Abstract
Proper skeletal muscle function is dependent on spatial and temporal control of gene expression in multinucleated myofibers. In addition, satellite cells, which are tissue-specific stem cells that contribute critically to repair and maintenance of skeletal muscle, are also required for normal muscle physiology. Gene expression in both myofibers and satellite cells is dependent upon nuclear proteins that require facilitated nuclear transport. A unique challenge for myofibers is controlling the transcriptional activity of hundreds of nuclei in a common cytoplasm yet achieving nuclear selectivity in transcription at specific locations such as neuromuscular synapses and myotendinous junctions. Nucleocytoplasmic transport of macromolecular cargoes is regulated by a complex interplay among various components of the nuclear transport machinery, namely nuclear pore complexes, nuclear envelope proteins, and various soluble transport receptors. The focus of this review is to highlight what is known about the nuclear transport machinery and its regulation in skeletal muscle and to consider the unique challenges that multinucleated muscle cells as well as satellite cells encounter in regulating nucleocytoplasmic transport during cell differentiation and tissue adaptation. Understanding how regulated nucleocytoplasmic transport controls gene expression in skeletal muscle may lead to further insights into the mechanisms contributing to muscle growth and maintenance throughout the lifespan of an individual.
1. Introduction
Skeletal muscle is a very plastic tissue that readily undergoes changes in mass and function in response to aging, injury, and disease. Such changes in muscle can impact breathing, locomotion, and metabolism and affect motility and lifespan. Proper skeletal muscle function is dependent on spatial and temporal control of gene expression mediated by proteins, such as transcription factors, that require facilitated transport to enter the nuclei. The subcellular localization of these regulatory proteins must be tightly controlled because altered import or export could result in aberrant muscle function.
Proper muscle function is dependent on myofibers, which are multinucleated cells containing many hundreds of nuclei distributed along the length of the cell in a common cytoplasm. Alongside each myofiber are adult muscle stem cells, called satellite cells, that lay beneath the basal lamina surrounding each myofiber. These satellite cells are normally quiescent but in response to muscle damage they are activated to begin proliferating and undergo differentiation and eventual fusion with each other or existing myofibers to repair muscles in a process called myogenesis. Myogenesis can be modeled in vitro by culturing myoblasts, the progeny of satellite cells, and inducing them to differentiate into multinucleated myotubes by changes in culture media. Myogenesis both in vivo and in vitro requires the coordinate activation and repression of many genes. Numerous nuclear proteins are required for proper gene expression and the nuclear repertoire of these proteins is very different along the myogenic continuum of quiescent satellite cells to mature postmitotic myofibers. How satellite cells differentially regulate nucleocytoplasmic transport of these nuclear proteins critical for regulating gene expression during quiescence, activation, and differentiation is unknown. In addition, how a myofiber with hundreds of nuclei coordinates and regulates nucleocytoplasmic transport is not clear.
2. Nuclear Envelope
The nuclear envelope of eukaryotic cells provides separation of the genetic material and transcriptional machinery within the nucleus from the translational machinery in the cytoplasm enhancing regulation of gene expression. The nuclear envelope is comprised of two lipid membrane bilayers, the outer nuclear membrane which is contiguous with the endoplasmic reticulum and the inner nuclear membrane which faces the nucleoplasm (Hetzer and Wente, 2009). The inner nuclear membrane contains integral transmembrane proteins that interact with lamins within a nuclear lamina meshwork of intermediate-type V filaments that lines the inner nuclear membrane. The nuclear lamina contributes to nuclear envelope stability and provides a platform for proteins involved in chromatin anchoring, DNA replication, and gene transcription (Kind and van Steensel, 2010).
Several studies have revealed a critical role for inner nuclear envelope proteins in regulating the expression of muscle-specific genes during muscle differentiation (Datta et al., 2009; Huber et al., 2009; Liu et al., 2009; Ostlund et al., 2009). For example, loss of function or mutation of lamins or lamin-associated inner nuclear membrane proteins can result in tissue-specific diseases which are referred to as nuclear envelopathies (Holaska, 2008; Mattout et al., 2006). These diseases encompass a wide range of clinical phenotypes with different envelopathies affecting different tissues including muscle. Mutations in the nuclear envelope transmembrane protein emerin are associated with Emery-Dreifuss muscular dystrophy (Manilal et al., 1996), while mutations in lamin A/C lead to two muscular dystrophies, Emery-Dreifuss muscular dystrophy and limb-girdle muscular dystrophy 1B (Muchir et al., 2000). Emerin sequesters β-catenin and Lim domain protein, LMO7, at the inner nuclear periphery to regulate their participation in gene transcription; therefore, disease-causing mutations in emerin may disrupt access of these proteins to the transcriptional machinery (Holaska et al., 2006; Markiewicz et al., 2006). In addition, nuclear envelope transmembrane proteins, termed NETs, have been identified, a subset of which are hypothesized to have skeletal muscle-specific roles since they are highly expressed in skeletal muscle tissue compared with other mouse tissues (Chen et al., 2006; Schirmer et al., 2003). Specific roles for NETs appear to exist in signaling pathways during muscle differentiation (Datta et al., 2009; Huber et al., 2009; Liu et al., 2009). For example, during differentiation of C2C12 cells, a mouse muscle cell line (Yaffe and Saxel, 1977), depletion of NET25 led to elevated mitogen-activated kinase (MAPK) signaling which delayed myogenesis (Huber et al., 2009). In contrast, depletion of NET39 accelerated myogenesis through diminished mammalian target of rapamycin (mTOR) signaling and increased insulin-like growth factor 2 (IGF-2) production (Liu et al., 2009). Together, these studies expose a crucial role for nuclear envelope proteins in regulating gene expression during muscle differentiation. Mutations in nuclear envelope transmembrane proteins may alter signaling at the nuclear envelope and may contribute to the altered gene expression observed in laminopathies affecting skeletal muscle. Further studies will likely uncover functional roles for other nuclear envelope proteins in regulating skeletal muscle gene expression.
3. Nuclear Pore Complexes
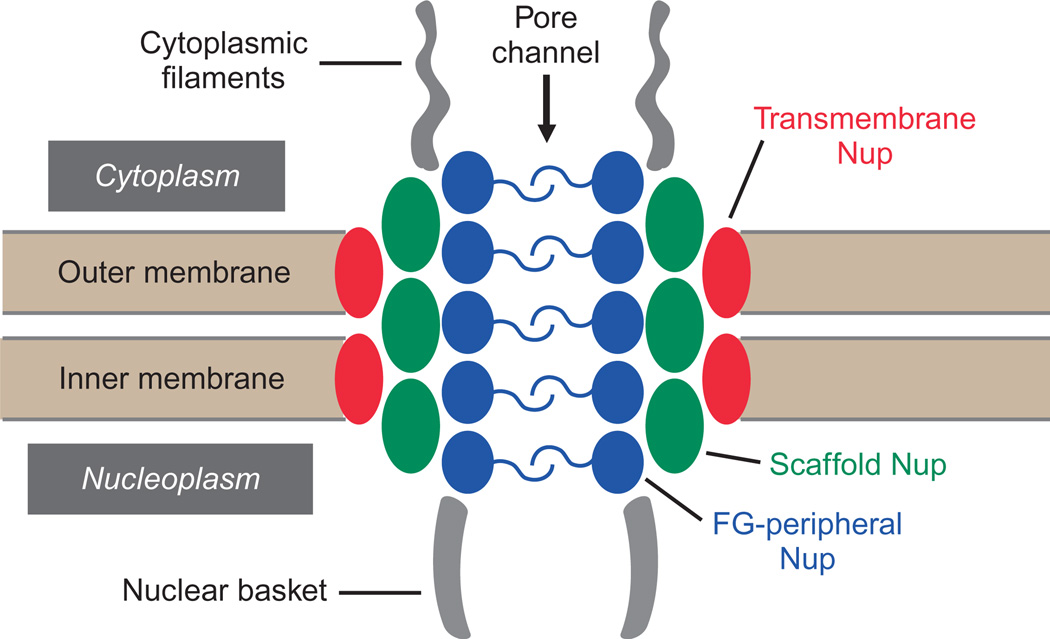
The nuclear envelope is perforated by nuclear pore complexes (NPCs) which fuse the outer nuclear membrane and inner nuclear membrane together to create channels for nucleocytoplasmic transport (Fig. 10.1; Lim and Fahrenkrog, 2006). NPCs are multiprotein suprastructures (~50 MDa) which provide channels for the nucleocytoplasmic exchange of ions and macromolecules (Alber et al., 2007). While smaller ions and molecules can diffuse through the NPC, molecules larger than ~40 kDa require a targeting signal and a soluble transport receptor to mediate transport through the NPC (Freitas and Cunha, 2009; Rabut et al., 2004; Weis, 2003). The NPC is comprised of ~30 types of nucleoporins or Nups, many of which are present in multiple copies, consistent with the eightfold symmetry of the NPC (Frenkiel-Krispin et al., 2010). Based on the current model of the NPC, Nups are categorized as scaffold, transmembrane, or peripheral Nups (Fig. 10.1; Fernandez-Martinez and Rout, 2009). Scaffold Nups, also termed core Nups, provide structure to the NPC core by forming a cage-like scaffold, while transmembrane Nups located at the nuclear envelope–NPC interface, function in NPC biogenesis and nuclear envelope anchoring (Strambio-De-Castillia et al., 2010). Peripheral Nups within the NPC function in cargo transport, chromatin anchoring, and gene transcription. Peripheral Nups lining the pore channel contain phenylalanine–glycine (FG) repeats that extend into the channel to function in NPC permeability and mediate facilitated transport of macromolecules (Strambio-De-Castillia et al., 2010). The physical mechanics of NPC permeability and transport are still unclear with several models proposing varying arrangements of FG-Nups during transport; however, the functional role of individual FG-Nups in mediating transport and regulating different transport pathways has been well established (Walde and Kehlenbach, 2010).
