SUMMARY
Minimal hammerhead ribozymes have been characterized extensively by static and time-resolved crystallography as well as numerous biochemical analyses, leading to mutually contradictory mechanistic explanations for catalysis. We present the 2.2 Å resolution crystal structure of a full-length Schistosoma mansoni hammerhead ribozyme that permits us to explain the structural basis for its 1000-fold catalytic enhancement. The full-length hammerhead structure reveals how tertiary interactions occurring remotely from the active site prime this ribozyme for catalysis. G-12 and G-8 are positioned consistent with their previously suggested roles in acid-base catalysis, the nucleophile is aligned with a scissile phosphate positioned proximal to the A-9 phosphate, and previously unexplained roles of other conserved nucleotides become apparent within the context of a distinctly new fold that nonetheless accommodates the previous structural studies. These interactions permit us to explain the previously irreconcilable sets of experimental results in a unified, consistent, and unambiguous manner.
INTRODUCTION
Hammerhead ribozymes are small self-cleaving RNAs, first discovered in viroids and satellite RNAs of plant viruses (Forster and Symons, 1987; Prody et al., 1986), that catalyze a specific phosphodiester bond isomerization reaction in the course of rolling-circle replication. More recently, hammerhead-encoding sequences have been identified in the DNA genomes of schistosomes and other nonviral species. The hammerhead ribozyme can be truncated to a minimal, catalytically active motif consisting of three base-paired stems flanking a central core of 15 mostly invariant nucleotides (Haseloff and Gerlach, 1989; Ruffner et al., 1990; Symons, 1997; Uhlenbeck, 1987). The conserved central bases, with few exceptions, are essential for the hammerhead ribozyme’s catalytic activity. Most of the biochemical and structural studies aimed toward understanding the catalytic mechanism (Blount and Uhlenbeck, 2005; McKay, 1996; Scott, 1999a; Wedekind and McKay, 1998) have employed the minimal construct.
Since its discovery, the hammerhead ribozyme has been the subject of numerous biochemical and biophysical investigations, yet the relationship between the structure of the hammerhead RNA and its catalytic activity has been a subject of major and increasing controversy (Blount and Uhlenbeck, 2005) since the first structures appeared (Pley et al., 1994; Scott et al., 1995). Two competing and seemingly mutually exclusive hypotheses (Murray et al., 1998b; Wang et al., 1999) have emerged regarding the extent of the conformational change required to activate catalysis in the ribozyme; each has now accumulated a substantial body of work to support it, but a compelling case for excluding either set of data has never been attained.
The original hammerhead ribozyme crystal structures were obtained from minimal sequences with non-cleavable substrate analogs. The first structure (Pley et al., 1994) incorporated an all-DNA substrate analog, and the second structure (Scott et al., 1995) replaced the cleavage-site nucleophile with an inert 2′-O-methyl ether linkage in an otherwise all-RNA construct. In both of these structures, the attacking nucleophile, the 2′ oxygen of the cleavage-site ribose, must rotate at least 90° to mount an in-line attack upon the scissile phosphate. Hence, a conformational change in the minimal hammerhead structures is required to account for the previously observed inversion at the scissile phosphate (Koizumi and Ohtsuka, 1991; Slim and Gait, 1991; van Tol et al., 1990) indicative of an inline attack mechanism. Subsequent time-resolved crystallographic analyses conducted with hammerhead RNAs with active, unaltered nucleophiles have revealed a series of localized cleavage-site conformational changes (Dunham et al., 2003; Murray et al., 1998b; Scott et al., 1996) as well as the structure of an enzyme-product complex (Murray et al., 2000). These structures appeared to represent at least a subset of the conformational changes required for catalysis. Cleavage of the crystallized minimal hammerhead RNA sequence was faster and more extensive within the crystal than in solution (Murray et al., 2002), suggesting that the global fold must somehow correspond to the catalytically active form of the ribozyme.
Many of the biochemical experiments performed in solution, in contrast, appear to indicate that much larger-scale conformational changes than those observed in the crystal structure analyses must take place before or accompanying catalysis (Blount and Uhlenbeck, 2005; Heckman et al., 2005; Suzumura et al., 2000; Wang et al., 1999). The experimental results most strikingly incompatible with the crystallographic analyses include the apparent proximity between the A-9 and scissile phosphates. These phosphates are almost 20 Å apart in the minimal hammerhead crystal structures. When both are substituted with phosphorothioates, a single bridging divalent soft metal ion such as Cd2+ rescues catalysis (Blount and Uhlenbeck, 2005; Wang et al., 1999). In addition, a chemical crosslinking study (Heckman et al., 2005) and an NMR analysis (Simorre et al., 1997) each imply that U-4 and U-7 are in closer proximity in the active hammerhead ribozyme than is observed in the crystal structures of the minimal hammerhead. The universally assumed strict requirement for divalent metal ions for hammerhead ribozyme catalysis was refuted (Murray et al., 1998a), yet controversy has remained (O’Rear et al., 2001) over whether magnesium is required for the chemical step of the self-cleavage reaction or whether, when present, it plays a more ancillary (and nonessential) structural role (Scott, 1999b).
Recently, sequence-specific stabilizing interactions capable of enhancing the reaction rate by a factor of almost 1000 were found to exist between the more distal parts of stems I and II of many naturally occurring hammerhead ribozymes (Canny et al., 2004; De la Peña et al., 2003; Khvorova et al., 2003). The nucleotides implicated in these distal contacts are absent in the minimal hammerhead sequences. The tertiary interactions that occur only in the full-length ribozyme sequences (Forster and Symons, 1987; Prody et al., 1986) may in fact mediate switching between nuclease and ligase activities during satellite RNA virus replication. The hypothesis that the full-length hammerhead ribozyme might therefore stabilize a catalytically active conformation that is evanescent in the minimal hammerhead compelled us to obtain its three-dimensional structure.
RESULTS AND DISCUSSION
A 63 nucleotide hammerhead ribozyme composed of enzyme and substrate-analog strands was prepared based upon a well-characterized and highly catalytically active sequence derived from the Schistosoma hammerhead RNA (Canny et al., 2004; Khvorova et al., 2003). The substrate-analog strand incorporated a 2′-OMe C-17 at the active site to prevent cleavage and two nonessential 5Br-U residues for MAD phasing, as well as a nonessential deoxy-C at the 3′ end to enhance synthetic yield, in an otherwise all-RNA construct. The sequence used for crystal structure determination, containing five base-pair switches in nonessential regions, is given in Figure 1. These changes, introduced to enhance crystallization, were shown not to disrupt catalytic activity (see the Supplemental Data available with this article online).
Figure 1. The Sequence and the Structure.
