Abstract
Fur is a transcriptional regulator involved in iron-dependent control of gene expression in many bacteria. In this work we analyzed the phenotype of a fur mutant in Sinorhizobium meliloti, an α-proteobacterium that fixes N2 in association with host plants. We demonstrated that some functions involved in high-affinity iron transport, siderophore production, and iron-regulated outer membrane protein expression respond to iron in a Fur-independent manner. However, manganese-dependent expression of the MntABCD manganese transport system was lost in a fur strain as discerned by constitutive expression of a mntA::gfp fusion reporter gene in the mutant. Thus, Fur directly or indirectly regulates a manganese-dependent function. The data indicate a novel function for a bacterial Fur protein in mediating manganese-dependent regulation of gene expression.
The intracellular concentrations of transition metals such as Fe, Mn, Co, Cd, Ni, and Zn are precisely controlled through the regulation of its transport across the membrane (21, 22). The transport of iron is the best studied transition metal uptake system in bacteria. As no relevant efflux systems are known for iron, it has been hypothesized that core regulation must be at the acquisition level. Bacteria have evolved highly efficient mechanisms to acquire this metal in naturally occurring iron-restricted conditions. Transport of ferric siderophores and heme are two widespread strategies for iron acquisition when the metal is limiting. In gram-negative bacteria, siderophore- or heme-based iron transport systems comprise three main components: iron-repressed outer membrane proteins involved in recognition and transport of the iron-containing compounds; the TonB-ExbB-ExbD complex to couple the proton motive force with outer membrane transport events; and ATP-binding cassette (ABC) transporters present in the inner membrane. ABC transporters involved in high-affinity iron acquisition systems are also present in gram-positive bacteria. Since these bacteria do not possess an outer membrane, lipoproteins analogous to periplasmic binding proteins play a role similar to that of iron-regulated outer membrane proteins (2, 5).
More recently, the physiological significance of manganese homeostasis has received attention (19, 22). Neither specific outer membrane proteins nor specific chelators for manganese (as siderophore for iron) seem to be required for manganese acquisition. Bacterial manganese transport systems identified so far comprise: the natural resistance-associated macrophage protein (NRAMP)-type transporters (mntH); the ABC-type transporters mntABC, scaABC, and sitABC; and the P-type ATPase transporter mntA (mntP) found only in Lactobacillus plantarum (23). The SitABC/D operon was originally reported as being involved in iron uptake, but recent studies have clearly defined this system as a manganese transporter (24, 34). Accordingly, Que and Helmann (40) and Kehres and Maguire (25) proposed to rename this operon as MntABC/D.
Fur is a global regulator of iron homeostasis in numerous bacteria (8, 17). This protein senses the intracellular ferrous ion concentration, through the formation of a Fur-Fe2+ complex, which in turn interacts with specific DNA targets in the promoters of iron-repressed genes. Presumptive fur genes have been found in the genome of some gram-positive bacteria and in almost all gram-negative bacteria.
The Fur family comprises the classical Fur, Zur (a Zn uptake regulator), PerR (an oxidative stress response regulator), and Irr (an iron response regulator involved in heme biosynthesis) (6, 11, 14, 32). In Bacillus subtilis, Staphylococcus aureus, Escherichia coli, and Salmonella enterica, a different metalloprotein, MntR, has been described as a central regulator of manganese homeostasis (18, 24, 33, 39). In B. subtilis MntR has a dual role: under high-manganese conditions it represses the expression of the mntH and mntABCD genes, but under low-manganese conditions it acts as a positive regulator of the MntABCD transporter (39). In E. coli and S. enterica, the expression of mntH is repressed by manganese and iron. MntR mediates manganese response in both bacteria, whereas MntR and Fur are involved in iron response of mntH gene in E. coli (24, 33). To our knowledge no MntR homologs have been found in α-proteobacteria.
One of the most relevant physiological differences between iron and manganese is that while iron is potentially toxic since it is able to catalyze a Fenton-type reaction producing highly noxious reactive oxygen species (O2−, H2O2, ·OH), manganese contributes to the detoxification of these reactive oxygen species. These facts led to the proposal that cells have to ensure not only specific intracellular levels for each metal but also an accurate balance between these two metals. Therefore, it is not surprising to find sophisticated mechanisms of coordination of iron sensing, manganese sensing and oxidative stress response such as the ones reported between PerR/MntR/Fur in S. aureus and B. subtilis or OxyR/MntR/Fur in S. enterica (18, 22, 30).
Sinorhizobium meliloti is an α-proteobacterium that belongs to the Rhizobiaceae family. Bacteria known generically as rhizobia can establish symbiotic associations with host plants to fix nitrogen within the plant. Bona fide Fur homologs have been identified in two genera of rhizobia: Bradyrhizobium (FurBj) and Rhizobium (FurRl) (13, 46). FurBj is able to complement an E. coli fur mutant and binds to a canonical DNA binding element called Fur box (13). Moreover, the rate of Fe uptake is higher in a Bradyrhizobium japonicum furBj mutant than in the wild-type strain when cells are grown in high-iron medium indicating that iron uptake is normally repressed by Fur (13). Unexpectedly, in Rhizobium leguminosarum iron regulation of some iron uptake genes is mediated by RirA rather than by Fur (43, 46). Besides, in silico studies indicate that Fur is absent in the Mesorhizobium loti genome. These facts suggest that the pattern of iron uptake regulation is not the same in all genera of rhizobia.
