SUMMARY
Loading of the ring-shaped Mcm2-7 replicative helicase around DNA licenses eukaryotic origins of replication. During loading, Cdc6, Cdt1 and the origin-recognition complex (ORC) assemble two heterohexameric Mcm2-7 complexes into a head-to-head double hexamer that facilitates bidirectional replication initiation. Using multi-wavelength single-molecule fluorescence to monitor the events of helicase loading, we demonstrate that double-hexamer formation is the result of sequential loading of individual Mcm2-7 complexes. Loading of each Mcm2-7 molecule involves the ordered association and dissociation of distinct Cdc6 and Cdt1 proteins. In contrast, one ORC molecule directs loading of both helicases in each double hexamer. Based on single-molecule FRET, arrival of the second Mcm2-7 results in rapid double-hexamer formation that anticipates Cdc6 and Cdt1 release, suggesting Mcm-Mcm interactions recruit the second helicase. Our findings reveal the complex protein dynamics that coordinate helicase loading and indicate that distinct mechanisms load the oppositely oriented helicases that are central to bidirectional replication initiation.
INTRODUCTION
Eukaryotic DNA replication must occur faithfully each cell cycle to maintain genomic stability. Initiation of replication occurs at genomic sites called origins. To ensure that no origin initiates replication more than once per cell cycle, the cell restricts the DNA loading and activation of the Mcm2-7 replicative helicase to distinct cell cycle stages (Siddiqui et al., 2013). Importantly, helicase loading (also known as pre-RC formation) licenses origins of replication by establishing the correct architecture for helicase activation and bidirectional replication initiation.
Three helicase-loading proteins direct Mcm2-7 loading: the origin recognition complex (ORC), Cdc6 and Cdt1 (reviewed in Yardimci and Walter, 2014). ORC binds origins of replication and recruits Cdc6 at the M/G1 transition. Cdc6-bound ORC recruits Mcm2-7 in complex with Cdt1 to origin DNA. In an ATP-hydrolysis-dependent reaction, recruited Mcm2-7 complexes are loaded around the origin DNA (Coster et al., 2014; Kang et al., 2014). Helicase loading requires opening and closing of the toroidal Mcm2-7 ring between the Mcm2 and Mcm5 subunits (Bochman and Schwacha, 2008; Costa et al., 2011; Samel et al., 2014). The product of helicase loading is a pair of tightly interacting Mcm2-7 complexes that encircle the double-stranded origin DNA in a head-to-head conformation, with staggered Mcm2/5 gates (Costa et al., 2014; Evrin et al., 2009; Remus et al., 2009; Sun et al., 2014).
Although the structure of the double-hexamer product of helicase loading is clear, important questions remain about how the helicase-loading proteins achieve this outcome. In particular, the mechanisms that load the first and second Mcm2-7 complex in opposite orientations are unclear (reviewed in Yardimci and Walter, 2014). Do the two Mcm2-7 complexes associate and load simultaneously or in an ordered fashion? Do the same or different ORC and Cdc6 proteins load each Mcm2-7 complex? To address these questions we have developed single-molecule assays to monitor helicase loading.
Single-molecule studies are a powerful tool to address questions of stoichiometry and dynamics during DNA replication events. Studies of this type have led to important insights including the dynamics and number of DNA polymerases acting at the replication fork (reviewed in Stratmann and van Oijen, 2014). Extending these approaches to replication initiation has the potential for additional discovery. Unlike current ensemble helicase loading assays, which can only detect events that survive multiple washes, single-molecule approaches readily detect short-lived interactions during cycles of enzymatic function. Single-molecule approaches also allow stoichiometric determinations that are difficult with ensemble helicase loading assays due to DNA-to-DNA asynchrony and heterogeneity. Finally, although multi-step reactions are frequently asynchronous, post-hoc synchronization of single-molecule data allows precise kinetic analysis of reaction pathways.
We have developed single-molecule assays that monitor the DNA association of eukaryotic helicase-loading proteins using colocalization single-molecule spectroscopy (CoSMoS) (Friedman et al., 2006; Hoskins et al., 2011). We show that the two Mcm2-7 hexamers are recruited and loaded in separate events that require distinct Cdc6 and Cdt1 molecules. In contrast, one ORC molecule directs loading of both Mcm2-7 complexes present in a double hexamer. Consistent with distinct mechanisms loading the two hexamers, we observe kinetic differences between events associated with loading the first and second helicase. By combining CoSMoS with fluorescence resonance energy transfer (FRET), we demonstrate that formation of the Mcm2-7 double-hexamer interface precedes dissociation of Cdc6 and Cdt1, suggesting interactions with the first Mcm2-7, rather than ORC, drive recruitment of the second helicase. Our observations reveal both the complex protein coordination required to assemble Mcm2-7 double hexamers and the mechanisms ensuring the two Mcm2-7 molecules are loaded in the opposite orientations required for bidirectional replication initiation.
RESULTS
A single-molecule assay for helicase loading
To develop a single-molecule assay for eukaryotic helicase loading we used CoSMoS to monitor the origin-DNA association of the proteins required for this process (ORC, Cdc6, Cdt1, Mcm2-7). First, we immobilized origin-containing DNA by coupling it to microscope slides. We determined the location of surface-attached DNA on the slide using a DNA-coupled fluorophore (Fig. 1A). We monitored associations of one or two proteins (labeled with distinguishable fluorophores) with origin DNA using colocalization of the protein- and DNA-associated fluorophores (Fig. S1A). Fluorescent labeling of ORC, Cdc6, Cdt1 and Mcm2-7 was accomplished using a SNAP-tag or sortase-mediated coupling of fluorescent peptides (Gendreizig et al., 2003; Popp et al., 2007). In each case, the fluorescent tags did not interfere with protein function in ensemble helicase-loading reactions (Fig. S1B). After imaging the locations of slide-coupled DNA molecules, purified ORC, Cdc6, and Cdt1/Mcm2-7 were added (one or two of which were fluorescently labeled) and the location of each DNA molecule was continuously monitored for labeled protein colocalization in one-second intervals for 20 minutes.
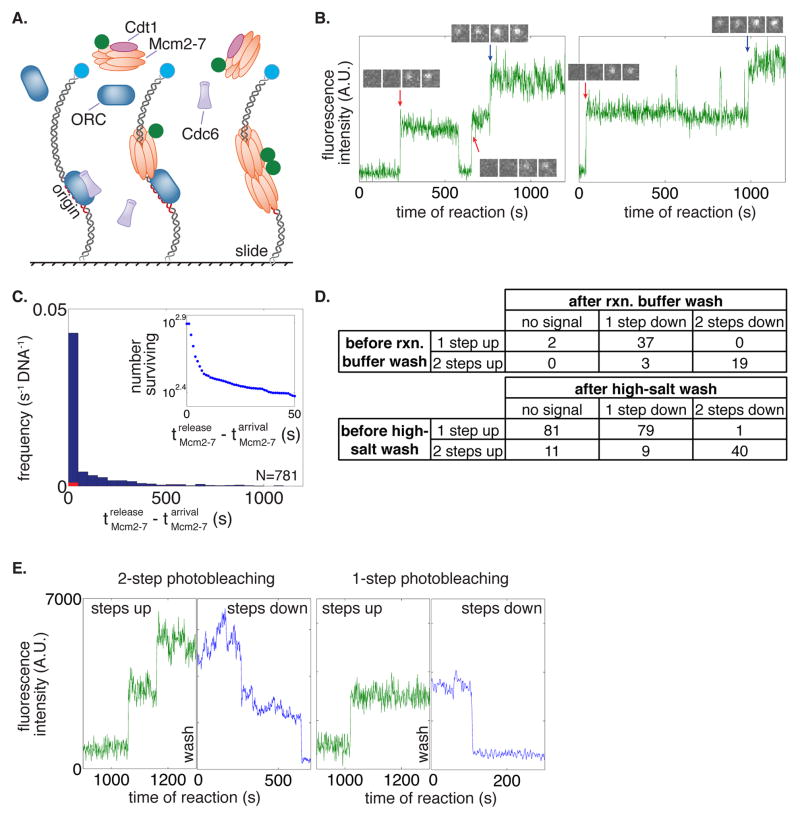
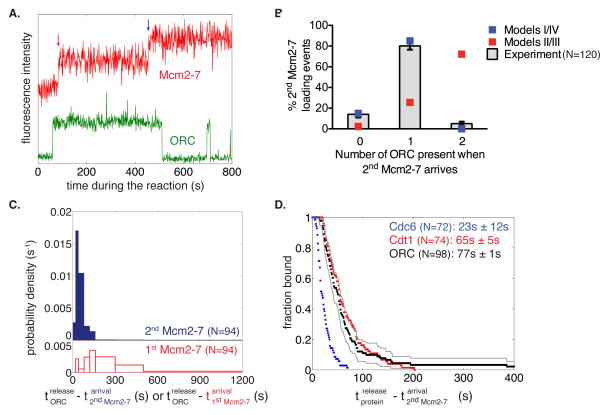
Figure 1. Mcm2-7 hexamers associate with and are loaded on DNA in a one-at-a-time manner.
