Abstract
Chromophore-assisted laser inactivation (CALI) is a technique whereby engineered proteins and dye molecules that produce substantial amounts of reactive oxygen species upon absorption of light are used to perturb biological systems in a spatially and temporally defined manner. CALI is an important complement to conventional genetic and pharmacological manipulations. In this review, we examine the applications of CALI to cell biology and discuss the underlying photochemical mechanisms that mediate this powerful technique.
Introduction
Although analysis of loss-of-function phenotypes leads to important clues about protein function, long-term global protein inactivation via gene knockout or RNA interference (RNAi) can lead to compensatory changes at the molecular level that are difficult to detect or control. Use of pharmacological inhibitors is limited by a lack of subcellular spatial resolution and transport across the plasma membrane. By contrast, instantaneous and spatially precise protein inactivation through chromophore-assisted laser inactivation (CALI) [1,2], in principle, avoids these issues. In CALI, a small dye conjugated to an antibody, the protein of interest or a fluorescent protein fused to the target protein is introduced or expressed in the cell. An area of interest is then irradiated with an intense beam of light. Absorption of light by the chromophore leads to the production of highly reactive free radicals, including reactive oxygen species (ROS) that, in turn, inactivate the proximate protein(s). Because the free radical species generated during CALI are short-lived, the damage radius has been reported to be limited to proteins that are immediately adjacent to the irradiated chromophore [3,4]. Given that inactivation can be accomplished within short timescales (often <1 s) and that the inactivating light beam can be directed to small regions within a single cell, the technique provides a spatially and temporally controlled loss-of-function tool for cell and developmental biology that complements other loss-of-function methodologies.
There are several important precursors to the development of CALI. Hematoporphyrin therapy for certain cancers, in which tumor-localized hematoporphyrin is irradiated and toxic singlet-oxygen released, is essentially a CALI effect [5]. Light-induced damage to fluorescently labeled chromosomes and cytoskeletal components has been reported at various times. This includes damage to chromosomes following laser irradiation of bound dyes [6], breakage of actin filaments labeled with 5-iodoacetamido fluorescein that creates additional barbed ends supporting more filament growth [7] and breakup of microtubules incorporating tubulin conjugated to different fluorescent dyes upon excitation with intensities typical of those employed in fluorescence microscopy [8]. More recently, researchers observed that fluorescently labeled stress fibers that looked normal in the light microscope exhibited ‘fraying’ in correlative electron microscopy (G. Borisy, personal communication). Despite these deleterious effects, these observations indicated that photochemically produced reactants, properly harnessed, can function as selective loss-of-function tools with high temporal and spatial precision. In this review, we summarize applications of CALI to cell biology and discuss the photochemical mechanisms underlying this methodology.
Development of CALI and its applications to cell biology
CALI requires that a chromophore, which acts as a photosensitizer, is placed immediately adjacent to the target protein. Over the years, there have been several creative strategies to achieve this. CALI was originally developed to employ the dye malachite green as a chromophore after several dyes were surveyed as suitable candidates [1]. Malachite green was conjugated to a target-specific non-function-blocking antibody that was microinjected into the cell or bound to cell-surface determinants [2]. In subsequent years, other dyes – usually xanthene-based – have also been used because they are better generators of ROS. Thus, researchers used fluorescein-labeled antibodies to inactivate β-galactosidase in vitro and β1 integrins in vivo [9], and employed CALI of fluorescein isothiocyanate (FITC) antibodies to the nuclear Ki-47 protein to demonstrate its role in nucleolar rRNA synthesis [10].
More recently, two methodologies have been described using membrane-permeable fluorescent ligands that bind to genetically encoded target sequences. These strategies avoid the need to generate non-function-blocking antibodies that are specific for a single target, which are not always available. They also avoid having to use microinjection for cytoplasmic targets, which can introduce a background of labeled but unbound antibody. The first method uses the biarsenical dyes ReAsH (resorufin-based arsenical hairpin binder) and FlAsH (fluorescein-based arsenical hairpin binder). ReAsH and FlAsH have an intrinsic high-affinity interaction with the tetracysteine motif Cys-Cys-Pro-Gly-Cys-Cys (Kd in the pM or lower range). ReAsH-based CALI was used to inactivate connexin 43 that was expressed as a tetracysteine fusion, and loss-of-function was monitored electrophysiologically by the decrease in intracellular coupling mediated by gap junctions [11]. FlAsH was demonstrated to label synaptogamin-I into which the tetracysteine motif was engineered and expressed in synaptogamin-I null Drosophila mutants. CALI of dissected larvae that had been labeled with FlAsH impaired neurotransmitter release from the neuromuscular junction by interfering with the synaptic vesicle post-docking fusion mechanism [12].
An alternative targeting strategy was subsequently developed that exploited the high-affinity interaction between FKBP12 (F36V), a protein that binds to the natural ligand FK506, and its synthetic ligand for FKBP12 (SLF′) (Kd ~10 pM) [13]. In this methodology, the protein target is expressed as a fusion with FKB12 and the cell is incubated with fluorescein-labeled SLF′. SLF is membrane permeable and thus avoids the need to microinject any reagents. It was demonstrated that CALI of fluorescein-labeled SLF′ targeted to β-galactosidase could inactivate this enzyme both in vitro and when expressed in fibroblasts.
