Abstract
Mitochondrial function is paramount to energy homeostasis, metabolism, signaling, and apoptosis in cells. Mitochondrial complex V (ATP synthase), a molecular motor, is the ultimate ATP generator and a key determinant of mitochondrial function. ATP synthase catalyzes the final coupling step of oxidative phosphorylation to supply energy in the form of ATP. Alterations at this step will crucially impact mitochondrial respiration and hence cardiac performance. It is well established that cardiac contractility is strongly dependent on the mitochondria, and that myocardial ATP depletion is a key feature of heart failure. ATP synthase dysfunction can cause and exacerbate human diseases, such as cardiomyopathy and heart failure. While ATP synthase has been extensively studied, essential questions related to how the regulation of ATP synthase determines energy metabolism in the heart linger and therapies targeting this important mechanism remain scarce. This review will visit the main findings, identify unsolved issues and provide insights into potential future perspectives related to the regulation of ATP synthase and cardiac pathophysiology.
Keywords: ATP synthase, mitochondria, cardiac hypertrophy, heart failure, energy metabolism, mPTP
Introduction
Mitochondrial complex V or ATP synthase is an enzyme complex that works as a molecular machine to generate and hydrolyze ATP in cells in the last step of the mitochondrial respiratory process. Therefore, ATP synthase plays pivotal roles not only in maintaining the cellular energy state, but also in determining mitochondrial respiratory function. Cardiac contraction and relaxation are energy demanding processes that depend on mitochondrial function and efficient ATP production/reservation. Dysregulation of ATP synthase activity should have major impacts on mitochondrial respiration and hence cardiac performance. Mitochondrial energy disturbances are involved in cardiac pathological development [1]. For example, myocardial ATP depletion is a key issue of heart failure [1-3]. Therefore, further research on the regulation of the mitochondrial ATP synthase in the heart may help discover novel therapeutic strategies for the treatment of cardiac disorders. This review will discuss the current knowledge of ATP synthase regulation in the heart, the potential challenges in the field, and the potential perspectives on the translational potential of the related research.
ATP synthase: a molecular machine that makes ATP
ATP synthase, also known as F1F0-ATP synthase or complex V, is the key energy generator for most life forms on earth. This large, mitochondrial protein complex is bound to the inner mitochondrial membrane along with the other respiratory chain complexes I-IV. The enzyme functions through a reversible rotary complex, whereby the direction of its rotation determines the synthesis or hydrolysis of ATP. In order to generate ATP from ADP and Pi, ATP synthase rotates using the driving force of an electrochemical potential built up in the intermembrane space by the I-IV respiratory chain complexes (see review [4,5]. Conversely, ATP synthase can spin in the reverse direction and hydrolyze ATP to pump protons back to the intermembrane space to maintain membrane potential. ATP synthase consists of two distinct subcomplexes with complementary functions. The Fo complex contains transmembrane subunits that transport protons from the intermembrane space and the F1 is a peripheral complex in the matrix, which catalyzes nucleotide binding for ATP synthesis [6-9]. F0 and F1 are connected through two stalk-like subunits: a central rotor shaft and a peripheral stator [6]. In the mammalian mitochondrial enzyme, F1 is composed of three copies each of subunits α and β, and one each of subunits γ, δ and ε. F0 consists of a subunit c ring (comprising 12 copies) and one copy each of subunits a, b, d, h (F6) and the O subunit or oligomycin sensitivity conferring protein (OSCP). A number of additional subunits (e, f, g, i/j, k and A6L) are associated with F0, although their precise locations within the complex remain unknown [10-13]. Protons accumulated in the intermembrane space are driven through a channel in F0, which causes rotation of the c-ring along with the attached central stalk. Subsequently, rotation of subunit γ, within the F1-α3β3 hexamer provides energy for ATP synthesis at the catalytic sites (located in each of the three β subunits, at the interface with an adjacent α subunit) [14]. The rotation of the F1 hexamer (α3β3) may enable the Interconversion of the states relative to the γ-subunit [6].
Despite the accumulation of this detailed knowledge of ATP synthase constituents in the past two decades, the roles of ATP synthase associated proteins in this crucial enzyme complex remain understudied, especially in the in vivo context. So far, most of the genetic investigations related to in the vitro function of ATP synthase have been conducted on either cultured cells or yeast. The exciting development in molecular genetics that enable genetic manipulations to be done with relative ease will open new doors for the further in vivo study of the function and regulation of this most important enzyme complex in our body.
The regulation of ATP synthase activity
The in-depth understanding of how the ATP synthase works to generate and hydrolyze ATP is well known, but the underlying mechanism of how ATP synthase is regulated remains obscure. Current literatures propose multi-levels of regulating mechanisms for ATP synthase activity, which primarily rely on elements directly involved in its operation. Among these elements are ADP, Mg2+, Pi, ATP, and others, such as anions [15-17]. The heart is known to be an energy-demanding organ for contraction/relaxation and ion transport [18], but the mechanisms that enable it to alter rapidly the ATP level to meet the fluctuating demand remain unclear. In general, the mitochondrial ATP synthase is regulated to maintain its steady and dynamic states of capacity.
Steady state regulation
Given that the ATP synthase plays such a pivotal role in cellular function, it is essential for maintaining the constitutive expression of key components of this enzyme complex. Based on current literatures, it appears that transcriptional, post-transcriptional, and protein assembling regulations determine the steady state of the ATP synthase activity (Figure 1).
Figure 1.
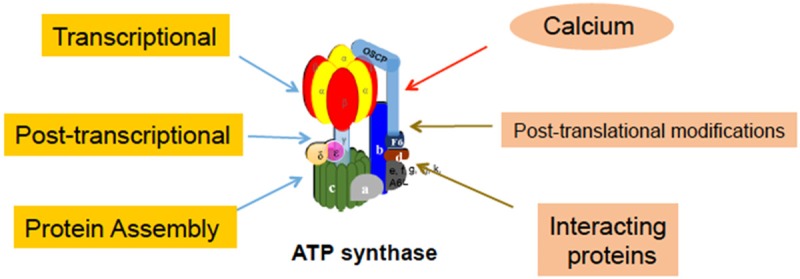
Summary of ATP synthase activity regulation. The steady state of mitochondrial ATP synthase activity is regulated at the transcriptional, post-transcriptional and protein assembly levels and the dynamic state of mitochondrial ATP synthase activity is regulated by calcium transient, post-translational modifications and interacting proteins.
Transcriptional regulation
Transcriptional regulation of metabolisms is essential in controlling the rate of metabolism in response to various physiological and pathological cues. Transcription factors, such as many nuclear receptors, are among the key transcriptional regulators of metabolic pathways [19]. Because genes for enzymes of oxidative phosphorylation are thought to be housekeeping genes that are transcribed constitutively [20], the transcriptional regulation of the component proteins of ATP synthase has been limited. In general, changes of ATP synthase content or activity appear to occur preferentially at the protein levels. However, mutations that mostly lead to the deficiency of the enzyme have been identified on genes encoding ATP synthase component proteins. Clinical cases with nuclear genetic defects of mitochondrial ATP synthase, such as the mitochondrial DNA ATP6 and the nuclear ATP12 genes [21,22], have been reported. They are characterized by early onset, lactic acidosis, 3-methylglutaconic aciduria, hypertrophic cardiomyopathy, and encephalopathy, followed by premature death [23]. On the other hand, it appears that the transcripts of ATP synthase components could be regulated by common transcription factors, such as peroxisome proliferator activator receptor δ (PPARδ) [24,25] and estrogen related receptors (ERRs) [26]. Such regulation could lead to the co-activation of peroxisome proliferator activator receptor γ [26] as part of the overall metabolic responses under different circumstances. Intuitively, ATP synthase transcripts are expressed at different tissue-specific levels with higher levels found in skeletal muscle and heart and lower levels in other tissues [27]. Specifically, in vitro studies have demonstrated that the transcriptional expression of ATP synthase components is controlled by various transcriptional regulation factors. For example, ATP factor 1 (ATPF1), which is present in human HeLa nuclei, plays a critical role in transcriptional activation of the α subunit of the ATP synthase [28]. The same group further illustrated that upstream stimulatory factor 2 (USF2) [29-31] and the transcription factor Yin Yang 1 (YY1) promotes transcription expression of the α subunit [32]. To further exemplify, it has been shown that hypoxia suppresses the transcript expression of the subunit e of ATP synthase. Therefore, ATP synthase could also be regulated at the transcriptional level by oxygen availability [33].
