Abstract
Soft selective sweeps represent an important form of adaptation in which multiple haplotypes bearing adaptive alleles rise to high frequency. Most statistical methods for detecting selective sweeps from genetic polymorphism data, however, have focused on identifying hard selective sweeps in which a favored allele appears on a single haplotypic background; these methods might be underpowered to detect soft sweeps. Among exceptions is the set of haplotype homozygosity statistics introduced for the detection of soft sweeps by Garud et al. (2015). These statistics, examining frequencies of multiple haplotypes in relation to each other, include H12, a statistic designed to identify both hard and soft selective sweeps, and H2/H1, a statistic that conditional on high H12 values seeks to distinguish between hard and soft sweeps. A challenge in the use of H2/H1 is that its range depends on the associated value of H12, so that equal H2/H1 values might provide different levels of support for a soft sweep model at different values of H12. Here, we enhance the H12 and H2/H1 haplotype homozygosity statistics for selective sweep detection by deriving the upper bound on H2/H1 as a function of H12, thereby generating a statistic that normalizes H2/H1 to lie between 0 and 1. Through a reanalysis of resequencing data from inbred lines of Drosophila, we show that the enhanced statistic both strengthens interpretations obtained with the unnormalized statistic and leads to empirical insights that are less readily apparent without the normalization.
Keywords: Haplotype statistics, selective sweeps, Drosophila melanogaster
Introduction
A selective sweep, the process whereby beneficial mutations at a locus that contribute to the fitness of an organism rise in frequency to become prevalent in a population, can occur through two main mechanisms that leave distinct genomic signatures (Pritchard et al., 2010; Cutter and Payseur, 2013; Messer and Petrov, 2013). A relatively new adaptive allele can proliferate so that the single haplotype on which it has occurred reaches a high frequency, resulting in a signature of a “hard” selective sweep (Maynard Smith and Haigh, 1974; Kaplan et al., 1989; Kim and Stephan, 2002). Alternatively, a mutation that arises de novo multiple times or exists as standing genetic variation on several haplotype backgrounds before the onset of positive selection can increase in frequency; in these cases, multiple favored haplotypes have relatively high frequencies, generating a signature of a “soft” selective sweep (Hermisson and Pennings, 2005; Przeworski et al., 2005; Pennings and Hermisson, 2006a). Soft sweeps can provide an effective mechanism for natural selection and might explain a sizeable fraction of selective events in many systems (Orr and Betancourt, 2001; Innan and Kim, 2004; Pritchard et al., 2010; Messer and Petrov, 2013).
Most statistical methods that have been designed to detect selective sweeps from patterns of genetic polymorphism search for patterns expected under a hard-sweep model, such as the presence of a single common haplotype (Hudson et al., 1994), high haplotype homozygosity (Depaulis and Veuille, 1998; Sabeti et al., 2002; Voight et al., 2006), high-frequency derived variants and related features of site-frequency spectra (Tajima, 1989; Braverman et al., 1995; Fay and Wu, 2000; Nielsen et al., 2005), or local loss of variation near a putative selected site (Maynard Smith and Haigh, 1974; Begun and Aquadro, 1992; Kim and Stephan, 2002). Many methods that search for patterns expected with hard sweeps, however, can be less well suited to the problem of identifying soft sweeps (Pennings and Hermisson, 2006b; Teshima et al., 2006; Cutter and Payseur, 2013). Therefore, current genomic scans for selective sweeps might be limited in their ability to uncover an important class of adaptive events.
Recently, it has been shown that statistics based on haplotype homozygosity can identify both hard and soft sweeps from population-genomic data (Ferrer-Admetlla et al., 2014; Garud et al., 2015). Garud et al. (2015) developed a haplotype homozygosity statistic, H12, relying on the principle that in a soft sweep, the most frequent haplotype might not predominate in frequency, and instead, multiple frequent haplotypes might be present. In terms of frequencies pi ≥ 0 for i = 1, 2, 3, . . . with and p1 ≥ p2 ≥ p3 ≥ . . ., Garud et al. (2015) defined H12 as
| (1) |
This statistic calculates homozygosity by combining the two largest haplotype frequencies into a single value and then computing a haplotype homozygosity. Garud et al. (2015) determined that H12 has reasonable power to detect both hard and soft sweeps, applying the statistic to Drosophila population-genomic data and identifying abundant signatures of natural selection.
To determine whether the genomic regions with the highest values of H12 were compatible with either a hard-sweep or soft-sweep pattern, Garud et al. (2015) examined a second statistic, H2/H1, a ratio of a haplotype homozygosity H2 that excludes the most frequent haplotype and a haplotype homozygosity H1 that includes this haplotype:
| (2) |
| (3) |
For high values of H12, hard sweeps are expected to produce relatively low values of H2/H1 because they produce a single high-frequency haplotype (very high p1, low p2). Soft sweeps, on the other hand, produce multiple high-frequency haplotypes (high p1, p2, and perhaps others), and are expected to produce higher values of H2/H1.
Garud et al. (2015) found that this two-step process—identification of regions with high H12 followed by examination of H2/H1—could both detect selective sweeps in general and distinguish hard and soft sweeps. As we will show, however, a complication in the approach is that the permissible range of H2/H1 varies with the value of H12. Thus, the magnitude of H2/H1 that might be regarded as indicative of a soft or hard sweep can depend on the associated values of H12. This potential difference in interpretations for values of H2/H1 as a function of H12 can present a particular challenge when comparing H2/H1 at multiple loci with a wide range of H12 values.