S. meliloti 1021 genome carries three fur-like putative genes: fur, irr, and zur. Iron-dependent repression of hemin and rhizobactin 1021 receptors has been described previously, but the regulator involved in this response is not known (3, 28). Concerning Mn acquisition, the presence of one ABC transporter (sitABC, renamed here as mntABC) and one presumptive NRAMP-type transporter (Sma1115) were identified by physiological or bioinformatic studies (35).
In this work we study the regulation of the expression of the hemin receptor ShmR and manganese transport system MntABCD in wild-type and fur mutant backgrounds. We found that S. meliloti Fur protein (FurSm) is involved in manganese regulation of mntA but not in iron regulation of shmR. Moreover, fur mutants are not affected in siderophore production or in the expression of some iron-regulated outer membrane proteins.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
Bacteria and plasmids used in this study are listed in Table 1. S. meliloti strains were grown at 30°C in tryptone-yeast extract medium (TY) (4) or in defined minimal medium (M3) (3). Metal chelated medium was obtained by supplementation with ethylenediamine-di-o-hydroxyphenylacetic acid (EDDHA) (TYE or M3E). Micromolar concentrations of added EDDHA are indicated throughout this work as subscripts. FeCl3 and MnCl2 were added at a final concentration of 50 μM as indicated. E. coli strains were grown at 37°C in Luria broth. When required kanamycin (50 μg/ml), ampicillin (50 μg/ml), spectinomycin (100 μg/ml), or streptomycin (100 μg/ml) was added to the medium. Gentamicin at 20 or 40 μg/ml was used for E. coli or S. meliloti, respectively, grown in solid medium.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| 1021 | Wild type, Str | 29 |
| 242 | Wild type, Str | 9 |
| Mf1 | 1021 furSm::Ω cassette | This work |
| Mf2 | 242 furSm::Ω cassette | This work |
| 1021(pShm) | 1021 containing plasmid pShm | This work |
| 242 (pShm) | 242 containing plasmid pShm | This work |
| 1021 (pMan) | 1021 containing plasmid pMan | This work |
| 242 (pMan) | 242 containing plasmid pMan | This work |
| Mf1 (pShm) | Mf1 containing plasmid pShm | This work |
| Mf2 (pShm) | Mf2 containing plasmid pShm | This work |
| Mf1 (pMan) | Mf1 containing plasmid pMan | This work |
| Mf2 (pMan) | Mf2 containing plasmid pMan | This work |
| 1021(pOT) | 1021 containing plasmid pOT1 | This work |
| 242(pOT) | 242 containing plasmid pOT1 | This work |
| Mf1(pLAFur,pMan) | Mf1 containing both plasmids pLAFur and pMan | This work |
| E. coli DH5α | supE44 ΔlacU169(φ80lacZΔM15)hsdR17 recA1 gyrA96 thi-1 relA1 | 15 |
| Plasmids | ||
| pBluescriptSK (pBSK) | Cloning vector, Apr | Stratagene |
| pΩ45 | Plasmid containing Ω Spr Str cassette | 8 |
| pWS233 | Suicide vector in rhizobia, Gmr | 41 |
| pOT1 | Wide-host-range gfp-UV promoter-probe plasmid, Gmr, derivative of pBBR1 | 1 |
| pOT2 | Wide-host-range gfp-UV promoter-probe plasmid, Gmr, derivative of pBBR1 | 1 |
| pRK2013 | Helper plasmid, Kmr | 10 |
| pFurSm | 1.25-kb PCR fragment carrying the furSm gene and flanking regions cloned in pBSK | This work |
| pBSKfurSmΩ | 0.8-kb furSm ends and flanking regions carrying an Ω cassette in place of furSm gene cloned in pBSK | This work |
| pWSfurSmΩ | pWS233 carrying the 0.8-kb fragment containing an Ω cassette in place of fursm | This work |
| pShmR | pBSK with a 2.8-kb PCR fragment containing shmR, its presumptive promoter, and flanking regions | This work |
| pShm | pOT1 with a 650-bp HindIII, SalI fragment containing the presumptive shmR promoter, Gmr | This work |
| pMan | pOT2 with a 750-bp KpnI, PstI fragment containing the presumptive mntA promoter, Gmr | This work |
| pLAFR3 | Low-copy-number, broad-host-range cosmid derivative of pRK290 (IncP1) PLAFR3 containing furSm and its presumptive promoter | 42 |
| pLAFur | This work |
Phylogenetic analysis.
