Abstract
Biogenic amines regulate a variety of behaviors. Their functions are predominantly mediated through G-protein-coupled 7-transmembrane domain receptors (GPCR), 16 of which are predicted to exist in the genome sequence of the nematode Caenorhabditis elegans. We describe here the expression pattern of several of these aminergic receptors, including two serotonin receptors (ser-1 and ser-4), one tyramine receptor (ser-2), and two dopamine receptors (dop-1 and dop-2). Moreover, we describe distinct but partially overlapping expression patterns of different splice forms of the ser-2 tyramine receptor locus. We find that each of the aminergic receptor genes is expressed in restricted regions of the nervous system and that many of them reveal significant overlap with the expression of regulatory factors of the LIM homeobox (Lhx) gene family. We demonstrate that the expression of several of the biogenic amine receptors is abrogated in specific cell types in Lhx gene mutants, thus establishing a role for these Lhx genes in regulating aspects of neurotransmission. We extend these findings with other cell fate markers and show that the lim-4 Lhx gene is required for several but not all aspects of RID motor neuron differentiation and that the lim-6 Lhx gene is required for specific aspects of RIS interneuron differentiation. We also use aminergic receptor gfp reporter fusions as tools to visualize the anatomy of specific neurons in Lhx mutant backgrounds and find that the development of the elaborate dendritic branching pattern of the PVD harsh touch sensory neuron requires the mec-3 Lhx gene. Lastly, we analyze a mutant allele of the ser-2 tyramine receptor, a target of the ttx-3 Lhx gene in the AIY interneuron class. ser-2 mutants display none of the defects previously shown to be associated with loss of AIY function.
Keywords: LIM homeobox genes, Biogenic amine receptors, C. elegans, Tyramine
Introduction
LIM homeobox (Lhx) genes code for regulatory factors involved in nervous system development (Bach, 2000; Hobert and Westphal, 2000; Jessell, 2000; Shirasaki and Pfaff, 2002). Expression of these genes can often be found in postmitotic neurons, suggesting a role for Lhx genes in determining and maintaining the differentiated state of neurons (Hobert and Westphal, 2000). Whereas the function of Lhx genes in some neuronal cell types has been elucidated, the limited availability of cell fate markers has hindered an analysis of Lhx gene function in many other neuronal cell types. Further complicating the matter, Lhx genes can act in a manner highly dependent on cellular context. For example, the Apterous-type ttx-3 Lhx gene acts as a regulator of all known subtype-specific characters of one specific C. elegans interneuron class but does not appear to affect the differentiation of other neuron classes in which ttx-3 is normally expressed (Altun-Gultekin et al., 2001). For this reason, the function of any given Lhx gene must be painstakingly assessed for each individual neuron class without drawing any assumptions from other neuron classes.
Besides the context-dependent action of individual transcription factors, another theme emerging from the study of transcriptional control of neuronal development is the progressively restricted action of hierarchically ordered transcription factor cascades. Early acting transcription factors affect the overall generation of a neuron, while later acting transcription factors affect the adoption of all known subtype characteristics without affecting the pan-neuronal properties of a neuron. Even later acting factors determine only a very limited subset of neuronal subtype characteristics. Different types of Lhx genes have been found to act at each of these distinct steps in neuronal development. For example, the vertebrate Lhx factor, Isl1, is required for motor neuron generation (Pfaff et al., 1996). The C. elegans mec-3 and ttx-3 genes are necessary for the adoption of all subtype but not pan-neuronal characteristics of various neuron classes (Altun-Gultekin et al., 2001; Duggan et al., 1998; Way and Chalfie, 1988), whereas the lim-6 Lhx gene is required for the expression of only a restricted subset of motor neuron features (Hobert et al., 1999) (O.H., unpublished observations).
To determine at what step a given transcription factor acts requires the availability of tools to assess how a given neuron subtype is affected by the loss of that transcription factor. Such tools may, for example, allow for the visualization of the anatomical or physiological properties of a neuron. Perhaps best suited for the analysis of transcription factor function, however, are molecular tools, that is, cell fate markers that reflect neuron subtype-specific gene expression profiles. The activity of transcription factors acting late in neuronal differentiation can be monitored by the expression of genes that determine specific functional properties of a neuron, such as ion channels, neurotransmitter synthesizing enzymes, or neurotransmitter receptors. In this paper, we describe a set of such terminal differentiation markers, namely G-protein-coupled neurotransmitter receptors, and use them to assess the roles that LIM homeobox genes play in the differentiation of specific neural subtypes. Given the established role of biogenic amines in the regulation of a variety of behaviors in the adult nervous system (e.g., Blenau and Baumann, 2001), we concentrated on metabotropic, G-protein-coupled neurotransmitter receptors (GPCR) predicted to have biogenic amine specificity.
Metabotropic receptors for the biogenic amines serotonin, dopamine, and tyramine have been described in C. elegans on a pharmacological level (Olde and McCombie, 1997; Rex and Komuniecki, 2002; Suo et al., 2002). However, their expression patterns have not previously been reported. These receptors are presumed to mediate the roles that both serotonin and dopamine were shown to have in modulating a variety of C. elegans behaviors, such as egglaying and locomotory behavior (Mendel et al., 1995; Sawin et al., 2000; Schafer and Kenyon, 1995; Segalat et al., 1995; Weinshenker et al., 1995). In addition, serotonin is involved in regulating mating behavior, feeding behavior, and the metabolic state of a worm (Avery and Horvitz, 1990; Loer and Kenyon, 1993; Sze et al., 2000). Localization of the receptors for serotonin and dopamine may provide important insights into the nature of the neuronal circuits that underlie the execution of these behaviors. The expression pattern analysis of serotonin, dopamine, and tyramine receptors that we present here is aimed at providing the first glimpses into this direction.
In contrast to serotonin and dopamine, the role of tyramine in C. elegans has not been reported to date. Tyramine is the chemical precursor to octopamine, a biogenic amine regulating a variety of behaviors in invertebrates and thought to be the invertebrate nervous system counterpart to norepinephrine (Roeder, 1999). While initially thought to simply be an octopamine precursor, the molecular cloning of high-affinity tyramine receptors in flies (Saudou et al., 1990) and in vertebrates (Borowsky et al., 2001) as well as the specific effect of tyramine on cocaine sensitization in flies (McClung and Hirsh, 1999) suggests tyramine functions as a neuroactive substance. Furthermore, a hypomorphic allele of the Drosophila tyramine receptor revealed a function for this protein in olfactory behavior and in modulating signaling at the neuromuscular junction (Kutsukake et al., 2000; Nagaya et al., 2002). We present here an expression pattern analysis of a worm tyramine receptor, its various splice forms, and characterize a putative null allele of the receptor.
Materials and methods
Sequence analysis
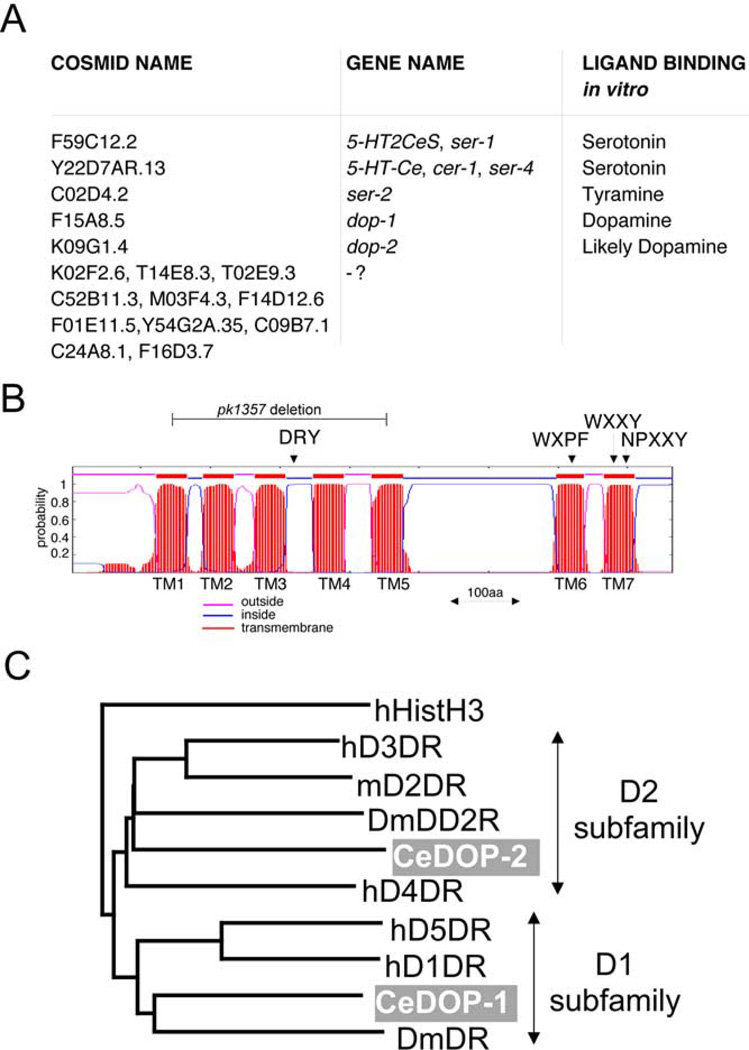
Biogenic amine receptors were identified in reiterative BLASTp searches (Altschul et al., 1990) using the Genbank BLAST server at http://www.ncbi.nlm.nih.gov/blast/ and the C. elegans genome BLAST server at the Sanger Center at http://www.sanger.ac.uk/Projects/C_elegans/blast_server.shtml. The pairwise sequence alignment was evaluated with the blosum62 substitution matrix. Reiterative BLAST searches identified 16 biogenic amine receptors with a characteristic 7-trans-membrane topology and specific sets of conserved amino acids (Gether, 2000) (Fig. 1A and B). These 16 proteins are all consistently picked up as the first 16 hits in BLAST searches of the Sanger Center C. elegans genome database using invertebrate and vertebrate biogenic amine receptors as the query sequence. Conversely, these 16 predicted proteins consistently pick up only biogenic amine receptors as first hits when they are used as the query sequence in Genbank BLAST searches. The next most similar group of proteins retrieved by BLAST using these 16 proteins as query are the GAR proteins (GAR-1/C15B12.5, GAR-2/F47D12.1, and GAR-3/C53A5.12), which are muscarinic acetylcholine receptors (Hwang et al., 1999). Aminergic receptor sequences were aligned with ClustalX v1.81, and a phylogenetic tree was constructed with NJPlot (using the neighbor joining method) and PAUP4.0b10 (using parsimony analysis). Transmembrane prediction was conducted by using the TMHMM Server v2.0 at http://ww-w.cbs.dtu.dk/services/TMHMM/ (Krogh et al., 2001).
Fig. 1.
The complete set of predicted biogenic amine receptors in the C. elegans genome. (A) Biogenic amine receptors retrieved through reiterative BLAST searches are shown. Ligand binding was assessed by in vitro binding assays as described in the respective references mentioned in the text. We infer dopamine as a likely ligand for DOP-2 given its similarity to the D2-dopamine receptor family (C). (B) Transmembrane prediction of a representative biogenic amine receptor, SER-2A, using a hidden Markov model (TMHMM) (Krogh et al., 2001). Characteristic motifs thought to be involved in biogenic amine binding (WXPF and WXXY) (Roth et al., 1997), receptor activation (DRY), and receptor desensitization/internalization (NPXXY) (Gether, 2000) are present in all putative biogenic amine receptors listed in (A). The position of the ser-2 deletion allele, pk1357 (described later in the text), is also shown. (C) The C. elegans genome contains two dopamine receptors, F15A8.5/ DOP-1 and K09G1.4/DOP-2 (gray shading), which are representative of the D1 and D2 dopamine receptor subfamilies. D1- and D2-type dopamine receptor sequences were retrieved from GenBank and assembled into a phylogenetic tree with the distance-based neighbor-joining method using ClustalX. A similar clustering of DOP-1 and DOP-2 can be observed with parsimony analysis using PAUP (data not shown). Human histamine receptor H3 was used as an outlier. “h” stands for human, “m” for mouse, and “Dm” for Drosophila. GenBank Accession nos. are: DmDD2R: AAN15955.1, hD3DR: AAB08750.1, hD5DR: AAN01276.1, hD4DR: AAB59386.1, DmDR: AAF55030.2, hD1DR: AAB26273.1, CeDOP-1: AAO91737.1, mD2DR: NP_034207.1, CeDOP-2: NP_505478.2, hHistH3: AAD38151.1
Strains
The wild-type strain used is the N2 Bristol isolate. LIM homeobox null mutant strains used in this study are: ttx-3(mg158), ttx-3(ot22) (Altun-Gultekin et al., 2001), lim-6(nr2073) (Hobert et al., 1999), lim-4(ky403) (Sagasti et al., 1999), egl-38(n578)mec-3(n3197), mec-3(u298), mec-3(e1338) (Way and Chalfie, 1988), and lin-11(n389); him-5(e1467)V (Ferguson and Horvitz, 1985). RIS cell fate markers are nuIs1 [Is(glr-1::gfp; lin-15)] (Hart et al., 1995), otIs11 [Is(zig-5::gfp; rol-6)] (Aurelio et al., 2002), and otIs39 [Is(unc-47Δ::gfp; lin-15)] (Aurelio et al., 2003). RID cell fate markers are lqIs4 [Is(ceh-10::gfp; lin-15)] (a gift from E. Lundquist), otIs33 [Is(kal-1::gfp; pBX] (Bu¨low et al., 2002), and otIs12 [Is(zig-5::gfp; rol-6)] (Aurelio et al., 2002). Markers for the AIY interneuron class are mgIs18 [Is(ttx-3::gfp; lin-15)] (Altun-Gultekin et al., 2001) and otIs121 [Is(ttx-3::rfp; unc-122::gfp] (see below).
