Abstract
Context
Gliomas, particularly glioblastomas, are among the deadliest of human tumors. Gliomas emerge through the accumulation of recurrent chromosomal alterations, some of which target yet-to-be-discovered cancer genes. A persistent question concerns the biological basis for the coselection of these alterations during gliomagenesis.
Objectives
To describe a network model of a cooperative genetic landscape in gliomas and to evaluate its clinical relevance.
Design, Setting, and Patients
Multidimensional genomic profiles and clinical profiles of 501 patients with gliomas (45 tumors in an initial discovery set collected between 2001 and 2004 and 456 tumors in validation sets made public between 2006 and 2008) from multiple academic centers in the United States and The Cancer Genome Atlas Pilot Project (TCGA).
Main Outcome Measures
Identification of genes with coincident genetic alterations, correlated gene dosage and gene expression, and multiple functional interactions; association between those genes and patient survival.
Results
Gliomas select for a nonrandom genetic landscape—a consistent pattern of chromosomal alterations—that involves altered regions (“territories”) on chromosomes 1p, 7, 8q, 9p, 10, 12q, 13q, 19q, 20, and 22q (false-discovery rate–corrected P<.05). A network model shows that these territories harbor genes with putative synergistic, tumor-promoting relationships. The coalteration of the most interactive of these genes in glioblastoma is associated with unfavorable patient survival. A multigene risk scoring model based on 7 landscape genes (POLD2, CYCS, MYC, AKR1C3, YME1L1, ANXA7, and PDCD4) is associated with the duration of overall survival in 189 glioblastoma samples from TCGA (global log-rank P=.02 comparing 3 survival curves for patients with 0–2, 3–4, and 5–7 dosage-altered genes). Groups of patients with 0 to 2 (low-risk group) and 5 to 7 (high-risk group) dosage-altered genes experienced 49.24 and 79.56 deaths per 100 person-years (hazard ratio [HR], 1.63; 95% confidence interval [CI], 1.10–2.40; Cox regression model P=.02), respectively. These associations with survival are validated using gene expression data in 3 independent glioma studies, comprising 76 (global log-rank P=.003; 47.89 vs 15.13 deaths per 100 person-years for high risk vs low risk; Cox model HR, 3.04; 95% CI, 1.49–6.20; P=.002) and 70 (global log-rank P=.008; 83.43 vs 16.14 deaths per 100 person-years for high risk vs low risk; HR, 3.86; 95% CI, 1.59–9.35; P=.003) high-grade gliomas and 191 glioblastomas (global log-rank P=.002; 83.23 vs 34.16 deaths per 100 person-years for high risk vs low risk; HR, 2.27; 95% CI, 1.44–3.58; P<.001).
Conclusions
The alteration of multiple networking genes by recurrent chromosomal aberrations in gliomas deregulates critical signaling pathways through multiple, cooperative mechanisms. These mutations, which are likely due to nonrandom selection of a distinct genetic landscape during gliomagenesis, are associated with patient prognosis.
Malignant gliomas, with disproportionately high morbidity and mortality,1 are among the most devastating of human tumors. Particular genomic alterations are fundamental to both their formation and their malignant progression.2, 3 Although genomic instability (Box) lends a dynamic character to the human glioma genome, gliomas harbor recurrent chromosomal alterations (Box).2,4,5 Chromosomal alterations presumably exert their tumor-promoting effect on glioma cells by modifying the expression or function of distinct genes, which map to those alterations,6 so as to deregulate growth factor signaling and survival pathways. For many chromosomal alterations, the biologically relevant target genes remain to be discovered.
Box.
Glossarya
- Chromosomal alteration
is an irregularity in the number or structure of chromosomes, usually in the form of a gain (duplication), loss (deletion), exchange (translocation), or alteration in sequence (inversion) of genetic material.
- Chromosomal rearrangement
encompasses several different classes of events: deletions, duplications, inversions, insertions, and translocations. Each of these events can be caused by breakage of DNA double helices in the genome at 2 different locations, followed by a rejoining of the broken ends to produce a new chromosomal arrangement of genes, different from the gene order of the chromosomes before they were broken. Insertions and translocations can involve the exchange of genetic material between nonhomologous chromosomes.
- Cytogenetic bands
correspond to an alternating dark and light banding pattern of chemically stained chromosomes and provide a tool by which abnormalities in chromosomes from diseased cells can be identified.
- Gene dosage
refers to the copy number for a specific gene determined in analytic approaches that do not assess single cells but describe the average copy number profile of a complex tumor in which some cell populations may harbor copy number alterations of a distinct gene and some may not.
- Genome-wide gene dosage profile
is a gene-by-gene representation of gene copy numbers across the whole genome displayed in genome order along the chromosomes.
- Genomic instability
is a biological process consisting of chromosomal rearrangements and duplications. These phenotypes are often seen in the karyotype of cancer cells, where there is an imbalance between the mechanisms of cell-cycle control and mutation rates within aberrant genes.
- Interactome
is the whole set of molecular interactions in cells, most notably protein-protein interactions and protein-DNA interactions.
- Landscape gene
is a gene mapping to a chromosomal region that is part of a consistent pattern of chromosomal alterations.
- Nonrandom genetic landscape
is a consistent pattern of distinct chromosomal alterations. Nonrandomness refers to the significant co-occurrence of these alterations and not to their recurrence frequency. A random alteration may well recur in gliomas and, thus, represent a biological consistency, but there is no statistically recognizable pattern of its recurrence with other alterations.
Oncogenic research on gliomas has focused on the tumor-promoting or tumor-suppressive function of target genes within individual chromosomal alterations.7 However, these alterations do not exist in isolation, nor do single genes account for gliomagenesis. Rather, there may be mechanistic links to genes at other, coincident alterations. To the extent that a set of coincident alterations confers a survival advantage, cells possessing it will be selected by somatic evolution.
A major challenge in cancer research is to distinguish genetic changes that directly drive the disease process from “passenger” mutations, which are neutral to the process and have been accumulated by chance.8 Herein, we take a systems biology approach to this complexity by integrating multidimensional genomic data and functional interactions among genes. The goal is to reduce the complexity and dimensionality of these data to provide biological insights that may translate into improved management of high-grade gliomas. Our central hypothesis is that such an approach reveals a nonrandom genetic landscape (Box)—a consistent pattern of distinct chromosomal alterations—in gliomas, which facilitates gliomagenesis in a cooperative fashion. Using more than 500 glioma samples—including multidimensional data from The Cancer Genome Atlas Pilot Project (TCGA), which aims to discover major cancer-causing genome alterations—we delineate the relationships of tumor-promoting genes in gliomas. At the molecular level, these relationships explain the selection of such a complex genetic landscape during gliomagenesis, and at the clinical level, they impart prognostic insight and suggest therapeutic strategy.
METHODS
Tumor Samples
Forty-five fresh-frozen glioma specimens of varying morphology were collected at Stanford University under Stanford Institutional Review Board–approved guidelines. Written informed consent was obtained from all patients. Specimens were analyzed by a neuropathologist to confirm the histological diagnosis and the presence of vital tumor tissue without excessive contamination (<10%) by normal brain and tumor necrosis. Genome-wide gene dosage (Box)—the copy number for a specific gene—data were generated as previously reported4 using sex-matched reference DNA (Promega, Madison, Wisconsin) and a Stanford human complementary DNA (cDNA) micro-array containing 41 421 Integrated Molecular Analysis of Genomes and Their Expression (IMAGE) Consortium cDNA clone objects9 (hereafter referred to as clones) corresponding to 27 290 different UniGene cluster IDs (Stanford Functional Genomics Facility, Stanford, California). Corresponding genome-wide expression profiles for these tumors and pooled normal human brain RNA (Stratagene, La Jolla, California) were previously reported.10,11 Raw data for gene dosage and expression profiling are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE1991 and GSE2223, respectively, and at the Stanford Microarray Database (SMD) (http://smd.stanford.edu/). Agilent Human Genome CGH Microarray 244A gene dosage data, Affymetrix HT Human Genome U133A Array Plate Set gene expression data, and clinical data for 219, 188, and 207 glioblastomas, respectively, were obtained from the Open-Access and Controlled-Access Data Tiers Portal (http://tcga-data.nci.nih.gov/tcga/findArchives.htm) of TCGA (http://cancergenome.nih.gov/index.asp) on National Human Genome Research Institute approval. At the time of data retrieval from TCGA, there was incomplete overlap in the types of data available for each sample. Alignment of sample identifiers yielded 189 samples with both gene dosage and clinical data, 175 samples with gene expression and clinical data, 175 samples with gene dosage and gene expression data, and 172 samples with all 3 data types. Affymetrix HG-U133A gene expression data and clinical data from 3 high-grade glioma/glioblastoma studies made public at Gene Expression Omnibus between March 8, 2006, and October 10, 2008, were obtained from the University of Texas M. D. Anderson Cancer Center (MDA)12 (GSE4271), the University of California at Los Angeles (UCLA)13 (GSE4412), and Lee et al14 (GSE13041), comprising 76, 70, and 191 samples, respectively.