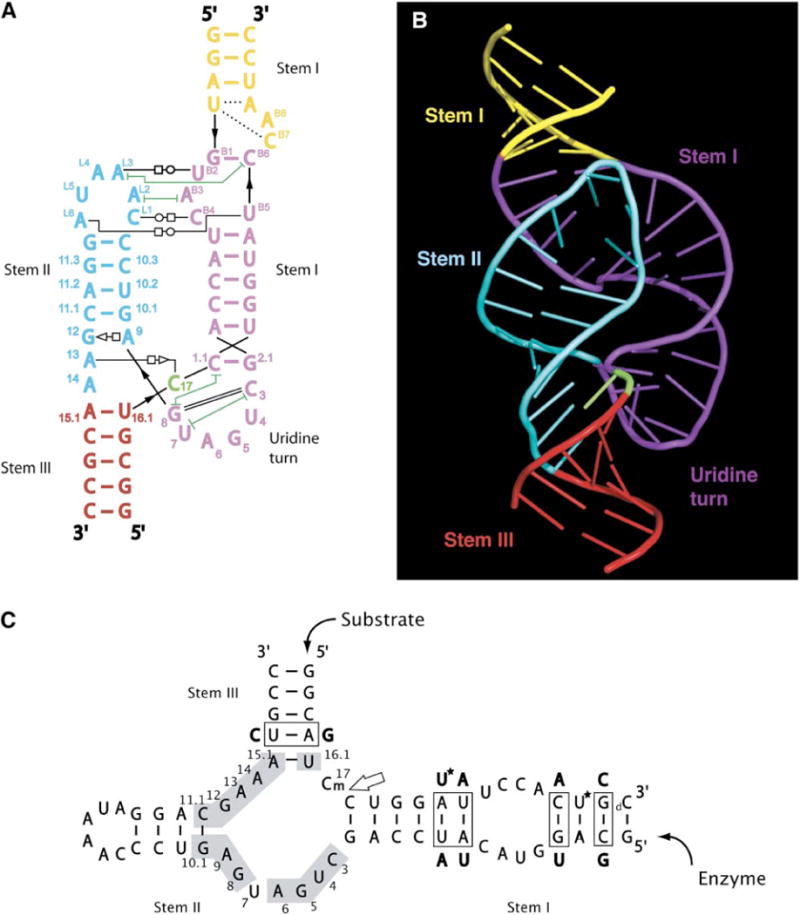
The secondary structure of the full-length hammerhead ribozyme (A) is oriented and color coded to complement the three-dimensional structural representation (B). Canonical base-pairing interactions are shown as colored lines, and other hydrogen bonding interactions are shown as thin black lines having various annotations according to the following key (Yang et al., 2003): open circle next to open square = Watson-Crick/Hoogsteen; open square next to open triangle = Hoogsteen/sugar edge; dashed line = single hydrogen bond; green lines with T termini = nonadjacent base stacking. Thick black lines indicate backbone continuity where the sequence has been separated for diagrammatic clarity. The sequence used for structural determination, and its derivation from the Schistosoma hammerhead ribozyme, is shown in (C). The conserved nucleotides are numbered using the standard convention and are shown on a gray background. The large arrow points to the scissile bond. Base-pair switches from the wild-type sequence, introduced to aid crystallization, are boxed, and the base substitutions we employed are indicated adjacent to the boxes. Two 5-bromo-uridine substitutions, used for MAD phasing a single crystal, are indicated by asterisks. The lowercase m signifies a 2′-O-methyl modification at C-17 to prevent cleavage. The lowercase d designates a deoxynucleotide used at the 3′ end of the substrate to increase synthetic yields. Figure 1B and all subsequent molecular model figures were made with PyMOL (http://pymol.sourceforge.net/).
The full-length Schistosoma hammerhead ribozyme formed monoclinic (C2) crystals having one molecule per asymmetric unit with a solvent content of 59.9% (Table S1). An unambiguous Br-phased 2.2 Å resolution MAD map permitted tracing of the entire molecule prior to refinement. The crystal lattice is stabilized by stacking interactions between helical ends of the molecules, van der Waals interactions between adjacent helices, and two intermolecular nucleotide interactions described below.
Global Fold
The full-length Schistosoma hammerhead structure presented here is 50% larger than a typical minimal hammerhead. The region of this structure corresponding to the minimal motif is similar to the minimal hammerhead fold—three helices form a junction resembling a lowercase γ in which stem II aligns coaxially with stem III. However, the remaining portion of the Schistosoma ribozyme, composed of the extended stem I and II, rejoins in an inverted lowercase γ, creating a helical bubble-like structure. This has an effect of stacking the terminal section of stem I coaxially upon stems II and III while distorting the lower part of stem I.
Tertiary Contacts between Stems I and II
One of the most prominent features of the full-length hammerhead ribozyme structure is the stem II loop/stem I bulge interaction that appears to induce the structural organization of the catalytic core. The loop/bulge interaction is composed of an intricate network of inter-helical non-canonical base pairs and stacks interdigitating the stem II loop into stem I (Figure 1 and Figure 2), kinking stem I in such a way as to coaxially align its distal helix on top of the stem II-stem III coaxial arm. The bulge and loop nucleotides are also involved in the stabilization of the crystal lattice. The B-8 adenosine participates in crystal contacts by stacking onto the B-8 adenine of the adjacent molecule while, on the opposite side of the molecule, N3 of the L-4 adenosine forms a stabilizing hydrogen bond with the 2′ oxygen of G-11.3.
Figure 2. Stereo Diagrams of the Full-Length Hammerhead Ribozyme and the Experimentally Phased MAD Electron Density Map.
