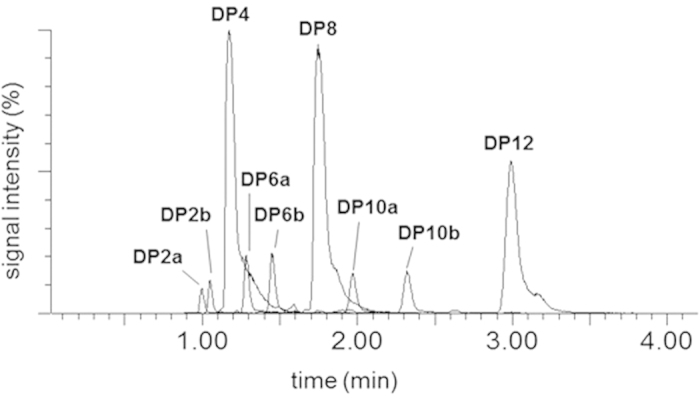
UPLC-TOF MS (ESI+) analysis of a MeOH extract of B. cereus strain F3876/87 with peak assignment of two didepsipeptide methyl esters with m/z 240.1212 (DP2a; C10H19NO4Na, +0.1 ppm) and m/z 254.1368 (DP2b; C11H21NO4Na, +2.0 ppm), the tetradepsipeptide methyl ester with m/z 453.2576 (DP4; C21H38N2O7Na, -1.5 ppm), two hexadepsipeptide methyl esters with m/z 624.3472 (DP6a; C29H51N3O10Na, +0.8 ppm) and m/z 638.3629 (DP6b; C30H53N3O10Na, +0.9 ppm), the octadepsipeptide methyl ester (DP8; m/z 837.4806; C40H70N4O13Na, -4.7 ppm), two decadepsipeptide methyl esters with m/z 1008.5733 (DP10a; C48H83N5O16Na, +0.7 ppm) and m/z 1022.5889 (DP10b; C49H85N5O16Na, +0.8 ppm), and the dodecadepsipeptide methyl ester (DP12) with m/z 1207.6941 (C58H100N6O19Na, +1.0 ppm).
Table 1. Chromatographic and Mass Spectrometric Data Oligodepsipeptides (DP2a/b – DP12) and their Methyl and Ethyl esters Released from B. cereus Cultures upon Extraction with MeCN/H2O (70/30, v/v), MeOH, and EtOH, Respectively.
| No.a | Peptides (R=H) and Peptide Esters (R=Me, Et)b |
UPLC-TOF MS data |
|||||||
|
free peptidesc |
methyl esterd |
ethyl estere |
13C-labeled ethyl esterf |
||||||
| RTg(min) | ECh (EM/AM; var., ppm)i | RTg (min) | ECh (EM/AM; var., ppm)i | RTg(min) | ECh (EM/AMh; var., ppm)i | RTg(min) | ECh (EM/AMh; var., ppm)i | ||
| DP2a | D-O-Leu-D-Ala-O-R | 0.97 | C9H17NO4Na (226.1055/226.1055; -0.1) | 1.00 | C10H19NO4Na (240.1212/240.1211; +0.1) | 1.05 | C11H21NO4Na (254.1368/254.1377; +3.5) | — | — |
| DP2b | L-O-Val-L-Val-O-R | 0.99 | C10H19NO4Na (240.1212/240.1211; -0.1) | 1.05 | C11H21NO4Na (254.1368/254.1373; +2.0) | 1.12 | C12H23NO4Na (268.1525/268.1537; +4.5) | — | — |
| DP4 | L-O-Val-L-Val-D-O-Leu-D-Ala-O-R | 1.10 | C19H34N2O7Na (425.2264/425.2265; +0.2) | 1.17 | C20H36N2O7Na (439.2420/439.2420; -0.1) | 1.24 | C21H38N2O7Na (453.2577/453.2569; -1.8) | 1.24 | 12C1913C2H38N2O7Na (455.2644/455.2645; -1.8) |
| DP6a | D-O-Leu-D-Ala-(L-O-Val-L-Val-D-O-Leu-D-Ala)-O-R | 1.14 | C28H49N3O10Na (610.3316/610.3323; +1.1) | 1.28 | C29H51N3O10Na (624.3472/624.3477; +0.8) | 1.35 | C30H53N3O10Na (638.3629/638.3628; -0.2) | 1.35 | 12C2813C2H53N3O10Na (640.3696/640.3688; -1.2) |
| DP6b | (L-O-Val-L-Val-D-O-Leu-D-Ala)-L-O-Val-L-Val-O-R | 1.22 | C29H51N3O10Na (624.3472/624.3472; -0.3) | 1.45 | C30H53N3O10Na (638.3629/638.3635; +0.9) | 1.52 | C31H55N3O10Na (652.3785/652.3777; -1.2) | 1.52 | 12C2713C4H55N3O10Na (656.3919/656.3916; -0.5) |
| DP8 | (L-O-Val-L-Val-D-O-Leu-D-Ala)2-O-R | 1.43 | C38H66N4O13Na (809.4524/809.4530; +0.7) | 1.75 | C39H68N4O13Na (823.4681/823.4688; +0.9) | 1.78 | C40H70N4O13Na (837.4837/837.4798; -4.7) | 1.78 | 12C3613C4H70N4O13Na (841.4971/841.4961; -1.2) |
| DP10a | D-O-Leu-D-Ala-(L-O-Val-L-Val-D-O-Leu-D-Ala)2-ORj | 1.44 | C47H81N5O16Na (994.5576/994.5580; +0.4) | 1.97 | C48H83N5O16Na (1008.5733/1008.5740; +0.7) | 1.95 | C49H85N5O16Na (1022.5889/1022.5871; -1.8) | 1.94 | 12C4513C4H85N5O16Na (1026.6023/1026.6012; -1.1) |
| DP10b | (L-O-Val-L-Val-D-O-Leu-D-Ala)2-L-O-Val-L-Val-O-R | 1.54 | C48H83N5O16Na (1008.5733/1008.5735; +0.2) | 2.32 | C49H85N5O16Na (1022.5889/1022.5897; +0.8) | 2.26 | C50H87N5O16Na (1036.6046/1036.6031; -1.4) | 2.21 | 12C4413C6H87N5O16Na (1042.6247/1042.6244; -0.3) |
| DP12 | (L-O-Val-L-Val-D-O-Leu-D-Ala)3-O-R | 1.78 | C57H98N6O19Na (1193.6784/1193.6787; +0.3) | 2.99 | C58H100N6O19Na (1207.6941/1207.6953; +1.0) | 2.71 | C59H102N6O19Na (1221.7097/1221.7050; -3.8) | 2.68 | 12C5313C6H102N6O19Na (1227.7299/1227.726; -3.1) |
aCompound number of peptides detected; bSequence of open-chain peptides and peptide esters determined my MSn analysis; c,d,e Corresponding peptides and esters released upon treatment of B. cereus cell pellets with MeCN/H2O (70/30, v/v), MeOH, and EtOH, respectively; f 13C-Labeled ethyl esters released of strain F4810/72 cultivated in MOD-medium supplemented with 13C1-valine;17,19 g Retention time on RP-18 UPLC; h Elemental composition of analyte; i Exact mass (EM, calcd) and accurate mass (AM) of analyte’s [M+Na]+ ion determined by means of TOF MS.
