Background: NF-κB essential modulator (NEMO) is the key regulator of NF-κB signaling, but how it performs this regulation is poorly understood.
Results: NEMO is a highly flexible protein that shows ligand-specific conformational change.
Conclusion: Unanchored polyubiquitin binding controls NEMO conformational state and functional activity.
Significance: Our findings provide a detailed mechanistic understanding of NEMO activity and develop the non-degradative role of free polyubiquitin.
Keywords: adaptor protein, allosteric regulation, biophysics, conformational change, polyubiquitin chain, NEMO, high pressure
Abstract
The NF-κB essential modulator (NEMO) is the master regulator of NF-κB signaling, controlling the immune and nervous systems. NEMO affects the activity of IκB kinase-β (IKKβ), which relieves the inhibition of the NF-κB transcriptional regulation machinery. Despite major effort, there is only a very sparse, phenomenological understanding of how NEMO regulates IKKβ and shows specificity in its large range of molecular interactions. We explore the key molecular interactions of NEMO using a molecular biophysics approach, incorporating rapid-mixing stopped-flow, high-pressure, and CD spectroscopies. Our study demonstrates that NEMO has a significant degree of native structural disorder and that molecular flexibility and ligand-induced conformational change are at the heart of the molecular interactions of NEMO. We found that long chain length, unanchored, linear polyubiquitin drives NEMO activity, enhancing the affinity of NEMO for IKKβ and the kinase substrate IκBα and promoting membrane association. We present evidence that unanchored polyubiquitin achieves this regulation by inducing NEMO conformational change by an allosteric mechanism. We combine our quantitative findings to give a detailed molecular mechanistic model for the activity of NEMO, providing insight into the molecular mechanism of NEMO activity with broad implications for the biological role of free polyubiquitin.
Introduction
The NF-κB transcriptional regulation pathway is one of the most important signaling pathways in human biology, with an essential role in the immune and nervous systems (1). Activation of the NF-κB pathway occurs via a large range of signaling inputs. However, the canonical pathway presents a point of convergence, centered on the activity of a complex comprising at least IκB kinase-β (IKKβ)3 and the regulatory protein NF-κB essential modulator (NEMO), termed IKK (2, 3). In this pathway, IKK is considered to be responsible for phosphorylation of IκBα, inducing proteasomal degradation. IκBα sequesters the NF-κB complex in the cytoplasm, and its degradation allows the NF-κB complex to enter the nucleus to support expression of proinflammatory or anti-apoptotic genes (1). NEMO increases IKKβ activity in response to NF-κB canonical pathway stimuli and is one of very few non-redundant components of NF-κB signaling (4). NEMO is therefore thought of as the gatekeeper of NF-κB signaling owing to its role as an adaptor of the many diverse signaling inputs feeding into the NF-κB signaling pathway and is a crucial player in healthy human biology (1, 5). NEMO regulates IKKβ potentially by promoting autophosphorylation/TAK1 phosphorylation of the activation loop, enhancing enzyme activity (6–8), or by ensuring the specificity of IKKβ for IκBα putatively by facilitating the recruitment of IκBα to the kinase (5). However, despite a large amount of research, the molecular mechanism for NEMO regulation of IKKβ remains equivocal.
NEMO binds to unanchored (free) polyubiquitin chains via a specific domain, UBAN (ubiquitin binding to ABIN and NEMO). The UBAN domain has been found to bind linear (Met1) and Lys11-, Lys48-, and Lys63-linked polyubiquitin, but with far higher affinity for the linear linked chains (9–11). The biological role of free polyubiquitin chains represents a new field of study, but already non-covalent polyubiquitin binding is key to understanding IKK activation (8). NEMO activity requires binding of free polyubiquitin (7, 8, 10). IL-1 and TNF do not induce activation of IKKβ in cells expressing NEMO mutations that prevent interaction with Met1- and Lys63-linked polyubiquitin (12, 13). Evidence from single-domain constructs suggests that free polyubiquitin binding induces NEMO conformational change (11). However, there is no evidence for ligand-induced conformational change with the full-length human protein or what conformational change might achieve.
