Abstract
BACKGROUND & AIMS
Defects in colonic epithelial barrier defenses are associated with ulcerative colitis (UC). The proteins that regulate bacterial clearance in the colonic epithelium have not been completely identified. The chromosome-associated protein D3 (dCAP-D3), regulates responses to bacterial infection. We examined whether CAP-D3 promotes bacterial clearance in human colonic epithelium.
METHODS
Clearance of Salmonella or adherent-invasive Escherichia coli LF82 was assessed by gentamycin protection assays in HT-29 and Caco-2 cells expressing small hairpin RNAs against CAP-D3. We used immunoblot assays to measure levels of CAP-D3 in colonic epithelial cells from patients with UC and healthy individuals (controls). RNA sequencing identified genes activated by CAP-D3. We analyzed the roles of CAP-D3 target genes in bacterial clearance using gentamycin protection and immunofluorescence assays and studies with pharmacologic inhibitors.
RESULTS
CAP-D3 expression was reduced in colonic epithelial cells from patients with active UC. Reduced CAP-D3 expression decreased autophagy and impaired intracellular bacterial clearance by HT-29 and Caco-2 colonic epithelial cells. Lower levels of CAP-D3 increased transcription of genes encoding SLC7A5 and SLC3A2, whose products heterodimerize to form an amino acid transporter in HT-29 cells following bacterial infection; levels of SLC7A5–SLC3A2 were increased in tissues from patients with UC, compared with controls. Reduced CAP-D3 in HT-29 cells resulted in earlier recruitment of SLC7A5 to Salmonella-containing vacuoles, increased activity of mTORC1, and increased survival of bacteria. Inhibition of SLC7A5–SLC3A2 or mTORC1 activity rescued the bacterial clearance defects of CAP-D3– deficient cells.
CONCLUSIONS
CAP-D3 downregulates transcription of genes that encode amino acid transporters (SLC7A5 and SLC3A2) to promote bacterial autophagy by colon epithelial cells. Levels of CAP-D3 protein are reduced in patients with active UC; strategies to increase its levels might restore mucosal homeostasis to patients with active UC.
Keywords: condensin, inflammatory bowel disease, innate immunity, dysbiosis
Introduction
Ulcerative colitis (UC) is one type of inflammatory bowel disease (IBD) which results in chronic mucosal inflammation, tissue destruction, and severe pain 1. The incidence of UC is increasing worldwide, with lifelong drug treatment or surgery as the only therapeutic options 2. While the initiating event and causes of disease relapse are poorly understood, there are several factors shown to have major roles in disease etiology, including genetics, the environment, the immune system, and the microbiome 3, 4.
The bacterial contribution to UC is well established. During active UC in humans and mouse colitis models, bacteria penetrate the mucus layer and come into close contact with colonic epithelial cells, and this has been observed even prior to inflammation 5-9. In addition, prior enteric infections are a risk factor for the development of UC 10, 11.
Autophagy is a cellular defense mechanism against intracellular bacterial infection 12, 13. The intracellular pathogen, Salmonella enterica serovar typhimurium (Salmonella), invades cells and creates specialized vacuoles for replication 14. Often during this process, this Salmonella-containing Vacuole (SCV) becomes damaged and targeted for autophagic clearance through a cascade of events resulting in the recruitment of LC3 (microtubule associated protein light chain 3) 15, 16. The membrane-bound form of LC3 (LC3-II) is a component of a multi-lamellar membrane that engulfs the damaged SCV, forming an autophagosome which fuses with lysosomes to kill trapped bacteria. Salmonella can escape autophagy through recruitment and activation of mTORC1 on the SCV 17. Other invasive bacteria, including the Crohn’s Disease-associated adherent-invasive E. coli (AIEC) strain, LF82, are also targeted by autophagy, but are able to replicate in patients with defective autophagy pathways18, 19. These studies underscore the importance of understanding the proteins and pathways involved in promoting bacterial clearance through autophagy.
Previously, our lab demonstrated dCAP-D3 (Chromosome Associated Protein- D3) to be a novel player in the innate immune response against bacterial infection in the model organism, Drosophila melanogaster 20. dCAP-D3 is a subunit of the Condensin II complex, a complex which promotes chromatin condensation at the beginning of mitosis. Recently, additional roles for the human Condensin II complex outside of mitosis have been characterized, including regulation of transcription 20-22. In Drosophila, dCAP-D3 directly upregulates genes required for innate immunity following systemic bacterial infection and its expression is necessary for the fly’s ability to clear bacteria and survive infection 20.
Given that intestinal dysbiosis is linked to multiple disease states, including UC, we investigated whether human CAP-D3 also regulated bacterial defenses in colon epithelial cells. We found that CAP-D3 protein levels are significantly decreased in colonic epithelial cells isolated from resected tissues of patients with active UC. When we tested HT-29 and Caco-2 human colon adenocarcinoma cell lines expressing inducible shRNAs targeting CAP-D3 expression, we discovered that decreased CAP-D3 expression impaired autophagy induction and decreased clearance of Salmonella or AIEC LF82. RNA-sequencing of mRNAs from control and CAP-D3 deficient cells revealed that large, neutral amino acid transporters, including two members of a heterodimeric, bidirectional transporter, SLC7A5 and SLC3A2, were negatively regulated by CAP-D3. These transporters were also found to be elevated in epithelial cells from patients with UC. In CAP-D3-deficient cells, increased SLC7A5 expression resulted in its earlier localization to SCVs, resulting in recruitment of mTORC1 and autophagy repression. These studies are the first to identify CAP-D3 as a regulator of antimicrobial responses and innate immunity in the gut epithelium, and link CAP-D3 to regulation of mTORC1 activity. Our findings suggest that methods to control the levels of CAP-D3 may have therapeutic benefit for patients with active UC.
Materials and Methods
Cell lines
HT-29 cells were cultured in RPMI 1640 media with 10% fetal bovine serum (FBS; Gibco), and 1% penicillin/streptomycin. Caco-2 cells were cultured in DMEM media, with sodium pyruvate, 10% FBS, and 1% penicillin/streptomycin. Cells were treated with the SLC3A2/SLC7A5 inhibitors, 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH, 20mM, Sigma) or D-phenylalanine (25 mM, Sigma) 1h prior or 3h prior (respectively) to infection and maintained during the infection. Cells were treated with the mTORC1 agonist rapamycin (25mg/ml in DMSO, LC Labs) 1h prior to infection and maintained during the infection.