Although these strategies described have increased the utility and general applicability of CALI, the need for exogenously added labeling reagents that require the washing out of excess unbound chromophore introduces the risk of non-specific dye-binding and potential collateral photodamage during irradiation. In such cases, non-transfected cell lines that exhibit background staining after incubation with the labeling reagent cannot be fruitfully used. In addition, dyes such as FlAsH and ReAsH and their antidotes can produce toxicity in some cells (D. Piston, personal communication). Specificity can also be problematic when the FKBP12 (F36V) mutant is fused to target proteins, as described. Although the mutant FKBP12 is recognized by no known proteins except the synthetic ligand conjugated to fluorescein (FL-SLF′), cells incubated with this ligand exhibited appreciable non-specific inactivation [13].
A new class of CALI reagents, termed environment-sensitive photosensitizers (ESPers) has recently been developed [14]. One of the problems with soluble CALI dyes that bind to molecules of interest is the off-target effects, in which damaging singlet-oxygen is produced by photoexcited reagents away from the target site. To obviate this deleterious effect, compounds have been developed that consist of a CALI chromophore, a targeting ligand and a polarity-sensitive electron-donor moiety, all covalently conjugated. The electron donor quenches singlet-oxygen production by the excited CALI-chromophore in aqueous environments; however, when this new class of CALI reagent is bound to a hydrophobic environment of a target receptor, singlet-oxygen generation occurs and inactivates the receptor. Using this new strategy, the inactivation of the inositol (1,4,5)-trisphosphate [Ins(1,4,5)P3] receptor in DT40 cells was demonstrated by monitoring the inhibition of calcium release from the endoplasmic reticulum (ER) into the cytoplasm upon Ins(1,4,5)P3 stimulation.
The potential problems caused by labeling the protein of interest with exogenous reagents can be completely avoided by using genetically encoded fluorescent protein fusions. In both single photon and multiphoton excitation applications, enhanced green fluorescent protein (EGFP), a widely used fluorescent protein, has been shown to be an effective CALI chromophore. Using cells expressing EGFP-α-actinin, CALI was employed to demonstrate that α-actinin provides a mechanical link between stress fibers and focal adhesions [15]. Furthermore, EGFP-connexin 43 expressed in HeLa cells could be inactivated by two-photon excitation with the result that electrical coupling between cells was abrogated [16]. The same study also showed that two-photon CALI of EGFP-Aurora B disrupts mitosis; Aurora B is a kinase essential for mitosis. In another study, CALI of EGFP-capping protein (EGFP-CP) increased uncapped actin filaments, resulting in enhanced filament growth and the formation of numerous protrusive structures [17] (Figure 1). CALI of EGFP-Mena in Ena (also known as VASP)-deficient cells was also shown to stabilize lamellipodial protrusions, in a manner completely consistent with earlier genetically based sequestration experiments [18,19]. Although many fluorescent proteins are weak singlet-oxygen producers, an efficient genetically encoded ROS generator, killer red [20], results in cell lethality at high illumination doses; at lower doses, killer red fusions with pleckstrin homology domains leads to dissociation of the domain from the plasma membrane, indicating specific CALI action. Overall, several different molecules have been shown to be adequate CALI reagents, as assessed by the production of distinct loss-of-function phenotypes. These results beg the question of how, photochemically speaking, the selective inactivation occurs.
Figure 1.

CALI of EGFP-CP induces the formation of dorsal ruffles and filopodia. (a) Fluorescence (left) and differential interference contrast (DIC) (right) images of Rat2 fibroblasts expressing EGFP-CP before CALI; the irradiated area is indicated with a black circle (diameter: 24 μm). (b) Fluorescence immediately after CALI (left) and DIC (middle) and dorsal surface F-actin (right) (stained by Alexa Fluor 568-labeled phalloidin) 3 min after CALI. Numerous actin-rich dorsal ruffles and filopodia are locally formed after irradiation.
Mechanisms of CALI
In this section, we discuss the photochemical agents that mediate CALI, what is known about their mode of action, and the key issue of collateral damage occurring during CALI.