Fur and Irr sequences were obtained from GenBank for all the species considered in this study. Phylogenetic trees based on amino acid sequences of Fur and Irr were inferred with maximum-parsimony and neighbor-joining (NJ) (p distance matrix) analyses, using MEGA2 (25). Confidence in topologies was assessed using bootstrapping (100 replicates). Trees were rooted using the Fur-like sequence of M. loti.
Primers.
For amplification of the 1.25-kb fragment containing the furSm gene of S. meliloti 1021 and flanking regions, the following pair of primers (F1 and F2) were used: 5′-ATATTCGGGAAGGGCTTCAG-3′ (forward primer) and 5′-TGGAAGAACCTTTCGAACCA-3′ (reverse primer). An inverted PCR was performed to amplify furSm ends and flanking regions contained in pBSK using primers P3 and P4 (5′-AACTCGGCTATGACCTGGTG-3′ and 5′-ATCCGATTCTTGCTCTGGCTC-3′, respectively) as forward primer and as reverse primer.
The 2.8-kb S. meliloti 1021 DNA fragment containing shmR was amplified by using as forward primer 5′-ATTCGTCTCGCTCCGTAAAA-3′ (S5) and as reverse primer 5′-AGAAACGCGACGATCAAAAT-3′ (S6).
Cloning and mutagenesis.
Digestion with restriction endonucleases, DNA ligation, and minipreparation of plasmid DNA were performed essentially as described by Sambrook et al. (40). DNA fragments containing furSm or shmR genes were obtained from PCR amplification of total DNA isolated from S. meliloti strain 1021, and the blunt-end fragments were ligated with EcoRV-digested pBluescript SK. Cloning was confirmed by restriction mapping and sequencing.
To replace the furSm gene with an omega cassette, the following strategy was used. An inverted PCR was performed with Pfu polymerase and primers complementary to furSm extremities (about 20 to 40 bp each) using pFursm as target DNA. The PCR-amplified fragment contained linear pFurSm without the internal part (360 bp) of furSm. This PCR product was ligated with the cassette obtained by digestion of pΩ45 with SmaI. The resulting pBSKfurSmΩ construction was digested to confirm the correct insertion of the cassette. Fragments obtained by EcoRI/SalI double digestion of pBSKfurSmΩ and of pWS233, were ligated. The new plasmid was mobilized into S. meliloti strains 1021 and 242 by triparental mating by using DH5α(pRK2013) as a helper strain (10). Spr Gms colonies were selected and genomic DNAs were isolated to check gene replacement by PCR using F1 and F2 primers.
Plasmid pLAFur for complementation of fur mutations was generated by cloning the 1,248-bp EcoRI/HindIII fragment of pFurSm containing wild-type furSm gene in pLAFR3 digested with EcoRI and HindIII.
Reporter fusions.
A 650-bp HindIII, SalI fragment of pShmR containing the putative promoter of shmR was subcloned in the appropriate orientation, upstream of the gfp reporter gene of pOT1, a low-copy-number plasmid able to replicate in rhizobia (1). The new plasmid was named pShm. Plasmid pMan was constructed by ligation of a 750-bp KpnI, PstI fragment of pFurSm containing the presumptive promoter of mntA with KpnI-, PstI-digested pOT2 plasmid. Cloning was confirmed by restriction mapping. Plasmids obtained were transferred into S. meliloti wild-type and mutant strains by triparental mating using pRK2013 as a helper plasmid.
GFP-UV expression.
Qualitative green fluorescent protein-ultraviolet (GFP-UV) expression of cultures grown on solid M3 or on TY medium was evaluated by visualization of plates under UV light. For quantitative measurements of fluorescence of GFP-UV in cultures, 10 ml of M3, M3 plus 50 μM FeCl3, M3 plus 50 μM MnCl2, or M3 plus 100 μM EDDHA was supplemented with gentamicin at 15 μg/ml, and cultures were inoculated with ca. 108 washed cells of 1021(pOT), 1021(pShm), 1021(pMan), Mf1(pShm), Mf1(pMan), 242(pOT), 242(pShm), 242(pMan), Mf2(pShm), and Mf2(pMan) strains. One hundred and fifty microliters of a 3-day-old culture (of about 5 × 108 CFU/ml) were transferred to microtiter plates (four wells per condition), and fluorescence was evaluated with a 960 plate reader (FLUOstar OPTIMA; BMG Labtechnologies) using a 390-nm (10-nm bandwidth) excitation filter and 520-nm emission filter (10-nm bandwidth). Cell optical density at 620 nm (OD620) was measured. Quantitative relative fluorescence was determined according the method of Allaway et al. (1) as fluorescence emission at 520 nm/OD620.
Siderophore production and outer membrane protein expression.