Isolation of ser-2 cDNAs
cDNA was isolated by PCR from mixed stage and embryonic cDNA libraries using gene-specific primers and vector primers. So as to avoid bias, cDNA was isolated using a two-part approach. To isolate the 5′ end of the cDNA, a nested PCR was carried out. The first reaction 5′ primer, M13-Reverse, corresponded to vector sequence 5′ of the cDNA insert, whereas the nested 5′ primer was T3, also upstream of the cDNA insertion site. The first reaction 3′ primer (5′-CTAAGGTTGCGCACTCATTC) is complementary to the end of the ser-2 gene. The nested 3′ primer (5′-CTCTTTTCTCCTTCGCCAC) is complementary to sequence at the start of the tenth exon. To isolate the 3′ end of the gene, a similar strategy was employed. 3′ first and second reaction vector primers were M13-Forward and T7, respectively. The first reaction 5′ primer corresponded to sequence in exon 2 (5′-ATGGTGTTACGAGCCATCG). The nested reaction 5′ primer used (5′-ATGACCTCTAT-GAAACCGCC) is found in exon 2, 74 nucleotides downstream of the first reaction 5′ primer. Given a predicted cDNA size of approximately 1500 base pairs, all bands larger than 750 base pairs were cloned into the pCR-XL-TOPO vector (Invitrogen product # K4700-10). A total of 38 candidates containing inserts were randomly selected and sequenced using 5′ and 3′ vector primers.
In order to determine whether there was a correlation between the inclusion of exon 5A and promoter choice, the following experiments were performed. Ninety-four pCR-XL-TOPO transformants containing cDNA inserts were randomly selected and screened by PCR using primers specific to exons 1a (5′-TGCTAAGACATCATGTTCCG) and 5a (5′-CCCTCGCCATAGTTAGATTG). No product was detectable, although a similarly sized product using primers specific to exons 1a and 6 was observed. In addition, a different set of 94 randomly selected transformants was selected to determine which promoter was used to drive expression of exon 5a–containing cDNAs. Specifically, the M13-Reverse vector primer and the exon 5a primer were used to obtain 12 cDNAs out of the 94 screened. These 12 PCR products were sequenced with respect to the 5′-most end of the cDNA. Two were found to contain exon 1bc, 10 contained exon 1d, whereas none were identified as having exon 1a sequence.
Generation of reporter gene fusions and transgenic strains
All three ser-2 reporter constructs (see Fig. 3A) were generated by using a PCR fusion protocol, using pPD95.75 as a template for green fluorescent protein (gfp) (Hobert, 2002). For all gfp fusion primers listed, gfp vector sequence is indicated in lowercase, and gene-specific sequence is indicated in uppercase. The primers for the respective fusions are: ser-2prom1 5′upstream: 5′-GGACTCCCTCCTTGTATGGC (no nested primer was used); ser-2prom1 3′primer: 5′-cccggggatcctcTA-GAGGTCATGGGAATGTCC; ser-2prom2 5′upstream (OH346): 5′-GTCTGGTAAGTTGAACATGGAGTC; ser-2prom2 5′upstream nested (OH347): 5′-ATTTTATGACTT-TCACTAGAAATG; ser-2prom2 3′ primer (OH345): 5′ -agtcgacctgcaggcatgcaagctCATTTTTGCAAATTACTTGA-GGC; ser-2prom3 5′upstream (OH349): 5′-CATGCT-TGTTCTAGTGATCAAAATTG; ser-2prom3 5′upstream nested (OH350): 5′-GTAAAAGTTTAGTAAATTAACTGC; and ser-2prom3 3′ primer (OH348): 5′-agtcgacctgcaggcatg-caagctCATTATGTGTTGTGATGTCACAA.
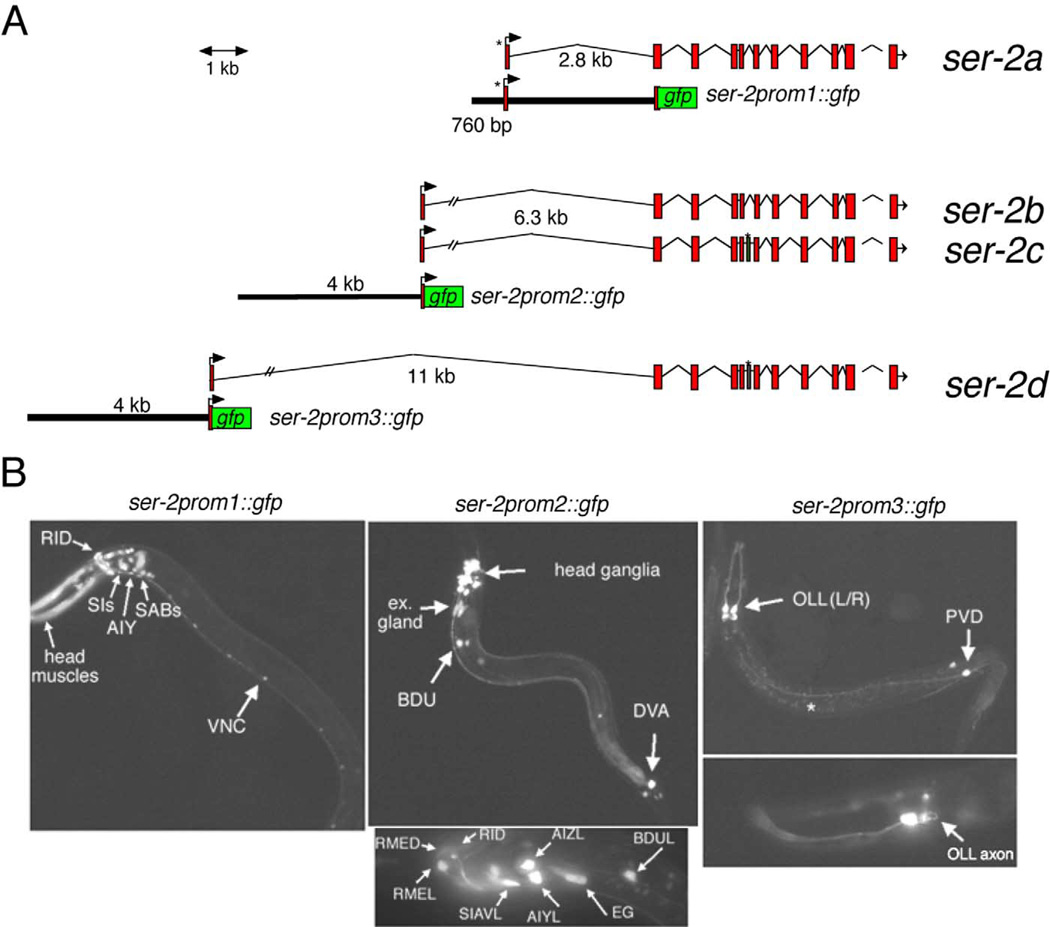
Fig. 3.
Expression pattern of ser-2reporter gene fusions. (A) Reporter gene constructs. (B) Expression of gfp in transgenic worms. Anterior is always to the left. Reporter transgenes are: ser-2prom1::gfp = otls107, ser-2prom2::gfp = otEx536, ser-2prom3::gfp = otIs138. Animals were scored during larval stages. Expression in the AIY interneuron class was confirmed by colabeling with otIs121 (see Materials and methods). The white star indicates nonspecific gut autofluorescence.
ser-2prom1::gfp was injected into lin-15 mutant animals with lin-15(+) to generate adEx1450, which was chromosomally integrated to yield otIs107. ser-2::prom2::gfp transgenic lines were generated both with rol-6 as an injection marker in an N2 wild-type background (otEx448) as well as with pBX as an injection marker in a pha-1(e2123) mutant background (otEx536, otEx537). All transgenic lines showed similar expression patterns. ser-2prom3::gfp was injected with rol-6 as an injection marker into an N2 wild-type background (otEx449–456). All transgenic lines showed similar expression patterns. otEx449 was chromosomally integrated to yield transgene otIs138, which was twice backcrossed. A translational fusion of the whole ser-2 locus to gfp was created by using an in vivo recombination technique. Specifically, two overlapping PCR fragments, one containing the 5′ part of a locus, the other containing the remainder of the locus PCR-fused to gfp, were coinjected into the worm. Recombination of these two fragments via the homologous region [which had been previously observed using rol-6 and unc-68 as test cases (Maryon et al., 1998; Mello et al., 1991)] leads to the expression of a full-length ser-2::gfp fusion.
The dop-1prom1::gfp fusion was created by PCR fusion with primers that range from −3.46 kb to −354 bp of the corrected ATG (see main text). The sequences of the primers are: dop-1prom1 5′upstream (oBJ1): 5′-TCAAG-CACTTTGTGAACACG; dop-1prom1 5′upstream nested (oBJ2): 5′-TAGGATGGGCGACCTAGGATG; and dop-1prom1 3′primer (oBJ3): 5′-gtcgacctgcaggcatcgaagctTTT-GATTTTTTTAAACCAG. The PCR product was injected into pha-1(e2123ts) along with pBX as an injection marker. Array names are otEx233–236.
The dop-1prom2::gfp fusion was created by using the same set of upstream primers as in dop-1prom1::gfp but using a 3′ primer that extends six nucleotides 3′ to the corrected ATG. The sequence is: 5′-gcaggcatgcaagctATCGTTCATC-GAATTTCTGG The PCR product was injected into pha-1(e2123ts) along with pBX as an injection marker. Array names are otEx1043–1045.
The dop-1trans::gfp fusion was created by overlap extension PCR using the upstream primer sequence (5′-TCCCTCCCATTCAAAGTGTTCAC) located 4.1 kb upstream of the predicted ATG, and the downstream primer (5′ -catacctttgggtcctTTGGCAGAAGAGTTTGCGTATCC) 4.6 kb downstream of the ATG within the seventh exon of the gene. No nested primers were used for this reaction. The purified GFP fusion product was coinjected with lin-15(+) into lin-15(n765ts) worms to generate adEx1647.
The dop-2::gfp fusion was created by subcloning 4.4 kb of genomic DNA directly upstream of the predicted ATG start codon into a gfp expression vector and was kindly provided by Gisela Sandoval. DNA was injected into pha-1(e2123ts) along with pBX as an injection marker to create transgene otEx980.
The ser-4::gfp fusion was created by PCR fusion. The upstream primer (5′-TTCCCCCCCTCTGACGACTACT) is located 4.1 kb upstream of the predicted ATG, and the downstream primer (5′-tgggtcctttggccaatccAGAAGTCA-CAGACCACGAC) is 3.7 kb downstream of the ATG within the third exon of the gene. No nested primers were used for the fusion reaction. The transgene array is adEx1616.
ser-1::gfp was created by PCR fusion of a 9.4-kb genomic fragment to gfp and encompasses 1.7 kb of sequences upstream of the ATG start codon. The resulting transgenic line was chromosomally integrated (kindly provided by Stephen Nurrish).
Reporter fusions for the remaining, uncharacterized biogenic amine receptor genes contain the genomic regions described below. A slight variation of the gfp fusion protocol described above was used such that nested primers were not used and a slightly different primer in the 3′ UTR of gfp was utilized (T. Niacaris, unpublished observations).
C09B7.1::gfp was created by using the upstream primer (5′-CGGGCCTGTGAAATGTCAACT) located 2.2 kb upstream of the predicted start site and the downstream primer (5′ccaatcccGGGGATCCACATGCCGAAAGACTT) 6.5 kb downstream of the ATG within the fourth exon of 7. The construct was injected into lin-15(n765ts) mutants with lin-15(+) to generate adEx1641.
K02F2.6::gfp was created by using the upstream primer (5′-TTTCCCCTTCATTCATTTTCCGT) located 4.1 kb upstream of the predicted start site and the downstream primer (5′ -ggccaatcccggGGATCGAATTGCCGTTGAG) 2.1 kb downstream of the ATG within the seventh exon of 7. The construct was injected into lin-15(n765ts) mutants with lin-15(+) to generate adEx1644.
T02E9.3::gfp was created by using the upstream primer (5′-GTCGCTTTCTTTCCCATCTTGTT) located 4.0 kb upstream of the predicted start site and the downstream primer (5′ -ccaatcccggggATCCTTCCTGTTCAGCCATTTTT) 3.5 kb downstream of the ATG within the seventh exon of 7. The construct was injected into lin-15(n765ts) mutants with lin-15(+) to generate adEx1646.
F14D12.6::gfp was created by using the upstream primer (5′-CGTATGTCCAAGAGCCCGCTA) located 4.0 kb upstream of the predicted start site and the downstream primer (5′ -ttggccaatcCCGGGTTGATACCAGAGTTACCTG) 1.8 kb downstream of the ATG within the eighth exon of 9. The construct was injected into lin-15(n765ts) mutants with lin-15(+) to generate adEx1649.
F01E11.5::gfp was created by using the upstream primer (5′-TTTGAAAATCTCTCGCTGATCCG) located 0.3 kb upstream of the predicted start site and the downstream primer (5′ -cggggatcctctagagTCGACTTGCCCACTACTTTCG) 7.7 kb downstream of the ATG within the ninth exon of 13. The construct was injected into lin-15(n765ts) mutants with lin-15(+) to generate adEx1648.
M03F4.3::gfp was created by using the upstream primer (5′-CGCTCCATATCGCTTGTAAGC) located 0.2 kb upstream of the potential start site and the downstream primer (5′-cggggatcctctagagTCGTCCAAAGCGAGCCG) 3.8 kb downstream of the ATG within the second exon of 11. The construct was injected into lin-15(n765ts) mutants with lin-15(+) to generate adEx1519.
The last two reporter gene fusions only contain a short region upstream of the ATG. This is due to a change in gene structure predictions since the time at which primers were designed. Current predictions of gene structure are based on recently identified expressed sequence tags (http://www.wormbase.org).
Microscopy and cell identifications
Animals were mounted and analyzed with a Zeiss Axioplan 2 microscope equipped with Nomarski filters and a fluorescent light source. Images were collected with a CCD camera and the Openlab Software package, with the exception of Fig. 5F, which was collected with a confocal microscope.