Epithelial cell isolation from resected patient colon tissues
Epithelial cells were isolated from de-identified, colonic tissue collected from individuals undergoing colonic resection (Cleveland Clinic Tissue Procurement Service; IRB protocol#05-205)23.
Characterization of disease activity in patient tissue samples
Disease activity of obtained samples was determined by 1) review of de-identified pathology reports; 2) microscopic examination by a pathologist using the six-grade classification system described in 24; 3) observance of neutrophils inside the crypt lumen in S100A12 immunostained tissue sections. S100A12 had previously been shown to stain the activated neutrophils inside of colonic crypts in patients with active CD25. S100A12 staining correlated well with other measures of disease activity in both CD and UC tissues used in our studies.
Bacterial infections and gentamycin protection assays
Confluent cells were treated for 48h with 1mM IPTG prior to infection to induce shRNA expression. Infections with Salmonella enterica serovar Typhimurium SL1344 or AIEC LF82 were performed at a multiplicity of infection of 10 as previously described 26, 27. Assays were performed in triplicate in at least three independent experiments.
RNA-sequencing of cellular mRNAs
Total cellular RNA was isolated as described in 20. Briefly, TRIzol (Invitrogen) was used to harvest total RNA from tissues and cells according to manufacturer’s protocol. Samples were DNase treated and run on RNAeasy columns (Qiagen). RNA samples from 3 independent experiments including timepoints taken at 0, 0.5 and 7 hours post-infection were analyzed on a bioanalyzer for quality; one of the 0.5 hour post-infection samples in the control and in the CAP-D3 shRNA expressing cells was excluded at this time due to poor RNA purity. Directional, cDNA libraries made from cellular mRNAs were generated from the other 16 samples using the Illumina TruSeq RNA library kit and sequenced (paired-end sequencing of 100 bp reads) in the Genomics Core at the University of Chicago on an Illumina HiSeq2500, according to standard Illumina protocols. Raw data files from RNA-sequencing experiments have been deposited under the NCBI GEO accession number GSE62520.
qRT-PCR
RNA isolation and qRT-PCR were performed as described in 20. qRT-PCR reactions were run on a Roche Lightcycler 480. Three independent experiments were performed in all cases and results were averaged together. Primers are listed in Supplemental data.
Immunoblotting
Protein harvesting and immuoblotting was performed as described in 28 with more detail in Supplemental Methods.
Immunofluorescence
Immunofluorescence experiments were performed as described in 28 with more detail in Supplemental Methods.
Results
CAP-D3 promotes the clearance of intracellular bacteria through autophagy
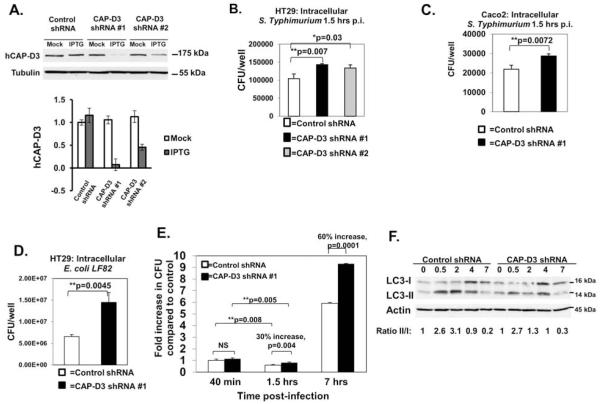
Based on our previous findings that dCAP-D3 was necessary for efficient clearance of bacteria in the model organism, Drosophila, we asked whether this function was conserved in human cells 20. For these studies, two human colon epithelial cell lines, HT-29 and Caco-2, were transduced with lentivirus expressing IPTG inducible non-target shRNA as a control or IPTG inducible CAP-D3 shRNA. Results show that CAP-D3 shRNA expression reduces CAP-D3 levels by more than 50% (Figure 1A). These cell lines were infected with the intracellular pathogen, Salmonella (Figures 1B and 1C) or with AIEC LF82 (Figure 1D), and gentamycin protection assays were performed. Results show significant increases in viable, intracellular Salmonella and viable, intracellular AIEC LF82 in cells expressing CAP-D3 shRNA in comparison to cell lines expressing control shRNA. These results indicate that CAP-D3 is involved in the intracellular clearance of bacteria in human intestinal epithelial cells. Additionally, the findings demonstrate a conserved role for CAP-D3 in anti-microbial defense, which can target bacteria associated with IBD pathogenesis.
Figure 1. CAP-D3 promotes bacterial autophagy in human colon epithelial cells.
A) Immunoblot showing reduced expression of CAP-D3 in HT-29 cells treated with IPTG to induce expression CAP-D3 shRNAs in comparison to cells treated with PBS (mock) and cells expressing control (non-target) shRNA. CAP-D3 protein levels were normalized to tubulin and quantified in the bottom panel. Gentamycin protection assays of Salmonella infected HT-29 (B) or Caco-2 (C) cell lines at 1.5 hours post-infection, and of AIEC LF82 in HT-29 cells (D) demonstrate significant increases in bacteria in cells expressing CAP-D3 shRNAs. Mean±SD. E) Gentamycin protection assays performed in Salmonella-infected HT-29 cells expressing CAP-D3 shRNA or control shRNA show enhanced bacterial survival in CAP-D3 deficient cells beginning after 40 minutes. Bacterial survival graphed relative to CFU/well of control shRNA expressing cells at the earliest timepoint. Mean±SD. F) Immunoblots demonstrate lower ratios of LC3-II/LC3-I protein in Salmonella infected HT-29 cells expressing CAP-D3 shRNA. Protein levels normalized to actin, then quantified relative to uninfected controls. The experiment shown is representative of three independent immunoblots.
To better understand the role of CAP-D3 in promoting bacterial clearance, gentamycin protection assays were performed over a timecourse and viable intracellular Salmonella loads were assayed at various timepoints post-infection. At the earliest timepoint (40 min), equivalent amounts of bacteria were recovered from HT-29 cells expressing control and CAP-D3 shRNAs. However, bacterial loads became significantly higher in CAP-D3 deficient cells at 1.5 and 7 hours post-infection (Figure 1E). These results suggest that increased bacterial entry is not the cause of increased bacterial loads in the CAP-D3 deficient cells.
