ABSTRACT
Bordetella fimbriae (FIM) are generally considered to function as adhesins despite a lack of experimental evidence supporting this conclusion for Bordetella pertussis and evidence against a requirement for FIM in adherence of Bordetella bronchiseptica to mammalian cell lines. Using B. bronchiseptica and mice, we developed an in vivo adherence assay that revealed that FIM do function as critically important adhesins in the lower respiratory tract. In the first few days postinoculation, FIM-deficient B. bronchiseptica induced a more robust inflammatory response than wild-type bacteria did, suggesting that FIM, like filamentous hemagglutinin (FHA), allow B. bronchiseptica to suppress the innate immune response to infection. Localization analyses indicated that FIM are required for efficient attachment to airway epithelium, as bacteria lacking FIM localized to alveoli. FHA-deficient bacteria, in contrast, localized to airways. Bacteria unable to produce both FIM and FHA localized to alveoli and caused increased inflammation and histopathology identical to that caused by FIM-deficient bacteria, demonstrating that lack of FIM is epistatic to lack of FHA. Coinoculation experiments provided evidence that wild-type B. bronchiseptica suppresses inflammation locally within the respiratory tract and that both FHA and FIM are required for defense against clearance by the innate immune system. Altogether, our data suggest that FIM-mediated adherence to airway epithelium is a critical first step in Bordetella infection that allows FHA-dependent interactions to mediate tight adherence, suppression of inflammation, and resistance to inflammatory cell-mediated clearance. Our results suggest that mucosal antibodies capable of blocking FIM-mediated interactions could prevent bacterial colonization of the lower respiratory tract.
IMPORTANCE
Although fimbriae (FIM) have been shown to be important mediators of adherence for many bacterial pathogens, there is surprisingly little experimental evidence supporting this role for Bordetella fimbria. Our results provide the first demonstration that Bordetella FIM function as adhesins in vivo, specifically to airway epithelium. Furthermore, our results suggest that FIM mediate initial interactions with airway epithelial cells that are followed by tight filamentous hemagglutinin (FHA)-mediated binding and that together, FIM and FHA allow Bordetella to suppress inflammation, leading to prolonged colonization. Given the shortcoming of the current acellular component pertussis (aP) vaccine in preventing colonization, these findings suggest that generation of antibodies capable of blocking FIM-mediated adherence could potentially prevent Bordetella colonization.
INTRODUCTION
The “classic” or mammalian Bordetella species, which include Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica, are Gram-negative bacteria that cause respiratory infections in mammals (1). B. bronchiseptica colonizes the nasopharynx and trachea in a broad range of hosts, including rabbits, rats, mice, and occasionally humans, often resulting in persistent, asymptomatic infections (2). Phylogenetic analyses indicate that Bordetella pertussis, the causative agent of whooping cough (pertussis), evolved from a B. bronchiseptica-like ancestor, narrowing its host range to humans exclusively and typically causing acute respiratory disease, particularly in infants and young children (3, 4). Widespread use of a whole-cell pertussis (wP) vaccine in the 1950s led to a rapid decrease of pertussis morbidity and mortality. Safety concerns, however, led to the replacement of the wP vaccine with acellular component pertussis (aP) vaccines (5). aP vaccines contain pertussis toxin (PTX) and one or more of the putative adhesins, filamentous hemagglutinin (FHA), fimbria (FIM), or pertactin (PRN). Coinciding with the switch to using only aP vaccines, cases of pertussis in the United States and other countries have increased steadily since the 1990s (6–8). Given the reemergence of this disease, it is important to better understand the mechanisms utilized by Bordetella to colonize and persist in the respiratory tract.
Despite differences in host range and disease-causing propensity, B. bronchiseptica and B. pertussis are extremely similar and produce a nearly identical set of virulence factors. One such virulence factor is a type I pilus system, typically called fimbria (FIM) in Bordetella. The putative chaperone, usher, and tip adhesin are encoded by the fimBCD genes, respectively, and are required for fimbrial biogenesis (9). Most Bordetella strains characterized produce FIM composed of either Fim2 or Fim3 as the major fimbrial subunit (10). The structural genes fim2 and fim3 are not linked to each other or to the fimBCD operon (10). Additional major fimbrial subunit-encoding genes have been identified, including fimX, fimN, and fimA (11–13). The fimA gene, located immediately 5′ to the fimBCD operon, is a pseudogene in B. pertussis (13). Although most aP vaccines contain the major fimbrial subunits, Fim2 and Fim3, whether antibodies against these proteins contribute to protection against colonization or disease is unknown.
Because B. pertussis is a human-specific pathogen that does not readily infect laboratory animals, we have been using B. bronchiseptica with its natural hosts to understand the contribution of specific virulence factors to Bordetella infection (14–16). The amino acid sequences of the FimD proteins produced by B. pertussis (Tohama I) and B. bronchiseptica (RB50) are 95% identical, and the major fimbrial subunits, Fim2 and Fim3, are 73% and 94% identical, respectively (9, 17, 18). It is likely that the fimbriae produced by B. bronchiseptica and B. pertussis play similar, if not identical, roles during infection, and we hypothesize that information gleaned from studies using B. bronchiseptica and natural-host animal models will be applicable to B. pertussis.
Although fimbriae have been shown to be important mediators of adherence for many bacterial pathogens, there is surprisingly little experimental evidence supporting this role for Bordetella FIM. A B. pertussis strain containing an insertion mutation in fimD was defective for adherence to adherent monocytes in vitro (19). However, as this strain is also defective for FHA production, the contribution of FIM alone could not be determined (9, 19). We previously constructed a ΔfimBCD strain of B. bronchiseptica that does not produce fimbria of any type and is unaltered for FHA production. Unexpectedly, this strain did not differ from wild-type (WT) bacteria in its ability to adhere to various epithelial and macrophage cell lines in vitro (20). However, a B. pertussis strain defective for both FIM and FHA had reduced adherence to baboon trachea explants, and FIM-defective B. bronchiseptica had reduced adherence to rabbit trachea explants (21, 22), suggesting that FIM may be important for adherence specifically to ciliated respiratory epithelial cells. Although studies have been conducted to identify host cell receptors for FIM (23–25), these experiments used purified fimbrial subunits and nonciliated cell lines and whether the interactions identified reflect those that occur with native FIM in vivo is unknown.
Using a colonization model in which rats are inoculated with a small-volume inoculum into the tip of the nose, we showed that FIM and FHA are necessary for B. bronchiseptica to colonize the lower respiratory tract, specifically the trachea (20, 26). When inoculated directly into the tracheas of rats, FIM-deficient bacteria were unable to persist in the trachea, but they colonized and persisted in the nasal cavity (20). These data suggest that FIM are required to adhere to tracheal tissue, to resist mucociliary clearance, and/or to avoid clearance by the innate immune system. Our lab and others have used a large-volume, intranasal inoculation mouse model to investigate the host response to Bordetella infection. This inoculation method presumably deposits bacteria throughout the nose, trachea, and lungs of the animal. Using this model, our lab has shown that FHA is necessary for bacterial persistence in the lower respiratory tract and that FHA-deficient bacteria induce a more robust inflammatory response than WT bacteria do (27–29). These data suggest that FHA is involved in suppressing the host immune response to aid bacterial persistence. On the basis of the similar tracheal colonization defect of ΔfhaB and ΔfimBCD mutants in rats, we hypothesized that FIM may also contribute to immune modulation and bacterial persistence in the lower respiratory tract, and we set out to test this hypothesis.
RESULTS
Fimbriae are required for adherence in vivo.