Genomic Data Processing
Gene dosage data deposited into the SMD were background subtracted and included for downstream analysis if the reference channel intensity was at least 2.5 times the observed background and Pearson correlation was greater than 0.6 for the sample and reference channels. Locally weighted least squares (LOWESS) normalization was performed using SNOMAD (http://pevsnerlab.kennedykrieger.org/snomad.htm) and the Institute for Genomic Research TM4 Microarray Data Analysis System (TIGR MIDAS) (http://www.tm4.org/midas.html). The GoldenPath Human Genome Assembly (http://genome.ucsc.edu) was used to map fluorescence ratios of the arrayed human cDNAs to chromosomal positions. Circular binary segmentation (CBS)15 from the R package “DNA-copy” (http://www.r-project.org/) was used to estimate segmented regions of equal dosage along each chromosome using 33 245 clones that passed initial data normalization and filtering. To increase the number of estimated segmented regions obtained by CBS and thereby the sensitivity of the algorithm, the α level was set to .05 rather than the default of .01. Segmented gene dosage values were deemed changed compared with normal human reference DNA if they fell outside of the ±3-SD range of all segmented log2 values of control (glioma and normal brain) self-to-self hybridizations, resulting in classification as copy number “gain” for gene dosage values >0.2135 and “loss” for gene dosage values <−0.2135. Agilent Human Genome CGH Microarray 244A data from TCGA were background corrected using subtraction then replacement of negative or zero values with half the minimum of the positive corrected values on the array, then were LOWESS normalized using the “limma” package for R. Data from TCGA were similarly processed as described above using CBS via the “snapCGH package” for R. Gene dosage segments were classified as chromosomal gain or loss if the absolute value of the predicted dosage was more than 0.75 times the interquartile range of the difference between observed and predicted values for each region. Gene expression data deposited into the SMD were background subtracted and included for downstream analysis if the reference channel intensity was at least 1.5 times the observed background. LOWESS normalization was performed using SNOMAD and TIGR MIDAS. Preprocessed gene expression data obtained from TCGA,2 MDA,12 UCLA,13 and Lee et al14 were normalized as previously described.
Gene Dosage Data Analysis
We developed a new permutation-based approach, which calculates the probabilistic fit for the coincidence of distinct chromosomal alterations. Gene-by-gene CBS dosage data were averaged for each of 802 chromosomal cytogenetic bands (Box) corresponding to standard chromosomal high-resolution banding, based on an International System for Human Cytogenetic Nomenclature (ISCN) 850 chromosome ideogram.16 For each tumor (t) and cytoband (i), measurements x[t,i] were scored +1 for gain, −1 for loss, and 0 otherwise (ie, unchanged). Scores were mapped into a data matrix (x) with tumors and cytobands ordered as rows and columns, respectively. The association of alterations within each possible combination of 2 cytobands (i, j) was assessed based on computing the score S[i,j] as follows:
where n[i,j] indicated the number of tumors with both i and j altered, and x[t,i]*x[t,j] scores +1 if cytobands [i, j] for tumor t are both amplified or both deleted, –1 if one is amplified and the other deleted, and 0 otherwise (ie, both unchanged or only one cytoband altered). One thousand permutations were applied within the columns of the data matrix to estimate the false-discovery rate (FDR). Associations of pairs of cytobands with an FDR <0.05 were considered significant. The same approach was applied to the gene dosage data of individual candidate genes.
Gene Dosage and Expression Integration
For the Stanford samples, gene dosage and gene expression data (expression in >80% of tumors) were available for 15 608 clones that mapped to 10 351 genes, 7509 of which had either losses or gains in at least 2 tumors. For the latter, signal-to-noise (s2n) ratios were computed to assess the influence of dosage alteration for each gene on its transcript,17–19 defined by the difference of the means (m) of expression levels in the groups of gene dosage–altered (mc) and –unaltered (mn) samples, divided by the sum of standard deviations (s) of expression levels in both groups (sc and sn, respectively). The above data matrix x was transformed such that gene dosage change (c) was assigned g for gain, l for loss, and n otherwise. The significance of the s2n ratio for each gene was estimated by randomly permuting the gene alteration labeling vector 10 000 times. We further determined corresponding FDR-adjusted q values using the “qvalue” package for R (default parameters),20 allowing for a more straightforward interpretation of statistical significance in the context of multiple hypothesis testing. False-discovery rate estimation was performed by setting fixed cutoffs and estimating the FDR by permutations as previously described.20 These computations were done separately for l vs n and g vs n. For clones with alterations in both directions, s2n ratio and corresponding q value for the more recurrent change were chosen. Similar s2n calculations and permutation-based P value computations were conducted to test and validate associations between gene dosage and gene expression for selected candidate genes using TCGA gene dosage and expression data. Due to the intentional selection of these genes for hypothesis testing in TCGA data, FDR was controlled using the method of Benjamini and Hochberg,21 a limiting case of q value correction.
Gene Set Enrichment Analyses and Modeling of Gene Interactions
Gene set enrichment analyses, using hypergeometric testing for the set of genes with significant gene dosage–gene expression relationships, were executed using Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Mountain View, California) to identify overrepresented biological functions and signaling pathways from the IPA database for this gene set with right-tailed P <.05. Ingenuity Pathways Analysis was also used to identify direct and indirect physical (protein-protein) and functional interactions between genes with significant gene dosage–gene expression relationships. An initial gene set was first overlaid onto the set of all cataloged IPA interactions and “focus genes” required in the IPA algorithm were identified as the subset having direct interactions with other genes in the initial set. The specificity of a gene’s interactions was calculated as the percentage of its interactions with other significant genes in the initial set. Genes with the highest specificity of connections were used for the initiation and growth of biological networks with a maximum of 35 genes. Networks of highly interconnected genes were identified by statistical likelihood as follows:
where N is the number of genes in the genomic network, of which G are focus genes, for a network of s genes, f of which are focus genes; and C(N,s) is the binomial coefficient. The score reflects the negative logarithm of the P value that signifies the likelihood of the focus genes in a network being found together as a result of random chance. We merged the top-scoring networks using a “force-direct layout algorithm” to form a composite network representing the underlying biology of the process. Gene set enrichment analyses for the composite network were computed as above. For purposes of further investigation, genes with connectivity (ie, number of interactions with other genes) above the 95% quantile of the overall distribution of gene connectivity in the composite network were deemed network “hub” genes. Genes with tumor-related biological functions and interactions with hub genes were deemed “hub-interacting” genes. For the interaction of the hub genes with each other and with the hub-interacting genes, the mode of functional interaction was integrated with the direction of gene dosage change of each gene constituent of a gene pair to determine the presence of a cooperatively tumorigenic relationship. The BioGRID protein interaction repository22 was used to confirm high connectivity of candidate genes yielded by the IPA analyses by providing an experimentally based data source for physical protein interactions. The entire set of interactions observed in humans (release 2.0.51; March 26, 2009) was downloaded and the number of interactions (or connectivity) observed for all proteins represented in the data set was compared with the number of interactions observed for the candidate genes yielded by the IPA analyses using a Wilcoxon rank-sum test.
Survival Analysis and Multigene Predictor
Survival curves were estimated by the Kaplan-Meier product-limit method, and survival distributions between groups were compared using the log-rank test. Univariate and multivariate Cox proportional hazards regression analyses were performed with overall survival as the dependent variable, and the proportional hazards assumption was tested using interactions with time. To create a multigene predictor model, we first selected predictor genes based on their statistical significance at P<.05 in univariate Cox models relating continuous gene dosage data to survival in the TCGA data set. To combine data for the 7 detected genes, we used a schema according to the presence of alterations of these genes in individual tumors. Based on the number of alterations we assigned tumors to a high-risk group (≥5 of 7 genes altered in the direction associated with gliomagenesis) and a low-risk group (≤2 genes altered). The remainder of the tumors were considered moderate-risk. For the data sets that only included gene expression data without gene dosage data, we used transcript levels above or below their respective medians within a data set to reflect possible chromosomal gain or loss. For genes whose deletion or amplification demonstrated association with survival data at the genetic level in the TCGA analysis, we assigned an “alteration call” to denote potential chromosomal loss or gain if the transcript level was below or above the median. We created low-, moderate-, and high-risk groups for the transcript data based on the number of alteration calls using the same criteria as for the genetic data.