Intrinsic protein structural disorder is the lack of a fixed three-dimensional structure and is increasingly recognized as a functional feature of the native state of proteins and enzymes. For example, disorder has emerged as a key concept required to understand protein-protein interactions (14, 15), allostery (16), ligand promiscuity (16), enzyme activity (17), and cell signaling (18, 19). Structural disorder can give rise to a range of atypical molecular mechanisms, including specificity for multiple interaction partners (16), high-specificity low-affinity binding (20), and fast association and dissociation rates (19). Structurally disordered proteins are best described thermodynamically in the context of a multidimensional free energy landscape, comprising energetic “hills” and “valleys” that mark the thermodynamic boundaries to different conformational states. Disordered and highly dynamic proteins are considered to have a relatively “rugged” free energy landscape, occupying a broad equilibrium of conformational states that are able to interconvert on rapid timescales (21), i.e. proteins comprising a significant degree of structural disorder are highly plastic, and their functional activity is therefore potentially tunable (22). The role of structural disorder is increasingly being recognized for proteins involved in signaling pathways, as the atypical molecular mechanisms arising from structural disorder are able to account for much of the complexity of signaling interactions.
Experimental Procedures
Protein Expression and Purification
The NEMO expression plasmid was purchased from Addgene (catalog no. 11966) and comprises full-length human NEMO in the pET-28a expression vector with an N-terminal His6 tag for purification (13). The plasmid was transformed into a BL21 derivative Escherichia coli cell line, T7 Express (New England Biolabs), and induced overnight with 0.5 mm isopropyl β-d-thiogalactopyranoside at 25 °C. The NEMO protein was purified from lysed cells via nickel affinity chromatography. We purified NEMO from the soluble fraction and achieved high purity primarily by massively overloading the nickel affinity resin and incubating with 1 mm ATP for a prolonged period of time. The incubation with ATP served to remove tightly bound DnaK. Using this method, we typically recovered ∼20% of the total expressed NEMO protein. Purified protein was dialyzed extensively into the required buffer using spectroscopic grade buffer components. IKKβ and IκBα peptides were commercially synthesized by GenScript with a purity of >98%. Polyubiquitin was purchased from Enzo Life Sciences (catalogue no. BML-UW0825-0001).
Static Fluorescence and High-pressure Measurements
Fluorescence emission spectra were monitored on a PerkinElmer Life Sciences LS 50B fluorescence spectrophotometer connected to a circulating water bath. Excitation and emission slit widths were selected to give the best signal-to-noise ratio, typically 5 nm. Spectra were the average of a minimum of three recorded spectra. All experiments were performed in 50 mm Tris-Cl, 50 mm NaCl, and 5 mm DTT (pH 8) unless stated otherwise. High-pressure measurements were made using a high-pressure cell (ISS, Champaign, IL) with a custom fiber optic mounted onto the fluorometer. The cell was connected to a circulating water bath for pressure/temperature (p/T) dependence experiments. In all experiments, the corresponding background signal attributable to emission arising from the buffer components or ligand was subtracted from the data sets.
Stopped-flow Fluorescence and CD Measurements
Stopped-flow fluorescence emission transients were monitored on a Hi-Tech Scientific thermostated stopped-flow apparatus (TgK Scientific, Bradford on Avon, United Kingdom). Typically, three to five transients were recorded for each reported measurement. Transients were fit as described below. CD measurements were made on a thermostated Chirascan spectrophotometer (Applied Photophysics, Leatherhead, UK) using a high-quality quartz cell with a 0.1-mm path length. NEMO and ligands were incubated with 50 μm 8-anilinonaphthalene-1-sulfonic acid (ANS), and the effective ANS concentration did not vary on mixing.
Liposome Preparation and Binding Assay
l-α-Phosphatidylethanolamine and l-α-phosphatidylcholine mixed liposomes were prepared at a 50:50 ratio. 30 mg of total lipid was dissolved in 2 ml of a 1:1 methanol/chloroform mixture. The solvent was evaporated under a nitrogen stream, and residual chloroform was removed by freeze-drying for 4 h, followed by resuspension in buffer (50 mm Tris-Cl (pH 8.0), 50 mm NaCl, and 5 mm DTT) to 1 mg/ml and hydration by vigorous agitation for 1 h. Resuspended lipids were sonicated in a Decon FS100 Frequency Sweep water bath for 90 min and collected by centrifugation at 16,000 × g for 10 min at 4 °C in an Eppendorf F45-30-11 rotor. The supernatant was discarded, and the liposomes were resuspended again in buffer to 1 or 2 mg/ml and used immediately. NEMO in 10 mm Tris-SO4, 20 mm NaCl, and 2 mm β-mercaptoethanol was spun at 16,000 × g for 10 min at 4 °C before mixing, followed by incubation at room temperature for 30 min, and then spun at 20,000 × g for 15 min at 4 °C. The supernatant was discarded, and pellets were resuspended in SDS loading buffer before boiling at 90 °C for 5 min. Samples were analyzed by SDS-PAGE. Protein complexes were incubated at room temperature for 30 min with the liposomes and then pelleted by centrifugation. SDS-PAGE analysis was performed on the pelleted samples under the specific conditions reported in the figure legends.