To investigate the contribution of FIM to adherence to respiratory epithelium in the context of natural infection, we developed an in vivo adherence assay. We inoculated mice intranasally with 50 µl phosphate-buffered saline (PBS) containing 7.5 × 104 CFU of bacteria, euthanized the mice 30 min later, cannulated the tracheas, performed bronchoalveolar lavage (BAL) with 1 ml PBS, and determined the number of CFU recovered. When BAL was not performed, we recovered equivalent numbers of CFU for all strains tested, approximately 3.0 × 104 CFU, indicating that all mice received similar inocula (Fig. 1A). This number also represents the maximum number of CFU recoverable from the lungs using this inoculation protocol. When we inoculated mice with wild-type (WT) bacteria and then performed BAL, we recovered approximately 7.5 × 102 CFU in the BAL fluid (BALF) (Fig. 1A), corresponding to ~1% of the recoverable CFU, which we calculated as the mean CFU recovered by BAL divided by the mean CFU recovered from the lungs when BAL was not performed (Fig. 1B). We homogenized and plated the postlavage lungs and recovered approximately 5 × 104 CFU (Fig. 1A), corresponding to ~75% of the recoverable CFU (Fig. 1B). Therefore, for WT bacteria, almost all of the recoverable bacteria remained in the lungs following BAL, presumably because they adhered tightly to respiratory epithelium.
FIG 1 .
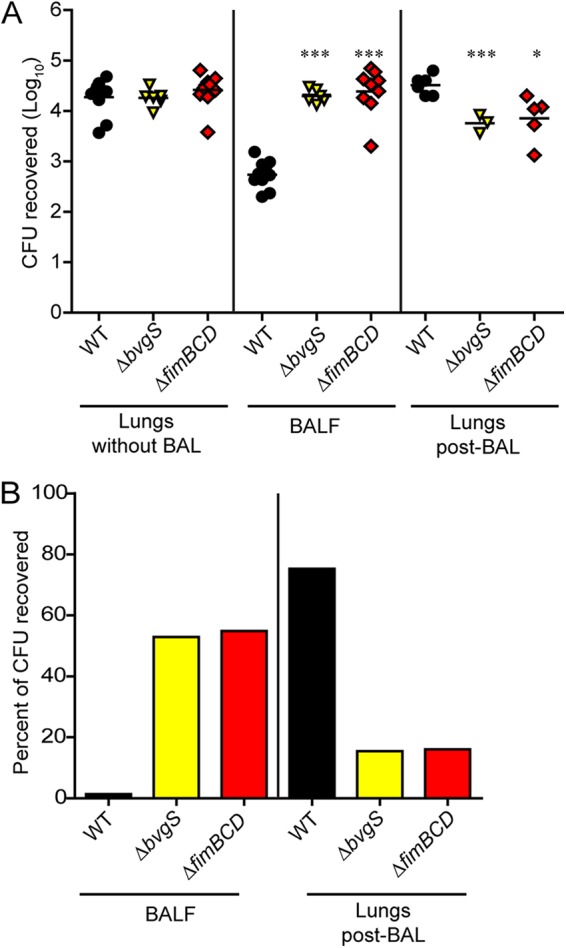
(A) Mice were inoculated intranasally with approximately 7.5 × 104 CFU of WT or mutant bacteria. The numbers of CFU recovered from lungs for which bronchoalveolar lavage (BAL) was not performed, from BAL fluid (BALF), and from lungs after BAL (post-BAL) are shown. Each symbol represents the value for an individual mouse, and the black bar shows the mean for the group of mice. Mean values for mutant bacteria that are significantly different from the mean value for WT bacteria are indicated by asterisks as follows: *, P < 0.05; ***, P < 0.001. (B) Data shown as the percentage of the recoverable CFU, which is calculated as the mean number of CFU recovered from either BALF or lungs post-BAL divided by the mean number of CFU recovered from lungs for which BAL was not performed for each strain. Values are means for two independent experiments.
When we inoculated mice with a strain defective for production of all known protein virulence factors (ΔbvgS), we recovered approximately 3 × 104 CFU in the lavage fluid, corresponding to ~55% of the recoverable CFU, while we recovered 7 × 103 CFU in the post-BAL lung homogenate, corresponding to ~15% of the recoverable CFU (Fig. 1A and B). Maximum recovery of avirulent bacteria by BAL, therefore, is about 55% of the recoverable CFU in this assay (compared with 1% for WT bacteria). When we inoculated mice with a strain deficient for production of FIM (ΔfimBCD), the CFU recovered by BAL was ~55% of the inoculum, and ~15% of the inoculum was recovered in the post-BAL lung homogenate, indicating that FIM-deficient bacteria are as defective for adherence as ΔbvgS bacteria are (Fig. 1A and B). We conclude from these results that FIM contribute substantially to adherence to mouse respiratory tissue. Moreover, as FIM-deficient B. bronchiseptica was not defective for adherence to a variety of cell lines in vitro (20), these results suggest that FIM are required specifically for adherence to respiratory epithelium and perhaps to ciliated respiratory epithelium.
We also inoculated mice with strains deficient in production of FHA (ΔfhaB), pertactin (Δprn), both FHA and FIM (ΔfimBCD ΔfhaB), adenylate cyclase toxin (ΔcyaA), or the type 3 secretion system (ΔbscN) and measured adherence. We recovered ~3 × 104 CFU in BALF from mice inoculated with either ΔfhaB or ΔfimBCD ΔfhaB bacteria, which is nearly identical to the number recovered from FIM-deficient bacteria. In contrast, Δprn mutants adhered similarly to WT bacteria, supporting in vitro evidence that pertactin is not necessary for adherence (30), and as expected, bacteria defective for adenylate cyclase toxin and the type 3 secretion system also adhered similarly to WT bacteria (data not shown). These results indicate that both FIM and FHA are required for bacterial adherence to mouse respiratory epithelium within the first hour of infection. The fact that the number of CFU of ΔfimBCD and ΔfhaB mutant bacteria recovered from BALF was similar to that of the ΔbvgS strain suggests that FIM and FHA are the two main, if not only, factors that mediate adherence to respiratory tissue and that they function interdependently, i.e., both are required.
Fimbriae are required for persistence in the lower respiratory tract.
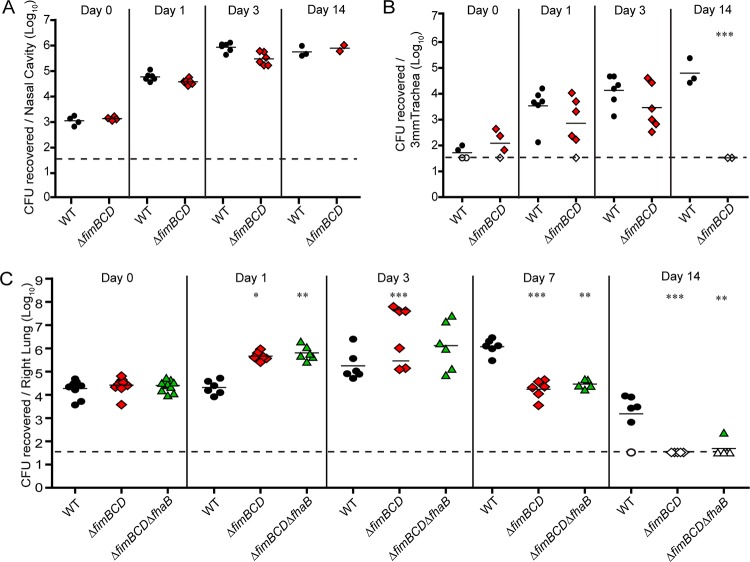
We inoculated 6-week-old BALB/c mice intranasally with 7.5 × 104 CFU of either wild-type, ΔfimBCD, or ΔfimBCD ΔfhaB B. bronchiseptica. For mice inoculated with WT or ΔfimBCD mutant bacteria, we determined bacterial burden in the nasal cavity, trachea, and right lung lobes at various times postinoculation (p.i.). For mice inoculated with the ΔfimBCD ΔfhaB mutant, we determined bacterial burden only in the lungs. CFU recovered from tissues harvested 1 h p.i. (day 0) were similar among all animals, indicating that consistent inoculation between bacterial strains and replicates occurred (Fig. 2A to C). There was no difference in the number of CFU recovered from the nasal cavities of animals inoculated with WT or ΔfimBCD bacteria at any time p.i. (Fig. 2A). We also recovered similar numbers of CFU of WT and ΔfimBCD bacteria from the tracheas 1 and 3 days p.i. However, 14 days p.i., no ΔfimBCD mutants were recovered from the tracheas, while the number of WT bacteria in the trachea remained high at this time point (Fig. 2B). Similar to what we have observed in rats (20), therefore, fimbriae are required for persistence in the tracheas of mice.