Statistical Calculations and Data Visualization
Unless otherwise specified, all statistical calculations were performed using R software, version 2.8.1, and packages from Bioconductor, release 2.3 (http://www.bioconductor.org/). Paired and unpaired t test, Wilcoxon rank-sum test, and 2-way contingency table analysis based on Pearson χ2 test were used as appropriate. Odds ratios in the 2-way contingency table analysis were computed according to the equation (a/b)/(c/d), where a and b indicate the number of landscape genes (Box)—genes mapping to the nonrandom genetic landscape—with and without significant gene-dosage effect, respectively; and c and d indicate the number of nonlandscape genes with and without significant gene dosage effect, respectively. Corresponding 95% confidence intervals (CIs) for the estimated parameters were computed on “constant χ2 boundaries” as detailed elsewhere.23 Unsupervised hierarchical clustering was done in Cluster (http://rana.lbl.gov/EisenSoftware.htm),24 and 2-way complete linkage clustering based on Pearson correlation as the distance metric was applied. Two-class, unpaired Significance Analysis of Microarrays (SAM; version 3.02; http://www-stat.stanford.edu/~tibs/SAM/)25 was used to assess the weight of individual genes in driving the clustering. TreeView (http://rana.lbl.gov/EisenSoftware.htm) and Caryoscope (http://dahlia.stanford.edu:8080/caryoscope/index.html) software were used to map gene dosage and s2n ratios as a function of human genome order to an ISCN-850 human chromosome ideogram.
RESULTS
Patients
Forty-five patients treated at Stanford University between April 5, 2001, and April 19, 2004, constituted the initial molecular discovery set. Diagnoses included 26 glioblastomas, 5 astrocytic gliomas (grades I-III), 8 oligodendrogliomas (including 3 anaplastic oligodendrogliomas), and 6 anaplastic oligoastrocytomas according to World Health Organization classification.26 Two hundred nineteen glioblastoma samples collected between July 26, 1989, and November 23, 2007, and profiled as part of TCGA constituted a molecular validation set and a clinical training set. Corresponding clinical data were available for 207 of the 219 patients (77 females and 130 males), of which 192 were dead and 15 were alive at last follow-up. Mean patient age was 55.8 (SD, 15.1) years. Median follow-up was 50.6 (range, 1.1–503.4) weeks. A sample population of 76 high-grade glioma patients (25 females and 51 males) treated at MDA,12 whose data were made public on March 15, 2006, constituted a first clinical validation set. Patients included 55 with glioblastomas and 21 with anaplastic astrocytomas. Mean patient age was 45.8 (SD, 12.9) years. Sixty-two patients were dead and 14 were alive at last follow-up. Median follow-up was 93.0 (range, 3.0–477.0) weeks. A sample population of 70 high-grade glioma patients (43 females and 27 males) treated at UCLA,13 made public on March 8, 2006, constituted a second clinical validation set. Patients included 47 glioblastomas, 8 anaplastic astrocytomas, 9 anaplastic oligodendrogliomas, and 6 anaplastic oligoastrocytomas. Mean patient age was 45.6 (SD, 15.0) years. Forty-four patients were dead and 26 were alive at last follow-up. Median follow-up was 63.2 (range, 1.0–359.0) weeks. A unified sample population of 191 glioblastoma patients (74 females and 117 males) from multiple institutions,14 made public on October 10, 2008, constituted a third clinical validation set. Mean patient age was 53.8 (SD, 13.6) years. One hundred seventy-six patients were dead and 15 were alive at last follow-up. Median duration of follow-up was 55.6 (range, 1.0–479.0) weeks.
Modeling of a Nonrandom Genetic Landscape in Gliomas
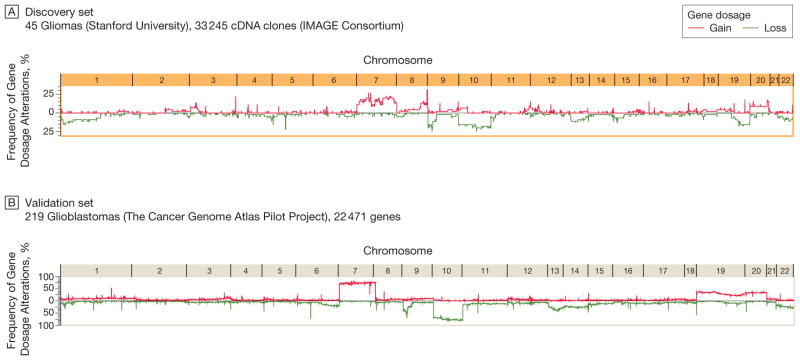
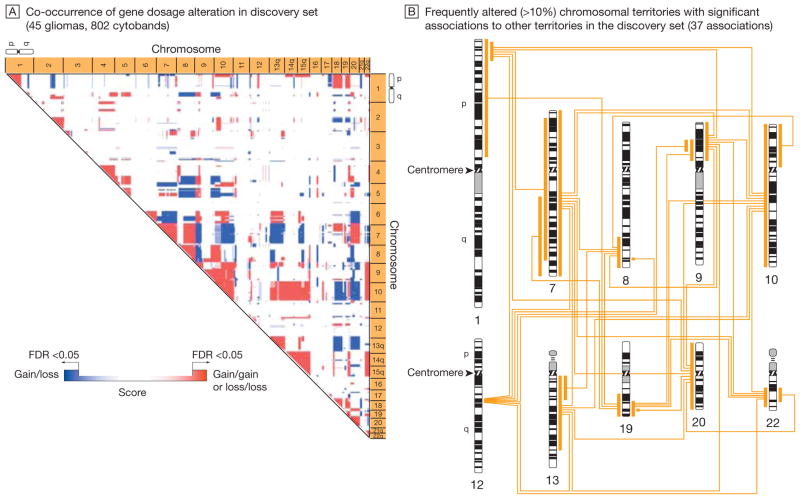
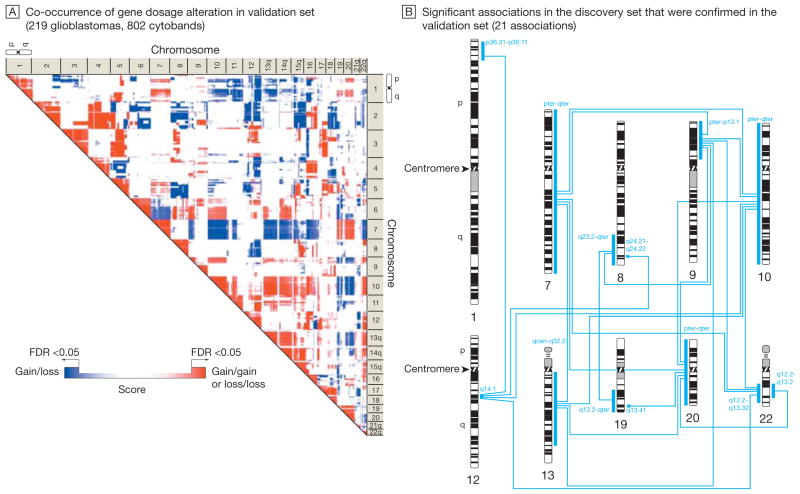
Figure 1 shows the genome-wide gene dosage profiles (Box)—a gene-by-gene representation of gene copy numbers across the whole genome—for the Stanford (45 tumors) and TCGA (219 tumors) data sets according to genome position. The profiles highlight a similar pattern of recurrent chromosomal alterations, including gains and losses (Figure 1), and their boundaries throughout the glioma genome (eFigure 1, available at http://www.jama.com). We modeled the association of chromosomal alterations with each other. Figure 2A shows a genome-wide association matrix for 802 chromosomal cytogenetic bands (cytobands; Box)—subregions of a chromosome visible microscopically after special staining—in the 45 Stanford tumors. Numerous significant (FDR<0.05) cytoband-cytoband associations are identified across the human glioma genome (Figure 2A). Filtering of those associations for more than 10% frequency identifies an initial genetic landscape of aneuploidy (–, losses; +, gains) involving 10 networking regions (or territories) on chromosomes 1p-, 7+, 8q+, 9p-, 10-, 12q+, 13q-, 19q-, 20+, and 22q- displaying 37 associations (Figure 2B). An independent validation analysis of 219 TCGA glioblastomas using the same permutation-based approach reveals a very similar association matrix (Figure 3A), confirming 9 (90%) of 10 chromosomal territories (except for the long arm of chromosome 1p) and 21 (56.8%) of 37 territorial associations (Figure 3B). The chromosomal territories confirmed in this validation analysis are hereafter referred to as a (TCGA-validated) genetic landscape.