In a line of work separate from the use by Garud et al. (2015) of homozygosity-based soft sweep statistics, Rosenberg and Jakobsson (2008) and Reddy and Rosenberg (2012) analyzed the properties of homozygosity statistics in relation to the frequency of the most frequent allele, identifying upper and lower bounds on homozygosity given the frequency of the most frequent allele. This work, along with related work on other statistics (Long and Kittles, 2003; Hedrick, 2005; Jost, 2008; Vanliere and Rosenberg, 2008; Maruki et al., 2012; Jakobsson et al., 2013), seeks to understand mathematical bounds on population-genetic statistics, so that their application and interpretation can be suitably informed by the mathematical constraints on their numerical values.
Here, to facilitate the interpretation of the statistics of Garud et al. (2015) and to enhance comparisons among values of these statistics at loci with different haplotype homozygosities, we use a result from Rosenberg and Jakobsson (2008) to determine the upper and lower bounds on H2/H1 as a function of H12. The upper bound provides a basis for normalization of H2/H1 to produce a statistic with the same range, from 0 to 1, irrespective of the value of H12. Using the upper bound and the new normalized statistic, we reexamine Drosophila data analyzed by Garud et al. (2015), demonstrating that the upper bound, (H2/H1)max, and the normalized statistic, (H2/H1)′, enable improved insights regarding soft selective sweeps on the basis of genetic polymorphism data.
Theory
Our goal is to determine the maximum of H2/H1 given the value of H12, for 0 < H12 ≤ 1. For convenience, we denote Z = H2/H1. We denote the desired upper bound by Zmax.
For generality in our description, we consider “alleles” at a locus. These distinct “alleles” can be viewed as representing distinct haplotypes at a specific location in the genome; the assumption is that a set of distinct genetic types is considered, representing perhaps distinct haplotypes or distinct alleles in the traditional sense, and the sum of the frequencies of the types is 1.
We sort alleles in descending order of frequency, so that p1 > 0 and p1 ≥ p2 ≥ p3 ≥ ... ≥ 0. The number of alleles is left unspecified, and it can be arbitrarily large; thus, . For our mathematical analysis, we consider parametric allele frequencies; that is, the pi are treated as known frequencies in a population rather than values estimated from samples. The mathematical setting follows Rosenberg and Jakobsson (2008).
We let M = p1 + p2. Because p1 > 0, M, H12, and H1 are all strictly positive. By analogy with H1 and H2, denote . Thus, by eq. 1,
| (4) |
The upper bound on H2/H1 given H12
We proceed in two main steps. First, for fixed H12 and fixed M, we determine the maximum of Z as a function of p1. Next, we identify the value of M that maximizes Z. This pair of steps constructs the set of allele frequencies that generates the maximal Z at fixed H12. A graphical overview of the argument appears in Figure 1.
Figure 1.
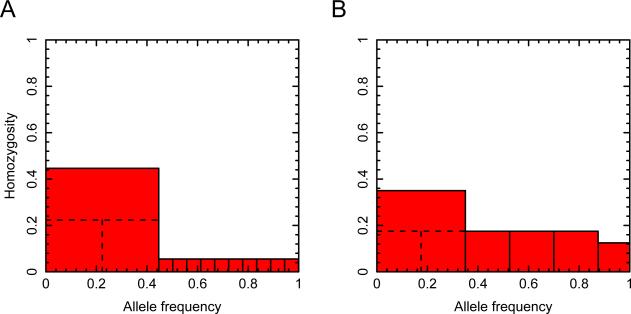
A geometric illustration of the argument for finding the upper bound on H2/H1 as a function of H12. In both panels, the unit interval (x-axis) is partitioned into components representing allele frequencies. H12 is represented by the sum of the areas of the red shaded regions, each indicating a squared frequency; the largest red square indicates (p1 + p2)2 or M2. (A) Step 1: for fixed H12 and fixed M, H2/H1 is maximal when p2 = p1. The maximal H2/H1 requires p1 to be as small as possible, but p1 ≥ p2 by definition; at the maximal H2/H1, p1 and p2 are equal. (B) Step 2: allowing M to vary while keeping H12 fixed, H2/H1 is maximal when M is as small as possible. At the maximum for H2/H1, M is reduced to the point where p1 and as many subsequent alleles as possible have identical frequency, and at most one remaining allele of smaller frequency completes the unit interval. In both panels, H12=0.23. Part A uses for M and for each of 10 additional alleles. The dashed lines illustrate the choice of p2 = p1 = M/2. Part B achieves the maximum of H2/H1 = 221/270 ≈ 0.8185 (eq. 12), with M = 0.35.
Maximizing Z for fixed H12 and fixed M
Because and p2 = M – p1, H2 can be rewritten
| (5) |
Note that by eq. 4, for fixed H12 and fixed M, H3 is constant. Because M = p1 + p2, p1 ≥ p2, and p1 > 0, it follows that M/2 ≤ p1 ≤ M. Treated as a function of p1, on the interval [M/2, M], (M – p1)2 + H3 is decreasing.
Using eq. 5, Z = H2/H1 can be written
| (6) |
In eq. 6, for fixed H12 and fixed M, is increasing in p1 and (M – p1)2 + H3 is decreasing. The ratio is therefore increasing in p1, so that the entire expression for Z decreases with p1. It is therefore maximized when p1 is minimized—in other words, when p1 = p2 = M/2. The maximal Z for fixed H12 and fixed M is
| (7) |
It remains to maximize Z by finding the value of M that maximizes eq. 7 for fixed H12. By rewriting eq. 7 as Z = 1 – M2/(4H12 – 2M2), it can be seen that for fixed H12, as M increases, M2 increases, 4H12 – 2M2 decreases, and Z decreases. Thus, for fixed H12, the maximal Z, treated as a function of M, occurs when M isas small as possible.