FIG 2 .
(A to C) Bacterial burden in respiratory tissues from mice inoculated with WT or mutant bacteria. Each symbol represents the value for an individual animal, and the black bar represents the mean for the group. The horizontal dashed line represents the lower limit of detection. Values are means from at least two independent experiments. Mean values for mutant bacteria that are significantly different from the mean value for WT bacteria are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We recovered approximately 1-log-unit-more CFU of ΔfimBCD orΔfimBCD ΔfhaB bacteria than WT bacteria from the lungs 1 day p.i. (Fig. 2C). At 3 days p.i., mice inoculated with ΔfimBCD bacteria split into two distinct groups. One group had significantly higher bacterial burden compared to the burden in mice inoculated with WT bacteria and were moribund. The bacterial burden of the second group was similar to the burden of mice inoculated with WT bacteria, and these mice showed no signs of respiratory distress. This “bimodal” phenotype at 3 days p.i. is similar to what has been observed in mice inoculated with ΔfhaB bacteria (27–29). For mice inoculated with ΔfimBCD ΔfhaB mutants, the bacterial burden was not clearly bimodal but spread between 105 and 107 bacteria at 3 days p.i. By 7 days p.i., the burdens of both the ΔfimBCD bacteria and ΔfimBCD ΔfhaB bacteria were significantly lower than the burden of WT bacteria, and the mutants were undetectable by 14 days p.i.
The persistence defect of ΔfimBCD and ΔfimBCD ΔfhaB bacteria was similar to that of ΔfhaB bacteria, indicating that FIM, like FHA, are required for bacterial persistence in the lower respiratory tract (27–29). These data suggest that, like FHA, FIM may be involved in suppressing inflammation.
Fimbriae are required to modulate the innate immune response in mice.
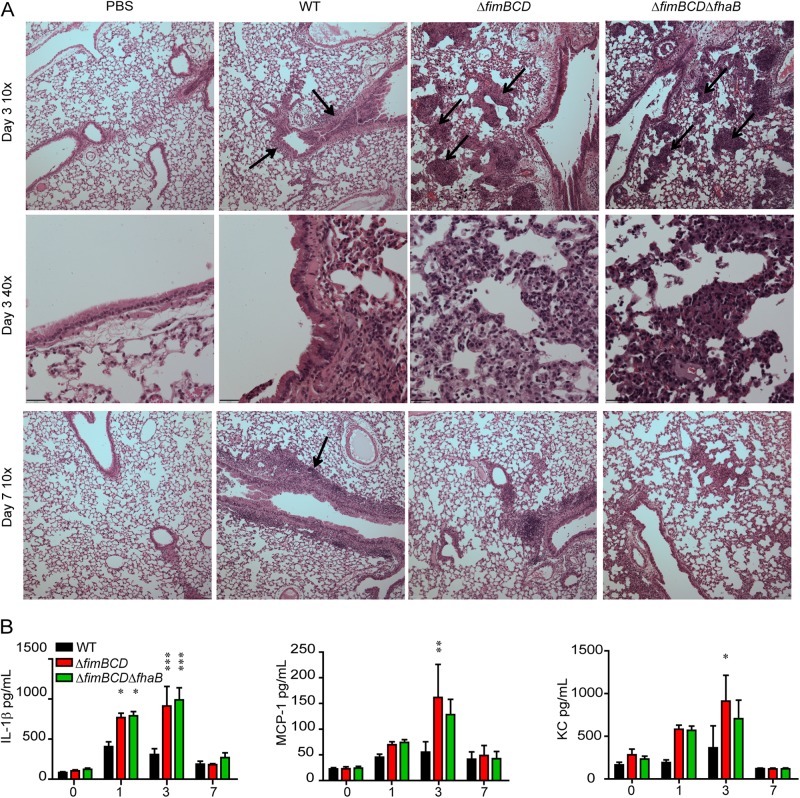
We examined hematoxylin-and-eosin (H&E)-stained lung sections to evaluate inflammation in both the major airways and alveoli during infection (Fig. 3A). The lungs of mice inoculated with only PBS appeared healthy, with little, if any, evidence of inflammation at any time point p.i. The lungs of mice inoculated with WT bacteria contained cellular infiltrates around the major airways (Fig. 3A, black arrows), but the alveoli and alveolar spaces were free of any signs of inflammation at 3 days p.i. The cellular infiltrates around major airways persisted to 7 days p.i. in mice inoculated with WT bacteria (Fig. 3A, black arrow), coinciding with the high bacterial burden at this time point. The lungs of mice inoculated with ΔfimBCD bacteria displayed some cellular infiltration around the major airways but also showed substantial visually distinct cellular infiltrate within the alveolar spaces at 3 days p.i. (Fig. 3A, black arrows). This histopathology pattern, which was present in the lungs of mice with either high or low bacterial burdens at day 3 p.i., differed dramatically from that of the lungs of mice inoculated with FHA-deficient bacteria, which showed increased cellular infiltrate primarily around the major airways and no patches of cellular infiltration in alveoli (27). By 7 days p.i., the patches of inflammatory cell recruitment in alveoli of ΔfimBCD mutant-inoculated mice was absent, and there was decreased cellular infiltrate around the major airways, corresponding with the decreased bacterial burden at this time point. The lungs of mice inoculated with ΔfimBCD ΔfhaB bacteria appeared similar to those of mice inoculated with ΔfimBCD bacteria at both time points p.i., with cellular infiltration evident around the major airways as well as distinct cell recruitment within the alveolar spaces (Fig. 3A, black arrows) at 3 days p.i. At 7 days p.i., there were fewer inflammatory cells present, coinciding with decreased bacterial burden.
FIG 3 .
(A) Hematoxylin-and-eosin-stained 5-μm lung sections at ×10 and ×40 magnification. Black arrows indicate areas of cellular infiltrate. (B) Cytokine and chemokine levels in lung homogenates of animals inoculated with WT bacteria or with ΔfimBCD or ΔfimBCD ΔfhaB mutant bacteria. The y axis shows time postinoculation (in days). Values are means ± standard errors (SE) (error bars) from at least two independent experiments. Mean values for mutant bacteria that are significantly different from the mean value for WT bacteria are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We also measured cytokine and chemokine levels in right lung homogenates in enzyme-linked immunosorbent assays (ELISAs) (Fig. 3B). Interleukin-1β(IL-1β) levels were significantly increased in the lungs of mice inoculated with ΔfimBCD or ΔfimBCD ΔfhaB bacteria compared to WT bacteria 1 and 3 days p.i. These differences did not correlate with bacterial burden, as IL-1β levels were increased in all animals inoculated with the mutant bacteria, even those with lower bacterial burdens at day 3 p.i. Moreover, monocyte chemotactic protein 1 (MCP-1) and neutrophil chemokine CXCL1 (CXC chemokine ligand 1) (KC [keratinocyte-derived chemokine]) levels were significantly increased in the lungs of mice inoculated with ΔfimBCD bacteria compared to the lungs of mice inoculated with WT bacteria 3 days p.i. MCP-1 and KC levels in mice inoculated with ΔfimBCD ΔfhaB mutant bacteria were higher than in mice inoculated with WT bacteria, but the differences were not statistically significant. Other cytokines, such as interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), interleukin-10, -12p70, -22, and -23 (IL-10, IL-12p70, IL-22, and IL-23) were measured, and no significant difference was found between mice inoculated with ΔfimBCD or ΔfimBCD ΔfhaB bacteria compared to those inoculated with WT bacteria at any time point.