Results and Discussion
NEMO Is Highly Structurally Disordered in the Native State
Based on the combined results of a range of structure prediction algorithms, the NEMO amino acid sequence predicts major regions of structural disorder comprising 49% of the sequence space (Fig. 1). These predicted regions are found essentially between the solved crystal and NMR structures of truncated NEMO variants (Fig. 1), suggesting that the failure to date to crystallize the full-length human protein is due to a significant fraction of disordered content.
FIGURE 1.
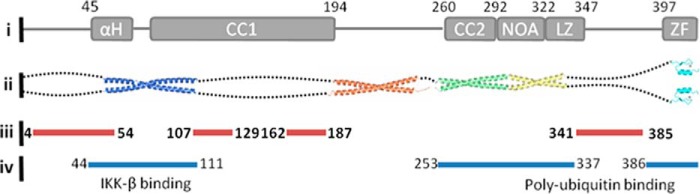
Summary of NEMO structural biology and predicted disordered structure content. i, NEMO domain map. αH, α-helix; CC1 and CC2, coiled coils 1 and 2, respectively; NOA, NEMO-optineurin-ABIN; LZ, leucine zipper; ZF, zinc finger. ii, solved NEMO structures deposited in the Protein Data Bank (codes 3BRV, 3CL3, 2ZVN, and 2JVX). iii, predicted structurally disordered regions of NEMO based on consensus between PONDR-FIT, ANCHOR, IUPred, and SPINE-D. Only regions of >5 amino acids are shown for clarity. iv, binding regions for IKKβ and polyubiquitin.
We experimentally determined the secondary structure composition using far-UV CD spectroscopy (Fig. 2A). The relative proportion of secondary structural content was determined by fitting the experimentally derived data using the CONTIN algorithm via the DichroWeb server (Fig. 2B). From this analysis, NEMO is composed primarily of α-helix (48%) and random coil (41%), with minor fractions of β-sheet (1.5%) and turn (9.5%) content. The percentage of structural disorder in NEMO predicted by far-UV CD correlates with the percentage predicted from the amino acid sequence (Fig. 1): 41 and 49%, respectively. We explored the temperature stability of NEMO secondary structure content via thermal denaturation (Fig. 2, A (inset) and B). We fit the CD signal at 222 nm to Equation 1 for a simple two-state transition,
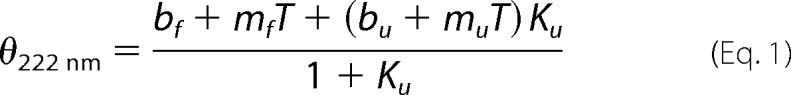 |
where
where m and b are the slope and intercept of the folded (f) and unfolded (u) baseline, respectively; Tm is the melting temperature; and ΔH is the van 't Hoff enthalpy of unfolding at Tm. Fitting the data shown in Fig. 2A (inset) gives Tm = 49.9 ± 1.5 °C and ΔH = 114.8 ± 18.48 kJ/mol. We note that in the region where we conducted our biophysical studies (10–25 °C), we found little if any secondary structural change (1–2%) (Fig. 2B) and no evidence for protein unfolding. Disordered proteins typically display higher melting temperatures compared with ordered structures (23), so our relatively large Tm value can be seen as additional evidence for the significant degree of NEMO structural disorder.
FIGURE 2.
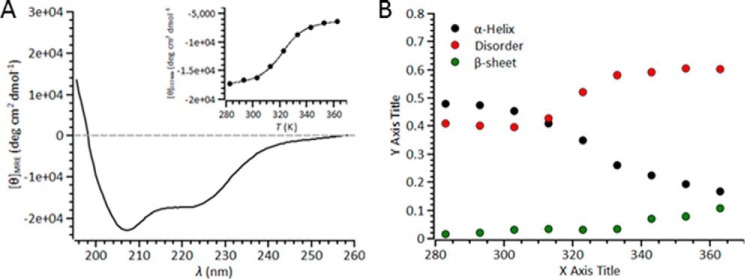
CD spectrum showing that NEMO has a large percentage of disordered structure content. A, far-UV CD spectra of NEMO. The data shown are the raw averaged data from 10 spectra, with the corresponding buffer spectra subtracted. The inset shows the temperature dependence of the signal at 222 nm, fit to Equation 1. B, results of fitting far-UV CD data (contin set 7). The variation in turn content is not shown for clarity. Conditions were 20 mm Tris-SO4 (pH 8), 50 mm NaCl, 2 mm β-mercaptoethanol, and 6 μm NEMO. MRE, mean residue ellipticity; deg, degrees.