(A) Schematic for the single-molecule helicase-loading assay. Alexa488-labeled (blue circle) 1.3kb origin-DNAs were coupled to microscope slides. Purified ORC, Cdc6, and Cdt1/Mcm2-7 (at least one fluorescently labeled, Mcm2-7 in this illustration) were incubated with slide-coupled DNA and colocalization of the fluorescently-labeled protein with the DNA was monitored.
(B) Mcm2-7 complexes sequentially associate with origin DNA. Plots display the Mcm2-74SNAP549 fluorescence intensity recorded at two representative DNA molecules. Insets show fluorescence images (4 × 1 s) taken during the sequential association of first (red arrow) and second (blue arrow) Mcm2-7.
(C) Mcm2-7 dwell times on DNA have a multiexponential distribution. Mcm2-7 dwell times were plotted as a histogram. Combined data from first and second Mcm2-7 associations are included; vertical axis represents the number of dwells of the specified duration per s per DNA molecule. Red bars are results from a separate experiment using mutant origin DNA. Inset shows the distribution of Mcm2-7 dwell times on DNA molecules as a semilogarithmic cumulative survival plot; only a portion of the entire plot is shown to emphasize that the distribution has at least two exponential components.
(D) Mcm2-7 associates with DNA one at a time. The number of associations present at standard protein concentrations before a reaction-buffer (top) or high-salt-buffer (0.5 M NaCl; bottom) wash is compared to the number of fluorophores that are detected by photobleaching immediately after the wash.
(E) Two representative traces before and after a high-salt wash and photobleaching. Reactions were washed twice with a high-salt buffer and imaged at higher laser power in the absence of an oxygen scavenging system until all fluorophores were photobleached. Traces of Mcm2-74SNAP549 associations during the reaction (green) are plotted adjacent to photobleaching steps after a high-salt wash (blue).
Multiple observations indicated that Mcm2-7-DNA colocalizations represented events of helicase loading (Table S1, Movies S1, S2 and S3). First, colocalizations of Mcm2-7 with the DNA was dramatically reduced in the absence of ORC or Cdc6, proteins required for helicase loading (Yardimci and Walter, 2014). Second, stable association (>30 s) of Mcm2-7 was dependent on the presence of the ORC DNA binding site (the ARS-consensus sequence, ACS). Third, ORC, Cdc6, origin DNA and ATP hydrolysis were each required to form Mcm2-7 molecules that were resistant to a high-salt wash (Table S1), a biochemical test for loaded helicases encircling dsDNA independently of helicase-loading proteins (Donovan et al., 1997; Randell et al., 2006).
Mcm2-7 association and loading occurs in a one-at-a-time manner
Our initial studies monitored Mcm2-7 association with origin DNA. We performed CoSMoS helicase-loading experiments using Mcm2-7 containing SNAP-tagged Mcm4 labeled with 549 fluorophore (Mcm2-74SNAP549, Fig. 1) and unlabeled ORC, Cdc6 and Cdt1. Over the course of 20 minutes we observed both single- and double-stepwise increases in Mcm2-7-associated fluorescence intensity at origin DNAs (Fig. 1B, S1C). Mcm2-7 dwell time distributions were multi-exponential with many short-lived (<30 s) and fewer longer-lived (>30 s) relative increases in fluorescent intensity, suggesting at least two distinct types of Mcm2-7 association with the DNA (Fig. 1C).
There are two possible explanations for the multiple stepwise increases in DNA-colocalized Mcm2-7-coupled fluorescence. The simplest interpretation of this data is that Mcm2-7 hexamers associate with origin-DNA in a one-at-a-time manner, with multiple hexamers accumulating over time. Alternatively, it was possible that each increase in fluorescence was due to the simultaneous association of a Mcm2-7 multimer (e.g. a pre-formed dimer of two Mcm2-7 hexamers). To distinguish between these possibilities, we used photobleaching to count the number of DNA-associated Mcm2-7 hexamers. To this end, we first observed Mcm2-74SNAP549 associations with DNA and then washed the surface-tethered DNA molecules with reaction buffer, removing unbound proteins. Then, to promote photobleaching, we increased laser excitation power and removed oxygen scavengers. Comparison of the number of Mcm2-74SNAP549 photobleaching steps after the wash with the number of association steps that accumulated before the wash showed no single-step increase in fluorescence before the wash resulted in a two-step photobleaching afterward (Fig. 1D, top). We confirmed that loss of fluorescence was due to photobleaching and not dissociation of Mcm2-7 by observing previously non-illuminated microscope fields of view. These data eliminate models in which multiple Mcm2-7 complexes are recruited simultaneously. We conclude that Mcm2-7 association occurs in a one-at-a-time manner.
We next asked whether loading of salt-resistant Mcm2-7 hexamers around origin-DNA occurred sequentially or simultaneously. We used the same photobleaching assay (described above) except a high-salt wash was used to remove any incompletely-loaded Mcm2-7 complexes prior to photobleaching. If loading of both Mcm2-7 hexamers occurs simultaneously, we should observe only even numbers of high-salt resistant hexamers. In contrast, if loading occurs sequentially, we should observe even and odd numbers of high-salt resistant hexamers. At low protein concentrations we observed both one- and two-step photobleaching events (Fig. 1D, bottom, 1E). Roughly half (79/160) of all single Mcm2-7-associated fluorophores that colocalized with origin DNA before the high-salt wash were high-salt resistant, and 67% (40/60) of the double Mcm2-7-associated fluorophores were high-salt resistant. When we increased protein concentrations, we also observed DNA molecules with three and four origin-dependent, high-salt resistant Mcm2-7 complexes (Fig. S1D), indicating that more than one double-hexamer loading event occurred at a single origin.
We considered the possibility that the apparent colocalization of odd numbers of loaded Mcm2-7 complexes was due to incomplete fluorescent labeling of Mcm2-7. For example, a single salt-resistant Mcm2-7-associated fluorophore could be the result of loading two Mcm2-7 complexes, only one of which is fluorescently labeled. To address this possibility, we purified Mcm2-7 complexes that were labeled on two subunits with different fluorophores (Mcm2-74SNAP549/7SORT649). Because the SNAP-tag and sortase labeling approaches are independent of each other, we could use single-molecule imaging to determine the efficiency of each labeling protocol (79% for SNAP and 77% for sortase). This labeling protocol also increased the proportion of Mcm2-7 complexes that have at least one coupled fluorophore to 95%. Using the measured labeling efficiencies, we determined the number of high-salt resistant Mcm2-7 complexes with no more than one of each fluorophore that would be expected if only double hexamers were loaded (Fig. S1E, Model II). Assays with Mcm2-74SNAP549/7SORT649 yielded single, salt-resistant fluorophores in a proportion that is inconsistent with this model. Instead our data is consistent with a model where both single and double hexamers are loaded (in a 52:48 ratio based on our data; Fig. S1E, Model I). We conclude that Mcm2-7 complexes are both recruited and loaded onto origin DNA in a sequential manner.