Taken together, these data indicate that FIM, like FHA, are required to suppress inflammation during infection. The dramatically different histopathology, however, suggests that FIM and FHA may play different roles in pathogenesis, and the fact that the ΔfimBCD ΔfhaB double mutant induced a histopathology pattern similar to that induced by the ΔfimBCD mutant indicates that the ΔfimBCD mutation is epistatic to the ΔfhaB mutation.
Fimbriae do not complement “in trans” during coinoculation.
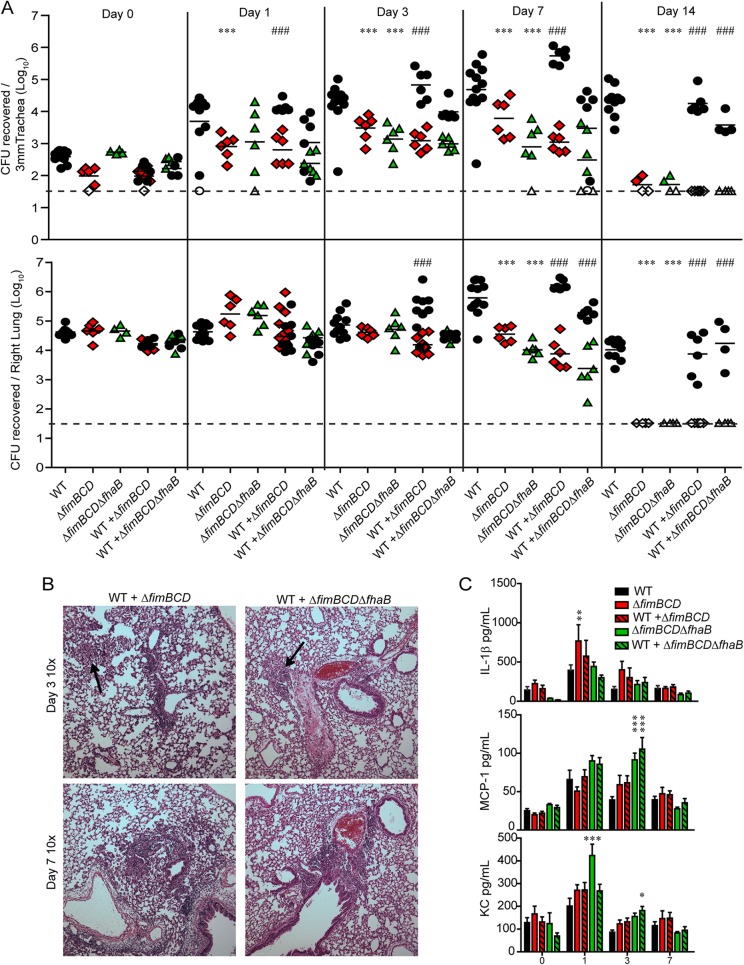
Our data here and from previous studies suggest that WT B. bronchiseptica is able to suppress the initial inflammatory response to infection, contributing to decreased pathology and increased bacterial persistence (27–29). We have previously shown that when mice are coinoculated with WT and FHA-deficient bacteria, the level of inflammation in the lungs is less than that induced by inoculation with the ΔfhaB mutant alone, and the ΔfhaB mutant persists longer than when inoculated into mice in the absence of the WT strain, suggesting that FHA-producing WT bacteria are able to complement “in trans” (29). We hypothesized that WT B. bronchiseptica would similarly be able to rescue FIM-deficient bacteria from inflammation-mediated clearance. To test this hypothesis, we inoculated 6-week-old mice with 1.5 × 105 CFU of WT, ΔfimBCD, or ΔfimBCD ΔfhaB bacteria alone or a mixture of 7.5 × 104 CFU each of WT and ΔfimBCD bacteria or WT and ΔfimBCD ΔfhaB bacteria (so that the total number of CFU in each inoculum was 1.5 × 105) and determined the bacterial burden.
Strikingly, the numbers of mutant and WT bacteria recovered from the trachea and lungs from coinoculated animals were similar to those from animals inoculated with WT or mutant bacteria alone (Fig. 4A); the presence of WT bacteria did not improve the persistence of the mutant bacteria, and the presence of mutant bacteria did not lead to increased clearance of WT bacteria. These data indicate that, unlike the case with FHA, WT bacteria cannot complement ΔfimBCD or ΔfimBCD ΔfhaB bacteria in trans, further supporting the hypothesis that FIM and FHA contribute differently to infection.
FIG 4 .
(A) Bacterial burden in respiratory tissue from mice inoculated with WT or mutant bacteria alone or coinoculated with WT and mutant bacteria. Each symbol represents the value for an individual animal, and the black bar represents the mean of the group of animals. The horizontal dashed line represents the lower limit of detection. (B) Hematoxylin-and-eosin-stained 5-µm lung sections at a magnification of ×10. Arrows indicate areas of cellular infiltration. (C) Cytokine and chemokine levels in lung homogenates of animals inoculated with WT bacteria or ΔfimBCD or ΔfimBCD ΔfhaB mutant bacteria. Values are means (A) or means ± SE (C) for at least two independent experiments. Mean values for single mutant strains that are significantly different from the mean value for WT bacteria are indicated by asterisks as follows: * = P < 0.05; **, P < 0.01; ***, P < 0.001. Mean values for each strain in coinoculated animals that are significantly different from each other are indicated (###).
Lung sections from mice coinoculated with WT and either mutant strain appeared similar to lung sections of mice inoculated with ΔfimBCD or ΔfimBCD ΔfhaB bacteria alone: there was cell recruitment around the major airways as well as in the alveolar spaces (Fig. 4B, black arrows), indicating that the presence of WT bacteria cannot prevent the histopathology seen in lungs inoculated with FIM-deficient bacteria (Fig. 4B). In general, levels of IL-1β, KC, and MCP-1 were higher in coinoculated mice than in mice inoculated with WT bacteria alone, although most differences were not statistically significant (Fig. 4C).
These data indicate that the presence of WT bacteria in the lungs did not increase the survival of FIM-deficient bacteria and did not prevent cellular infiltrate from entering alveolar space. A possible explanation for the inability of WT bacteria to complement FIM-deficient bacteria in trans is that WT and FIM-deficient bacteria localize differently in this model.
FIM-deficient bacteria localize differently than WT and FHA-deficient bacteria in the lower respiratory tract.