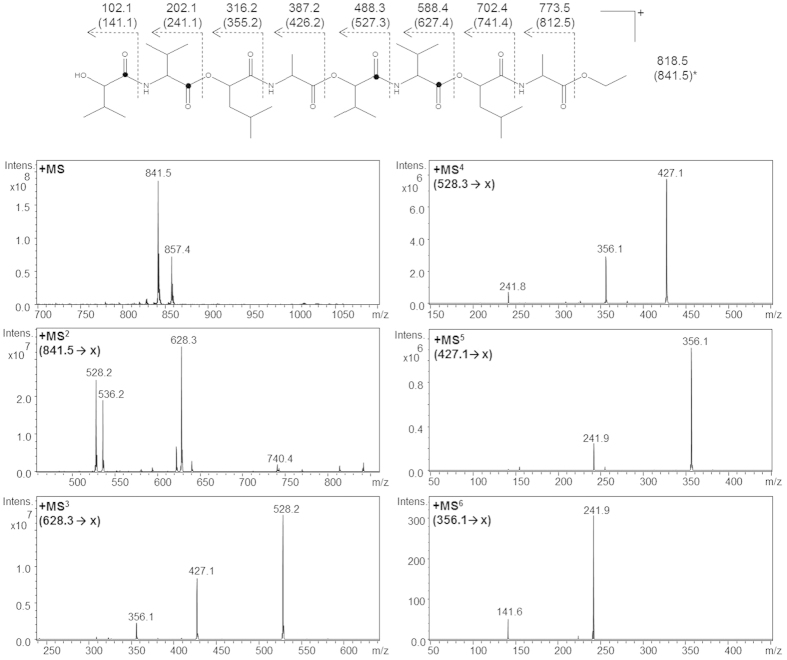
These findings were further confirmed by a 13C-labeling experiment. Cultivation of strain F4810/72 in MOD-medium supplemented with 13C1-valine revealed 13C6-labeled cereulide17,18,19 as well as the ethyl esters of the 13C2-labeled tetradepsipeptide (DP4), a 13C2- and a 13C4-labeled hexadepsipeptide (DP6a/b), a 13C4-labeled octadepsipeptide (DP8), a 13C4- and a 13C6-labeled decadepsipeptide (DP10a/b), as well as a 13C6-labeled dodecadepsipeptide (DP12) released from the cell pellet upon ethanol extraction (Table 1). To confirm the constitutional arrangement of the amino acids and hydroxycarboxylic acids in the target depsipeptides, sequence analyses were performed by MSn-experiments. Using the 13C4-octadepsipeptide ethyl ester as an example, MS fragmentation of the pseudomolecular ion (m/z 841.5, [M+Na]+) revealed specific fragment ions for the hexadepsipeptide L-O-Val-L-Val-D-O-Leu-D-Ala-L-O-Val-L-Val (m/z 841.5 [M+Na]+ → 628.3, 528.2 [M+K]+) released after cleavage of D-O-Leu-D-Ala (Fig. 5). After isolation of the ion m/z 628.3, the recorded MS3 spectrum showed specific fragment ions indicating the cleavage of the amino acid valine (m/z 628.3 [M+K]+ → 528.2, [M+K]+) or the dipeptide L-O-Val-L-Val (m/z 628.3 [M+K]+ → 427.1 [M+K]+) to release the tetradepsipeptide L-O-Val-L-Val-D-O-Leu-D-Ala which could also be detected in the MS4 spectrum after isolation of the fragment ion m/z 528.2. Further fragmentation of the ion m/z 427.1 ([M+K]+ → m/z 356.1 → m/z 241.9; MS5 spectrum) and m/z 356.1 (M+K]+ → m/z 241.9 → m/z 141.6; MS6 spectrum), respectively, revealed a L-O-Val-L-Val-D-O-Leu-D-Ala sequence of the tetradepsipeptide and unequivocally confirmed two 13C-labeled carbon atoms in α-hydroxy isovaleric acid (L-O-Val) and valine (L-Val), respectively (Fig. 5). Similar MSn experiments enabled the sequence elucidation of the other peptides and peptide esters (Table 1, and Supplementary Information). Most intriguingly, the amino acid sequences found for the open-chain tetra- and octadepsipeptides were L-O-Val-L-Val-D-O-Leu-D-Ala and (L-O-Val-L-Val-D-O-Leu-D-Ala)2 and did not match with D-O-Leu-D-Ala-L-O-Val-L-Val and (D-O-Leu-D-Ala-L-O-Val-L-Val)2 proposed as intermediary depsipeptides in cereulide biosynthesis8.
Figure 5.
MSn sequencing of the 13C4-octadepsipeptide ethyl ester (upper masses: calculated, lower masses given in parenthesis: measured [M+K]+ ions); •: 13C-labeling of α-hydroxy isovaleric acid and valine.
As the identified di-, hexa-, and decadepsipeptides are not in line with the proposed biosynthetic assembly of three ready-made tetradepsipeptides8,9, these findings prompted us to ask whether the candidate depsipeptide esters are released from chemically unstable NRPS-bound PCP-S-ester or TE-O-ester intermediates of cereulide biosynthesis upon transesterification with ethanol, or are formed as artifacts by esterification of free depsipeptides or as cereulide degradation products released upon ethanolysis. In a first set of experiments, the B. cereus cell pellet was exhaustively extracted with MeCN to separate cereulide, followed by an extraction of the cereulide-free residue with MeOH instead of EtOH. UPLC-TOF MS analysis confirmed cereulide as the major compound in the MeCN extract, whereas the ions m/z 439.2420, 823.4681 and 1207.6941, corresponding to candidate peptide methyl esters, were detected in the MeOH extract (Table 1). UPLC-TOF MS analysis of methanolic or ethanolic solutions containing the synthesized dipeptides D-O-Leu-D-Ala and L-O-Val-L-Val, or purified cereulide did not reveal even trace amounts of the candidate peptide methyl or ethyl esters after incubation for 4 days at room temperature (data not shown). These data show the low reactivity of the free depsipeptides and confirm the chemical stability of cereulide as reported earlier20,21,22.