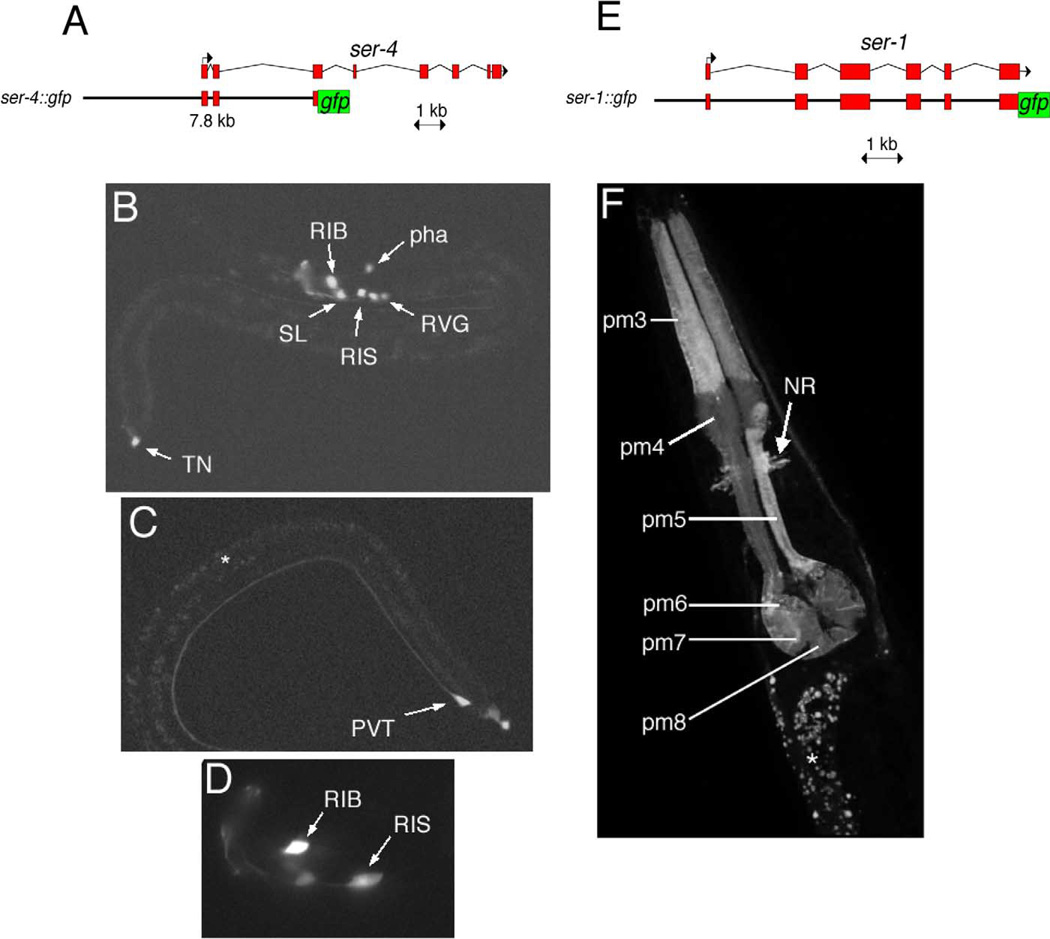
Fig. 5.
Expression of ser-4::gfp and ser-1::gfp. (A) Reporter gene construct for ser-4. (B) Overview of expression of ser-4::gfp (adEx1616) in a mid-larval stage animal. RIB, RIS = head neuron classes. SL = sublateral neurons. pha = pharyngeal neuron. RVG = retrovesicular ganglion. TN = tail neuron (either DVA or DVC). (C) Expression in the PVT tail neuron. (D) Higher magnification view of the two head neuron classes that express ser-4::gfp most consistently, RIB and RIS. All animals are at mid larval stages. (E) Reporter gene construct for ser-1. (F) The ser-1::gfp fusion is mainly expressed in the pharynx. The white arrow points to the axon bundle of the nerve ring (note that this reporter fusion contains the ser-1 coding region and may hence be transported to axons). Expression in the pharyngeal muscle group pm4 is consistently weaker than in other muscles. The white star indicates nonspecific gut autofluorescence.
Identifications of cells that express gfp reporter gene fusions were confirmed by cell position and axon morphology. As a landmark for relative cell position, amphid sensory neurons were filled with the red fluorescent dye, DiI (Hedgecock et al., 1985). For example, expression of ser-4::gfp in RIB was confirmed by the stereotyped location of the GFP-positive cell sandwiched between the DiI-filled ASH and ASJ neurons. All three neurons send an axon through the amphid commissure (red and green labeled). Expression of gfp transgenes in the AIY interneuron class was examined by crossing the respective gfp strains with a chromosomally integrated transgene, otIs121, which expresses red fluorescent protein (rfp) exclusively in AIY. The colocalization of gfp and rfp signals was then assessed. The otIs121 transgenic strains express dsRed (Clontech, Inc.) under the control of an AIY-specific promoter fragment from the ttx-3 gene (construct kindly provided by Zeynep Altun). A coelomocyte-specific gfp reporter (unc-122::gfp) was used as an injection marker to generate the strain.
Phenotypic analysis of Lhx mutants
All Lhx mutants described here are null alleles of the respective genes. Their effects on the expression of cell fate markers and/or on axon morphology were assessed by crossing either extrachromosomal or chromosomally integrated reporter gene arrays into the respective Lhx null mutant background. This allowed for a comparison of the same array in wild-type and mutant backgrounds.
Isolation of a ser-2 deletion allele
A previously described PCR-based deletion library screening approach was used (Jansen et al., 1997). Primer pairs were chosen to span a genomic region of approximately 3.5 kb. The sequence of the primer upstream of the predicted start was 5′-AAATTCGTAAAGGCGCTATTAC (ser-2 5A-outer), whereas that of the primer downstream of the predicted stop was 5′-TGCGACATGATTCACAGAC (ser-2 3A-outer). The sequence of the nested primer upstream of the start was 5′-CCAGGAAGGAAATAGTT-GAGAC (ser-2 5A-nested). The sequence of the nested primer downstream of the stop was 5′-TGCGACATGAT-TCACAGAC (ser-2 3A-nested). A 1442-bp deletion was retrieved from the Plasterk lab Millennium Deletion Library.
Behavioral assays
Behavioral assays were performed according to the following references: Odortaxis, attraction (diacetyl, benzaldehyde, isoamyl alcohol, thiazole): Bargmann et al., 1993. Odortaxis, repulsion (nonanone): Troemel et al., 1997. Odor-induced modulation of reversal behavior, thrashing, reversal frequency, body bends/time: Tsalik and Hobert, 2003. Chemotaxis: Wicks et al., 2000. Thermotaxis: Altun-Gultekin et al., 2001. Copper barrier choice assay: Ishihara et al., 2002. Maze Assay: Brockie et al., 2001b. Basal/ enhanced slowing response: Sawin et al., 2000. Amplitude of locomotion: Sagasti et al., 1999. Dauer formation was measured in a daf-7(e1372) mutant background as described in Hobert et al., 1997. Harsh touch: Way and Chalfie, 1989.
Adaptations to diacetyl and benzaldehyde were performed as follows: Well-fed animals were washed free from food and placed on a 2% agar-containing assay plate, which contained 5 agar plugs placed on the inside of the lid. The agar plugs were saturated with the specified odorant and worms were left to adapt for 1 h. Controls were treated in parallel, except no odorant was added to the agar plugs. After the adaptation period, worms were subjected to odortaxis assays according to previously published techniques (Bargmann et al., 1993).
Defecation was scored by first picking individual young adult worms onto a standard NGM plate coated with OP50 bacteria. The time between expulsion steps was then measured for four cycles for each of four worms of each genotype. Controls were tested in parallel. The experimenter was blind to the genotypes of the test and control animals.
Egg laying was assessed by picking late-stage L3′s onto an OP50-coated NGM plate. These larvae were left to develop for 24 h. Different NGM plates were coated with either 200 µl of octopamine (Sigma-Aldrich) for a final concentration of 16 mg/ml or with an equal volume of water and each was allowed to dry. At this point, 2 adult worms were picked onto each OP50-coated NGM plate (containing octopamine or water) and allowed to lay eggs. The total number of eggs laid by the 2 worms on each plate was scored at specified time intervals. The data presented in Fig. 10E represent the total number of eggs laid per plate averaged across 5 independent samples for a total of 10 worms of each genotype.
Fig. 10.
ser-2 mutant animals show normal locomotory and sensory behaviors. (A) ser-2 is expressed in primary interneurons that were shown to be required for sensory processing (Mori and Ohshima, 1995; Tsalik and Hobert, 2003; White et al., 1986). (B) Normal axon morphology of AIY interneurons in wild-type and ser-2(pk1357) adult animals, as visualized with the mgIs18 transgene (lateral view). (C–E) Results of a representative set of behavioral assays testing ser-2 mutant animals. For a complete list, see Table 1. Behaviors shown here measure various aspect of locomotory behavior [basal and enhanced slowing response, (C) Sawin et al., 2000], chemosensory behavior [maze assay, (D) Brockie et al., 2001b] and egg-laying behavior in the presence or absence of octopamine (E). Numbers above bars in (C) indicate the number of worms at that set of conditions. The maze assay in (D) represents a more complex chemical environment than the originally described olfactory assays (Bargmann et al., 1993) as it incorporates volatile attraction (“X” in schematic denotes diacetyl) and soluble chemical repulsion (line in schematic designate upper barrier). Results from fifteen maze assays for the wild-type and eight for ser-2(pk1357) are reported. Each assay represents more than 200 worms/assay plate. Data in (E) represents the total number of eggs laid by two adult worms/plate as a function of time averaged across five assay plates under each set of conditions and for each genotype. Error bars represent standard error of the mean.
Results
Biogenic amine receptors in the C. elegans genome
To characterize the role that various Lhx proteins play in neuronal differentiation, we first had to identify and characterize a set of molecular tools reflective of neuronal sub-type-specific gene expression. We therefore searched for biogenic amine receptors in the C. elegans genome sequence using reiterative BLAST searches with known vertebrate and invertebrate metabotropic biogenic amine-receptors, namely serotonin, dopamine, histamine, octopamine, and tyramine receptors. We identified 16 predicted C. elegans proteins with a 7-transmembrane topology that share homology to these receptors and contain characteristic motifs involved in biogenic amine binding, G-protein coupling, and desensitization/internalization (Fig. 1A and B; see Materials and methods for details on the search). A previous analysis of the unfinished C. elegans genome sequence had identified a largely overlapping, though less complete set of proteins (Bargmann, 1998).
Pharmacological and ligand binding properties were described for 4 of these 16 receptors, demonstrating that Y22D7AR.13/5-HT-Ce/CER-1/SER-4 (from here on referred to as SER-4, the CGC approved name) and F59C12.2/5-HT2Ce/SER-1 (from here on referred to as SER-1) are serotonin receptors (Hamdan et al., 1999; Olde and McCombie, 1997), whereas SER-2 is a tyramine receptor (Rex and Komuniecki, 2002) and DOP-1 is a dopamine receptor (Suo et al., 2002) (Fig. 1A).
Sequence comparisons did not allow us to unambiguously group the other putative biogenic amine receptors into specific subfamilies, thus making their ligand binding properties difficult to predict. One exception is the K09G1.4 protein, which together with DOP-1, shares the highest sequence similarity to dopamine receptors. Along with DOP-1, it is consistently picked up as one of the first two hits when BLASTing the C. elegans genome database with known vertebrate dopamine receptors (P value in the 1e-40 to 1e-43 range, depending on which vertebrate dopamine receptor is used). Due to primary sequence features and their choice of downstream targets, dopamine receptors can be divided into two subfamilies, the D1- and the D2-sub-family (Vallone et al., 2000). Whereas the DOP-1 receptor falls into the D1-like subfamily, we find that K09G1.4 is of the D2-like subfamily of dopamine receptors (Fig. 1C). We named this receptor DOP-2.
We have focused our analysis on the biogenic amine receptors SER-1, SER-4, SER-2, and DOP-1, whose ligand properties have been characterized in vitro (Hamdan et al., 1999; Olde and McCombie, 1997; Rex and Komuniecki, 2002; Suo et al., 2002). In addition, due to its strong primary sequence affinity to the dopamine receptor class, we have also included DOP-2 in our analysis.
The tyramine receptor locus ser-2 produces several unusual transcripts
cDNAs for the ser-1, ser-4, and dop-1 loci have been previously isolated (Hamdan et al., 1999; Olde and McCombie, 1997; Suo et al., 2002). One nearly complete EST clone from Y. Kohara’s collection (yk1356c04), which extends 5′ into the first exon of the predicted gene, defines the gene structure of dop-2 (Fig. 2A). Our own analysis of transcripts from the ser-2 locus, however, differed from that of previously described ser-2 transcripts, whose isolation was biased due to PCR primer choice (Rex and Komuniecki, 2002). We found that the ser-2 locus is large and complex coding for several splice variants (Fig. 2B). There are four distinct features of these splice variants.
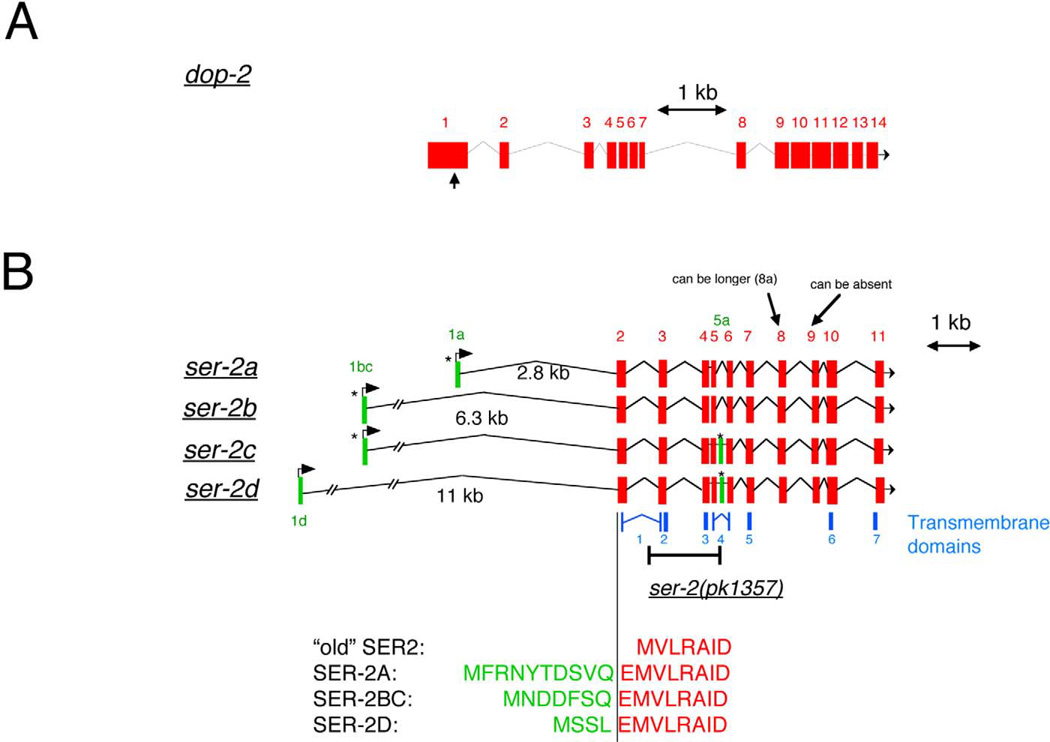
Fig. 2.
dop-2 and ser-2 cDNAs. (A) Predicted gene structure of the dop-2 locus, confirmed with the near-complete EST clone yk1356c04 (start of clone indicated by an arrow in the first exon; extends to 3′ end of gene). (B) Gene structure of the ser-2 locus as deduced by PCR-mediated analysis of transcripts from a cDNA library. ser-2a and ser-2b are the most prevalent splice forms (13/19 and 4/19 independently isolated library clones, respectively). We also detected transcripts in which the third to last exon, coding for part of a large intracellular loop, is missing (data not shown). Previously described ser-2 cDNAs incorrectly started at the second exon due to bias in primer choice during PCR amplification (“OLD SER-2”; Rex and Komuniecki, 2002). The predicted ATG start codon of the first exons in ser-2a and ser-2bc are preceded by an in-frame stop codon (indicated by “*”).