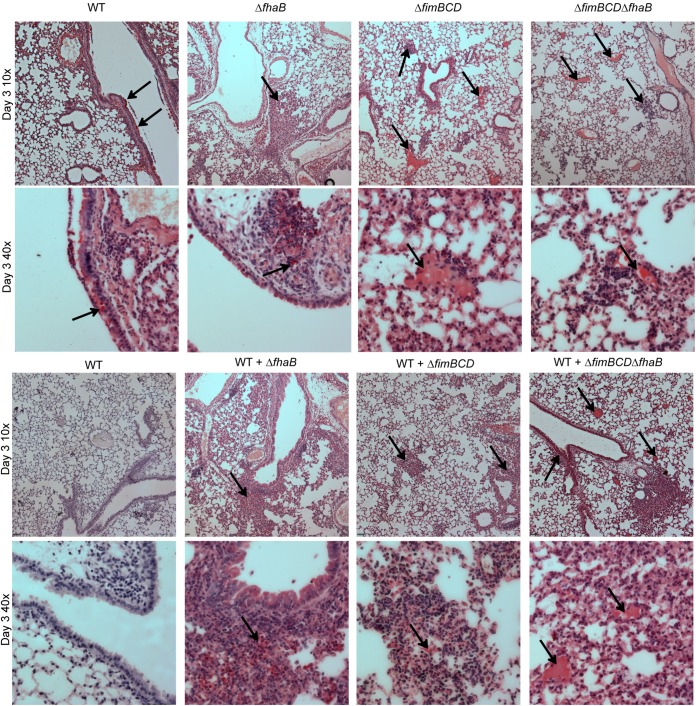
To determine the location of WT and mutant bacteria in the lungs, we inoculated mice with 7.5 × 104 CFU of WT or mutant bacteria and then sacrificed the mice 3 days p.i. and prepared the left lung lobe for sectioning. We then performed immunohistochemistry using serum from a rabbit chronically infected with WT B. bronchiseptica as the primary antibody, goat anti-rabbit conjugated to alkaline phosphatase (AP) as the secondary antibody, and naphthol red as the AP substrate. The lung sections were then counterstained with hematoxylin, which stains nuclei dark purple. The lungs of mice inoculated with WT bacteria showed red staining around the ciliated epithelium of the major airways (Fig. 5, black arrows), while control lungs, which were not incubated with rabbit serum, did not have any red staining (Fig. 5). This staining pattern indicates that WT bacteria localize to the major airways during infection.
FIG 5 .
Lung sections 3 days p.i. from mice inoculated with WT or mutant bacteria (A) or coinoculated with WT and mutant bacteria (B). Immunohistochemistry was performed using convalescent rabbit serum from a rabbit infected with WT bacteria and goat anti-rabbit secondary antibody conjugated to alkaline phosphatase, incubated with naphthol red substrate. Red staining (black arrows) indicate alkaline phosphatase activity. Tissue was counterstained with hematoxylin. Pictures were taken at magnifications of ×10 and ×40. Control indicates no primary antibody staining.
The lungs of mice inoculated with FHA-deficient bacteria appeared similar to the lungs of mice inoculated with WT bacteria, with noticeable red staining around the major airways, both on the ciliated epithelium and in the cellular infiltrate beneath the epithelial cells (Fig. 5A, black arrows), but very little red staining in the alveolar space, suggesting that FHA-deficient bacteria localize similarly to WT bacteria. In contrast, the lungs of mice inoculated with FIM-deficient bacteria showed a dramatically different staining pattern. These lungs had red staining throughout the alveolar space (Fig. 5A, black arrows). The lungs of mice inoculated with ΔfimBCD ΔfhaB double mutant bacteria appeared similar to the lungs of mice inoculated with FIM-deficient bacteria. These results indicate that FIM-producing bacteria localize to major airways and bronchioles, while FIM-deficient bacteria localize predominantly to alveoli, suggesting that FIM mediate attachment specifically to ciliated epithelia, which line bronchi and bronchioles. Without FIM, many, if not most, bacteria bypass the ciliated epithelium and are deposited in alveoli.
The lungs of mice coinoculated with WT and FHA-deficient bacteria had red staining around the major airways (Fig. 5B, black arrows), but very little red staining in alveolar space, similar to the lungs of mice inoculated with WT or FHA-deficient bacteria alone. In contrast, the lungs of mice coinoculated with WT and either ΔfimBCD or ΔfimBCD ΔfhaB bacteria had red staining in the major airways and distinct staining in alveolar space (Fig. 5B, black arrows). These data suggest that even during coinoculation, WT bacteria and FHA-deficient bacteria localize primarily to major airways, while FIM-deficient bacteria are delivered primarily to alveoli. Furthermore, these results provide a possible explanation for why coinoculation with WT bacteria can rescue FHA-deficient bacteria but not FIM-deficient bacteria: immune suppression mediated by WT bacteria occurs locally in the major airways and does not affect bacteria in alveolar space because the WT bacteria do not gain access to this location.
We conducted an in vivo adherence assay using an equal mixture of WT and FIM-deficient bacteria (data not shown). The numbers of CFU recovered were similar to those from mice inoculated with WT or FIM-deficient bacteria alone. This result provides further evidence that WT and FIM-deficient bacteria function independently in the respiratory tract.
DISCUSSION
Bordetella FIM are generally considered to function as adhesins despite there being no reports of adherence studies using B. pertussis strains defective only for production of FIM. Studies of B. bronchiseptica also failed to provide convincing evidence that Bordetella FIM function as adhesins, as FIM-deficient bacteria showed no defect in adherence assays using a variety of nonciliated cell lines (20), and this strain was only modestly defective in adherence to ciliated tracheal explants (22). Our results therefore provide the first demonstration that Bordetella FIM are important adhesins, and they indicate that fimbriae mediate adherence specifically to airway epithelium. Our results also show that FIM and FHA work together, playing equally important roles in allowing Bordetella to suppress inflammation, leading to prolonged colonization.
A murine model in which large numbers of bacteria are delivered intranasally in a large volume has been used by our group and others to study respiratory infection by Bordetella (16, 27–29, 31–34). It has been presumed that this inoculation method deposits bacteria evenly throughout the respiratory tract. Within the trachea, bronchi, and bronchioles, bacteria must overcome mucociliary clearance through tight adherence. The bacteria also interact with or stimulate sentinel innate immune cells, such as alveolar macrophages and dendritic cells within the lower respiratory tract, which in turn stimulate the initial inflammatory response characterized by the recruitment of phagocytic cells, predominately neutrophils, to the site of infection. In this model, both bacterial burden and cellular infiltrate peak at about 7 days postinoculation. Bacterial load then decreases, with clearance from the lungs occurring over the next 2 to 3 weeks and requiring adaptive immunity (16). For B. pertussis, the bacteria are cleared from the entire respiratory tract by about 30 to 40 days postinoculation, while for B. bronchiseptica, the bacteria are cleared from the lower respiratory tract but persist in the nasal cavity indefinitely (16).
Our newly developed in vivo adherence assay and bacterial localization analyses showed that WT B. bronchiseptica bacteria are not distributed evenly throughout the lungs following high-dose, large-volume inoculation. Instead they localize predominately to airway epithelium. This localization requires FIM, as FIM-deficient mutants bypassed the ciliated epithelium and localized to the alveoli. In the alveoli, FIM-deficient bacteria did not adhere tightly enough to resist bronchoalveolar lavage, despite producing wild-type levels of FHA. It is possible that FHA receptors are not present on alveolar pneumocytes. Alternatively, tight adherence may require FIM-mediated interactions that induce changes in either bacterial or host cells. FHA-deficient bacteria, in contrast, localized to the airways, presumably due to fimbrial attachment to ciliated epithelium, but these FIM-mediated interactions alone were insufficient to resist bronchoalveolar lavage. Our data therefore suggest a model in which adherence of Bordetella is a two-step process requiring both FIM and FHA. In this model, fimbriae mediate initial interactions to ciliated epithelia, and this critical first step then allows FHA to mediate tighter adherence to these cells.
