DNA methylation patterns in the developing and adult mammalian brain point to a role in synaptic development and maturation.
The methylation of DNA in mammalian genomes regulates gene expression, guiding differentiation and maintaining cellular identity within tissues. However, it may have a distinct function in the brain. On page XX of this issue, Lister et al. (1) present a comprehensive analysis of DNA methylation and hydroxymethylation at single-base resolution in the mammalian frontal cortex. The authors chart out striking postnatal alterations in neuronal methylation profiles that occur as synapses develop and are refined, from the fetal to adult stage. The patterns suggest that DNA methylation is important in the maturation of neurons in the developing brain.
DNA methylation represses gene expression in all mammalian cells. The methylation of cytosines in the context of cytosine-guanine dinucleotides (mCG) is a stable repressive mark on DNA. However, the Tet family of enzymes converts methylcytosine to hydroxymethylcytosine (hmC), an oxidized form that can be demethylated (2, 3). hmC is enriched in stem cells and neurons, suggesting that they might be particularly susceptible to changes in DNA methylation state (2, 4). Intriguingly, stem cells and brain tissue also contain substantial cytosine methylation outside of the CG context [mCH, where H is adenine (A), thymine (T), or cytosine (C)], which is rare in most somatic cells (5, 6). Although the role of hmC and mCH in stem cells has been extensively investigated at base-pair resolution (5, 7), there has been limited examination of DNA methylation at high-resolution in the brain.
Using high-throughput sequencing to profile the mouse and human cortex from the fetal to adult stage, Lister et al. revealed a conserved, genome-wide increase in mCH amounts in the brain after birth. Although the period during which mCH accumulates differs between mice and humans (several weeks versus several years, respectively), the increase coincides with the peak of synaptogenesis and synaptic pruning in the brain for each organism. Thus, the acquisition of mCH may be linked to neuronal maturation. High amounts of mCH were found in neurons compared to low amounts in nonneuronal (glial) cells. Although the average percentage methylation detected in neurons at CH is quite low (~2–6%) and quite high at CG (~80%), because CG is rare in mammalian genomes relative to CH, a similar number of total mCH and mCG events occur in neurons. Indeed, in adult human neurons, the total number of mCH sites surpasses that of mCG sites. Rather than a minor addition to the methylation modifications in the genome (“methylome”), mCH is likely a major substrate for gene regulation in the maturing brain.
hmC builds up in neurons with a timing similar to that of mCH (8), raising the possibility that, in addition to occurring at CG, hydroxymethylation also takes place at CH in the brain. Lister et al. used another high-throughput sequencing method (9) to profile hmC at single-base resolution in mouse fetal and adult frontal cortex. Surprisingly, even though hmC and mCH both accumulate as the brain matures, hydroxymethylation occurs almost exclusively in the hmCG context.
To better understand how methylation contributes to transcriptional regulation, Lister et al. performed integrated analysis of hmC, mCG, and mCH profiles, messenger RNA expression, and epigenomic datasets from the brain. The authors observed an expected enrichment of mCG and mCH at genes and distal regulatory elements correlating with transcriptional repression, whereas hmC is deposited across active genes (6, 8). Previously undescribed patterns of methylation in the brain also were uncovered, such as enrichment of mCH at genes that escape X-chromosome inactivation in females. Lister et al. also noted that, while glial genomes contain low global mCH, mCH is enriched at genes that are repressed in either neurons or glia, indicating that mCH may enforce cell-type–specific transcription in these related cell lineages. Further, the authors detected thousands of developmental-stage– and cell-type–specific methylation sites at putative enhancers. Thus, some inactive enhancers in the fetal brain are marked with hydroxymethylation, which likely leads to subsequent demethylation and activation in the adult.
The findings of Lister et al. pave the way for examining how the brain-specific methylome is established. For example, the enzyme(s) that generates high amounts and mCH in neurons have not yet been described. A strong candidate is Dnmt3a, a methyltransferase that peaks in expression in neurons during the period of mCH deposition, and displays methylation activity on CH dinucleotides (10). Because hmC and mCH profiles display conservation and reproducibility across individuals, they likely result from regulated processes. However, it is unclear to what extent the patterns of hmC and mCH in neurons result from sequence-directed targeting mechanisms or if they occur secondarily, guided by accessible chromatin structure or transcriptional activity.