Dihydroxamate siderophore production was evaluated by the ferric perchlorate method (7). For these studies, cultures of wild-type and mutant strains were grown in liquid M3 minimal medium with different metal availability. Stationary-phase cultures were centrifuged at 10,000 × g for 10 min at 4°C, supernatants were mixed with equal volumes of 10 mM FeCl3 and 0.2 M HClO4, and the absorbance at 450 nm was measured after 10 min of incubation. Deferoxamine mesylate (Desferal; Novartis Co.), a trihydoxamate siderophore, was used as a standard. Values were recorded as micromolar concentrations of rhizobactin 1021 of a supernatant corresponding to a cell culture of OD620 equal to 1. Independent assays were performed at least three times. Outer membrane preparations were carried out as previously described (3). Briefly, cells obtained from 50 ml of saturated cultures were disrupted by three passages through a French pressure cell at 25,500 lb/in2 and incubated for 90 min with a solution containing 0.4 mg of DNase per ml, 0.4 mg of RNase per ml, and 10 mg of lysozyme per ml and for an additional 60 min with 0.75% N-laurylsarcosine at room temperature. Cell debris were removed by centrifugation, and the outer membrane enriched supernatant was centrifuged for 2 h at 60,000 × g. Outer membrane proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver stained.
Plant assays.
S. meliloti 1021 and 242 mutants were screened for their symbiotic phenotype on alfalfa plants. Bacteria were grown to early stationary phase on TY-liquid medium. Washed cells were used to inoculate Medicago sativa cv. Creola plants at a final concentration of 5 × 106 CFU per plant. Alfalfa plants were grown aseptically in 15 ml of N-free Jensen medium (45) solidified with agar at 15 g/liter. Plants were maintained at 21 ± 2°C in a controlled light room with a photoperiod of 12 h. At least five tubes (two plants per tube) were used for each strain's inoculation. The complete experiment was independently repeated twice. Plant dry weights 2 months after inoculation were determined.
RESULTS
In silico studies.
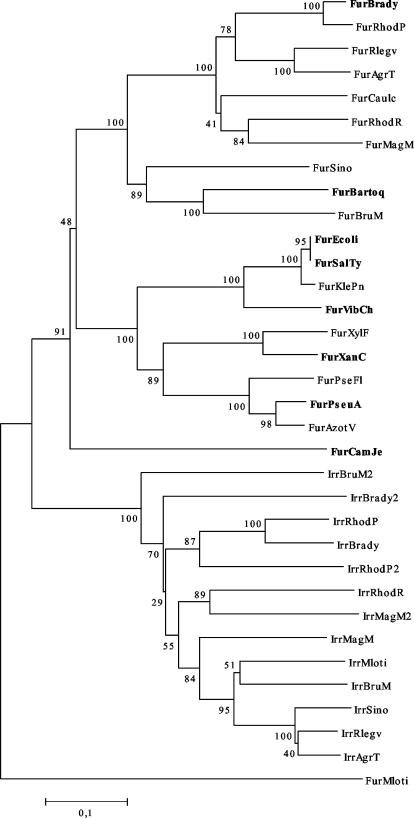
The S. meliloti 1021 genome reveals a presumptive fur gene upstream of the mntABC operon, but in the opposite orientation. We wanted to compare the sequence similarity between the Fur protein deduced from the S. meliloti genome and other presumptive proteins that have been assigned in different genomes as classical Fur proteins. A phylogenetic tree of Fur proteins and the related Irr protein, clearly differentiate these proteins in two well-defined groups (Fig. 1). This organization indicates that S. meliloti Fur (FurSm) protein is more related to Fur proteins from δ-proteobacteria or ɛ-proteobacteria than to a Fur-like protein from the same α-proteobacterium group. Although only the NJ tree is shown; the tree obtained by maximum parsimony with a heuristic search factor of 2 shows the same overall topology. The tree also shows that α-proteobacterium Fur proteins, with the exception of M. loti Fur, cluster separately from the δ-proteobacteria and the ɛ-proteobacterium examined, which is consistent with the apparent divergent evolutionary trajectories of these bacteria.
FIG. 1.
Phylogenetic analysis of Irr and Fur protein sequences. Trees were constructed by NJ based on p distances using MEGA2. The bootstrap values are shown on the branches and indicate the number of times out of 100 replications. M. loti Fur sequence (FurMloti NP_104436) was used as an outgroup to root the tree. Fur protein sequences: FurBrady (B. japonicum NC_004463.1), FurRhodP (Rhodopseudomonas palustris NZ_AAAF01000001.1), FurRlegv (R. leguminosarum bv. viciae O07315), FurAgrT (Agrobacterium tumefaciens NP_531060.1), FurCaulc (Caulobacter crescentus NP_418876.1), FurRhodR (Rhodospirillum rubrum |ZP_00013164.1), FurMagM (Magnetospirillum magnetotacticum ZP_00052636.1), FurSino (S. meliloti NP387131), FurBartoq (Bartonella quintana AAL04498), FurBruM (Brucella melitensis AAB81452), FurEcoli (E. coli K-12 P06975), FurSalTy (S. enterica serovar Typhimurium NP459678), FurKlePn (Klebsiella pneumoniae P45599), Fur VibCh (Vibrio cholerae P33087), FurXylF (Xylella fastidiosa NP779572), FurXanC (Xanthomonas campestris NP_636842), FurPseFl (Pseudomonas fluorescens O68563), FurPseAu (Pseudomonas aeruginosa Q03456), FurAzotV (Azotobacter vinelandii AAN03807), FurCamJe (Campylobacter jejuni P48796). Irr protein sequences: IrrBruM2 (B. melitensis NP_540872.1), IrrBrady2(B. japonicum NP_767856.1), IrrRhodP (R. palustris ZP_00010023.1), IrrBrady (B. japonicum NP_767408.1), IrrRhodP2 (R. palustris ZP_00011887.1), IrrRhodR (R. rubrum ZP_00013142.1), IrrMagM2(M. magnetotacticum ZP_00054559.1), IrrMagM (M. magnetotacticum ZP_00052390.1), IrrMloti (M.loti NP_106209.1), IrrBruM (B. melitensis AAO89498), IrrSino (S. meliloti NP384355), IrrRlegv (R. leguminosarum bv. viciae CAD37806), IrrAgrT (A. tumefaciens C58 E97377). Experimentally verified Fur proteins (13, 16, 26, 27, 31, 36, 44) are indicated in boldface type.