The minimal value of M given H12
We have shown that maximizing Z for fixed H12 and M requires p1 = p2 = M/2, and hence, using the descending order of the allele frequencies, p3 ≤ M/2. We have also shown that maximizing Z for fixed H12 over all possible M requires us to find the minimal M permissible for fixed H12. This problem can be solved with a known result. We first ignore the trivial case of H12 = 1, for which the maximal Z has M = 1, p1 = p2 = 1/2, H1 = 1/2, H2 = 1/4, and Zmax = 1/2.
By eq. 4, minimizing M for fixed H12 amounts to maximizing H3. Lemma 3 of Rosenberg and Jakobsson (2008) obtains the maximal sum of squares for a set of nonnegative numbers in a non-increasing sequence, each of which lies below the same specified constant, and whose sum is specified. In our case, the sequence is , the entries are bounded above by M/2, and their sum is 1 – p1 – p2 = 1 – M.
Applying the lemma, we obtain
| (8) |
where K = ⌈(1 – M)/(M/2)⌉ = ⌈2/M⌉ – 2 and ⌈x⌉ denotes the smallest integer greater than or equal to x; in the application of the lemma, K gives the number of nonzero numbers in the sequence that achieves the maximum. Equality is achieved if and only if ⌈2/M⌉ – 3 alleles (in addition to alleles 1 and 2) have frequency M/2, and one allele has frequency (1 – M) – (⌈2/M⌉ – 3)(M/2) = 1 – (⌈2/M⌉ – 1)(M/2).
The minimal M is obtained by substituting the upper bound from eq. 8 for H3 in eq. 4 and solving for M. The equation that must be solved is
| (9) |
Note that K is currently considered a function of M, equaling ⌈2/M⌉ – 2. However, we can instead determine the value of K as a function of H12, so that eq. 9 becomes a simple quadratic equation in M. To solve eq. 9 for M at a given H12, we must find the value of K—the number of alleles of nonzero frequency (not including alleles 1 and 2)—that applies for the given value of H12.
We break the unit interval (0, 1) into disjoint intervals [2/I, 2/(I – 1)) for integers I ≥ 3. On the interval [2/I, 2/(I – 1)) for M, K = I – 2. Inserting K = I – 2 into eq. 9, for M in this interval, the minimal M in terms of H12 is obtained by solving
| (10) |
for M. Thus, identifying the value of K in terms of H12 for use in eq. 9 amounts to finding the value of I in terms of H12 for use in eq. 10.
The right-hand side of eq. 10 is monotonically increasing on the interval [2/I, 2/(I – 1)), as it is a concave-up parabola in M with minimum at M = 2(I – 1)/(I2 – I + 2) = 2/[I + 2/(I – 1)] < 2/I. The vertex of the parabola lies to the left of the left endpoint of the interval, M = 2/I, so that on [2/I, 2/(I – 1)), the parabola is increasing.
At the left endpoint M = 2/I, H12 = (I + 2)/I2, and at the right endpoint M = 2/(I – 1), H12 = (I + 1)/(I – 1)2. Consequently, because H12 increases as a function of M on the interval [2/I, 2/(I – 1)), for this interval, H12 lies in [(I + 2)/I2, (I + 1)/(I – 1)2).
As a strictly monotonic continuous function from [2/I, 2/(I – 1)) to [(I + 2)/I2, (I + 1)/(I – 1)2), H12 is invertible as a function of M. Treated as a function of M in (0, 1), I satisfies 2/I ≤ M < 2/(I – 1); similarly, as a function of H12 in (0, 1), I satisfies (I + 2)/I2 ≤ H12 < (I + 1)/(I – 1)2. In other words, given H12, I must be equal to the smallest integer for which (I + 2)/I2 ≤ H12.
Solving this inequality, either or . The latter root is negative and can be discarded as I ≥ 3. The smallest integer that satisfies the former inequality is
| (11) |
We can now complete the solution for the minimal M as a function of H12: this minimum is a solution to eq. 10 when eq. 11 is used for I. The equation has two roots; the smaller root is smaller than 2/I, and therefore lies outside the interval [2/I, 2/(I – 1)) in which M must fall when H12 satisfies eq. 11. The minimal M therefore equals the larger root.
The formula for Zmax
Compiling the steps we have completed, we have that as a function of H12,
| (12) |
where M is the larger root of eq. 10,
| (13) |
and I satisfies eq. 11. The formula for Zmax holds for all H12 in (0, 1]; in the H12 = 1 case that we initially discarded, eq. 12 gives the correct value Zmax = 1/2. Zmax is reached when I – 1 alleles each have frequency M/2 and one allele has frequency 1 – (⌈2/M⌉ – 1)(M/2).