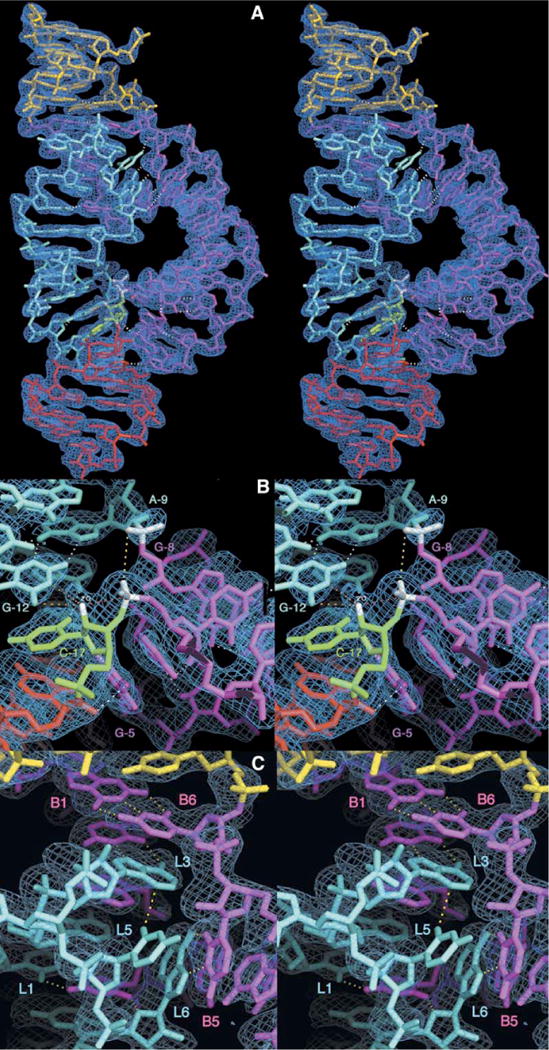
(A) Refined atomic model of the full-length Schistosoma mansoni hammerhead ribozyme superimposed upon the initial 2.2 Å resolution MAD electron density map contoured at 1.25 rmsd. The color scheme follows that introduced in Figure 1. Hydrogen bonds that mediate tertiary interactions between stems I and II and within the active site are shown as dotted white lines. (B) Closeup of the active site with the same electron density map. Several of the nucleotides implicated in catalysis, including G-12, G-8, A-9, and C-17, are labeled. In addition to the white hydrogen bonds, the yellow dotted line indicates the 4.3 Å distance between the A-9 and scissile phosphates, and the red dotted line indicates the direction of in-line attack of the 2′-O of C-17 upon the adjacent scissile phosphate. The nucleophile and the scissile phosphate, as well as the A-9 phosphate, are highlighted in white. The orange dotted lines represent contacts or hydrogen bonds that may be active in catalysis. The extra bit of density surrounding the 2′-O of C-17 corresponds to the 2′-CH3 bound to the 2′-O to prevent the cleavage reaction from taking place. The methyl group has been omitted from the figure to emphasize the additional electron density but is included in the coordinates that have been deposited in the Protein Data Bank (ID code 3ZP8).
(C) Closeup of the stem II loop/stem I bulge interaction, showing a hydrogen bonding network summarized in Figure 1A.
More importantly, however, the tertiary contacts between the loop and bulge regions induce structural changes affecting the catalytic core. This interaction imparts a severe bend upon, and partial unwinding of, stem I, compared to the more canonical A form stem I helix observed in minimal hammerhead crystal structures. These distortions appear to accommodate G-8 and U-7 in the catalytic pocket and in turn stabilize the rearrangement of the augmented stem II helix that enables G-8 to form the Watson-Crick base pair with C-3 in the catalytic pocket. Concurrently, an overwinding or right-handed twist of stem II positions the conserved G-12, A-13, and A-14 precisely against the catalytic-site C-17, helping to lock the latter in a catalytically active conformation in which C-17 is oriented for in-line attack (Figure 1 and Figure 2).
New Tertiary Contacts within the Active Site
The structure of the invariant uridine turn and cleavage site of the full-length hammerhead ribozyme differs significantly from that of the minimal hammerhead. In the full-length hammerhead ribozyme, the invariant nucleotides implicated in catalysis form base pairings and extensive hydrogen bonding networks in a rather compact arrangement, whereas in the minimal hammerhead structures, these same nucleotides are distributed widely across the three-helix junction, making very few contacts with one another (Figure 3). The cleavage-site base, C-17, resides outside of the uridine turn in the full-length structure, positioned almost perpendicular to G-5 and A-6, whose exocyclic functional groups are oriented toward the ribose. This is most clearly visible in Figure 2B, where G-5 and A-6 appear to be wedged between C-17 and stem I, splaying apart C-17 and C-1.1. As with the minimal hammerhead uridine turn, G-5 and A-6 stack upon one another, but, unlike the minimal hammerhead, the bases in the full-length ribozyme are both oriented toward the exposed ribose of C-17 and are involved in an extensive hydrogen bonding network that helps to position C-17. The resulting orientation of C-17 brings the attacking 2′ oxygen within about 3 Å of the scissile phosphorus and within 17° of perfect in-line geometry, indicating that the hammerhead ribozyme is closely approaching the expected trigonal bipyramidal transition-state phosphate configuration in which the attacking nucleophile and leaving-group oxygens occupy the axial positions of a pentacoordinated oxyphosphorane. G-8 now occupies the position of C-17 observed in the minimal hammerhead structures, forming a Watson-Crick base pair with C-3 of the uridine turn. U-7 (rather than U-4 in the minimal hammerhead structure) now stacks beneath C-3. U-7 and U-4 thus reside close together in space but do not hydrogen bond to one another (Figure 3B).
Figure 3. Side-by-Side Comparison of the Minimal and Full-Length Hammerhead Ribozyme Structures.
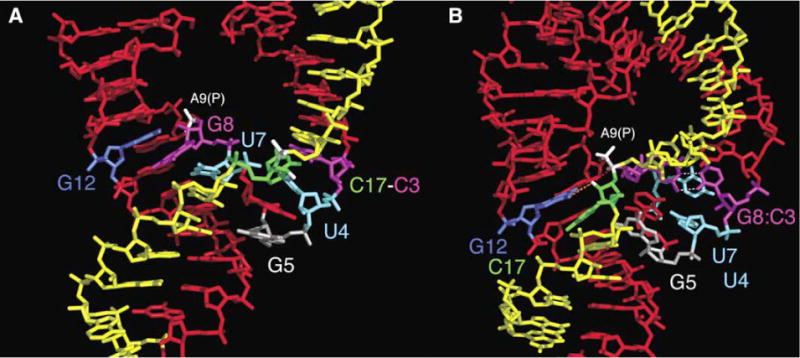
Several dramatic changes induced by the loop/bulge tertiary interaction between stems I and II reshape the active site of the full-length hammerhead ribozyme (B) relative to that of the minimal hammerhead (A). The most radical of these are the formation of a base pair between G-8 and C-3 (magenta) and the complete reorientation of the cleavage-site base, C-17 (green). As a result, the adjacent scissile phosphate (highlighted in white) approaches within 4.3 Å of the A-9 phosphate (also in white). The position of G-12 (blue) is similar in both structures, but in the full-length hammerhead, it interacts with the cleavage-site ribose (orange dotted line), whereas G-8 (magenta) rotates from the position occupied in stem II of the minimal structure to pair with C-3 (magenta), creating an extension of stem I. Similarly, U-7 (cyan) in the full-length hammerhead rotates into the space occupied by the base of U-4 (cyan). G-5 (silver) in the uridine turn helps to position C-17 (green) in the full-length structure by forming a hydrogen bond to its furanose oxygen. The differences are illustrated dynamically with an adiabatic morphing between the minimal and full-length active-site structures (see Movie S1 in the Supplemental Data).
The endocyclic N1 of G-12 is within hydrogen bonding distance of the 2′ oxygen attacking nucleophile of C-17, permitting us to suggest that G-12, when deprotonated, may serve as a general base that initiates the cleavage reaction. The 2′-OH of G-8 appears to hydrogen bond to the 5′ oxygen of the scissile phosphate, suggesting its possible involvement in general-acid-catalysis-mediated leaving-group charge stabilization in the cleavage reaction.