Expression of iron-repressed outer membrane proteins and siderophore production respond to iron in a Fur-independent manner.
Since Fur has been described as a central iron-sensing metalloregulator, we wanted to test its role in the regulation of some iron-repressed functions in Sinorhizobium. For this purpose we constructed a furSm knockout mutant in S. meliloti by allelic exchange of the furSm gene for a spectinomycin resistance cassette. The expected replacement by the omega cassette of the furSm genes of S. meliloti strains 1021 and 242 was confirmed by PCR using primers F1 and F2, described in Materials and Methods. A unique PCR-amplified fragment of 1,250 bp was obtained for 1021 and 242 wild-type S. meliloti strains, and a unique fragment of about 3,100 bp was obtained for Mf1 and Mf2 mutants.
Previously, Battistoni et al. (3) showed that S. meliloti strain 242 expresses three outer membrane proteins with estimated molecular masses of 80, 82, and 92 kDa in response to iron limitation. We compared the induction of these proteins in S. meliloti strain 242 and in the fur mutant strain Mf2 grown in iron-supplemented or iron-chelated medium. Expression of the outer membrane proteins was iron repressed in both the wild-type and fur strains, indicating that the iron responsiveness was Fur independent (data not shown).
Under iron-restricted conditions S. meliloti strains 1021 and 242 are able to synthesize rhizobactin 1021, a dihydroxamate-type siderophore (34; E. Fabiano, unpublished data). The presence of this siderophore in culture supernatants was visualized by the development of a reddish color in acidic medium with a ferric perchlorate reagent (7). The rhizobactin 1021 production was estimated for cell cultures of an OD620 equal to 1 (micromolar concentration/OD620). As shown in Table 2, when the fur mutant strain Mf1 and parent strain S. meliloti 1021 were grown in M3E100 iron-chelated medium, rhizobactin 1021 production was 135 ± 30 μM and 120 ± 4 μM rhizobactin/OD620, respectively. Siderophore production of cultures grown on 50 μM FeCl3 supplemented M3 medium were less than 30 μM/OD620. Similar results were obtained for S. meliloti 242 and Mf2 strains. These results indicate that FurSm is not involved in iron regulation of siderophore production in S. meliloti.
TABLE 2.
Effect of Mn2+ and Fe3+ in siderophore production of S. meliloti wild-type strains and fur mutants
| S. meliloti strain | Mean rhizobactin 1021 concn ± SD (nM) of cultures grown ina:
|
||||
|---|---|---|---|---|---|
| M3Fe50 | M3 | M3Mn50 | M3E100 | M3E100Mn50 | |
| 1021 | 22 ± 1 | 61 ± 6 | 58 ± 4 | 120 ± 24 | 144 ± 32 |
| Mf1 | 24 ± 8 | 78 ± 8 | 68 ± 9 | 135 ± 30 | 106 ± 18 |
| 242 | 33 ± 4 | 109 ± 10 | 95 ± 10 | 173 ± 18 | 132 ± 30 |
| Mf2 | 26 ± 1 | 94 ± 10 | 70 ± 20 | 160 ± 42 | 160 ± 38 |
Concentration of rhizobactin 1021 in the supernatant was estimated under the assumption that 1 mol of the trihydroxamate deferoxamine mesylate is equivalent to 1.5 mol of the dihydroxamate rhizobactin 1021. Results were normalized to an optical density of 1. The data shown are based on three separate experiments.
ShmR is a hemin-binding outer membrane protein expressed in iron-restricted cultures of S. meliloti. In order to study factors involved in the regulation of this protein, we studied the fluorescence produced by S. meliloti 1021 and 242 wild-type strains, as well as by the fur mutants containing pShm, a pOT1 derived plasmid that carried the presumptive shmR promoter upstream the gfp-UV reporter gene. As shown in Fig. 2 and Fig. 3, fluorescence corresponding to the expression of the green fluorescent protein was clearly observed in S. meliloti wild-type strains containing pShm when they are grown on iron-limited medium. However, the addition of 50 μM FeCl3 repressed GFP-UV expression. This genetic approach confirms previous biochemical observations showing that ShmR expression is iron-regulated (3). Interestingly, the same iron-dependent regulation of GFP-UV expression was also observed with the fur mutants containing pShm. No significant differences were detected between S. meliloti 1021(pShm) and Mf(pShm) strains, indicating that FurSm is not involved in iron regulation of ShmR expression. Similar results were obtained with S. meliloti strain 242.