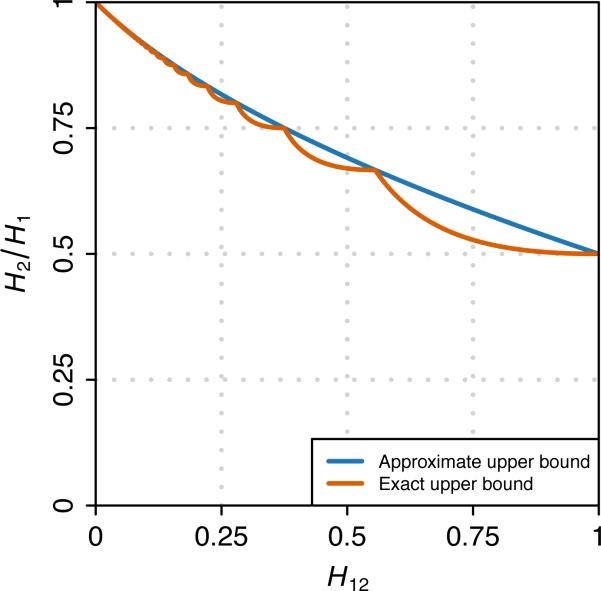
Figure 2 plots eq. 12 as a function of H12 over the unit interval. A piecewise structure of the upper bound Zmax is visible, reflecting the fact that at points H12 = (I + 2)/I2 for integers I ≥ 3, the value of I as a function of H12 changes, and Zmax is not differentiable. Zmax approaches a limiting value of 1 as H12 approaches 0, and it declines monotonically to a value of 1/2 at H12 = 1.
Figure 2.
The upper bound on H2/H1 as a function of H12. The exact upper bound is given by eq. 12, and the approximate upper bound is given by eq. 15.
An approximation to Zmax
It is convenient to consider a simple approximation to Zmax by examining the points H12 = (I + 2)/I2 for integers I ≥ 3. At these points, applying eqs. 11-13,
| (14) |
M = 2/I, and Zmax = (I – 1)/I. Eqs. 11-13 simplify because Zmax is achieved when I alleles each have frequency 1/I, unlike for other H12 < 1, where one nonzero frequency differs from the others.
We can approximate Zmax by finding a function Ymax that satisfies
at the points specified by integers I ≥ 3 and using this function to interpolate across all values of H12. When H12 = (I + 2)/I2 for integers I ≥ 3, I satisfies eq. 14, and
Multiplying the numerator and denominator of this equation by , we have
| (15) |
This approximate bound agrees with the strict bound Zmax at points H12 = (I + 2)/I2 for integers I ≥ 3, and it matches Zmax at the endpoints of the unit interval. In Figure 2, it can be seen that Ymax provides a reasonable approximation to Zmax over the entire interval.
Not only is Ymax an approximation to the strict upper bound Zmax, Ymax ≥ Zmax for H12 in (0, 1], so that Ymax is itself an upper bound. To prove this result, using eqs. 15 and 12, we have
The denominator is positive, as 2H12 – M2 = M2 + 2H3 > 0. It remains to show that
Squaring both sides, Ymax(H12) – Zmax(H12) ≥ 0 if
As H12 is positive, Ymax(H12) – Zmax(H12) ≥ 0 if H12 lies in the closed interval bounded by the roots of the quadratic term in brackets, or (M2 – M)/2 and (M2 + M)/2. Because 0 < M ≤ 1, the smaller root is at most 0, and H12 ≥ (M2 – M)/2 always holds. It thus suffices to prove H12 ≤ (M2 + M)/2.
Recalling eq. 4, we must show that H3 ≤ (M – M2)/2. Eq. 8 provides a maximum on H3 in terms of M; substituting this maximum for H3, we have H3 ≤ (M – M2)/2 if
This last inequality is true by definition of K = ⌈2/M⌉ – 2, as 2/M – 2 ≤ K < 2/M – 1 implies KM + M – 2 < 0 and KM + 2M – 2 ≥ 0. We can therefore conclude that H3 ≤ (M – M2)/2, and hence H12 ≤ (M2 + M)/2, and Ymax(H12) – Zmax(H12) ≥ 0 for H12 in (0, 1].
The lower bound on H2/H1 given H12
It is straightforward to show that for any H12 in (0, 1], H2/H1 can get arbitrarily close to 0. For H12 = 1, we set p1 = 1 – ε and p2 = ε for a small ε > 0. Then H2/H1 = ε2/[(1 – ε)2 + ε2], which approaches 0 as ε → 0. Otherwise, we construct a scenario with one frequent allele and K rare alleles, and demonstrate that H2/H1 → 0 as K → ∞.
Suppose and for large K. Frequency p1 is large and the remaining frequencies are small. In this case,
The denominator has higher degree in K than the numerator, so that limK → ∞(H2/H1) = 0.
The mean range of H2/H1 given H12
Determining the mean of the range of Z, treated as a function of H12 over the unit interval, can provide a sense of the magnitude of the constraint placed by H12 on Z. For a statistic with a larger mean range, a greater proportion of the unit interval can be achieved, and the statistic is less constrained than is one with a smaller mean range.
Because Ymax(H12) ≥ Zmax(H12), the simpler Ymax can assist in evaluating the mean size of the range of Z. As the minimum Z approaches 0 for all H12 in (0, 1], the size of the range for Z is simply Zmax. On the unit interval, Ymax has mean
| (16) |
and therefore, the mean Zmax is smaller than 17/24. This mean exceeds 1/2, as the minimal Zmax for H12 in (0, 1], at H12 = 1, is 1/2. Numerical integration of eq. 12 to obtain the mean Zmax gives
| (17) |
This result illustrates that the mean across the unit interval for H12 of the error in the approximation of Zmax by Ymax is small, approximately 0.708 – 0.684 = 0.024. Further, although the range of Z is constrained, the mean range over all values of H12 in (0, 1] is larger than corresponding mean constraints in other contexts involving homozygosity, Fst, the r2 statistic for linkage disequilibrium, and the frequency of the most frequent allele (Rosenberg et al., 2003; Rosenberg and Jakobsson, 2008; Vanliere and Rosenberg, 2008; Reddy and Rosenberg, 2012; Jakobsson et al., 2013; Edge and Rosenberg, 2014).