In a second set of experiments, a 13C-labeling study was performed to confirm that the open-chain depsipeptides are not formed from ready-made cereulide upon enzymatic cleavage. A Bacillus cereus culture was incubated in the presence of 13C6-cereulide for up to 40 h and 13C6-cereulide as well as non-labeled cereulide were analyzed by means of UPLC-TOF-MS. The data clearly showed that cereulide is produced de-novo as expected while the spiked 13C6-labeled cereulide did not decline significantly over time, thus demonstrating that cereulide is not enzymatically degraded by B. cereus. This is well in line with the observation that none of the 13C-labelled tetra-, octa- and depsipeptide ethyl esters were detectable in the 13C6-cereulide-spiked incubations by means of LC-TOF MS. These findings exclude the open-chain depsipeptides as cereulide degradation products and further strengthen the proposed release of the target depsipeptide esters by transesterification of biosynthetic intermediates like NRPS-bound PCP-S-esters and TE-O-esters, respectively.
Quantitative B. cereus strain screening for cereulide productivity and NRPS-bound depsipeptide intermediates
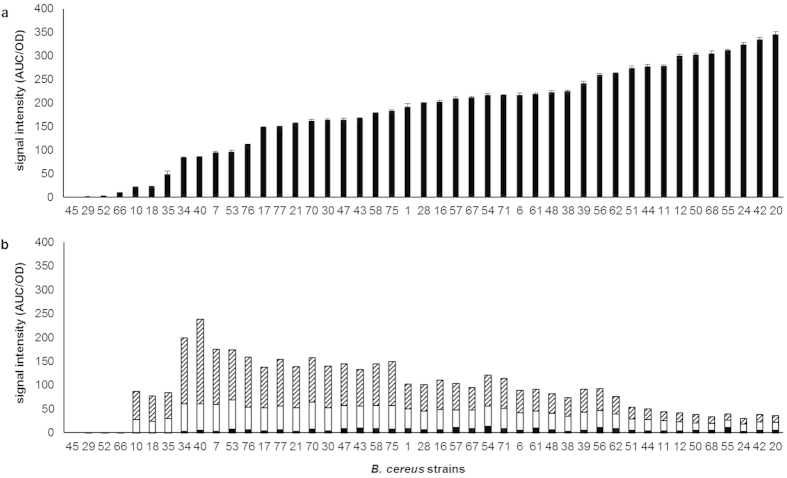
A selection of 42 B. cereus strains adjusted in optical density (OD) were analyzed by UPLC-TOF MS for cereulide and the major tetra- (DP4), octa- (DP8), and dodecadepsipeptide ethyl esters (DP12) released upon ethanol extraction of the cell pellet17. No/low producing strains (no. 45, 29, 52, 66) neither produced elevated levels of cereulide, nor could depsipeptide intermediates be detected in significant amounts (Fig. 6). Intriguingly, cereulide productivity in medium to high producers seem to be inversely correlated with the total abundance of released tetra-, octa-, and dodecadepsipeptide esters measured as a proxy for NRPS-bound depsipeptides. Moreover, with decreasing cereulide productivity higher amounts of tetradepsipeptide esters were liberated when compared to the octa- and dodecadepsipeptide esters (Fig. 6). These data are first evidence that cereulide high producers rapidly oligomerize the precursor depsipeptides followed by subsequent cyclization, whereas less productive strains seem to accumulate NRPS-bound intermediates at the stage of tetradepsipeptides, only slowly converting them to higher oligomers.
Figure 6.
UPLC-ESI-TOF MS analysis of (a) cereulide and (b) the ethyl esters of tetradepsipeptide (dashed bar), octadepsipeptide (white bar) and dodecadepsipeptide (black bar) in 42 B. cereus strains.
Gene sequence analyses
At the C-terminus of the enniatin synthetase (Esyn) gene cluster two PCP domains are located, one catalyzing ester bonds and thereby performing peptide elongation and cyclisation, and another one acting as a waiting position23,24. A similar mechanism can be ruled out for cereulide synthesis, since gene sequence analysis did not reveal any evidence for duplicated PCP domains in CesA or CesB (data not shown). A second alternative might be that cereulide synthesis follows the classical collinear principle of NRPS product assembly including partial hydrolysis of the preformed tetradepsipetides, mediated by condensation domain reversibility. For instance, VibFC2 and VibH are not only mediating condensation in vibriobactin synthesis but also catalyze hydrolysis of single monomers by back reaction25. However, the latter mechanism is not supported by our experimental findings for two reasons: first, only didepsipeptides and a multiple thereof were found by means of UPLC-TOF MS and not even traces of depsipeptide intermediates showing uneven numbers of monomers were detectable; second, a controlled partial hydrolysis is contradictory to the observed occurrence of two (DP6a/b) instead of one hexadepsipeptide and two (DP10a/b) instead of one decadepsipeptide, respectively (Table 1).
Besides their well-known pivotal role in peptide bound formation, C domains have been reported to catalyze ester bound formation and may also have dual functions by acting as epimerization and condensation domain26,27,28. Very recently, an unprecedented function of a C domain was reported for beta lactam antibiotics of the nocardicin family. The C5 domain of nocardicin NRPS (NocB) not only fulfills its canonical function by forming the corresponding amide bond in the growing peptide chain, but also mediates cyclization of the beta lactam ring29. Thus, the spectrum of additional functional activities reported for C domains in non-ribosomal multi enzyme complexes is still growing and may significantly contribute to the diversification of naturally synthetized peptide products. As shown in Fig. 7, primary sequence analysis of the Ces C domains revealed that CesB_C3 belongs to the L C domain class (LCL) while CesA_C2 belongs to the D C domain class (DCL)30, which is in full agreement with the stereochemistry of cereulide. CesA_C1 shows some characteristic features of LCL, albeit, based on the depsipeptide structure and previous stereochemical experiments, one would rather expect a DCL8. A similar deviation from the expected C class was reported for NocB29. The C5 domain of NocB shows DCL characteristics, despite receiving an L-peptide (Fig. 7). Comparison of Ces C domains to C domain sequences in the natural product domain seeker database NaPDoS31 revealed a distinct clustering of CesA_C1 and CesB_C3 while CesA_C2 was grouping with dual C domains and DCL (data not shown).
Figure 7.