Figure 10.1.
Schematic illustrating the relative location of various Nups within the NPC. The NPC resides within the nuclear envelope, a bilipid membrane comprised of an inner and outer nuclear membrane. The NPC has cytoplasmic filaments that extend into the cytoplasm and a nuclear basket that extends into the nucleoplasm. Peripheral Nups containing FG repeats line the pore channel to function in NPC permeability and the facilitated transport of macromolecules. Transmembrane Nups localize to the nuclear envelope–NPC interface, while scaffold Nups reside between transmembrane and peripheral Nups.
NPCs within the nuclear envelope display variability in density and distribution between cell types and even within a single cell (Hetzer and Wente, 2009). For example, NPC density differs between Xenopus oocytes (>50 NPCs/mm2) and C2C12 muscle cells (5 NPCs/mm2) by 10-fold (D’Angelo et al., 2009; Hetzer and Wente, 2009), meanwhile a 50% increase in NPC density was observed as mouse embryonic stem cells differentiated into cardiomyocytes (Perez-Terzic et al., 2007). In addition, differences in NPC density across the nuclear envelope have been observed in Saccharomyces cerevisiae which suggests nuclear transport may be spatially regulated across the nuclear envelope (Winey et al., 1997). NPC nucleoporin composition also varies between cell types where multiple Nups involved in nucleocytoplasmic transport display differential expression between tissues (Hetzer and Wente, 2009; Smitherman et al., 2000; Tran and Wente, 2006). In skeletal muscle, the transcripts for numerous Nups are upregulated during satellite cell activation suggesting that an increase in NPC biogenesis occurs in proliferating satellite cells (Fukada et al., 2007; Pallafacchina et al., 2010). Meanwhile, Tetrahymena thermophila, a binucleated ciliated protozoa expresses a subset of Nups that differentially localize to either the macronucleus or the micronucleus to regulate transport of cargoes involved in nucleus-specific functions (Malone et al., 2008). Variations in NPC density, distribution, and nucleoporin composition between cell types or nuclei sharing a common cytoplasm, suggest that NPCs and nucleoporins are regulated to accommodate for ever changing demands on nuclear transport in both mono- and multinucleated cells.
In proliferating eukaryotic cells, new NPCs are formed during mitosis and interphase which allows for regular replacement of Nups (Doucet and Hetzer, 2010). Therefore, de novo assembly of NPCs would not be predicted to occur in postmitotic cells. Experiments examining the synthesis of scaffold and peripheral Nups in postmitotic C2C12 myotubes, revealed that some peripheral Nups, such as NUP153 and NUP50, are continuously synthesized, whereas scaffold Nups, such as NUP107 and NUP160, are transcriptionally downregulated in myotubes and are consequently not replaced (D’Angelo et al., 2009). Accumulation or loss of damaged scaffold Nups in nondividing cells, such as muscle, could result in dysfunctional NPCs and loss of integrity of the nucleocytoplasmic barrier. Indeed, oxidized scaffold Nups in neuronal nuclei from aged rats are associated with leakage of cytoplasmic proteins into the nucleus (D’Angelo et al., 2009). These studies have significant implications for the functional integrity of the NPC in quiescent satellite cells and postmitotic multinucleated muscle cells. Further studies are required to determine if Nups in NPCs become damaged in skeletal muscle and whether the nucleocytoplasmic barrier is altered and contributes to muscle dysfunction during disease and aging.
4. Nuclear Import Pathways
Nuclear transport is a process whereby proteins or other macromolecules traverse the NPC either by directly interacting with peripheral FG-Nups within the NPC channel or by binding to import or export receptors that mediate transport through the NPC (Walde and Kehlenbach, 2010). The majority of nuclear transport receptors are karyopherin family members termed importins, transportins, or exportins which mediate transport across the NPC in an energy-dependent manner (Tran and Wente, 2006; Weis, 2003). Transport receptors tend to be divided into importins, which bind a nuclear localization signal (NLS) within a cargo protein to target it into the nucleus and exportins, which bind a nuclear export sequence (NES) within a cargo protein to target it for export to the cytoplasm (Cook et al., 2007; Kalderon et al., 1984a,b; Kutay and Guttinger, 2005; Lange et al., 2007). Karyopherin transport receptors fall within two families, the karyopherin alpha (KPNA) family and the karyopherin beta family (Wagstaff and Jans, 2009). Increasing evidence suggests that both these karyopherin families have critical roles in controlling the nuclear import and export of key proteins involved in genetic reprogramming and cell adaptation in a large number of cell types and tissues (Kohler et al., 1999; Quensel et al., 2004; Talcott and Moore, 2000; Yasuhara et al., 2007). Below we describe the mechanisms of karyopherin-dependent nuclear transport and detail what is currently known about these transport receptors and pathways in skeletal muscle.
4.1. Classical nuclear import: Karyopherin alpha family
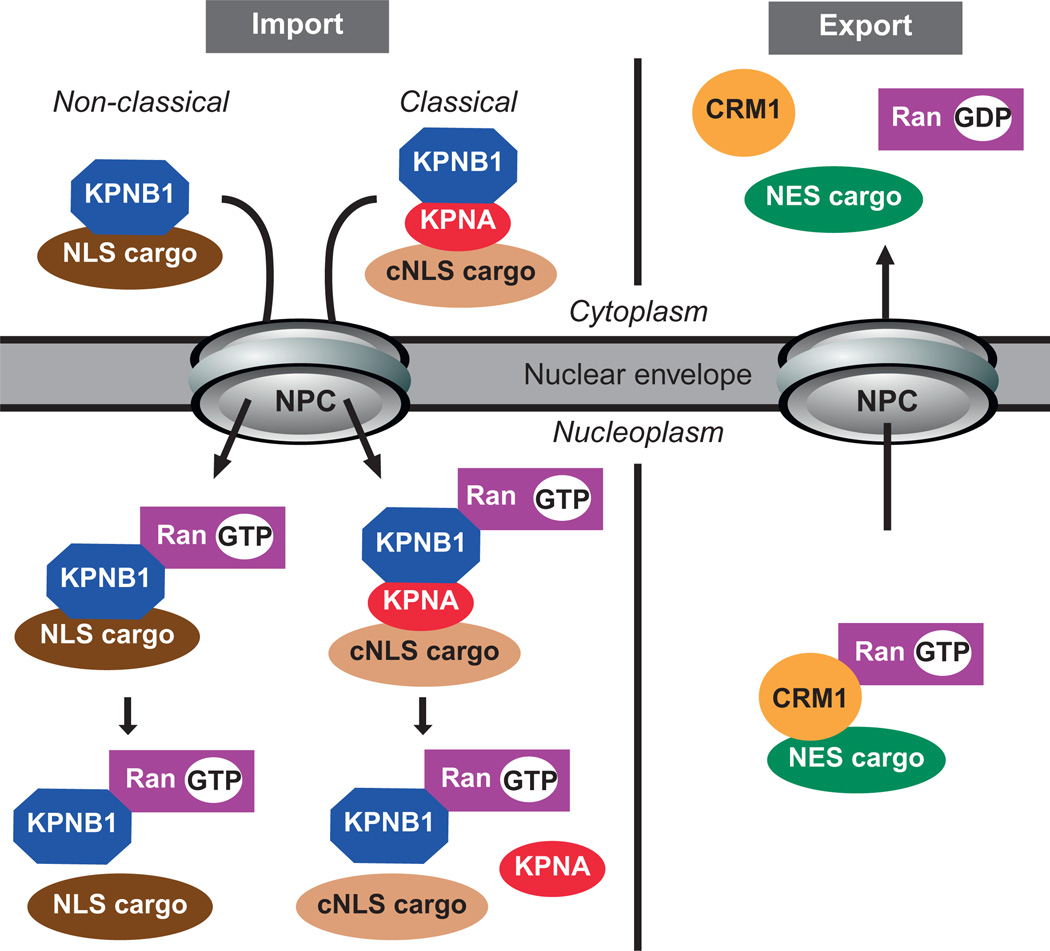
Classical nuclear import, which is the best characterized of the nuclear transport pathways, is an active process that depends on KPNA and beta family members as well as a classical nuclear localization signal sequence (cNLS) defined by a string of basic residues contained within a protein (Hodel et al., 2001; Kalderon et al., 1984b; Robbins et al., 1991). In Mus musculus, 35–55% of nuclear proteins may depend on classical nuclear import for nuclear targeting as determined using a bioinformatics approach (Marfori et al., 2010). KPNAs recognize two types of cNLS, a monopartite signal comprised of a single string of basic amino acid residues or a bipartite signal containing two strings of basic variable residues flanking a 10–12 amino acid linker region (Kalderon et al., 1984b; Robbins et al., 1991). The prototypical sequence for the monopartite is the SV40 large-T antigen sequence, PKKKRKV, and for the bipartite, the nucleoplasmin sequence KRPAATKKAGQAKKKK (Hodel et al., 2001; Kalderon et al., 1984a; Robbins et al., 1991). Classical nuclear protein import is mediated by a heterotrimeric complex of KPNA, which recognizes and binds the cNLS signal within a cargo protein, and karyopherin beta1 (KPNB1), which binds KPNA and interacts with FG-Nups within the NPC to mediate nuclear import (Fig 10.2; Matsuura et al., 2003; Matsuura and Stewart, 2005). Once in the nucleus, a small GTPase, Ran-GTP, binds KPNB1, triggering disassembly of the trimeric complex and subsequent cargo release (Cook et al., 2007). Disassembly of the import complex is also facilitated by CAS, the export receptor for KPNA and another member of the karyopherin family (Kutay et al., 1997), and the nucleoporin, NPAP60L (Ogawa et al., 2010). Upon cargo release, KPNA is recycled back to the cytoplasm by CAS, while KPNB1 is returned to the cytoplasm in complex with Ran- GTP (Hood and Silver, 1998; Kutay et al., 1997). Thus, classical nuclear import cycles and directionality depend upon the GTPase, Ran, which facilitates assembly and disassembly of transport complexes (Cook et al., 2007). Ran-GTP, but not Ran-GDP, triggers cargo release in the nucleus upon binding to KPNB1, therefore, the directionality of import is driven by the presence of Ran-GTP in the nucleus and Ran-GDP in the cytoplasm (Lonhienne et al., 2009). Ran-GTP levels are maintained in the nucleus through the nuclear import of Ran-GDP (Ribbeck et al., 1998) and conversion to Ran-GTP by the nuclear localized Ran guanine nucleotide exchange factor (Ran GEF; Cook et al., 2007; Smith et al., 1998). Meanwhile, in the cytoplasm, Ran-GTP is hydrolyzed to Ran-GDP by the Ran GTPase-activating protein (Ran GAP).