Distinct Cdc6 and Cdt1 molecules load the first and second Mcm2-7
We investigated the number of Cdt1 and Cdc6 molecules required for helicase loading and their relative times of DNA association. Both proteins are essential for loading but show little or no association with DNA in bulk assays (Coster et al., 2014; Kang et al., 2014), suggesting that their protein and/or DNA associations during helicase loading are transient. To detect these associations, we simultaneously monitored the binding of two different protein pairs labeled with distinguishable fluorophores: either Cdt1SORT549 with Mcm2-74SNAPJF646 or Cdc6SORT549 with Mcm2-74SNAPJF646. The associations of both fluorophores with origin-DNA were monitored simultaneously, revealing relative times of arrival and departure for the two molecules in each pair.
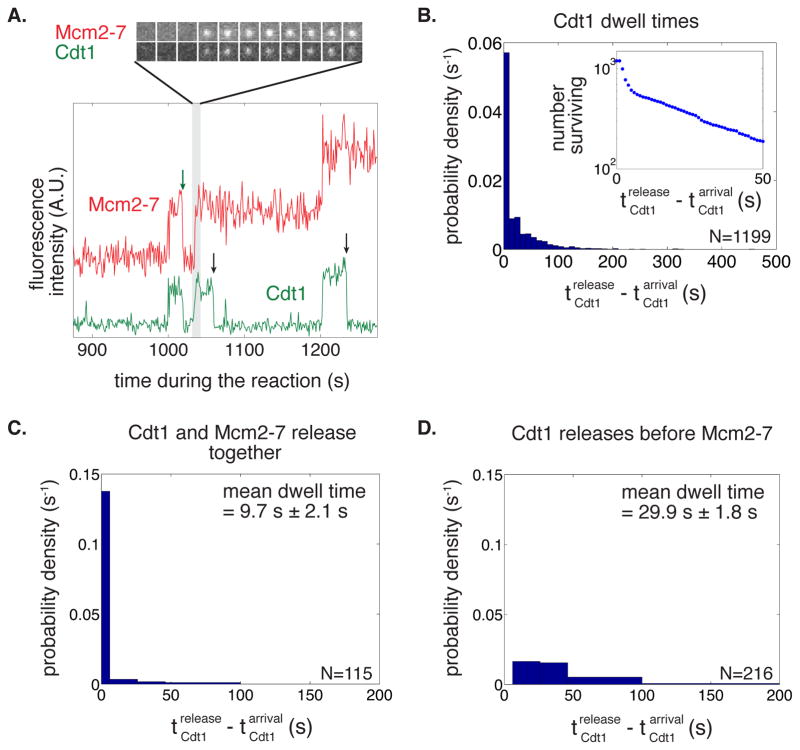
Consistent with being recruited to origins as a complex, we typically observed that Cdt1 and Mcm2-7 associated with origin DNA simultaneously (Fig. 2A, S2A-C). Uncommon instances where Cdt1 or Mcm2-7 are seen associating separately (Cdt1 alone: 11.4%, Mcm2-7 alone: 18.6%) are likely caused by incomplete dye labeling of the other protein because the frequencies of these events are similar to the fractions of unlabeled Mcm2-7 or Cdt1 (14% and 20%, respectively). Like Mcm2-7, Cdt1 dwell times followed a multi-exponential distribution, indicating the presence of at least two types of Cdt1-containing complexes on the DNA (Fig. 2B). Consistent with this interpretation, we identified two classes of Mcm2-7/Cdt1 dwell-time and departure behaviors. In many instances Cdt1 and Mcm2-7 were released simultaneously (i.e. within 1 s, see Fig. S2B and S2C). This release pattern occurs whether or not the DNA molecule already had an associated Mcm2-7. These associations were typically short-lived (Fig. 2C) and represent non-productive binding events. Interestingly, these events were less frequent if the Mcm2-7/Cdt1 was the second (29%) rather than the first (53%) to arrive at the DNA. In the remaining cases, Cdt1 was typically longer-lived (Fig. 2D) and was released from origin DNA by itself, leaving behind an associated Mcm2-7. Clearly, only instances when Cdt1 is released independently of Mcm2-7 can be on the pathway for double-hexamer formation. Because Cdt1-associated fluorophore photobleaching was much slower than Cdt1 dissociation (Fig. S2D; Table S2), nearly all loss of fluorescent colocalization was due to dissociations, not photobleaching.
Figure 2. Distinct Cdt1 molecules load the first and the second Mcm2-7 hexamer.
(A) Cdt1 molecules arrive with Mcm2-7 but release quickly after the complex arrives. A representative two-color recording of Mcm2-74SNAPJF646 and Cdt1SORT549 fluorescence at an origin-DNA location is shown. The baseline of the red plot (Mcm2-7) is shifted up relative to the green plot (Cdt1) throughout the manuscript when two-color recordings are displayed together. The sequence of single-frame images of the Cdt1- and Mcm2-7-fluorescent spots illustrates the concurrent arrival of Cdt1 and Mcm2-7. Cdt1 release occurs either with (green arrow) or without (black arrows) concurrent Mcm2-7 release.
(B) Cdt1 dwell times on DNA have a multiexponential distribution. Cdt1 dwell times were plotted as a histogram. Inset shows semilogarithmic cumulative survival plot as in Fig. 1C.
(C–D) There are two types of Cdt1 release events.
(C) Histogram shows the duration of Cdt1 origin-DNA associations when Cdt1 releases with Mcm2-7. The mean dwell time ± standard error of the mean (s.e.m.) is reported.
(D) Histogram shows the duration of Cdt1 origin-DNA associations when Cdt1 releases before Mcm2-7. The mean dwell time ± s.e.m. is reported.
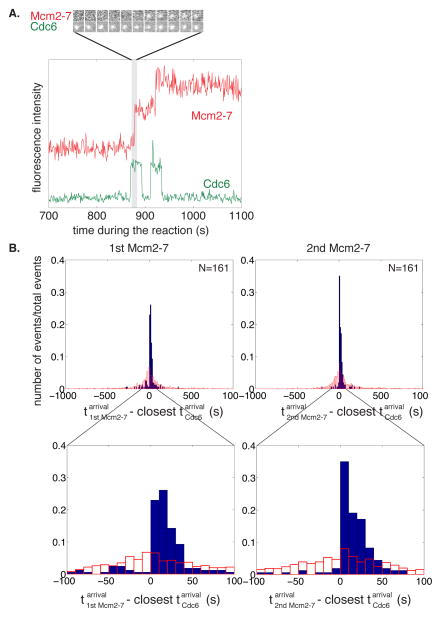
Like Cdt1, Cdc6 association with the DNA is dynamic with distinct molecules acting during loading of the first and second Mcm2-7 (Fig. 3A, S3A). Simultaneous analysis of Mcm2-7 and Cdc6 DNA association showed short Cdc6-DNA associations (mean lifetime 27.8 ± 1.5 s; Fig. S3B), a subset of which directed Mcm2-7 recruitment (35.8%, N=514, Fig. 3A, S3A). Cdc6 consistently anticipated Mcm2-7 arrival at the DNA (>85%, Fig. 3A, S3A). The remaining cases likely reflected the action of unlabeled Cdc6. We observed distinct Cdc6 proteins direct recruitment of the first and second Mcm2-7 with a similar rate constant (Fig. S3C). The high frequency of Cdc6 DNA associations led us to test and confirm that sequential binding of Cdc6 and Mcm2-7 was not coincidental for either Mcm2-7 loading event (Fig. 3B).