The amount of intracellular bacteria recovered in these assays actually decreased between 40 minutes and 1.5 hours in both CAP-D3 and control shRNA expressing cells. These results suggest that both cell lines are able to kill bacteria, but that CAP-D3 shRNA expressing cells have a killing defect. Therefore, we performed TUNEL assays on HT-29 cells expressing CAP-D3 or control shRNA to quantify the numbers of bacteria that exhibited damaged and degraded DNA, which might be indicative of dying bacteria. Significantly higher numbers of TUNEL positive bacteria were seen in control shRNA expressing cells at 7 hours post-infection (Supplementary Figure 1A and 1B). TUNEL staining also showed that there was minimal damage to epithelial cell DNAs following infection at the timepoints studied, suggesting that differences in the levels of epithelial cell death were not the reason for increased bacterial loads in CAP-D3 deficient cells (Supplementary Figure 1C).
One early cellular response to intracellular bacterial infections is the targeting of these pathogens for destruction through capture in an autophagosome12. To examine whether decreased CAP-D3 expression influenced autophagy induction in response to Salmonella infection, the conversion of the autophagy marker protein, LC3, from the cytosolic form (LC3-I) to the autophagosome-associated form (LC3-II) was assessed by immunoblot. Results demonstrate that the ratio of LC3-II to LC3-I is lower in CAP-D3 shRNA expressing cells compared to control shRNA expressing cells, with the greatest difference occurring at 2 hours post-infection (Figure 1F). These results suggest that CAP-D3 deficient cells are diminished in their capacity to clear intracellular pathogens due to autophagy.
CAP-D3 levels are significantly decreased in colon epithelial cells from patients with UC
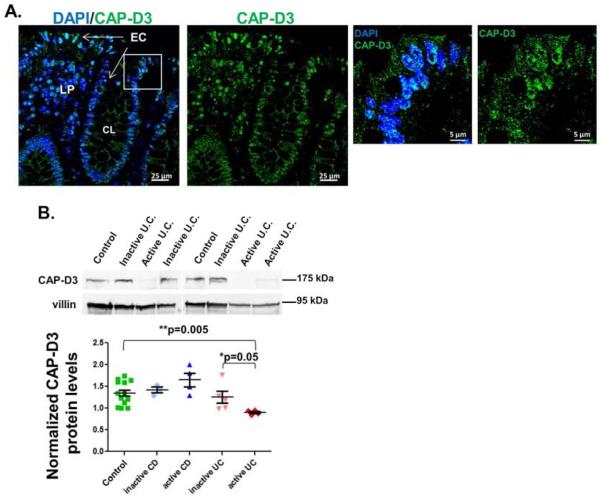
Given that bacterial invasion into the intestinal epithelium is believed to contribute to the pathogenesis of UC, we tested whether CAP-D3 levels are altered in the colonic epithelial cells of patients with IBD. Immunostaining for CAP-D3 expression in tissue sections from control patients indicated that CAP-D3 localized to both the nucleus and the cytoplasm of colonic epithelial cells (EC) (Figure 2A and Supplementary Table 1). Next, we profiled the CAP-D3 expression levels in epithelial cells isolated from colonic resections of patients with active or inactive IBD by immunoblot. Samples were classified as being active or inactive (see Methods and Supplemental Table 1). CAP-D3 protein was analyzed by immunoblot and normalized to protein levels of the epithelial cell specific marker, villin (Figure 2B). Comparison of CAP-D3 protein levels from all patient tissue samples, demonstrates that CAP-D3 levels are not significantly different between control patients, patients with active or inactive CD, and patients with inactive UC. However, the levels in patients with active UC were significantly lower. These findings suggest an inverse correlation between CAP-D3 levels and disease activity specifically in patients with UC.
Figure 2. CAP-D3 protein levels are decreased in colonic epithelial cells from patients with active UC.
A) Immunofluorescence analysis of CAP-D3 (green) and DAPI (blue) staining of colonic tissue from a control patient. B) Immunoblotting for CAP-D3 protein in epithelial cells isolated from resected colon tissue from control patients, patients with inactive CD, active CD, inactive UC, or active UC shows CAPD3 is significantly lower in patients with active UC. Protein levels normalized to villin, then quantified relative to a control sample. Mean±SEM.
Amino acid transporters, SLC7A5 and SLC3A2 are upregulated at early timepoints post-infection in CAP-D3 deficient cells and in CAP-D3 deficient tissues from patients with UC
To determine whether CAP-D3 regulates transcription of genes which promote bacterial clearance in colon epithelial cells, RNA-sequencing of total mRNAs in uninfected or Salmonella infected HT-29 cells expressing CAP-D3 or control shRNA was performed. Results showed CAP-D3 transcripts were decreased in CAP-D3 shRNA expressing cells by almost 2 log fold (Supplementary Table 2). Analysis of differentially expressed transcripts at 0.5 hours post-infection and at 7 hours post-infection revealed that the expression of many genes significantly changed in response to the presence of the bacteria (Supplementary Table 3). Upon comparison of differentially expressed gene sets in control shRNA vs. CAP-D3 shRNA expressing cells at 0.5 hours post-infection, 13 genes were found to be significantly upregulated and shared between the two cell lines (Supplementary Figure 2A). These included genes encoding the AP-1 transcription factors FOS, FOSB, and JUNB, as well as Immediate Early Response genes EGR1, EGR3 and IER2. AP-1 transcription factors and Immediate Early Response genes have previously been shown to be transiently upregulated in colon epithelial cells shortly after infection with Salmonella, thus verifying that our assays were performed correctly 29, 30. Our results corroborated the transient nature of induction of the Immediate Early Response genes, since most were seen to decrease at 7 hours post-infection (Supplementary Figure 2B and 2C).