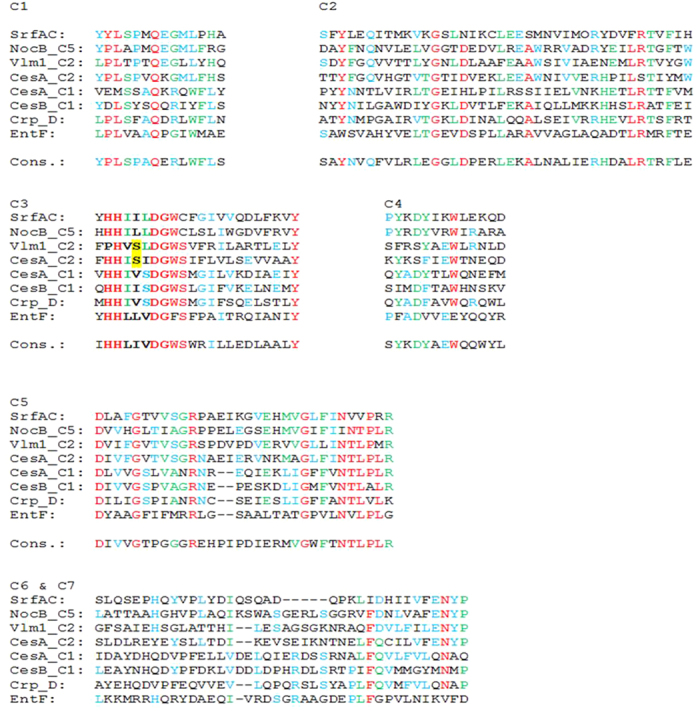
Alignment of the condensation (C) domain core motifs of Ces and core motifs from selected bacterial NRPS. SrfAC: terminal C domain from surfactin synthetase as representative of a DCL; NocB: C5 domain from the nocardicin synthetase as representative of a C domain with non-canonical functions; Vlm1: C2 domain in the valinomycin synthetase, which shows the same genetic architecture as the cereulide synthetase; Crp_D: C domain (M2) from cryptophycin synthetase catalyzing ester bound formation; EntF: C domain from enterobactin synthetase. According to literature42, the consensus sequence from Pfam database for core motifs C1 to C5 is depicted. The C3 core motif comprises the conserved catalytic motif HHxxxDG (printed in bold).
Remarkably, the core motifs of CesA_C2 show a relatively high homology to the second C domain in Vlm1 of the valinomycin NRPS although the overall similarities between the two NRPS is low (approx. 33%). Since formation of cereulide and valinomycin is thought to follow the same biosynthetic logic8, it is proposed that CesA_C2 and Vlm1_C2 act as ester synthases rather than as amide synthases. For this function, the location of the C domains, designated as C*, at the C terminus of CesA/Vlm1 might be of relevance. C domains are most commonly encoded in the same coding DNA sequence (CDS) as their acceptor molecules, upstream of the corresponding A domain, and there are only very few examples were a C domain is located in the C terminal proximitiy of the preceding CDS in NRPS32,33,34. For the C5 domain of NocB a unique amino acid located immediately upstream to the conserved HHxxxDG motif has been reported, which is crucial for the proper function of C529. The HHxxxDG motif promotes the condensation reaction and transfer of the intermediate to the downstream PCP, with the second His thought to be central for the catalytic activity of the C domain12. Interestingly, we also observed an unusual amino acid inserted the HHxxxxDG motif of CesA_C2 and Vlm1_C2 (Fig. 7). Intensive database searches of NRPS C domains revealed a serine at a position in CesA_C2 (C*) and Vlm_C2 were usually aliphatic amino acids are positioned (Fig. 7). Further structural and biochemical analyses, which are clearly beyond the scope of this study, will be necessary in the future to fully decipher the role of C* in cereulide assembly. However, the location of C* in the C terminus of CesA may facilitate a postulated function of this domain as esterification center for cereulide assembly.
Proposed biosynthesis of cereulide
Taking all the experimental data and bioinformatic analysis on the ces gene locus into account and considering previously published data8,9, the following modified mechanism for the biosynthesis of cereulide is proposed (Fig. 8a). Initially, the dipeptides D-O-Leu-D-Ala and L-O-Val-L-Val, both bound as a thioester at PCP, are generated independently by CesA and CesB. The proposed condensation domain C* (CesA_C2)8 transfers the hydroxy group of D-O-Leu-D-Ala-S-PCP to the thioester carbon of L-O-Val-L-Val-S-PCP to form an ester bond resulting in the PCP-bound tetradepsipeptide L-O-Val-L-Val-D-O-Leu-D-Ala-S-PCP (DP4)24, which is subsequently transferred to the serine hydroxy group of the CesB C-terminal TE domain (Fig. 8a). Catalyzed by the condensation domain C* (CesA_C2), a second L-O-Val-L-Val-D-O-Leu-D-Ala-S-PCP, produced as described above, is then connected to the TE-bound tetradepsipeptide by esterification, thus forming the octadepsipeptidyl-O-TE intermediate (DP8) and, further on, the dodecadepsipeptide-O-TE (DP12) upon extension by a third L-O-Val-L-Val-D-O-Leu-D-Ala unit prior to macrolactonization to the cyclic twelve residue product cereulide (Fig. 8a). It is therefore proposed that the C* domain located at the C-terminus of CesA and the TE domain located at the C-terminus of CesB function as a cooperative esterification and depsipeptide elongation center (DP4 → DP8 → DP12), repeatedly recruiting the action of the C* domain prior to macrocyclization by the TE domain. It is tempting to speculate that a previously described internal promoter of yet unknown function may play a role in adjusting and fine tuning of the independent transcription of cesA and cesB, thereby decoupling CesA and CesB to a certain extend11. Alternative modular expression of the polyketide synthetase pikAIV has been reported to be involved in the generation of two macrolactones differing in size while a potential contribution of alternative gene expression to the structural diversification of NRPS products still needs to be deciphered35.
Figure 8.