Normalized statistics
Because H2/H1 can approach 0 for any H12, a normalization of H2/H1 to lie in [0, 1] need only be concerned with the upper bound on H2/H1. We can therefore define exact and approximate normalizations of Z at given values of H12 as follows:
| (18) |
| (19) |
The denominators of these equations are computed using eqs. 12 and 15, respectively.
Application to data
We illustrate the bounds on H2/H1 as functions of H12 by reexamining two Drosophila melanogaster data sets studied by Garud et al. (2015), each containing fully sequenced genomes of inbred lines generated from samples taken in North Carolina. First, we consider the Drosophila Genetic Reference Panel (DGRP) data set consisting of sequences of 145 inbred lines (Mackay et al., 2012). Next, we examine the Drosophila Population Genomic Panel (DPGP) consisting of 40 strains. We consider these two data sets generated with different samples both to show an example use of the upper bounds and to demonstrate how inferences from samples with differing numerical patterns in H12 and H2/H1 can be viewed as comparable.
DGRP data
We first consider the DGRP data set studied by Garud et al. (2015). As a consequence of inbreeding, the DGRP genomes are largely homozygous. On each of the four autosomal arms, Garud et al. (2015) examined haplotypes within analysis windows of 400 single-nucleotide polymorphisms (SNPs, ~10kb). Because low recombination rates can result in high haplotype homozygosities, Garud et al. (2015) excluded analysis windows overlapping 100 kb tracts measured by Comeron et al. (2012) to have recombination rates lower than 5 × 10–7 centimorgans per base pair (cM/bp). To classify haplotypes within windows, Garud et al. (2015) assigned the 400-SNP haplotypes into groups according to exact sequence identity. If a haplotype with missing data matched multiple haplotypes at all genotyped sites in the analysis window, then the haplotype was randomly assigned to one of these classes. In the DGRP data set, all heterozygous sites in a strain were treated as missing data. Examining all 4,013,703 segregating sites across the 145 strains, 0.7% heterozygous sites per base pair per strain and 4.2% missing data per base pair per strain were observed. If a haplotype could not be conclusively assigned based on the information at non-missing data sites, then the haplotype was randomly assigned to a haplotype class that matched at all other sites; across all analysis windows and strains, 18% of assignments to haplotype classes used this method of random assignment. Windows were incremented by 50 SNPs, so that consecutive windows overlapped by 350 SNPs.
Each window has a haplotype frequency distribution across the 145 lines, enabling computations of H12, H1, and H2. To avoid inflating the number of selective events inferred in a genomic region, Garud et al. (2015) grouped together consecutive windows as belonging to the same “peak” if the H12 values in all of the grouped windows were above a critical H12 value calculated under a neutral demographic model. They assigned H12 and H2/H1 values to individual peaks by using the values calculated in the analysis window with the largest H12 within a peak. Garud et al. (2015) focused on the 50 peaks with the largest H12 values, none of which possessed two or more windows sharing the same highest H12 value. The top three peaks coincided with the loci Ace, Cyp6g1, and CHKov1, prominent cases of adaptation previously discovered by detailed focused analyses (Daborn et al., 2001; Catania et al., 2004; Menozzi et al., 2004; Aminetzach et al., 2005; Karasov et al., 2010; Schmidt et al., 2010; Magwire et al., 2011).
Effect of normalization in the DGRP data
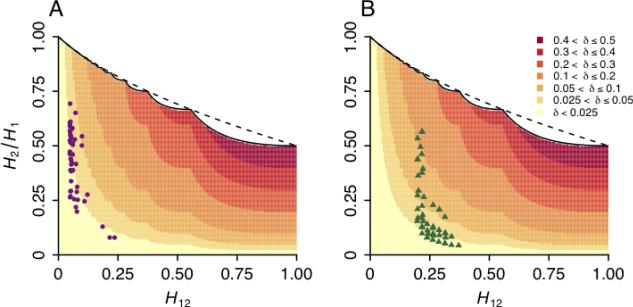
We assessed the effect of the application of Z′ to H12 and H2/H1 values calculated for the top 50 peaks in the DGRP data set. To do so across the full range of possible values for (H12, H2/H1), we first calculated the change δ = Z′ – Z in H2/H1 produced by the normalization. For any value of H12, as H2/H1 increases, δ also increases, reflecting the monotonicity of the upper bound on H2/H1 with increasing H12 (Figure 3A). The maximal δ of 1/2 is achieved when H12 = 1 and H2/H1 = 1/2.
Figure 3.
The effect of the application of Z′ on H2/H1 values in data from Drosophila. The shaded regions show the change δ in H2/H1 values after applying the normalization, where δ = Z′ – Z. Overlaid are points representing the top 50 windows for H12 in Drosophila melanogaster genome scans. (A) Drosophila Genetic Reference Panel (DGRP) data. (B) Drosophila Population Genomic Panel (DPGP) data. The solid line shows the exact upper bound on H2/H1 (eq. 12), and the dashed line shows the approximate upper bound (eq. 15).
Overlaid in Figure 3A are the H12 and H2/H1 values from the 50 top peaks in the DGRP data set. The values of H12 generally lie below 0.25, with most values occurring near 0.1. The values of H2/H1 span a wide range, with most (H12, H2/H1) combinations lying in a region of the space where δ is between 0.025 and 0.05.