Because of the general lack of in vivo information, the significance of transcriptional regulation of the mitochondrial ATP synthase on the development of myocardial pathophysiology is not clear. However, the human cases of ATP synthase deficiency have clearly manifested how crucial it is to maintain an optimal level of ATP synthase in the body, especially for preserving the normal function of the heart.
Post-transcriptional regulation
The ATP synthase is also regulated at the post-transcriptional level by controlling translation of the enzyme complex. For instance, the expression of its catalytic subunit (β subunit) is stringently controlled at post-transcriptional levels. Micro-RNA plays an important role in regulating the translation of the β subunit. Willers IM et al showed that miR-127-5p represses the β subunit translation by inhibiting the 3’UTR of the β subunit mRNA of human ATP synthase [34].
The regulation of protein assembly
The assembly of F1 hexamer structure requires two specialized chaperones, Atp11p and Atp12p in yeast [35], which bind transiently to the α and β subunits. In the absence of Atp11p and Atp12p, the hexamer is not formed, and the α and β subunits precipitate as large insoluble aggregates [35-38]. This appears to be the case in humans too [39]. Mutants lacking the α and β subunits of F (1), or the Atp11p and Atp12p chaperones that promote F (1) assembly, have normal levels of the bicistronic ATP8/ATP6 mRNAs, but fail to synthesize Atp6p and Atp8p [40]. Another recent study showed that the INA complex facilitates assembly of the peripheral stalk of the mitochondrial F1F0-ATP synthase [41]. Additionally, it has been shown that OSCP plays a key role in the biogenesis of ATP synthase [42]. It is likely that p53 interacts with OSCP to facilitate the assembly and stabilization the ATP synthase complex [43].
Dynamic regulation
Because changes in cellular energy demand occur instantly and fluctuate rapidly, flux through mitochondrial ATP synthase must also change to maintain cellular ATP levels. Direct regulations at the level of ATP synthase appear to occur in mitochondria of these cells. ATP synthase activity increases in rat cardiomyocytes subjected to high-energy demand (beating, positive inotropic substances) and decreases in anoxic cells [44-46]. Several dynamic regulatory elements have been shown to act at the level of the ATP synthase (Figure 1).
Regulation of ATP synthase activity by mitochondrial calcium (Ca2+)
Mitochondrial Ca2+ transients occur during the contractile/relaxation cycle and are translated into overall rise in mitochondrial ATP production to keep pace with the functional demand [47]. Therefore, mitochondrial Ca2+ plays crucial roles in the regulation of the ATP synthase activity. However, the molecular mechanisms underpinning the direct regulation of calcium on the ATP synthase remain obscure. Early studies based on purified protein showed Ca2+ might regulate the ATP synthase via a protein named calcium binding ATPase inhibitor (CaBI) [15,16,24]. CaBI is reported to be a 6.3 kD protein, which interacts with ATP synthase in a Ca2+ dependent manner [48,49]. In vitro protein treatment of purified CaBI on extracted ATP synthase promotes ATP synthesis and inhibits ATP hydrolysis [49]. Neverthless, as of today, very little is known as no specific gene that encodes for this elusive protein has been identified. Recently, a report showed that the β subunit of the ATP synthase binds Ca2+, but with unknown effects [50]. In addition, Protein kinase Cδ, which is a Ca2+ signaling protein, can regulate ATP synthase by binding to the d subunit of the F0 sector [51,52]. It has also been reported that S100A1 is an F1 interacting protein in the mitochondria and promotes ATP synthesis in a Ca2+ -dependent manner [53]. Despite these advancements, our understanding of how mitochondrial Ca2+ directly regulates the ATP synthase activity remain poor, due partly to the technical difficulties involved in measuring acute alteration of ATP contents in different cellular compartments, especially in the mitochondria. Recent developments using fluorescent markers for the real time measurement of mitochondrial Ca2+ in cultured cells [54] provide new tools for the field to gain better insight toward solving at least part of the puzzle.
Regulation of ATP synthase activity by post-translational modifications
Not surprisingly, posttranslational modifications (PTM) of ATP synthase play important roles in the regulation of ATP synthase activity. Evidence of direct regulations by post-translational modifications on key subunits of ATP synthase has been surprisingly limited [55-58]. However, more research has been emerging, including reports on several post-translational modifications on various subunits of the ATP synthase complex (see review [59]). Human ATP synthase β is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle [57]. Further, the β-subunit of the ATP synthase is phosphorylated following myocardial preconditioning in rabbit myocytes [60]. In a study using a 32P γ subunit labeling strategy, Hopper et al observed that the γ subunit of ATP synthase subunit was phosphorylated [61]. Additionally, Ko et al employed Phospho-tyrosine antibodies to confirm that the platelet-derived growth factor (PDGF) induced phosphorylation of the δ subunit [62]. This group also used 32P labeling to show that the δ subunit could be differentially phosphorylated in vitro by mitochondrial extracts that had been isolated from either untreated NIH3T3 cells or from PDGF-treated NIH3T3 cells. However, further studies are required to understand how phosphorylation of a specific subunit of ATP synthase alters its function.
Using a proteomic approach, Wang et al uncovered several oxidative stress-related protein modifications occurring on ATP synthase in failing dyssynchronous hearts, which can be corrected by the cardiac resynchronization therapy (CRT), a clinically effective treatment for failing dyssynchronous hearts [63]. Multiple oxidative posttranslational modifications occur at a selective Cysteine in ATP synthase α subunit, which may act as a redox sensor modulating ATP synthase function [63]. The role of oxidative posttranslational modifications in the regulation of the ATP synthase complex is most frequently discussed in the context of heart failure and its possible clinical treatment [64]. Most recently, epigenetic regulation of ATP synthase has also been uncovered. Several subunits of the ATP synthase complex contain lysine modifications, including methylation and acetylation. For example, SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress [65]. However, it remains unknown if these posttranslational modifications also occur in cardiomyocytes and the in vivo context. Further investigations are needed to explore the translational potential of targeting the post-translational modification of ATP synthase.
Regulation of ATP synthase activity by its interacting proteins
Some proteins that are associated with ATP synthase, but not considered to be subunits, are also often involved in the regulation of mitochondrial ATP synthase. Table 1 has summarized the ATP synthase interacting proteins in the literatures and their potential function in controlling the enzyme’s activity.
Table 1.
Interacting proteins of the mitochondrial ATP Synthase
| Interactions | Functions | Citations | |
|---|---|---|---|
| IF1 | F1α, F1β | ATP synthase dimer formation | [100,117] |
| Cyclophilin D | OSCP, subunit d, and subunit b | mPTP | [88,118,119] |
| Bcl-xL | F1α, F1β | Membrane potential, mPTP and apoptosis | [91,120] |
| p53 | OSCP | Apoptosis, mPTP | [43,121,122] |
| S100A1 | F1α, F1β | Increase of ATP synthase activity | [53] |
| Factor B | F1α, OSCP | Component for ATP synthase complex formation | [82,123] |
| Strap | F1β | Modulator for cellular energy metabolism | [124] |
| PKCδ | subunit d | Inhibited ATP synthase activity | [86,87] |
Shown is a list of well-characterized interacting proteins of the mitochondrial ATP synthase along with information of the corresponding interacting subunits of ATP synthase and the related functions.