Figure 3. Distinct Cdc6 molecules recruit and load the first and the second Mcm2-7 hexamer.
(A) Distinct Cdc6 molecules anticipate each Mcm2-7 association. A representative fluorescence intensity record for Mcm2-74SNAPJF646 and Cdc6SORT549 at origin-DNA. Images of the Cdc6- and Mcm2-7-associated fluorescent spots show Cdc6 binds before the arrival of the first Mcm2-7 complex.
(B) Cdc6 association anticipates binding of the first and second Mcm2-7. Full histogram (top) and expanded view (bottom) of Mcm2-7 arrival time minus the closest Cdc6 arrival time on the same DNA molecule (blue bars). Data is separated into Mcm2-7 complexes arriving at the DNA first (left) or second (right). In >85% of the observations the difference was greater than zero, indicating that Cdc6 arrived before Mcm2-7; in the remaining <15%, Mcm2-7 arrived before Cdc6 (likely due to an unlabeled Cdc6 molecule). Red bars show a control analysis in which each Mcm2-7 arrival time was paired with the closest Cdc6 arrival time on a different, randomly selected DNA molecule. The randomized control does not show the prominent peak at differences between 0 and +50 s indicating the sequential association of Cdc6 and Mcm2-7 was not coincidental.
Release of Cdc6 and Cdt1 is sequential during helicase loading
We next asked whether helicase loading led to a defined order of Cdc6 and Cdt1 release. We took two approaches to address this question: (1) we performed experiments in which Cdc6 and Cdt1 were labeled with different fluorophores, and (2) we compared the times of Cdc6 and Cdt1 release relative to the time of the corresponding Mcm2-7 association in the previously described double-labeled experiments (Mcm2-74SNAPJF646 with either Cdt1SORT549 or Cdc6SORT549).
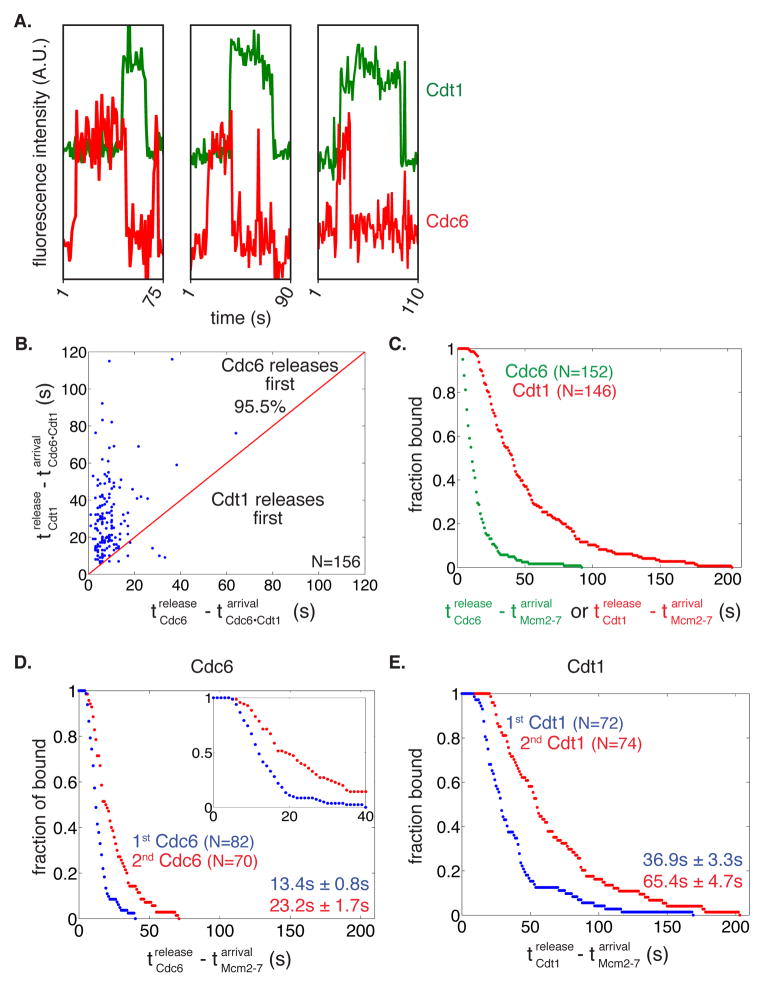
When Cdc6 and Cdt1 were labeled in the same experiment, we consistently saw Cdc6 associating with and releasing from origin-DNA before Cdt1 (Fig. 4A, S4A). Because only non-productive Cdt1-DNA interactions had dwell times less than 6 s (see Fig. 2C), we excluded these molecules from our analysis. Cdc6SORT649 is released before Cdt1SORT549 in >95% of cases when Cdt1 and Cdc6 were co-localized on a DNA (Fig. 4B). When the fluorophores coupled to the proteins were swapped (Cdc6SORT549 and Cdt1SORT649) >90% of observations showed Cdc6 dissociates from origin-DNA before Cdt1 (Fig. S4B). This lower percentage is likely due to the higher photobleaching rate of the 649 dye (Table S2). These results suggest that Cdc6 is released prior to Cdt1 during helicase loading.
Figure 4. Cdc6 release occurs before Cdt1 release.
(A) Three representative fluorescence intensity records for Cdc6SORT649 and Cdt1SORT549 showing arrival and departure of Cdc6 before Cdt1.
(B) Release of Cdc6 anticipates Cdt1 release in a majority of cases. Time of Cdt1 release (y-axis) is plotted against time of Cdc6 release (x-axis, both times are measured from start of simultaneous presence of Cdc6 and Cdt1). The red line represents where points would fall if Cdc6 and Cdt1 released simultaneously. The fraction of measurements in which Cdc6 is released before Cdt1 is reported.
(C) Release of Cdc6 occurs before release of Cdt1 during double hexamer formation. Survival function for Cdc6SORT549 and Cdt1SORT549 dwell times after the first or second Mcm2-7 associates with origin DNA. The y-axis represents the fraction of Cdc6 or Cdt1 molecules that are still associated after the time represented on the x-axis.
(D-E) Cdc6 and Cdt1 release events are slower for the second versus the first Mcm2-7 loading events.
(D) The time of Cdc6 release after Mcm2-7 association is plotted for the first (blue) and second (red) Mcm2-7 association as a survival plot (the fraction of Cdc6 molecules that remain DNA-associated is plotted against time). Inset shows the first 40 s of the entire plot to emphasize the presence of a lag prior to DNA release. Numbers are mean release times ± s.e.m. for the first or second Mcm2-7-associated Cdc6 molecule.
(E) Cdt1 release after the first (blue) and second Mcm2-7 association (red) as a survival plot as described for 4D.
Because Mcm2-7 was unlabeled in the previous experiments, we did not know which of the Cdc6-Cdt1 DNA co-localization events directed double-hexamer formation. To address whether Cdc6 is released before Cdt1 during double-hexamer formation, we analyzed the time that each Cdc6 or Cdt1 spent on the DNA with Mcm2-7. Consistent with the Cdc6-Cdt1 double-labeling experiments, the average time between Mcm2-7 arrival and Cdc6 release is significantly shorter than the corresponding time before Cdt1 release (Fig. 4C). Both the Cdc6SORT549 and Cdt1SORT549 release times are >50-fold shorter than the fluorescent dye lifetimes calculated from photobleaching rates (Table S2), verifying that these are dissociation events and not due to photobleaching. We conclude that each Mcm2-7 loading event is associated with the ordered release of Cdc6 followed by Cdt1 from the DNA.