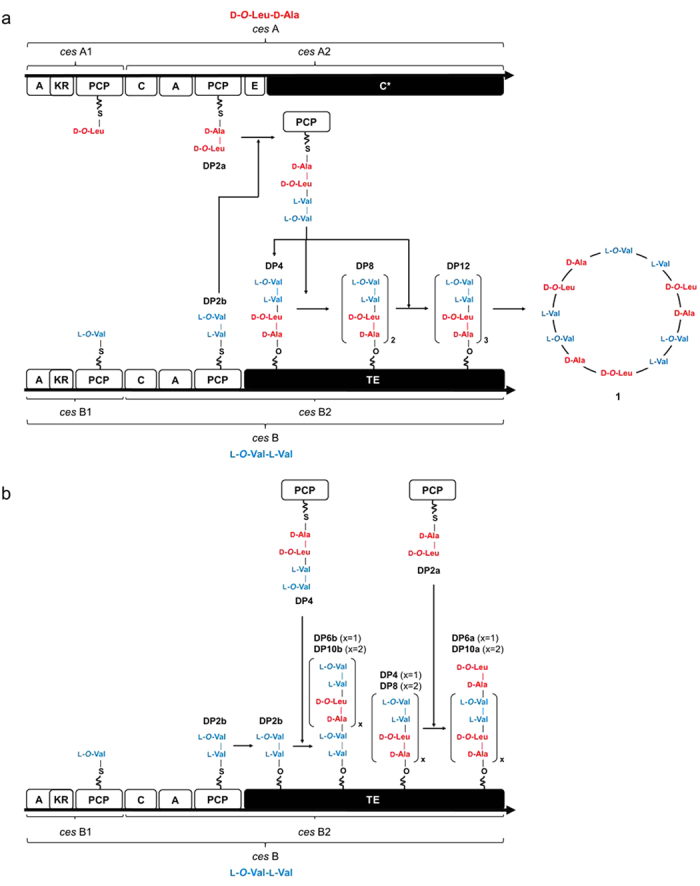
(A) Proposed biosynthesis of cereulide. The CesA and CesB modules of cereulide NRPS are made up the adenylation (A) domain, that recognize, select, and reduce α-ketocarboxylic acids to α-hydroxyacyl-S-pantetheinyl intermediates by means of a ketoreductase (KR) domain. The peptidyl carrier protein (PCP) captures the precursor adenylates as covalent thioesters via a phosphopantetheine linker. Dipeptide formation is catalyzed by a condensation (C) domain catalyzing the nucleophilic attack of the downstream PCP-bound acceptor monomer on the activated thioester of the upstream PCP-bound donor monomer intermediate, thus D-O-Leu-D-Ala (DP2a) and L-O-Val-L-Val (DP2b) are generated independently by CesA and CesB. The C* domain catalyzes the ester bond formation to reveal the PCP-bound tetradepsipeptide DP4 which is subsequently transferred to the CesB C-terminal TE domain. Again catalyzed by the C* domain, a second DP4 is then connected to the TE-bound tetradepsipeptide by esterification, thus affording DP8 and DP12 upon extension by a third DP4 unit prior to macrolactonization to cereulide (1). (B) As side products, hexa- and decadepsipeptide esters are generated by an incorrect transfer of DP2a onto DP4 and DP8 leading to the DP6a and DP10a, respectively. Transesterification of DP2b onto the TE domain, followed by C* domain catalyzed esterification with one or two units of DP4 gives rise to the DP6b and DP10b, respectively.
Although NRPS product assembly frequently seems to display a strict correlation between the enzymatic domain sequence of the megasynthase and the position of the amino acid building blocks in the peptide product, this colinearity rule is not strictly reinforced in nature36,37. Recently, the molecular mechanisms involved in the noncanonical biosynthesis of the thalassospiramide family of cyclic lipopeptides have been convincingly demonstrated to involve atypical NRPS biochemical features like module skipping and multimodule iteration38.
The proposed mechanism considering dipeptides rather than single monomers as the basic modules in tetradepsipeptide assembly and oligomerization of tetradepsipeptides to give octa- and dodecadepsipeptides is well in line with the higher abundance of tetra-, octa-, and dodecadepsipeptide esters released upon ethanolysis when compared to di-, hexa-, and decadepsipeptide esters (Fig. 6). The low abundance hexa- and decadepsipeptide esters are proposed to be generated by an incorrect transfer of D-O-Leu-D-Ala-S-PCP (DP2a) onto L-O-Val-L-Val-D-O-Leu-D-Ala-O-TE (DP4) and (L-O-Val-L-Val-D-O-Leu-D-Ala)2-O-TE (DP8) leading to the generation of the hexadepsipeptide O-Leu-D-Ala-(L-O-Val-L-Val-D-O-Leu-D-Ala)-O-TE (DP6a) and O-Leu-D-Ala-(L-O-Val-L-Val-D-O-Leu-D-Ala)2-O-TE (DP10a), respectively (Fig. 8b). Incorrect transesterification of L-O-Val-L-Val-S-PCP (DP2b) to the TE domain, followed by C* domain catalyzed esterification with one or two units of tetradepsipeptidyl-S-PCP gives rise to the TE-bound hexadepsipeptide (L-O-Val-L-Val-D-O-Leu-D-Ala)-L-O-Val-L-Val-O-TE (DP6b) and decadepsipeptide (L-O-Val-L-Val-D-O-Leu-D-Ala)2-L-O-Val-L-Val-O-TE (DP10b), respectively.
These newly proposed mechanisms also help to explain the biosynthesis of isocereulides A (2) to G (8). Formation of hexa- and decadepsipeptides via NRPS misoperation is key to form the isocereulides E, F, and G, whereas peptide assembly in the biosynthesis of isocereulides A-D corresponds well to that of cereulide differing that L-O-Val is substituted by L-O-Leu (isocereulide A), L-Val by L-Ala (isocereulide D), D-Ala by D-Ser (isocereulide C), and D-O-Leu by D-O-Val (isocereulide B) due to a relaxed substrate specificity of the A domains and/or an inefficient proof reading by CesT.
In summary, this study reveals novel insights into the biosynthesis of cereulide and isocereulides, highlighting dipeptides rather than single amino or hydroxy acids as the basic modules in tetradepsipeptide assembly. It is proposed that the C* domain located at the C-terminus of CesAand the TE domain located at the C-terminus of CesB works as a cooperative esterification and depsipeptide elongation center, repeatedly recruiting the action of the C* domain to oligomerize tetradepsipeptides to give octa- and dodecadepsipeptides prior to macrocyclization by the TE domain. Since the valinomycin synthetase Vlm shows the same genetic architecture as Ces, it is tempting to speculate that the novel mechanism of non-ribosomal peptide assembly proposed in this study is not a specificity of the cereulide synthetase but a more common logic for biosynthesis of ester bond containing NRPS products, thereby contributing to biodiversity of natural depsipeptide products. Further structural and functional studies will be necessary to decipher the exact mechanism of the unusual C* domain located in trans in the C-terminus of CesA. Such knowledge would not only contribute to a better understanding of the complex biosynthetic pathways involved in the natural generation of depsipeptides, but could also open new avenues for the development of novel pharmaceuticals.