About six decades ago, inhibitor factor 1 (IF1) was identified to be the first nuclear-encoded ATP-synthase interacting protein [66-68]. IF1 is an evolutionarily well conserved mitochondrial protein that interacts with the F1 sector of ATP synthase and is not considered a subunit of the mitochondrial ATP synthase [69,70]. Numerous studies confirmed that IF1 inhibits the ATP hydrolysis activity of the mitochondrial ATP-synthase [66,69-71]. Interestingly, IF1 is activated under acidic conditions, such as in myocardial ischemia [72,73]. ATP hydrolysis occurs when the electrochemical proton gradient across the mitochondrial inner membrance is lost (e.g., during hypoxic/ischemic conditions), and the enzyme reverses in an attempt to restore mitochondrial membrane potential [74,75]. Therefore, IF1 is a potential drug target for enhancing ATP reserves in the heart [69,70]. In fact, preclinical assessments on IF-1 mimetic compounds have shown promising results in animal studies [73,76].
Most of the early knowledge of IF1 is based on studies on bovine heart mitochondria and has shown that IF1 can respond rapidly to the energy state of the mitochondrial membrane [18,77]. IF1 interacts with the ATP synthase in mitochondria of many species, including the rat, even though its inhibitory function on ATP hydrolysis at least in the heart seems less effective in small animals than in large animals [78-80]. This conclusion appears to reduce the enthusiasm for studying IF1 in genetically manipulated mouse models. This view may hold some true at least in terms of IF1’s cardiac role. A recent study on an IF1 knockout mouse model showed no basal phenotype, although ATP hydrolysis was elevated at least in mitochondrial samples extracted from liver [81]. Whether ATPase activity is affected in the IF1 knockout heart remains unknown. As stated previously, IF1 is activated under acidic conditions, so further studies are warranted to test if the loss of IF1 in mice under myocardial ischemia would lose the capacity to prevent accelerated ATP depletion.
Other ATP synthase interacting proteins have been identified: factor B [82-85], which is essential for ATP synthesis and involved in the regulation of ATP synthase oligomerization; CaBI [48], which may upregulate the enzyme activity in response to higher levels of cytoplasmic Ca2+, and finally S100A1 [53], which improves catalytic efficiency in cardiac muscle. In recent years, a few more proteins have been shown to interact with and regulate ATP synthase. Nguyen et al showed that Protein kinase Cδ interacts with the d subunit of the F0 sector and inhibits ATPase activity [52,86,87]. It has also been reported that cyclophilin D, a member of the cyclophilin family of chaperones, can constitutively bind ATP synthase, thus slowing ATP synthesis and hydrolysis rates through interaction with the lateral stalk [88-90]. Additionally, recent studies have shown that the Bcl II family member, Bcl-xL, interacts with ATP synthase to inhibit ATPase activity in neurons [91]. Surprisingly, it was discovered that the tumor suppressor protein p53 is localized to the mitochondria and interacts with ATP synthase by binding to OSCP [43]. Furthermore, Stress-responsive activator of p300 (Strap), a p53 cofactor, interacts with the β subunit of ATP synthase to inhibit ATP synthase activity with similar potency as oligomycin and to induce apoptosis [92].
It has long been suggested that IF1 is the only naturally occurring, endogenous protein that interacts with the mitochondrial ATP synthase. However, it becomes clear that many more endogenous proteins must be involved in the regulation of mitochondrial ATP synthase. Further in-depth investigations are necessary to explore the significance of these newly emerging interacting proteins in regulating the enzyme activity of ATP synthase in vivo.
The role of ATP synthase in mitochondrial function
The mitochondrial ATP synthase certainly does not serve its sole function as an energy generator. By reversing ATP hydrolysis activity, ATP synthase plays a vital role in maintaining mitochondrial membrane potential [93] (Figure 2). IF1 is the first identified natural protein that prevents excess hydrolysis of ATP [69,70]. It has also been reported that ATP synthase is involved in mitochondrial protein import [94] and the mobilization of cytochrome c during apoptosis [95]. Further evidence implicates that ATP synthase dimerization, which is facilitated by IF1, plays an important role in forming mitochondrial cristae and the F0 c-ring itself or the dimerized ATP synthase may form the permeability transition pore (mPTP) (Figure 2). All of these should be crucial in maintaining mitochondrial function.
Figure 2.
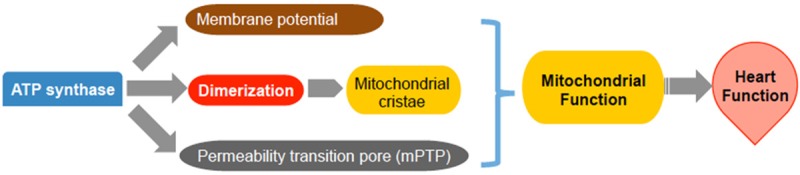
The roles of ATP synthase in mitochondrial function. The mitochondrial ATP synthase activity plays a key role in mitochondrial function and cardiac function in determining membrane potential, mitochondrial cristae and the opening of the mitochondrial permeability transition pore (mPTP).
ATP synthase is suggested to have a role in crista morphogenesis. A recent study suggests that ATP synthase contributes to the optimal supramolecular organization of the respiratory chain [96], and even the density of cristae structure [97,98]. ATP synthase occupancy rises correspondingly with the cellular demand for OXPHOS. Mitochondrial ATP synthases cluster as discrete domains that reorganize with the cellular demand for oxidative phosphorylation [99] and play a key role in cristae morphogenesis [98]. Other than inhibiting ATPase activity, IF1 may also regulate the oligomeric state of ATP synthase by facilitating the dimerization of ATP synthase via a molecular link between two F1 domains [100]. IF1 limits the apoptotic-signalling cascade by preventing mitochondrial remodeling and preserves cristae structure to limit apoptotic cell death signaling [101].
However, the role of IF1 in promoting the dimerization of ATP synthase and mitochondrial remodeling is still under debate. Studies on mitochondrial extracted from bovine heart showed that the ATP synthase dimer is a stable inactive structure and its formation is not mediated by IF1 binding [102]. A cell culture study in human HeLa cells could not confirm that IF1 overexpression facilitates mitochondrial cristae formation [103]. Similarly, the same group reported that in vivo IF1 knockout did not alter the morphology of mitochondrial cristae in various tissues of the IF1 knockout mice under basal condition [81]. It would be intriguing to further examine whether IF1 could help maintain mitochondrial morphology under hypoxic/ischemia conditions because of the acidic conditions required for IF1 activation. Therefore, further investigations that subject animals to IF1 overexpression and knockout under different pathological conditions may help resolve the above inconsistent observations.
The mitochondrial permeability transition, or MPT, refers to the increase in the permeability of the mitochondrial membranes to molecules of less than 1.5 KD, which results from the opening of a mitochondrial permeability transition pore (mPTP). The mPTP is a protein pore located at the inner membrane of the mitochondria under certain pathological conditions such as myocardial ischemia. The opening of mPTP leads to mitochondrial swelling and cell death of apoptosis or necrosis. Under physiological conditions, oxidative phosphorylation is responsible for ATP production with relatively low ROS production. In contrast, when mPTP is open, the mitochondria not only produces excessive reactive oxygen species (ROS), but also consumes ATP in a futile cycle of efforts to restore the membrane potential, thereby exacerbating cellular damage. However, the exact components of mPTP remain unknown. While cyclophilin D is thought to be an important regulatory protein in determining the opening of mPTP, other earlier identification of mPTP components based mostly on in vitro biophysical and biochemical investigations failed to prove their necessity for the proper function of mPTP, at least in various single gene knockout mouse models (see review [104]). In cyclophilin D knockout mice with myocardial ischemia/reperfusion injury, the extracted mitochondria show reduced mPTP opening and therefore show myocardial protective effects [105,106].