Figure 1.
Gene Dosage Alterations Across the Glioma Genome
Cartesian line diagrams depict the frequency of gene dosage alteration across a tumor population for genes mapped according to genome position along the chromosomes (X and Y chromosomes omitted). Gene dosage profiles were generated using circular binary segmentation change point analysis of microarray-based genomic hybridization profiles. Both the discovery and validation sets show consistently high frequencies of alterations involving chromosomes 1p, 7, 8q, 9p, 10, 12, 13, 19, 20, and 22. IMAGE indicates Integrated Molecular Analysis of Genomes and Their Expression Consortium.
Figure 2.
Model of a Nonrandom Genetic Landscape in Gliomas: Discovery Set
A permutation-based approach, which calculates the probabilistic fit for the coincidence of distinct chromosomal alterations, was applied to the gene dosage data in the discovery set from Stanford University. The gene dosage data generated by the circular binary segmentation change point algorithm were averaged according to 802 cytogenetic bands (cytobands)—subregions of a chromosome visible microscopically after special staining—that correspond to an International System for Human Cytogenetic Nomenclature (ISCN) 850 chromosome ideogram. The association map (A) indicates chromosomal regions (territories) with significant co-occurrence of gene dosage alteration (false-discovery rate [FDR]–corrected P<.05). Red scores denote the significant association of 2 chromosomal gains or 2 chromosomal losses; blue scores, the significant association of a gain and a loss event. The color gradation reflects a score that indicates the significance of cytoband-cytoband associations. The association map shows a consistent pattern of significant associations, which denotes a nonrandom genetic landscape in gliomas. B, Chromosomal territories that showed an alteration frequency of greater than 10% (involving chromosomes 1p, 7, 8q, 9p, 10, 12, 13, 19, 20, and 22) in the discovery set and their significant associations are mapped to a human ISCN-850 chromosome ideogram. (See interactive Figure 1 showing significant associations in the discovery and validation sets at http://www.jama.com.)
Figure 3.
Model of a Nonrandom Genetic Landscape in Gliomas: Confirmation by Validation Set
Permutation-based approach described in Figure 2 was applied to the validation set from The Cancer Genome Atlas Pilot Project (TCGA). The association map (A) indicates chromosomal regions (territories) with significant co-occurrence of gene dosage alteration (false-discovery rate [FDR]–corrected P<.05). Red scores denote the significant association of 2 chromosomal gains or 2 chromosomal losses; blue scores, the significant association of a gain and a loss event. The color gradation reflects a score that indicates the significance of cytoband-cytoband associations. The association map shows a consistent pattern of significant associations, which denotes a nonrandom genetic landscape in gliomas. B, Significant associations identified in the discovery set (Figure 2B) that were confirmed in the TCGA validation set. The blue bars and corresponding blue labels indicate chromosome territories with TCGA-confirmed significant associations to other territories (TCGA-validated nonrandom genetic landscape). (See interactive Figure 1 showing significant associations in the discovery and validation sets at http://www.jama.com.)
Gene Dosage Reflected in Gene Expression
We observed a significant (q<.10) gene dosage–gene transcript relationship for 1562 (15.1%) of 10 351 expressed genes in the 45 Stanford tumors, including genes implicated in gliomagenesis such as EGFR, MYC, MDM2, GBAS, PTEN, CDK6, MTAP, IGF1R, MXI1, WDR11, and FGFR2. Gene set enrichment analyses—which test for high representation of distinct biological processes and signaling pathways—revealed a significant (P<.05 by right-tailed Fisher exact test) enrichment of those genes for various tumor-promoting (and developmental) functions and pathways (eFigure 2).
We noted a widespread association of changes in gene dosage and expression across the glioma genome (eFigure 3). However, there was a greater likelihood for a gene dosage effect on the expression of genes mapping to the genetic landscape than on the expression of genes mapping to random genetic alterations (P<.001 by Pearson χ2 test; odds ratio, 7.7; 95% CI, 6.9–8.6) (eFigures 3 and 4). Within the genetic landscape, we noted a greater propensity for alterations to be hypomorphic, ie, to reduce gene expression, rather than hypermorphic, ie, to increase gene expression (for hypomorphic vs hypermorphic, odds ratio, 8.4; 95% CI, 7.3–9.6 vs 3.5; 95% CI, 2.8–4.2; P <.001 for both) (eFigures 3 and 4).
Highly Interacting Landscape Genes
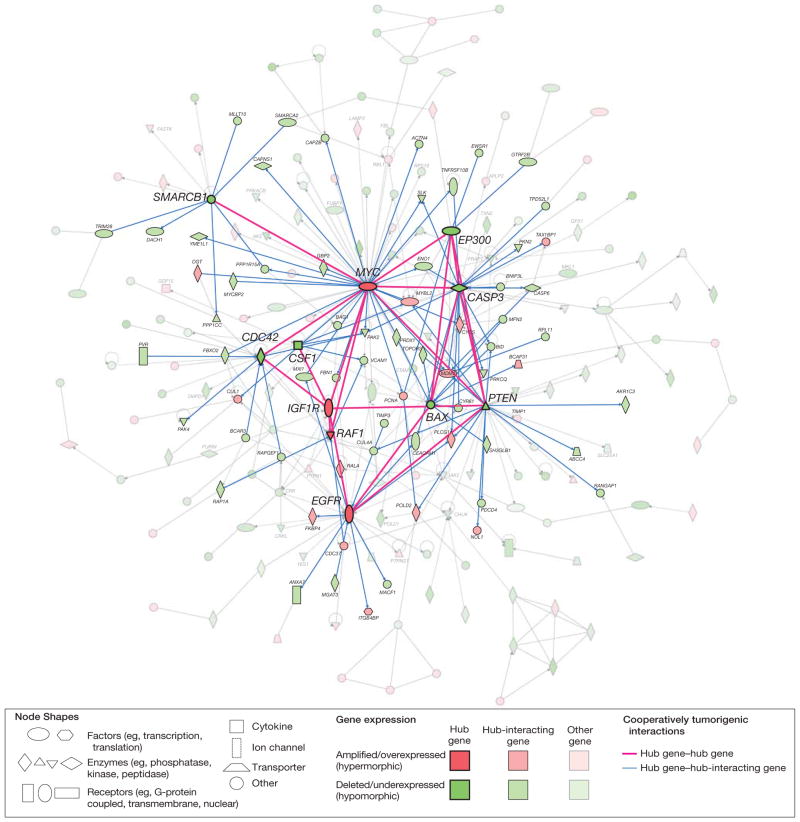
Genes with a role in gliomagenesis are more likely to function as part of a cooperative group or network rather than individual units.10,27,28 Our hypothesis that the characterized common genetic landscape alters biological networks to promote glioma formation prompted network modeling to identify the interaction of the genes affected by dosage effects in the glioma genome. This yielded a “node-and-edge” graph that maps 214 genes (represented as nodes) into a network of various physical (protein-protein) or functional interactions (represented as edges connecting the nodes) (Figure 4). The modeled interactions are both directed (well-defined information flow; eg, from a transcription factor to the gene it regulates) and undirected (eg, mutual binding relationships). Gene set enrichment analyses revealed a greater probability for over-representation of tumor-promoting (83.1% of genes) and developmental (29.5%) functions and pathways in the network than for all 1562 gene dosage–driven genes (eFigure 2).