Kinetic evidence for distinct mechanisms loading the first and second helicase
We reasoned that if loading of the first and second helicases occurred by different mechanisms, the time that Cdc6 and Cdt1 would spend associated with the first versus the second Mcm2-7 would differ. The resulting survival curves showed delays between arrival of Mcm2-7 and release of Cdc6 or Cdt1, suggesting that the release of both proteins involves multiple steps after Mcm2-7 recruitment. Although the order of Cdc6 and Cdt1 release remained the same, we found that the release times were significantly longer for the second Mcm2-7 loading event for both Cdc6 (p < 0.003, Fig. 4D) and Cdt1 (p < 0.001, Fig. 4E). These kinetic data suggest that loading of the first and second helicase occurs through distinct mechanisms.
A single ORC directs formation of the Mcm2-7 double hexamer
There are multiple models for the stoichiometry of ORC during helicase loading (Fig. S5A). One ORC molecule could direct both helicase loading events (Model I). Alternatively, two ORC molecules could be present throughout the loading reaction (Model II). Finally, it is possible that distinct ORC molecules direct each loading event but both ORC molecules are only present for the second loading event (Model III) or, like Cdc6 and Cdt1, each ORC is only present during loading of one Mcm2-7 (Model IV). To distinguish between these models, we performed CoSMoS with simultaneous labeling of ORC and Mcm2-7.
Initially, we fluorescently labeled ORC on the Orc1 subunit (ORC1SORT549) and observed associations with DNA in the presence of unlabeled Cdc6, Cdt1 and Mcm2-7. ORC DNA binding showed a broad distribution of dwell times (Fig. S5B, left panel). Consistent with the long-lived associations reflecting ORC binding to the ACS, mutation of this element resulted in >94% of ORC DNA associations being short-lived (<10 s, Fig. S5B, right panel). The associations of ORC are shorter than the calculated fluorescent dye lifetimes confirming that we are observing dissociations and not photobleaching (Fig. S5C, Table S2).
To identify ORC molecules involved in helicase loading, we simultaneously monitored ORC and Mcm2-7 DNA associations (Fig. 5A). As expected, ORC associates with DNA and Cdt1/Mcm2-7. Unlike Cdc6 and Cdt1, we consistently observed a single increase in ORC fluorescence that remained present continuously during recruitment of the first and second Mcm2-7 complexes (Fig. 5A, S5D).
Figure 5. A single ORC complex directs recruitment and loading of the first and second Mcm2-7 hexamer.
(A) Representative fluorescence intensity record for ORC1SORT549 and Mcm2-74SNAPJF646 at an origin-DNA location. Association of first and second Mcm2-7 are marked with red and blue arrows, respectively.
(B) A single ORC complex directs recruitment of two hexamers. The fraction (± s.e.) of DNA molecules observed to have zero, one or two ORC fluorophores bound when the second Mcm2-7 was recruited, is plotted (bars) together with the predicted number of associated fluorophores (red and blue squares) of different models (see Fig. S5A).
(C) ORC is released rapidly after recruitment of the second Mcm2-7 hexamer. Histograms showing the time between the association of the second Mcm2-7 and ORC release (top), or association of the first Mcm2-7 and ORC release (bottom).
(D) Release of Cdc6SORT549 (blue), Cdt1SORT549 (red), and ORC1SORT549 (black) after the association of the second Mcm2-74SNAPJF646 complex is plotted as a survival function. There are two ORC molecules that associate for >400s (1033.8s and 709.6s) that are not shown and disproportionately affect the mean dwell time. Grey lines represent a 95% confidence interval for the ORC dataset showing that there is no significant difference between Cdt1 and ORC release time distributions. Numbers in parentheses represent the mean release times ± s.e.m.
Because ORC multimers have been detected (Sun et al., 2012), we addressed whether ORC complexes dimerize in solution prior to DNA binding by counting the number of photobleaching steps associated with single increases in ORC-associated fluorescence (as was described for Mcm2-7). The large majority of cases were consistent with ORC binding as a single complex (67 of 69, Fig. S5E). These data confirmed that the single increases in ORC-associated fluorescence were due to single ORC molecules associating with origin-DNA during loading.
Although the majority of observations involved a single ORC directing loading of two Mcm2-7 hexamers, occasionally we observed the presence of multiple DNA-bound ORC molecules at the time of a Mcm2-7 association. To address which models for ORC function during helicase loading were possible, we counted the number of DNA-associated ORC molecules (by counting step-wise increases in ORC fluorescence) during the second Mcm2-7 hexamer association (Fig. 5B). Models II and III predict two ORC molecules bound to DNA when the second Mcm2-7 is recruited. In contrast to these models, we observed two ORC molecules associated during loading of the second hexamer only 5% of the time (as opposed to 70% predicted by Model II or III using the measured ORC labeling efficiency; 85%; see Supplemental Experimental Procedures). Instead, we observe a single ORC present during association of the second helicase 80% (96/120) of the time, very close to the percentage expected if a single ORC is responsible for loading the second Mcm2-7 (85%). To distinguish between models I and IV, we asked whether the same or different ORC molecules directed the first and second helicase-loading events. Consistent with model I, 94% (N=96) of observations showed a single ORC complex continuously present during both Mcm2-7 recruitment events. Thus, our data indicates one ORC molecule directs loading of both the first and the second Mcm2-7 hexamer (Model I).
Interestingly, in most traces where two Mcm2-7 associate with the DNA, we observed dissociation of ORC from origin DNA soon after binding of the second Mcm2-7 hexamer (see Fig. 5A, S5D). Plotting the times between the association of the second Mcm2-7 hexamer and ORC release (Fig. 5C, blue bars), we observed only one instance where ORC released from DNA in <15 s (13.1 s), followed by a short time interval (15 s – 90 s) during which 87% of the ORC complexes were released. The shape of this distribution suggests that, like Cdc6 and Cdt1, release of ORC is a multi-step process. In contrast, a much broader distribution was observed when ORC release was measured relative to DNA association of the first Mcm2-7 hexamer (Fig. 5C, red bars), suggesting ORC release is independent of this event. To investigate the order of ORC release relative to the other helicase-loading proteins, we compared the distribution of ORC, Cdc6 and Cdt1 dwell times after binding of the second Mcm2-7 complex (Fig. 5D), using data from two-color experiments with Mcm2-74SNAPJF646 and 549-labeled ORC, Cdt1 or Cdc6. Photobleaching of the 549-labeled proteins was insignificant relative to their observed dwell times (Table S2). Although there is a significant difference between release of Cdc6 and ORC (p < 0.001), we saw no significant difference in the distributions of Cdt1 and ORC release (Fig. 5D). Thus, loading of the first Mcm2-7 allows ORC retention, whereas loading of the second Mcm2-7 appears to induce the linked release of ORC and the second Cdt1.