As a result of these conformational rearrangements, the scissile phosphate adjacent to C-17 becomes positioned quite close to the A-9 phosphate. The closest nonbridging oxygen distance between the two phosphates is now 4 Å, rather than 20 Å as in the minimal hammerhead structure. A stereo view of the active site of the full-length hammerhead ribozyme, depicting these interactions and the experimentally phased electron density map that defines their positions at 2.2 Å resolution, is provided in Figure 2B.
The Full-Length Hammerhead Structure Corroborates Previous Experiments
The full-length hammerhead ribozyme structure enables us to explain experiments thought to be irreconcilable with crystal structures of the minimal ribozyme constructs.
The previously observed inversion of configuration of the scissile phosphate (Koizumi and Ohtsuka, 1991; Slim and Gait, 1991; van Tol et al., 1990) conclusively demonstrated that phosphodiester isomerization takes place via an in-line attack mechanism in which the 2′ oxygen of C-17 is the nucleophile. However, in the minimal hammerhead crystal structures (Pley et al., 1994; Scott et al., 1995), the cleavage-site nucleotide, C-17, was not observed to be in position for an in-line attack upon the scissile phosphate. In the full-length hammerhead ribozyme structure, by contrast, the attacking nucleophile is now unambiguously positioned for in-line attack (Figure 2B and Figure 4A).
Figure 4. Positioning of Nucleotide Functional Groups for Acid-Base Catalysis.
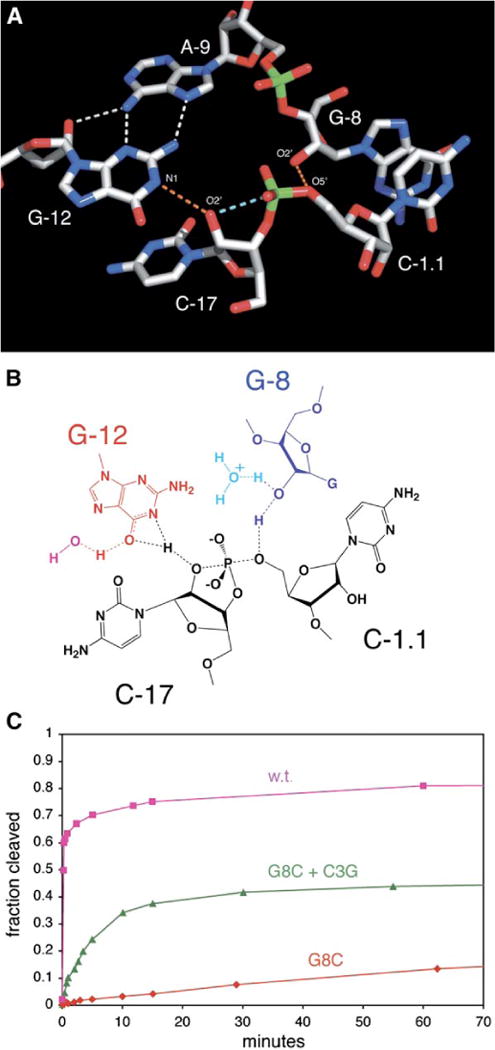
(A) Atoms are color coded as follows: carbon, gray; oxygen, red; nitrogen, blue; phosphorus, green. Hydrogen bonds are shown as white and orange dotted lines, and the light blue dotted line shows that the 2′ oxygen, the attacking nucleophile of C-17, is in line with the scissile phosphorus and leaving-group 5′ oxygen. The orange dotted lines indicate hydrogen bonds that are potentially active for acid-base catalysis.
(B) A hypothetical transition-state configuration involving G-12 (red) as a general base, G-8 (blue) as a general acid, and the substrate RNA (black). Specific-base catalysis via a water or hydroxide ion (magenta) may also play a role in this mechanism. Similarly, specific-acid catalysis from a water molecule or hydronium ion (cyan) avoids the need to invoke a fully charged alkoxide anion.
(C) The G8-C3 base pair is important for catalysis. The fraction of substrate cleaved as a function of time at pH 7.4 for the active sequence and mutations is shown. Pink squares represent the cleavage time course of the crystallographic construct (kobs = 50 min−1, a positive control). Red diamonds represent the activity of the G8C single mutant (kobs < 0.0001, a negative control), which decreases the activity about 500,000-fold. Green triangles represent the activity of the C3G + G8C double mutant (kobs = 0.22 min−1), which rescues the activity of the ribozyme, demonstrating the importance of the base pair.
The three invariant guanosines, G-5, G-8, and G-12, do not appear to make essential contacts in the minimal hammerhead structure, yet it has been shown that altering even a single functional group on any of these nucleotides abolishes catalysis (Blount and Uhlenbeck, 2005; Horton et al., 1998; McKay, 1996; Wedekind and McKay, 1998). The full-length ribozyme structure provides compelling reasons for such strict conservation. G-5 is observed to contribute to the hydrogen bonding network responsible for forming the specific binding pocket into which C-17 is positioned, while G-8 makes a Watson-Crick pair with C-3. The N1 of G-12 is within hydrogen bonding distance of the 2′ oxygen, suggesting that near the apparent pKa of the ribozyme, 8.5, it might be able to act (in a deprotonated form) as a general base that abstracts the 2′ proton at the active site. The essential (McKay, 1996) 2′-OH of G-8, similarly, is within hydrogen bonding distance of the leaving-group 5′ oxygen, suggesting that this secondary alcohol might be positioned to donate a proton to the primary 5′ alkoxide that forms as the cleavage reaction takes place, thus acting as a general-acid catalyst. The positions of G-12 and G-8 are in general agreement with a recently proposed mechanism for the involvement of these nucleotides in acid-base catalysis (Han and Burke, 2005). The observed pH-dependent sensitivity of G-8 to modifications (Han and Burke, 2005) is also consistent with the observed base-pairing interaction with C-3.
Chemical crosslinking (Heckman et al., 2005) and NMR (Simorre et al., 1997) studies have suggested that U-4 and U-7 are close in space, in contrast to what is observed in the minimal hammerhead ribozyme structure. In the full-length ribozyme structure, U-4 is rotated to create room for U-7 to stack under C-3, bringing U-7 within crosslinking distance of U-4.