Even though it has long been speculated that ATP synthase may play a key role in mitochondrial permeability transition [107], direct experimental proof has emerged only recently. Based on findings that the c subunit of the ATP synthase is required for MPT-driven mitochondrial fragmentation and cell death triggered by Ca2+ overload and oxidative stress, Bonora et al proved that the c subunit of the ATP synthase constitutes a critical component of the mPTP [108]. Another group further provided evidence that cyclosporine A binds to the OSCP and β-subunits of the ATP synthase, indirectly inhibiting the c-subunit (mPTP) pore by inducing a conformational change in ATP synthase that places F1 over the pore and its conductance [109]. Bcl-xL can be found within the c-subunit of the ATP synthase and is similar to mPTP [91]. The F1 prevents mPTP opening by being placed over the pore of a leak conductance within the c-subunit ring [91,109]. This model predicts that cyclophilin D, which is known to bind to OSCP [88], acts on the pore by facilitating the removal of the F1 from the c-subunit in a CsA-sensitive manner during pore opening [109]. On the other hand, Giorgio et al have shown that the binding of cyclophilin D to OSCP facilitates ATP synthase dimerization, which in turn becomes a conductance channel responsible for the opening of mPTP [110]. Apparently, these two models (Figure 3) may be interrelated, eg., the formation of ATP synthase dimer may eventually alter the c-subunit conductance. However, further investigations are needed to determine if the c-subunit conductance is dimer dependent. It is also likely that changes in ATP synthase activity are sufficient to alter the formation of ATP synthase related mPTP opening and the detailed correlations should be rigorously investigated.
Figure 3.
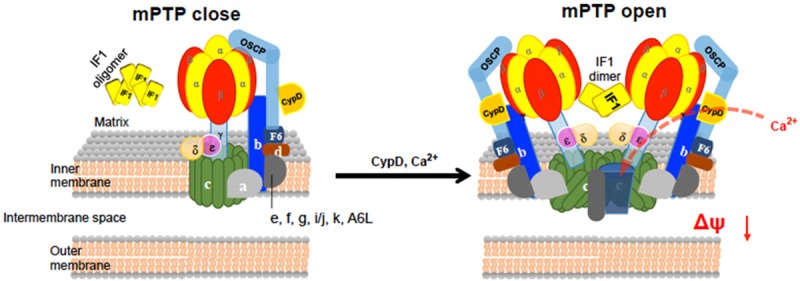
The schematic models of ATP synthase serving as the mitochondrial permeability transition (mPTP). The main mPTP regulator cyclophilin D (CypD) in mitochondrial matrix is a necessary mPTP component responding to stimuli to initiate mPTP opening upon its binding to ATP synthase. The c-ring of the ATP synthase F0 domain acts as the pore of the mPTP. Alternatively, the mitochondrial ATPase inhibitory factor 1 (IF1) dimer binds to the interface between α- and β-subunits of the ATP synthase F1 domain, inducing the dimerization of the F1F0-ATP synthase and forming a pore.
In general, emerging evidence supports a lasting speculation that ATP synthase acts as the key component of mPTP. However, it is apparent that the molecular and structural details remain scant. Further investigations are required to answer many unresolved questions. For instance, what conformational changes will enable the ATP synthase to act as an mPTP? What is the role of F1 in regulating the mPTP opening? Does the dimerization of ATP synthase consist of the mPTP or is it merely a required condition for the C-ring channel to work as one? Moreover, in vivo evidence is also needed to confirm the in vitro findings. Preclinical animal models that illustrate the role of ATP synthase serving as the key components of mPTP will be highly valuable for further development of therapeutic strategies targeting ATP synthase.
Targeting ATP synthase regulation as a therapeutic target for cardiac disorders
Diminished energy supply is a key factor contributing to both the initiation and progressive transition of congestive heart failure (CHF) [2,111]. The activity of electron transport-chain complexes and ATP synthase capacity are reduced in failing hearts [112-114]. The increased opening of mPTP is one main feature of hearts under ischemia/reperfusion. This impairment in ATP generation and mPTP opening further augments the release of ROS, which exacerbates damages of mitochondria and other important cellular structures. ATP depletion and dysfunctional mitochondria are crucial components of not only impaired contractile function, but also programmed cell death, leading to a remarkable net loss of functional myocardium. Therefore, the mitochondrial function or the energetics of the heart are integrally linked with the causes and phenotype of heart failure. There is a common consensus that improving the myocardial energetic state and preventing excessive mPTP opening should be a therapeutic goal in treating CHF.
Inhibiting the hydrolytic activity of ATP synthase during ischemia without interfering with the synthesis of ATP during normoxic condition is proposed to be therapeutic. Treatment with aurovertin and oligomycin, both inhibit similarly bi-direction of ATP synthase activity showed myocardial protective effects [115]. Similarly, IF-1 is a potential drug target because it is activated under acidic conditions, such as in myocardial ischemia, to enhance ATP reserves in the heart [69,70]. Preclinical assessments on IF-1 mimetic compounds did show promising results in animal studies [76,116]. Novel therapies targeting other ATP synthase interacting proteins are also possible to improve myocardial energetics and prevent mPTP opening and mitochondrial dysfunction.
Summary and conclusions
ATP synthase is a fascinating protein complex with an essential role in maintaining life. Its pivotal role is even more obvious in the most energy consuming heart. Several mechanisms are involved in maintaining the steady state activity of ATP synthase, thus determining mitochondrial function via its role in controlling cellular energetics, mitochondrial remodeling and mPTP opening (Figure 3). Novel therapies that can correct ATP synthase deficiency, energy depletion and mPTP opening in CHF are highly desired, but these must be first tested in animal models. Given that a majority of the current knowledge about the mitochondrial ATP synthase is based on protein biology, cellular biochemistry and in vivo yeast biology, it is highly desirable to gain insights into how this system works in animals and in humans. Studies on preclinical animal models will yield insights into the development of novel therapies targeting the mitochondrial ATP synthase.
Acknowledgements
This work was supported by a Basic Science Award (#7-12-BS-208) from the American Diabetes Association to Q.Y., a Grant-in-aid from American Heart Association to Q.Y., and a Post-doctoral Fellowship (13POST14180006) from the American Heart Association to Q.L.