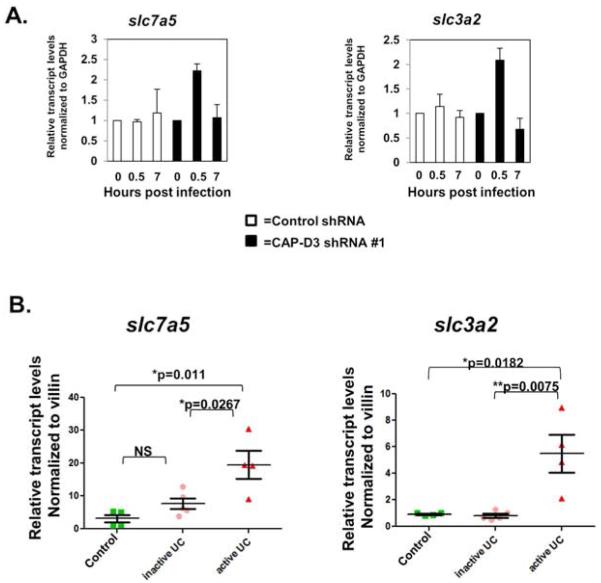
Gene ontology analysis of genes that were upregulated at 0.5 hours post-infection and unique to the CAP-D3 shRNA expressing cells, identified the most significantly upregulated group as “neutral amino acid transmembrane transporte(s)” (Supplementary Figure 3A). Closer inspection of the RNA-sequencing data identified the large neutral amino acid transporter genes, SLC7A5, SLC1A5, and SLC4A5 as being significantly upregulated in CAP-D3 shRNA expressing cells at 0.5 hours post-infection and then downregulated at 7 hours post-infection (Supplementary Figure 3B and 3C). The same effect was seen for the SLC3A2 gene which encodes an adaptor protein for several heterodimeric transporters, including SLC7A5. qRT-PCR confirmed that SLC7A5 and SLC3A2 transcripts were transiently upregulated in CAP-D3 shRNA expressing cells only (Figure 3A). Immunoblots examining protein expression for each gene demonstrated that SLC7A5 protein levels were modestly higher (20%) in CAP-D3 shRNA expressing cells starting at 2 hours post-infection (Supplemental Figure 3D). This small increase in protein levels could be due to the fact that only 20-30% of HT-29 cells are infected with Salmonella, and while larger amounts of SLC7A5 are actually made in the infected CAP-D3 deficient cells, they are diluted by the protein levels in the uninfected cells which make up the majority of the sample. In contrast, a sustained increase in SLC3A2 protein levels was seen in CAP-D3 shRNA expressing cells as compared to control shRNA expressing cells (Supplemental Figure 3E). Since we had found that CAP-D3 levels were significantly lower in colon epithelial cells isolated from patients with active UC (Figure 2B), we tested whether SLC7A5 and SLC3A2 transcripts might be upregulated in these cells. Excitingly, qRTPCR results demonstrated significantly higher transcript levels for both genes in colon epithelial cells from patients with active UC in comparison to cells from control patients or patients with inactive UC (Figure 3B). These results suggest that CAP-D3 is necessary to repress transcription of amino acid transporter genes immediately following bacterial infection.
Figure 3. Salmonella infection increases SLC7A5-SLC3A2 gene expression in CAP-D3 deficient cells.
A) qRT-PCR for transcript levels of SLC7A5 and SLC3A2, in Salmonella-infected HT-29 cells normalized to GAPDH transcript levels demonstrate increases in CAP-D3 shRNA expressing cells. B) qRT-PCR for transcript levels of SLC3A2 and SLC7A5 in colonic epithelial cells isolated from resected patient tissues demonstrate increases in patients with active UC compared to controls and patients with inactive UC. Transcript levels were normalized to villin transcript levels and then to the transcript levels found in a single control patient sample.
SLC7A5 localizes to the SCV earlier, and its activity is necessary for enhanced bacterial survival in CAP-D3 deficient cells
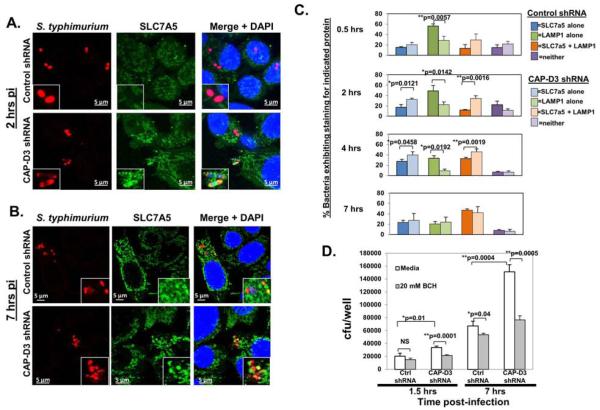
SLC7A5 and SLC3A2, together, comprise a bidirectional, heterodimeric, amino acid transporter for large neutral amino acids 31. SLC7A5-SLC3A2 is upregulated in response to stress with a subsequent positive regulation of the mTORC1 complex, which contains the mammalian target of rapamycin (mTOR) and is a known inhibitor of autophagy 32-35. Additionally, the activity of SLC7A5-SLC3A2 has been previously shown to be necessary for the ability of bacteria to escape autophagy 36. Therefore, to test whether the increased levels of SLC7A5-SLC3A2 observed in CAP-D3 deficient cells might result in the inhibition of autophagy, we first examined the localization pattern of SLC7A5 during Salmonella infection. Surprisingly, results showed that SLC7A5 appears to localize to bacteria containing vacuoles (Figure 4A and 4B). More bacteria containing vacuoles stained positive for SLC7A5 between 0.5 and 4 hours in CAP-D3 shRNA expressing cells compared to control shRNA expressing cells (Figure 4C). Following entry into a host cell, Salmonella can reside within an endosome or in the cytoplasm. The endosome housing the bacterium is called the Salmonella-containing vacuole or SCV, and, within 1-2 hours, acquires markers of a late endosome/lysosome (LEL), including LAMP1 14. To determine whether SLC7A5 was in fact localizing to bacteria-containing LELs, or intermediate SCVs, immunofluorescence assays were performed with antibodies to LAMP1 and SLC7A5. Results demonstrated a greater number of bacteria colocalizing with both SLC7A5 and LAMP1 in CAP-D3 shRNA expressing cells at earlier timepoints post-infection (Figure 4C, Supplementary Figure 4, and Supplementary Table 4). Conversely, the number of bacteria staining positive for LAMP1 alone was less in CAP-D3 deficient cells compared to controls. By 7 hours post-infection, there was no difference in SLC7A5/LAMP1 colocalization at bacterial cells between CAP-D3 deficient or control cells. Finally, to test whether the function of SLC7A5 was responsible for the enhanced bacterial survival in the CAP-D3 deficient cells, infected cells were treated with two different inhibitors of SLC7A5-SLC3A2 activity, 2-amino-2-norbornanecarboxylic acid (BCH) and D-phenylalanine. Both molecules have previously been shown to inhibit L-leucine uptake by SLC7A5-SLC3A2, and we verified that this is true for HT-29 cells, as well (Supplementary Figure 5A)34, 37-39. Gentamycin protection assays demonstrated increased numbers of viable Salmonella in CAP-D3 shRNA expressing cells at both 1.5 and 7 hours post-infection (Figure 4D). Excitingly, both BCH and D-phenylalanine treatment of these cells completely rescued the increased bacterial levels to those seen in the control shRNA expressing cells at both 1.5 and 7 hours post-infection (Figure 4D and Supplementary Figure 5B). BCH and D-phenylalanine treatment of control shRNA expressing cells had no significant effects on bacterial numbers at the 1.5 hour timepoint, but did decrease the numbers at the later timepoint. Taken together, these data suggest that 1) Salmonella infection of colon epithelial cells results in SLC7A5 localization to LELs/SCVs and 2) that the earlier localization and activity of SLC7A5 in CAPD3 deficient cells is required for the enhanced bacterial survival.