Photoactive agents
CALI-induced photodamage is photodynamic, meaning that, in addition to the CALI reagent acting as a photosensitizer, the process involves light and oxygen. In CALI, target proteins are inactivated by ROS. These are generated when chromophores that have become excited through light absorption react with oxygen in the environment. The toxicity of ROS is a consequence of their ability to oxidize proteins, lipids, DNA and carbohydrates [21]. In most cases, the predominant ROS causing CALI is thought to be singlet oxygen (1O2; molecular oxygen in an excited state) because only it is generated by energy transfer from an excited sensitizer chromophore to oxygen [22]. In this process, a small amount of energy (94 kJ mol−1, ~0.98 eV) delivered to molecular oxygen in the triplet ground state (3O2) results in 1O2, which has both valence electrons in the same orbital with spins paired. The 1O2 has a lifetime in solution of 1–10 μs in the absence of molecules in the environment with which it can react [23]. A higher excited state for 1O2 oxygen exists but it has an even shorter lifetime, leaving little time for chemical reaction. Excited state chromophores produced by absorption of light can also stimulate electron-transfer reactions occurring from molecular oxygen, resulting in the formation of other ROS such as superoxide anion. However, these processes are usually characterized by lower forward rate constants [23], leading to the assumption that 1O2 is the predominant ROS that mediates CALI. Nevertheless, evidence exists that CALI can be mediated by other ROS or by 1O2 acting in concert with other ROS, particularly for malachite green and killer red (see later).
Indeed, detailed studies have demonstrated that singlet oxygen is the photoactive agent for CALI mediated by fluorescein [9,24] and fluorescein derivatives [13]. Using several inhibitors of photodamage and D2O and oxygen perfusion to increase any oxygen-dependent effects, it was concluded that singlet oxygen was the likely photoactive agent in CALI mediated by ReAsH and FlaSH [11]. It was also concluded, by comparing the inhibition caused by azide, superoxide dismutase and mannitol [20], that 1O2 was the predominant agent mediating killer red CALI. ROS species, including 1O2, are also known to cause the green fluorescent protein (GFP)-mediated photo-oxidation of diaminobenzidine to an osmophilic polymer for electron microscopic localization [25] and are involved in EGFP-based CALI (M. McLean et al., unpublished). Azide and edaravone, both ROS scavengers, inhibit EGFP CALI of connexin 43 [16]. In contrast to the mechanisms mediated by 1O2 described here, CALI produced by exciting malachite green is mediated by the OH radical. This conclusion was based on the effects of different scavengers and the fact that manipulation of oxygen content and singlet-oxygen lifetime had no effect [4]. In the case of killer red, evidence for some damage occurring via superoxide anions in addition to that mediated by 1O2 was obtained [20].
Use of elevated oxygen content, D2O and free radical scavengers that react with photogenerated ROS provide only indirect evidence for the photoactive species involved. It is therefore difficult to discriminate unambiguously between different ROS [26]; presumably the yields for the different species depend somewhat on the environment of the chromophore and the local oxygen concentration. In the case of fluorescent proteins, it has been shown unambiguously that photobleaching of GFP produces 1O2; researchers used an electron spin resonance (ESR) assay, in which 1O2 reacts with an ESR probe, TEMPO, to alter its spectrum. In this case, photobleaching was reduced by the 1O2 quenchers azide and β carotene [27].
Once 1O2 (and possibly other ROS) is produced, it can either react with cellular targets including proteins, nucleic acids and lipids, or be physically quenched by reaction with other compounds in the cellular environment. The reactions with proteins predominate [23] and are of immediate interest for CALI. 1O2 can oxidize side chains of amino acid residues including histidine, tyrosine, tryptophan, methionine and cysteine, leading to both intramolecular and protein protein cross-linking and aggregation, loss of enzyme and other functional activities and protein peroxide formation [23], as depicted in Figure 2. Direct peptide backbone breakage induced by oxidation seems to be less likely [23]. Thus, 1O2 reactions with susceptible amino acid residues represent potential damage pathways that result in CALI. The most detailed information to date was obtained by Yan et al. [28], who showed that oxidative photodamage to illuminated FlAsH-labeled calmodulin results in sulfoxide formation on surface methionine residues, which disrupts the binding interface between calmodulin and its partners. No dependence on the distance between FlAsH and particular methionines was found, indicating that reaction with rather than diffusion of 1O2 was rate-limiting. In addition, reactive histidine intermediates were formed, leading to potential cross-linking to binding partners [28].
Figure 2.
Potential photodamage pathways in CALI.
Relative production of ROS by different chromophores
CALI depends on the effectiveness of the chromophore in generating ROS. Small molecule dyes of the xanthene family are much better in this respect than most fluorescent proteins [11,29]. Presumably, because fluorescent proteins (FPs) appear in organisms exposed to sunlight, evolution would require that FPs do not produce abundant ROS upon illumination [30]. GFP is often referred to as ‘light in a can’ because the chromophore is surrounded by and influenced by the β-can structure of the protein with ‘lids’ at either end [31]. This structure hinders interaction between the fluorophore and molecular oxygen, the source for singlet-oxygen production. Presumably, this is the reason that it takes much more light to produce a demonstrable CALI effect when EGFP, rather than small organic dyes, is employed as a chromophore [15,17].