Figure 10.2.
Basic model of karyopherin-mediated nuclear import and export pathways. The nuclear import pathway involves the import receptors karyopherin alpha (KPNA) and/or KPNB1 which can recognize proteins containing a classical (cNLS) or nonclassical (NLS) nuclear localization signal, respectively. The nuclear import of a cNLS-containing cargo involves both KPNA and KPNB1 import receptors, since KPNA recognizes the cNLS motif in the cargo protein and then KPNB1 mediates translocation of the import complex through the NPC by interacting with FG-Nups within the NPC. Once in the nucleus, Ran-GTP binding to KPNB1 results in the dissociation of the import complex and cargo release into the nucleus. The nuclear export pathway consists of an obligate trimeric complex consisting of the exportin (CRM1 for classical NES-containing cargo), export cargo, and Ran-GTP. Translocation of the complex through the NPC is mediated by interaction between the exportin and FG-Nups. Once in the cytoplasm, the hydrolysis of Ran-GTP to Ran-GDP results in the dissociation of the export complex and subsequent cargo release.
KPNB1 is the sole member of the karyopherin beta family to participate in classical nuclear import with KPNA (Liu and Liu, 2007). KPNB1 and other karyopherin beta family members participate in nonclassical nuclear import which involves either direct NLS binding or the use of non-KPNA adaptor proteins. Modeling studies for cNLS import reveal that the addition of the KPNA adaptor to the transport cycle, as opposed to direct protein import by KPNB1 alone, reduces import efficiency (Riddick and Macara, 2007); however, the loss of import efficiency is offset by the addition of multiple KPNA adaptor paralogs that allows for additional points of control over the nuclear localization of cNLS proteins.
Saccharomyces cerevisiae contains a single, essential KPNA import receptor. Six KPNA paralogs are found in mouse: KPNA1, KPNA2, KPNA3, KPNA4, KPNA6, and KPNA7 (Hu et al., 2010; Tsuji et al., 1997). Seven KPNA paralogs exist in human with which the mouse homologues share 90% amino acid identity (Kelley et al., 2010; Kohler et al., 1997, 1999; Tsuji et al., 1997). Confusion regarding KPNA protein nomenclature between species exists in the literature. While KPNA gene names between human and mouse are consistent, protein nomenclature using the terms importin alpha or importins does not match between these two species (Table 10.1). In this chapter, we refer to KPNA paralogs using the KPNA/kpna nomenclature, instead of importin alpha, to minimize confusion when discussing KPNA paralog function. KPNA paralogs in mouse and human are categorized into three subtypes based on percentage of amino acid identity. Mouse subtypes are Subtype S: KPNA1 and KPNA6; Subtype P: KPNA2; and Subtype Q: KPNA3 and KPNA4. Subtype members share 80–90% amino acid identity, whereas different subtypes share 40–50% amino acid identity (Tsuji et al., 1997). All KPNA paralogs function as nuclear import receptors, but paralogs may differ in their cNLS binding affinities and/or specificities for cNLS proteins (Hodel et al., 2001; Kohler et al., 1999; Quensel et al., 2004; Talcott and Moore, 2000; Timney et al., 2006; Yasuhara et al., 2007). For example, human regulator of chromosome condensation 1, RCC1, which is the Ran GEF, depends solely on KPNA4 to access to the nucleus, while other cNLS proteins, such as RNA Helicase A, may utilize multiple KPNA paralogs to access the nucleus, but may have preference for one paralog over another (Aratani et al., 2006; Quensel et al., 2004). In silico experiments suggest that the rate of nuclear import of a cNLS cargo is limited by the levels of KPNA and Ran (Riddick and Macara, 2005).
Table 10.1.
Karyopherin alpha paralogs in different organisms
| Homo sapiens | Mus musculus | Drosophila melanogaster | Saccharomyces cerevisiae | |||||
|---|---|---|---|---|---|---|---|---|
| Subtype | Gene name | Importin designation |
Gene name |
Importin designation |
Gene name | Importin designation |
Gene name |
Importin designation |
| S | KPNA1 | alpha5 | Kpna1 | alpha1 | Kap-alpha1 | alpha1 | Srp1 | Kap60 |
| KPNA5 | alpha6 | – | – | – | – | – | – | |
| KPNA6 | alpha7 | Kpna6 | alpha6 | – | – | – | – | |
| P | KPNA2 | alpha1 | Kpna2 | alpha2 | Pen | alpha2 | – | – |
| KPNA7 | alpha7 | Kpna7a | alpha7a | – | – | – | – | |
| Q | KPNA3 | alpha4 | Kpna3 | alpha3 | – | – | – | – |
| KPNA4 | alpha3 | Kpna4 | alpha4 | Kap-alpha3 | alpha3 | – | – | |
KPNA family members are categorized into three subtypes, S, P, and Q based on amino acid sequence homology.
Saccharomyces cerevisiae has a single karyopherin (SRP1), while Homo sapiens, Mus musculus, and Drosophila melanogaster each contain multiple karyopherin paralogs. The gene name and importin alpha designation are given for each species to clarify confusion regarding karyopherin/importin designations between species. A dash (–) indicates the absence of a karyopherin homologue in that species.
Placement of recently discovered murine KPNA7 into subtype P is tentative.
The steady-state levels of different KPNA paralogs can vary both among tissue types and within a single tissue during differentiation suggesting distinct roles for individual KPNA paralogs in importing key factors required for cell function and differentiation (Goldfarb et al., 2004; Mason and Goldfarb, 2009; Okada et al., 2008; Poon and Jans, 2005). For example, during mouse spermatogenesis, KPNA paralogs are expressed with unique cellular and temporal expression profiles at discrete stages of development (Hogarth et al., 2006). In contrast, during neural differentiation of mouse embryonic stem cells in vitro, the steady-state levels of one KPNA paralog increase, while that of another paralog decrease, thereby allowing for differential nuclear import of transcription factors involved in maintaining either the undifferentiated or differentiated state (Yasuhara et al., 2007). This KPNA paralog switching was proposed by the authors of this study as a general mechanism that enables cells to coordinate differentiation by controlling the subcellular localization of transcription factors. In support of the subtype switching model, KPNA steady-state profiles in multiple differentiating human and mouse cell types are characterized by increases in expression of one KPNA paralog with concomitant decreases in another paralog (Kamei et al., 1999; Kohler et al., 1997, 2002; Okada et al., 2008); however, in skeletal muscle cells, an increase in the steady-state levels of all five Kpnas was observed during differentiation (Hall et al., unpublished data). The increase in all Kpnas may suggest an overall increase in demand for nuclear import during skeletal muscle differentiation. These studies suggest that the role of KPNAs in cell differentiation may differ between cell types that express different cNLS-containing cargo proteins.
Additional evidence for the nonredundant roles of individual KPNA paralogs in cellular physiology stems from loss-of-function experiments in model organisms. Kpna1 null Drosophila melanogaster developed normally but displayed defects in gametogenesis resulting in sterility in both males and females (Ratan et al., 2008). Similarly, male and female sterility occurred in Kpna2 null flies (Mason et al., 2002). The sterility in females could be rescued only by Kpna2 transgenes, whereas the sterility in males could be rescued by Kpna1, Kpna2, or Kpna3 transgenes suggesting distinct requirements for KPNA2 in male and female gametogenesis (Mason et al., 2002). In contrast, Kpna3 null flies displayed defects throughout development, whereas late stages of development and photoreceptor development could only be rescued with Kpna3 but not Kpna1 or Kpna2 transgenes (Mason et al., 2003). RNAi experiments in Caenorhabditis elegans demonstrated that KPNA3 but not KPNA2 is required for oocyte development (Geles and Adam, 2001). Together, these results in genetic model organisms support the notion that KPNA paralogs have evolved distinct functions in different cell types during development.
Loss-of-function experiments in vitro also provide support for distinct roles of KPNA paralogs in controlling cell proliferation and differentiation. During neural differentiation of mouse embryonic stem cells, depletion of KPNA1 by RNAi-mediated knockdown resulted in accelerated neural differentiation, while loss of KPNA5 delayed differentiation (Yasuhara et al., 2007). In primary mouse muscle cells, the nonredundant roles for individual paralogs were revealed by siRNA experiments in which KPNA1 knockdown increased myoblast proliferation but KPNA2 knockdown decreased proliferation (Hall et al., unpublished data). In contrast, no proliferation defect was observed with KPNA4 knockdown. KPNAs import negative regulators of proliferation, such as Rb and p27Kip1 in other cell types (Hu et al., 2005; Shin et al., 2005), so results of these RNAi experiments could suggest that KPNA1 imports a negative regulator of proliferation during myogenesis. Contrary to these findings, in Hela cells, RNAi-mediated knockdown of each KPNA, including KPNA1, resulted in a decrease in cell proliferation, which suggests that the role of individual KPNAs in cell proliferation may differ between cell types (Kohler et al., 2002). Contrary to the results obtained in vitro, a recent study examining a Kpna1 null mice revealed normal development of brain and other tissues, however, KPNA1 may have a different role in proliferation in the brain or compensation by other KPNAs may have occurred during development (Shmidt et al., 2007). Further evidence for the nonredundant roles of KPNA paralogs in skeletal muscle comes from knockdown experiments where depletion of KPNA2, but not KPNA1 or KPNA4, resulted in reduced myotube size and decreased ability of muscle cells to migrate (Hall et al., unpublished data). The small myotube phenotype observed may be due to the reduced import of multiple cNLS-containing proteins involved in regulating cell migration. Indeed, a similar migration defect was also observed upon loss of KPNA2 in a lung cancer cell line (Wang et al., 2010). These studies suggest that myoblast proliferation and myotube growth rely on specific KPNA paralogs to regulate the nuclear import of key factors involved in proliferation and myotube growth during myogenesis.