References
- 1.Sinatra ST. Metabolic cardiology: an integrative strategy in the treatment of congestive heart failure. Altern Ther Health Med. 2009;15:44–52. [PubMed] [Google Scholar]
- 2.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 3.Gupta A, Akki A, Wang Y, Leppo MK, Chacko VP, Foster DB, Caceres V, Shi S, Kirk JA, Su J, Lai S, Paolocci N, Steenbergen C, Gerstenblith G, Weiss RG. Creatine kinase-mediated improvement of function in failing mouse hearts provides causal evidence the failing heart is energy starved. J Clin Invest. 2012;122:291–302. doi: 10.1172/JCI57426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida M, Muneyuki E, Hisabori T. ATP synthase--a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 5.Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 7.van Raaij MJ, Abrahams JP, Leslie AG, Walker JE. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc Natl Acad Sci U S A. 1996;93:6913–6917. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Ko Y, Delannoy M, Ludtke SJ, Chiu W, Pedersen PL. Mitochondrial ATP synthasome: three-dimensional structure by electron microscopy of the ATP synthase in complex formation with carriers for Pi and ADP/ATP. J Biol Chem. 2004;279:31761–31768. doi: 10.1074/jbc.M401353200. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Saxena AK, Simcoke WN, Garboczi DN, Pedersen PL, Ko YH. Mitochondrial ATP synthase. Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism. J Biol Chem. 2006;281:13777–13783. doi: 10.1074/jbc.M513369200. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 11.Karrasch S, Walker JE. Novel features in the structure of bovine ATP synthase. J Mol Biol. 1999;290:379–384. doi: 10.1006/jmbi.1999.2897. [DOI] [PubMed] [Google Scholar]
- 12.Rubinstein JL, Walker JE, Henderson R. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 2003;22:6182–6192. doi: 10.1093/emboj/cdg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubinstein JL, Dickson VK, Runswick MJ, Walker JE. ATP synthase from Saccharomyces cerevisiae: location of subunit h in the peripheral stalk region. J Mol Biol. 2005;345:513–520. doi: 10.1016/j.jmb.2004.10.060. [DOI] [PubMed] [Google Scholar]
- 14.Noji H, Yasuda R, Yoshida M, Kinosita K Jr. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 15.Zharova TV, Vinogradov AD. Energy-linked binding of Pi is required for continuous steady-state proton-translocating ATP hydrolysis catalyzed by F0. F1 ATP synthase. Biochemistry. 2006;45:14552–14558. doi: 10.1021/bi061520v. [DOI] [PubMed] [Google Scholar]
- 16.Zharova TV, Vinogradov AD. Requirement of medium ADP for the steady-state hydrolysis of ATP by the proton-translocating Paracoccus denitrificans Fo. F1-ATP synthase. Biochim Biophys Acta. 2006;1757:304–310. doi: 10.1016/j.bbabio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal B. A role for anions in ATP synthesis and its molecular mechanistic interpretation. J Bioenerg Biomembr. 2011;43:299–310. doi: 10.1007/s10863-011-9358-3. [DOI] [PubMed] [Google Scholar]
- 18.Harris DA, Das AM. Control of mitochondrial ATP synthesis in the heart. Biochem J. 1991;280:561–573. doi: 10.1042/bj2800561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 20.Kagawa Y, Ohta S. Regulation of mitochondrial ATP synthesis in mammalian cells by transcriptional control. Int J Biochem. 1990;22:219–229. doi: 10.1016/0020-711x(90)90333-x. [DOI] [PubMed] [Google Scholar]
- 21.Houstek J, Klement P, Floryk D, Antonicka H, Hermanska J, Kalous M, Hansikova H, Hout’kova H, Chowdhury SK, Rosipal T, Kmoch S, Stratilova L, Zeman J. A novel deficiency of mitochondrial ATPase of nuclear origin. Hum Mol Genet. 1999;8:1967–1974. doi: 10.1093/hmg/8.11.1967. [DOI] [PubMed] [Google Scholar]
- 22.Houstek J, Pickova A, Vojtiskova A, Mracek T, Pecina P, Jesina P. Mitochondrial diseases and genetic defects of ATP synthase. Biochim Biophys Acta. 2006;1757:1400–1405. doi: 10.1016/j.bbabio.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Houstek J, Kmoch S, Zeman J. TMEM70 protein - a novel ancillary factor of mammalian ATP synthase. Biochim Biophys Acta. 2009;1787:529–532. doi: 10.1016/j.bbabio.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Ding Y, Yang KD, Yang Q. The role of PPARdelta signaling in the cardiovascular system. Prog Mol Biol Transl Sci. 2014;121:451–473. doi: 10.1016/B978-0-12-800101-1.00014-4. [DOI] [PubMed] [Google Scholar]
- 25.Yang Q, Li Y. Roles of PPARs on regulating myocardial energy and lipid homeostasis. J Mol Med (Berl) 2007;85:697–706. doi: 10.1007/s00109-007-0170-9. [DOI] [PubMed] [Google Scholar]
- 26.Schilling J, Kelly DP. The PGC-1 cascade as a therapeutic target for heart failure. J Mol Cell Cardiol. 2011;51:578–583. doi: 10.1016/j.yjmcc.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierce DJ, Jordan EM, Breen GA. Structural organization of a nuclear gene for the alpha-subunit of the bovine mitochondrial ATP synthase complex. Biochim Biophys Acta. 1992;1132:265–275. doi: 10.1016/0167-4781(92)90160-2. [DOI] [PubMed] [Google Scholar]
- 28.Vander Zee CA, Jordan EM, Breen GA. ATPF1 binding site, a positive cis-acting regulatory element of the mammalian ATP synthase alpha-subunit gene. J Biol Chem. 1994;269:6972–6977. [PubMed] [Google Scholar]
- 29.Breen GA, Jordan EM. Upstream stimulatory factor 2 activates the mammalian F1F0 ATP synthase alpha-subunit gene through an initiator element. Gene Expr. 1998;7:163–170. [PMC free article] [PubMed] [Google Scholar]
- 30.Breen GA, Jordan EM. Transcriptional activation of the F(1)F(0) ATP synthase alpha-subunit initiator element by USF2 is mediated by p300. Biochim Biophys Acta. 1999;1428:169–176. doi: 10.1016/s0304-4165(99)00061-6. [DOI] [PubMed] [Google Scholar]
- 31.Breen GA, Jordan EM. Upstream stimulatory factor 2 stimulates transcription through an initiator element in the mouse cytochrome c oxidase subunit Vb promoter. Biochim Biophys Acta. 2000;1517:119–127. doi: 10.1016/s0167-4781(00)00269-4. [DOI] [PubMed] [Google Scholar]
- 32.Breen GA, Vander Zee CA, Jordan EM. Nuclear factor YY1 activates the mammalian F0F1 ATP synthase alpha-subunit gene. Gene Expr. 1996;5:181–191. [PMC free article] [PubMed] [Google Scholar]
- 33.Levy FH, Kelly DP. Regulation of ATP synthase subunit e gene expression by hypoxia: cell differentiation stage-specific control. Am J Physiol. 1997;272:C457–465. doi: 10.1152/ajpcell.1997.272.2.C457. [DOI] [PubMed] [Google Scholar]
- 34.Willers IM, Martinez-Reyes I, Martinez-Diez M, Cuezva JM. miR-127-5p targets the 3’UTR of human beta-F1-ATPase mRNA and inhibits its translation. Biochim Biophys Acta. 2012;1817:838–848. doi: 10.1016/j.bbabio.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Ackerman SH. Atp11p and Atp12p are chaperones for F(1)-ATPase biogenesis in mitochondria. Biochim Biophys Acta. 2002;1555:101–105. doi: 10.1016/s0005-2728(02)00262-1. [DOI] [PubMed] [Google Scholar]
- 36.Ludlam A, Brunzelle J, Pribyl T, Xu X, Gatti DL, Ackerman SH. Chaperones of F1-ATPase. J Biol Chem. 2009;284:17138–17146. doi: 10.1074/jbc.M109.002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickova A, Potocky M, Houstek J. Assembly factors of F1FO-ATP synthase across genomes. Proteins. 2005;59:393–402. doi: 10.1002/prot.20452. [DOI] [PubMed] [Google Scholar]
