Abstract
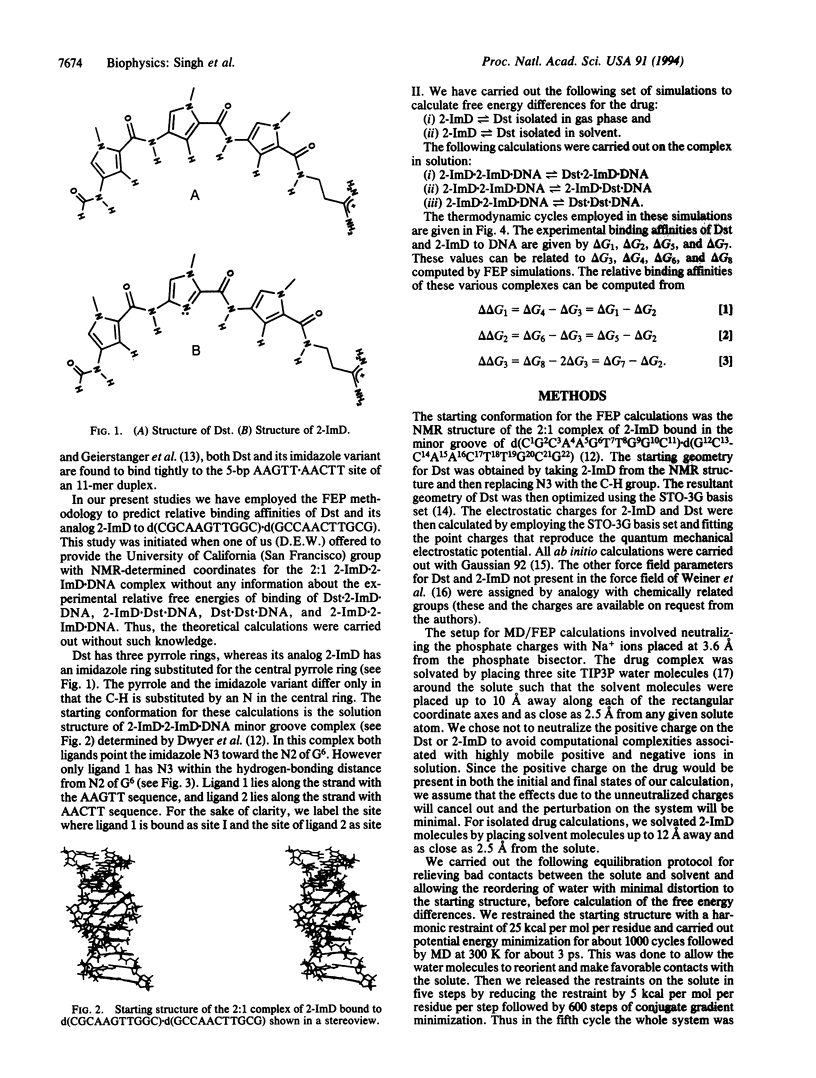
We report here an effort to use molecular dynamics/free energy perturbation methodology to calculate relative binding affinities of two related drugs to DNA. Specifically, we focus on the relative binding free energies of distamycin (Dst) and its analog, 2-imidazoledistamycin (2-ImD), to d(CGCAAGTTGGC).d(GCCAACTTGCG). The pyrrole (Dst) and the imidazole variant (2-ImD) differ only in that the C-H is substituted by an N in the central ring. The starting conformation for these calculations was the previously determined solution structure of two 2-ImD molecules in the minor groove of the above 11-residue DNA. In this complex both the ligands have the imidazole nitrogen (N3) oriented toward the amino group of G6. However only ligand 1 (site I) has N3 within the hydrogen bonding distance from N2 amino proton of G6. We have calculated the difference in free energy of binding of 2-ImD versus Dst in three different cases by mutating 2-ImD-->Dst reversibly. In the first case ligand 1 (site I) is mutated, in the second case ligand 2 (site II) is mutated, and in the third case both the ligands are mutated. These calculations show that at site I Dst has weaker binding affinity than 2-ImD by 0.7 kcal/mol, at site II Dst has stronger binding affinity than 2-ImD by 2.9 kcal/mol, and when occupying both site I and site II, Dst binds with greater affinity than 2-ImD by 1.8 kcal/mol. Recent experimental findings agree semiquantitatively (within 1 kcal/mol) with the calculations presented here. Hence the methodology presented here can be used to predict relative binding energies of two or more closely related molecules to DNA.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bash P. A., Singh U. C., Brown F. K., Langridge R., Kollman P. A. Calculation of the relative change in binding free energy of a protein-inhibitor complex. Science. 1987 Jan 30;235(4788):574–576. doi: 10.1126/science.3810157. [DOI] [PubMed] [Google Scholar]
- Boehncke K., Nonella M., Schulten K., Wang A. H. Molecular dynamics investigation of the interaction between DNA and distamycin. Biochemistry. 1991 Jun 4;30(22):5465–5475. doi: 10.1021/bi00236a020. [DOI] [PubMed] [Google Scholar]
- Cieplak P., Rao S. N., Grootenhuis P. D., Kollman P. A. Free energy calculation on base specificity of drug--DNA interactions: application to daunomycin and acridine intercalation into DNA. Biopolymers. 1990 Mar-Apr;29(4-5):717–727. doi: 10.1002/bip.360290406. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton S., Rudolph B., Lybrand T., Singh U. C., Shafer R., Brown S., Kollman P., Case D. A., Andrea T. A combined 2D-NMR and molecular dynamics analysis of the structure of the actinomycin D: d(ATGCAT)2 complex. J Biomol Struct Dyn. 1989 Apr;6(5):929–969. doi: 10.1080/07391102.1989.10506524. [DOI] [PubMed] [Google Scholar]
- Ferguson D. M., Radmer R. J., Kollman P. A. Determination of the relative binding free energies of peptide inhibitors to the HIV-1 protease. J Med Chem. 1991 Aug;34(8):2654–2659. doi: 10.1021/jm00112a048. [DOI] [PubMed] [Google Scholar]
- Feuerstein B. G., Pattabiraman N., Marton L. J. Molecular dynamics of spermine-DNA interactions: sequence specificity and DNA bending for a simple ligand. Nucleic Acids Res. 1989 Sep 12;17(17):6883–6892. doi: 10.1093/nar/17.17.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago F., Richards W. G. Netropsin binding to poly[d(IC)].poly[IC)] and poly[d(GC].poly[d(GC)]: a computer simulation. Mol Pharmacol. 1990 Mar;37(3):341–346. [PubMed] [Google Scholar]