Recruitment of a Second Mcm2-7 Results in Rapid Double Hexamer Formation
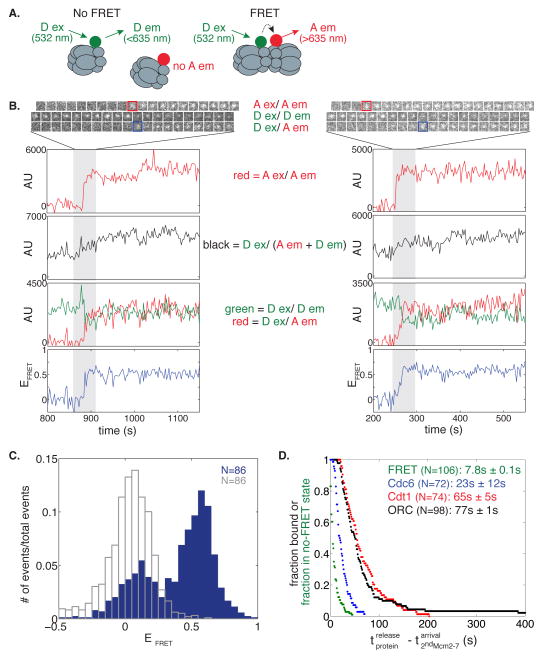
The interactions that drive recruitment of the second Mcm2-7 remain unclear (Yardimci and Walter, 2014). To gain insight into this event, we used fluorescence resonance energy transfer (FRET)-CoSMoS (Crawford et al., 2013) to detect the proximity of the Mcm7 N-terminal domains upon double-hexamer formation (Costa et al., 2014; Sun et al., 2014). To this end we labeled the Mcm7 subunit in separate preparations of Mcm2-7 with either 549 (Mcm2-77SORT549 - donor) or 649 (Mcm2-77SORT649 - acceptor) fluorophore (Fig. 6A). When mixed in an equimolar ratio, the differently labeled Mcm2-7 should be in the same double hexamer ~50% of the time, and those molecules should exhibit substantial FRET efficiency (EFRET) because the Mcm7 N-terminal regions are in close proximity in the double hexamer (Sun et al., 2014). We alternated between 633 and 532 nm laser excitation to monitor both arrival of each Mcm2-7 and EFRET. Importantly, when Mcm2-77SORT549 and Mcm2-77SORT649 were sequentially recruited to the origin DNA (in either order) we observed rapid development of a high EFRET ≈ 0.53 state (Fig. 6B and 6C, blue bars; Fig. S6). A second peak at EFRET ≈ 0.02 was also observed in the absence of acceptor (Fig. 6C, unfilled grey bars) and thus represents state(s) with no detectable FRET. Consistent with the detected FRET signal occurring as a consequence of double-hexamer formation, the high EFRET state was stable for hundreds of seconds and 95% (55/58) of the complexes that exhibited EFRET ≈ 0.53 were high-salt resistant.
Figure 6. Double hexamer formation occurs quickly upon recruitment of the second Mcm2-7 hexamer.
(A) When the two fluorophores (green circle = Dy549, red circle = Dy649) are not associated, excitation of the donor (D ex) will only yield emission from the donor (D em). However when the two fluorophores are in close proximity, we observe acceptor emission (A em) upon D ex, and a weaker D em signal. Wavelengths represent laser excitation and emissions that were monitored.
(B) Representative fluorescence records for experiments using a mixture of Mcm2-77SORT549 and Mcm2-77SORT649 showing FRET upon arrival of the second Mcm2-7. Red squares highlight when Mcm2-77SORT649 associates with DNA (Mcm2-77SORT549 is already present), and blue squares highlight when FRET occurs. Images and records of fluorescence intensity for D ex/D em (Mcm2-77SORT549), A ex/A em (Mcm2-77SORT649), total emission (D ex / (D em + A em), and FRET (D ex/A em) are shown together with calculated EFRET.
(C) Histogram of EFRET is plotted for times when a single Mcm2-77SORT549 and a single Mcm2-77SORT649 are present (blue bars) or when only Mcm2-77SORT549 is associated with the DNA (unfilled grey bars). The histogram displays the first ten consecutive EFRET measurements after arrival of the second Mcm2-7 for 86 DNA molecules (the same number of molecules and time points were used for the control). EFRET data below −0.5 was excluded from the plot (3/860 signal points and 17/860 control points).
(D) Double-hexamer formation anticipates Cdc6, Cdt1 and ORC release. Survival after the association of the second Mcm2-7 complex of the no-FRET state (green) and of DNA-bound Cdc6SORT549 (blue), Cdt1SORT549 (red), and ORC1SORT549 (black). Mean times ± s.e.m. until FRET increase and ORC, Cdt1, Cdc6 release are reported for comparison.
To determine when double-hexamer formation occurs relative to binding of the second Mcm2-7, we compared the time of FRET formation to the time of arrival of the second Mcm2-7 (Fig. 6D). We found the mean time between recruitment of the second Mcm2-7 hexamer until formation of FRET was 7.8 ± 0.1 s. This time is significantly shorter than release of Cdc6 after arrival of the second Mcm2-7 hexamer (23.2 ± 1.7s, p < 0.001), indicating that formation of the N-terminal-to-N-terminal interactions anticipates, and is therefore independent of, Cdc6 and Cdt1 release (Fig. 6D).
DISCUSSION
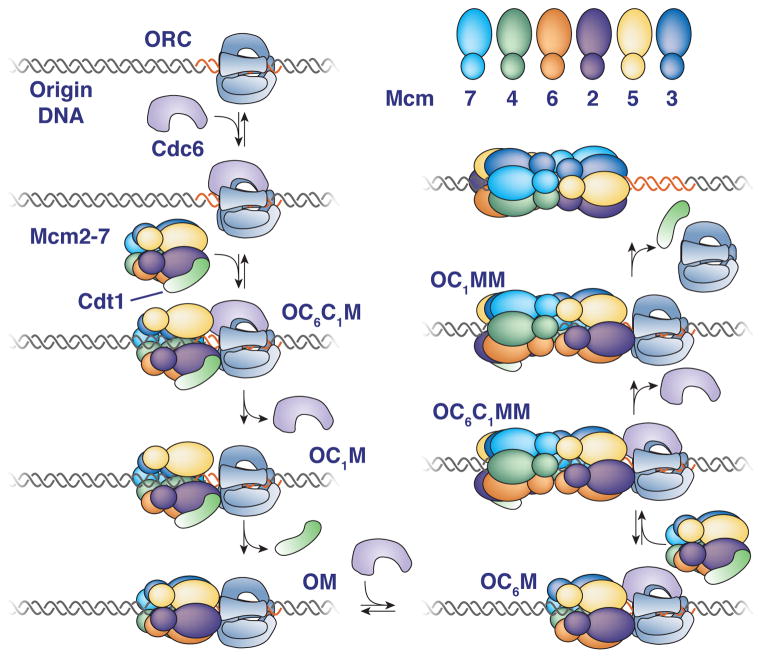
By determining precise protein/DNA stoichiometry and real-time dynamics, the single-molecule observations of helicase loading described here provide important insights into this event. Together, our findings strongly support a model in which the first and second helicase are loaded by distinct mechanisms and the second Mcm2-7 complex is recruited through interactions with the first. Accordingly, we propose a new model for helicase loading that is consistent with our current data and is described below (Fig. 7).
Figure 7. Proposed model for helicase loading.
Proteins present are indicated adjacent to each illustration (O=ORC, C6=Cdc6, C1=Cdt1, M=Mcm2-7). Reversible steps that are observed are indicated. See text for details.
Recruitment and loading of Mcm2-7 helicases occur in a one-at-a-time manner
Monitoring associations in real-time reveals sequential recruitment and loading of Mcm2-7 helicases to origin-DNA. One-at-a-time recruitment is consistent with an initial complex containing a single Mcm2-7 associated with the three helicase-loading proteins (Sun et al., 2013) and ensemble assays that show temporal separation of Mcm2-7 recruitment (Fernández-Cid et al., 2013). Recent structural observations indicate that the Mcm2/5 gates, which must open to provide DNA access to the Mcm2-7 central channel (Samel et al., 2014), are staggered in the double hexamer (Costa et al., 2014; Sun et al., 2014). Concerted Mcm2-7 loading would require alignment of the two Mcm2/5 gates to allow simultaneous DNA entry into the central channels of both hexamers. In contrast, sequential Mcm2-7 loading can readily accommodate the formation of a staggered-gate double-hexamer structure.