FIG. 2.
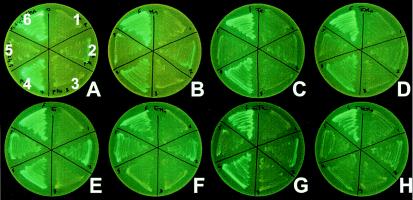
GFP-UV expression of shmR-gfp and mntA-gfp transcriptional fusions in S. meliloti 242 wild-type strain and in a fur mutant (Mf2). Strains used were 242(pOT) (sectors 1), 242(pShm) (sectors 2), 242(pOT) (sectors 3), 242(pMan) (sectors 4), Mf2(pShm) (sectors 5), and Mf2(pMan) (sectors 6). Growth media shown are M3 (A), M3Mn50 (B), M3Fe50 (C), M3Fe50Mn50 (D), M3E100 (E), M3E100Mn50 (F), M3E100Fe50 (G), and M3E100Mn50Fe50 (H).
FIG. 3.
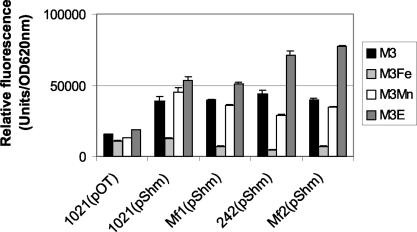
In vivo expression of shmR promoter in S. meliloti 1021 and S. meliloti 242 and in the fur mutants Mf1 and Mf2. Cultures were grown in M3 defined medium supplemented with 50 μM FeCl3 (M3Fe), 50 μM MnCl2 (M3Mn), or 100 μM EDDHA (M3E). Specific fluorescence was determined in arbitrary fluorescence units per units of OD620. The data shown represent the average + standard deviation (error bars) of four measures from one assay. The complete assay was repeated twice.
The expression of the mntA gene involved in manganese acquisition is regulated by manganese in a Fur-dependent manner.
We previously showed that the mntABC operon is involved in manganese acquisition in S. meliloti 242 (35). This operon is adjacent to the furSm gene but in the opposite orientation in the S. meliloti 1021 genome. Here, we asked whether mntA expression could be regulated by Fur. A fragment containing the presumptive mntA promoter was cloned upstream of the gpf reporter gene in the plasmid pOT2 generating pMan. As shown in Fig. 2 and 4, S. meliloti wild-type strains containing pMan plasmid fluoresced in minimal or rich medium without added manganese. The addition of 50 μM MnCl2 to the medium inhibited, but did not abolish, fluorescence expression. These results indicate that the mntA gene responds to manganese and that this control is at the transcriptional level. S. meliloti fur mutants, Mf1 and Mf2, containing the reporter plasmid pMan expressed GFP-UV strongly in minimal and rich medium regardless of manganese addition. However, when the wild-type fur gene was introduced in trans in Mf1(pMan) strain, manganese-dependent expression of MntA was restored to the same level as in the wild-type strain (Table 3). Thus, the defect in the fur gene is the cause of the observed phenotype. Taken together, these results clearly show that the FurSm protein regulates manganese-dependent mntA repression.
FIG. 4.
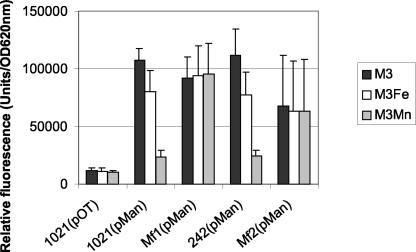
In vivo expression of mntA promoter in S. meliloti 1021 and S. meliloti 242 and in the fur mutants Mf1 and Mf2. Cultures were grown in M3 defined medium supplemented with 50 μM FeCl3 (M3Fe) or 50 μM MnCl2 (M3Mn). Specific fluorescence was determined in arbitrary fluorescence units per units of OD620. The data shown represent the average + standard deviation (error bars) of four measures from three independent assays.
TABLE 3.
fur gene complements in trans the Mn-derepressed phenotype of Mf1
| Strain | Relative fluorescencea (mean ± SD)
|
|
|---|---|---|
| M3 | M3Mn | |
| 1021(pMan) | 107 ± 10 | 23 ± 6 |
| Mf1(pMan) | 91 ± 18 | 95 ± 26 |
| Mf1(pLAFur,pMan) | 75 ± 16 | 24 ± 10 |
Relative fluorescence is expressed as arbitrary units (103) per unit of OD620. Data are based on three separate experiments.