Classical nuclear import has been implicated in transporting cargoes across large distances or from specific sites in the cell to the nucleus. This phenomenon has been most extensively studied in neurons (Lai et al., 2008; Mikenberg et al., 2007; Thompson et al., 2004), but emerging evidence supports the presence of such spatial signaling in skeletal muscle. In rodent hippocampal neurons, classical nuclear import mediates the transport of cNLS cargoes from the synapse to the nucleus upon receptor activation (Thompson et al., 2004). In skeletal muscle cells, KPNA mediates the nuclear import of myopodin, an actin bundling protein (Faul et al., 2007), which has been shown to shuttle between the sarcomeric Z-disc and the nucleus in a differentiation and stress-dependent manner (Weins et al., 2001). However, the role of myopodin nucleocytoplasmic shuttling during cell differentiation and stress is unclear. In contrast, a Z-disc-associated protein with known nuclear function is muscle limb protein (MLP), which acts as a mechanosensor in rat cardiomyocytes (Boateng et al., 2009). Loss of cNLS-dependent import of MLP results in disarranged sarcomeres. Another cargo that undergoes similar shuttling is serum response factor (SRF), a transcription factor required for skeletal muscle growth that shuttles between the sarcomere and the nucleus (Li et al., 2005). The nuclear import of SRF occurs via KPNA1/KPNB1 of the classical nuclear import pathway (McConville et al., 2010). These findings suggest that nucleocytoplasmic import has a significant role in transmitting signals from sarcomeres to the nucleus in response to stimuli that induce muscle cell remodeling to adapt to cellular stress. Further studies should shed light on the role of classical nuclear import in overcoming the unique spatial challenges of signaling in a multinucleated muscle cell.
4.2. Karyopherin beta family members mediate import and export
Karyopherin beta family members comprise the majority of nuclear transport receptors which includes karyopherin beta import receptors (importins or transportins) involved in protein import and exportins involved in protein export (Cook and Conti, 2010; Cook et al., 2007). Similar to KPNA-dependent nuclear import, the karyopherin beta nuclear import pathway is dependent upon energy and the Ran gradient for directionality (Lonhienne et al., 2009). Karyopherin beta import receptors bind directly to nonclassical NLSs in cargo proteins to mediate import (Fig. 10.2) or may use a non-KPNA adaptor protein for NLS cargo recognition (Cook et al., 2007; Kutay and Guttinger, 2005; Lange et al., 2007). Currently, only karyopherin beta2-dependent cargoes have defined NLSs termed PY-NLS, while cargoes depending on other karyopherin beta family members do not have recognizable amino acid sequences that comprise NLS motifs (Marfori et al., 2010).
In total, 14 karyopherin beta import receptors exist in S. cerevisiae, while humans have over 19 karyopherin betas (Chook and Suel, 2010). Karyopherin beta family members display protein homology ranging from 15% to 20% and function in the import of distinct sets of proteins, RNAs, and Nups (Chook and Suel, 2010; Marfori et al., 2010). KPNB1, the best characterized member of the karyopherin family, is one of four essential karyopherin beta family members in S. cerevisiae (Chook and Suel, 2010). In contrast to classical nuclear import, KPNB1-dependent nuclear import does not occur through a conserved receptor–cargo binding conformation which provides KPNB1 with the flexibility to bind a wide variety of cargoes containing unique classes of NLS signals (Fiserova et al., 2009; Marfori et al., 2010).
Several studies provide evidence that karyopherin beta family members have critical roles in regulating the cellular localization of different NLS proteins involved in myogenesis and neuromuscular junction physiology (Giagtzoglou et al., 2009; Higashi-Kovtun et al., 2010; Mosca and Schwarz, 2010; van der Giessen and Gallouzi, 2007). In C2C12 myoblasts, the nuclear import of the RNA-binding protein HuR depends upon the karyopherin beta family member transportin-2 (van der Giessen and Gallouzi, 2007). HuR is a RNA-binding protein involved in regulating the stability of mRNA transcripts encoding MyoD and Myogenin which are myogenic transcription factors required for differentiation (Figueroa et al., 2003; van der Giessen and Gallouzi, 2007). During differentiation, cleavage of HuR prevents its nuclear import by transportin-2 which results in the stabilization of MyoD and Myogenin mRNAs and enhancement of myogenesis (Mazroui et al., 2008). Several studies using a Drosophila model system have uncovered roles for karyopherin beta family members in regulating the import of proteins involved in postsynaptic membrane development and neurotransmitter release at the neuromuscular junction (Giagtzoglou et al., 2009; Higashi-Kovtun et al., 2010; Mosca and Schwarz, 2010). Wingless signaling at the neuromuscular junction causes cleavage and release of the C terminus of Frizzled2 (Fz2-C), which is then imported into the nucleus by KPNA2/KPNB1, or by the karyopherin beta family member karyopherin-beta11 (Mosca and Schwarz, 2010). In Drosophila mutants lacking either KPNA2 or karyopherin-beta11, a reduction in the nuclear import of Fz2-C and defects in the postsynaptic membrane were observed suggesting that multiple transport pathways are required for membrane development at the neuromuscular junction. Another karyopherin beta involved in neuromuscular junction physiology is Drosophila karyopherin beta13 which controls neurotransmitter release and intracellular Ca2+ levels at the neuromuscular junction (Giagtzoglou et al., 2009). These data suggest that karyopherin beta family members regulate myogenesis, neuromuscular development and neurotransmitter release by importing a variety of proteins, including RNA-binding proteins and cell-surface receptor components.
Karyopherin beta family members also play roles in facilitated nuclear export of proteins to the cytoplasm (Cook and Conti, 2010; Wente and Rout, 2010). The best understood export pathway is the recognition of a classical nuclear export signal (NES) by the karyopherin beta, CRM1 (Fig. 10.2). The classical NES signal consists of a short string of hydrophobic leucine-rich residues, which are difficult to identify because they share sequence similarity with the hydrophobic cores of most proteins (Cook et al., 2007). Prototypical sequences for an NES are the cyclin D NESs, RFLSLEPL, and TPTDVRDVDI as well as the mitogen-activated protein kinase kinase (MAPKK) NES, LQKKLEELEL (Kutay and Guttinger, 2005; Poon and Jans, 2005). Export receptors or exportins recognize and bind export cargoes while bound to the GTPase, Ran-GTP, in an obligate trimeric complex (Cook et al., 2007; Kutay and Guttinger, 2005).
As with facilitated nuclear import, directionality of export is driven by the compartmentalization of Ran-GTP in the nucleus and Ran-GDP in the cytoplasm. At least six exportin genes are found in mouse and human (Okada et al., 2008). These exportins facilitate the nuclear export of a variety of cargoes with some exportins displaying different specificity for individual cargoes. Similar to NLS-mediated import, NES-containing proteins may be exported by a single exportin or may utilize multiple exportins (Okada et al., 2008). For example, the essential yeast exportin, CSE1, or CAS in vertebrates, has one export cargo, KPNA of the classical nuclear import pathway (Cook et al., 2007), while the best characterized exportin, CRM1(XPO1 or exportin-1), mediates export of at least 10 different classical NES-containing proteins (Cook et al., 2007; Shen et al., 2010; Wada et al., 1998). Exportin family members also facilitate the export of tRNAs and pre-miRNAs (Cook et al., 2007; Lund et al., 2004). Together, these studies suggest exportins may have differing roles in the export of a wide variety of cargo proteins and RNAs required for proper cell function.
Nucleocytoplasmic shuttling via CRM1-dependent export and KPNA-dependent import coordinate the nuclear steady-state levels of proteins critical to muscle cell biology. For example, the nuclear localization of the transcription factor NF-κB is controlled by the subunit p65 which mediates both import and export of the complex through binding to KPNA/KPNB1 or CRM1, respectively (Micheli et al., 2010; Zerfaoui et al., 2010). The nuclear accumulation of p65 NF-κB suppresses MyoD transcription (Guttridge et al., 2000) therefore modulation of nuclear import or export of p65 could control p65 NF-kB activity, MyoD expression, and ultimately myogenesis. The nuclear import and export of the forkhead box transcription factor FOXO3a is critical for skeletal muscle atrophy (Sandri et al., 2004). In C2C12 cells, the nuclear import of Foxo3a was observed upon inhibition of the phosphatidylinositol 3-kinase, PI3K/Akt pathway, while nuclear export of FOXO3a was observed upon activation of the stress-activated protein kinase (SAPK) pathway (Clavel et al., 2010). Control over the cellular localization of FOXO3a by two different signaling pathways may provide global control over atrophy by regulating the transcription of genes, such as Atrogin-1, that are involved in skeletal muscle atrophy (Clavel et al., 2010). Control over the nucleoctyoplasmic shuttling of cargo proteins by different transport pathways may provide global control over gene expression during myogenesis.