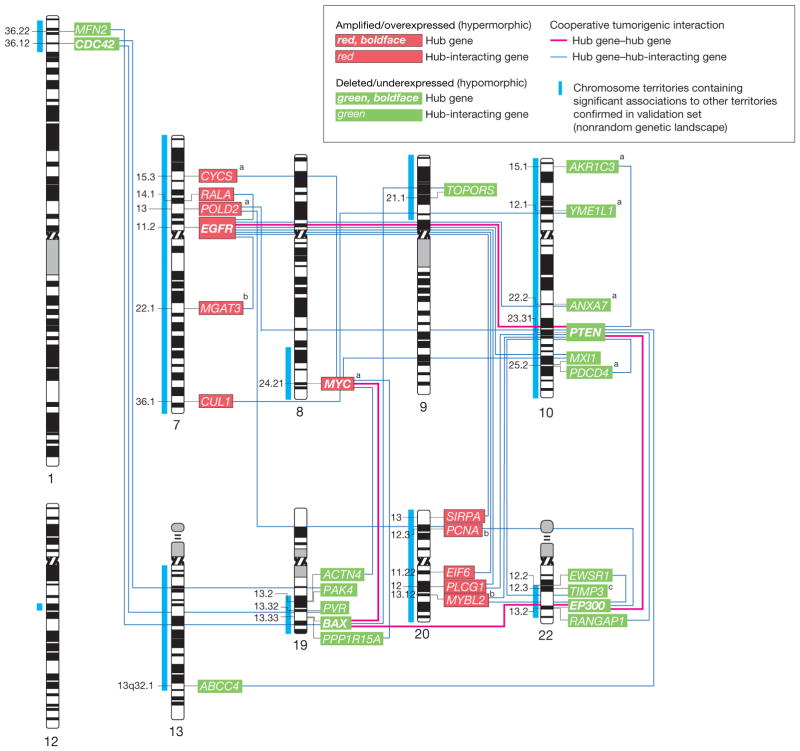
Figure 4.
Complex Network of Glioma Gene Interactions Highlighting Hub Gene–Hub Gene and Hub Gene–Hub-Interacting Gene Interactions
Graphical representation of the interactions and networking of 214 glioma genes. The 214 genes represent the subset of all 1562 genes with significant gene dosage–transcript relationships (false-discovery rate–adjusted q<.10) that map to the top-scoring subnetworks identified in a network modeling approach using Ingenuity Pathway Analysis. These subnetworks were merged using a force-directed layout algorithm to form a composite network representing the underlying biology of the process. Genes are represented as nodes using various shapes that represent the functional class of the gene product. The interactions of genes with tumor-related biological functions and fulfilling the criterion of a network hub (11 genes) or interacting with hub genes (hub-interacting gene; 92 genes) are highlighted. Hub gene refers to a gene that shows a number of interactions with other genes (the hub-interacting genes or other hub genes) that is above the 95% quantile of the overall distribution of the number of interactions of all genes in the network (eFigure 5A). Cooperatively tumorigenic interactions are interactions for which integration of the mode of interaction for a hub-hub (solid red lines) or hub–hub-interacting gene pair (blue lines) with the direction of gene dosage–gene expression change (gain or loss) reveals an effect on this interaction that synergistically promotes tumorigenesis. Paired t testing comparing the number of cooperative vs noncooperative interactions for 11 hub genes with each other and 92 hub-interacting genes with tumor-related functions reveals a significant prevalence (P=.003) of cooperatively tumorigenic interactions (125 [71.8%] of all 174 interactions) (eFigure 4). (See interactive Figure 2 of the complete network at http://www.jama.com.)
The organization of genes and proteins into functional networks likely reflects cellular function.27 We studied the genes with high connectivity (a large number of incident edges), as their alteration may affect many other genes in the network; such highly influential genes should be preferred therapeutic targets. We identified 11 highly connected hub genes (Figure 4 and eFigure 5A), all of which have a tumor-promoting function and notably included genes with a known biological role in gliomagenesis, such as EGFR, MYC, PTEN, and BAX. Genes that interact functionally with the hub genes are also likely to fundamentally influence the network. Analysis of the reciprocal functional relationship of the 11 hub genes with each other and with 92 hub-interacting genes with a tumor-related function disclosed 125 cooperatively tumorigenic gene-gene relationships (Figure 4). A matched-pair analysis of the number of putatively cooperative vs nonco-operative interactions for those genes revealed a significant prevalence (125 [71.8%] of all 174 interactions; P = .003 by paired t test) of cooperatively tumorigenic relationships in the network (eFigure 6).
We found that 6 (54.5%) of 11 hub genes and 52 (56.5%) of 92 tumor-related hub-interacting genes mapped to the TCGA-validated genetic landscape. We tested the nonrandomness of coalteration of the individual hub and hub-interacting genes mapping to the genetic landscape (the landscape hub and hub-interacting landscape genes, constituting together the landscape genes) by applying the same approach used initially to model a genome-wide genetic landscape. We then integrated the association data with the interaction data of our network model (Figure 4) to identify significant interactions for the landscape genes; that is, physical (protein-protein) and functional interactions between genes whose gene dosage alterations are significantly associated (FDR<0.05) (eFigure 5, B and C). This analysis revealed 37 significant interactions involving all 6 (100%) landscape hub genes—CDC42, EGFR, MYC, PTEN, BAX, and EP300—and 31 (59.6%) of the 52 hub-interacting landscape genes (eFigure 5, B and C). Thirty-one (83.7%) of these 37 significant interactions demonstrated a cooperatively tumorigenic relationship (Figure 5). Mapping of these relationships to a human chromosome ideogram reveals that the involved genes are broadly dispersed throughout the TCGA-validated landscape (Figure 5).
Figure 5.
Networking of 31 Hub and Hub-Interacting Genes With Cooperatively Tumorigenic Relationships That Map to the TCGA Validated Landscape
Human chromosome ideogram representation of the networking of 31 hub and hub-interacting genes that map to The Cancer Genome Atlas Pilot Project (TCGA)–validated genetic landscape and possess cooperatively tumorigenic relationships. These cooperatively tumorigenic relationships emerge through the mode of interaction between the genes and their significant coalteration. Gene dosage–gene expression integration for 30 of the 31 genes with combined availability of gene dosage and expression data in TCGA confirmed significant gene dosage effects on transcription for 27 (90.0%) of 30 genes (Benjamini-Hochberg false-discovery rate–corrected P<.05). Labels indicate the cytogenetic band mapping of the genes.
aGenes with a significant (P<.05) relationship between gene dosage and duration of patient survival in the TCGA data set based on Cox proportional hazards regression analysis.
bGenes without confirmed significant gene dosage–gene expression relationship in the TCGA data set.
cTIMP3 is embedded within an intron of the SYN3 gene. Gene dosage information for TIMP3 was only indirectly available through a probe mapping to SYN3 in TCGA. Integration of gene dosage information using this probe and TIMP3 transcript information revealed a significant (P<.05) gene dosage–transcript relationship.
To validate a significant gene-transcript correlation for the 31 landscape genes with cooperatively tumorigenic relationships, we integrated corresponding gene dosage and expression profiles in TCGA using a statistical approach similar to that above. Combined gene dosage and gene expression data were available for 30 of the 31 genes in TCGA’s data set. This analysis confirmed significant gene dosage effects on expression for 27 (90.0%) of the 30 genes, excluding MGAT3, MYBL2, and PCNA (Benjamini-Hochberg FDR-corrected P<.05) (Figure 5).
Finally, we confirmed high connectivity for 25 of the 27 validated landscape genes (MFN2 and ABCC4 did not have interactions reported at the time of analysis) using BioGRID, a general repository for protein-protein interactions. For 8445 available genes (the entire set of human protein interactions) with at least 1 interaction reported in humans, there were 30 386 gene-gene pairs with at least 1 reported interaction between them. The median number of reported interactions for the entire data set was 3 (range, 1–202). For the 25 validated landscape genes available for analysis, the median number of reported interactions was 12 (range, 1–108). A Wilcoxon rank-sum test comparing the number of observed interactions yielded P<.001, confirming the significantly higher interconnectivity of the TCGA-validated landscape genes than of the entire set of human protein interactions.