ANS is an environmentally sensitive, small-molecule, extrinsic fluorescent dye that binds non-covalently to solvent-exposed non-polar cavities in proteins (24). Upon binding, the quantum yield of the dye increases, and the emission spectrum shows a significant blue shift. These spectral changes are broadly considered to reflect the exposure of hydrophobic residues and a molten globule-like state of the protein (24–27). Molten globules are considered to have some folded character but also a significant degree of structural disorder, representing a third class of protein structure between tightly folded and unfolded. We found that under native conditions (low temperature, no denaturant), NEMO bound to ANS (Fig. 3A). The spectra exhibited a significant increase in ANS emission and a blue shift, giving 513.5 to 465 nm (emission maxima) and 515 to 480 nm (center of spectral mass) for ANS and NEMO + ANS, respectively (Fig. 3A). As such, these data potentially suggest that NEMO adopts a native molten globule-like state. This is consistent with both structure prediction (Fig. 1) and our CD data (Fig. 2), which suggest a nearly equal fraction of α-helical and disordered content.
FIGURE 3.
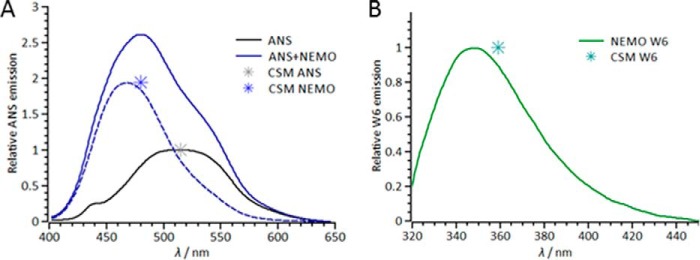
Fluorescence probes of NEMO conformational state. A, fluorescence emission spectra attributable to ANS emission in the presence (solid blue line and dashed blue line (deconvolved spectrum)) and absence (solid black line) of NEMO. B, fluorescence emission spectrum of the single NEMO Trp residue (Trp6 (W6)). The center of spectral mass (CSM) is shown as an asterisk at the emission maxima. Conditions were 50 mm Tris-Cl (pH 8), 50 mm NaCl, 5 mm DTT, and 6 μm NEMO at 10 °C.
Tryptophan fluorescence emission is a common reporter of the conformational state of proteins, being sensitive to the degree of solvent exposure but also proximity to the peptide bond (28). NEMO has a single native Trp residue (position 6) located at the N terminus near the IKKβ-binding region and predicted to be in a region of structural disorder (Fig. 1). By inference, Trp6 therefore reports on the conformational state of the IKKβ-binding region. The emission spectrum of Trp6 (Fig. 3B) is consistent with an extended, solvent-exposed Trp residue, having a heavily red-shifted center of spectral mass (359 nm) and emission maximum (348.5 nm). This is consistent with the inference that Trp6 is in a solvent-exposed, disordered environment.
Our CD and fluorescence data combined therefore suggest that, in our hands, NEMO is a native molten globule protein with a “core” of folded (primarily helical) structure and a significant degree of disordered content. We suggest that our preparations reflect the native state of NEMO primarily because they are from the soluble fraction, are very thermally stable, and bind to known ligands (see below). Formally assigning a protein as a native molten globule is challenging. In the case of NEMO, we argue this is warranted, given that, in this state, NEMO binds to functionally relevant ligands, and this induces NEMO conformational change (see below).
Probing the NEMO Free Energy Landscape
The extrinsic (ANS) and intrinsic (Trp6) fluorescence probes described above are potentially excellent metrics of native state NEMO conformational change. To demonstrate this, one can alter the conformational state of the protein and observe if there is a change in fluorescence reporter signal. Hydrostatic pressure is a powerful tool that can be used to perturb the equilibrium of protein conformational states, acting on a pre-existing equilibrium without altering the internal energy of the system. Moreover, the thermodynamic conjugate of pressure is volume, meaning that this physical perturbation connects directly to protein structure and conformational change (29).