5. Identifying Classical Nuclear Import-Dependent Cargoes
Identifying the specific cargo proteins that are transported via various nucleocytoplasmic transport pathways is key for understanding the regulatory networks that govern cell function. Here we focus on how cNLS-dependent cargoes are identified since this pathway is the best characterized nuclear transport pathway; however, many of the challenges in cargo identification presented here apply also to other receptor-mediated transport pathways. In Mus musculus, 30–55% of nuclear proteins are predicted to depend on classical nuclear import (Marfori et al., 2010). However, only a few proteins with key functional roles in muscle are known to contain a functional cNLS, such as NOTCH (Huenniger et al., 2010) and NFATc2 (Okamura et al., 2000). While bioinformatics approaches exist to identify classical nuclear import signal sequences within cargoes, these putative cargoes still require functional testing to ensure that such signals actually mediate transport via this pathway.
Putative cNLS motif sequences within proteins can be identified with prediction software (Cokol et al., 2000; Horton et al., 2007; Kosugi et al., 2009; Nguyen Ba et al., 2009). The consensus sequence for the monopartite cNLS has been characterized in both structural and thermodynamic studies where the first residue is a lysine followed by a second and fourth basic residue as follows: K(K/R)X(K/R) (Conti and Kuriyan, 2000; Fontes et al., 2000; Hodel et al., 2001). The consensus sequence for the bipartite cNLS has also been characterized as KRX10-12KRRK (Fontes et al., 2003). Small deviations from the consensus sequence may increase or decrease KPNA-cargo binding affinity, while large deviations likely result in failed import because KPNA-cargo binding is either too weak or too strong for efficient cargo import and release (Lange et al., 2007). A drawback to cNLS prediction algorithms is that linear sequence is analyzed and these algorithms do not identify nonlinear synthetic cNLS signals created through intra- or interprotein interactions. For example, signal transducer and activator of transcription (STAT1) forms a homodimeric complex in which each dimer contributes basic resides to form a functional synthetic cNLS that is not detected by current cNLS algorithms (Fagerlund et al., 2002). While cNLS prediction models identify consensus sequences, functional studies meeting several criteria must be performed before a cNLS is deemed functional (Lange et al., 2007). A cNLS is functional if it is both necessary and sufficient for import of the cargo protein and import of the cargo depends upon the classical nuclear import machinery (Lange et al., 2007).
An alternative approach to identifying cNLS-dependent cargoes is a candidate-based approach which involves transport receptor loss-of-function experiments, whereby a phenotype observed upon receptor depletion may offer hints to potential cargo. A candidate-based approach may prove difficult since the phenotypes observed during depletion of transport receptors are likely combinatorial due to the altered nuclear transport of many cargo proteins that are involved in regulating a large number of genes. Overall, identifying cargoes dependent upon classical nuclear import receptors will be critical to understanding the role of nucleoctyoplasmic transport in regulating cell function and fate in skeletal muscle.
6. Remodeling of the Nuclear Transport Machinery
Alterations in global nucleocytoplasmic transport provide another layer of control over gene expression. Global changes in the efficiency or rate of nuclear transport can occur through alterations or remodeling of key components of the nuclear transport machinery. For example, altering the expression or localization of karyopherin transport receptors, Ran and/or Ran-associated proteins, or Nups results in changes in transport efficiency (Hodel et al., 2001, 2006; Riddick and Macara, 2005; Timney et al., 2006; Wagstaff and Jans, 2009). A muscle cell could alter nucleocytoplasmic transport to adjust for changes in demand for nuclear transport over a wide range of cellular conditions such as cell quiescence, proliferation, differentiation, stress, aging, and disease.
Experimental evidence indicates that steady-state levels for different components of the nuclear transport machinery can vary during myogenesis. Microarray analyses suggest that satellite cell entry into the cell cycle is marked by global remodeling of the nuclear transport machinery (Fukada et al., 2007; Pallafacchina et al., 2010). A wide variety of mRNAs that encode components of the nuclear transport machinery, such as nucleoporins and various karyopherin transport receptors, were increased in proliferating satellite cells in vivo compared to quiescent satellite cells. This widespread upregulation of the nuclear transport machinery may be functionally required to allow for rapid changes in gene expression associated with the myogenic lineage progression of satellite cells. Nuclear pore composition can influence stem cell differentiation as evidenced from studies of NUP-133-deficient epiblast and embryonic stem cells in mice which differentiated inefficiently along the neural lineage (Lupu et al., 2008). Nuclear transport machinery remodeling has also been observed during muscle differentiation, however the extent and type of remodeling differs among muscle cell types. In mouse primary muscle cells, the steady-state levels of all five KPNA import receptors increased during skeletal muscle differentiation suggesting an increase in demand for nuclear import during differentiation (Hall et al., unpublished data). The differentiation of mouse embryonic stem cells into a cardiac lineage resulted in the downregulation of karyopherins, exportins, Nups- and Ran-related proteins, while an increase in NPC density was observed along with the expansion of individual NPC diameter suggesting an increase in demand for nuclear transport during cardiac differentiation (Perez-Terzic et al., 2007). In contrast, during the differentiation of C2C12s, the steady-state levels of Nup proteins and overall density of NPCs remained constant (D’Angelo et al., 2009). Differences in nuclear transport machinery remodeling appear to be cell type-dependent and suggest that remodeling of the nuclear transport machinery is a key process that controls global nucleocytoplasmic transport during cellular differentiation. Characterizing the functional role of members of the nuclear transport machinery during skeletal muscle proliferation and differentiation will be essential to understanding the role of nucleocytoplasmic transport as a driver of muscle cell differentiation and function.
Remodeling of the nuclear transport machinery has also been observed during cellular response to chemical or mechanical stress in multiple cell types. Remodeling of the nuclear transport machinery to reduce or block transport during cellular stress may allow a cell to globally “pause” gene signaling pathways in order to redirect gene expression to respond to a particular cellular stress. Cellular stress, such as oxidative stress, can inhibit both import and export transport receptors and reduce the levels, localization and posttranslation modifications of several Nups involved in nuclear export (Crampton et al., 2009; Kodiha et al., 2008; Miyamoto et al., 2004). In vascular smooth muscle cells (VSMCs), remodeling of the nuclear import machinery was observed during exposure to ceramide, a antiproliferative sphingolipid implicated in the final stage of atherosclerotic plaque formation. Ceramide treatment of cultured VSMCs resulted in reduced cell proliferation and inhibition of classical nuclear import due to mislocalization of KPNA and CAS to the cytoplasm (Faustino et al., 2008). Nuclear transport machinery remodeling during mechanical stress was also observed during stretching of VSMCs which results in smooth muscle cell hyperplasia and hypertrophy (Richard et al., 2007). Mechanical stretching of VSMCs in vitro resulted in an increase in nuclear import and the steady-state protein levels of Nups, along with alterations in MAPK signaling. Further studies are required to determine whether remodeling of the nuclear transport machinery also occurs with oxidative or mechanical stress in skeletal muscle cells and what role it may play in controlling the nuclear localization of critical proteins necessary for cellular responses to these physiologic perturbations.
Aging and disease are also associated with changes in the nuclear transport machinery. In myocardial microvascular endothelial cells and human fibroblasts, a reduction in expression of KPNA and therefore, classical nuclear import occurred with cellular aging (Ahluwalia et al., 2010; Pujol et al., 2002). Another study using Hela cells expressing a mutant form of Lamin A responsible for the premature aging disease, Hutchinson Gilford progeria syndrome, revealed a reduction in nuclear import efficiency along with alterations in the localization of NUP62, NUP153, and the exportin CRM1 (Busch et al., 2009), suggesting that remodeling of the nuclear import machinery may have a role in disease pathology. In contrast, human cardiomyocytes from patients with heart failure, displayed an increase in karyopherin import receptors, exportins, Ran regulators, and Nups as well as differences in NPC configuration and morphology as compared to healthy cardiomyocytes (Cortes et al., 2010) suggesting that remodeling may occur as a disease response. These findings suggest that changes in the nuclear transport machinery may occur either as result of aging and disease or may occur in response to disease to restore tissue function. Given the extensive loss of skeletal muscle mass that can occur with aging or disease, further studies are warranted to determine if remodeling of the nuclear transport machinery also occurs with age or disease in skeletal muscle and whether nucleocytoplasmic communication is impaired as a consequence.
7. Challenges in Studying Nucleocytoplasmic Transport in Multinucleated Cells
The basic mechanics for nucleocytoplasmic transport of proteins and RNA identified to date and described in this review have almost exclusively been examined in cells with a single nucleus. Skeletal muscle is the only permanent multinucleated cell type in the body and constitutes ~50% of body mass; yet, how these cells spatially and temporally regulate and coordinate nucleocytoplasmic transport among hundreds of nuclei is unknown.
Spatial and temporal regulation of nucleocytoplasmic transport between nuclei likely occurs within a single myofiber since transcriptional activity of specific gene loci can differ among nuclei within the same myofiber. Such differences in transcription could arise from specific regional requirements for cell function. Myonuclei at the neuromuscular junction express transcripts for subunits of the acetylcholine receptor at much higher levels than nonsynaptic nuclei leading to the accumulation of the acetylcholine receptor protein at the neuromuscular junction (Burden, 1993; Fontaine and Changeux, 1989; Sanes et al., 1991; Simon et al., 1992) and thereby facilitating coordinated neuronal activation of muscle contraction. In addition, the myonuclei located at the myotendinous junction in stretched myofibers express the transcript for sarcomeric myosin heavy chain at higher levels than other myonuclei (Dix and Eisenberg, 1990), thereby enhancing sarcomere addition and cell growth at the ends of myofibers in response to muscle stretching. Less clear are the reasons why myonuclei distributed along the length of a myofiber and at times even right next to each other exhibit differences in the transcription of both endogenous genes and transgenes (Newlands et al., 1998). Transcriptional differences among nuclei also occur in myotubes in vitro (Berman et al., 1990; Su et al., 1995). The molecular mechanisms responsible for such nuclear diversity in transcriptional activity in skeletal muscle are unknown. Differences in transcriptional activity among nuclei in a common cytoplasm also occur in other cell types and organisms. For example, only a subset of nuclei in multinucleated mouse osteoclasts or human placental syncytiotrophoblasts is transcriptionally active (Ellery et al., 2009; Youn et al., 2010). In binucleated Tetrahymena thermophila, the macronucleus is transcriptionally active, whereas the micronucleus is transcriptionally inert (Karrer, 2000). Further, nuclei in the syncytial blastoderm of Drosophila and the syncytial germ line of C. elegans are also transcriptionally distinct (Burden, 1993). The molecular mechanisms responsible for the transcriptional differences among nuclei in these other cell types and organisms are not fully elucidated.