DPGP data
Our second example considers the DPGP data set that was also studied by Garud et al. (2015). The DPGP data set (Mackay et al., 2012) consists of 40 of the original 145 inbred lines in the DGRP data set, sequenced and assembled separately from the DGRP data (www.dpgp.org).
In the DPGP data set, considering all 2,337,358 segregating sites across the 40 lines, there were 1.2% heterozygous sites per base pair per strain, and the missing data rate was 7.5%. With this reduced sample size compared to the DGRP data—and hence, with both shorter distances over which haplotypes become unique and faster computation times—Garud et al. (2015) measured H12 values in shorter overlapping analysis windows of 100 SNPs incremented by 1 SNP. The treatment of haplotypes and missing data proceeded in the same manner as in the DGRP analysis. In this scan, averaging across lines, haplotypes with missing data were clustered with other haplotypes matching at all other positions at a lower rate of 2.7%.
As in the DGRP analysis, Garud et al. (2015) identified the 50 peaks with the highest H12. This analysis produced a distinct but overlapping set of high-H12 windows as the DGRP top 50 peaks, again recovering known cases of adaptation at Ace, Cyp6g1, and CHKov1.
Effect of normalization in the DPGP data
As in our analysis of the DPGP data, we assessed the effect of the application of Z′ to high-H12 peaks in the DPGP data set. Figure 3B plots the (H2/H1, H12) values for the top 50 peaks in the DPGP data. In comparison to those seen in the DGRP data set, the H12 values in the DPGP data are generally greater, and the H2/H1 values lower. As a consequence, the points in the DPGP data lie in a region of the space in which normalization has a greater effect, often with δ > 0.05.
Comparison of DGRP and DPGP
Garud et al. (2015) compared the positions of the top 50 peaks in the DPGP data set according to H12 with the positions of the top 50 peaks in the DGRP data set to determine if the same selective events were identified in the two data sets. To do so, Garud et al. (2015) overlapped the edge coordinates of the peaks in the two data sets, where the edge coordinates of each peak correspond to the positions of the first SNP of the first analysis window and the last SNP of the last analysis window within a peak. An overlap was defined as a non-empty intersection of the two genomic regions defining the boundaries of the two peaks, one from one data set and one from the other. Garud et al. (2015) found that 16 DPGP peaks overlapped 13 DGRP peaks, 10 of which were among the top 15 peaks in the DGRP scan. In three cases, two DPGP peaks overlapped one DGRP peak because multiple non-overlapping peaks in the DPGP data were in the same region as a DGRP peak. These multiple proximate peaks in the DPGP data set might have been part of the same selective events.
Jointly considering the DGRP and DPGP data sets, different sample depths and analysis window sizes can result in different distributions of H12 and H2/H1 values, and thus, in different inferences about selection. As a consequence, although several H12 peaks overlap in the DGRP and DPGP scans, the H12 and H2/H1 values for the top peaks differ between the two data sets. This result complicates the comparison of the selection signals obtained between the two data sets. Application of our normalization, however, can facilitate a meaningful comparison of the H12 and H2/H1 values measured in different data sets that potentially uncover the same selective events.
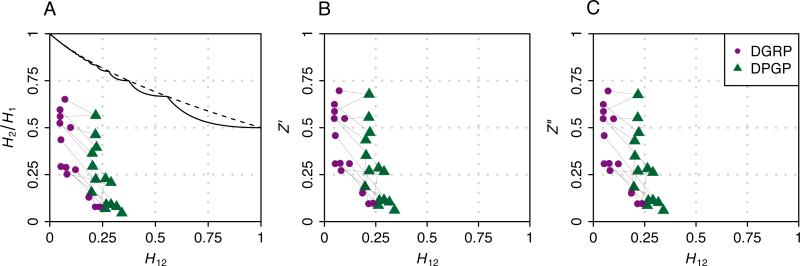
We applied the Z′ and Z″ normalizations to overlapping peaks in the two data sets. Figure 4A shows that prior to normalization, the H2/H1 values for DGRP exceed those of DPGP, as was seen previously in the plots of all 50 windows in Figure 3. However, after normalization, the distributions of H2/H1 values for the two scans are comparable despite the differences in H12. We quantified this change with a paired two-tailed Wilcoxon signed-rank test, testing the null hypothesis that the distributions of H2/H1 values in the DGRP and DPGP data are the same before and after application of Z′ and Z″. Because 16 peaks in the DPGP data set overlap 13 peaks in the DGRP set, where three pairs of DPGP peaks each overlap unique peaks in the DGRP data, we removed one of the overlapping peaks from each pair in order to perform a paired test. We applied this procedure eight times to account for every possible combination of discarded peaks, finding that in all cases, before application of Z′ or Z″, H2/H1 was greater in the DGRP data than in the DPGP data (P = 0.0473, averaged across the eight choices). After application of Z′ and Z″, however, the comparison of DGRP and DPGP did not produce a significant difference (P = 0.1946 and P = 0.1781 for Z′ and Z″, respectively, averaged across the eight choices). Thus, because normalization reduces the difference in H2/H1 values between corresponding peaks in the DGRP and DPGP data, the normalization suggests that differences in H2/H1 for corresponding peaks are attributable largely to the different values of H12 in the two data sets rather than to genuine differences in the biological signals that the two data sets provide.
Figure 4.