CALI versus photobleaching
It would be expected that fluorescence, photobleaching and CALI are proportional to the number of trips through the excited state per unit of time with the relative efficiencies for each process characterized by their respective quantum yields. The ROS generated by the excited dye leads to both CALI and dye bleaching, but usually at different rates, which results in different quantum yields for the two processes. However, intricacies exist that makes understanding the processes complicated. For example, the excited state of the dye can provide access to a high-energy reaction pathway for bleaching that does not involve interaction with oxygen, or an interaction that does not result in 1O2 formation. Even when the chromophore seems to be substantially bleached, 1O2 production can still occur [20]. Thus, perfect correlation of the amount of fluorescence bleaching and the amount of CALI is probably not to be expected; rather, the bleaching and CALI pathways can be separate. Indeed, there is evidence that ‘bleaching does not necessarily lead to inactivation of protein function’ [16]. For fluorescent proteins, it is likely that the ability of the chromophore to induce CALI of the target protein will depend on the spatial disposition of the chromophore with respect to the target and the ability of photogenerated ROS to readily escape the fluorescent protein structure to inactivate the target protein. The latter factor will be a determinant in the rates of photobleaching versus CALI; for example, it is conceivable that if ROS cannot escape the fluorescent protein, the bleaching rates would be large, whereas CALI would be insignificant.
Researchers have reported that the flourescein-mediated CALI process seems to be distinct from the fluorophore photobleaching mechanism [24]. In contrast to frequently encountered mechanisms of 1O2 production and dye bleaching that are mediated via the triplet state of the dye, CALI affected by soluble fluorescein proceeded, via 1O2 production, from the first excited singlet state of the dye as a one photon process. It should be noted, however, that strong evidence exists for fluorescein triplet-state-mediated production of 1O2 that results in CALI [28].
For GFP, there is evidence that the triplet state exists and that this state leads to generation of ROS for CALI [32,33]. Non-light-emitting (dark) excited states other than the triplet state exist in GFP and its mutants, complicating description of the photophysical dynamics [33 35]. Observations from studies in which the interval between pulses of excitation was varied, provide support for an active GFP triplet state [36]. It was found that optimum fluorescence brightness was achieved when the separation between pulses of excitation energy was approximately a microsecond, the duration of a typical triplet state. It was speculated that, by enabling the long-lived triplet state, which is more susceptible to photochemical reactions, to relax between pulses as opposed to keeping it populated by more frequent pulses less photochemical bleaching occurs, resulting in an order of magnitude increase in the total yield of fluorescent photons from fluorophores such as GFP or Atto532. Finally, recent studies show that 1O2, as identified by an infrared phosphorescence emission spectrum, is produced from the triplet state of GFP and from the triplet state of model compounds that simulate the chromophore of GFP [32]. From this work, a triplet state lifetime of ~25 μs is deduced, which is approximately consistent with earlier fluorescence correlation spectroscopic studies [33-35]. This long lifetime is suggested to be as a result of the β-can structure limiting access of oxygen to the excited chromophore, which is consistent with more efficient singlet-oxygen production from the unprotected model chromophore than from EGFP, as noted earlier.
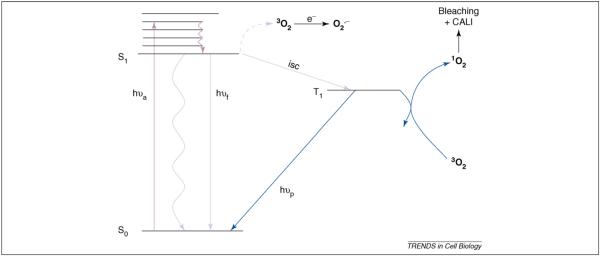
Various reaction pathways that can lead to CALI are depicted in a simplified Jablonski energy-level diagram (Figure 3) to provide a speculative model. In this scheme, excitation of the fluorescent protein or dye is followed by vibrational relaxation, the dropping of the molecule to its lowest excited singlet state for several nanoseconds. From this state, the excited chromophore can emit light, bleach, generate ROS, stimulate electron transfer from ground-state molecular oxygen, 3O2, to produce superoxide anions or undergo intersystem crossing to the triplet state. However, because the triplet state is long-lived, most photochemistry, including ROS production, subsequent bleaching and CALI, proceeds from it. It should be noted that, in general, the fluorescence pathway denoted by hnf, can be destroyed via a selective bleaching mechanism that leaves the chromophore intact and still capable of mediating CALI.
Figure 3.
ROS production in terms of simplified energy-level diagram. Excitation (denoted by absorption of a photon, hυa) occurs to one of several higher vibronic states followed by rapid internal conversion (compact squiggly arrow) resulting in the molecule dropping to the lowest excited singlet state (S1) in which it exists for several nanoseconds. From this state, several fates are possible: the excited chromophore can emit (denoted by emission of a photon, hυf), transfer its energy to the solvent without emission (extended squiggly arrow), stimulate electron transfer from 3O2 (depicted as the broken line from S1 to 3O2) to produce superoxide anion (O2•−) or undergo intersystem crossing (isc) to the triplet manifold. (Note, to simplify the diagram, generation of 1O2 from the chromophore single state is not depicted.) The triplet state is long lived and, for this reason, much photochemistry proceeds from it, including ROS production; emission from the triplet state is called phosphorescence (denoted by emission of a photon, hυp).