Following adherence, the ability of Bordetella to influence the innate immune response is evident in the first 3 or 4 days postinoculation. We have previously shown a role for FHA in modulating the innate immune response, as FHA-deficient B. bronchiseptica bacteria were hyperinflammatory compared to WT bacteria (27, 29). Here, we showed that FIM-deficient mutants were similar to FHA-deficient mutants in bacterial burden and in inducing high levels of proinflammatory cytokines and chemokines during the first 3 days of infection, suggesting that FIM also contribute to suppression of inflammation (Fig. 2). The lungs of mice inoculated with FHA- or FIM-deficient bacteria, however, displayed strikingly different histopathology. Unlike FHA-deficient bacteria, which caused increased cellular infiltrate around the bronchioles, FIM-deficient bacteria caused increased cellular infiltrate in the alveoli. This difference correlates with the different localizations of these strains in the lungs. It also indicates that FHA alone is insufficient to modulate the inflammatory response in the alveoli, since FIM-deficient bacteria that localize to this site produce FHA. Furthermore, cytokine levels and histopathology from mice inoculated with ΔfimBCD ΔfhaB bacteria were similar to those of mice inoculated with ΔfimBCD bacteria. These data indicate that the lack of FIM is epistatic to the lack of FHA; without FIM, the presence or absence of FHA did not change the outcome of infection, underscoring the importance of FIM for FHA-mediated interactions.
In this murine model, bacterial burden and inflammatory infiltrate in the lungs both peak at about 7 days postinoculation. Subsequent to this time point, FHA- and FIM-deficient bacteria are cleared rapidly, while WT bacteria persist for approximately three more weeks (16, 29). Rapid clearance of the FIM and FHA mutant bacteria may reflect a decreased ability of these mutants to resist killing by inflammatory cells. However, because these mutants induce a hyperinflammatory environment, it is also possible that their rapid clearance is due primarily, if not solely, to the increased numbers and/or activation of recruited inflammatory cells. Our lab has shown that mice coinoculated with WT and FHA-deficient bacteria exhibit less inflammation than mice inoculated with FHA-deficient bacteria alone. Furthermore, in coinoculated animals, there was increased persistence of FHA-deficient bacteria than in animals inoculated with only the ΔfhaB mutant (29). We hypothesized that the increased persistence of ΔfhaB bacteria in coinoculated animals resulted from FHA-producing WT bacteria suppressing inflammation. We also hypothesized that the decreased ability of FHA-deficient mutants compared to WT bacteria to persist in coinoculated animals indicated a decreased ability of FHA-deficient bacteria to resist phagocytic cells. Our new results show that WT and FHA-deficient bacteria colocalize in the major airways, supporting the hypothesis that mutant bacteria benefit from local immune suppression mediated by WT bacteria and further supporting the hypothesis that FHA-deficient bacteria are unable to resist clearance by inflammatory cells, even if the activation state of these cells is suppressed by the presence of WT bacteria.
In contrast, WT bacteria did not improve the survival of FIM-deficient bacteria, which were cleared as rapidly from mice coinoculated with WT bacteria as from mice inoculated with FIM-deficient bacteria alone (Fig. 3). The inability of WT bacteria to complement “in trans” the ΔfimBCD mutant (and the ΔfimBCD ΔfhaB mutant) may be due simply to differences in localization, which would suggest that FHA-mediated suppression of inflammation occurs locally within the major airways. Rapid clearance of the ΔfimBCD and ΔfimBCD ΔfhaB mutant bacteria likely reflects both an inability to suppress inflammation locally and an inability to resist the recruited phagocytic cells. Together, our data indicate that once FIM-mediated interactions localize WT bacteria to the ciliated epithelium, FHA-mediated interactions provide both localized immune suppression and protection against immune-mediated clearance.
We have been using B. bronchiseptica and rabbits, rats, and mice to investigate how specific virulence factors and the regulation of virulence factor-encoding genes contribute to pathogenesis (20, 27, 29, 30). Because these animals are natural hosts for B. bronchiseptica, the results obtained from these studies are biologically relevant, i.e., we are confident in our conclusions that FHA and FIM are required for lower respiratory tract colonization of these hosts by B. bronchiseptica. However, B. bronchiseptica infection of these rodents does not result in the same course of infection or disease characteristics as B. pertussis infection of humans, and therefore, B. bronchiseptica infection of rabbits, rats, or mice should not be considered models of human pertussis. Nonetheless, we have demonstrated previously that the genes encoding FHA, adenylate cyclase, and BvgAS in B. pertussis can substitute for the corresponding genes in B. bronchiseptica during infection (15, 28, 30), suggesting that these virulence factors and regulatory system perform the same function for these two bacterial species during infection of their respective hosts. In our current study, we did not determine whether the genes encoding FIM in B. pertussis could substitute for their homologs in B. bronchiseptica. However, our results are consistent with those of a report showing that a B. pertussis strain defective for both FHA and FIM was defective for persistence and caused increased cellular infiltrate in the alveoli of mice, a study in which the authors concluded that FIM were important for modulating the immune response (35). We hypothesize, therefore, that B. pertussis FIM are required for adherence to airway epithelium and for FHA-mediated immunomodulation during human infection. Testing this hypothesis and determining whether and how these factors contribute to disease characteristics unique to human pertussis may be achievable with the recently developed baboon model (36).
Given the reemergence of pertussis and the increasing reports of shortcomings of the acellular vaccine (36–38), it is important to reevaluate vaccine design and immunization route. While FIM2 and FIM3 are components of the acellular vaccine and induce an antibody response, the contribution and method of action of this response to bacterial clearance are unclear. Our results suggest that antibodies that block FIM-mediated attachment may prevent bacterial attachment and colonization in the lower respiratory tract, but whether the current aP vaccine or natural immune response generates antibodies capable of performing this function is uncertain. As Bordetella is primarily an extracellular respiratory pathogen, generation of mucosal IgA antibodies capable of blocking bacterial adherence could potentially prevent Bordetella colonization. Most studies focus primarily on serum IgG responses following vaccination, so further characterization of the antibody response may be warranted.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (42). Our protocol was approved by the University of North Carolina IACUC (10-134, 12-307, and 13-238). All animals were properly anesthetized for inoculations, monitored regularly, and euthanized when moribund, and efforts were made to minimize suffering
Growth media and bacterial strains.
Escherichia coli was grown in lysogeny broth (LB) or on LB agar (1.5%) at 37°C. Wild-type (WT) Bordetella bronchiseptica RB50 and mutant derivatives were grown at 37°C on Bordet-Gengou (BG) agar again (Becton Dickinson Microbiology Systems) supplemented with 7.5% defibrinated sheep blood (Colorado Serum Co., Denver, CO) or in Stainer-Scholte (SS) broth with 100 mg/ml (2,6-O-dimethyl)-β-cyclodextrin. When necessary, media were supplemented with streptomycin (Sm) (20 µg/ml), gentamicin (Gm) (30 µg/ml), kanamycin (50 µg/ml), or diaminopimelic acid (DAP) (300 µg/ml).
Construction and cloning of plasmids were accomplished in E. coli DH5α. Plasmids were introduced into B. bronchiseptica via mating with E. coli RHO3. In-frame markerless deletion mutations were made using the pSS4245 allelic exchange system. pUC18-based plasmids were utilized to deliver genes encoding gentamicin resistance (aacC1) and kanamycin resistance (nptII) to the attTn7 site via transposase-mediated insertion. Detailed descriptions of each strain are given in Table 1.
TABLE 1 .