H12 and H2/H1 values calculated in overlapping peaks in the DGRP and DPGP data sets before normalization and after the application of Z′ and Z″. Corresponding points for the DGRP and DPGP data sets are connected by lines. Note that because the 16 DPGP peaks overlap 13 DGRP peaks, three DGRP points are each connected to a pair of DPGP points. Also, two pairs of DPGP points with different chromosomal locations have the same (H12, H2/H1) coordinates. (A) Unnormalized H2/H1 values. Overlaid are the exact upper bound (solid) and the approximate upper bound (dashed) as given by eq. 12 and eq. 15. (B) Values of Z′ (eq. 18). (C) Values of Z″ (eq. 19).
Note that normalization can in principle change the rank order of peaks for a given data set, as a lower H2/H1 at a higher H12 can be shifted after normalization above a higher H2/H1 at a lower H12. In our examples with the DGRP and DPGP data sets, however, relatively few reorderings of peaks took place upon normalization. We calculated a Spearman rank correlation coefficient to quantify the difference in rank order of Z and Z′ values and Z and Z″ values for the overlapping peaks in the DGRP and DPGP data sets, and in all four calculations (DGRP Z to Z′, DGRP Z to Z″, DPGP Z to Z′, DPGP Z to Z″), the correlation coefficient exceeded 0.999.
Discussion
Statistical methods for detecting selective sweeps from genomic data have enabled the identification of cases of adaptation in multiple organisms. Many statistics have been developed to identify hard selective sweeps, and recent attention has now also focused on detecting soft sweeps (Messer and Neher, 2012; Peter et al., 2012; Fu and Akey, 2013; Messer and Petrov, 2013; Vitti et al., 2013; Ferrer-Admetlla et al., 2014; Jensen, 2014; Wilson et al., 2014). Garud et al. (2015) recently proposed the haplotype homozygosity statistics H12 and H2/H1 to discover both hard and soft selective sweeps and to differentiate whether top candidates for selection have signatures of hard or of soft sweeps. They applied their method to two Drosophila population-genomic data sets, DGRP and DPGP, recovering known cases of adaptation as well as finding new candidates.
In this paper, we have shown that the permissible range of H2/H1 values is dependent on their associated H12 values, and that therefore, the interpretation of H2/H1 in distinguishing hard and soft sweeps can be challenging when comparing H2/H1 values across loci with a broad distribution of H12 values. To facilitate interpretation of H2/H1 values measured in scans with a wide range of H12 values, we developed approximate and exact normalizations Z′ and Z″ that can be applied to H2/H1. The application of the statistics Z′ and Z″ to data has the greatest impact for H2/H1 values with high associated H12 values (>0.5).
We illustrated the use of the new bounds and normalizations using data from Drosophila. Garud et al. (2015) compared the H12 peaks in the DGRP and DPGP data sets, finding that 13 DGRP peaks overlapped 16 DPGP peaks. However, the overlapping H12 peaks in the two data sets had significantly different H2/H1 values despite presumably reflecting the same selective events. In applying Z′ and Z″ to the H2/H1 values observed at the highest, overlapping H12 peaks in the two data sets, we found that the comparison of distributions of H2/H1 values observed in the two scans did not produce a significant difference after normalization. Thus, the differences in distributions of H12 and H2/H1 across data sets might be attributable to differences in sample sizes and analysis window sizes in the two scans rather than to differences in biological signal. Indeed, the two data sets differed in a number of ways that could have generated higher H12 values on average for DPGP compared to DGRP. DPGP had a smaller sample size; in evaluating H12 from a finite sample of size n ≥ 2, eq. 1 has a minimum of (n + 2)/n2, which is greater for smaller n. H12 was also applied to DPGP in smaller analysis windows; decreasing the window size increases the probability of haplotype identity, thus increasing measures of homozygosity.
Our work on the relationship between H12 and H2/H1 parallels other studies (Long and Kittles, 2003; Rosenberg et al., 2003; Hedrick, 2005; Rosenberg and Jakobsson, 2008; VanLiere and Rosenberg, 2008; Maruki et al., 2012; Reddy and Rosenberg, 2012; Jakobsson et al., 2013; Edge and Rosenberg, 2014) in obtaining bounds on population-genetic statistics. A shared feature common to these studies is that in each study, unexpected or counterintuitive bounds are identified that are informative for sensible interpretation. As in some of these studies, however, our calculations consider an unspecified number of haplotypes K. If we instead required that K be specified as a finite constant, it would not be possible to reach the lower bound of 0 on H2/H1 because the lower bound is obtained from a limiting scenario with large numbers of low-frequency alleles. The difference in bounds between arbitrary-K and finite-K cases can for some statistics be nontrivial, especially for small K (Reddy and Rosenberg, 2012); for future work, it will be of interest to determine the magnitude of the effect on the H2/H1 bounds of fixing the value of K.
The proposed normalizations, Z′ and Z″, offer an improvement in the interpretation of the H12 and H2/H1 statistics proposed by Garud et al. (2015). Further simulation-based investigation of the influence on H12 and H2/H1 of such variables as haplotype window sizes and sample sizes will be important for continuing to clarify the behavior of the statistics in models of selective sweeps. Nevertheless, as shown in our Drosophila example, the normalization of H2/H1 in data sets of varying sample sizes and SNP densities can help with the interpretation of selection scans, especially as data for testing population-genomic hypotheses become increasingly available in a variety of organisms.