Methods
Chemicals
The following chemicals were purchased for cultivation and extraction of bacterial cultures: MeOH and EtOH (Mallinckrodt Baker B.V., Deventer, Holland), pepton tryptone, yeast extract (Oxoid Hamphsire, England), NaCl (Carl Roth, Karlsruhe, Germany), and D-(+)-glucose monohydrate (Fluka, Sigma Aldrich, Steinheim, Germany). H2O for chromatography was purified with an integral-5 system (Millipore, Schwalbach, Germany), solvents used were of HPLC or LC-MS grade (J.T. Baker, Deventer, Holland). Purified reference material of cereulide (1) and isocereulide A-G (2–8) was prepared as reported recently17,18. The dipeptides D-O-Leu-D-Ala and L-O-Val-L-Val were synthesized following a literature procedure18. Cultivation of strain F4810/72 in MOD-medium supplemented with 13C1-valine was performed as reported recently18,19.
Bacterial Strains and Growth Conditions
Five B. cereus strains were used in this study: the emetic reference strain F4810/72 and the foodborne intoxication strain WSBC 1092539, both recently classified as a cereulide high producers, and the cereulide low to medium producers F3876/87 isolated from a patient’s vomit and E07395/2 and WSBC 10942, both isolated from B. cereus intoxicated foods17. LB broth (100 mL) supplemented with D-glucose (0.2%) was inoculated with an overnight pre-culture (103 cfu/mL) and, then, incubated at 24 °C in baffled flasks (500 mL) whilst rotary shaking (150 rpm). After incubation for 24 h, the culture of strain F3876/87 was autoclaved (15 min, 121 °C) and centrifuged (7800 × g, 121 °C, 12 min; Sigma 3-18 K), pellets from culture portions (100 mL) were frozen in liquid nitrogen and stored at -20 °C until further use.
Comparative UPLC-TOF MS Profiling of B. cereus Strains WSBC 10925 and F3876/87
Bacterial pellets, harvested from cultures (100 mL) of strains WSBC 10925 and F3876/87, respectively, were extracted with EtOH (10 mL) by shaking at room temperature for 15 h, the extract was centrifuged twice (7800 × g, 20 °C, 12 min), the supernatant was centrifuged (18600 × g, 20 °C, 5 min) and membrane filtered (0.2 μm; Phenomenex, Aschaffenburg, Germany) to remove remaining cells and cell debris. Aliquots (3 μL) of ethanolic cell pellet extracts were analysed by means of UPLC-TOF MS using the following solvent gradient for chromatography (0.4 mL/min, 45 °C): starting with a mixture (10/90, v/v) of water and methanol, the methanol content was increased to 100% within 8 min and, then, kept constant for 1 min.
Mass Spectrometric Screening of Solvent Extractables from Strain F3876/87
Bacterial pellets, prepared from F3876/87 liquid cultures (100 mL) by centrifugation, were extracted with 10 mL of MeOH, MeCN, or MeCN/H2O (70/30, v/v), respectively, by shaking overnight (15 h) at room temperature. The individual solvent extracts were centrifuged (7800 × g; 20 °C, 12 min), membrane-filtered (0.2 μm; PTFE; Phenomenex, Aschaffenburg, Germany) to remove remaining cell debris, made up to 10 mL with MeOH and, then, analyzed by means of UPLC-TOF MS using the following chromatographic conditions (0.4 mL/min, 40 °C): starting with a mixture of H2O/MeOH (10/90, v/v), the MeOH content was kept constant for 1 min at 90%, increased to 100% within 8 min, kept constant for 1 min, decreased within 0.1 min to 90% MeOH, followed by re-equilibration for 0.4 min.
Isolation and Structure Determination of a Candidate Tetradepsipeptide Ethyl Ester (m/z 453) and Octadepsipeptide Ethyl Ester (m/z 837), Respectively
The combined EtOH extracts obtained from cultures (100 mL) of strain F4810/72 were concentrated to about 25 mL in vacuum and, after dilution with H2O (1/10, v/v), aliquots (5 mL) were separated by means of preparative RP-HPLC on a PrepStar system (Varian, Darmstadt, Germany) consisting of two HPLC-pumps (Model SD-1), a two-wavelength UV detector (Prostar 325), a fraction collector (Model 701), and equipped with a 250 × 21.2 mm, 4 μm, 90A, Jupiter Proteo column (Phenomenex) as the stationary phase. Monitoring the effluent at 220 nm, chromatography (flow rate: 18.0 mL/min) was performed starting with a MeOH/H2O mixture (85/15, v/v) for 1 min, increasing the MeOH content to 100% within 10 min, holding for 10 min, followed by a decrease of the MeOH content to 85% within 1 min and re-equilibration for 2 min at 85% MeOH. A total of 14 HPLC fractions were collected, separated from solvent in vacuum, the residue of each fraction was suspended in H2O (10 mL), freeze-dried twice and, then, screened for the target compounds showing the pseudomolecular ions m/z 453 and 837, respectively, by means of UPLC-TOF MS using the following chromatographic conditions (0.3 mL/min, 40 °C): starting with a mixture of H2O/MeOH (40/60, v/v), the MeOH content was kept constant for 1 min at 60%, increased to 90% within 8 min, increased to 95% within 10 min, increased to 100% within 1 min, kept constant for 1 min, decreased within 1 min to 60% MeOH, followed by re-equilibration for 1 min. UPLC-TOF MS analyses revealed the candidate tetradepsipeptide ethyl ester (m/z 453) and the octadepsipeptide ethyl ester (m/z 837) in HPLC fraction 3.
Further purification was done by means of analytical HPLC on a 250 × 4.6 mm, 4 μm, 90 A, Jupiter Proteo column (Phenomenex). Monitoring the effluent at 220 nm, chromatography (flow rate: 1.0 mL/min) was started with a mixture of H2O/MeOH (40/60, v/v), the MeOH content was kept constant for 1 min at 60%, increased within 20 min to 80%, increased within 5 min to 86%, increased within 10 min to 90%, increased within 1 min to 100%, kept constant for 3 min, decreased within 1 min to 60% MeOH, followed by re-equilibration for 4 min. After removing the solvent in vacuum, the candidate tetradepsipeptide ethyl ester (m/z 453; 0.4 mg) and the octadepsipeptide ethyl ester (m/z 837; 0.3 mg) obtained were suspended in water (3 mL) and freeze-dried. Aliquots (~0.1 mg) of the target compounds were dissolved in methanolic KOH solution (1.2 mol/L, 80% MeOH), incubated at 50 °C for 2 h, then adjusted to pH 5.0, and analyzed by means of UPLC-TOF MS in the negative electrospray mode. Chromatography was performed on a 2.1 × 150 mm, 1.7 μm, BEH C18 column (Waters) at 45 °C using aqueous HCOOH (0.1% in H2O) as solvent A and MeCN containing 0.1% HCOOH as solvent B and the following gradient (flow rate: 0.4 mL/min): For dipeptide analysis in the hydrolysates, chromatography started with 1% solvent B for 2.5 min, which was increased to 95% within 5.5 min, hold for 1.5 min and, then, decreased again to 1% within 0.5 min. For TOF MS analysis, scan time for the MSe method (centroid) was set to 0.1 sec, high-resolution negative ionization mode and the following ion source parameters were selected: capillary voltage (-2.5 kV), sampling cone (50 V), source temperature (150 °C), desolvation temperature (450 °C), cone gas (10 L/h) and desolvation gas (850 L/h). For the MSe method, the second MS scan function used a transfer collision energy ramp from 20 to 40 eV.