We could not observe a decrease in fluorescence from the mntA-gfp fusion on plates supplemented with 50 μM FeCl3. However, quantification of fluorescence in the liquid assay revealed a modest repression upon the addition of FeCl3. This repression was not observed in the fur mutants. Thus, Fe has a modest effect on mntA expression that is dependent on Fur.
Expression of the iron-regulate ShmR outer membrane protein and siderophore production are not repressed by manganese.
In order to evaluate if manganese could functionally mimic the effect of iron in regulation of some iron-repressed function, shmR expression and siderophore production were determined in liquid medium supplemented with 50 μM MnCl2. As shown in Table 2 no significant differences were obtained in rhizobactin 1021 concentration in manganese-supplemented medium (M3 versus M3Mn50 and M3E100 versus M3E100Mn50). Conversely, addition of 50 μM FeCl3 to M3 medium produced a clear repression on siderophore production.
In vivo expression of the shmR promoter was evaluated, and the results obtained are shown in Fig. 3. Comparison of GFP-UV fluorescence obtained for 1021(pShm) and Mf1(pShm) cultures grown in M3 or M3Mn50 demonstrate that this promoter is regulated by iron but not by manganese. Similar results were obtained for 242(pShm) and Mf2(pShm) (data not shown). Therefore, Mn2+ cannot substitute for Fe2+ in regulation of shmR.
Symbiotic phenotype of fur mutants.
No significant differences could be detected in plant dry weight or visualization of nodules formed between plants inoculated with wild-type strains or fur mutant strains (data not shown). These results indicate that FurSm expression is not essential for symbiosis with alfalfa in the conditions assayed here.
DISCUSSION
To determine the role of FurSm in S. meliloti, we have carried out a phenotypic study of fur knockout mutants of two S. meliloti strains: S. meliloti 1021 and S. meliloti 242. A Fur homolog was identified from the genome sequence of S. meliloti 1021. Sequence alignments and phylogenetic analysis strikingly assign FurSm into the group of classical Fur proteins (Fig. 1). Interestingly, we found that mntA expression is repressed by manganese and that this repression requires a furSm functional gene. Previously we showed that the MntABCD system is involved in manganese uptake in S. meliloti 242 (35). Fur-mediated regulation of manganese acquisition has been reported in E. coli and S. enterica though this regulator represses mntH gene in an iron-dependent way (24, 33). Our studies do not reveal whether FurSm indirectly regulates manganese-responsive genes or whether it is itself a manganese-sensing regulator. In that sense it is possible that Fur metal specificity would have changed during evolution, as seems to be the case of some DtxR homologs (Tro, Sca, and MntR) (20, 37, 39). Due to the redox properties of Fe, many in vitro DNA-binding studies of Fur are carried out with manganese, indicating that this metal could occupy the iron-binding site. Fur protein sequence comparisons show that the amino acids predicted to be implicated in iron binding are conserved (36). Nonetheless, subtle changes in other positions could lead to relevant changes in metal affinity of the binding site. In view of previous observations of Guedon and Helmann (12), we cannot rule out the possibility that the balance of transition metals differs between bacterial species, and this fact could determine metal regulator selectivity in a particular bacterial background. In that sense, it is interesting that Kehres et al. (24) reported that the intracellular Mn concentration in Enterobacteriaceae could reach levels as high as 10 mM, whereas iron content is usually in the range of some hundred micromolar. Therefore, it is possible that Fur responds to Mn2+ even if it has a higher affinity for Fe2+.
Our results indicate that FurSm appears not to be a global regulator of iron acquisition in S. meliloti. Iron-dependent expression of outer membrane proteins and siderophore production were not affected in S. meliloti fur mutants. In a different genus of rhizobia, Bradyrhizobium, the FurBj protein is involved in the repression of iron uptake under high-iron conditions (13). However, in R. leguminosarum high-affinity iron uptake systems are iron regulated in a Fur-independent manner, indicating the existence of a different mechanism of regulation (46). Surprisingly, no typical fur homolog could be identified in the M. loti genome. Our results together with these previous observations point out that at least in some rhizobial species repression of iron-regulated genes is not exclusively mediated by Fur. In R. leguminosarum a new regulator, RirA, has been described as being involved in iron homeostasis. A rirA homolog (aau3) is also present in the S. meliloti genome, although its function has not been studied.
Acknowledgments
We are very grateful to Federico Battistoni and Francisco Noya for critical suggestions.
E.F. was supported by PEDECIBA and by an ASM International Fellowship to work in the MRO lab. R.P. was supported by CSIC, and M.R.O. was supported by NSF grant MCB-0089928.