Simultaneous substitutions of the pro-R phosphate nonbridging oxygens of A-9 and the scissile phosphate in the hammerhead ribozyme (Wang et al., 1999) appear to create a binding site for a single soft metal ion (such as Cd2+), yet these phosphates are almost 20 Å from one another in the minimal hammerhead ribozyme structures (Pley et al., 1994; Scott et al., 1995). In the full-length ribozyme structure, these nonbridging oxygens are within about 4 Å of one another and could potentially bind a single divalent metal ion without further rearrangement. Phosphorothioate metal rescue has in fact been observed for individual substitutions of these sites in the full-length hammerhead at low ionic strength (Osborne et al., 2005). However, no Mg2+ ions were observed in the crystal structure, in which a low concentration of Mg2+ and a high concentration of NH4+ are present. Under low-ionic-strength conditions, divalent metal ions appear to stimulate folding of the full-length ribozyme (Kim et al., 2005; Osborne et al., 2005; Penedo et al., 2004) and might help to stabilize the active fold by minimizing electrostatic repulsion. In the presence of molar concentrations of NH4+ ions, however, divalent cations are not required for catalysis in the full-length hammerhead sequence crystallized, in accordance with our previous findings on various minimal hammerhead constructs (Murray et al., 1998a).
How Does the Full-Length Hammerhead Activate Catalysis?
Based on the newly observed interactions listed above, the stem II loop/stem I interactions stabilize or constrain a conformation within the active site that appears to position G-12 for general-base catalysis. G-8, in pairing with C-3, appears poised to supply a 2′ proton to the leaving group as a general-acid catalyst. In addition, the activesite conformation stabilized by the distal tertiary interactions orients the 2′ oxygen attacking nucleophile of C-17 such that it becomes aligned with the scissile phosphate and leaving-group 5′ oxygen of the cleavage site. These arrangements are most clearly seen in Figure 4A.
General-Base Catalysis and G-12
The N1 of G-12 is almost certainly protonated in the crystal structure of the full-length hammerhead ribozyme and is within approximate hydrogen bonding distance (3.5 Å) of the 2′-O of C-17. Because C-17 has been modified with a 2′-OMe to prevent the cleavage reaction, there is no 2′-H to be abstracted. If G-12 functions as a general base, as its positioning and previous results (Han and Burke, 2005) suggest, it is most likely that the 2′ proton would be abstracted by a deprotonated N1. The pKa of G-12 would need to shift from about 9.4 to 8.5 to be consistent with the apparent pKa of the hammerhead ribozyme. Substitution of G-12 with inosine (pKa 8.7), 2,6-diaminopurine (pKa 5.1), or 2-aminopurine (pKa 3.8) shifts the reaction-rate profile in a manner consistent with G-12′s suggested role in general-base catalysis (Han and Burke, 2005). However, a formal negative charge on N1 seems unlikely, and the possibility that a tautomeric fluctuation participates in this catalytic scheme may also be considered. In our crystal structure, the exocyclic O6 oxygen of G-12 is 3.3 Å from the 2′-O of C-17. Ordinarily, the ketolic form dominates well in excess of 99%, but a transient tautomeric fluctuation might allow the 2′-H of C-17 to be abstracted by N1, whose labile proton resides on O6 until it is abstracted by an ambient water molecule or hydroxide ion. As the 2′ proton is removed, the covalent bond between the 2′-O nucleophile and the prealigned scissile phosphorus atom could then proceed to form. This mechanism in a sense involves specific base catalysis, invoking a proton relay, and possesses the merit of not requiring a full negative charge to accumulate on either the N1 or the O6 of G-12. A possible mechanistic scheme wherein a water molecule abstracts the enolic proton from G-12, shown in Figure 4B, uses the overwhelming propensity to return to the keto tautomeric form to ensure that the 2′ oxygen does not recapture a proton but instead forms a bond with the adjacent phosphorus.
General-Acid Catalysis and G-8
Donation of a proton is required to counter the accumulating negative charge on the 5′ leaving group of the hammerhead cleavage reaction, a primary alkoxide. Previous biochemical analyses (Han and Burke, 2005) have implicated G-8 as a possible general acid in the hammerhead ribozyme cleavage reaction. The simplest explanation for this observation, in the absence of structural information, is that the proton on the N1 of G-8 is labile and can be donated to the 5′ oxygen leaving group in the cleavage reaction as it is displaced from the scissile phosphate. However, in the crystal structure, the base of G-8 is paired with C-3, and it is the 2′-OH of G-8 that is observed to hydrogen bond rather tightly (3.18 Å) to the 5′-O leaving group (Figure 4A). Thus, if G-8 functions as a general acid in the cleavage reaction, either it must break its pairing with C-3 and reorient toward the leaving group or it is the 2′-OH of G-8 that is the relevant functional group in acid catalysis. To test between these hypotheses, we switched the bases within the pair to C-8 and G-3. Both G-8 and C-3 are known to be invariant; changing either nucleotide abolishes activity in the minimal hammerhead (McKay, 1996). However, we observed (Figure 4C) that the G8C + C3G double mutant (kobs = 0.22 min−1 at pH 7.5) rescues the deleterious effects of the single G8C mutation (kobs < 0.0001 at pH 7.5), demonstrating that the base pairing, when reestablished, restores catalysis. This result, combined with the repeated observation that the 2′-OH of G-8 is required for hammerhead ribozyme activity (McKay, 1996), in turn permits us to suggest that the ribose 2′-OH of G-8 is indeed the active functional group in hammerhead ribozyme acid catalysis. Although it is clear that a secondary alcohol would readily donate a proton to a primary alkoxide, the reverse reaction seems less plausible. A more obviously reversible reaction might again involve partial proton dissociation in the transition state, accompanied by a water assisting as a specific-acid catalyst. Such a scheme is presented in the hypothetical transition-state structure depicted in Figure 4B. Han and Burke (2005) observe that G-8, but not G-12, mutations partially inhibit ribozyme folding, consistent with the G-8/ C-3 base pair.
Transition-State Stabilization and the Role of Various Solvent Cations
An excess of negative charge is predicted to accumulate in the pentacoordinated oxyphosphorane transition state. A transition-state stabilization interaction that would mask the electrostatic repulsive force between the oxyphosphorane and the A-9 phosphate would likely contribute to catalytic enhancement. The molar concentration of ammonium cations in the crystal environment provides sufficient charge screening to enable the full-length hammerhead ribozyme to fold into the observed conformation. Our observation that the full-length hammerhead ribozyme is active in molar concentrations of nonmetallic monovalent ammonium ions, both in the absence of and when supplemented with Mg2+, indicates that any generic high concentration of positive charge is sufficient to stabilize the hammerhead ribozyme transition state (data not shown). A divalent metal ion, if present in a low-ionicstrength solution, might help to stabilize the close approach between the A-9 and the scissile phosphates.
In an effort to identify any specific divalent metal-ion binding sites, two additional experiments, in which either 50 mM Mg2+ or 10 mM Mn2+ was soaked into full-length hammerhead ribozyme crystals, were performed. No Mg2+ ions in the 50 mM soak were observed near the active site, and a single, low-occupancy Mn2+ site similar to the binding mode observed in the minimal hammerhead constructs (but notably not bridging the two phosphates) was observed in a 2.0 Å resolution data set (data not shown). The potential involvement of G-12 and G-8 in acid-base catalysis and the ability of the RNA to fold into a structure that is unambiguously poised for catalysis in the presence of ammonium ion, together with our observation that no metal ions appear in the active site, support the hypothesis that the hammerhead ribozyme possesses all of the chemical constituents that are strictly required for the self-cleavage reaction (Murray et al., 1998a; Scott, 1999b).