Incubation of B. Cereus in the Presence of 13C6-Cereulide
Aliquots (19.8 mL) of LB medium (including 0.2% glucose) were mixed with an ethanolic solution (0.2 mL) of 13C6-cereulide (10 μg/mL) and, then, inoculated with the B. cereus strain F4801/72 (103 KBE/mL). Before (time point: 0 h) and after incubation for 12, 20, and 40 h at 24 °C whilst shaking (150 rpm), aliquots (2.0 mL) were taken, autoclaved (15 min, 121 °C), pellets collected by centrifugation (13000 rpm, 4 min), and extracted with EtOH (1 mL) by shaking overnight (15 h) at room temperature. After centrifugation (7800 × g; 12 min, each), the supernatants were membrane-filtered (0.2 μm; PTFE; Phenomenex), made up to 1 mL with EtOH and, then, directly used for UPLC-TOF-MS analysis.
Mass Spectrometry
High-resolution mass spectrometric analyses (UPLC-ESI-TOF MS) were performed on a Waters Synapt G2-S HDMS spectrometer coupled to an Acquity UPLC core system (Waters, Milford, MA, USA) consisting of a binary solvent manager, sample manager and a 2 × 150 mm, 1.8 μm, HSS T3 C18 column (Waters, Manchester, UK). Data processing was performed by using MassLynx 4.1 SCN 851 (Waters, Manchester, UK) and the elemental composition tool for determining the accurate mass. MS analyses were performed in the positive ESI and high resolution mode with the instrument setting for the capillary voltage (+2.5 kV), sampling cone (50 V), source temperature (120 °C), desolvation temperature (450 °C), cone gas (10 L/h), and desolvation gas (850 L/h) given in parenthesis. All data were lock mass corrected on the pentapeptide leucine enkephaline (Tyr-Gly-Gly-Phe-Leu, m/z 556.2771, [M+H]+ and m/z 554.2615, [M-H]–) in a solution (2 ng/μL) of MeCN/0.1% HCO2H (1/1, v/v). Scan time for the lock mass was set to 0.3 s, an interval of 10 s and 3 scans to average with a mass window of ±0.5 Da. Calibration of the Synapt G2-S in the range from m/z 50 to 1300 was performed using a solution of sodium formate (5 mmol/L) in 2-propanol/H2O (9/1, v/v). The UPLC and Synapt G2-S systems were operated with MassLynx software (Waters, Manchester), data processing and analysis were performed using MarkerLynx XSTM software. The raw data obtained from UPLC-TOF MS analysis were processed with MarkerLynx XS using ApexTrack peak integration to detect chromatographic peaks. Marker intensity threshold was set to 2000 cps, mass window was 0.02 Da, retention time window was 0.1 s, and data were de-isotoped.
MSn-experiments were performed on a Bruker Daltonics HCTultra PTM Discovery SystemTM (Bruker Daltonics Inc., Billerica, MA, USA) using direct sample infusion via a syringe pump (4 μL/min; kd scientific, USA) and manual MSn (m/z 50-1300) using the following instrument parameters: ultra-scan mode (26.000 m/z/sec), max. accumulation time (300 ms), isolation width (m/z 4), collision amplitude (1 V; ramp: 30-200%). The following source parameters were applied: capillary voltage (+4 kV), end plate offset (-500 V), nebulizer (30 psi), dry gas (8 L/min), dry temperature (300 °C), skimmer (40 V), capillary exit (166 V). Data acquisition and processing were done by using Bruker Daltonics Data Analysis Version 3.4 (Bruker Daltonics Inc.).
Bioinformatic Analysis of the Cereulides Synthetase Gene Locus
The previously sequenced cereulide synthetase gene locus ces9 was derived from the NCBI database (Acc.No. NG_036207) and subjected to bioinformatics analysis. Sequence similarity searches were performed using the Basic Local Aligment Search Tools BLASTP40. The software packages ClustalX and TREECON were used for sequence alignments and cluster analysis41,42. To search for conserved sequence domains the online software tool Pfam43 was employed and analysis of the condensation domain function was carried out using the database NaPDos31. The C domain core motifs were identified according to Rausch et al.30
Additional Information
How to cite this article: MARXEN, SANDRA et al. Depsipeptide Intermediates Interrogate Proposed Biosynthesis of Cereulide, the Emetic Toxin of Bacillus cereus. Sci. Rep. 5, 10637; doi: 10.1038/srep10637 (2015).
Supplementary Material
Acknowledgments
We are grateful to Sofie Lösch and Ines Otte for technical assistance. This research project (project AiF 16845 N) was supported by the German Ministry of Economics and Technology (via AiF) and the FEI (Forschungskreis der ährungsindustrie e.V., Bonn).
Footnotes
Author Contributions S.M., A.R. and E.F. performed bacteria growth and extraction experiments. Depsipeptide isolation and structure determination was done by S.M., T.D.S. and T.H. M.E-S. carried out the bioinformatic analysis of the ces gene cluster. T.D.S, G.L., S.S. and M.E-S have conceptualized and supervised the study. T.H. has conceptualized, supervised and acted as overall study director.