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Molecular cloning strain | 30 |
| RH03 | Conjugation strain; Kms; DAP auxotroph | 39 |
| B. bronchiseptica strains | ||
| RB50 | “Wild-type” Bordetella bronchiseptica complex I strain | 14 |
| RB50 Kmr | RB50 containing constitutively expressed nptII inserted via a pUC18-based plasmid at the attTn7 site | This study |
| RB63 | RB50 containing deletion of the fimBCD operon | 20 |
| RB63 Gmr | RB63 containing constitutively expressed aacC1 inserted via a pUC18-based plasmid at the attTn7 site | This study |
| SP5 | RB50 containing an in-frame deletion of codons 227 to 756 of prn | 22 |
| RBX9f | RBX9 with a deletion mutation of the fimA-fhaB intergenic region | 31 |
| RB515 | RB50 containing a deletion in cyaA | 27 |
| WD3 | RB50 with an in-frame deletion of bscN | 40 |
| RB54 | RB50 Bvg− phase-locked variant with an in-frame deletion of bvgS | 14 |
| AS16 | RB63 with a mutation of the fhaB-MCD | This study |
| AS16 Gmr | AS16 containing constitutively expressed aacC1 inserted via a pUC18-based plasmid at the attTn7 site | This study |
| Plasmids | ||
| pSS4245 | pBR322-based allelic exchange plasmid; Apr Kmr | 30 |
| pTsS3 | Tn7 transposase expression vector containing tnsABCD; Apr | 41 |
| pUC18-miniTn7 | Transposition vector; Apr Kmr | 41 |
Intranasal mouse inoculation.
Bacteria were grown overnight in Stainer-Scholte broth with 100 mg/ml (2,6-O-dimethyl)-β-cyclodextrin. When necessary, media were supplemented with streptomycin (20 µg/ml), gentamicin (30 µg/ml), or kanamycin (50 µg/ml). Six-week-old BALB/c mice from Jackson Laboratories (Bar Harbor, ME) were inoculated intranasally with 7.5 × 104 or 1.5 × 105 CFU of B. bronchiseptica in 50 μl PBS. Mice were inoculated with strains RB50, RB63, RBX9F, RB54, SP5, and AS16 or a mixture of strains RB50 and RB63 or strains RB50 and AS16. The right lung lobes, tracheae, and nasal cavities were harvested from mice at specific time points postinoculation (p.i.) in 1 ml PBS. Tissue was homogenized, serial dilutions were plated on BG agar, and the number of CFU were determined.
Cytokine and histological analyses.
Using lung homogenates, the cytokine and chemokine responses to infection were measured using ELISA kits (R&D Systems). Homogenates were diluted 1:10, and then IL-1β, KC, MCP-1 and IL-17 cytokines and chemokines were measured following the manufacturer’s instructions. The cytokine concentrations were calculated using standard curve data for each cytokine. Absorbance was determined using a Molecular Devices plate reader and analyzed by Softmax Pro software (Molecular Devices). To prepare histology slides, left lung lobes were harvested at the time points indicated in the figures and inflated with 10% formalin. The Animal Histopathology Core Lab then embedded the tissue in paraffin, sectioned the tissue so that the tissue section was 5 µm thick, and then stained the tissue with hematoxylin and eosin (H&E). Lung sections were examined at the Microscopy Services Laboratory using bright-field imaging on an Olympus BX61 microscope at magnifications of ×10 and ×40.
In vivo adherence.
Bacteria were grown as described above, and 6-week-old BALB/c mice were inoculated with 7.5 × 104 CFU of bacteria. Thirty minutes p.i., the mice were euthanized, the tracheae were cannulated, and bronchoalveolar lavage was performed with 1 ml PBS to determine adherence. The right and left lung lobes were then excised and homogenized in 1 ml PBS to determine CFU remaining post-BAL. Serial dilutions of both BAL and homogenate were plated on BG agar to determine the number of CFU recovered.
Bacterial localization.
Six-week-old BALB/c mice were inoculated as described above. Lung tissue was harvested 3 days p.i., and the left lobe was inflated with 10% formalin. The Animal Histopathology Core Lab then embedded the tissue in paraffin and provided unstained 5-μm lung sections. The tissue sections were incubated with serum from a rabbit chronically infected with B. bronchiseptica RB50 bacteria and then incubated with goat anti-rabbit secondary antibody conjugated to alkaline phosphatase (Promega). The tissue sections were then incubated with alkaline phosphatase substrate naphthol red (Sigma), counterstained with hematoxylin (Sigma), and examined on an Olympus Bx61 microscope at magnifications of ×10 and ×40.
Statistical analysis.
Statistical analysis was performed using Prism 5.0 software from GraphPad Software, Inc. Statistical significance was determined using unpaired Student’s t test or analysis of variance (ANOVA). Figures were generated using Adobe Illustrator CS6 (Adobe Systems, Inc.).
ACKNOWLEDGMENTS
This work was supported by the NIH Research Project Grant Program (grants AI43986 and AI094991 to P.A.C.) from the National Institutes of Health (NIH).
We thank members of our laboratory for many helpful discussions and Bill Goldman for advice on sample preparation for the microscopy studies. Tissue preparation and sectioning were performed by the University of North Carolina Animal Histopathology Core Lab.
Footnotes
Citation Scheller EV, Melvin JA, Sheets AJ, Cotter PA. 2015. Cooperative roles for fimbria and filamentous hemagglutinin in Bordetella adherence and immune modulation. mBio 6(3):e00500-15. doi:10.1128/mBio.00500-15.