Nuclear proteins are localized to some nuclei and not others within a myofiber which further suggests differential nuclear targeting occurs in skeletal muscle. One such protein is endonuclease G, which is a mitochondrial protein that can translocate to nuclei and induce DNA fragmentation and apoptosis independent of caspase. At the initiation of disuse muscle atrophy, endonuclease G translocates to a subset of myofiber nuclei (Dupont-Versteegden et al., 2006) and may serve as a means to control the loss of myonuclei commonly observed in disuse atrophy without cell death. Other examples of such proteins are specific nuclear envelope proteins which are more highly concentrated in synaptic myonuclei compared to nonsynaptic nuclei. These include Syne-1, Syne-2, and nesprin-1α which appear to participate in nuclear localization and/or anchoring (Apel et al., 2000; Grady et al., 2005; Puckelwartz et al., 2010; Zhang et al., 2007). Unequal nuclear localization of proteins is also observed in cultured myotubes. These include transcription factors with important roles in muscle differentiation and growth such as NFAT5 (O’Connor et al., 2007), NFATc1 (Abbott et al., 1998), and MyoD (Ferri et al., 2009), the growth inhibitory protein, myostatin (Artaza et al., 2002) as well as MYO18B (Salamon et al., 2003), an unconventional myosin heavy chain with an unidentified role in muscle physiology. The mechanisms that govern differential protein targeting among neighboring myonuclei in vivo and in vitro are unknown but may be related to nucleus-specific transport mechanisms and merit further study.
Studies in Tetrahymena provide potential clues for how nucleus-specific transport mechanisms may be regulated in multinucleated myofibers. Certain nuclear proteins in Tetrahymena are selectively accumulated in either macro- or micro-nuclei (White et al., 1989). Analyses of GFP-labeled KPNA proteins revealed that 9 of the 13 KPNA proteins localized exclusively to the micronucleus suggesting that nucleus-specific transport systems must exist (Malone et al., 2008). Further studies demonstrated that the NPCs of macronuclei and micronuclei contain unique subsets of FG-containing nucleoporins which are responsible for this nuclear selectivity (Iwamoto et al., 2009; Malone et al., 2008). Interestingly, homologs of NUP98 contributed to nuclear selectivity: two NUP98 homologs localized exclusively to macronuclei, whereas the other two exclusively localized to micronuclei. Specific structural components of the NUP98 homologs were functionally required for the nuclear selectivity as shown by chimeric protein experiments (Iwamoto et al., 2009). The NUP98 homologs that localized to the macronucleus contained amino acid repeats of GLFG, whereas homologs that localized to the micronucleus lacked GLFG and instead contained novel NIFN repeats. These results suggest that structural alterations of the NPC can contribute to nucleus-specific protein transport in a multinucleated cell. Such structural alterations may modulate the interaction of karyopherin transport receptors with specific components of the NPC and consequently alter nuclear accumulation of proteins. Immunofluorescence analyses of primary mouse myotubes in vitro reveal that the steady-state levels of KPNA2 differ among nuclei supporting the hypothesis that specific karyopherin transport receptors may also undergo selective nuclear targeting in skeletal muscle as in Tetrahymena (Hall et al., unpublished data). Further studies are required to define the contribution of NPC composition and karyopherin transport receptors to nuclear differences in transcription and protein content in skeletal muscle.
8. Summary
Nucleocytoplasmic transport plays a key regulatory role in cellular physiology. While much is known about facilitated nuclear transport in other cell types, the study of nucleocytoplasmic transport in skeletal muscle is still in its infancy. Multinucleated myofibers are faced with unique challenges compared to most other mammalian cell types in controlling the function of hundreds of nuclei in a common cytoplasm. Although a fair bit is known about nuclear envelope proteins in skeletal muscle because of their association with several muscular dystrophies, very little is known about the NPC or karyopherin transport receptors. Further knowledge about the nuclear transport machinery is needed in skeletal muscle to enhance our understanding of how gene expression is controlled in normal, aged, and diseased muscle as well as to provide insight into satellite cell biology.
ACKNOWLEDGMENTS
G. K. P. is supported by National Institute of Health grants AR051372, AR052730, AR047314, and NS059340.
Abbreviations
- cNLS
classical nuclear localization signal
- NLS
nuclear localization signal
- Kpna
karyopherin alpha
- kpnb1
karyopherin beta1
- NPC
nuclear pore complex
- Nup
nucleoporin
REFERENCES
- Abbott KL, Friday BB, Thaloor D, Murphy TJ, Pavlath GK. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell. 1998;9:2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahluwalia A, Narula J, Jones MK, Deng X, Tarnawski AS. Impaired angiogenesis in aging myocardial microvascular endothelial cells is associated with reduced importin alpha and decreased nuclear transport of HIF1 alpha: Mechanistic implications. J. Physiol. Pharmacol. 2010;61:133–139. [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 2000;275:31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- Aratani S, Oishi T, Fujita H, Nakazawa M, Fujii R, Imamoto N, Yoneda Y, Fukamizu A, Nakajima T. The nuclear import of RNA helicase A is mediated by importin-alpha3. Biochem. Biophys. Res. Commun. 2006;340:125–133. doi: 10.1016/j.bbrc.2005.11.161. [DOI] [PubMed] [Google Scholar]
- Artaza JN, Bhasin S, Mallidis C, Taylor W, Ma K, Gonzalez-Cadavid NF. Endogenous expression and localization of myostatin and its relation to myosin heavy chain distribution in C2C12 skeletal muscle cells. J. Cell. Physiol. 2002;190:170–179. doi: 10.1002/jcp.10044. [DOI] [PubMed] [Google Scholar]
- Berman SA, Bursztajn S, Bowen B, Gilbert W. Localization of an acetylcholine receptor intron to the nuclear membrane. Science. 1990;247:212–214. doi: 10.1126/science.1688472. [DOI] [PubMed] [Google Scholar]
- Boateng SY, Senyo SE, Qi L, Goldspink PH, Russell B. Myocyte remodeling in response to hypertrophic stimuli requires nucleocytoplasmic shuttling of muscle LIM protein. J. Mol. Cell. Cardiol. 2009;47:426–435. doi: 10.1016/j.yjmcc.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden SJ. Synapse-specific gene expression. Trends Genet. 1993;9:12–16. doi: 10.1016/0168-9525(93)90066-Q. [DOI] [PubMed] [Google Scholar]
- Busch A, Kiel T, Heupel WM, Wehnert M, Hubner S. Nuclear protein import is reduced in cells expressing nuclear envelopathy-causing lamin A mutants. Exp. Cell Res. 2009;315:2373–2385. doi: 10.1016/j.yexcr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Chen IH, Huber M, Guan T, Bubeck A, Gerace L. Nuclear envelope transmembrane proteins (NETs) that are up-regulated during myogenesis. BMC Cell Biol. 2006;7:38. doi: 10.1186/1471-2121-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Suel KE. Nuclear import by karyopherin-betas: Recognition and inhibition. Biochim. Biophysic. Acta. 2010 doi: 10.1016/j.bbamcr.2010.10.014. [E pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel S, Siffroi-Fernandez S, Coldefy AS, Boulukos K, Pisani DF, Derijard B. Regulation of the intracellular localization of Foxo3a by stress-activated protein kinase signaling pathways in skeletal muscle cells. Mol. Cell. Biol. 2010;30:470–480. doi: 10.1128/MCB.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Cook AG, Conti E. Nuclear export complexes in the frame. Curr. Opin. Struct. Biol. 2010;20:247–252. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cortes R, Rosello-Lleti E, Rivera M, Martinez-Dolz L, Salvador A, Azorin I, Portoles M. Influence of heart failure on nucleocytoplasmic transport in human cardiomyocytes. Cardiovasc. Res. 2010;85:464–472. doi: 10.1093/cvr/cvp336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton N, Kodiha M, Shrivastava S, Umar R, Stochaj U. Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol. Biol. Cell. 2009;20:5106–5116. doi: 10.1091/mbc.E09-05-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Guan T, Gerace L. NET37, a nuclear envelope transmembrane protein with glycosidase homology, is involved in myoblast differentiation. J. Biol. Chem. 2009;284:29666–29676. doi: 10.1074/jbc.M109.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Eisenberg BR. Myosin mRNA accumulation and myofibrillogenesis at the myotendinous junction of stretched muscle fibers. J. Cell Biol. 1990;111:1885–1894. doi: 10.1083/jcb.111.5.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Hetzer MW. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 2010;119:469–477. doi: 10.1007/s00412-010-0289-2. [DOI] [PubMed] [Google Scholar]
- Dupont-Versteegden EE, Strotman BA, Gurley CM, Gaddy D, Knox M, Fluckey JD, Peterson CA. Nuclear translocation of EndoG at the initiation of disuse muscle atrophy and apoptosis is specific to myonuclei. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R1730–R1740. doi: 10.1152/ajpregu.00176.2006. [DOI] [PubMed] [Google Scholar]