Figure 4. SLC7A5 localizes to Salmonella-containing vacuoles early in infection and its transporter activity is essential for enhanced bacterial survival in CAP-D3 deficient cells.
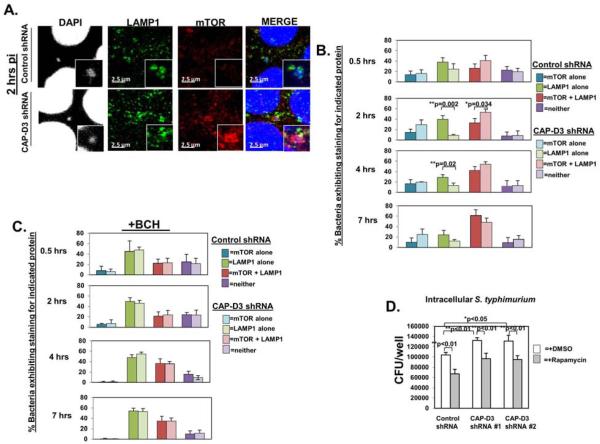
A-B) Immunofluorescence analysis of Salmonella (red) and SLC7A5 (green) and bacteria nuclei (DAPI-blue) in HT-29 cells demonstrates SLC7A5 localization to bacteria earlier (2 hours post-infection) in CAP-D3 deficient cells. C) Quantification of the percentage of SLC7A5+, LAMP1+, SLC7A5+/LAMP1+ Salmonella in HT-29 cell lines at various times during infection. D) The SLC7A5-SLC3A2 inhibitor, BCH, rescues the increased numbers of Salmonella in CAP-D3-deficient cells at 1.5h post-infection in gentamycin protection assays.
SLC7A5 is required for earlier mTORC1 localization to the SCV in CAP-D3 deficient cells
Upregulation of SLC7A5 expression has been shown to activate mTORC1 32. Increased amino acid levels lead to mTORC1 localization to the lysosome where it is subsequently activated and attenuates autophagy 40, 41. At early timepoints post-infection with Salmonella in HeLa cells, an amino acid starvation response was shown to occur, causing mTOR to localize away from LELs/SCVs and then return once amino acid levels had returned to normal 17. Recently, this relocalization of mTOR to the SCV was shown to be dependent on the activities of SLC7A5, SLC3A2 and SLC1A5 36. Given that our results showed SLC7A5 activity to be necessary for the enhanced bacterial survival in CAP-D3 shRNA expressing cells (Figure 4D), we tested whether SLC7A5 mediates mTOR localization to the LEL/intermediate SCV, leading to its activation and inhibition of autophagy. Immunofluorescence analysis of LAMP1 and mTOR colocalization to SCVs demonstrated more bacteria staining positive for both proteins, together, in CAPD3 shRNA expressing cells at 2 hours post-infection, as compared to control shRNA expressing cells (Figure 5A, 5B and Supplementary Table 5). In fact, the majority of LAMP1 positive SCVs in CAP-D3 shRNA expressing cells also stained for mTOR at this timepoint, whereas, this did not occur in the control shRNA expressing cells until 7 hours post-infection. Treatment of Salmonella infected control and CAPD3 shRNA expressing cells with BCH decreased numbers of bacteria staining positive for both mTOR and LAMP1 in the CAP-D3 shRNA expressing cells at 2 hours post-infection (Figure 5C and Supplementary Table 6). BCH treatment, which did not kill epithelial cells (Supplementary Figure 6) also increased numbers of SCVs staining positive for LAMP1 alone in both cell lines (Supplementary Table 6). Finally, treatment of Salmonella infected cells with an inhibitor of mTORC1 activity, rapamycin, behaved similarly to BCH treatment and reduced the enhanced bacterial numbers in CAP-D3 shRNA expressing cells to the levels seen in the control shRNA expressing cells at 1.5 hours post-infection (Figure 5D). These results suggest that the major underlying cause of enhanced bacterial survival in the CAP-D3 deficient cells is SLC7A5-mediated mTORC1 activation and attenuation of autophagy.
Figure 5. SLC7A5-dependent mTOR localization to Salmonella-containing vacuoles and mTORC1 activity are required for enhanced Salmonella survival in CAP-D3 deficient cells.
A) Immunofluorescence analysis of LAMP1 (green) and mTOR (red) and bacterial nuclei (DAPI-blue) in HT-29 cells at 2 hours post-infection demonstrates increased colocalization in CAP-D3 deficient cells. B) Quantification of the percentage of mTOR+, LAMP1+, mTOR+/LAMP1+ Salmonella in HT-29 cell lines various times post-infection. C) Quantification as in (B) but in cells treated with BCH. D) The mTORC1 inhibitor, rapamycin, rescues the increased numbers of Salmonella in HT-29 cell lines at 1.5h post-infection in gentamycin protection assays.
Discussion
The intestinal epithelium is an extremely important barrier separating the immense numbers of bacteria in the gut lumen from the underlying tissues and immune cells. Perturbations to the epithelium, including increased bacterial infiltration and survival in intestinal epithelial cells can result in excessive inflammation and are thought to play a major role in the pathogenesis of IBD. Our studies have uncovered a completely novel anti-microbial role for the CAP-D3 protein in human colon epithelial cells which may have implications for patients with IBD. Results described here show that in normal colonic epithelial cells, the majority of LELs housing Salmonella bacteria do not contain mTOR at their surface during the early hours post-infection, and are targeted for autophagy by the epithelial cell (Figure 6A). By 7 hours post-infection, mTOR has now localized to the late SCV, become activated (as part of mTORC1) and is able to block autophagy. This timecourse is very similar to what has been published for Salmonella infection of HeLa cells 17. Interestingly, immediately following infection of colonic epithelial cells deficient for CAP-D3, there is an increase in transcription of genes involved in amino acid transport, including SLC7A5 and SLC3A2, which complex to form a bidirectional, heterodimeric Leucine transporter. We observed that in this cell type, SLC7A5 localizes to SCVs, and its upregulation in CAP-D3 deficient cells results in a faster accumulation of SLC7A5 protein at the surface of the LEL/intermediate SCV. The activity of SLC7A5-SLC3A2 is required to increase localization of mTOR to the SCV, resulting in mTORC1 activation and less bacteria being targeted for autophagy in CAP-D3 deficient cells (Figure 6B).