REFERENCES
- 1.Melvin JA, Scheller EV, Miller JF, Cotter PA. 2014. Bordetella pertussis pathogenesis: current and future challenges. Nat Rev Microbiol 12:274–288. doi: 10.1038/nrmicro3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodnow RA. 1980. Biology of Bordetella bronchiseptica. Microbiol Rev 44:722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diavatopoulos DA, Cummings CA, Schouls LM, Brinig MM, Relman DA, Mooi FR. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog 1:e45. doi: 10.1371/journal.ppat.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J, Zhang Y, Buboltz AM, Zhang X, Schuster SC, Ahuja U, Liu M, Miller JF, Sebaihia M, Bentley SD, Parkhill J, Harvill ET. 2012. Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13:545. doi: 10.1186/1471-2164-13-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preston A, Maskell DJ. 2002. A new era of research into Bordetella pertussis pathogenesis. J Infect 44:13–16. doi: 10.1053/jinf.2001.0933. [DOI] [PubMed] [Google Scholar]
- 6.Poland GA. 2012. Pertussis outbreaks and pertussis vaccines: new insights, new concerns, new recommendations? Vaccine 30:6957–6959. doi: 10.1016/j.vaccine.2012.09.084. [DOI] [PubMed] [Google Scholar]
- 7.Fisman DN, Tang P, Hauck T, Richardson S, Drews SJ, Low DE, Jamieson F. 2011. Pertussis resurgence in Toronto, Canada: a population-based study including test-incidence feedback modeling. BMC Public Health 11:694. doi: 10.1186/1471-2458-11-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong JY. 2010. Update on pertussis and pertussis immunization. Korean J Pediatr 53:629–633. doi: 10.3345/kjp.2010.53.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willems RJ, van der Heide HG, Mooi FR. 1992. Characterization of a Bordetella pertussis fimbrial gene cluster which is located directly downstream of the filamentous haemagglutinin gene. Mol Microbiol 6:2661–2671. doi: 10.1111/j.1365-2958.1992.tb01443.x. [DOI] [PubMed] [Google Scholar]
- 10.Willems R, Paul A, van der Heide HG, ter Avest AR, Mooi FR. 1990. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J 9:2803–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kania SA, Rajeev S, Burns EH, Odom TF, Holloway SM, Bemis DA. 2000. Characterization of fimN, a new Bordetella bronchiseptica major fimbrial subunit gene. Gene 256:149–155. doi: 10.1016/S0378-1119(00)00360-7. [DOI] [PubMed] [Google Scholar]
- 12.Pedroni P, Riboli B, de Ferra F, Grandi G, Toma S, Aricò B, Rappuoli R. 1988. Cloning of a novel pilin-like gene from Bordetella pertussis: homology to the fim2 gene. Mol Microbiol 2:539–543. doi: 10.1111/j.1365-2958.1988.tb00061.x. [DOI] [PubMed] [Google Scholar]
- 13.Boschwitz JS, van der Heide HG, Mooi FR, Relman DA. 1997. Bordetella bronchiseptica expresses the fimbrial structural subunit gene fimA. J Bacteriol 179:7882–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter PA, Miller JF. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun 62:3381–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez de Tejada G, Miller JF, Cotter PA. 1996. Comparative analysis of the virulence control systems of Bordetella pertussis and Bordetella bronchiseptica. Mol Microbiol 22:895–908. [DOI] [PubMed] [Google Scholar]
- 16.Harvill ET, Cotter PA, Miller JF. 1999. Pregenomic comparative analysis between Bordetella bronchiseptica RB50 and Bordetella pertussis Tohama I in murine models of respiratory tract infection. Infect Immun 67:6109–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooi FR, van der Heide HG, ter Avest AR, Welinder KG, Livey I, van der Zeijst BA, Gaastra W. 1987. Characterization of fimbrial subunits from Bordetella species. Microb Pathog 2:473–484. [DOI] [PubMed] [Google Scholar]
- 18.Willems RJ, Geuijen C, van der Heide HG, Matheson M, Robinson A, Versluis LF, Ebberink R, Theelen J, Mooi FR. 1993. Isolation of a putative fimbrial adhesin from Bordetella pertussis and the identification of its gene. Mol Microbiol 9:623–634. [DOI] [PubMed] [Google Scholar]
- 19.Hazenbos WL, van den Berg BM, van’t Wout JW, Mooi FR, van Furth R. 1994. Virulence factors determine attachment and ingestion of nonopsonized and opsonized Bordetella pertussis by human monocytes. Infect Immun 62:4818–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattoo S, Miller JF, Cotter PA. 2000. Role of Bordetella bronchiseptica fimbriae in tracheal colonization and development of a humoral immune response. Infect Immun 68:2024–2033. doi: 10.1128/IAI.68.4.2024-2033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Funnell SG, Robinson A. 1993. A novel adherence assay for Bordetella pertussis using tracheal organ cultures. FEMS Microbiol Lett 110:197–203. [DOI] [PubMed] [Google Scholar]
- 22.Edwards JA, Groathouse NA, Boitano S. 2005. Bordetella bronchiseptica adherence to cilia is mediated by multiple adhesin factors and blocked by surfactant protein A. Infect Immun 73:3618–3626. doi: 10.1128/IAI.73.6.3618-3626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazenbos WL, Geuijen CA, van den Berg BM, Mooi FR, van Furth R. 1995. Bordetella pertussis fimbriae bind to human monocytes via the minor fimbrial subunit FimD. J Infect Dis 171:924–929. [DOI] [PubMed] [Google Scholar]
- 24.Hazenbos WL, van den Berg BM, Geuijen CW, Mooi FR, van Furth R. 1995. Binding of FimD on Bordetella pertussis to very late antigen-5 on monocytes activates complement receptor type 3 via protein tyrosine kinases. J Immunol 155:3972–3978. [PubMed] [Google Scholar]
- 25.Geuijen CA, Willems RJ, Mooi FR. 1996. The major fimbrial subunit of Bordetella pertussis binds to sulfated sugars. Infect Immun 64:2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotter PA, Yuk MH, Mattoo S, Akerley BJ, Boschwitz J, Relman DA, Miller JF. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect Immun 66:5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henderson MW, Inatsuka CS, Sheets AJ, Williams CL, Benaron DJ, Donato GM, Gray MC, Hewlett EL, Cotter PA. 2012. Contribution of Bordetella filamentous hemagglutinin and adenylate cyclase toxin to suppression and evasion of interleukin-17-mediated inflammation. Infect Immun 80:2061–2075. doi: 10.1128/IAI.00148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julio SM, Inatsuka CS, Mazar J, Dieterich C, Relman DA, Cotter PA. 2009. Natural-host animal models indicate functional interchangeability between the filamentous haemagglutinins of Bordetella pertussis and Bordetella bronchiseptica and reveal a role for the mature C-terminal domain, but not the RGD motif, during infection. Mol Microbiol 71:1574–1590. doi: 10.1111/j.1365-2958.2009.06623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inatsuka CS, Julio SM, Cotter PA. 2005. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci U S A 102:18578–18583. doi: 10.1073/pnas.0507910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inatsuka CS, Xu Q, Vujkovic-Cvijin I, Wong S, Stibitz S, Miller JF, Cotter PA. 2010. Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infect Immun 78:2901–2909. doi: 10.1128/IAI.00188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason E, Henderson MW, Scheller EV, Byrd MS, Cotter PA. 2013. Evidence for phenotypic bistability resulting from transcriptional interference of bvgAS in Bordetella bronchiseptica. Mol Microbiol 90:716–733. doi: 10.1111/mmi.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connelly CE, Sun Y, Carbonetti NH. 2012. Pertussis toxin exacerbates and prolongs airway inflammatory responses during Bordetella pertussis infection. Infect Immun 80:4317–4332. doi: 10.1128/IAI.00808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scanlon KM, Gau Y, Zhu J, Skerry C, Wall SM, Soleimani M, Carbonetti NH. 2014. Epithelial anion transporter pendrin contributes to inflammatory lung pathology in mouse models of Bordetella pertussis infection. Infect Immun 82:4212–4221. doi: 10.1128/IAI.02222-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfe DN, Karanikas AT, Hester SE, Kennett MJ, Harvill ET. 2010. IL-10 induction by Bordetella parapertussis limits a protective IFN-gamma response. J Immunol 184:1392–1400. doi: 10.4049/jimmunol.0803045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandebriel RJ, Hellwig SMM, Vermeulen JP, Hoekman JHG, Dormans JAMA, Roholl PJM, Mooi FR. 2003. Association of Bordetella pertussis with host immune cells in the mouse lung. Microb Pathog 35:19–29. doi: 10.1016/S0882-4010(03)00087-1. [DOI] [PubMed] [Google Scholar]
- 36.Warfel JM, Zimmerman LI, Merkel TJ. 2014. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc Natl Acad Sci U S A 111:787–792. doi: 10.1073/pnas.1314688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tartof SY, Lewis M, Kenyon C, White K, Osborn A, Liko J, Zell E, Martin S, Messonnier NE, Clark TA, Skoff TH. 2013. Waning immunity to pertussis following 5 doses of DTaP. Pediatrics 131:e1047–e1052. doi: 10.1542/peds.2012-1928. [DOI] [PubMed] [Google Scholar]
- 38.Witt MA, Katz PH, Witt DJ. 2012. Unexpectedly limited durability of immunity following acellular pertussis vaccination in preadolescents in a North American outbreak. Clin Infect Dis 54:1730–1735. doi: 10.1093/cid/cis287. [DOI] [PubMed] [Google Scholar]
- 39.López CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 75:6496–6503. doi: 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuk MH, Harvill ET, Miller JF. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol Microbiol 28:945–959. doi: 10.1046/j.1365-2958.1998.00850.x. [DOI] [PubMed] [Google Scholar]
- 41.Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 42.National Research Council. 2011. Guide to the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]