Comparing the Hammerhead to Other Self-Cleaving Ribozymes
The hammerhead, hepatitis delta virus (HDV), and hairpin ribozymes each catalyze the same phosphodiester isomerization reaction. However, despite some superficial similarities, each of these ribozymes deploys a unique strategy for affecting catalysis.
The HDV ribozyme, for instance, is an obligate metalloenzyme, requiring magnesium for catalysis under all conditions (Ferre-D’Amare et al., 1998; Ke et al., 2004; Murray et al., 1998a; Nakano et al., 2001, 2003; Scott, 1999b). In the uncleaved structure, C-75 is located nearest to the cleavage-site 2′-O, and a divalent metal ion appears to make a through-water contact to the 5′-O leaving group. C-75 is therefore believed to abstract the 2′-H in the (unobserved) transition state, thus functioning as a general base in the cleavage reaction, and the hydrated divalent metal ion is thought to be the general acid, supplying a proton from a coordinated water to the cleavage-site 5′-O as a negative charge develops. Thus, in sharp contrast to the hammerhead and hairpin ribozymes, a pyrimidine nucleotide base appears to play the role of a general base in the cleavage reaction, and a hydrated metal ion appears to play the role of the general acid.
Unlike the HDV ribozyme, the hammerhead and hairpin ribozymes are not metalloenzymes (Curtis and Bartel, 2001; Murray et al., 1998a; O’Rear et al., 2001). All ribozymes were once thought to be metalloenzymes (Dahm et al., 1993; Dahm and Uhlenbeck, 1991; Pyle, 1993) that use hydrated metal ions (Dahm et al., 1993) or inner-sphere-coordinated metal ions (Lott et al., 1998) in catalysis. The hammerhead and hairpin, rather than simply functioning as passive scaffolds for metal-ion binding (Yarus, 1993), are now seen to have structures in which the nucleotide functional groups, as previously suggested (Murray et al., 1998a; Scott, 1999b), are clearly positioned for acid-base catalysis.
The first structure of the hairpin ribozyme (Rupert and Ferre-D’Amare, 2001) revealed that the N1 of G-8 in the hairpin ribozyme active site is within hydrogen bonding distance of the cleavage-residue 2′ oxygen, suggesting its role as the base in the cleavage reaction, and the subsequent transition-state analog structure (Rupert et al., 2002) revealed an adenosine (A-38) positioned near the 5′ oxygen, suggesting its role as the acid. The exocyclic amines of G-8, A-38, and A-9 were also observed to hydrogen bond to the nonbridging oxygens, presumably helping to disperse the excess negative charge that accumulates in the transition state.
In contrast to the hairpin, the uncleaved complex of the full-length hammerhead ribozyme reveals contacts with both the attacking nucleophile (the 2′-O) and the leaving group (the 5′-O). G-12 in the hammerhead appears to play the same role as G-8 in the hairpin, i.e., that of a general base in the cleavage reaction, whereas a protonated N1 of A-38 in the hairpin ribozyme transition-state analog structure appears to play the role of the hammerhead’s G-8 2′-OH, i.e, that of a general acid in the cleavage reaction.
The uncleaved hammerhead structure, unlike the hairpin ribozyme, does not make contacts to the scissile phosphate nonbridging oxygen atoms, suggesting that dispersal of the accumulating negative charge, as well as screening of the electrostatic repulsion between the A-9 and scissile phosphates, is accomplished via nonspecific cation interactions, where ammonium, other monovalent cations, or divalent metal ions can supply a high concentration of positive charge. This is consistent with the observation that the hammerhead ribozyme requires that a greater concentration of monovalent cations be present for full activity than the hairpin ribozyme does (Murray et al., 1998a; Scott, 1999b), and it also explains why the hammerhead ribozyme appears to require divalent cations under low-ionic-strength conditions.
How (and Why) Does the Minimal Hammerhead Function?
Despite dramatic differences within the active site, the minimal hammerhead ribozyme can cleave in the crystal (Murray et al., 1998b, 2000, 2002; Scott et al., 1996). The pH dependence is similar to that observed in solution, and the same cleavage products are obtained (Murray et al., 2002). The rate of cleavage for the minimal hammerhead ribozyme is greater in the crystal than in solution under otherwise identical conditions (Murray et al., 1998b, 2002), approaching the rate observed for the most active minimal hammerhead sequence, HH-α1 (Clouet-d’Orval and Uhlenbeck, 1997). The extent of substrate cleavage in the crystal is about 95%, whereas the extent of cleavage for the same RNA in solution is only about 70%, indicating that the crystal lattice is apparently doing more to enhance than inhibit the cleavage reaction (Murray et al., 2002). Crystal-lattice contacts restrict the positions of the distal ends of stems I, II, and III (Dunham et al., 2003), making it difficult to reconcile this and the accompanying 5-fold cleavage-rate enhancement with a large-scale change in the global fold of the minimal hammerhead RNA.
The solution to this conundrum is that the original, minimal hammerhead fold is approximated within the global fold of the full-length hammerhead ribozyme, as shown in Figure 5. This is consistent with observations made in solution (Penedo et al., 2004). Therefore, cleavage would not be prevented in a crystal lattice that confines the RNA to the observed minimal hammerhead fold but simultaneously permits rearrangement of the active site, at least transiently, into that observed in the full-length structure. Tethering stem I to stem II within the minimal hammerhead crystals has the effect of permitting a pre-cleavage conformational change to occur yet prevents catalytic turnover (Dunham et al., 2003). This result is consistent with the need for stem I to unwind and undergo the comparatively small helical conformational changes required for cleavage to occur, such as those that appear by comparing the pre-cleavage and post-cleavage hammerhead folds with the analogous section of the full-length hammerhead structure, as seen in Figures 5C, 5E, and 5D, respectively.
Figure 5. Aspects of the Full-Length Hammerhead Ribozyme Fold.