First, a set of 3 different first exons can be utilized. Each of the 3 different splice variants produces a slightly different N terminus (Fig. 2B). Second, an additional internal exon, termed 5a, is included in some transcripts. Interestingly, the presence of an in-frame stop codon in exon 5a produces a shortened and presumably nonfunctional protein. Third, exon 8 uses 2 distinct splice donor sites to splice to exon 9. The 2 splice forms differ by 23 amino acids, which are found in the third intracellular loop. This alternative splicing has been previously reported (Rex and Komuniecki, 2002). Fourth, exon 9 is absent in several transcripts, and similar to the alternative splice forms involving exon 8, shortens the third intracellular loop.
Whereas the functional consequences of differential splicing of exons 8 and 9 are difficult to predict, the inclusion of exon 5a will produce an obvious functional consequence, that is, the production of a truncated protein. Intriguingly, the inclusion of exon 5a appears to be coupled to the choice of the first exon. Using PCR, we found that if exon 1a is utilized, the resulting protein is always full-length and never contains the internal stop codon. In contrast, if exon 1d is utilized, a truncated protein is always produced. If exon 1bc is utilized, both types of protein, truncated and nontruncated, are produced. The functional relevance of such different transcripts is discussed in a later section.
Expression pattern of reporter gene fusions to the ser-2 tyramine receptor locus
We next examined the expression pattern of ser-2, dop-1, dop-2, ser-1, and ser-4. To this end, we made use of reporter gene fusions in which putative regulatory elements of the respective loci are fused to the gene coding for green fluorescent protein (gfp) (Chalfie et al., 1994). This sort of reporter gene analysis has in the past served as a useful first approximation of endogenous gene expression profiles of neuronally expressed gene families (e.g., Aurelio et al., 2002; Brockie et al., 2001a; Nathoo et al., 2001; Salkoff et al., 2001; Troemel et al., 1995).
We generated and analyzed three distinct transcriptional fusions to each alternative first exon of the ser-2 locus. We previously reported that a fragment of the ser-2 locus encompassing the 5′ upstream regulatory region, exons 1a and 2 as well as the first intron, fused in-frame to gfp (ser-2prom1::gfp) is expressed in the AIY interneuron class and a set of unidentified neurons (Altun-Gultekin et al., 2001). We have now identified these neurons as head and tail interneuron classes, namely AVHL/R, AUAL/R, AIYL/R, RICL/R, SABVL/R, RID, RIAL/R, SABD, SDQ, CANL/R, DA9, LUAL/R, ALNL/R, and PVCL/R (Fig. 3). In addition to its expression in neurons, ser-2prom1::gfp is also expressed in pharyngeal cells (NSM neurons and pm1/6 muscles) and in head muscles (data not shown). In males, expression can be observed in posterior dorsal and ventral body wall muscles, the male-specific diagonal muscles, and several posterior neurons likely to be CP neurons (data not shown). We confirmed that no more transcriptional regulatory information is contained within intronic regions by generating a fusion of gfp to the full coding genomic ser-2 locus using an in vivo recombination technique (see Materials and methods). Transgenic animals expressing such a construct show an expression pattern similar to the one observed with the ser-2prom1::gfp construct (data not shown).
Expression using the upstream regulatory regions of exon 1bc (ser-2prom2::gfp) is mostly restricted to the AIYL/R, AIZL/R, RID, DVA, BDUL/R, SIADL/R, and SIAVL/R interneurons (Fig. 3). Less consistent expression is observed in PVT. In addition, expression is observed in the RMEL/R motor neurons. Outside the nervous system, expression can be observed in the excretory gland cells. The expression of ser-2prom2::gfp in AIY and RID is notable since the ser-2prom1::gfp expression construct, which encompasses a distinct set of upstream regulatory sequences (Fig. 3A), is also expressed in these two cell types. Therefore, the expression of ser-2a and ser-2b/ser-2c splice forms in the same cell types (AIY and RID neuron class), relies on separable cell-specific regulatory elements. Furthermore, we have previously shown that, in spite of their apparent morphological and functional differences, the AIY interneurons and the RID motor neuron share a substantial set of terminal differentiation characteristics (see Fig. 11) (Altun-Gultekin et al., 2001).
Fig. 11.
LIM homeobox genes and neuronal differentiation. The effects of disrupting an Lhx gene (red) on the expression of cell fate markers and axogenesis in various neuronal cell types is schematically depicted. Biogenic amine receptors presented in this paper are in blue. In those cases where no effect is observed, other factors (“?”) must regulate expression of the marker and/or axogenesis. Dark shading indicates that the Lhx gene is required to adopt all known subtype-specific features of a neuron; light gray shading indicates that the Lhx gene is required for the adoption of several but not all subtype characters of a neuron; no shading indicates that the Lhx gene has no known effect on differentiation. See Hobert et al. (1999) for lim-6 function in RME and Altun-Gultekin et al. (2001), for ceh-10 and ttx-3 function in AIY. Not shown in this summary are the effect of mec-3 on all known light-touch sensory neuron markers (Zhang et al., 2002) and the limited effect of ttx-3 on other neuron classes (Altun-Gultekin et al., 2001). We could not assess axogenesis of the AIZ interneurons since too many other head neurons are labeled with the ser-2prom2::gfp reporter, obscuring the AIZ axons. The vertebrate homologs of the Lhx genes are: ttx-3: Lhx2/9, lin-11: Lhx1/5, lim-6: Lmx1.1/2, lim-4: Lhx6/8 (Hobert and Westphal, 2000). The molecular identity of the terminal differentiation genes are: unc-17= ACh transporter; sra-11 and C50F7.1= orphan 7TMRs; glr-1= glutamate receptor; zig-5= secreted 2-lg domain protein; ceh-10 and ceh-23= homeobox genes; unc-47/unc-25= GABA transporter/synthesizing enzyme; hen-1 and kal-1= secreted signaling/matrix proteins (Altun-Gultekin et al., 2001). Note that AIY and RID share a striking homology with respect to gene expression profiles. Five of six cell-fate markers in RID are also expressed in AIY (underlined gene names). However, the gene regulatory hierarchy is very different in these neurons. In AIY, the ceh-10 homeobox gene is required for the expression of all known cell-specific characters (Altun-Gultekin et al., 2001), but in RID it does not appear to regulate the cell-fate markers that we tested (lim-4, ser-2a; data not shown). Moreover, Lhx genes expressed in AIY and RID have differential importance in both cell types. The AIY-expressed ttx-3 Lhx gene affects all known AIY properties, whereas the RID-expressed lim-4 Lhx gene affects only a restricted subset of RID properties. These points illustrate the context dependency of transcription factor action. Lastly, it is interesting to note that RID can be made to appear more AIY-like through the ectopic expression of ttx-3 (Altun-Gultekin et al., 2001), suggesting that lim-4 and ttx-3 act in a strictly nonredundant manner.
The upstream regulatory region of the third splice form, containing exon 1d, drives expression exclusively in two sensory neuron classes, OLL(L/R) and PVD(L/R) (Fig. 3). The PVD neuron class will be described in more detail in a later section.
Taken together, we infer from the reporter gene constructs that the ser-2 gene is expressed in a restricted set of sensory, inter- and motor neurons. The best characterized ser-2-expressing interneuron classes are AIY, AIZ, and RIA, which process a variety of sensory modalities (see Fig. 10A). While the respective upstream regulatory regions of the three alternative first exons drive expression in largely, though not entirely, distinct sets of cells, it is possible that the expression of the endogenous ser-2 splice variants may be the combined sum of regulatory information provided by all three regulatory regions. Testing such a possibility would require exon 1a-, 1bc-, and 1d-specific probes (either antibody or in situ probes), which are currently not available. No matter the results such an analysis would produce, the available data demonstrate the presence of distinct regulatory regions preceding distinct and alternative first exons.
Expression pattern of reporter gene fusions to the dopamine receptor loci, dop-1 and dop-2
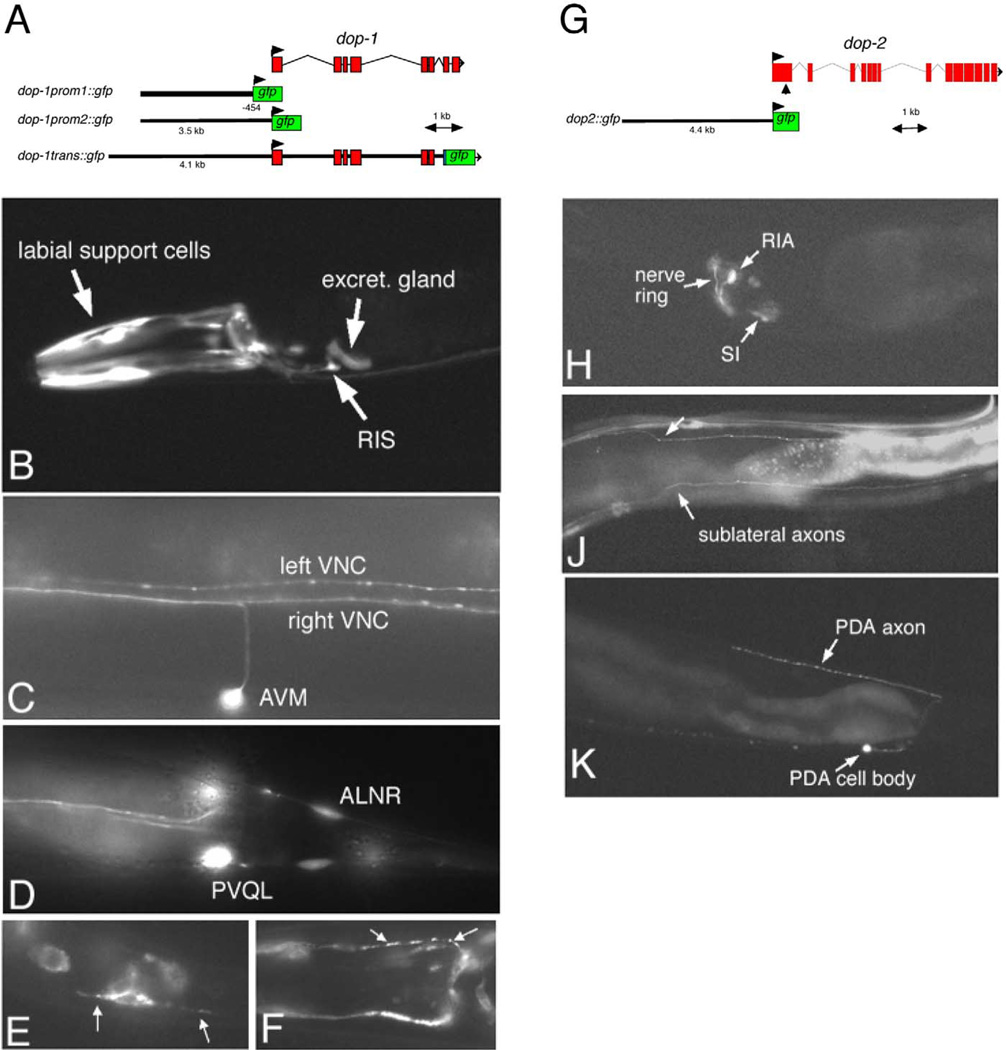
In contrast to the ser-2::gfp reporter fusions, a dop-1::gfp reporter gene fusion has a more restricted expression pattern in the nervous system in larval and adult animals. Within the head ganglia, dop-1prom1::gfp is consistently expressed only in the RIS interneuron class (Fig. 4). Only weak and inconsistent expression can be observed in other unidentified head neurons. Consistent and strong expression can be observed in the excretory gland cells and head muscles as well as in several labial and amphid sensory neuron support cells (sheath/socket cells) (Fig. 4A). In the midbody and tail region, reporter gene expression is evident in the AVM and ALM touch sensory neurons, in the ALN and PLN neuron classes and in the PVQ interneurons, which extend axons along the left and right ventral nerve cord, respectively (Fig. 4C and D). Reporter gene fusions that contain more genomic sequence (dop-1prom2::gfp and dop-1trans::gfp; see Fig. 4A) show expression in similar sets of cells. In addition, dop-1prom2::gfp expression is observed in additional sets of unidentified head neurons. A dop-1trans::gfp reporter protein, which contains dop-1 coding sequences, is as expected, localized to the plasma membrane and has a punctate appearance along axons and muscle arms (Fig. 4E and F).
Fig. 4.
Expression of dop-1::gfp and dop-2::gfp. (A) Reporter gene constructs for dop-1. dop-1prom1::gfp and dop-1prom2::gfp show largely similar patterns of expression. (B–D) dop-1prom1::gfp (otEx233). (E, F) dop-1trans::gfp (adEx1647). (B) Lateral view of the head region. (C) Ventral view of a region in the anterior half of an adult animal, showing axons in the left and right ventral nerve cord fascicles. (D) Ventral view of the tail ganglia. The PVQR cell body is out of focus. (E, F) Localized expression of a dop-1 translational fusion (white arrows) in tail neuron axons (E) and head muscle arms (F). (G) Reporter gene construct for dop-2. (H) Lateral view of the head region of adult dop-2prom::gfp-expressing animals (otEx980). Expression is strongest and most consistent in RIA(L/R) but less consistent in sublateral interneurons (SIA and/or SIB; labeled “SI”). Inconsistent expression of gfp in additional neurons can often be observed. (J) Sublateral interneurons send characteristic axons along sublateral tracts (white arrows). (K) Lateral view of the tail region, showing expression of dop-2prom2::gfp in the PDA neuron, which characteristically sends an axon from the ventral to the dorsal nerve cord.