References
- Ehling-Schulz M., Fricker M., Scherer S. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 48, 479–487 (2004). [DOI] [PubMed] [Google Scholar]
- Stenfors Arnesen L. P., Fagerlund A., Granum P. E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606 (2008). [DOI] [PubMed] [Google Scholar]
- Agata N. et al. A novel dodecadepsipeptide, cereulide, isolated from Bacillus cereus causes vacuole formation in HEp-2 cells. FEMS Microbiol. Lett. 121, 31–34 (1994). [DOI] [PubMed] [Google Scholar]
- Suwan S. et al. Structure of cereulide, a cyclic dodecadepsipeptide toxin from Bacillus cereus and studies on NMR characteristics of its alkali metal complexes including a conformational structure of the K+ complex. J. Chem. Soc. Perkin Trans. 1, 765–775 (1995). [Google Scholar]
- Tomoda H. et al. New cyclodepsipeptides, enniatins D, E and F produced by Fusarium sp. FO-1305. J. Antibiot. 45, 1207–1215 (1992). [DOI] [PubMed] [Google Scholar]
- Xu Y. et al. Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genet. Biol. 46, 353–364 (2009). [DOI] [PubMed] [Google Scholar]
- Ehling-Schulz M. et al. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71, 105–113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magarvey N. A., Ehling-Schulz M., Walsh C. T. Characterization of the cereulide NRPS α-hydroxy acid specifying modules: Activation of α–keto acids and chiral reduction on the assembly line J. Am. Chem. Soc. 128, 10698–10699 (2006). [DOI] [PubMed] [Google Scholar]
- Ehling-Schulz M. et al. Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6, 20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A. et al. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1 J. Bacteriol. 189, 52–64 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dommel M. K. et al. Identification of the main promoter directing cereulide biosynthesis in emetic Bacillus cereus and its application for real-time monitoring of ces gene expression in foods. Appl Environ Microbiol. 76, 1232–1240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber S. A., Marahiel M. A. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105, 715–738 (2005). [DOI] [PubMed] [Google Scholar]
- Belshaw P. J., Walsh C. T., Stachelhaus T. Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284, 486–489 (1999). [DOI] [PubMed] [Google Scholar]
- Clugston S. L., Sieber S. A., Marahiel M. A., Walsh C. T. Chirality of peptide bond-forming condensation domains in nonribosomal peptide synthetases: the C5 domain of tyrocidine synthetase is a (D)C(L) catalyst. Biochemistry 42, 12095–12104 (2003). [DOI] [PubMed] [Google Scholar]
- Strieker M., Tanovic A., Marahiel M. A. Nonribosomal peptide synthetases: structures and dynamics. Curr. Opin. Struct. Biol. 20, 234–240 (2010). [DOI] [PubMed] [Google Scholar]
- Koglin A., Walsh C. T. Highlight: structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 26, 987–1000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark T. et al. Mass spectrometric profiling of Bacillus cereus strains and quantitation of the emetic toxin cereulide by means of stable isotope dilution analysis and HEp-2 bioassay. Anal. Bioanal. Chem. 405, 191–201 (2013). [DOI] [PubMed] [Google Scholar]
- Marxen S. et al. Chemodiversity of cereulide, the emetic toxin of Bacillus cereus. Anal. Bioanal. Chem. 407, 2439–2453 (2015). [DOI] [PubMed] [Google Scholar]
- Bauer T. Stark T., Hofmann T., Ehling-Schulz M. Development of a stable isotope dilution analysis (SIDA) for the quantification of the Bacillus cereus toxin cereulide in foods. J. Agric. Food Chem. 58, 1420–1428 (2010). [DOI] [PubMed] [Google Scholar]
- Melling J., Capel B. J. Characteristics of Bacillus cereus emetic toxin. FEMS Microbiol. Lett. 4, 133–135 (1978). [Google Scholar]
- Shinagawa K., Konuma H., Sekita H., Sugii S., Emesis of rhesus monkeys induced by intragastric administration with the HEp-2 vacuolation factor (cereulide) produced by Bacillus cereus. FEMS Microbiol. Lett. 130, 87–90 (1995). [DOI] [PubMed] [Google Scholar]
- Rajkovic A. et al. Heat resistance of Bacillus cereus emetic toxin, cereulide. Lett. Appl. Microbiol. 46, 536–541 (2008). [DOI] [PubMed] [Google Scholar]
- Hornbogen T., Glinski M., Zocher R. Biosynthesis of depsipeptide mycotoxins in Fusarium. Eur. J. Plant Pathol. 108, 713–718 (2002). [Google Scholar]
- Glinski M., Urbanke C., Hornbogen T., Zocher R. Enniatin synthetase is a monomer with extended structure: evidence for an intramolecular reaction mechanism. Arch. Microbiol. 178, 267–273 (2002). [DOI] [PubMed] [Google Scholar]
- Balibar C. J., Walsh C. T. From thioesters to amides and back: condensation domain reversibility in the biosynthesis of vibriobactin. Chembiochem. 9, 42–45 (2008). [DOI] [PubMed] [Google Scholar]
- Balibar C. J., Vaillancourt F. H., Walsh C. T. Generation of D amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem. Biol. 11, 1189–1200 (2005). [DOI] [PubMed] [Google Scholar]
- Zaleta-Rivera K. et al. A bidomain nonribosomal peptide synthetase encoded by FUM14 catalyzes the formation of tricarballylic esters in the biosynthesis of fumonisins. Biochemistry 45, 2561–2569 (2006). [DOI] [PubMed] [Google Scholar]
- Ding Y., Rath C. M., Bolduc K. L., Håkansson K., Sherman D. H. Chemoenzymatic synthesis of cryptophycin anticancer agents by an ester bond-forming non-ribosomal peptide synthetase module. J. Am. Chem. Soc. 133, 14492–14495 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli N. M., Long D. H., Townsend C. A. β-Lactam formation by a non-ribosomal peptide synthetase during antibiotic biosynthesis. Nature , in press, 10.1038/nature14100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C., Hoof I., Weber T., Wohlleben W., Huson D. H. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evolutionary Biology 7, 78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N. et al. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7, e34064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Fiss E., Jacobs W. R. Jr. Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J. Bacteriol. 180, 4676–4685 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duitman E. H. et al. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96, 13294–13299 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S. et al. Paenilamicin: structure and biosynthesis of a hybrid nonribosomal peptide/polyketide antibiotic from the bee pathogen Paenibacillus larvae. Angew. Chem. Int. Ed. 53, 10821–10825 (2014). [DOI] [PubMed] [Google Scholar]
- Xue Y., Sherman D.H. Alternative modular polyketide synthase expression controls macrolactone structure. Nature. 403, 571–575 (2000). [DOI] [PubMed] [Google Scholar]
- Wenzel S. C., Müller R. Formation of novel secondary metabolites by bacterial multimodular assembly lines: deviations from textbook biosynthetic logic. Curr. Opin. Chem. Biol. 9, 447–458 (2005). [DOI] [PubMed] [Google Scholar]
- Lane A. L., Moore B. S. A sea of biosynthesis: marine and natural products meet the molecular age. Nat. Prod. Rep. 28, 411–428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. C. et al. Biosynthetic multitasking facilitates thalassospiramide structural diversity in marine bacteria. J. Am. Chem. Soc. 135, 1155–1162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling-Schulz. M. et al. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151, 183–197 (2005). [DOI] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., De Wachter R. Construction of evolutionary distance trees with TREECON for windows: accounting for variation in nucleotide substitution rate among sites. Comput. Appl. Biosci. 13, 227–230 (1997). [DOI] [PubMed] [Google Scholar]
- Finn R. D. et al. Pfam: the protein families database. Nucleic. Acids Res. 42, database issue, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.