Radius of CALI action and collateral damage
Although the CALI technique has been used to inactivate varied protein targets, skepticism remains about the specificity of this approach and its general applicability, mainly because of the potential for unintended collateral damage. This is a legitimate concern and, in some cases, such damage has been demonstrated (as cited in the introduction). During the development of the fluorescence recovery after photobleaching (FRAP) technology, it was demonstrated that fluorescein-labeled red cell-membrane proteins could be cross-linked by photobleaching of the dye [37]. This question was addressed for fluorescein-CALI and researchers provided compelling evidence by using sensitive transport assays that the inactivation radius for fluorescein extends beyond its immediate target [38]. An α-bungarotoxin site was introduced into the glutamine receptor, a G-protein-coupled receptor (GPCR) that modulates N-type calcium channels in neurons, so that it could be labeled with fluorescein α-bungarotoxin, enabling CALI to be performed. It was found that CALI significantly affected calcium transport modulated by a glutamate receptor, as expected. However, if the α-bungarotoxin site was engineered into other transporters unrelated to glutamate receptor, modulation of N-type calcium channels was also reduced. Moreover, even voltage-dependent but receptor-independent modulation of N-type calcium transport could be disrupted by CALI of a non-GPCR labeled with fluorescein α-bungarotoxin. In addition, during CALI of FITC antibodies to Ki-67 nuclear protein, this protein was extensively cross-linked and a portion of the associated upstream binding factor (UBF) was also found to be cross-linked to Ki-67 [10].
In other cases, notably involving EGFP-CALI, controls indicate that collateral damage was not important to the observed effects. For example, in the case of stress fiber detachment from focal adhesions mediated by CALI of EGFP-α-actinin, CALI of EGFP-focal adhesion kinase (FAK), another focal adhesion localized protein, no detachment of stress fibers occurred [15]. For CALI of EGFP-capping protein (CP), when EGFP-CP was ectopically expressed over the light-insensitive endogenous CP, no protrusive phenotype was observed. It was only when the endogenous protein was removed from the cell that the CALI loss-of-function occurred. This provided the best possible control for non-specific CALI effects, because only the protein of which the concentration was manipulated could have been mediating the effect. Additionally, CALI of EGFP-CP and EGFP-Mena resulted in positive subcellular changes that mimicked the previously known loss-of-function phenotypes of these proteins, making it highly unlikely that CALI-induced changes were the result of non-specific photodamage [17].
Collateral damage presumably depends on the distance the ROS can diffuse before it is inactivated or quenched. In Table 1, we present some estimates of distances the photoproduced ROS must travel to have the observed effect. Jay and co-workers, in pioneering work on malachite green, concluded from the average intermolecular separation of quencher at the concentration needed to achieve half-maximal quenching, that the OH radical moved over distances of 1–3 nm to effect CALI, indicating that the damage radius is small [4]. They also demonstrated that malachite-green-mediated CALI of the T-cell receptor in situ is restricted spatially within the multisubunit complex [39]. Using quenchers and defined spacers between the fluorophore and the target enzyme, other researchers concluded that 1O2, the production of which was sensitized by fluorescein excitation, had a damage radius of ~4 nm [9]. CALI of β–galactosidase (β-Gal) in vitro indicates that the motionally averaged distance between chromophore and site of action is in the order of 5 nm or less (D. Humphrey et al., unpublished). This is based on structural models of a β-Gal monomer showing that the distance between the N terminus and the active site is ~2.5 nm and that between the C terminus and the active site is ~3.5 nm. In addition, the distance from the chromophore to the ‘lid’ of the β-can of EGFP is ~1.5 nm along the axis of the can. Indeed, the effectiveness of CALI drops markedly comparing N-and C-terminal fusions of EGFP to β-Gal, a distance of ≈ 1 nm. These results indicate that the damage radius for CALI is of the order of 5 nm or less and is consistent with the lower range of singlet-oxygen diffusion distances (see Ref. 11 and references therein). The structure of CP-β [40] and the location of EGFP at its C terminus indicates that ROS generated by excitation of EGFP must travel ~6 nm to inactivate actin binding unless an intra- or intermolecular cross-linking process, operative at shorter ROS diffusion distances, results in occlusion of the actin binding site. As mentioned earlier, ROS photosensitized by fluorescein must travel at least 5 nm to achieve collateral damage of other transport channels [38].
Table 1.
Estimated radii of spherical ROS-induced photodamage zones
| CALI target | Photochemical intermediary | Range of action | Refs |
|---|---|---|---|
| Malachite green β-galactosidase | OH-radical (not from triplet state) | 1–3 nm | [39] |
| EGFP-β-galactosidase | ROS | Several nm | D. Humphrey et al., unpublished |
| EGFP-CP | ROS | ~6 nm | [17] |
| Fluorescein | 1O2 (produced from singlet state of dye) | ~4 nm | [9,24] |
| Fluorescein α-bungarotoxin–Ca2+
channel |
Probably 1O2 | Collateral damage to other transporters implies range of action ≥ 5 nm |
[38] |
For reference, 1O2 has a lifetime of 3 μs in water and 65 μs in D2O, giving diffusion distances of 130 nm and 605 nm, respectively, if a diffusion coefficient for 1O2 of ~10−5 cm2 s−1 is assumed. Primarily owing to proximate reactive amino acid residues in the cellular milieu, the 1O2 lifetime is reduced to ~200ns in the cytoplasm [26,41]. During this lifetime, 1O2 could diffuse ~30 nm, indicating a large damage radius with considerable potential for collateral damage. (Note that a recent study indicates the lifetime of 1O2 in cytosolic environments approaches that in water meaning that it could diffuse longer distances before being quenched, which indicates that it is a selective rather than a broadly reactive species [42].) However, when the 1O2 generator is conjugated to proteins of interest, presumably the effective diffusion distance that determines the damage radius is reduced even further by an abundance of reactive sites proximate to the site of generation [28].