Although the profiles point to regulatory roles for mC and hmC in the brain, the mechanisms by which these marks are read out to affect gene expression await elucidation. It will be essential to understand which DNA binding factors are recruited to mCG, mCH, and hmCG to mediate their regulatory functions. Multiple proteins differentially bind to unmethylated, methylated, and hydroxymethylated DNA (11, 12). In neurons, high hmCG amounts at some CG sites could block proteins from binding, whereas mCH may provide an array of new binding sites in the genome. Notably, the methyl-DNA–binding protein MeCP2 accumulates in the brain with similar timing to mCH and hmC (13). Thus, MeCP2 may bind to the additional mCH sites in neurons to repress genes. Because disruption of MeCP2 leads to the neurological disorder Rett syndrome (14), it will be important to determine how mCH affects the function of this protein.
Could the epigenomic rearrangements uncovered by Lister et al. participate in the development and maturation of synaptic connections? Because the frontal cortex develops postnatally in concert with input from the environment, it is possible that neuronal-specific DNA methylation contributes to sensory input–dependent changes in gene expression and synaptic development. Alternatively, high amounts of mCH and hmC may facilitate the generation of diverse neuronal subtypes; acquisition of the neuronal-specific methylome coincides with the final stages of neuronal differentiation that give rise to the many neuronal subtypes of the brain. The propensity of mCH to mark cell-type–specific genes and the differential methylation of enhancers suggest that the methylation profile of a neuron may dictate its specific gene expression profile and functions within the context of a neural circuit. Future studies should determine whether epigenomic rearrangements affect neuronal cell-type diversity and/or sensory-dependent development in the brain.
Figure 1. A distinct neuronal methylome.
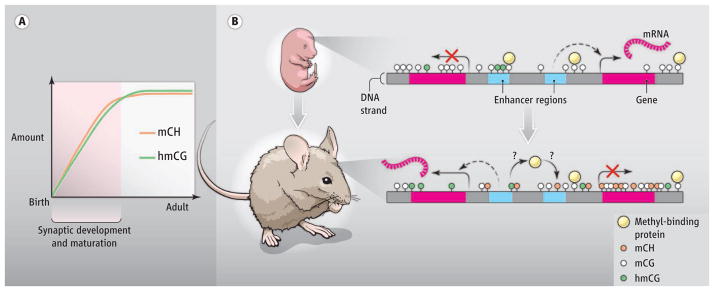
(A) High mCH and hmCG accumulate in mammalian neurons postnatally, coinciding with the period of active synapse development and maturation. (B) A schematic of the same genomic region in fetal and adult brain shows that mCH builds up in adult neurons and is enriched at repressed genes and enhancers. hmCG associates with active genes and can premark enhancers in the fetal brain that will become demethylated and active in the adult. Differential affinity to hmCG and mCH may alter the binding profile of methyl-DNA binding proteins and thereby control distinct neuronal transcription.
References
- 1.Lister R, et al. Science. 2013;341:XXX. doi: 10.1126/science.1237905. [DOI] [Google Scholar]
- 2.Tahiliani M, et al. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo JU, Su Y, Zhong C, Ming GL, Song H. Cell Cycle. 2011;10:2662. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriaucionis S, Heintz N. Science. 2009;324:929. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lister R, et al. Nature. 2009;462:315. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie W, et al. Cell. 2012;148:816. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziller MJ, et al. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szulwach KE, et al. Nat Neurosci. 2011;14:1607. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, et al. Cell. 2012;149:1368. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsahoye BH, et al. Proc Natl Acad Sci USA. 2000;97:5237. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. Cell. 2012;151:1417. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spruijt CG, et al. Cell. 2013;152:1146. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Skene PJ, et al. Mol Cell. 2010;37:457. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amir RE, et al. Nat Genet. 1999;23:185. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]