Landscape Gene Alterations Associated With Patient Outcome
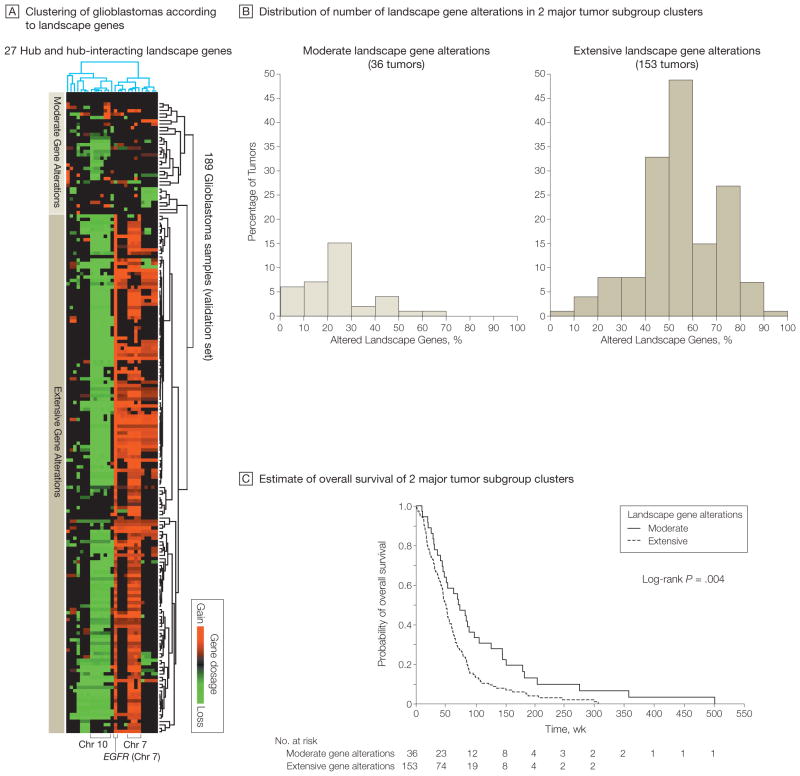
Unsupervised hierarchical clustering—which assigns objects (herein, patient and genes) into groups (called clusters) so that objects from the same cluster are more similar to each other than are objects from different clusters—of 189 glioblastomas with gene dosage and clinical data in TCGA based on the 27 TCGA-validated landscape genes identified 2 major subgroups with a significant difference in their mean number of landscape gene alterations (P<.001 by unpaired t-test): one consisting of 153 tumors showing substantial alterations of the landscape genes (mean, 54.7% [SD, 16.1%]) and one consisting of 36 tumors showing far fewer alterations (mean, 24.3% [SD, 15.5%]) (Figure 6, A and B). Supervised group predictor analysis revealed highest group-predictive power for altered genes on chromosomes 7 and 10.
Figure 6.
Subgroups of Glioblastomas With Distinct Landscape Gene Profiles Associated With Patient Survival
A, Unsupervised hierarchical clustering (2-way complete linkage clustering based on Pearson correlation as the distance metric) of 189 of 219 glioblastomas from The Cancer Genome Atlas Pilot Project (TCGA) (with availability of corresponding survival data) according to the 27 landscape genes in Figure 5 with a TCGA-validated gene dosage–expression relationship. Rows on heat map represent patient samples; columns represent the 27 genes. Two major tumor clusters with distinct gene dosage alteration patterns were identified (moderate and extensive). Supervised group predictor analysis revealed highest group-predictive power for altered genes on chromosomes 7 and 10. B, Distribution of the number of landscape gene alterations among the 2 major clustering subgroups identified by the hierarchical clustering in panel A. The groups show a significant difference in mean number of landscape gene alterations (P<.001 by unpaired t test). C, Kaplan-Meier estimates of overall survival of the 2 major clustering subgroups indicated in panels A and B. Median follow-up was 73.1 (range, 10.6–503.4) weeks for the group with modest alterations and 47.0 (range, 1.1–307.4) weeks for the group with extensive alterations.
We hypothesized that the group of tumors with extensive landscape gene alterations might be those that are biologically more aggressive and, hence, have a worse prognosis. A 2-class model confirmed a significant difference in overall survival between the 2 groups (median duration of survival of 51 vs 72 weeks), such that tumors with extensive alterations demonstrated a comparably unfavorable outcome (hazard ratio [HR], 1.75; 95% CI, 1.19–2.59; log-rank P=.004) (Figure 6C). Median duration of follow-up was 73.1 (range, 10.6–503.4) weeks for the group with modest alterations and 47.0 (range, 1.1–307.4) weeks for the group with extensive alterations. Tumors with wide-spread landscape gene alterations had a 2-year survival rate of 12.9% and 82.34 deaths per 100 person-years vs a 2-year survival rate of 32.0% and 48.31 deaths per 100 person-years for tumors with modest alterations.
Multigene Predictor Model
To determine the potential utility of a concise molecular profile in estimating the prognosis of an individual glioblastoma patient, we assessed the survival association of gene dosage for each of the 27 validated landscape genes in TCGA glioblastomas. Cox proportional hazard regression indicated statistically significant survival associations for 7 landscape genes, including the DNA polymerase delta 2, small subunit (POLD2) (P = .01) and cytochrome C, somatic (CYCS) (P = .01) genes on chromosome 7, the MYC oncogene on chromosome 8 (P=.03), and the aldoketo reductase family 1, member C3 (AKR1C3) (P = .002), YME1-like 1 (YME1L1) (P=.006), annexin A7 (ANXA7) (P=.008), and programmed cell death 4 (neoplastic transformation inhibitor) (PDCD4) (P=.01) genes on chromosome 10. Each of the 7 genes has a biological function whose alteration might contribute to tumorigenesis. The avian myelocytomatosis homologue (MYC) is a major oncogene10; ANXA7 is a candidate tumor suppressor gene in breast and prostate cancer29–33 and its protein product is a phosphorylation target of the epidermal growth factor receptor (EGFR) oncoprotein34; AKR1C3 and PDCD4 have been implicated in colorectal cancer35,36 and their expression is regulated by phosphatase and tensin homolog (PTEN)37; YME1L1 has been related to cancer development and progression38 and has been identified as an MYC-responsive gene,39 as has CYCS.40 The protein encoded by POLD2 is a subunit of the DNA polymerase delta complex, which also contains the protein product of the landscape gene PCNA; it is involved in DNA replication and repair and its expression is regulated by PTEN.37
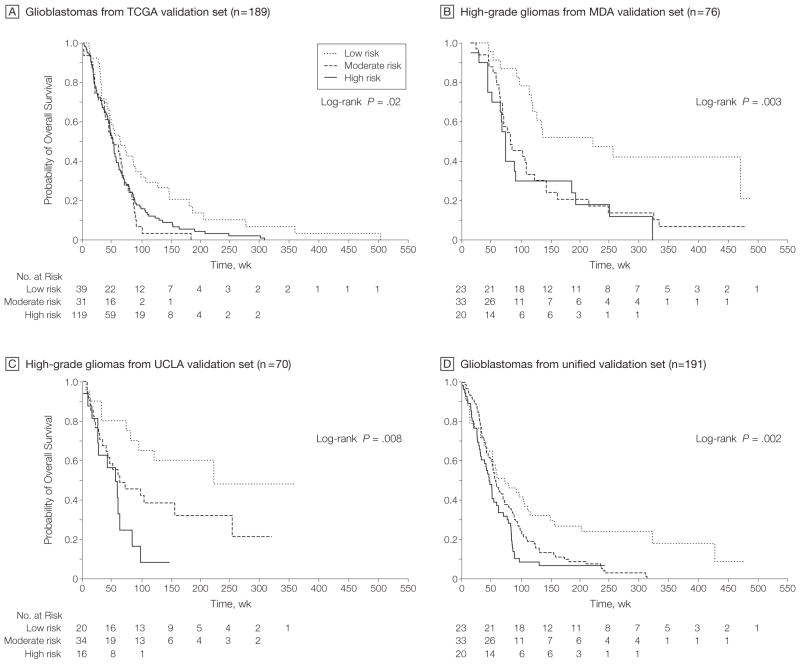
In the TCGA training set of 189 glioblastomas, the combined 7-gene predictor model assigned 119 tumors to a high-risk group (≥5 of 7 genes altered) with a median duration of follow-up of 49.3 (range, 2.4–307.4) weeks, 39 to a low-risk group (≤2 genes altered) with a median follow-up of 62.9 (range, 10.6–503.4) weeks, and 31 to a moderate-risk group (all others) with a median follow-up of 51.0 (range, 1.1–183.1) weeks. There was a significant difference in overall survival for the 3 groups (log-rank P = .02; Cox model HR, 1.63; 95% CI, 1.10–2.40 [P=.02] comparing high-risk vs low-risk groups and HR, 1.87; 95% CI, 1.13–3.10 [P = .01] comparing moderate-risk vs low-risk groups; 49.24, 92.66, and 79.56 deaths per 100 person-years for low, moderate, and high-risk groups, respectively) (Figure 7A). Similar multigene models using gene expression data revealed significant differences in survival for the MDA and UCLA validation sets (Figure 7, B and C). In the MDA set, 23, 33, and 20 patients were assigned to the low-, moderate-, and high-risk groups and had median follow-up times of 175 (range, 33–477) weeks, 70 (range, 12–467) weeks, and 62 (range, 3–311) weeks, respectively. The low-, moderate-, and high-risk groups demonstrated 15.13, 41.10, and 47.89 deaths per 100 person-years, respectively. A log-rank test for all 3 curves yielded P=.003 and Cox modeling yielded an HR of 3.04 (95% CI, 1.49–6.20; P = .002) comparing high-risk vs low-risk groups and an HR of 2.52 (95% CI, 1.33–4.78; P=.005) comparing moderate-risk vs low-risk groups (Figure 7B). In the UCLA set, 20, 34, and 16 patients were assigned to the low-, moderate-, and high-risk groups and had median follow-up times of 128.5 (range, 6–359) weeks, 58.5 (range, 8–320) weeks, and 49 (range, 1–147) weeks, respectively. The low-, moderate-, and high-risk groups demonstrated 16.14, 38.41, and 83.43 deaths per 100 person-years, respectively. A log-rank test for all 3 curves yielded P = .008 and Cox modeling yielded an HR of 3.86 (95% CI, 1.59–9.35; P=.003) comparing high-risk vs low-risk groups and an HR of 2.12 (95% CI, 0.97–4.63; P=.06) comparing moderate-risk vs low-risk groups, respectively (Figure 7C). In the multi-institutional validation set of 191 glioblastomas, 43, 92, and 56 patients were assigned to the low-, moderate-, and high-risk groups and had median follow-up times of 73 (range, 1–479), 57.5 (range, 8–313), and 47 (range, 1–242) weeks, respectively. The low-, moderate-, and high-risk groups demonstrated 34.16, 63.49, and 83.23 deaths per 100 person-years, respectively. A log-rank test for all 3 curves yielded P = .002 and Cox modeling yielded an HR of 2.27 (95% CI, 1.44–3.58; P<.001) comparing high-risk vs low-risk groups and an HR of 1.72 (95% CI, 1.14–2.59; P = .01) comparing moderate-risk vs low-risk groups (Figure 7D), confirming the robustness of the multigene predictor model in glioblastoma at the expression level. Although gene dosage was prognostic in TCGA tumors, we did not find an association between mRNA expression and survival in the TCGA data set. Heterogeneity of patient characteristics and treatment (tumor samples collected over nearly 2 decades at varying institutions) and the fact that gene transcription is more dynamic and fluctuant than gene dosage may explain the disparity in findings in the TCGA group at the genetic and transcript levels.