Although high-salt resistant single hexamers have been detected after artificially closing the Mcm2/5 gate (Samel et al., 2014), previous studies have not detected single loaded (high-salt-resistant) Mcm2-7 complexes in unperturbed helicase-loading reactions (Evrin et al., 2009; Kang et al., 2014; Remus et al., 2009). This difference may be due to the higher protein concentrations used in these ensemble reactions. Alternatively, the high-salt-resistant single hexamers may be less stable than the double hexamers resulting in their loss during sample preparation for chromatography or EM. Indeed, a higher percentage of double hexamers showed high-salt resistance relative to single hexamers (74% versus 49%; see Fig. 1D). The high-salt wash is effective in the single-molecule assay setting, however, as this treatment efficiently releases incompletely loaded Mcm2-7 formed in the absence of ATP hydrolysis (Table 1, ATPγS).
Ordered release of Cdc6 and Cdt1 molecules during double-hexamer loading
Our studies provide insights into Cdc6 and Cdt1 function during helicase loading. Previously, robust DNA association of these proteins was only observed when helicase-loading reactions were arrested at an early ATP-dependent step. We found that the initial ORC-Cdc6-Cdt1-Mcm2-7 (OC6C1M) complex has two possible fates (Fig. 7, left): (i) simultaneous release of Mcm2-7 and Cdt1 or (ii) sequential release of Cdc6 and Cdt1 with retention of Mcm2-7. The former is most likely the reversal of the initial Mcm2-7/Cdt1 association whereas the latter pathway leads to sequential formation of OC1M and OM complexes and Mcm2-7 loading. Based on this distinction, we propose that release of Cdt1 independent of Mcm2-7 is coupled to successful helicase loading (illustrated as closing of the Mcm2/5 gate, Fig. 7). Consistent with this hypothesis, treatments (e.g. ATPγS) or mutations (e.g. Mcm2-7 ATPase mutations, Coster et al., 2014; Kang et al., 2014) that lead to Cdt1 retention prevent helicase loading. We note that other times of ring closure (and opening) than those illustrated in the model are possible.
Electron microscopic (EM) and ensemble assays suggest the existence of helicase loading intermediates with ORC-Cdc6-Mcm2-7 (OC6M) and ORC-Cdc6-Mcm2-7-Mcm2-7 (OC6MM, Sun et al., 2014). Our findings suggest that the OC6M complex is a short-lived intermediate formed prior to recruitment of the second Mcm2-7/Cdt1 complex rather than being formed by release of Cdt1 from the OC6C1M (Fernández-Cid et al., 2013). We do not see evidence of an OC6MM complex during helicase loading and there is no direct evidence that Cdc6 is present in the 2-D class averages used in these studies (Sun et al., 2014). Given their relatively lower resolution, these studies could have detected either the OC6C1MM or OC1MM complexes that we observe (Fig. 7, right). Our previous studies found an intermediate with two Cdt1 complexes that is not detected in the current studies (Takara and Bell, 2011). During efforts to reconcile these findings, we found the Mcm2-7 protein used in the previous studies contained a non-lethal mutation in the C-terminus of Mcm3 that is predicted to inhibit Cdc6 interactions (Frigola et al., 2013). We suspect that this mutant enhanced dependence on other interactions leading to the detection of two Cdt1 associations.
Loading of the first and second Mcm2-7 occurs by distinct mechanisms
In addition to answering a long-standing question about ORC function, our data indicating that one ORC molecule directs Mcm2-7 double-hexamer formation strongly suggests that different mechanisms direct loading of the first and second Mcm2-7. EM studies suggest that during helicase loading ORC interacts with the C-terminal end of the first Mcm2-7 on adjacent DNA (Sun et al., 2014; 2013). Assuming this configuration, direct recruitment of the second Mcm2-7 complex by the same ORC would load the two Mcm2-7 molecules in a head-to-tail fashion (Fig. S7, top). Even if ORC had a second binding site for Mcm2-7 on its opposite side, a similar direct interaction with Mcm2-7 could not load two Mcm2-7 complexes with adjacent N-terminal domains (Fig. S7, bottom). Further evidence in favor of distinct mechanisms loading the first and second Mcm2-7 include: (i) the two loading events show different Cdc6, Cdt1 and ORC release kinetics; (ii) Cdt1 associated with the second loading event shows an increased propensity to release without Mcm2-7.
We considered the possibility that a second ORC binds DNA in the opposite orientation and loads the second helicase by the same mechanism as the first. Several observations argue against this model. First, because we do not consistently detect a second ORC during recruitment of the second Mcm2-7, the average dwell time for this second ORC would have to be below our detection limit (~0.5 s). This limit is >10-fold shorter than the average dwell time observed for ORC on non-origin DNA (Fig. S5B). Second, in contrast to a model in which a short-lived second ORC directs loading of the second Mcm2-7, the Cdc6 protein associated with loading the second Mcm2-7 is easily detected (23.2 s average dwell time, Fig. 4D). Third, even at diffusion-limited binding rates the sequential association of Cdc6 and Mcm2-7/Cdt1 with such a short-lived ORC is improbable. Finally, experiments showing that soluble ORC is not required for helicase loading if ORC is pre-loaded onto DNA (Bowers et al., 2004; Fernández-Cid et al., 2013; Duzdevich et al, 2015) are not consistent with a role for a short-lived second ORC.
Recruitment of the Second Mcm2-7
Instead of ORC and Cdc6 directly recruiting the second Mcm2-7/Cdt1 complex, our findings suggest that interactions involved in stabilizing the Mcm2-7 double hexamer mediate the recruitment of the second Mcm2-7/Cdt1. We detect these interactions prior to Cdc6 or Cdt1 release (Fig. 6), suggesting that formation of double hexamer interactions anticipates loading of the second helicase. Recent EM studies of a complex between one ORC and a head-to-head Mcm2-7 double hexamer are consistent with this hypothesis (Sun et al., 2014). Because FRET is not observed immediately upon recruitment of the second Mcm2-7, an intervening event (e.g. a Mcm2-7 conformational change or ATP hydrolysis) may be required to bring the Mcm7 N-terminal domains into close proximity. We do not know which parts of the Mcm2-7 N-terminal domains drive the proposed interactions. For simplicity, the model (Fig. 7) illustrates interactions consistent with those observed in EM studies of Mcm2-7 double hexamers (Costa et al., 2014; Sun et al., 2014). One argument against a model in which Mcm2-7 N-terminal domains drive recruitment of the second Mcm2-7 is the observation that a C-terminal mutation in Mcm3 that interferes with recruitment of the first Mcm2-7 also inhibits recruitment of the second Mcm2-7 (Frigola et al., 2013). This mutant has additional defects in Mcm2-7 ATP hydrolysis, however, which could explain a loading defect for the second Mcm2-7 (Coster et al., 2014; Kang et al., 2014; Sun et al., 2014).
Because purified Mcm2-7 complexes do not show affinity for one another in solution (Evrin et al., 2009), the first Mcm2-7 must be altered to facilitate interactions with a second Mcm2-7. A likely possibility is that ORC and Cdc6 alter the conformation of the first Mcm2-7 to facilitate these interactions (shown as separation of the Mcm2/Mcm5 N-terminal regions, Sun et al, 2013). In support of a role for Cdc6, although we observe an ORC-Mcm2-7 (OM) intermediate after the first loading event, this complex is unable to recruit a second Mcm2-7 until a second Cdc6 protein associates (OC6M).
The model for helicase loading presented here has several advantageous features. Because Cdc6 ATPase activity is required to remove incorrectly or incompletely loaded Mcm2-7 (Coster et al., 2014; Frigola et al., 2013; Kang et al., 2014), the use of different Cdc6 proteins to load the first and second Mcm2-7 would allow each event to be evaluated separately. More importantly, the use of Mcm2-7 N-terminal domain interactions to recruit the second Mcm2-7 ensures the establishment of a head-to-head double hexamer. This conformation is the first step in the establishment of bidirectional replication initiation and could be essential for initial DNA melting. Finally, the retention of ORC after the first loading event coupled with the release of ORC after the second loading event has the advantage of promoting the formation of double hexamers while inhibiting repeated loading of single hexamers.