Reporter gene fusions to dop-2 produce gfp expression exclusively in a small set (< 10) of neurons in the head and tail ganglia of larval and adult hermaphrodites (Fig. 4H). The strongest and most consistent expression is observed in the RIA(L/R) interneuron pair. In addition, expression of variable intensity and penetrance can be observed in at least a subset of the sublateral interneurons (SIA and SIB) which send characteristic axons along sublateral tracks (Fig. 4J) and in the unilateral RID neuron (data not shown). In the tail, a single neuron, the PDA neuron, shows dop-2::gfp expression (Fig. 4K).
Expression pattern of reporter gene fusions to serotonin receptor loci, ser-1 and ser-4
Expression of a ser-4::gfp reporter gene construct is restricted to a few neurons in the nervous system (Fig. 5). Most consistent expression can be observed in the RIB and RIS head interneuron classes. Less consistent expression is observed in a pharyngeal neuron, a pair of sublateral interor motorneurons, rarely in a pair of neurons in the retrove-sicular ganglion, the PVT tail neuron, and either the DVA or DVC tail interneuron (Fig. 5).
ser-1::gfp is expressed in most pharyngeal muscles (Fig. 5). ser-1::gfp was not obviously detectable in pharyngeal neurons, but it is possible that weak expression may be obscured by the strong pharyngeal muscle gfp expression. Faint fluorescence can be observed surrounding the isthmus of the pharynx and may be ascribed to the neuropil of the nerve ring.
Expression pattern of other biogenic amine receptors
We also generated translational gfp fusions to the biogenic amine receptor loci M03F4.3, T02E9.3, F14D12.6, F01E11.5, K02F2.6, and C09B7.1. Since we did not confirm the gene structure predictions by cDNA analysis and since the nature of the ligands for these receptors has neither been examined in vitro nor can be easily predicted by sequence alignment and phylogenetic analysis, we refrained from engaging in a detailed expression pattern analysis of transgenic animals expressing these reporters. However, we noted that each one of these reporter gene constructs showed a very restricted expression pattern almost exclusively in a subset of head and tail neurons (data not shown).
Overlap of aminergic receptor and LIM homeobox gene expression
The expression of each of the biogenic amine reporter gene fusions, described in detail above, shows significant overlap with the expression of different Lhx gene classes (Hobert et al., 1997, 1998, 1999; Sagasti et al., 1999; Way and Chalfie, 1989) (see Fig. 11). This observation allowed us to address the following questions. First, how do the respective Lhx genes affect biogenic amine receptor expression? Second, since some of the aminergic receptor reporter genes readily visualize the anatomy of individual neuron classes, we could ask whether mutations in the Lhx genes affect the anatomy of the neurons in which they are normally expressed. In the next sections, we will describe how null mutations in five distinct classes of Lhx genes (ttx-3, lim-4, lin-11, lim-6, mec-3) affect the aminergic receptor expression profiles and/or the axon morphologies of eight different neuron classes (AIY, AIZ, RID, RIS, RME, PVD, PVQ, DVA).
ttx-3 affects AIY interneuron differentiation
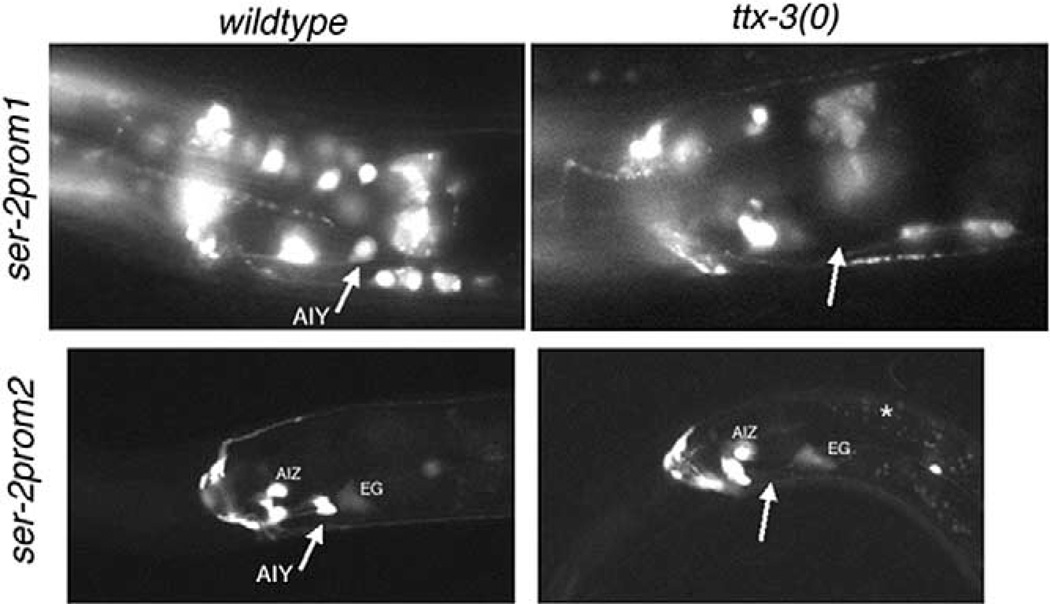
We have previously shown that expression of an extra-chromosomal ser-2prom1 reporter gene array in the AIY interneuron class depends on the presence of a functional ttx-3 Lhx gene (Altun-Gultekin et al., 2001). Using a chromosomally integrated reporter array, we confirmed this finding (1/17 animals shows wild-type expression) (Fig. 6). In addition, we tested whether transcription of the ser-2b/ ser-2c splice forms, which contain a regulatory element for AIY expression that is independent of that for the ser-2a splice form (Fig. 2), is also under the control of ttx-3. By crossing the ser-2prom2::gfp reporter array into ttx-3 null mutant animals, we indeed found this to be the case (0/17 animals show wild-type expression) (Fig. 6). ttx-3 is thus required for the expression of all indicated ser-2 splice forms in AIY. These results are consistent with previous evidence that ttx-3 affects all known subtype-specific features of the AIY interneuron class (Altun-Gultekin et al., 2001).
Fig. 6.
ser-2 reporter gene fusions require ttx-3 for correct expression in AIY. Expression of the ser-2prom1::gfp (otIs107) transgene (upper panel) and the ser-2prom2::gfp (otEx536) transgene (lower panel) in wild-type and ttx-3(mg158) mutant animals. Whereas the respective transgenes are always expressed in AIY in wild-type animals, they fail to be expressed in ttx-3(mg158) mutant animals (otIs107: 16/17 animals show no expression; otEx536: 17/17 animals show no expression). The white star indicates nonspecific gut autofluorescence.
lim-4 affects RID motor neuron differentiation
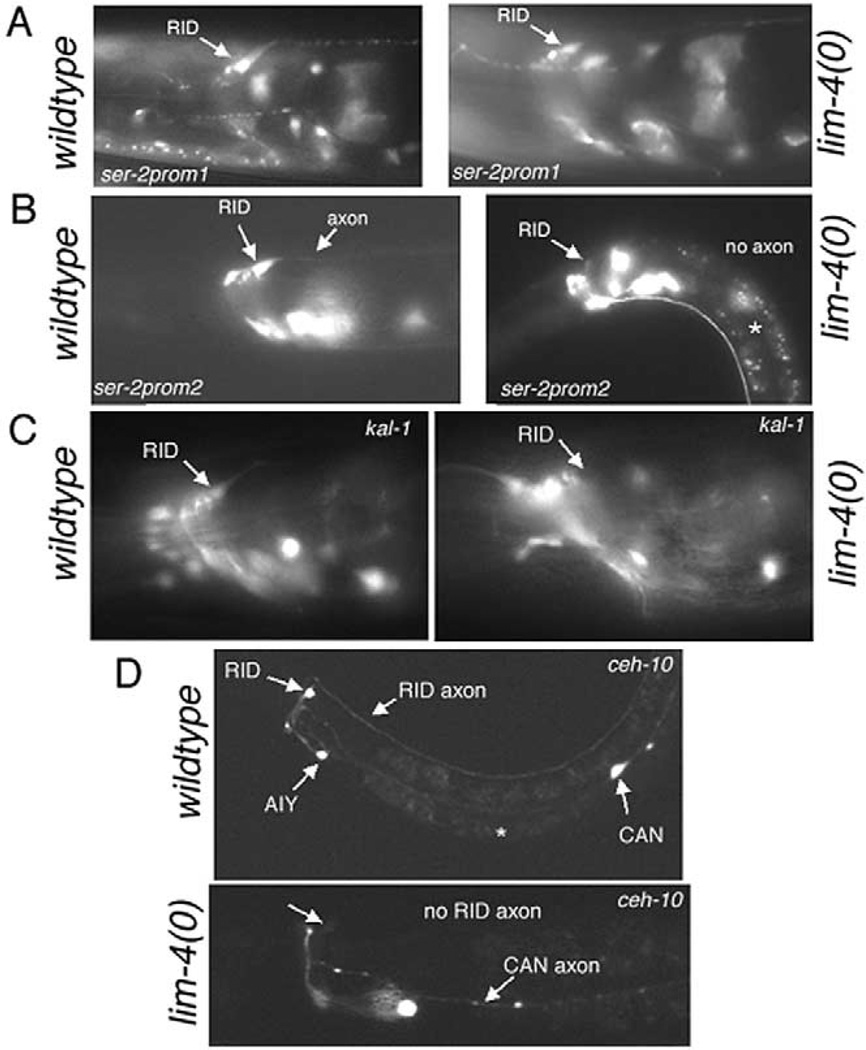
The expression of two splice forms of ser-2 also overlaps with the expression of another Lhx gene, the Lhx6/ 8-type Lhx gene lim-4. This overlap can be observed in the unilateral RID motorneuron, which sends an axon along the dorsal nerve cord. Interestingly, we found that lim-4 regulates expression only one of the two distinct RID-expressing ser-2 regulatory elements. ser-2prom1::gfp expression (monitoring the ser-2a splice form) in RID is largely unaffected in lim-4 null mutants (24/30 animals with normal expression), whereas the RID-regulatory element in ser-2prom2::gfp (monitoring expression of ser-2b/ser-2c) is severely affected in lim-4 null mutants (0/42 with normal expression) (Fig. 7A and B). These findings prompted us to assess the differentiated state of RID in lim-4 null mutants in greater detail. To this end, we made use of four more RID cell fate markers, namely the kal-1 gene (Bu¨low et al., 2002), the ceh-10 gene (Svendsen and McGhee, 1995), the zig-5 immunoglobulin gene (Aurelio et al., 2002), and a novel putative neuropeptide receptor, C50F7.1 (A.S.W. and O.H., unpublished observations). Reporter gene fusions revealed significant RID differentiation defects in lim-4 mutant animals (Fig. 7C and D). A kal-1::gfp reporter construct shows defective expression of the transgene in 65% (n = 34) of animals examined (Fig. 7C). A ceh-10::gfp reporter gene fusion is never appropriately expressed in RID (Fig. 7D; n = 27). A zig-5::gfp reporter gene fusion showed reduced or absent expression in RID in 100% of animals examined (n = 33; data not shown). In contrast, expression of a C50F7.1::gfp fusion gene shows no defect in lim-4 mutants (95% of animals tested show normal expression; n = 19; data not shown). The RID-expressing reporter constructs that are not affected by lim-4, namely ser-2prom1::gfp and C50F7.1::gfp, allowed us to visualize RID motor axon outgrowth in the dorsal nerve cord, which we found to be unaffected by loss of lim-4 (Fig. 7A).
Fig. 7.
lim-4 affects RID differentiation. (A) Expression of ser-2prom1::gfp (otIs107) in wild-type and lim-4(ky403) mutant animals. Expression levels in RID and axon anatomy of RID are largely unaffected. 24/30 animals show normal expression levels and axon anatomy; 6/30 had slightly dimmed expression. In many cases, the cell body was obscured by strong head muscle staining and expression in RID had to be assessed by gfp levels in the axon rather than cell body; wild-type control: 34/34 animals show strong expression. (B) Expression of ser-2prom2::gfp (otEx536) in wild-type and lim-4(ky403) mutant animals. Whereas the transgene is always expressed in RID of wild-type animals (n= 42), it is significantly downregulated in lim-4(ky403) mutants (0/42 animals show expression comparable to wild-type, 36/42 have no expression in RID; 6/42 have very faint expression in a cell that is in a similar position to RID; the figure panel shows an example of the latter case). The white star indicates nonspecific gut autofluorescence. (C) Expression of kal-1::gfp (otIs33) is affected in RID by lim-4 null mutations (12/34 animals show wild-type expression levels; 42/42 wild-type animals show expression in RID). (D) Expression of ceh-10::gfp (lqIs4) is affected in RID in lim-4 null mutant animals (0/27 animals show wild-type expression levels; 43/43 wild-type animals show expression in RID).
Taken together, lim-4 regulates some but not all aspects of RID differentiation (ser-2b/c, kal-1, zig-5, and ceh-10 expression) (see Fig. 11). At this point, it is unclear whether LIM-4 binds to cis regulatory elements of the ser-2, kal-1, zig-5, and ceh-10 loci or if it acts indirectly through intermediate factors to affect their expression.
lim-6 affects RIS interneuron differentiation
The usefulness of the ser-2 reporter gene fusions to assess cellular differentiation in Lhx gene mutant backgrounds prompted us to utilize our aminergic receptor probes to assess the state of cellular differentiation in other Lhx gene mutants and to ask whether it is a common theme of Lhx gene function to regulate biogenic amine receptor expression, either directly or indirectly.