Acknowledgments
We thank Doc Edge, Arbel Harpak, Rajiv McCoy, Pleuni Pennings, Dmitri Petrov, and Ben Wilson for helpful comments. Support was provided by NIH grant R01 HG005855.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aminetzach YT, Macpherson JM, Petrov DA. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science. 2005;309:764–767. doi: 10.1126/science.1112699. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W. The hitchhiking e ect on the site frequency spectrum of DNA polymorphisms. Genetics. 1995;140:783–796. doi: 10.1093/genetics/140.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania F, Kauer MO, Daborn PJ, Yen JL, ffrench-Constant RH, et al. World-wide survey of an Accord insertion and its association with DDT resistance in Drosophila melanogaster. Molecular Ecology. 2004;13:2491–2504. doi: 10.1111/j.1365-294X.2004.02263.x. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genetics. 2012;8:e1002905. doi: 10.1371/journal.pgen.1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nature Reviews Genetics. 2013;14:262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P, Boundy S, Yen J, Pittendrigh B, ffrench-Constant R. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Molecular Genetics and Genomics. 2001;266:556–563. doi: 10.1007/s004380100531. [DOI] [PubMed] [Google Scholar]
- Depaulis F, Veuille M. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Molecular Biology and Evolution. 1998;15:1788–1790. doi: 10.1093/oxfordjournals.molbev.a025905. [DOI] [PubMed] [Google Scholar]
- Edge MD, Rosenberg NA. Upper bounds on FST in terms of the frequency of the most frequent allele and total homozygosity: the case of a specified number of alleles. Theoretical Population Biology. 2014;97:20–34. doi: 10.1016/j.tpb.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Molecular Biology and Evolution. 2014;31:1275–1291. doi: 10.1093/molbev/msu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Akey J. Selection and adaptation in the human genome. Annual Review of Genomics and Human Genetics. 2013;14:467–489. doi: 10.1146/annurev-genom-091212-153509. [DOI] [PubMed] [Google Scholar]
- Garud NR, Messer PW, Buzbas EO, Petrov DA. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genetics. 2015;11:e1005004. doi: 10.1371/journal.pgen.1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. A standardized genetic di erentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ. Evidence for positive selection in the superoxide dismutase (sod) region of Drosophila melanogaster. Genetics. 1994;136:1329–1340. doi: 10.1093/genetics/136.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kim Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10667–10672. doi: 10.1073/pnas.0401720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Edge MD, Rosenberg NA. The relationship between FST and the frequency of the most frequent allele. Genetics. 2013;193:515–528. doi: 10.1534/genetics.112.144758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JD. On the unfounded enthusiasm for soft selective sweeps. Nature Communications. 2014;5:5281. doi: 10.1038/ncomms6281. [DOI] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure di erentiation. Molecular Ecology. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The “hitchhiking e ect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T, Messer PW, Petrov DA. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genetics. 2010;6:e1000924. doi: 10.1371/journal.pgen.1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Kittles RA. Human genetic diversity and the nonexistence of biological races. Human Biology. 2003;75:449–471. doi: 10.1353/hub.2003.0058. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM. Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genetics. 2011;7:e1002337. doi: 10.1371/journal.pgen.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruki T, Kumar S, Kim Y. Purifying selection modulates the estimates of population di erentiation and confounds genome-wide comparisons across single-nucleotide polymorphisms. Molecular Biology and Evolution. 2012;29:3617–3623. doi: 10.1093/molbev/mss187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking e ect of a favourable gene. Genetical Research. 1974;23:23–35. [PubMed] [Google Scholar]
- Menozzi P, Shi MA, Lougarre A, Tang ZH, Fournier D. Mutations of acetylcholinesterase which confer insecticide resistance in Drosophila melanogaster populations. BMC Evolutionary Biology. 2004;4:4. doi: 10.1186/1471-2148-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Neher RA. Estimating the strength of selective sweeps from deep population diversity data. Genetics. 2012;191:593–605. doi: 10.1534/genetics.112.138461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Petrov DA. Population genomics of rapid adaptation by soft selective sweeps. Trends in Ecology and Evolution. 2013;28:659–669. doi: 10.1016/j.tree.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, et al. Genomic scans for selective sweeps using SNP data. Genome Research. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Betancourt AJ. Haldane's sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps II—molecular population genetics of adaptation from recurrent mutation or migration. Molecular Biology and Evolution. 2006a;23:1076–1084. doi: 10.1093/molbev/msj117. [DOI] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genetics. 2006b;2:e186. doi: 10.1371/journal.pgen.0020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM, Huerta-Sanchez E, Nielsen R. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genetics. 2012;8:e1003011. doi: 10.1371/journal.pgen.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Current Biology. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- Reddy SB, Rosenberg NA. Refining the relationship between homozygosity and the frequency of the most frequent allele. Journal of Mathematical Biology. 2012;64:87–108. doi: 10.1007/s00285-011-0406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Jakobsson M. The relationship between homozygosity and the frequency of the most frequent allele. Genetics. 2008;179:2027–2036. doi: 10.1534/genetics.107.084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. American Journal of Human Genetics. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genetics. 2010;6:e1000998. doi: 10.1371/journal.pgen.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima KM, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Research. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanLiere JM, Rosenberg NA. Mathematical properties of the r2 measure of linkage disequilibrium. Theoretical Population Biology. 2008;74:130–137. doi: 10.1016/j.tpb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annual Review of Genetics. 2013;47:97–120. doi: 10.1146/annurev-genet-111212-133526. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biology. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BA, Petrov DA, Messer PW. Soft selective sweeps in complex demographic scenarios. Genetics. 2014;198:669–684. doi: 10.1534/genetics.114.165571. [DOI] [PMC free article] [PubMed] [Google Scholar]