EXPERIMENTAL PROCEDURES
Protein Purification and Labeling
Wild-type Mcm2–7/Cdt1 and ORC complexes were purified as described previously (Kang et al., 2014). Wild-type Cdc6 was purified as described in Frigola et al., 2013. We used a variety of protein fusions to fluorescently label ORC (Ubiquitin-GGG-Flag at the N-terminus of Orc1), Cdc6 (GST-SUMO-GGG tag at the N-terminus), and Cdt1/Mcm2-7 (Ubiquitin-GGG-Flag at the N-terminus of Mcm7 or Cdt1, and/or a SNAP-tag (NEB) at the N-terminus of Mcm4). The Ubiquitin (in vivo) and GST-SUMO (using Ulp1 protease) fusions were removed to reveal three N-terminal glycines required for sortase labeling. Sortase was used to couple fluorescently labeled peptide (DY549P1- or DY649P1-CHHHHHHHHHLPETGG; referred to as SORT549 and SORT649 respectively in the manuscript) to the N-terminus of these proteins. SNAP-Surface549 (NEB, SNAP549 in the manuscript) or SNAP-Janelia Fluor 646 (SNAPJF646; Grimm et al., 2015) was coupled to SNAP-tagged Mcm2-7 (See Supplemental Experimental Procedures for these purification protocols). For Sortase labeling, peptide-coupled proteins were separated from uncoupled proteins using Complete-His-Tag Resin (Roche).
Single-Molecule Microscopy
The micro-mirror total internal reflection (TIR) microscope used for multiwavelength single-molecule using excitation wavelengths 488, 532, and 633 nm has been previously described (Friedman and Gelles, 2012; Friedman et al., 2006). Biotinylated AlexFluor488-labeled 1.3kb-long DNA molecules containing an origin were coupled to the surface of a reaction chamber through streptavidin. Briefly, the chamber surface was cleaned and derivatized using a 200:1 ratio of silane-NHS-PEG and silane-NHS-PEG-biotin (see Supplemental Experimental Procedures). We identified DNA molecule locations by acquiring 4–7 images with 488 nm excitation at the beginning of the experiment. Unless otherwise noted, helicase loading reactions contained 0.25nM ORC, 1nM Cdc6 and 2.5nM Cdt1/Mcm2-7. Reaction buffer was as previously described (Kang et al., 2014) except without any glycerol and with the addition of 2 mM dithiothreitol, 2 mg/ml bovine serum albumin (EMD Chemicals; La Jolla, CA), and an oxygen scavenging system (glucose oxidase/catalase) to minimize photobleaching (Friedman et al., 2006). After addition of protein to the DNA-coupled chamber, frames of one second duration were acquired according to the following protocol: (1) a single image frame visualizing the DNA positions (488 excitation), (2) 60 frames monitoring both the 549 and 649 fluorophores (simultaneous 532 and 633 excitation) and (3) a computer-controlled focus adjustment (using a 785 nm laser). This cycle was repeated roughly 20 times in the course of an experiment (~20 mins). Chambers were then washed with either three chamber volumes of reaction buffer or two volumes of the same buffer with 0.5M NaCl in place of 300mM K-glutamate and one volume reaction buffer. For photobleaching, laser power(s) were increased and one or multiple fluorophores were imaged simultaneously until no visible spots remained. Typically, photobleaching was also examined in a second field of view that was not imaged during the loading reaction.
FRET experiments
The conditions for monitoring FRET were similar to the other experiments, with a few exceptions. Typical reactions contained 0.75 nM ORC, 3nM Cdc6, 5nM Cdt1/Mcm2-77SORT549 and 5nM Cdt1/Mcm2-77SORT649. DNA was imaged before and immediately after adding the reaction to the slide but not throughout the experiment. The imaging protocol alternated between 1 s frames with the 532 laser on and 1 s frames with the 633 laser on over 20-30 minutes. Apparent EFRET was calculated as described (Crawford et al., 2013).
Supplementary Material
Acknowledgments
We are grateful to members of the Bell laboratory for useful discussions. We thank Daniel Duzdevich and Eric C. Greene for comments on the manuscript and helpful discussions. We thank Jonathan B. Grimm and Luke D. Lavis for graciously providing the Janelia Fluors. This work was supported by NIH grants GM52339 (S.P.B.). S.T. was supported in part by an NIH Pre-Doctoral Training Grant (GM007287). L.J.F. and J.G. were supported by NIH R01 GM81648 and a grant from the G. Harold and Leila Y. Mathers Foundation. S.P.B. is an investigator with the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
S.T. designed and conducted experiments with feedback from L.J.F., J.G. and S.P.B. S.T. and L.J.F. analyzed data. N.A.I developed labeling strategies and S.T., and N.A.I. generated proteins. S.T. and S.P.B. composed the paper with input from all authors, and S.P.B. and J.G. directed the project.
References
- Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JCW, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Renault L, Swuec P, Petojevic T, Pesavento JJ, Ilves I, MacLellan-Gibson K, Fleck RA, Botchan MR, Berger JM. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. eLife. 2014;3:e03273. doi: 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster G, Frigola J, Beuron F, Morris EP, Diffley JFX. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell. 2014;55:666–677. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. Proc Natl Acad Sci USA. 2013;110:6783–6788. doi: 10.1073/pnas.1219305110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duzdevich D, Warner MD, Ticau S, Ivica NA, Bell SP, Greene EC. The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level. Mol Cell. 2015;58:483–494. doi: 10.1016/j.molcel.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cid A, Riera A, Tognetti S, Herrera MC, Samel S, Evrin C, Winkler C, Gardenal E, Uhle S, Speck C. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell. 2013;50:577–588. doi: 10.1016/j.molcel.2013.03.026. [DOI] [PubMed] [Google Scholar]
- Friedman LJ, Gelles J. Mechanism of Transcription Initiationat an Activator-Dependent Promoter Defined by Single-Molecule Observation. Cell. 2012;148:679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys J. 2006;91:1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, Remus D, Mehanna A, Diffley JFX. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreizig S, Kindermann M, Johnsson K. Induced protein dimerization in vivo through covalent labeling. J Am Chem Soc. 2003;125:14970–14971. doi: 10.1021/ja037883p. [DOI] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Meth. 2015 doi: 10.1038/nmeth.3256. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Warner MD, Bell SP. Multiple Functions for Mcm2–7 ATPase Motifs during Replication Initiation. Mol Cell. 2014;55:655–665. doi: 10.1016/j.molcel.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- Randell JCW, Bowers JL, Rodríguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samel SA, Fernandez-Cid A, Sun J, Riera A, Tognetti S, Herrera MC, Li H, Speck C. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28:1653–1666. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K, On KF, Diffley JFX. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol. 2013;5:a012930. doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann SA, van Oijen AM. DNA replication at the single-molecule level. Chem Soc Rev. 2014;43:1201. doi: 10.1039/c3cs60391a. [DOI] [PubMed] [Google Scholar]
- Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H. Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes Dev. 2014;28:2291–2303. doi: 10.1101/gad.242313.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Evrin C, Samel SA, Fernández-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kawakami H, Zech J, Speck C, Stillman B, Li H. Cdc6-Induced Conformational Changes in ORC Bound to Origin DNA Revealed by Cryo-Electron Microscopy. Structure. 2012;20:534–544. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takara TJ, Bell SP. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2–7 helicases. EMBO J. 2011;30:4885–4896. doi: 10.1038/emboj.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Walter JC. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol. 2014;21:20–25. doi: 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.