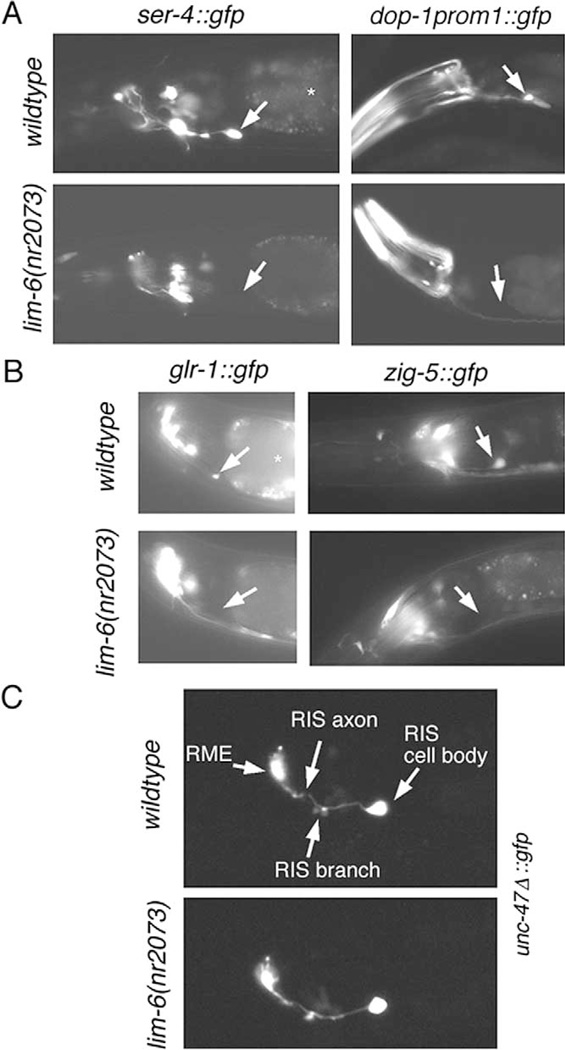
The sole neuron that expresses the dop-1prom1::gfp reporter in head ganglia, the unilateral RIS interneuron (Fig. 4), also expresses the lim-6 Lhx gene (Hobert et al., 1999). Similarly, a ser-4 reporter gene construct is also expressed in RIS (Fig. 5). By crossing each reporter transgene into a lim-6 null mutant background, we found that dop-1 expression in RIS requires lim-6 (Fig. 8A). lim-6 also significantly affects the expression of the ser-4 serotonin in RIS (Fig. 8A). We examined whether lim-6 affects RIS differentiation on a broader level and examined the expression of two additional RIS cell fate markers, the glr-1 glutamate receptor (Hart et al., 1995; Maricq et al., 1995) and the zig-5 Ig domain-containing gene (Aurelio et al., 2002). Reporter gene fusions for both genes fail to be appropriately expressed in lim-6 mutants (Fig. 8B).
Fig. 8. lim-6 affects RIS differentiation. (A–C) Expression of various reporter transgenes in wild-type and lim-6(nr2073) null mutants. The white arrow points to the unilateral RIS interneuron. The white star indicates nonspecific gut autofluorescence. The penetrance of defects in (A) and (B) are as follows: ser-4::gfp (adEx1616) 96% on in wild-type (n= 24), 30% on in lim-6(nr2073) (n= 27). dop-1prom1::gfp(otEx233) 91% on in wild-type (n= 33), 20% on in lim-6(nr2073) (n= 25). glr-1::gfp (nuIs1) 100% on in wild-type (n= 21), 9% on in lim-6(nr2073) (n= 33). zig-5::gfp (otls11) 100% on in wild-type (n= 25), 3% on in lim-6(nr2073) (n= 31). (C) The RIS interneuron is generated and its axon morphology (including its characteristic branch; White et al., 1986) is normal in lim-6 null mutants (40/40 animals). The reporter transgene used, otIs39, is a deletion fragment of the unc-47::gfp reporter and is expressed within the head ganglia in RME(L/R), RIS, and weakly in AVL.
We have previously demonstrated that an unc-25::gfp reporter for the GABA-synthesizing enzyme glutamic acid decarboxylase also fails to be appropriately expressed in lim-6 null mutants (Hobert et al., 1999). However, RIS is not completely undifferentiated or even absent in lim-6 mutants since expression of the unc-47 GABA transporter is unaffected in lim-6 mutants (Fig. 8C) (Hobert et al., 1999). With a newly generated transgene, otIs39 (an unc-47::gfp derivative), we were now able to assess RIS anatomy in lim-6 mutant animals and found no gross defects (Fig. 8C). Taken together, we have provided evidence that lim-6 mutations severely affect the differentiation of the RIS interneuron without affecting its generation, survival, or overall anatomy. LIM-6 may directly bind the regulatory elements upstream of the dop-1, ser-4, glr-1, zig-5, and unc-25 loci or may regulate the expression of another factor, which then regulates the expression of these genes.
ser-2 and dop-1 as cell fate markers to assess neuronal differentiation in other Lhx mutant backgrounds
The lin-11 Lhx1/3-type gene was previously reported to be expressed in either DVA or DVC (Sarafi-Reinach et al., 2001). Due to position and overlap of expression of lin-11::gfp and ser-2prom2::gfp reporters, we could now unambiguously identify the lin-11-expressing cell as DVA. lin-11 had previously been shown to affect the differentiation of a variety of neurons (Hobert et al., 1998; Sarafi-Reinach et al., 2001), but its function in several other neuron types, such as DVA, had remained obscure. We addressed the effect of completely eliminating lin-11 activity on DVA development by crossing a ser-2prom2::gfp transgene (otEx536) into lin-11(n389) null mutants and found that expression of ser-2prom2 in DVA is unaffected. Moreover, DVA axonal anatomy appears indistinguishable from that of wild-type (data not shown). These observations allow us to conclude that lin-11 does not affect the generation or gross differentiation of this neuron.
Removal of the AIZ interneuron class causes behavioral defects similar, though less severe, to those seen in lin-11 null mutants (Hobert et al., 1998; Mori and Ohshima, 1995). Given the profound effects of the ttx-3 Lhx gene on the differentiation of the AIY interneuron class, which is presynaptic to AIZ, it was conceivable that, in lin-11 mutants, the AIZ neuron would be similarly undifferentiated. However, we find that ser-2prom2::gfp expression in the AIZ interneurons is unaffected in lin-11 null mutants (data not shown). Thus, although loss of lin-11 appears to disrupt the functionality of the AIZ interneuron class, it does not completely disrupt its differentiation program.
dop-1 reporter gene expression in the PVQ tail interneurons allowed us to assess whether the development of these neurons requires the lin-11 Lhx gene, which is also expressed in PVQ (Hobert et al., 1998). dop-1 expression, axon morphology, and placement of PVQ were unaffected in lin-11(n389) null mutants (data not shown).
The lim-6 Lhx gene is coexpressed with ser-2prom2 in the RMEL/R motor neurons. Consistent with the lack of effect of loss of lim-6 on other RME(L/R) markers (Hobert et al., 1999), we find that ser-2prom2 is correctly expressed in RMEL/R in lim-6 null mutants (data not shown).
Taken together, using aminergic receptor expression as a way to assess cellular differentiation in a variety of genetic backgrounds, we found that several distinct types of Lhx genes have no impact on the overall generation or morphology of specific neuronal cell types (see Fig. 11).
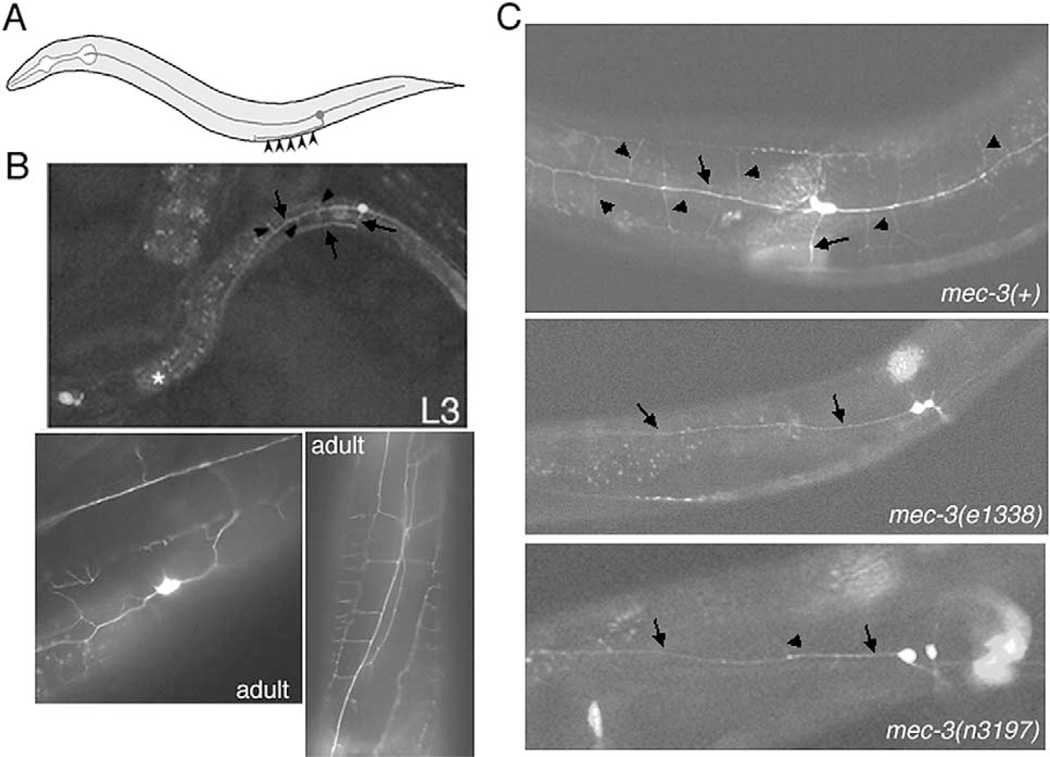
The dendritic branching pattern of the PVD harsh touch sensory neuron requires mec-3
The ser-2prom3::gfp reporter allowed us to visualize unusual aspects of PVD(L/R) anatomy. These neurons were not completely traced in a previous electron microscopical reconstruction of the nervous system, although it was noted that the postembryonically generated PVD(L/R) cells each project an anteriorly and posteriorly directed dendrite with no synaptic output and a ventrally directed axon that joins the ventral nerve cord to synapse with command interneurons (White et al., 1986) (Fig. 9A). The ser-2prom3::gfp reporter extends these previous findings by revealing that the anteriorly and posteriorly projecting dendrites display an extensive branching pattern, leading to an almost web-like structure covering most of the animal (Fig. 9B). This morphology can be observed in midlarval stages when the reporter gene construct starts to be expressed and appears to be further elaborated upon maturation of the animal. We observe this morphology in seven independent transgenic lines. Since this unusual PVD morphology has also recently been observed with antibody staining against a PVD-expressed antigen (Halevi et al., 2002), we consider it unlikely that this morphology is a reporter gene-induced artifact. The highly branched morphology of PVD may be a critical aspect of its function as a harsh touch sensory neuron, which was previously characterized through laser ablation studies (Way and Chalfie, 1989).
Fig. 9.
mec-3 affects dendritic branching of the PVD sensory neurons. (A, B) The ser-2prom3::gfp reporter gene fusion reveals the complex morphology of the PVD harsh touch sensory neurons. (A) Morphology of the main axons of PVD as determined by White et al. (1986). Arrowheads indicate the sites of PVD synaptic output to command interneurons. (B) Upper panel: Morphology of PVD visualized with ser-2prom3::gfp in a mid-larval stage animal. Main axons are labeled with arrows (note the anteriorly, posteriorly, and ventrally directed axons). Additional branches, not previously reported, are indicated with arrowheads. Middle and lower panels: Two examples of PVD morphology in adult animals. The branching pattern has become more elaborate. Similar morphologies have been observed in seven independent transgenic lines (otEx449-456). (C) PVD morphology is disrupted in mec-3 mutant animals. Upper panel: Adult wild-type control animals. PVD is visualized with the otIs138 transgene. Arrows point to main axon shaft, arrowheads to axon branches. Middle and lower panels: PVD morphology in the two recessive mec-3 loss-of-function alleles e1338 and n3197. 0/25 e1338 animals show the extensive braching patterns of wildype animals; 1/23 n3197 animals show the branching pattern; 22/23 animals show either no branching or just occasional short branches (arrowhead). The weak mec-3 allele, u298, shows a somewhat reduced elobration of dendritic branches, which we found difficult to acurately quantify.
The mec-3 Lhx gene, a founding member of the Lhx gene family, is required for the adoption of all known differentiation features of the AVM, PVM, ALM, and PLM light touch neurons (Way and Chalfie, 1988). In addition to those neurons, mec-3 is also expressed in the PVD harsh touch neurons. Furthermore, the harsh touch sensory defect observed by PVD ablation is phenocopied in mec-3 mutants (Way and Chalfie, 1989). The effect of mec-3 on PVD function was, however, not previously understood on a molecular or cellular level. The ser-2prom3::gfp construct allowed us to visualize the generation and correct positioning of the cell body and its axonal and dendritic processes in mec-3 mutants. We observed that mec-3 does not affect the adoption of these cell fate characters (Fig. 9C). However, we found a striking effect of loss of mec-3 function on the elaborate dendritic branching pattern of PVD. In the two strong loss-of-function alleles, n3197 and e1338, the dendritic branching is virtually absent (Fig. 9C). We hypothesize that impaired harsh touch defects in mec-3 mutants may at least be partly caused by failure of PVD to establish an intact dendritic branching pattern.
Isolation and characterization of a ser-2 null allele
Having established potential sites of biogenic amine receptor expression, we next attempted to elucidate their functional roles. We focused our efforts on the ser-2 gene due to its intriguing sites of neuronal expression. Specifically, ser-2 reporter gene constructs are expressed in a set of interneuron classes (AIY, AIZ, and RIA) whose synaptic connectivities to various sensory neuron classes suggest that these cells may play a role in processing sensory information (Fig. 10A) (White et al., 1986). Consistent with this notion, all three neuron classes have been shown to have a prominent role in thermotaxis behavior (Mori and Ohshima, 1995). More recently, we implicated the AIY and AIZ interneuron classes in controlling locomotory behavior and the response to defined chemo- and odorsensory cues (Tsalik and Hobert, 2003). Lastly, the disruption of the ttx-3 Lhx gene, which we have shown here to cause defects in the expression of several ser-2 reporter genes in the AIY interneuron class, leads to strong defects in all known AIY interneuron-mediated behaviors (Tsalik and Hobert, 2003).
These observations prompted us to test whether ser-2 may have a role in any of the AIY-, AIZ-, and RIA-mediated behaviors and/or other behaviors potentially regulated by other ser-2-expressing cells. To this end, we isolated animals from a mutagen-induced library containing a deletion in the ser-2 locus. The mutant allele retrieved, pk1357, contains a 1442-bp deletion that removes exon 2 through exon 4 of the ser-2 coding sequence and causes a 128-amino-acid in-frame deletion encompassing parts of transmembrane domain 1, all of transmembrane domains 2 and 3, and parts of transmembrane domain 4 (Figs. 1B and 2B). The predicted protein product of ser-2(pk1357) only contains four transmembrane regions, possibly leading to an inversion of intra- and extracellular domains. The deletion also encompasses several critical sites necessary for proper receptor functioning, including a conserved negatively charged aspartate residue at position 127 (numbering from SER-2A) involved in ligand binding, as well as the highly conserved DRY motif required for G-protein coupling (Rex and Komuniecki, 2002; Wess, 1998). In addition, the i1 loop is absent, which is also critical for receptor/G-protein coupling in numerous GPCR’s of this class (Wess, 1998). Taken together, it appears likely that ser-2(pk1357) represents a strong loss-of-function if not a null allele.