Overview of factors influencing CALI in a cellular context
It is clear that the factors affecting the efficacy of CALI are twofold. First, the properties of the chromophore (fluorophore) itself and its immediate environment determine the quantum efficiency for 1O2 and other ROS generation (Figure 3). In general, bleaching of the fluorophore and chromophore might or might not be parallel phenomena. However, by definition, CALI will cease when the chromophore is completely transformed into a species that no longer absorbs the wavelengths of light being used. Second, the extent of CALI, as mediated by ROS, depends on the local environment (Figure 2). Direct inactivation via photogenerated ROS attacking an active site or binding interface is essentially an intramolecular phenomenon and, therefore, not directly dependent on overall protein concentration. Indirect inactivation occurs because the photogenerated ROS oxidize surface residues so that proximate proteins become cross-linked. Clearly, this effect depends on protein concentration and is likely to be a key factor in the crowded intracellular environment. Because multiple inactivation pathways are potentially accessible in the cellular milieu, CALI might well be more efficient in cells than in vitro. This might explain why several studies on cells have reported distinct inactivation phenotypes. However, endogenous cytoplasmic agents such as glutathione might reduce ROS and thus protect against CALI to some extent. Some experimental considerations for successful CALI experiments are given in Box 1.
Box 1. Special considerations for successful CALI experiments.
Desirable properties of CALI chromophores
First, the chromophore should bind extremely tightly to the target to avoid ambiguities in what is inactivated. In this respect, the fluorescent proteins, – given that they are covalently attached to the target have an advantage over small dyes that diffuse to their targets after introduction into the cell. Second, the probe must be an efficient sensitizer for ROS production. The small xanthene based dyes, such as FlaSH or ReAsH, have a great advantage in that they are excellent sensitizers, meaning that much lower irradiation doses can be applied to achieve CALI. Thus, often CALI can be achieved in a reasonable time using arc lamp sources as opposed to focused laser beams.
Surmounting the effect of the endogenous protein
When a modified protein is ectopically introduced into a cell, it is present in amounts in excess of the background of endogenous cellular proteins. In a worst case scenario, the light insensitive endogenous material can fully compensate for a CALI induced loss of function. This has been demonstrated in the case of EGFP CALI of CP [17]. The success of this experiment was absolutely dependent upon removing the endogenous protein; with it present, no CALI effect was observed. In other cases, however, loss of function could be achieved with the endogeneous protein present, with varying degrees of completion. This is a factor that should be addressed on a case by case basis. It should be noted that valuable controls can be performed with mutant proteins where the activity to be inactivated by CALI is already knocked out. Irradiation of such a protein fused to GFP provides a good test for non specific effects.
Diffusion of target molecules during subcellular perturbations
When perturbations are meant to be localized, special consideration needs to be given to the intracellular mobility of the target molecule. This is because spatial localization of inactivated or activated mobile molecules can be eroded by diffusion out of the irradiated region. The magnitude of this effect depends on the diffusion coefficient of the target molecule in the cytoplasm or on the plasma membrane, and on the size of the irradiated region. Roughly speaking, the time for a localized inactivation profile to dissipate is proportional to the square of the illuminated region dimension divided by the diffusion coefficient. CALI of actin binding proteins is instructive in this regard. In the case of a actinin at the junction between stress fibers and focal adhesions, a local effect could be readily achieved by focusing a small laser beam (~2 μm) on the junction because, under these conditions, the time for fluorescence recovery after CALI (photobleaching), which is dominated by exchange with a actinin bound to the junction, is in the order of minutes [15]. By contrast, to achieve CALI of EGFP CP, the laser beam had to be expanded to cover a substantial portion of the cell [17]. Diffusion of CP in the cytoplasm is fast owing to rapid dissociation from f actin in cells [44], so inactivated CP in smaller illuminated regions was quickly replaced by fully functional protein. Consequently, the characteristic null CP phenotype could not develop. Thus, it is advisable to perform a FRAP measurement on the target molecule before attempting subcellular CALI to design an experimental routine that takes the diffusion and/or exchange properties of the target molecule into account. Without this knowl edge, such an experiment might be impossible, failing despite the fact that CALI has occurred efficiently.