REFERENCES
- 1.Allaway, D., N. A. Schofield, M. E. Leonard, L. Gilardoni, T. M. Finan, and P. S. Poole. 2001. Use of differential fluorescence induction and optical trapping to isolate environmentally induced genes. Environ. Microbiol. 3:397-406. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, F. Rodríguez-Quiñones. 2002. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Battistoni, F., R. Platero, R. Durán, C. Cerveñansky, J. Battistoni, A. Arias, and E. Fabiano. 2002. Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti. Appl. Environ. Microbiol. 68:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., and M. Braun. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 529:78-85. [DOI] [PubMed] [Google Scholar]
- 6.Bsat, N., A. Herbig, L. Casillas-Martínez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues; identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 7.Carson, K. C., S. Holliday, A. R. Glenn, and M. J. Dilworth. 1992. Siderophore and organic acid production in root nodule bacteria. Arch. Microbiol. 157:264-271. [DOI] [PubMed] [Google Scholar]
- 8.Escolar, L., J. Pérez-Martín, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabiano, E., P. R. Gill, F. Noya, P. Bagnasco, L. De La Fuente, and A. Arias. 1995. Siderophore-mediated iron acquisition mutants in Rhizobium meliloti 242 and its effect on the nodulation kinetic of alfalfa nodules. Symbiosis 19:197-211. [Google Scholar]
- 10.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guedon, E., and J. D. Helmann. 2003. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 48:495-506. [DOI] [PubMed] [Google Scholar]
- 13.Hamza, I., R. Hassett, and M. R. O′Brian. 1999. Identification of a functional fur gene in Bradyrhizobium japonicum. J. Bacteriol. 181:5843-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamza, I., Z. Qi, N. D. King, and M. R. O′Brian. 2000. Fur-independent regulation of iron metabolism by Irr in Bradyrhizobium japonicum. Microbiol. 146:669-676. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Hantke, K. 1984. Cloning of the repressor protein gene of iron regulated system in E. coli K-12. Mol. Gen. Genet. 197:337-341. [DOI] [PubMed] [Google Scholar]
- 17.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 18.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44:1269-1286. [DOI] [PubMed] [Google Scholar]
- 19.Horsburgh, M. J., S. J. Wharton, M. Karavolos, and S. J. Foster. 2002. Manganese: elemental defence for a life with oxygen? Trends Microbiol. 10:496-501. [DOI] [PubMed] [Google Scholar]
- 20.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 21.Jakubovics, N. S., and H. F. Jenkinson. 2001. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology 147:1709-1718. [DOI] [PubMed] [Google Scholar]
- 22.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J. Bacteriol. 184:3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kehres, D. G., and M. E. Maguire. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27:263-290. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 26.Lam, M. S., C. M. Litwin, P. A. Carroll, and S. B. Calderwood. 1994. Vibrio cholerae fur mutation associated with loss of repressor activity: implications for the structural-functional relationships of fur. J. Bacteriol. 176:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loprasert, S., R. Sallabhan, S. Atichartpongkul, and S. Mongkolsuk. 1999. Characterization of a ferric uptake regulator (fur) gene from Xanthomona campestris pv. phaseoli with unusual primary structure, genome organization, and expression patterns. Gene 239:251-258. [DOI] [PubMed] [Google Scholar]
- 28.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. Ó Cuív, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterisation of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 31.Park, S. Y., K. L. Kelminson, A. K. Lee, P. Zhang, R. E. Warner, D. H. Rehkopf, S. B. Calderwood, J. E. Koehler. Identification, characterization, and functional analysis of a gene encoding the ferric uptake regulation protein in Bartonella species. J. Bacteriol. 183:5751-5755. [DOI] [PMC free article] [PubMed]
- 32.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 33.Patzer, S. I., and K. Hantke. 2001. Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J. Bacteriol. 183:4806-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persmark, M., P. Pittman, J. S. Buyer, B. Schwyn, P. R. Gill, and J. B. Neilands. 1993. Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. J. Am. Chem. Soc. 115:3950-3956. [Google Scholar]
- 35.Platero, R. A., M. Jaureguy, F. J. Battistoni, and E. R. Fabiano. 2003. Mutations in sitB and sitD genes affect manganese-growth requirements in Sinorhizobium meliloti. FEMS Microbiol. Lett. 218:65-70. [DOI] [PubMed] [Google Scholar]
- 36.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 37.Posey, J. E., J. M. Hardham, S. J. Norris, and F. C. Gherardini. 1999. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. USA 96:10887-10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prentki, P. and, H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 39.Que, Q., and J. D. Helmann. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454-1466. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Selbitschka, W., S. Niemann, and A. Pühler. 1993. Construction of gene replacement vectors for Gram-bacteria using a genetically modified sacRB gene as a positive selection marker. Appl. Microbiol. Biotechnol. 38:615-618. [Google Scholar]
- 42.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todd, J. D., M. Wexler, G. Sawers, K. H. Yeoman, P. S. Poole, and A. W. B. Johnston. 2002. RirA, an iron-responsible regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148:4059-4071. [DOI] [PubMed] [Google Scholar]
- 44.Van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. I.B.P. handbook N15. Blackwell, Oxford, United Kingdom.
- 46.Wexler, M., J. D. Todd, O. Kolade, D. Bellini, A. M. Hemmings, G. Sawers, and A. W. B. Johnston. 2003. Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum. Microbiology 149:1357-1365. [DOI] [PubMed] [Google Scholar]