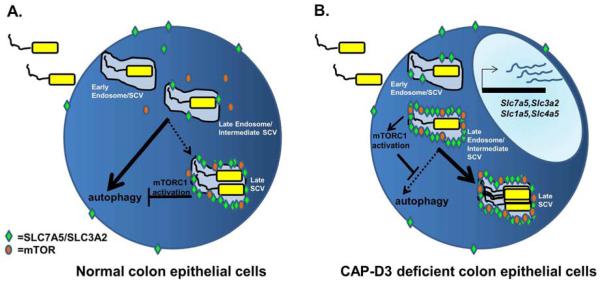
Figure 6. Possible model of how CAP-D3 deficiency leads to enhanced Salmonella survival.
A) In normal, colonic epithelial cells, Salmonella is endocytosed into an early endosome which becomes the early SCV (Salmonella-Containing Vacuole). The early endosome/SCV will acquire LAMP-1 (and other proteins) and become a Late Endosome/Lysosome/Intermediate SCV. The majority of Intermediate SCVs at this point will be targeted for autophagy since mTOR is not localized to the SCV and mTORC1 activity is low. A small number of Intermediate SCVs, however, will have begun to sequester existing SLC7a5 protein to their membranes and this could allow them to recruit mTOR, increase mTORC1 activity and block autophagy; this Late SCV would then escape destruction, and bacteria could replicate. In CAP-D3 deficient colonic epithelial cells, the burst of SLC7A5-SLC3A2 transcription occurring immediately after infection leads to increased levels of protein, allowing more Intermediate SCVs to be decorated with SLC7A5 earlier, thereby recruiting and activating mTORC1 more quickly and blocking autophagy sooner. This quicker version of events, leads to a larger number of SCVs escaping autophagy and enhances bacterial survival. This working model could also apply to other bacteria that invade epithelial cells and are typically cleared by autophagy, including AIEC LF82.
Current reports have associated the increased expression of SLC7A5-SLC3A2 with mTORC1 activation 42, 43. This amino acid sensing mechanism of mTORC1 activation takes place on lysosomal membranes44. Activation involves several factors, including plasma membrane uptake of Leucine43, the leucyltRNA synthetase (LRS)45 and the scaffolding protein p6234. The SLC7A5-SLC3A2 transporter is an obligate exchanger and it requires an intracellular amino acid (usually Glutamine) to be exchanged for the uptake of Leucine46. Therefore the coordinated expression of amino acid transporters can regulate amino acid flux that can modulate intracellular signaling, including mTORC1. Interestingly, expression of these proteins (SLC7A5, SLC3A2, LRS, p62) and several other amino acid transporters increases under conditions that induce the eIF2α/ATF4 transcriptional reprogramming of stressed cells46, 47. This reprogramming also includes expression of genes involved in increasing autophagy,48 thus further assisting the positive effects of amino acid uptake on mTORC1 signaling. The eIF2α/ATF4 signaling pathway is known to be involved in the development of IBD49, 50. However, the regulation of expression and function of amino acid transporters and their relationship to the regulation of autophagy in IBD and specifically in pathogen penetration, is not well understood. Interestingly, SLC3A2 (also known as CD98) expression increases during the progression of Dextran Sodium Sulfate (DSS) induced colitis in mice, and knockdown of SLC3A2 in murine intestinal epithelial cells provides resistance to DSS induced colitis51, 52. Considering that SLC3A2 interacts with many membrane proteins, including heterodimeric transporters, our studies introduce a novel concept in IBD pathology, the deregulation of amino acid uptake by heterodimeric transporters that require SLC3A2.
While mTORC1 activation on LELs has been previously described 41, our data showing the localization of SLC7A5 to SCVs has not been shown. Currently, we cannot definitively say whether activity of SLC7A5-SLC3A2 at the plasma membrane or at the LEL/SCV is responsible for the localization of mTORC1 to the LEL/SCV. Previous studies demonstrated that inhibition of clathrin-mediated endocytosis prevents mTOR localization to the SCV in HeLa cells, suggesting that intracellular vesicles do play a major role in the targeting of mTOR 36. The fact that a population of SLC7A5 protein localizes to SCVs which stain negative for LAMP1 at early timepoints post-infection argues that SLC7A5 localization is driven by bacterial infection and, in fact, a portion of the SLC3A2 extracellular domain has been shown to directly interact with Citrobacter rodentium and Enteropathogenic Escherichia coli (EPEC) proteins in in vitro surface plasmon resonance experiments 53. Future studies will determine if SLC7A5 localization at LELs is a more general nutrient sensing mechanism of mTORC1 activation or if it is uniquely linked to bacterial infection of colon epithelial cells.
The novel antibacterial function for CAP-D3 becomes extremely significant in light of our data showing that CAP-D3 levels are decreased in patients with active ulcerative colitis (Figure 2). The lower levels of CAP-D3 in the epithelium of these patients, in combination with another event such as a decrease in the protective mucus layer or change in the gut environment to favor expansion of a pathobiont may have contributed to the disease pathogenesis observed in these individuals. While no GWAS associations have been identified for CAP-D3 and IBD, it is important to remember that CAP-D3 is part of a multi-subunit complex, the Condensin II complex, with essential functions during the cell cycle. The phosphorylation of CAP-D3 has been shown to be necessary for the mitotic-specific functions of the Condensin II complex in ensuring efficient cellular division, and it therefore stands to reason that null, germline mutations in CAP-D3 would most likely result in early lethality 54. Whether or not the involvement of CAP-D3 in bacterial clearance is dependent on the Condensin II holocomplex requires further study. The manner by which CAP-D3 regulates transcription of amino acid transporter genes is also still under investigation; ChIP-sequencing of CAP-D3 binding sites will be required to determine if and how CAP-D3 binds to amino acid transporter gene loci. It is clear, however that the antimicrobial effects of CAP-D3 may be broad in nature, since our data shows that CAP-D3 promotes clearance of not only Salmonella, but AIEC LF82, as well. Studies involving other types of bacteria, including Gram positive bacteria as well as commensal organisms whose numbers have been shown to be greater in patients with IBD will help to answer this question.