- Ellery PM, Cindrova-Davies T, Jauniaux E, Ferguson-Smith AC, Burton GJ. Evidence for transcriptional activity in the syncytiotrophoblast of the human placenta. Placenta. 2009;30:329–334. doi: 10.1016/j.placenta.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund R, Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J. Biol. Chem. 2002;277:30072–30078. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- Faul C, Dhume A, Schecter AD, Mundel P. Protein kinase A, Ca2+/calmodulin-dependent kinase II, and calcineurin regulate the intracellular trafficking of myopodin between the Z-disc and the nucleus of cardiac myocytes. Mol. Cell. Biol. 2007;27:8215–8227. doi: 10.1128/MCB.00950-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino RS, Cheung P, Richard MN, Dibrov E, Kneesch AL, Deniset JF, Chahine MN, Lee K, Blackwood D, Pierce GN. Ceramide regulation of nuclear protein import. J. Lipid Res. 2008;49:654–662. doi: 10.1194/jlr.M700464-JLR200. [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Rout MP. Nuclear pore complex biogenesis. Curr. Opin. Cell Biol. 2009;21:603–612. doi: 10.1016/j.ceb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri P, Barbieri E, Burattini S, Guescini M, D’Emilio A, Biagiotti L, Del Grande P, De Luca A, Stocchi V, Falcieri E. Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. J. Cell. Biochem. 2009;108:1302–1317. doi: 10.1002/jcb.22360. [DOI] [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell. Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- Fontaine B, Changeux JP. Localization of nicotinic acetylcholine receptor alpha-subunit transcripts during myogenesis and motor endplate development in the chick. J. Cell Biol. 1989;108:1025–1037. doi: 10.1083/jcb.108.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J. Biol. Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- Freitas N, Cunha C. Mechanisms and signals for the nuclear import of proteins. Curr. Genomics. 2009;10:550–557. doi: 10.2174/138920209789503941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J. Mol. Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Fukada SI, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Geles KG, Adam SA. Germline and developmental roles of the nuclear transport factor importin alpha3 in C. elegans. Development. 2001;128:1817–1830. doi: 10.1242/dev.128.10.1817. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N, Lin YQ, Haueter C, Bellen HJ. Importin 13 regulates neurotransmitter release at the Drosophila neuromuscular junction. J. Neurosci. 2009;29:5628–5639. doi: 10.1523/JNEUROSCI.0794-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: A multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc. Natl. Acad. Sci. USA. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-κB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR. Border control at the nucleus: Biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi-Kovtun ME, Mosca TJ, Dickman DK, Meinertzhagen IA, Schwarz TL. Importin-beta11 regulates synaptic phosphorylated mothers against decapentaplegic, and thereby influences synaptic development and function at the Drosophila neuromuscular junction. J. Neurosci. 2010;30:5253–5268. doi: 10.1523/JNEUROSCI.3739-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J. Biol. Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- Hodel AE, Harreman MT, Pulliam KF, Harben ME, Holmes JS, Hodel MR, Berland KM, Corbett AH. Nuclear localization signal receptor affinity correlates with in vivo localization in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:23545–23556. doi: 10.1074/jbc.M601718200. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Calanni S, Jans DA, Loveland KL. Importin alpha mRNAs have distinct expression profiles during spermatogenesis. Dev. Dyn. 2006;235:253–262. doi: 10.1002/dvdy.20569. [DOI] [PubMed] [Google Scholar]
- Holaska JM. Emerin and the nuclear lamina in muscle and cardiac disease. Circ. Res. 2008;103:16–23. doi: 10.1161/CIRCRESAHA.108.172197. [DOI] [PubMed] [Google Scholar]
- Holaska JM, Rais-Bahrami S, Wilson KL. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum. Mol. Genet. 2006;15:3459–3472. doi: 10.1093/hmg/ddl423. [DOI] [PubMed] [Google Scholar]
- Hood JK, Silver PA. Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Kemp BE, Jans DA. Kinetic properties of nuclear transport conferred by the retinoblastoma (Rb) NLS. J. Cell. Biochem. 2005;95:782–793. doi: 10.1002/jcb.20439. [DOI] [PubMed] [Google Scholar]
- Hu J, Wang F, Yuan Y, Zhu X, Wang Y, Zhang Y, Kou Z, Wang S, Gao S. Novel importin-alpha family member Kpna7 is required for normal fertility and fecundity in the mouse. J. Biol. Chem. 2010;285:33113–33122. doi: 10.1074/jbc.M110.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol. Cell. Biol. 2009;29:5718–5728. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huenniger K, Kramer A, Soom M, Chang I, Kohler M, Depping R, Kehlenbach RH, Kaether C. Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cell. Mol. Life Sci. 2010;67:3187–3196. doi: 10.1007/s00018-010-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Mori C, Kojidani T, Bunai F, Hori T, Fukagawa T, Hiraoka Y, Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr. Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984a;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984b;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Yuba S, Nakayama T, Yoneda Y. Three distinct classes of the alpha-subunit of the nuclear pore-targeting complex (importin-alpha) are differentially expressed in adult mouse tissues. J. Histochem. Cytochem. 1999;47:363–372. doi: 10.1177/002215549904700310. [DOI] [PubMed] [Google Scholar]
- Karrer KM. Tetrahymena genetics: Two nuclei are better than one. Methods Cell Biol. 2000;62:127–186. doi: 10.1016/s0091-679x(08)61529-0. [DOI] [PubMed] [Google Scholar]
- Kelley JB, Talley AM, Spencer A, Gioeli D, Paschal BM. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010;11:63. doi: 10.1186/1471-2121-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol. 2010;22:320–325. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Kodiha M, Tran D, Qian C, Morogan A, Presley JF, Brown CM, Stochaj U. Oxidative stress mislocalizes and retains transport factor importin-alpha and nucleoporins Nup153 and Nup88 in nuclei where they generate high molecular mass complexes. Biochim. Biophys. Acta. 2008;1783:405–418. doi: 10.1016/j.bbamcr.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- Kohler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler M, Fiebeler A, Hartwig M, Thiel S, Prehn S, Kettritz R, Luft FC, Hartmann E. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell. Physiol. Biochem. 2002;12:335–344. doi: 10.1159/000067903. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Guttinger S. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Lai KO, Zhao Y, Ch’ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc. Natl. Acad. Sci. USA. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A, Olson EN. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl. Acad. Sci. USA. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Fahrenkrog B. The nuclear pore complex up close. Curr. Opin. Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Liu SM, Liu WM. Recent developments in the understanding of nuclear protein import. Protein Pept. Lett. 2007;14:723–733. doi: 10.2174/092986607781483895. [DOI] [PubMed] [Google Scholar]
- Liu GH, Guan T, Datta K, Coppinger J, Yates J, 3rd, Gerace L. Regulation of myoblast differentiation by the nuclear envelope protein NET39. Mol. Cell. Biol. 2009;29:5800–5812. doi: 10.1128/MCB.00684-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonhienne TG, Forwood JK, Marfori M, Robin G, Kobe B, Carroll BJ. Importin-beta is a GDP-to-GTP exchange factor of Ran: Implications for the mechanism of nuclear import. J. Biol. Chem. 2009;284:22549–22558. doi: 10.1074/jbc.M109.019935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev. Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Falkowska KA, Li AY, Galanti SE, Kanuru RC, LaMont EG, Mazzarella KC, Micev AJ, Osman MM, Piotrowski NK, Suszko JW, Timm AC, et al. Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila. Eukaryot. Cell. 2008;7:1487–1499. doi: 10.1128/EC.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manilal S, Nguyen TM, Sewry CA, Morris GE. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 1996;5:801–808. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Boden M, Kobe B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta. 2010 doi: 10.1016/j.bbamcr.2010.10.013. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Markiewicz E, Tilgner K, Barker N, van de Wetering M, Clevers H, Dorobek M, Hausmanowa-Petrusewicz I, Ramaekers FC, Broers JL, Blankesteijn WM, Salpingidou G, Wilson RG, et al. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 2006;25:3275–3285. doi: 10.1038/sj.emboj.7601230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Goldfarb DS. The nuclear transport machinery as a regulator of Drosophila development. Semin. Cell Dev. Biol. 2009;20:582–589. doi: 10.1016/j.semcdb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Mason DA, Fleming RJ, Goldfarb DS. Drosophila melanogaster importin alpha1 and alpha3 can replace importin alpha2 during spermatogenesis but not oogenesis. Genetics. 2002;161:157–170. doi: 10.1093/genetics/161.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Mathe E, Fleming RJ, Goldfarb DS. The Drosophila melanogaster importin alpha3 locus encodes an essential gene required for the development of both larval and adult tissues. Genetics. 2003;165:1943–1958. doi: 10.1093/genetics/165.4.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Lange A, Harreman MT, Corbett AH, Stewart M. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 2003;22:5358–5369. doi: 10.1093/emboj/cdg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout A, Dechat T, Adam SA, Goldman RD, Gruenbaum Y. Nuclear lamins, diseases and aging. Curr. Opin. Cell Biol. 2006;18:335–341. doi: 10.1016/j.ceb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Clair E, von Roretz C, Tenenbaum SA, Keene JD, Saleh M, Gallouzi IE. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp 32/PHAP-I regulation of apoptosis. J. Cell Biol. 2008;180:113–127. doi: 10.1083/jcb.200709030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville JF, Fernandes DJ, Churchill J, Dewundara S, Kogut P, Shah S, Fuchs G, Kedainis D, Bellam SK, Patel NM, McCauley J, Dulin NO, et al. Nuclear import of serum response factor in airway smooth muscle. Am. J. Respir. Cell Mol. Biol. 2010 doi: 10.1165/rcmb.2008-0393OC. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli L, Leonardi L, Conti F, Maresca G, Colazingari S, Mattei E, Lira SA, Farioli-Vecchioli S, Caruso M, Tirone F. PC4/Tis7/IFRD1 stimulates skeletal muscle regeneration and is involved in myoblast differentiation as a regulator of MyoD and NF-kB. J. Biol. Chem. 2010;286:5691–5707. doi: 10.1074/jbc.M110.162842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikenberg I, Widera D, Kaus A, Kaltschmidt B, Kaltschmidt C. Transcription factor NF-kappaB is transported to the nucleus via cytoplasmic dynein/dynactin motor complex in hippocampal neurons. PLoS ONE. 2007;2:e589. doi: 10.1371/journal.pone.0000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Saiwaki T, Yamashita J, Yasuda Y, Kotera I, Shibata S, Shigeta M, Hiraoka Y, Haraguchi T, Yoneda Y. Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J. Cell Biol. 2004;165:617–623. doi: 10.1083/jcb.200312008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ, Schwarz TL. The nuclear import of Frizzled2-C by Importinsbeta11 and alpha2 promotes postsynaptic development. Nat. Neurosci. 2010;13:935–943. doi: 10.1038/nn.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Bonne G, van der Kooi AJ, van Meegen M, Baas F, Bolhuis PA, de Visser M, Schwartz K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum. Mol. Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- Newlands S, Levitt LK, Robinson CS, Karpf AB, Hodgson VR, Wade RP, Hardeman EC. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: A simple hidden Markov model for nuclear localization signal prediction. BMC Bioinform. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RS, Mills ST, Jones KA, Ho SN, Pavlath GK. A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 2007;120:149–159. doi: 10.1242/jcs.03307. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Miyamoto Y, Asally M, Oka M, Yasuda Y, Yoneda Y. Two isoforms of Npap60 (Nup50) differentially regulate nuclear protein import. Mol. Biol. Cell. 2010;21:630–638. doi: 10.1091/mbc.E09-05-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N, Ishigami Y, Suzuki T, Kaneko A, Yasui K, Fukutomi R, Isemura M. Importins and exportins in cellular differentiation. J. Cell. Mol. Med. 2008;12:1863–1871. doi: 10.1111/j.1582-4934.2008.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Aramburu J, Garcia-Rodriguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptonal activity. Mol. Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- Ostlund C, Guan T, Figlewicz DA, Hays AP, Worman HJ, Gerace L, Schirmer EC. Reduction of a 4q35-encoded nuclear envelope protein in muscle differentiation. Biochem. Biophys. Res. Commun. 2009;389:279–283. doi: 10.1016/j.bbrc.2009.08.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Francois S, Regnault B, Czarny B, Dive V, Cumano A, Montarras D, Buckingham M. An adult tissue-specific stem cell in its niche: A gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4:77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Perez-Terzic C, Faustino RS, Boorsma BJ, Arrell DK, Niederlander NJ, Behfar A, Terzic A. Stem cells transform into a cardiac phenotype with remodeling of the nuclear transport machinery. Nat. Clin. Pract. 2007;4(Suppl. 1):S68–S76. doi: 10.1038/ncpcardio0763. [DOI] [PubMed] [Google Scholar]
- Poon IK, Jans DA. Regulation of nuclear transport: Central role in development and transformation? Traffic. 2005;6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Puckelwartz MJ, Kessler EJ, Kim G, Dewitt MM, Zhang Y, Earley JU, Depreux FF, Holaska J, Mewborn SK, Pytel P, McNally EM. Nesprin-1 mutations in human and murine cardiomyopathy. J. Mol. Cell. Cardiol. 2010;48:600–608. doi: 10.1016/j.yjmcc.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol G, Soderqvist H, Radu A. Age-associated reduction of nuclear protein import in human fibroblasts. Biochem. Biophys. Res. Commun. 2002;294:354–358. doi: 10.1016/S0006-291X(02)00492-8. [DOI] [PubMed] [Google Scholar]
- Quensel C, Friedrich B, Sommer T, Hartmann E, Kohler M. In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol. 2004;24:10246–10255. doi: 10.1128/MCB.24.23.10246-10255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Lenart P, Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Curr. Opin. Cell Biol. 2004;16:314–321. doi: 10.1016/j.ceb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Ratan R, Mason DA, Sinnot B, Goldfarb DS, Fleming RJ. Drosophila importin alpha1 performs paralog-specific functions essential for gametogenesis. Genetics. 2008;178:839–850. doi: 10.1534/genetics.107.081778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Gorlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard MN, Deniset JF, Kneesh AL, Blackwood D, Pierce GN. Mechanical stretching stimulates smooth muscle cell growth, nuclear protein import, and nuclear pore expression through mitogen-activated protein kinase activation. J. Biol. Chem. 2007;282:23081–23088. doi: 10.1074/jbc.M703602200. [DOI] [PubMed] [Google Scholar]
- Riddick G, Macara IG. A systems analysis of importin-{alpha}-{beta} mediated nuclear protein import. J. Cell Biol. 2005;168:1027–1038. doi: 10.1083/jcb.200409024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Macara IG. The adapter importin-alpha provides flexible control of nuclear import at the expense of efficiency. Mol. Syst. Biol. 2007;3:118. doi: 10.1038/msb4100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Salamon M, Millino C, Raffaello A, Mongillo M, Sandri C, Bean C, Negrisolo E, Pallavicini A, Valle G, Zaccolo M, Schiaffino S, Lanfranchi G. Human MYO18B, a novel unconventional myosin heavy chain expressed in striated muscles moves into the myonuclei upon differentiation. J. Mol. Biol. 2003;326:137–149. doi: 10.1016/s0022-2836(02)01335-9. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Johnson YR, Kotzbauer PT, Mudd J, Hanley T, Martinou JC, Merlie JP. Selective expression of an acetylcholine receptor-lacZ transgene in synaptic nuclei of adult muscle fibers. Development. 1991;113:1181–1191. doi: 10.1242/dev.113.4.1181. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science (New York, NY) 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Shen T, Liu Y, Contreras M, Hernandez-Ochoa EO, Randall WR, Schneider MF. DNA binding sites target nuclear NFATc1 to heterochromatin regions in adult skeletal muscle fibers. Histochem. Cell Biol. 2010;134:387–402. doi: 10.1007/s00418-010-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin I, Rotty J, Wu FY, Arteaga CL. Phosphorylation of p27Kip1 at Thr-157 interferes with its association with importin alpha during G1 and prevents nuclear re-entry. J. Biol. Chem. 2005;280:6055–6063. doi: 10.1074/jbc.M412367200. [DOI] [PubMed] [Google Scholar]
- Shmidt T, Hampich F, Ridders M, Schultrich S, Hans VH, Tenner K, Vilianovich L, Qadri F, Alenina N, Hartmann E, Kohler M, Bader M. Normal brain development in importin-alpha5 deficient-mice. Nat. Cell Biol. 2007;9:1337–1338. doi: 10.1038/ncb1207-1337. (author reply 1339). [DOI] [PubMed] [Google Scholar]
- Simon AM, Hoppe P, Burden SJ. Spatial restriction of AChR gene expression to subsynaptic nuclei. Development. 1992;114:545–553. doi: 10.1242/dev.114.3.545. [DOI] [PubMed] [Google Scholar]
- Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- Smitherman M, Lee K, Swanger J, Kapur R, Clurman BE. Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol. Cell. Biol. 2000;20:5631–5642. doi: 10.1128/mcb.20.15.5631-5642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: Bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Su X, Berman SA, Sullivan T, Bursztajn S. Myoblast and myotube nuclei display similar patterns of heterogeneous acetylcholine receptor subunit mRNA expression. J. Cell. Biochem. 1995;58:22–38. doi: 10.1002/jcb.240580105. [DOI] [PubMed] [Google Scholar]
- Talcott B, Moore MS. The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin alpha3/Qip. J. Biol. Chem. 2000;275:10099–10104. doi: 10.1074/jbc.275.14.10099. [DOI] [PubMed] [Google Scholar]
- Thompson KR, Otis KO, Chen DY, Zhao Y, O’Dell TJ, Martin KC. Synapse to nucleus signaling during long-term synaptic plasticity: A role for the classical active nuclear import pathway. Neuron. 2004;44:997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: Life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- van der Giessen K, Gallouzi IE. Involvement of transportin 2-mediated HuR import in muscle cell differentiation. Mol. Biol. Cell. 2007;18:2619–2629. doi: 10.1091/mbc.E07-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: A novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff KM, Jans DA. Importins and beyond: Non-conventional nuclear transport mechanisms. Traffic. 2009;10:1188–1198. doi: 10.1111/j.1600-0854.2009.00937.x. [DOI] [PubMed] [Google Scholar]
- Walde S, Kehlenbach RH. The part and the whole: Functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 2010;20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Wang CI, Wang CL, Wang CW, Chen CD, Wu CC, Liang Y, Tsai YH, Chang YS, Yu JS, Yu CJ. Importin subunit alpha-2 is identified as a potential biomarker for non-small cell lung cancer by integration of the cancer cell secretome and tissue transcriptome. Int. J. Canc. 2010 doi: 10.1002/ijc.25568. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Weins A, Schwarz K, Faul C, Barisoni L, Linke WA, Mundel P. Differentiation- and stress-dependent nuclear cytoplasmic redistribution of myopodin, a novel actin-bundling protein. J. Cell Biol. 2001;155:393–404. doi: 10.1083/jcb.200012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EM, Allis CD, Goldfarb DS, Srivastva A, Weir JW, Gorovsky MA. Nucleus-specific and temporally restricted localization of proteins in Tetrahymena macronuclei and micronuclei. J. Cell Biol. 1989;109:1983–1992. doi: 10.1083/jcb.109.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH, Jr, Mastronarde DN. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. A myogenic cell line with altered serum requirements for differentiation. Differentiation. 1977;7:159–166. doi: 10.1111/j.1432-0436.1977.tb01507.x. [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H, Yoneda Y. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat. Cell Biol. 2007;9:72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- Youn MY, Takada I, Imai Y, Yasuda H, Kato S. Transcriptionally active nuclei are selective in mature multinucleated osteoclasts. Genes Cells. 2010;15:1025–1035. doi: 10.1111/j.1365-2443.2010.01441.x. [DOI] [PubMed] [Google Scholar]
- Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, Liu T, Hans CP, Kim JG, Abd Elmageed ZY, Koochekpour S, Catling A, et al. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J. Immunol. 2010;185:1894–1902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]