Figure 7.
Multigene Risk Scoring Model in Malignant Gliomas
Patient assignment to low-, moderate-, and high-risk groups is based on risk scores generated by the number of gene dosage alterations (0–2, 3–4, and 5–7, respectively) of 7 landscape genes (POLD2, CYCS, MYC, AKR1C3, YME1L1, ANXA7, and PDCD4), each of which was individually linked to patient survival in Cox proportional hazard regression analyses in The Cancer Genome Atlas Pilot Project (TCGA). A, Kaplan-Meier estimates of overall survival for the 3 groups in 189 glioblastomas from TCGA) with available survival data. Median follow-up for the low-, moderate-, and high-risk groups was 62.9 (range, 10.6–503.4), 49.3 (range, 2.4–307.4), and 51.0 (range, 1.1–183.1) weeks, respectively. B, Estimates of overall survival in the University of Texas M. D. Anderson Cancer Center (MDA) validation set of 76 high-grade gliomas. Median follow-up for the low-, moderate-, and high-risk groups was 175 (range, 33–477) weeks, 70 (range, 12–467) weeks, and 62 (range, 3–311) weeks, respectively. C, Estimates of overall survival in the University of California, Los Angeles (UCLA) validation set of 70 high-grade gliomas. Median follow-up for the low-, moderate-, and high-risk groups was 128.5 (range, 6–359) weeks, 58.5 (range, 8–320) weeks, and 49 (range, 1–147) weeks, respectively. D, Estimates of overall survival in the unified validation set of 191 glioblastomas. Median follow-up for the low-, moderate-, and high-risk groups was 73 (range, 1–479), 57.5 (range, 8–313), and 47 (range, 1–242) weeks, respectively.
COMMENT
The pathobiology of human gliomas emerges through the actions of multiple genes and their interactions with each other. As the classic theories regarding glioma origin and gliomagenesis are currently being reappraised,41 we used a systems biology approach without regard to morphological subtypes to generate a model of a nonrandom genetic landscape in gliomas. Herein, nonrandomness describes a significant association of distinct chromosomal alterations. Our model proposes that this genetic landscape involves multiple reciprocal gene alterations, which cooperatively promote gliomagenesis and contribute to unfavorable patient prognosis, thus providing the selection pressure for its conservation.
Identification of the importance and mechanistic interplay of such gene alterations in gliomas prompts evaluation of their potential as therapeutic targets. The network context of a gene likely affects the efficacy of therapies that target its protein. The complexity of our landscape model helps explain the lack of therapeutic efficacy of strategies targeting single gene products. For example, targeting the oncoprotein EGFR alone has had limited clinical effect despite the prominence of EGFR in our model as a landscape hub gene.42 This paradox is resolved by the fact that the deregulation of EGFR in glioblastomas does not exist in isolation. But rather, the functional organization of EGFR, and other gene dosage–altered genes within the landscape, in a complex network of interactions predicts that monotherapeutic approaches will fail: extensive cross-talk between interacting genes and their pathways permits resistance to therapy. The topology of our landscape model suggests that molecular targeting of multiple (hub) genes will be more effective.
Our study has limitations. First, our landscape model reduces the biological complexity of human gliomas to the genetic level. However, the fundamental mechanisms of carcinogenesis include both genetic and epigenetic events. Future integration of genetics and epigenetics could better explain gliomagenesis. Second, our model assumes that cancer cells modify only pre-existing biological pathways. But, in complex diseases such as gliomas, genes may additionally interact in new pathways and networks. Therefore, exploration of gene networks without a priori assumptions may complement this network approach based on known gene interactions. We recognize that the connectivity (and, thus, the potential importance) of genes in a network like ours depends not only on biological reality but also on sampling, experimental conditions, and reporting of results; there may be other genes that significantly contribute to network behavior that our model has not addressed. Furthermore, the candidate gene-gene interactions that emerged from our model need to be further tested mechanistically in a brain tumor–specific context. And, finally, our assumption of nonrandom coselection of distinct chromosomal alterations may be invalidated by chromosomal rearrangement (Box) of parts between nonhomologous chromosomes in the genetic landscape. Recent evidence suggests that recurrent translocations might be more common in solid tumors than previously appreciated.43 However, physical rearrangements have not been reported for the chromosomal territories mapping to our TCGA-validated genetic landscape in gliomas, nor could they explain the frequent coselection of chromosomal loss and gain events within the landscape.
To date, therapeutic decisions for human gliomas are mainly guided by inexact and heterogeneous clinical and morphological measures. Risk estimation in glioblastomas, which share similar pathological characteristics but show substantial variation in outcome, is particularly challenging. This calls for better risk estimation strategies, which might improve treatment efficacy and quality of life. Such strategies have to take into account the key mechanisms contributing to glioblastoma pathogenesis.
Several gene sets have been proposed as multigene predictors of glioblastoma patient survival.5,13,44–46 Many of those sets have used so-called meta-genes—patterns of gene expression—for risk classification. Although meta-genes support the notion that, in a complex glioblastoma tumor, it is not a single gene but, rather, multiple genes that drive the disease process, the risk associations of metagenes are primarily correlative. While metagenes, by definition, constitute gene subsets identified without considering underlying mechanisms, some studies have shown that such metagenes can characterize clinically relevant molecular subtypes of high-grade gliomas.12–14 Recently, a consensus gene expression profile associated with patient outcome across independent data sets and an optimized 9-gene metagene predictor have been reported for high-grade gliomas.47
While the primary role of multi-gene predictor models would be to prospectively identify glioma patients with low vs high likelihood for a durable response to standard therapy, molecular predictor models that place genes into pathways and networks allow mechanistic insights that are crucial for developing novel molecularly targeted therapies. We show that a focused set of 7 landscape genes associates with patient survival across several different study populations with discrimination similar to that of correlative multigene predictors. These observations lay the foundation for future development of a mechanistically based molecular risk estimation model in glioblastomas and high-grade gliomas. In our study, the multigene predictor model associates with survival when taking into account either genetic or transcriptional information about those genes. The genetic model might lend itself better to translation into clinical practice for 2 reasons. First, while the expression of an individual gene can vary over short periods of time in cancers, gene dosage is more static and more amenable to predictive screens. Second, the gene expression model depends on risk groupings in which the transcript profile of an individual patient is related to the profiles of other patients, with a potential for demographic bias. In contrast, the genetic predictor allows risk estimation based on a patient’s individual biological profile by using a binary model (the gene is altered or it is not).