- 38.Lefebvre-Legendre L, Salin B, Schaeffer J, Brethes D, Dautant A, Ackerman SH, di Rago JP. Failure to assemble the alpha 3 beta 3 subcomplex of the ATP synthase leads to accumulation of the alpha and beta subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J Biol Chem. 2005;280:18386–18392. doi: 10.1074/jbc.M410789200. [DOI] [PubMed] [Google Scholar]
- 39.Hinton A, Gatti DL, Ackerman SH. The molecular chaperone, Atp12p, from Homo sapiens. In vitro studies with purified wild type and mutant (E240K) proteins. J Biol Chem. 2004;279:9016–9022. doi: 10.1074/jbc.M312631200. [DOI] [PubMed] [Google Scholar]
- 40.Rak M, Tzagoloff A. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc Natl Acad Sci U S A. 2009;106:18509–18514. doi: 10.1073/pnas.0910351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lytovchenko O, Naumenko N, Oeljeklaus S, Schmidt B, von der Malsburg K, Deckers M, Warscheid B, van der Laan M, Rehling P. The INA complex facilitates assembly of the peripheral stalk of the mitochondrial F1Fo-ATP synthase. EMBO J. 2014;33:1624–38. doi: 10.15252/embj.201488076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sangawa H, Himeda T, Shibata H, Higuti T. Gene expression of subunit c(P1), subunit c(P2), and oligomycin sensitivity-conferring protein may play a key role in biogenesis of H+-ATP synthase in various rat tissues. J Biol Chem. 1997;272:6034–6037. doi: 10.1074/jbc.272.9.6034. [DOI] [PubMed] [Google Scholar]
- 43.Bergeaud M, Mathieu L, Guillaume A, Moll UM, Mignotte B, Le Floch N, Vayssiere JL, Rincheval V. Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F(1)F0-ATP synthase. Cell Cycle. 2013;12:2781–2793. doi: 10.4161/cc.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das AM, Harris DA. Reversible modulation of the mitochondrial ATP synthase with energy demand in cultured rat cardiomyocytes. FEBS Lett. 1989;256:97–100. doi: 10.1016/0014-5793(89)81725-9. [DOI] [PubMed] [Google Scholar]
- 45.Das AM, Harris DA. Control of mitochondrial ATP synthase in heart cells: inactive to active transitions caused by beating or positive inotropic agents. Cardiovasc Res. 1990;24:411–417. doi: 10.1093/cvr/24.5.411. [DOI] [PubMed] [Google Scholar]
- 46.Das AM, Harris DA. Regulation of the mitochondrial ATP synthase in intact rat cardiomyocytes. Biochem J. 1990;266:355–361. doi: 10.1042/bj2660355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell CJ, Bright NA, Rutter GA, Griffiths EJ. ATP regulation in adult rat cardiomyocytes: time-resolved decoding of rapid mitochondrial calcium spiking imaged with targeted photoproteins. J Biol Chem. 2006;281:28058–28067. doi: 10.1074/jbc.M604540200. [DOI] [PubMed] [Google Scholar]
- 48.Yamada EW, Huzel NJ. The calcium-binding ATPase inhibitor protein from bovine heart mitochondria. J Biol Chem. 1988;183:11498–11503. [PubMed] [Google Scholar]
- 49.Yamada EW, Huzel NJ. Calcium-binding ATPase inhibitor protein of bovine heart mitochondria. Role in ATP synthesis and effect of Ca2+ Biochemistry. 1989;28:9714–9718. doi: 10.1021/bi00451a026. [DOI] [PubMed] [Google Scholar]
- 50.Hubbard MJ, McHugh NJ. Mitochondrial ATP synthase F1-beta-subunit is a calcium-binding protein. FEBS Lett. 1996;391:323–329. doi: 10.1016/0014-5793(96)00767-3. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen TT, Ogbi M, Yu Q, Fishman JB, Thomas W, Harvey BJ, Fulton D, Johnson JA. Modulation of the Protein Kinase C Interaction with the “d” Subunit of F1F0-ATP Synthase in Neonatal Cardiac Myocytes: Development of Cell-permeable, Mitochondrially Ta Rgeted Inhibitor and Facilitator PEP Tides. J Biol Chem. 2010;285:22164–22173. doi: 10.1074/jbc.M109.077578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen T, Ogbi M, Johnson JA. Delta protein kinase C interacts with the d subunit of the F1F0 ATPase in neonatal cardiac myocytes exposed to hypoxia or phorbol ester. Implications for F1F0 ATPase regulation. J Biol Chem. 2008;283:29831–29840. doi: 10.1074/jbc.M801642200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boerries M, Most P, Gledhill JR, Walker JE, Katus HA, Koch WJ, Aebi U, Schoenenberger CA. Ca2+-dependent interaction of S100A1 with F1-ATPase leads to an increased ATP content in cardiomyocytes. Mol Cell Biol. 2007;27:4365–4373. doi: 10.1128/MCB.02045-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prescott M, Lourbakos A, Bateson M, Boyle G, Nagley P, Devenish RJ. A novel fluorescent marker for assembled mitochondria ATP synthase of yeast. OSCP subunit fused to green fluorescent protein is assembled into the complex in vivo. FEBS Lett. 1997;411:97–101. doi: 10.1016/s0014-5793(97)00670-4. [DOI] [PubMed] [Google Scholar]
- 55.Hojlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase beta -subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J Biol Chem. 2003;278:10436–10442. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- 56.Reinders J, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, van der Laan M, Rehling P, Sickmann A, Pfanner N, Meisinger C. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol Cell Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Hojlund K, Yi Z, Lefort N, Langlais P, Bowen B, Levin K, Beck-Nielsen H, Mandarino LJ. Human ATP synthase beta is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle. Diabetologia. 2010;53:541–551. doi: 10.1007/s00125-009-1624-0. [DOI] [PubMed] [Google Scholar]
- 58.Kane LA, Youngman MJ, Jensen RE, Van Eyk JE. Phosphorylation of the F(1)F(o) ATP synthase beta subunit: functional and structural consequences assessed in a model system. Circ Res. 2010;106:504–513. doi: 10.1161/CIRCRESAHA.109.214155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kane LA, Van Eyk JE. Post-translational modifications of ATP synthase in the heart: biology and function. J Bioenerg Biomembr. 2009;41:145–150. doi: 10.1007/s10863-009-9218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marban E, Van Eyk JE. Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- 61.Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ko YH, Pan W, Inoue C, Pedersen PL. Signal transduction to mitochondrial ATP synthase: evidence that PDGF-dependent phosphorylation of the delta-subunit occurs in several cell lines, involves tyrosine, and is modulated by lysophosphatidic acid. Mitochondrion. 2002;1:339–348. doi: 10.1016/s1567-7249(01)00036-8. [DOI] [PubMed] [Google Scholar]
- 63.Wang SB, Foster DB, Rucker J, O’Rourke B, Kass DA, Van Eyk JE. Redox regulation of mitochondrial ATP synthase: implications for cardiac resynchronization therapy. Circ Res. 2011;109:750–757. doi: 10.1161/CIRCRESAHA.111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang SB, Murray CI, Chung HS, Van Eyk JE. Redox regulation of mitochondrial ATP synthase. Trends Cardiovasc Med. 2013;23:14–18. doi: 10.1016/j.tcm.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vassilopoulos A, Pennington JD, Andresson T, Rees DM, Bosley AD, Fearnley IM, Ham A, Flynn CR, Hill S, Rose KL, Kim HS, Deng CX, Walker JE, Gius D. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal. 2014;21:551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pullman ME, Monroy GC. A Naturally Occurring Inhibitor of Mitochondrial Adenosine Triphosphatase. J Biol Chem. 1963;238:3762–3769. [PubMed] [Google Scholar]