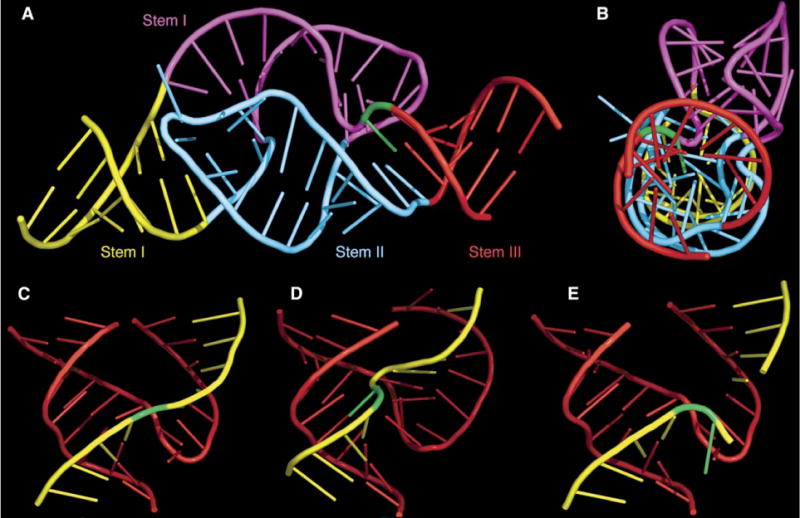
(A) Stems II and III are almost perfectly coaxial with the distal end of stem I. The remainder of stem I is quite curved and unwound.
(B) 90° rotation of the previous view, highlighting the coaxial stacking of all three stems as well as the profoundly distorted and unwound geometry of the remainder of stem I, highlighted in magenta.
(C) The region of the fold of the full-length hammerhead ribozyme (A) that corresponds to the previous, truncated hammerhead ribozyme structures. (D and E) Truncated precleavage (D) and postcleavage (E) hammerhead folds are shown for comparison. In each case, the enzyme strand is red; the substrate is yellow; and C-17, the cleavage-site nucleotide, is green.
Conclusions
The full-length hammerhead ribozyme structure differs significantly from those solved previously, presenting a refreshing look at how RNA structure induces hammerhead ribozyme catalysis. The Schistosoma ribozyme structure provides insight into how distant loop/bulge interactions affect the catalytic-core conformation by inducing changes in stem II while simultaneously unwinding stem I. Due to these changes, the catalytically important nucleotides become organized in such a manner as to align the catalytic-site C-17 very close to predicted active geometry. Moreover, we observe a fold familiar from the minimal hammerhead structures, yet, unlike before, it is now possible to formulate a structural explanation for the most challenging biochemical experiments thought to be incommensurate with the earlier structural data. In addition, the full-length hammerhead ribozyme structure reveals the potential roles of the invariant nucleotides G-8 and G-12 in acid-base catalysis.
EXPERIMENTAL PROCEDURES
RNA Synthesis
A full-length hammerhead ribozyme construct derived from the Schistosoma mansoni satellite DNA sequence (Ferbeyre et al., 1998) was selected based on its well-characterized enhancement of activity over the minimal hammerhead. The sequence was divided into separate enzyme and substrate strands similar to those previously published (Khvorova et al., 2003). Five nonessential base-pair switches were introduced into the full-length hammerhead construct in order to enrich the heterodimer population and thus facilitate its crystallization without compromising its activity. These changes were introduced outside of the loop, bulge, and invariant regions, aided by an RNA secondary structure prediction algorithm (Mathews et al., 1999, 2004). The 43 nucleotide enzyme strand sequence and the 2′ nucleotide substrate sequence are shown in Figure 1. The enzyme strand was in vitro transcribed from a DNA template containing a double-stranded T7 polymerase promoter and 2′-O-methyl modifications on the terminal two nucleotides. The enzyme DNA template, as well as the substrate RNA, was synthesized using an Expedite 8900. The substrate RNA contained a 2′-O-methylcytidine at the cleavage site (C-17), a deoxycytidine at the 3′ terminus, and two 5-bromo-uridines at positions 11 and 18. (See also Figure 1).
Crystallization
The enzyme and substrate were mixed in equimolar amounts in a solution containing 50 mM MES (pH 5.5), 1.5 mM EDTA. The complex was formed by incubating the mixture at 95° C for 2 min., at 65° C for 2 min., and finally at 27°C for 5 min. One millmolar MgCl2 was included in the mixture before the final incubation step. A 10 mg/ml concentration of RNA was used for the crystallization experiments. The reservoir solution contained 0.5 M (NH4)2SO4, 100 mM MES (pH 6.5), and 35% PEG 3350. After mixing the reservoir solution, the salt and PEG phases were allowed to separate, and only the salt phase was used for the drops. The crystals grew in hanging drops of 2 ml after 12 months of incubation at 28°C. The crystals were washed in the salt phase of the reservoir solution prior to cryofreezing.
Structure Determination and Refinement
The crystal structure was determined to 2.2 Å resolution using MAD phasing on crystals containing the double-bromine derivative substrate. Although three wavelengths were collected, data obtained using just the absorption peak and inflection energies yielded the best phases and thus were used for calculating the MAD map. The structure was subsequently refined using the peak data set (See Tables S1 and S2 in the Supplemental Data). Synchrotron data were collected at SSRL beamline 1–5. Mosflm and CCP4 crystallographic software (CCP4, 1994; Winn, 2003) were used for data processing (Table S1), followed by CNS (Brunger et al., 1998) for locating two bromine sites, MAD phasing, and simulated annealing refinement. Additional refinement was carried out using the CCP4 Refmac (Murshudov et al., 1997) package (Table S2). Model building was performed using O (Jones et al., 1991) and COOT (Emsley and Cowtan, 2004). Water positions were identified and tested within COOT. Figures were prepared using PyMOL (http://pymol.sourceforge.net/) and RNAView (Yang et al., 2003).
Subsequent to refinement, the structure was checked against the original MAD map, a composite-omit map generated in CNS (Brunger et al., 1998), and a minimally biased map calculated using EDEN (Somoza et al., 1995).
Activity Assays
Cleavage assays were performed using conditions similar to those previously published (Canny et al., 2004), except that the all-ribose enzyme was in >50-fold excess of the all-ribose 5′-labeled substrate. The enzyme concentration was 16.5 mM. The crystallographic construct assay conditions were 50 mM MES (pH 5.6) (Figure S1) or 50 mM Tris (pH 7.4) (Figure 4), 100 mM NaCl, 0.1 mM EDTA, 10 mM MgCl2 at 27°C. The assay conditions for the C3G + G8C double mutant, the G8C single mutant, and the positive control (Figure 4C) were 50 mM Tris (pH 7.4), 100 mM NaCl, 0.1 mM EDTA, 10 mM MgCl2 at 27° C. Prior to adding MgCl2, the enzyme and substrate (in the assay solution) were heated to 95° for 2 min, then 65° for 2 min, then allowed to equilibrate at 27° for 5 min. At this point, a sample was removed and designated as the zero time point. The reaction was initiated by adding the MgCl2, and the subsequent time point samples were collected.
Supplementary Material
Acknowledgments
We thank K. Lieberman, M. De la Peña, A. Khvorova, C. Dunham, and J. Burke as well as H.F. Noller, M. Ares, A. and H. Szöke, J. Nix, and the members of the Center for the Molecular Biology of RNA for helpful advice and discussions. Data collection was performed at the Stanford Synchrotron Radiation Laboratory, Stanford, CA, USA, and the Advanced Light Source, Berkeley, CA, USA. This research was funded by the NIH (R01 AI043393).