CONCLUSION
The current work provides a network model and biological rationale for the selection of a nonrandom genetic landscape in human gliomas. Coincident genetic alterations of multiple landscape genes may evolve rapidly under positive selection to provide multiple, synergistic mechanisms of dysregulation of critical signaling pathways toward gliomagenesis. A multigene predictor model incorporating 7 landscape genes demonstrates how molecular insights emerging from our integrative multidimensional analysis could translate into relevant clinical end points affecting the future management of gliomas.
Acknowledgments
Funding/Support: This work was supported by the State of Illinois Excellence in Academic Medicine Program (EAM Award 211 to Dr M. Bredel).
Role of the Sponsor: The funding organization had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
Footnotes
Definitions are in part adapted from the National Cancer Institute (NCI) Terminology Browser (http://nciterms.nci.nih.gov/NCIBrowser/Dictionary.do) using the NCI Thesaurus terminology.
Author Contributions: Dr M. Bredel had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Financial Disclosures: None reported.
Additional Information: eFigures 1 through 6 and interactive Figures 1 and 2 are available at http://www.jama.com. The results published herein are in part based on data generated by TCGA’s Pilot Project established by the National Cancer Institute and the National Human Genome Research Institute. Information about TCGA and the investigators and institutions who constitute TCGA’s research network can be found at http://cancergenome.nih.gov.
Study concept and design: M. Bredel, Sikic.
Acquisition of data: M. Bredel, Scholtens, Harsh, C. Bredel.
Analysis and interpretation of data: M. Bredel, Scholtens, Harsh, C. Bredel, Chandler, Renfrow, Yadav, Vogel, Scheck, Tibshirani, Sikic.
Drafting of the manuscript: M. Bredel.
Critical revision of the manuscript for important intellectual content: M. Bredel, Scholtens, Harsh, C. Bredel, Chandler, Renfrow, Yadav, Vogel, Scheck, Tibshirani, Sikic.
Statistical analysis: M. Bredel, Scholtens, Tibshirani.
Obtained funding: M. Bredel, Sikic.
Administrative, technical, or material support: Harsh, Chandler, Renfrow, Vogel.
Study supervision: M. Bredel, Sikic.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bredel M, Bredel C, Juric D, et al. High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer Res. 2005;65(10):4088–4096. doi: 10.1158/0008-5472.CAN-04-4229. [DOI] [PubMed] [Google Scholar]
- 5.Nigro JM, Misra A, Zhang L, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65(5):1678–1686. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 6.Chin K, Devries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452(7187):553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The IMAGE Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33(1):151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 10.Bredel M, Bredel C, Juric D, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65(19):8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 11.Stears RL, Getts RC, Gullans SR. A novel, sensitive detection system for high-density microarrays using dendrimer technology. Physiol Genomics. 2000;3(2):93–99. doi: 10.1152/physiolgenomics.2000.3.2.93. [DOI] [PubMed] [Google Scholar]
- 12.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Scheck AC, Cloughesy TF, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1(1):52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 16.Mitelman FE. ISCN 1995: An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: S Karger; 1995. [Google Scholar]
- 17.Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62(21):6240–6245. [PubMed] [Google Scholar]
- 18.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 19.Juric D, Bredel C, Sikic BI, Bredel M. Integrated high-resolution genome-wide analysis of gene dosage and gene expression in human brain tumors. Methods Mol Biol. 2007;377:187–202. doi: 10.1007/978-1-59745-390-5_12. [DOI] [PubMed] [Google Scholar]
- 20.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 22.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleiss JL. Statistical Methods for Rates and Proportions. 2. New York, NY: John Wiley & Sons; 1981. sect 5.6. [Google Scholar]
- 24.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson MR, Zhang B, Fang Z, Mischel PS, Horvath S, Nelson SF. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006;7:40. doi: 10.1186/1471-2164-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mischel PS, Cloughesy TF, Nelson SF. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci. 2004;5 (10):782–792. doi: 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 29.Srivastava M, Bubendorf L, Srikantan V, et al. ANX7, a candidate tumor suppressor gene for prostate cancer. Proc Natl Acad Sci U S A. 2001;98 (8):4575–4580. doi: 10.1073/pnas.071055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leighton X, Srikantan V, Pollard HB, Sukumar S, Srivastava M. Significant allelic loss of ANX7region (10q21) in hormone receptor negative breast carcinomas. Cancer Lett. 2004;210(2):239–244. doi: 10.1016/j.canlet.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava M, Bubendorf L, Raffeld M, et al. Prognostic impact of ANX7-GTPase in metastatic and HER2-negative breast cancer patients. Clin Cancer Res. 2004;10(7):2344–2350. doi: 10.1158/1078-0432.ccr-03-0278. [DOI] [PubMed] [Google Scholar]
- 32.Srivastava M, Torosyan Y, Raffeld M, Eidelman O, Pollard HB, Bubendorf L. ANXA7 expression represents hormone-relevant tumor suppression in different cancers. Int J Cancer. 2007;121(12):2628–2636. doi: 10.1002/ijc.23008. [DOI] [PubMed] [Google Scholar]
- 33.Torosyan Y, Dobi A, Naga S, et al. Distinct effects of annexin A7 and p53 on arachidonate lipoxygenation in prostate cancer cells involve 5-lipoxygenase transcription. Cancer Res. 2006;66(19):9609–9616. doi: 10.1158/0008-5472.CAN-06-1574. [DOI] [PubMed] [Google Scholar]
- 34.Furge LL, Chen K, Cohen S. Annexin VII and annexin XI are tyrosine phosphorylated in peroxovanadate-treated dogs and in platelet-derived growth factor-treated rat vascular smooth muscle cells. J Biol Chem. 1999;274(47):33504–33509. doi: 10.1074/jbc.274.47.33504. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa H, Liyanarachchi S, Davuluri RV, et al. Role of cancer-associated stromal fibroblasts in meta-static colon cancer to the liver and their expression profiles. Oncogene. 2004;23(44):7366–7377. doi: 10.1038/sj.onc.1208013. [DOI] [PubMed] [Google Scholar]
- 36.Yang HS, Matthews CP, Clair T, et al. Tumori-genesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26(4):1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsushima-Nishiu M, Unoki M, Ono K, et al. Growth and gene expression profile analyses of endometrial cancer cells expressing exogenous PTEN. Cancer Res. 2001;61(9):3741–3749. [PubMed] [Google Scholar]
- 38.Wan D, Gong Y, Qin W, et al. Large-scale cDNA transfection screening for genes related to cancer development and progression. Proc Natl Acad Sci U S A. 2004;101(44):15724–15729. doi: 10.1073/pnas.0404089101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo QM, Malek RL, Kim S, et al. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60(21):5922–5928. [PubMed] [Google Scholar]
- 40.Deng Y, Wu X. Peg3/Pw1 promotes p53-mediated apoptosis by inducing Bax translocation from cytosol to mitochondria. Proc Natl Acad Sci U S A. 2000;97(22):12050–12055. doi: 10.1073/pnas.97.22.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 42.Nicholas MK, Lukas RV, Jafri NF, Faoro L, Salgia R. Epidermal growth factor receptor-mediated signal transduction in the development and therapy of gliomas. Clin Cancer Res. 2006;12(24):7261–7270. doi: 10.1158/1078-0432.CCR-06-0874. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19) (q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 44.Rich JN, Hans C, Jones B, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65(10):4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- 45.Mischel PS, Shai R, Shi T, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22(15):2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 46.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. [PubMed] [Google Scholar]
- 47.Colman H, Zhang L, Sulman EP, et al. A multi-gene predictor of outcome in glioblastoma. Neuro Oncol. doi: 10.1093/neuonc/nop007. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]