Neurotransmitters including the biogenic amine serotonin were shown to be involved in neural differentiation (Lauder, 1993) and neuronal migration (Kindt et al., 2002). We tested whether neural development is affected in ser-2 null mutant animals, focusing on the AIY interneuron class, which normally expresses ser-2 and for which well-characterized tools for visualization are available. We find that loss of ser-2 does do not affect the generation of the AIY interneurons, their cell position, or their axodendritic anatomy (Fig. 10B). The ser-2prom1::gfp reporter, which labels many of the ser-2-expressing cells, appears to be normally expressed in ser-2 null mutants, demonstrating that ser-2-expressing cells are generated normally (data not shown).
We next subjected ser-2(pk1357) mutants to a variety of behavioral assays that measure sensory information processing, several of which we have previously shown to be affected by removal of ser-2-expressing cells (Tsalik and Hobert, 2003). We were unable to detect any significant behavioral defects in ser-2(pk1357) animals (Fig. 10C–E; Table 1). In Fig. 10C and D, we show representative examples of results in behavioral assays that represent highly sensitive tests for locomotory, odor- and chemosensory behavior. As a positive control, we tested ttx-3 mutant animals, in which the AIY interneuron class is disrupted and in which a specific set of behavioral defects can be observed (Tsalik and Hobert, 2003) (Table 1). The absence of locomotory defects and odorsensory defects contrasts the effects of tyramine reported in these behaviors in other invertebrates (Kutsukake et al., 2000; Roeder, 1999). In addition, since SER-2 also has appreciable affinity for octopamine (Rex and Komuniecki, 2002), which was previously shown to inhibit egg-laying behavior in worms (Horvitz et al., 1982), we assessed egg-laying behavior of ser-2(pk1357) mutants. We found that ser-2 mutants show normal egglaying behavior and respond to octopamine in a manner indistinguishable from wild-type (Fig. 10E). Octopamine has also been demonstrated to have a role in metabolic regulation in insects (Roeder, 1999) prompting us to assess fat-storage. Using nile red staining (Ashrafi et al., 2003) we observed no differences between ser-2 mutants and wild-type control animals (data not shown).
Table 1.
Summary of behavioral analysis of ser-2 null mutant animals
| Behavior testeda |
Behavior of ser-2(0) animalsb |
Behavior of ttx-3(0) animalsb |
|
|---|---|---|---|
| Sensory behaviors |
Odortaxis: attraction (diacetyl, benzaldehyde, isoamyl alcohol, thiazole) |
+ | − |
| Odortaxis: repulsion (nonanone) |
+ | + | |
| Adaptation (diacetyl, benzaldehyde) |
+ | − | |
| Odor induced modulation of reversal behavior |
+ | − | |
| Chemotaxis | + | + | |
| Thermotaxis | + | − | |
| Mechanosensation: harsh touchc |
+ | + | |
| Copper barrier choice assayd | + | − | |
| Maze Assay | + | − | |
| Locomotory behaviors |
Thrashing | + | − |
| Reversal frequency | + | − | |
| Body bends/time | + | − | |
| Basal/enhanced slowing response |
+ | + | |
| Amplitudee | + | + | |
| Others | Defecation | + | + |
| Male matingf | + | + | |
| Egg layingg | + | n.d. | |
| Dauer formationh | + | − |
References for behavioral assays are given in materials and methods.
“+” refers to ser-2(pk1357) or ttx-3(0) animals being indistinguishable from wild-type animals which were also tested in parallel. “−” refers to defects observed in ttx-3 mutants (data not shown and Tsalik and Hobert, 2003).
mec-3(e1338) (Way and Chalfie, 1989) served as the control for the harsh touch mechanosensation assay.
hen-1(tm501) (Ishiharaetal., 2002) served as the control for the copper barrier choice assay.
lim-4(ky403) (Sagasti et al., 1999) served as the control for the measure of amplitude.
This behavior was not tested in detail, but it was noted that ser-2 mutant males are capable of producing progeny when mated to wild-type animals.
Egg laying was tested in the absence and presence of octopamine (see text). n.d.: not determined.
Dauer formation was scored at 25°C in a daf-7(e1372) sensitized background.
SER-2 contains conserved residues whose mutagenesis has been shown to lead to constitutive, ligand-independent activity in related GPCR family members (Pauwels and Wurch, 1998; Wess, 1998). Introducing these mutations (using SER2A numbering, D155A in the DRY motif and A345E in the BBXXB motif) into SER-2 and expressing these constructs in the AIY neurons of wild-type animals yielded no obvious behavioral defects (data not shown). Taken together, we have been unable to detect a function for SER-2 using loss- and gain-of-function approaches.
Discussion
We reported here the in silico identification of biogenic amine receptors in the worm genome, an analysis of unusual splice variants of one of these receptors, the expression patterns of several of the receptors, and their use as markers to assess the fate of neuronal cell types in LIM homeobox gene mutants. Lastly, we assessed the consequences of a loss of the C. elegans tyramine receptor, ser-2.
Relation between aminergic receptor gene expression patterns and serotonin, dopamine and tyramine function and expression
What are the cells that produce serotonin, dopamine, and tyramine in C. elegans, and how do these cells relate to the sites of expression of the respective receptor? Furthermore, how does the site of receptor gene expression relate to the described functions of serotonin and dopamine? It is worth noting that the ensuing discussion relies on the analysis of reporter gene constructs, rather than endogenous transcripts. Although we cannot rule out the possibility that additional transcriptional regulatory elements exist that were not included in our reporter gene fusions, we nevertheless consider the results obtained from reporter genes as a reasonable, first approximation of the sites of endogenous gene expression.
The application of exogenous serotonin, serotonin-reuptake inhibitors, and the genetic analysis of mutants defective in serotonin biosynthesis revealed a complex picture for serotonin action. In brief, serotonin appears to regulate various aspects of egg-laying behavior, locomotory behavior, pharyngeal pumping, male mating behavior, and metabolic remodeling (Avery and Horvitz, 1990; Loer and Kenyon, 1993; Mendel et al., 1995; Sawin et al., 2000; Schafer and Kenyon, 1995; Segalat et al., 1995; Sze et al., 2000; Weinshenker et al., 1995). The sites of serotonin synthesis have been identified by using serotonin antibody staining and visualization of tryptophan hydroxylase reporters and include a rather limited set of sensory, inter- and motor neurons in the hermaphrodite, including the ADF sensory neurons, the AIM and RIH interneurons, the pharyngeal NSM neuron, and a subset of VC motor neurons (Desai et al., 1988; Sze et al., 2000). The expression of the ser-4 serotonin receptor does not correlate to the postsynaptic targets of these neurons. However, like other biogenic amines, serotonin can be humorally released to act on non-synaptically connected cells over some distance (Bunin and Wightman, 1999). Expression of ser-1 in pharyngeal muscle makes it a good candidate to be responsible for serotonin’s effect on pharyngeal pumping (Avery and Horvitz, 1990). However, a ser-1 mutant allele in which the C terminus of ser-1 is deleted shows no defects in pharyngeal function (T.N. and L.A., data not shown).
The effects and localization of dopamine have also been well documented. There are eight dopaminergic neurons in hermaphrodites which have been identified through FIF staining (Sulston et al., 1975) as well as by localization of tyrosine hydroxylase, cat-2, (Lints and Emmons, 1999), and a dopamine transporter, dat-1 (Nass et al., 2002). The expression patterns of the dop-1 and dop-2 receptors do not match with the postsynaptic targets of these neurons. However, as with ser-2, dopamine may act nonsynaptically, may be taken up by other cells and then rereleased, and/or other aminergic receptors (Fig. 1A) may be sensitive to it.
The octopamine producing enzyme, tyramine-β hydroxylase, is expressed exclusively in the RIC interneuron pair (M. Alkema and R. Horvitz, personal communication), making it likely that tyramine, the biosynthetic precursor to octopamine, is also produced in this pair of cells. In contrast to the neurotransmitter, expression of the ser-2 tyramine receptor is more widespread, making it possible that tyramine is produced in additional cells other than RIC. These issues will only be resolved upon the availability of tyramine-specific antibodies. The relatively broad expression of the tyramine receptor (~10% of all neurons in the nervous system) mirrors the broad expression of its fly, locust, and honeybee homologs (Arakawa et al., 1990; Blenau et al., 2000; Kutsukake et al., 2000; Saudou et al., 1990; Vanden Broeck et al., 1995).
Characterization of a worm tyramine receptor
The ser-2 locus displays intriguing splicing patterns. The choice of the first exon appears to determine which internal alternative splice site selection is made. Since our analysis suggests that each of the first exons is preceded by its own promoter, it appears that promoter choice determines the internal splicing pattern. Similar promoter/splice choice coupling events have been observed in only a few other instances, such as in the transcription and processing of the mammalian bcl-X locus and the fibronectin locus (Cramer et al., 1997; Pecci et al., 2001). In the latter case, the underlying molecular basis for such coupling lies in a promoter-dependent regulation of the activity of SR-type splicing factors (Cramer et al., 1999).
The functional output of alternative splicing in ser-2 exon 5a is the inclusion of a premature stop codon leading to the production of a truncated protein. In light of the previously recognized dimerization potential of G-protein coupled 7TMRs (Bouvier, 2001), it is possible to think of the truncated proteins as naturally occurring dominant negative forms of the receptor. Since the sites of expression of truncated and nontruncated transcripts overlap (e.g., the AIY and RID neurons express both ser-2a and ser-2b/ser-2c), it is interesting to speculate that the activity of the tyramine receptor within a single cell may be fine-tuned by differential regulation of the expression of one transcript versus the other. An indication for such differential regulation comes from our observation that the lim-4 transcription factor affects the expression of two RID-expressed splice forms (ser-2b/ser-2c), but not the third RID-expressed splice form (ser-2a).
We have described here a loss-of-function allele in the C. elegans tyramine receptor, ser-2. Using a variety of assays that measure the function of several of the neurons in which ser-2 is expressed, we found no significant consequences of loss of ser-2. This is in contrast to the odorsensory defects observed in Drosophila tyramine receptor mutants (Kutsukake et al., 2000; Nagaya et al., 2002). Also, the effects that tyramine has on the modulation of neuromuscular signaling in flies (Nagaya et al., 2002) are not mirrored by locomotory defects of any detectable type in ser-2 mutant animals. Our behavioral assays may lack the sensitivity to detect SER-2 function, SER-2 may not be the only tyramine receptor in C. elegans, and/or SER-2 may mediate aspects of neuronal circuit function of which we are not currently aware.
Function of Lhx genes in different tissue types
The expression of neurotransmitter receptors, such as the biogenic amine receptors described here, contributes to the subtype identity of a neuron and can be considered a hallmark of the terminal differentiation of a neuron. Probes that monitor the expression of these receptors, such as the reporter gene fusions that we describe here, can thus serve as valuable tools to assess the differentiated state of a neuron in specific mutant backgrounds. We have used these tools to address how specific types of Lhx genes affect the adoption of neuronal subtype identities. By showing that several distinct ser-2 transcriptional gene fusions fail to be appropriately expressed in the AIY interneuron class in ttx-3 mutant animals, we have extended our previous findings (Altun-Gultekin et al., 2001) that this Lhx gene affects all known subtype characteristics of the AIY interneuron class. A similar, though distinct scenario applies for the Lmx1-type lim-6 Lhx gene and the Lhx4-type lim-4 gene. With the help of biogenic amine receptor probes, we could show that lim-6 affects several but not all differentiated characteristics of the RIS interneuron class and that lim-4 affects several but not all differentiated characteristics of the RID motor neuron class. In contrast to ttx-3, which also severely affects AIY anatomy, neither lim-4 nor lim-6 mutants show obvious defects in the neuroanatomy of RID or RIS, respectively. We have thus defined two distinct scenarios for Lhx gene action. In one interneuron class, the Lhx gene ttx-3 affects the adoption of every subtype-specific aspect of an interneuron’s identity, including its axonal anatomy, but not its generation. In another interneuron class, the resident Lhx gene, lim-6, affects many but not all subtype-specific features of that interneuron. Similar to the lim-6 case, the lim-4 Lhx gene also appears to affect some but not all features of the RID neuron.
The cellular context-dependency of Lhx gene action is not only gene-specific but is also cell-specific. That is, different Lhx genes not only act distinctly in different cell types, but one Lhx gene may act distinctly in different cell types. We have previously shown that ttx-3 affects all known subtype characteristics of the AIY interneuron class, yet has little if any effect on the differentiation of other neuron classes that express ttx-3 (Altun-Gultekin et al., 2001). With the use of an aminergic receptor probe, we have obtained some preliminary evidence for an apparently similar theme for the mec-3 Lhx gene. mec-3 is required for the adoption of all subtype characteristics of the light touch sensory neurons, including the expression of all known touch neuron genes and the patterns of axon outgrowth of the light touch neurons (Way and Chalfie, 1988; Zhang et al., 2002). In contrast, our cell fate analysis of PVD in mec-3 mutants shows that mec-3 has no effect on the expression of the ser-2d splice form in PVD, nor does it have an effect on the extension of two dendrites along the length of the animal’s body and one axon in the ventral nerve cord. Intriguingly, however, mec-3 is required for one specific aspect of PVD differentiation, namely the elaboration of the neuron’s dendritic branching pattern, which may allow the neuron to fulfill its function as a harsh touch receptor. The apparently more restricted function of mec-3 in the PVD harsh touch sensory neurons versus the light touch neurons cannot be explained by the absence of its cofactor unc-86, which is also expressed in PVD, but may instead be due to the lin-4-mediated inhibition of the lin-14 transcription factor (Mitani et al., 1993). Thus, the extent of the effect that a given Lhx gene has on neuronal differentiation is dictated by the presence of specific cofactors, which together form a cell-type specific “transcription factor code.”