Functional recovery can occur as non irradiated molecules replace the inactivated species in the manipulated region. It is important to note that, although lateral transport of non irradiated material protein is obligatory in local functional exchange, additional reactions might be rate limiting in achieving complete phenotypic recovery. None theless, reversibility subsequent to exchange of irradiated for non irradiated target molecules provides an excellent indication that there has been bona fide photoregulation of the target.
From an experimental standpoint, for many fluorophores the quantum efficiencies for emission and even photobleaching are substantially higher than that for CALI. For example, in the case of EGFP-CP, confocal FRAP experiments demonstrated that bleaching did not produce the CALI-induced protrusive phenotype (E.V. et al., unpublished). In such cases, visualization and photobleaching of EGFP can be accomplished at doses where little CALI occurs, because the latter requires considerably higher laser power to produce an appreciable effect. The disadvantage is that it is difficult to perform rapid CALI with the more photostable fluorescent proteins using the low power lasers employed in conventional scanning confocal microscopes. Overall, however, this seems to be good news for cell and developmental biologists using both GFP fluorescence imaging and FRAP approaches.
Summary and outlook
Factors affecting CALI efficiency interplay in a complex fashion and depend on both photophysical and photochemical properties of the chromophore and the environment in which the chromophore finds itself. Thus, although in vitro screens might produce useful and important guidelines, the CALI efficiency ranking for different reagents may differ in a cellular environment. That said, screens can be designed to test fluorescent protein mutants and small molecular dyes for an optimum compromise between CALI and facile imaging [43]. Furthermore, small, highly specific molecules can be designed to bear both a CALI chromophore and a spectrally distinct fluorophore for imaging the location of the target protein without excessive bleaching. These might be based on, for example, the Halo® reagents (Promega; www.promega.com), which function as suicide inhibitors by covalently binding to the active site of a hydrolase fused to the target protein [43]. Using dyes that change absorption wavelengths upon binding to their target, it might even be possible to irradiate selectively only those dyes in proximity to the target.
Acknowledgements
We acknowledge helpful discussions with members of the Image and Photomanipulation Initiative of the Cell Migration Consortium and the support of the Consortium (NIH GM 64346 and NIH PO1-HL080166). We thank Jim Remington, Dave Piston, Petra Schwille, Tom Jovin, Vinod Subramaniam, Peter Devreotes and Harold Erickson for providing useful information for this review. We also thank Bing Yang for technical assistance in preparing this review.
References
- 1.Jay DG. Selective destruction of protein function by chromophore-assisted laser inactivation. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5454–5458. doi: 10.1073/pnas.85.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang FS, Jay DG. Chromophore-assisted laser inactivation (CALI): probing protein function in situ with a high degree of spatial and temporal resolution. Trends Cell Biol. 1996;6:442–445. doi: 10.1016/s0962-8924(96)40005-8. [DOI] [PubMed] [Google Scholar]
- 3.Linden KG, et al. Spatial specificity of chromophore assisted laser inactivation of protein function. Biophys. J. 1992;61:956–962. doi: 10.1016/S0006-3495(92)81902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao JC, et al. Chromophore-assisted laser inactivation of proteins is mediated by the photogeneration of free radicals. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2659–2663. doi: 10.1073/pnas.91.7.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dougherty TJ, et al. Photoradiation therapy for the treatment of malignant tumors. Cancer Res. 1978;38:2628–2635. [PubMed] [Google Scholar]
- 6.Rattner JB, Berns MW. Light and electron microscopy of laser microirradiated chromosomes. J. Cell Biol. 1974;62:526–533. doi: 10.1083/jcb.62.2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanni F, et al. Fluorescence photobleaching recovery in solutions of labeled actin. Biophys. J. 1981;35:351–364. doi: 10.1016/S0006-3495(81)84794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vigers GP, et al. Fluorescent microtubules break up under illumination. J. Cell Biol. 1988;107:1011–1024. doi: 10.1083/jcb.107.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck S, et al. Fluorophore-assisted light inactivation: a high-throughput tool for direct target validation of proteins. Proteomics. 2002;2:247–255. doi: 10.1002/1615-9861(200203)2:3<247::aid-prot247>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 10.Rahmanzadeh R, et al. Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis. Cell Prolif. 2007;40:422–430. doi: 10.1111/j.1365-2184.2007.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tour O, et al. Genetically targeted chromophore-assisted light inactivation. Nat. Biotechnol. 2003;21:1505–1508. doi: 10.1038/nbt914. [DOI] [PubMed] [Google Scholar]
- 12.Marek KW, Davis GW. Transgenically encoded protein photoinactivation (FlAsH-FALI): acute inactivation of synaptotagmin I. Neuron. 2002;36:805–813. doi: 10.1016/s0896-6273(02)01068-1. [DOI] [PubMed] [Google Scholar]