Footnotes
We dedicate this paper to Sir Aaron Klug on the occasion of his 80th birthday.
Supplemental Data
Supplemental Data include one figure, three tables, and one movie and can be found with this article online at http://www.cell.com/cgi/content/full/126/2/309/DC1/.
Accession Numbers
Coordinates and Fobs have been deposited in the Nucleic Acid Database under the Protein Data Bank ID code 3ZP8 (replaces 2GOZ).
References
- Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Clouet-d’Orval B, Uhlenbeck OC. Hammerhead ribozymes with a faster cleavage rate. Biochemistry. 1997;36:9087–9092. doi: 10.1021/bi9710941. [DOI] [PubMed] [Google Scholar]
- Curtis EA, Bartel DP. The hammerhead cleavage reaction in monovalent cations. RNA. 2001;7:546–552. doi: 10.1017/s1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm SC, Uhlenbeck OC. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Dahm SC, Derrick WB, Uhlenbeck OC. Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry. 1993;32:13040–13045. doi: 10.1021/bi00211a013. [DOI] [PubMed] [Google Scholar]
- De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham CM, Murray JB, Scott WG. A helical twistinduced conformational switch activates cleavage in the hammerhead ribozyme. J Mol Biol. 2003;332:327–336. doi: 10.1016/s0022-2836(03)00843-x. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Forster AC, Symons RH. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Han J, Burke JM. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Gerlach WL. Sequences required for self-catalysed cleavage of the satellite RNA of tobacco ringspot virus. Gene. 1989;82:43–52. doi: 10.1016/0378-1119(89)90028-0. [DOI] [PubMed] [Google Scholar]
- Heckman JE, Lambert D, Burke JM. Photocrosslinking detects a compact, active structure of the hammerhead ribozyme. Biochemistry. 2005;44:4148–4156. doi: 10.1021/bi047858b. [DOI] [PubMed] [Google Scholar]
- Horton TE, Clardy DR, DeRose VJ. Electron paramagnetic resonance spectroscopic measurement of Mn2+ binding affinities to the hammerhead ribozyme and correlation with cleavage activity. Biochemistry. 1998;37:18094–18101. doi: 10.1021/bi981425p. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Kim NK, Murali A, DeRose VJ. Separate metal requirements for loop interactions and catalysis in the extended hammerhead ribozyme. J Am Chem Soc. 2005;127:14134–14135. doi: 10.1021/ja0541027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M, Ohtsuka E. Effects of phosphorothioate and 2-amino groups in hammerhead ribozymes on cleavage rates and Mg2+ binding. Biochemistry. 1991;30:5145–5150. doi: 10.1021/bi00235a005. [DOI] [PubMed] [Google Scholar]
- Lott WB, Pontius BW, von Hippel PH. A two-metal ion mechanism operates in the hammerhead ribozyme-mediated cleavage of an RNA substrate. Proc Natl Acad Sci USA. 1998;95:542–547. doi: 10.1073/pnas.95.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DB. Structure and function of the hammerhead ribozyme: an unfinished story. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998a;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Murray JB, Terwey DP, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott WG. The structural basis of hammerhead ribozyme self-cleavage. Cell. 1998b;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]
- Murray JB, Szoke H, Szoke A, Scott WG. Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction. Mol Cell. 2000;5:279–287. doi: 10.1016/s1097-2765(00)80423-2. [DOI] [PubMed] [Google Scholar]
- Murray JB, Dunham CM, Scott WG. A pH-dependent conformational change, rather than the chemical step, appears to be rate-limiting in the hammerhead ribozyme cleavage reaction. J Mol Biol. 2002;315:121–130. doi: 10.1006/jmbi.2001.5145. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- Nakano S, Cerrone AL, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- O’Rear JL, Wang S, Feig AL, Beigelman L, Uhlenbeck OC, Herschlag D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne EM, Schaak JE, Derose VJ. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DM. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Breuning G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Ribozymes: a distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- Ruffner DE, Stormo GD, Uhlenbeck OC. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Rupert PB, Massey AP, Sigurdsson ST, Ferre-D’Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- Scott WG. Biophysical and biochemical investigations of RNA catalysis in the hammerhead ribozyme. Q Rev Biophys. 1999a;32:241–284. doi: 10.1017/s003358350000353x. [DOI] [PubMed] [Google Scholar]
- Scott WG. RNA structure, metal ions, and catalysis. Curr Opin Chem Biol. 1999b;3:705–709. doi: 10.1016/s1367-5931(99)00029-0. [DOI] [PubMed] [Google Scholar]
- Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Scott WG, Murray JB, Arnold JR, Stoddard BL, Klug A. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- Simorre JP, Legault P, Hangar AB, Michiels P, Pardi A. A conformational change in the catalytic core of the hammerhead ribozyme upon cleavage of an RNA substrate. Biochemistry. 1997;36:518–525. doi: 10.1021/bi9620520. [DOI] [PubMed] [Google Scholar]
- Slim G, Gait MJ. Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res. 1991;19:1183–1188. doi: 10.1093/nar/19.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza JR, Szoke H, Goodman DM, Beran P, Truckses D, Kim SH, Szoke A. Holographic methods in X-ray crystallography. IV. A fast algorithm and its application to macromolecular crystallography. Acta Crystallogr A. 1995;51:691–708. doi: 10.1107/s0108767395002315. [DOI] [PubMed] [Google Scholar]
- Suzumura K, Warashina M, Yoshinari K, Tanaka Y, Kuwabara T, Orita M, Taira K. Significant change in the structure of a ribozyme upon introduction of a phosphorothioate linkage at P9: NMR reveals a conformational fluctuation in the core region of a hammerhead ribozyme. FEBS Lett. 2000;473:106–112. doi: 10.1016/s0014-5793(00)01499-x. [DOI] [PubMed] [Google Scholar]
- Symons RH. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- van Tol H, Buzayan JM, Feldstein PA, Eckstein F, Bruening G. Two autolytic processing reactions of a satellite RNA proceed with inversion of configuration. Nucleic Acids Res. 1990;18:1971–1975. doi: 10.1093/nar/18.8.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Karbstein K, Peracchi A, Beigelman L, Herschlag D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- Wedekind JE, McKay DB. Crystallographic structures of the hammerhead ribozyme: relationship to ribozyme folding and catalysis. Annu Rev Biophys Biomol Struct. 1998;27:475–502. doi: 10.1146/annurev.biophys.27.1.475. [DOI] [PubMed] [Google Scholar]
- Winn MD. An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat. 2003;10:23–25. doi: 10.1107/s0909049502017235. [DOI] [PubMed] [Google Scholar]
- Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M. How many catalytic RNAs? Ions and the Cheshire cat conjecture. FASEB J. 1993;7:31–39. doi: 10.1096/fasebj.7.1.8422972. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