We also found that Lhx genes that have important functions in some cellular contexts (lim-6, lin-11) have limited if any effects on the differentiated state of the neuron in other cellular contexts (DVA, PVQ, AIZ, RME) (Fig. 11). Since AIZ is functionally disrupted in lin-11 mutants (Hobert et al., 1998), it is likely that lin-11 has other as of yet unrecognized targets in AIZ. In addition, we suspect that lim-6 and lin-11 also have unrecognized targets in RME, PVQ, and DVA. These apparently negative results with the aminergic receptor probes nevertheless make the relevant point that, in specific cellular contexts, Lhx genes only partially, if at all, affect neuronal differentiation. Even in those contexts where worm Lhx genes impinge on all subtype characteristics of a neuron (mec-3 in touch sensory neurons or ttx-3 in AIY interneurons), the neuron is still born and adopts pan-neuronal properties. Our current data thus support our previous speculation that it is a common theme of Lhx genes in C. elegans to act as postmitotic, identity-determining factors (Hobert and Ruvkun, 1998; Hobert and Westphal, 2000). Such a role is consistent with the postmitotic, identity-determining function of several Drosophila and vertebrate Lhx genes (Benveniste et al., 1998; Thor et al., 1999; Thor and Thomas, 1997; Thaler et al., 2002; Tsuchida et al., 1994), but contrasts the role of many other vertebrate Lhx genes, which have been implicated in earlier developmental events affecting neuron generation (Bulchand et al., 2001; Pfaff et al., 1996; Porter et al., 1997; Zhao et al., 1999). These earlier roles may reflect a vertebrate-specific and hence evolutionarily more recent cooption into novel contexts.
Acknowledgments
We thank Bob Johnston for generating dop-1prom1::gfp, Gisela Sandoval for dop-2::gfp, Erik Lundquist for lqIs4, Stephen Nurrish for ser-1::gfp, Zeynep Altun for ttx-3::rfp, Mark Alkema and Bob Horvitz for communicating the tbh-1 expression pattern, Millet Treinin for discussions on PVD morphology, Andy Fire for vectors, Ronald Plasterk and his lab for allowing us to use their deletion library and for assistance during the ser-2 screening procedure, Thomas Roeder for mentioning possible ser-2 functions and Piali Sengupta, Stefan Thor, and Sam Pfaff for reading the manuscript. This work was funded by grants from the NIH, the Sloan Foundation, the Searle Foundation, the Irma T. Hirschl Trust, the Klingenstein Foundation, and the Rita Allen Foundation (to O.H.).
Footnotes
Note added in proof. After acceptance of this paper, Suo et al. independently reported the characterization of DOP-2 and confirmed that DOP-2 is a D2-type dopamine receptor (Suo, S., Sasagawa, N., Ishiura, S., 2003. Cloning and characterization of a Caenorhabditis elegans D-2-type dopamine receptor, J. Neurochem., 86, 869–878).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Arakawa S, Gocayne JD, McCombie WR, Urquhart DA, Hall LM, Fraser CM, Venter JC. Cloning, localization, and permanent expression of a Drosophila octopamine receptor. Neuron. 1990;4:343–354. doi: 10.1016/0896-6273(90)90047-j. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Neuron. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Aurelio O, Boulin T, Hobert O. Identification of spatial and temporal cues that regulate postembryonic expression of axon maintenance factors in the C. elegans ventral nerve cord. Development. 2003;130:599–610. doi: 10.1242/dev.00277. [DOI] [PubMed] [Google Scholar]
- Aurelio O, Hall DH, Hobert O. Immunoglobulin-domain proteins required for maintenance of ventral nerve cord organization. Science. 2002;295:686–690. doi: 10.1126/science.1066642. [DOI] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J. Exp. Zool. 1990;253:263–270. doi: 10.1002/jez.1402530305. [DOI] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mech. Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Benveniste RJ, Thor S, Thomas JB, Taghert PH. Cell type-specific regulation of the Drosophila FMRF-NH2 neuropeptide gene by Apterous, a LIM homeodomain transcription factor. Development. 1998;125:4757–4765. doi: 10.1242/dev.125.23.4757. [DOI] [PubMed] [Google Scholar]
- Blenau W, Balfanz S, Baumann A. Amtyr1: characterization of a gene from honeybee (Apis mellifera) brain encoding a functional tyramine receptor. J. Neurochem. 2000;74:900–908. doi: 10.1046/j.1471-4159.2000.0740900.x. [DOI] [PubMed] [Google Scholar]
- Blenau W, Baumann A. Molecular and pharmacological properties of insect biogenic amine receptors: lessons from Drosophila melanogaster and Apis mellifera. Arch. Insect Biochem. Physiol. 2001;48:13–38. doi: 10.1002/arch.1055. [DOI] [PubMed] [Google Scholar]
- Borowsky B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S, Boyle N, Pu X, Kouranova E, Lichtblau H, Ochoa FY, Branchek TA, Gerald C. Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc. Natl. Acad. Sci. USA. 2001;98:8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat. Rev. Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 2001a;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Mellem JE, Hills T, Madsen DM, Maricq AV. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001b;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 2001;100:165–175. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- Bu¨low HE, Berry KL, Topper LH, Peles E, Hobert O. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc. Natl. Acad. Sci. USA. 2002;99:6346–6351. doi: 10.1073/pnas.092128099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunin MA, Wightman RM. Paracrine neurotransmission in the CNS: involvement of 5-HT. Trends Neurosci. 1999;22:377–382. doi: 10.1016/s0166-2236(99)01410-1. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Du H, Gu G, William CM, Chalfie M. Extracellular proteins needed for C. elegans mechanosensation. Neuron. 1996;16:183–194. doi: 10.1016/s0896-6273(00)80035-5. [DOI] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- Halevi S, McKay J, Palfreyman M, Yassin L, Eshel M, Jorgensen E, Treinin M. The C. elegans ric-3 gene is required for maturation of nicotinic acetylcholine receptors. EMBO J. 2002;21:1012–1020. doi: 10.1093/emboj/21.5.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Ungrin MD, Abramovitz M, Ribeiro P. Characterization of a novel serotonin receptor from Caenorhabditis elegans: cloning and expression of two splice variants. J. Neurochem. 1999;72:1372–1383. doi: 10.1046/j.1471-4159.1999.721372.x. [DOI] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hobert O, D’Alberti T, Liu Y, Ruvkun G. Control of neural development and function in a thermoregulatory network by the LIM homeobox gene lin-11. J. Neurosci. 1998;18:2084–2096. doi: 10.1523/JNEUROSCI.18-06-02084.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Regulation of interneuron function in the C elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Hobert O, Ruvkun G. A common theme for LIM homeobox gene function across phylogeny. Biol. Bull. 1998;195:377–380. doi: 10.2307/1543153. [DOI] [PubMed] [Google Scholar]
- Hobert O, Tessmar K, Ruvkun G. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development. 1999;126:1547–1562. doi: 10.1242/dev.126.7.1547. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Function of LIM homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Chalfie M, Trent C, Sulston JE, Evans PD. Serotonin and octopamine in the nematode Caenorhabditis elegans. Science. 1982;216:1012–1014. doi: 10.1126/science.6805073. [DOI] [PubMed] [Google Scholar]
- Hwang JM, Chang DJ, Kim US, Lee YS, Park YS, Kaang BK, Cho NJ. Cloning and functional characterization of a Caenorhabditis elegans muscarinic acetylcholine receptor. Receptors Channels. 1999;6:415–424. [PubMed] [Google Scholar]
- Ishihara T, lino Y, Mohri A, Mori I, Gengyo-Ando K, Mitani S, Katsura I. HEN-1, a secretory protein with an LDL receptor motif, regulates sensory integration and learning in Caenorhabditis elegans. Cell. 2002;109:639–649. doi: 10.1016/s0092-8674(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Jansen G, Hazendonk E, Thijssen KL, Plasterk RH. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kindt KS, Tam T, Whiteman S, Schafer WR. Serotonin promotes G(o)-dependent neuronal migration in Caenorhabditis elegans. Curr. Biol. 2002;12:1738–1747. doi: 10.1016/s0960-9822(02)01199-5. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kutsukake M, Komatsu A, Yamamoto D, Ishiwa-Chigusa S. A tyramine receptor gene mutation causes a defective olfactory behavior in Drosophila melanogaster. Gene. 2000;245:31–42. doi: 10.1016/s0378-1119(99)00569-7. [DOI] [PubMed] [Google Scholar]
- Lauder JM. Neurotransmitters as growth regulatory signals: role of receptors and second messengers. Trends Neurosci. 1993;16:233–240. doi: 10.1016/0166-2236(93)90162-f. [DOI] [PubMed] [Google Scholar]
- Lints R, Emmons SW. Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development. 1999;126:5819–5831. doi: 10.1242/dev.126.24.5819. [DOI] [PubMed] [Google Scholar]
- Loer CM, Kenyon CJ. Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J. Neurosci. 1993;13:5407–5417. doi: 10.1523/JNEUROSCI.13-12-05407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mech-anosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- Maryon EB, Saari B, Anderson P. Muscle-specific functions of ryanodine receptor channels in Caenorhabditis elegans. J. Cell Sci. 1998;111:2885–2895. doi: 10.1242/jcs.111.19.2885. [DOI] [PubMed] [Google Scholar]
- McClung C, Hirsh J. The trace amine tyramine is essential for sensitization to cocaine in Drosophila. Curr. Biol. 1999;9:853–860. doi: 10.1016/s0960-9822(99)80389-3. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel JE, Korswagen HC, Liu KS, Hajdu-Cronin YM, Simon MI, Plasterk RH, Sternberg PW. Participation of the protein Go in multiple aspects of behavior in C. elegans. Science. 1995;267:1652–1655. doi: 10.1126/science.7886455. [DOI] [PubMed] [Google Scholar]
- Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- Nagaya Y, Kutsukake M, Chigusa S, Komatsu A. A trace amine, tyramine, functions as a neuromodulator in Drosophila melanogaster. Neurosci. Lett. 2002;329:324. doi: 10.1016/s0304-3940(02)00596-7. [DOI] [PubMed] [Google Scholar]
- Nass R, Hall DH, Miller DM, 3rd, Blakely RD. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99:3264–3269. doi: 10.1073/pnas.042497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc. Natl. Acad. Sci. USA. 2001;98:14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olde B, McCombie WR. Molecular cloning and functional expression of a serotonin receptor from Caenorhabditis elegans. J. Mol. Neurosci. 1997;8:53–62. doi: 10.1007/BF02736863. [DOI] [PubMed] [Google Scholar]
- Pauwels PJ, Wurch T. Review: amino acid domains involved in constitutive activation of G-protein-coupled receptors. Mol. Neurobiol. 1998;17:109–135. doi: 10.1007/BF02802027. [DOI] [PubMed] [Google Scholar]
- Pecci A, Viegas LR, Baranao JL, Beato M. Promoter choice influences alternative splicing and determines the balance of isoforms expressed from the mouse bcl-X gene. J. Biol. Chem. 2001;276:21062–21069. doi: 10.1074/jbc.M008665200. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene lsl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Rex E, Komuniecki RW. Characterization of a tyramine receptor from Caenorhabditis elegans. J. Neurochem. 2002;82:1352–1359. doi: 10.1046/j.1471-4159.2002.01065.x. [DOI] [PubMed] [Google Scholar]
- Roeder T. Octopamine in invertebrates. Prog. Neurobiol. 1999;59:533–561. doi: 10.1016/s0301-0082(99)00016-7. [DOI] [PubMed] [Google Scholar]
- Roth BL, Shoham M, Choudhary MS, Khan N. Identification of conserved aromatic residues essential for agonist binding and second messenger production at 5-hydroxytryptamine2A receptors. Mol. Pharmacol. 1997;52:259–256. doi: 10.1124/mol.52.2.259. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 1999;13:1794–1806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salkoff L, Butler A, Fawcett G, Kunkel M, McArdle C, Paz-y-Mino G, Nonet M, Walton N, Wang ZW, Yuan A, Wei A. Evolution tunes the excitability of individual neurons. NeuroScience. 2001;103:853–859. doi: 10.1016/s0306-4522(01)00079-3. [DOI] [PubMed] [Google Scholar]
- Sarafi-Reinach TR, Melkman T, Hobert O, Sengupta P. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development. 2001;128:3269–3281. doi: 10.1242/dev.128.17.3269. [DOI] [PubMed] [Google Scholar]
- Saudou F, Amlaiky N, Plassat JL, Borrelli E, Hen R. Cloning and characterization of a Drosophila tyramine receptor. EMBO J. 1990;9:3611–3617. doi: 10.1002/j.1460-2075.1990.tb07572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Schafer WR, Kenyon CJ. A calcium-channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature. 1995;375:73–78. doi: 10.1038/375073a0. [DOI] [PubMed] [Google Scholar]
- Segalat L, Elkes DA, Kaplan JM. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Science. 1995;267:1648–1651. doi: 10.1126/science.7886454. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Sulston J, Dew M, Brenner S. Dopaminergic neurons in the nematode Caenorhabditis elegans. J. Comp. Neurol. 1975;163:215–226. doi: 10.1002/cne.901630207. [DOI] [PubMed] [Google Scholar]
- Suo S, Sasagawa N, Ishiura S. Identification of a dopamine receptor from Caenorhabditis elegans. Neurosci. Lett. 2002;319:13–16. doi: 10.1016/s0304-3940(01)02477-6. [DOI] [PubMed] [Google Scholar]
- Svendsen PC, McGhee JD. The C. elegans neuronally expressed homeobox gene ceh-10 is closely related to genes expressed in the vertebrate eye. Development. 1995;121:1253–1262. doi: 10.1242/dev.121.5.1253. [DOI] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contribute to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–239. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J. Neurobiol. 2003;56:178–197. doi: 10.1002/neu.10245. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Vallone D, Picetti R, Borrelli E. Structure and function of dopamine receptors. Neurosci. Biobehav. Rev. 2000;24:125–132. doi: 10.1016/s0149-7634(99)00063-9. [DOI] [PubMed] [Google Scholar]
- Vanden Broeck J, Vulsteke V, Huybrechts R, De Loof A. Characterization of a cloned locust tyramine receptor cDNA by functional expression in permanently transformed Drosophila S2 cells. J. Neurochem. 1995;64:2387–2395. doi: 10.1046/j.1471-4159.1995.64062387.x. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- Weinshenker D, Garriga G, Thomas JH. Genetic and pharmacological analysis of neurotransmitters controlling egg laying in C. elegans. J. Neurosci. 1995;15:6975–6985. doi: 10.1523/JNEUROSCI.15-10-06975.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wess J. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol. Ther. 1998;80:231–264. doi: 10.1016/s0163-7258(98)00030-8. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 2000;221:295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H. Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science. 1999;284:1155–1158. doi: 10.1126/science.284.5417.1155. [DOI] [PubMed] [Google Scholar]