- 67.van de Stadt RJ, de Boer BL, van Dam K. The interaction between the mitochondrial ATPase (F 1) and the ATPase inhibitor. Biochim Biophys Acta. 1973;292:338–349. doi: 10.1016/0005-2728(73)90040-6. [DOI] [PubMed] [Google Scholar]
- 68.Cintron NM, Pedersen PL. A protein inhibitor of the mitochondrial adenosine triphosphatase complex of rat liver. Purification and characterization. J Biol Chem. 1979;254:3439–3443. [PubMed] [Google Scholar]
- 69.Campanella M, Seraphim A, Abeti R, Casswell E, Echave P, Duchen MR. IF1, the endogenous regulator of the F(1)F(o)-ATPsynthase, defines mitochondrial volume fraction in HeLa cells by regulating autophagy. Biochim Biophys Acta. 2009;1787:393–401. doi: 10.1016/j.bbabio.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 70.Campanella M, Parker N, Tan CH, Hall AM, Duchen MR. IF(1): setting the pace of the F(1)F(o)-ATP synthase. Trends Biochem Sci. 2009;34:343–350. doi: 10.1016/j.tibs.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 71.Zanotti F, Gnoni A, Mangiullo R, Papa S. Effect of the ATPase inhibitor protein IF1 on H+ translocation in the mitochondrial ATP synthase complex. Biochem Biophys Res Commun. 2009;384:43–48. doi: 10.1016/j.bbrc.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 72.Rouslin W, Pullman ME. Protonic inhibition of the mitochondrial adenosine 5’-triphosphatase in ischemic cardiac muscle. Reversible binding of the ATPase inhibitor protein to the mitochondrial ATPase during ischemia. J Mol Cell Cardiol. 1987;19:661–668. doi: 10.1016/s0022-2828(87)80374-7. [DOI] [PubMed] [Google Scholar]
- 73.Grover GJ, Atwal KS, Sleph PG, Wang FL, Monshizadegan H, Monticello T, Green DW. Excessive ATP hydrolysis in ischemic myocardium by mitochondrial F1F0-ATPase: effect of selective pharmacological inhibition of mitochondrial ATPase hydrolase activity. Am J Physiol Heart Circ Physiol. 2004;287:H1747–1755. doi: 10.1152/ajpheart.01019.2003. [DOI] [PubMed] [Google Scholar]
- 74.Wu Q, Andrianaivomananjaona T, Tetaud E, Corvest V, Haraux F. Interactions involved in grasping and locking of the inhibitory peptide IF1 by mitochondrial ATP synthase. Biochim Biophys Acta. 2014;1837:761–72. doi: 10.1016/j.bbabio.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 75.Jennings RB, Reimer KA, Steenbergen C. Effect of inhibition of the mitochondrial ATPase on net myocardial ATP in total ischemia. J Mol Cell Cardiol. 1991;23:1383–1395. doi: 10.1016/0022-2828(91)90185-o. [DOI] [PubMed] [Google Scholar]
- 76.Grover GJ, Malm J. Pharmacological profile of the selective mitochondrial F1F0 ATP hydrolase inhibitor BMS-199264 in myocardial ischemia. Cardiovasc Ther. 2008;26:287–296. doi: 10.1111/j.1755-5922.2008.00065.x. [DOI] [PubMed] [Google Scholar]
- 77.Zanotti F, Raho G, Gaballo A, Papa S. Inhibitory and anchoring domains in the ATPase inhibitor protein IF1 of bovine heart mitochondrial ATP synthase. J Bioenerg Biomembr. 2004;36:447–457. doi: 10.1023/B:JOBB.0000047327.68173.9b. [DOI] [PubMed] [Google Scholar]
- 78.Rouslin W, Broge CW. IF1 function in situ in uncoupler-challenged ischemic rabbit, rat, and pigeon hearts. J Biol Chem. 1996;271:23638–23641. doi: 10.1074/jbc.271.39.23638. [DOI] [PubMed] [Google Scholar]
- 79.Rouslin W, Broge CW. Why the mitochondrial ATPase inhibitor IF1 fails to inhibit the mitochondrial ATPase in situ in fast heart-rate mammalian and avian hearts. Ann N Y Acad Sci. 1992;671:505–506. doi: 10.1111/j.1749-6632.1992.tb43842.x. [DOI] [PubMed] [Google Scholar]
- 80.Rouslin W, Broge CW. Isoform-independent heart rate-related variation in cardiac myofibrillar Ca(2+)-activated Mg(2+)-ATPase activity. Am J Physiol. 1996;270:C1271–1276. doi: 10.1152/ajpcell.1996.270.5.C1271. [DOI] [PubMed] [Google Scholar]
- 81.Nakamura J, Fujikawa M, Yoshida M. IF1, a natural inhibitor of mitochondrial ATP synthase, is not essential for the normal growth and breeding of mice. Biosci Rep. 2013;33 doi: 10.1042/BSR20130078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Belogrudov GI. Factor B is essential for ATP synthesis by mitochondria. Arch Biochem Biophys. 2002;406:271–274. doi: 10.1016/s0003-9861(02)00431-9. [DOI] [PubMed] [Google Scholar]
- 83.Belogrudov GI. Recent advances in structure-functional studies of mitochondrial factor B. J Bioenerg Biomembr. 2009;41:137–143. doi: 10.1007/s10863-009-9210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belogrudov GI, Hatefi Y. Factor B and the mitochondrial ATP synthase complex. J Biol Chem. 2002;277:6097–6103. doi: 10.1074/jbc.M111256200. [DOI] [PubMed] [Google Scholar]
- 85.Belogrudov GI, Tomich JM, Hatefi Y. ATP synthase complex. Proximities of subunits in bovine submitochondrial particles. J Biol Chem. 1995;270:2053–2060. doi: 10.1074/jbc.270.5.2053. [DOI] [PubMed] [Google Scholar]
- 86.Nguyen TT, Ogbi M, Yu Q, Johnson JA. Attenuation of the hypoxia-induced protein kinase Cdelta interaction with the ‘d’ subunit of F1Fo-ATP synthase in neonatal cardiac myocytes: implications for energy preservation and survival. Biochem J. 2010;429:335–345. doi: 10.1042/BJ20091927. [DOI] [PubMed] [Google Scholar]
- 87.Nguyen TT, Ogbi M, Yu Q, Fishman JB, Thomas W, Harvey BJ, Fulton D, Johnson JA. Modulation of the protein kinase Cdelta interaction with the “d” subunit of F1F0-ATP synthase in neonatal cardiac myocytes: development of cell-permeable, mitochondrially targeted inhibitor and facilitator peptides. J Biol Chem. 2010;285:22164–22173. doi: 10.1074/jbc.M109.077578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giorgio V, Bisetto E, Soriano ME, Dabbeni-Sala F, Basso E, Petronilli V, Forte MA, Bernardi P, Lippe G. Cyclophilin D modulates mitochondrial F0F1-ATP synthase by interacting with the lateral stalk of the complex. J Biol Chem. 2009;284:33982–33988. doi: 10.1074/jbc.M109.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chinopoulos C, Konrad C, Kiss G, Metelkin E, Torocsik B, Zhang SF, Starkov AA. Modulation of F0F1-ATP synthase activity by cyclophilin D regulates matrix adenine nucleotide levels. FEBS J. 2011;278:1112–1125. doi: 10.1111/j.1742-4658.2011.08026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chinopoulos C, Adam-Vizi V. Modulation of the mitochondrial permeability transition by cyclophilin D: Moving closer to F(0)-F(1) ATP synthase? Mitochondrion. 2012;12:41–5. doi: 10.1016/j.mito.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 91.Alavian KN, Li H, Collis L, Bonanni L, Zeng L, Sacchetti S, Lazrove E, Nabili P, Flaherty B, Graham M, Chen Y, Messerli SM, Mariggio MA, Rahner C, McNay E, Shore GC, Smith PJ, Hardwick JM, Jonas EA. Bcl-xL regulates metabolic efficiency of neurons through interaction with the mitochondrial F1FO ATP synthase. Nat Cell Biol. 2011;13:1224–1233. doi: 10.1038/ncb2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maniam S, Coutts AS, Stratford MR, McGouran J, Kessler B, La Thangue NB. Cofactor Strap regulates oxidative phosphorylation and mitochondrial p53 activity through ATP synthase. Cell Death Differ. 2015;22:156–163. doi: 10.1038/cdd.