Our studies have a number of clinical applications for patients with IBD. It is possible that by increasing the levels of CAP-D3 in the colon epithelium of patients with inactive UC, we could keep the numbers of invasive bacterial species at a minimum and possibly prevent associated immune signaling and flare ups. Additionally, the loss of Condensin II subunits, including CAP-D3, in cells of many different organisms has also been shown to result in lagging chromosomes, chromosome breaks and a subsequent lag in mitosis. This, combined with the fact that CAP-D3 binds to the classic tumor suppressor, pRB, suggests that CAP-D3 may act as a tumor suppressor, itself 28. Therefore, lower levels of CAP-D3 in the colon epithelium of patients with UC may not only lead to increased bacterial survival, but might also predispose a patient to genomic instability and subsequent tumor development. Currently, we do not know what regulates the levels of CAP-D3 expression, but this is now the focus of intense study and warrants investigations of CAP-D3 expression in mouse models of IBD, as well.
Supplementary Material
Supplementary Table 1. Information on de-identified patient tissues from which epithelial cells were isolated.
Supplemental Table 2. Analysis of differentially expressed genes in uninfected control shRNA and CAP-D3 shRNA expressing HT-29 cells.
Supplemental Table 3a. Analysis of genes that are differentially expressed in control shRNA expressing HT-29 cells as a result of infection.
Supplemental Table 3b. Analysis of genes that are differentially expressed in CAP-D3 shRNA expressing HT-29 cells as a result of infection.
Supplemental Table 4: Average percentage of bacteria/SCVs which stain positive for LAMP-1 and/or SLC7A5 over the infection timecourse in control and CAP-D3 shRNA
Supplemental Table 5: Average percentage of bacteria/SCVs which stain positive for LAMP-1 and/or mTOR over the infection timecourse in control and CAP-D3 shRNA
Supplemental Table 6: Average percentage of bacteria/SCVs which stain positive for LAMP-1 and/or mTOR over the infection timecourse in control and CAP-D3 shRNA + BCH
Acknowledgments
The authors thank the members of the Cleveland Clinic IBD club for their constructive comments and scientific input; John Peterson, Judy Drazba, Denise Hatala and Diane Mahovlic in the Lerner Research Institute Imaging Core for assistance with preparation of paraffin-embedded slides and in imaging experiments; Pieter Faber and the Genomics Core at the University of Chicago for generation of cDNA libraries and for sequencing; Madeleine Lemieux at Bioinfo.ca for analysis of RNA-sequencing data; Michael Weiner in the Lerner Research Institute Computing Services for assistance with packaging and transferring of data files; Eboni Ubinas and Tiffany Hollo in Tissue Procurement Services at the Cleveland Clinic for sample collection and de-identification; Dr. Deepa Patil in the Anatomical Pathology Department at the Cleveland Clinic for her expert opinions and initial consultation.
Grant support: This work was supported by a Chairman’s Innovative Research Award from the Lerner Research Institute (to MSL), National Institutes of Health research grants R01GM102400 (to MSL), R01DK082437 (to C.M.), R01DK050984 to C.F.), R01DK053307 and R37DK060596 (to MH), Department of Defense Peer Reviewed Medical Research Program grant PR110887 (to C.M.), and two separate grants from the Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the NIH and NIH roadmap for Medical Research (to MSL and to C.M). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: C.M. has received honoraria from Vertex Pharmaceuticals (Canada) Incorporated. The other authors have nothing to disclose.
Transcript Profiling: Raw data files from RNA-sequencing experiments have been deposited at the NCBI GEO repository under the data accession number GSE62520. Analyzed data files of all reads from all experiments are currently included in the manuscript as supplementary tables.
Author Contributions: ATS, CRH, CM, and MSL were responsible for study concepts and design. ATS, CRH, JRK, KPN, GW, TS, ES and MSL were responsible for acquisition of data. ATS, CRH, JRK, KPN, ED, YK, DK, MH, CD, BPR, CF, CM, and MSL were responsible for analysis and interpretation of data. ATS, CRH, MH, CF, CM, and MSL were responsible for drafting and revision of the manuscript.
References
- 1.Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med. 2011;365:1713–25. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 2.Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 3.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson ME, Gustafsson JK, Holmen-Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–91. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van der Sluis M, De Koning BA, De Bruijn AC, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–29. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Schultsz C, Van Den, Berg FM, Ten Kate FW, et al. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117:1089–97. doi: 10.1016/s0016-5085(99)70393-8. [DOI] [PubMed] [Google Scholar]
- 8.Swidsinski A, Loening-Baucke V, Herber A. Mucosal flora in Crohn’s disease and ulcerative colitis - an overview. J Physiol Pharmacol. 2009;60(Suppl 6):61–71. [PubMed] [Google Scholar]
- 9.Kleessen B, Kroesen AJ, Buhr HJ, et al. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034–41. doi: 10.1080/003655202320378220. [DOI] [PubMed] [Google Scholar]
- 10.Gradel KO, Nielsen HL, Schonheyder HC, et al. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137:495–501. doi: 10.1053/j.gastro.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Ternhag A, Torner A, Svensson A, et al. Short- and long-term effects of bacterial gastrointestinal infections. Emerg Infect Dis. 2008;14:143–8. doi: 10.3201/eid1401.070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–49. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamin JL, Sumpter R, Jr., Levine B, et al. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–34. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos-Morales F. Impact of Salmonella enterica Type III Secretion System Effectors on the Eukaryotic Host Cell. ISRN Cell Biology. 2012;2012:36. [Google Scholar]
- 15.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–9. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 16.Thurston TL, Wandel MP, von Muhlinen N, et al. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–8. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tattoli I, Sorbara MT, Vuckovic D, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–75. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–25. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 19.Lapaquette P, Glasser AL, Huett A, et al. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longworth MS, Walker JA, Anderssen E, et al. A shared role for RBF1 and dCAP-D3 in the regulation of transcription with consequences for innate immunity. PLoS Genet. 2012;8:e1002618. doi: 10.1371/journal.pgen.1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coschi CH, Ishak CA, Gallo D, et al. Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 2014;4:840–53. doi: 10.1158/2159-8290.CD-14-0215. [DOI] [PubMed] [Google Scholar]