- 13.Marks KM, et al. A general approach for chemical labeling and rapid, spatially controlled protein inactivation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9982–9987. doi: 10.1073/pnas.0401609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yogo T, et al. Selective photoinactivation of protein function through environment-sensitive switching of singlet oxygen generation by photosensitizer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:28–32. doi: 10.1073/pnas.0611717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajfur Z, et al. Dissecting the link between stress fibres and focal adhesions by CALI with EGFP fusion proteins. Nat. Cell Biol. 2002;4:286–293. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- 16.Tanabe T, et al. Multiphoton excitation-evoked chromophore-assisted laser inactivation using green fluorescent protein. Nat. Methods. 2005;2:503–505. doi: 10.1038/nmeth770. [DOI] [PubMed] [Google Scholar]
- 17.Vitriol EA, et al. Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6702–6707. doi: 10.1073/pnas.0701801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bear JE, et al. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 19.Bear JE, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 20.Bulina ME, et al. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez R, Redman R. Balancing the generation and elimination of reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3175–3176. doi: 10.1073/pnas.0500367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 23.Davies MJ. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003;305:761–770. doi: 10.1016/s0006-291x(03)00817-9. [DOI] [PubMed] [Google Scholar]
- 24.Horstkotte E, et al. Toward understanding the mechanism of chromophore-assisted laser inactivation evidence for the primary photochemical steps. Photochem. Photobiol. 2005;81:358–366. doi: 10.1562/2004-07-22-RA-240. [DOI] [PubMed] [Google Scholar]
- 25.Grabenbauer M, et al. Correlative microscopy and electron tomography of GFP through photooxidation. Nat. Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- 26.Gorman AA, Rodgers MA. Current perspectives of singlet oxygen detection in biological environments. J. Photochem. Photobiol. B. 1992;14:159–176. doi: 10.1016/1011-1344(92)85095-c. [DOI] [PubMed] [Google Scholar]
- 27.Greenbaum L, et al. Green fluorescent protein photobleaching: a model for protein damage by endogenous and exogenous singlet oxygen. Biol. Chem. 2000;381:1251–1258. doi: 10.1515/BC.2000.153. [DOI] [PubMed] [Google Scholar]
- 28.Yan P, et al. Fluorophore-assisted light inactivation of calmodulin involves singlet-oxygen mediated cross-linking and methionine oxidation. Biochemistry. 2006;45:4736–4748. doi: 10.1021/bi052395a. [DOI] [PubMed] [Google Scholar]
- 29.Surrey T, et al. Chromophore-assisted light inactivation and self-organization of microtubules and motors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4293–4298. doi: 10.1073/pnas.95.8.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giepmans BN, et al. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 31.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr. Opin. Struct. Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Jimenez-Banzo A, et al. Singlet oxygen photosensitization by EGFP and its chromophore HBDI. Biophys. J. 2008;94:168–172. doi: 10.1529/biophysj.107.107128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widengren J, et al. Photodynamic properties of green fluorescent proteins investigated by fluorescence correlation spectroscopy. Chem. Phys. 1999;250:171–186. [Google Scholar]
- 34.Haupts U, et al. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13573–13578. doi: 10.1073/pnas.95.23.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung G, et al. The role of dark states in the photodynamics of the green fluorescent protein examined with two-color fluorescence excitation spectroscopy. J. Phys. Chem. A. 2000;104:873–877. [Google Scholar]
- 36.Donnert G, et al. Major signal increase in fluorescence microscopy through dark-state relaxation. Nat. Methods. 2007;4:81–86. doi: 10.1038/nmeth986. [DOI] [PubMed] [Google Scholar]
- 37.Lepock JR, et al. Photoinduced crosslinking of membrane proteins by fluorescein isothiocyanate. Biochem. Biophys. Res. Commun. 1978;85:344–350. doi: 10.1016/s0006-291x(78)80048-5. [DOI] [PubMed] [Google Scholar]
- 38.Guo J, et al. Fluorophore-assisted light inactivation produces both targeted and collateral effects on N-type calcium channel modulation in rat sympathetic neurons. J. Physiol. 2006;576:477–492. doi: 10.1113/jphysiol.2006.113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao JC, et al. Chromophore-assisted laser inactivation of subunits of the T-cell receptor in living cells is spatially restricted. Photochem. Photobiol. 1995;62:923–929. doi: 10.1111/j.1751-1097.1995.tb09157.x. [DOI] [PubMed] [Google Scholar]
- 40.Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem. Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Baker A, Kanofsky JR. Quenching of singlet oxygen by biomolecules from L1210 leukemia cells. Photochem. Photobiol. 1992;55:523–528. doi: 10.1111/j.1751-1097.1992.tb04273.x. [DOI] [PubMed] [Google Scholar]
- 42.Hatz S, et al. Measuring the lifetime of singlet oxygen in a single cell: addressing the issue of cell viability. Photochem. Photobiol. Sci. 2007;6:1106–1116. doi: 10.1039/b707313e. [DOI] [PubMed] [Google Scholar]
- 43.Lee J, et al. A general system for evaluating the efficiency of chromophore-assisted light inactivation (CALI) of proteins reveals Ru(II) tris-bipyridyl as an unusually efficient “warhead”. Mol. Biosyst. 2008;4:59–65. doi: 10.1039/b712307h. [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi T, et al. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]