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei YH, Murphy MP. Qu-antitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur J Biochem. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- 94.Martin J, Mahlke K, Pfanner N. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J Biol Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- 95.Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, Korsmeyer SJ. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 96.Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kuhlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci U S A. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giraud MF, Paumard P, Soubannier V, Vaillier J, Arselin G, Salin B, Schaeffer J, Brethes D, di Rago JP, Velours J. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim Biophys Acta. 2002;1555:174–180. doi: 10.1016/s0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- 99.Jimenez L, Laporte D, Duvezin-Caubet S, Courtout F, Sagot I. Mitochondrial ATP synthases cluster as discrete domains that reorganize with the cellular demand for oxidative phosphorylation. J Cell Sci. 2014;127:719–726. doi: 10.1242/jcs.137141. [DOI] [PubMed] [Google Scholar]
- 100.Garcia JJ, Morales-Rios E, Cortes-Hernandez P, Rodriguez-Zavala JS. The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase. Biochemistry. 2006;45:12695–12703. doi: 10.1021/bi060339j. [DOI] [PubMed] [Google Scholar]
- 101.Faccenda D, Tan CH, Seraphim A, Duchen MR, Campanella M. IF1 limits the apoptotic-signalling cascade by preventing mitochondrial remodelling. Cell Death Differ. 2013;20:686–697. doi: 10.1038/cdd.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tomasetig L, Di Pancrazio F, Harris DA, Mavelli I, Lippe G. Dimerization of F0F1ATP synthase from bovine heart is independent from the binding of the inhibitor protein IF1. Biochim Biophys Acta. 2002;1556:133–141. doi: 10.1016/s0005-2728(02)00344-4. [DOI] [PubMed] [Google Scholar]
- 103.Fujikawa M, Imamura H, Nakamura J, Yoshida M. Assessing Actual Contribution of IF1, Inhibitor of Mitochondrial FoF1, to ATP Homeostasis, Cell Growth, Mitochondrial Morphology, and Cell Viability. J Biol Chem. 2012;287:18781–18787. doi: 10.1074/jbc.M112.345793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karch J, Molkentin JD. Is p53 the long-sought molecular trigger for cyclophilin D-regulated mitochondrial permeability transition pore formation and necrosis? Circ Res. 2012;111:1258–1260. doi: 10.1161/CIRCRESAHA.112.280990. [DOI] [PubMed] [Google Scholar]
- 105.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 106.Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- 107.Hunter DR, Haworth RA, Southard JH. Relationship between configuration, function, and permeability in calcium-treated mitochondria. J Biol Chem. 1976;251:5069–5077. [PubMed] [Google Scholar]
- 108.Bonora M, Bononi A, De Marchi E, Giorgi C, Lebiedzinska M, Marchi S, Patergnani S, Rimessi A, Suski JM, Wojtala A, Wieckowski MR, Kroemer G, Galluzzi L, Pinton P. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell Cycle. 2013;12:674–683. doi: 10.4161/cc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alavian KN, Beutner G, Lazrove E, Sacchetti S, Park HA, Licznerski P, Li H, Nabili P, Hockensmith K, Graham M, Porter GA Jr, Jonas EA. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc Natl Acad Sci U S A. 2014;111:10580–10585. doi: 10.1073/pnas.1401591111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Giorgio V, von Stockum S, Antoniel M, Fabbro A, Fogolari F, Forte M, Glick GD, Petronilli V, Zoratti M, Szabo I, Lippe G, Bernardi P. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci U S A. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ashrafian H, Redwood C, Blair E, Watkins H. Hypertrophic cardiomyopathy:a paradigm for myocardial energy depletion. Trends Genet. 2003;19:263–268. doi: 10.1016/S0168-9525(03)00081-7. [DOI] [PubMed] [Google Scholar]
- 112.Marin-Garcia J, Goldenthal MJ, Moe GW. Abnormal cardiac and skeletal muscle mitochondrial function in pacing-induced cardiac failure. Cardiovasc Res. 2001;52:103–110. doi: 10.1016/s0008-6363(01)00368-6. [DOI] [PubMed] [Google Scholar]
- 113.Quigley AF, Kapsa RM, Esmore D, Hale G, Byrne E. Mitochondrial respiratory chain activity in idiopathic dilated cardiomyopathy. J Card Fail. 2000;6:47–55. doi: 10.1016/s1071-9164(00)00011-7. [DOI] [PubMed] [Google Scholar]
- 114.Casademont J, Miro O. Electron transport chain defects in heart failure. Heart Fail Rev. 2002;7:131–139. doi: 10.1023/a:1015372407647. [DOI] [PubMed] [Google Scholar]
- 115.Rouslin W, Erickson JL, Solaro RJ. Effects of oligomycin and acidosis on rates of ATP depletion in ischemic heart muscle. Am J Physiol. 1986;250:H503–508. doi: 10.1152/ajpheart.1986.250.3.H503. [DOI] [PubMed] [Google Scholar]
- 116.Atwal KS, Wang P, Rogers WL, Sleph P, Monshizadegan H, Ferrara FN, Traeger S, Green DW, Grover GJ. Small molecule mitochondrial F1F0 ATPase hydrolase inhibitors as cardioprotective agents. Identification of 4-(N-arylimidazole)-substituted benzopyran derivatives as selective hydrolase inhibitors. J Med Chem. 2004;47:1081–1084. doi: 10.1021/jm030291x. [DOI] [PubMed] [Google Scholar]
- 117.Jackson PJ, Harris DA. The mitochondrial ATP synthase inhibitor protein binds near the C-terminus of the F1 beta-subunit. FEBS Lett. 1988;229:224–228. doi: 10.1016/0014-5793(88)80832-9. [DOI] [PubMed] [Google Scholar]
- 118.Chinopoulos C, Konrad C, Kiss G, Metelkin E, Torocsik B, Zhang SF, Starkov AA. Modulation of F0F1-ATP synthase activity by cyclophilin D regulates matrix adenine nucleotide levels. FEBS J. 2011;278:1112–1125. doi: 10.1111/j.1742-4658.2011.08026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chinopoulos C, Adam-Vizi V. Modulation of the mitochondrial permeability transition by cyclophilin D: moving closer to F(0)-F(1) ATP synthase? Mitochondrion. 2012;12:41–45. doi: 10.1016/j.mito.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 120.Chen YB, Aon MA, Hsu YT, Soane L, Teng X, McCaffery JM, Cheng WC, Qi B, Li H, Alavian KN, Dayhoff-Brannigan M, Zou S, Pineda FJ, O’Rourke B, Ko YH, Pedersen PL, Kaczmarek LK, Jonas EA, Hardwick JM. Bcl-xL regulates mitochondrial energetics by stabilizing the inner membrane potential. J Cell Biol. 2011;195:263–276. doi: 10.1083/jcb.201108059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maniam S, Coutts AS, Stratford MR, McGouran J, Kessler B, La Thangue NB. Cofactor Strap regulates oxidative phosphorylation and mitochondrial p53 activity through ATP synthase. Cell Death Differ. 2015;22:156–63. doi: 10.1038/cdd.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanadi DR, Pringle M, Kantham L, Hughes JB, Srivastava A. Evidence for the involvement of coupling factor B in the H+ channel of the mitochondrial H+-ATPase. Proc Natl Acad Sci U S A. 1984;81:1371–1374. doi: 10.1073/pnas.81.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zheng J, Ramirez VD. Purification and identification of an estrogen binding protein from rat brain: oligomycin sensitivity-conferring protein (OSCP), a subunit of mitochondrial F0F1-ATP synthase/ATPase. J Steroid Biochem Mol Biol. 1999;68:65–75. doi: 10.1016/s0960-0760(98)00161-7. [DOI] [PubMed] [Google Scholar]