- 22.Ono T, Yamashita D, Hirano T. Condensin II initiates sister chromatid resolution during S phase. J Cell Biol. 2013;200:429–41. doi: 10.1083/jcb.201208008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossmann J, Maxson JM, Whitacre CM, et al. New isolation technique to study apoptosis in human intestinal epithelial cells. Am J Pathol. 1998;153:53–62. doi: 10.1016/S0002-9440(10)65545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geboes K, Riddell R, Ost A, et al. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut. 2000;47:404–9. doi: 10.1136/gut.47.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foell D, Kucharzik T, Kraft M, et al. Neutrophil derived human S100A12 (EN-RAGE) is strongly expressed during chronic active inflammatory bowel disease. Gut. 2003;52:847–53. doi: 10.1136/gut.52.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homer CR, Kabi A, Marina-Garcia N, et al. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J Biol Chem. 2012;287:25565–76. doi: 10.1074/jbc.M111.326835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickerson KP, McDonald C. Crohn’s disease-associated adherent-invasive Escherichia coli adhesion is enhanced by exposure to the ubiquitous dietary polysaccharide maltodextrin. PLoS One. 2012;7:e52132. doi: 10.1371/journal.pone.0052132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longworth MS, Herr A, Ji JY, et al. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22:1011–24. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannemann S, Gao B, Galan JE. Salmonella modulation of host cell gene expression promotes its intracellular growth. PLoS Pathog. 2013;9:e1003668. doi: 10.1371/journal.ppat.1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruno VM, Hannemann S, Lara-Tejero M, et al. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor PM. Role of amino acid transporters in amino acid sensing. Am J Clin Nutr. 2014;99:223S–230S. doi: 10.3945/ajcn.113.070086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R, Zou Y, Mao D, et al. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J Cell Biol. 2014;206:173–82. doi: 10.1083/jcb.201403009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elorza A, Soro-Arnaiz I, Melendez-Rodriguez F, et al. HIF2alpha acts as an mTORC1 activator through the amino acid carrier SLC7A5. Mol Cell. 2012;48:681–91. doi: 10.1016/j.molcel.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 34.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18:524–33. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tattoli I, Philpott DJ, Girardin SE. The bacterial and cellular determinants controlling the recruitment of mTOR to the Salmonella-containing vacuole. Biol Open. 2012;1:1215–25. doi: 10.1242/bio.20122840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanai Y, Segawa H, Miyamoto K, et al. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98) J Biol Chem. 1998;273:23629–32. doi: 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- 38.Mastroberardino L, Spindler B, Pfeiffer R, et al. Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature. 1998;395:288–91. doi: 10.1038/26246. [DOI] [PubMed] [Google Scholar]
- 39.Sloan JL, Mager S. Cloning and functional expression of a human Na(+) and Cl(−)-dependent neutral and cationic amino acid transporter B(0+) J Biol Chem. 1999;274:23740–5. doi: 10.1074/jbc.274.34.23740. [DOI] [PubMed] [Google Scholar]
- 40.Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan X, Ross DD, Arakawa H, et al. Impact of system L amino acid transporter 1 (LAT1) on proliferation of human ovarian cancer cells: a possible target for combination therapy with anti-proliferative aminopeptidase inhibitors. Biochem Pharmacol. 2010;80:811–8. doi: 10.1016/j.bcp.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 43.Guan BJ, Krokowski D, Majumder M, et al. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem. 2014;289:12593–611. doi: 10.1074/jbc.M113.543215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng X, Liang Y, He Q, et al. Current models of mammalian target of rapamycin complex 1 (mTORC1) activation by growth factors and amino acids. Int J Mol Sci. 2014;15:20753–69. doi: 10.3390/ijms151120753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–24. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 46.Krokowski D, Han J, Saikia M, et al. A self-defeating anabolic program leads to beta-cell apoptosis in endoplasmic reticulum stress-induced diabetes via regulation of amino acid flux. J Biol Chem. 2013;288:17202–13. doi: 10.1074/jbc.M113.466920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J, Back SH, Hur J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–90. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.B’Chir W, Maurin AC, Carraro V, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–99. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huett A, Xavier RJ. Autophagy at the gut interface: mucosal responses to stress and the consequences for inflammatory bowel diseases. Inflamm Bowel Dis. 2010;16:152–74. doi: 10.1002/ibd.20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaser A, Adolph TE, Blumberg RS. The unfolded protein response and gastrointestinal disease. Semin Immunopathol. 2013;35:307–19. doi: 10.1007/s00281-013-0377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen HT, Dalmasso G, Torkvist L, et al. CD98 expression modulates intestinal homeostasis, inflammation, and colitis-associated cancer in mice. J Clin Invest. 2011;121:1733–47. doi: 10.1172/JCI44631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kucharzik T, Lugering A, Yan Y, et al. Activation of epithelial CD98 glycoprotein perpetuates colonic inflammation. Lab Invest. 2005;85:932–41. doi: 10.1038/labinvest.3700289. [DOI] [PubMed] [Google Scholar]
- 53.Charania MA, Laroui H, Liu H, et al. Intestinal epithelial CD98 directly modulates the innate host response to enteric bacterial pathogens. Infect Immun. 2013;81:923–34. doi: 10.1128/IAI.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abe S, Nagasaka K, Hirayama Y, et al. The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 2011;25:863–74. doi: 10.1101/gad.2016411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Information on de-identified patient tissues from which epithelial cells were isolated.
Supplemental Table 2. Analysis of differentially expressed genes in uninfected control shRNA and CAP-D3 shRNA expressing HT-29 cells.
Supplemental Table 3a. Analysis of genes that are differentially expressed in control shRNA expressing HT-29 cells as a result of infection.
Supplemental Table 3b. Analysis of genes that are differentially expressed in CAP-D3 shRNA expressing HT-29 cells as a result of infection.
Supplemental Table 4: Average percentage of bacteria/SCVs which stain positive for LAMP-1 and/or SLC7A5 over the infection timecourse in control and CAP-D3 shRNA
Supplemental Table 5: Average percentage of bacteria/SCVs which stain positive for LAMP-1 and/or mTOR over the infection timecourse in control and CAP-D3 shRNA
Supplemental Table 6: Average percentage of bacteria/SCVs which stain positive for LAMP-1 and/or mTOR over the infection timecourse in control and CAP-D3 shRNA + BCH