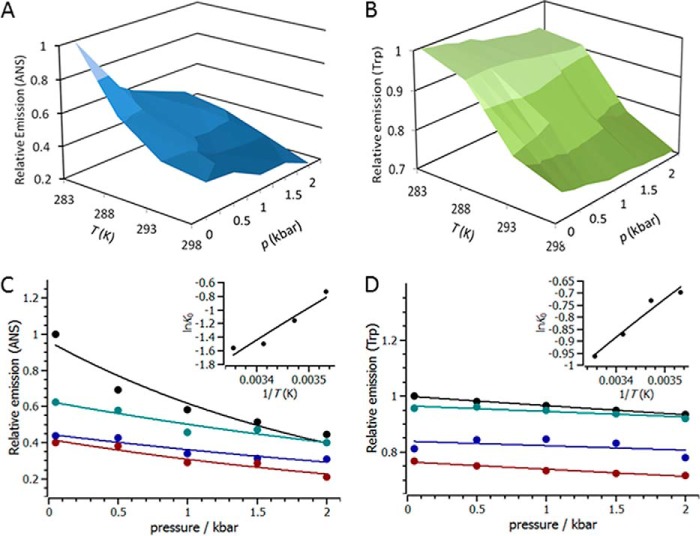
We turn to combined p/T dependence studies (30–32) to demonstrate the validity of these probes and also to explore whether NEMO is able to adopt a range of native equilibrium conformational states. Varying both pressure and temperature perturbs the equilibrium of protein conformational states but also allows thermodynamic information on this equilibrium to be extracted. Fig. 4 shows the p/T dependence of both the ANS (panel A) and Trp6 (panel B) signals across a limited p/T range. Based on our CD data (Fig. 2), the temperature range used is nondenaturing, with no significant secondary structural change, and we do not expect significant denaturation up to pressures of 2 kilobars.
FIGURE 4.
Pressure and temperature dependence of NEMO ANS (A and C) and Trp6 (B and D) emission. The specific conditions are given under “Experimental Procedures.” The relative ANS emission reflects the relative change in the ith conformational state, as given in Equation 3. The solid lines are the fits to Equation 3. The data are the pressure dependence at 10 °C (red), 15 °C (blue), 20 °C (green), and 25 °C (black). The insets show the temperature dependence of the extracted K0 values. The solid line is the fit to the van 't Hoff equation. Conditions were 50 mm Tris-Cl (pH 8), 50 mm NaCl, 5 mm DTT, and 6 μm NEMO.
In Fig. 4 (A and B), we see a significant decrease in the relative emission intensity of the fluorophores with both increasing pressure and temperature, i.e. by inference, the NEMO hydrophobic residues become more buried in the protein's interior across the pressure and temperature range, and Trp6 becomes more solvent-exposed. Based on our CD data, there was no significant change in secondary structure over the temperature range studied, so the observed conformational changes are most likely at the tertiary level. The pressure dependence data can be adequately fit to a simple physical model that implies a single transition between two states with changing hydrostatic pressure (Equation 3),
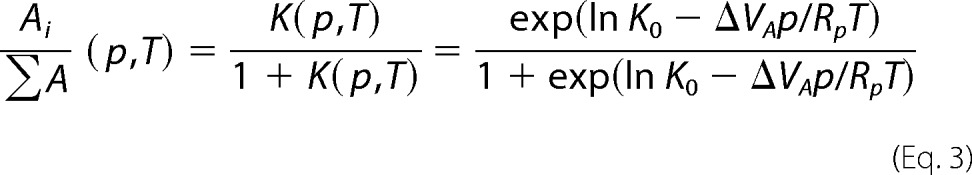 |
where Rp = 83.13 cm3/bar/mol/K when p is measured in bar, K0 is the equilibrium constant for the change in the relative population of the ith conformational state (A) extrapolated to 0 bar, and VA is the apparent difference in the volume associated with this equilibrium transition. Fits of Equation 3 to the pressure dependence data are shown in Fig. 4 (C and D). The temperature dependence of K0 is shown in the insets in Fig. 4 (C and D) and is given in Table 1. K0 can be fit to the van 't Hoff equation to give the enthalpy and entropy changes associated with NEMO conformational change (Fig. 4, C and D; and Table 1).
TABLE 1.
Parameters extracted from fitting combined NEMO p/T data for ANS and Trp6 emission
| ANS | Trp6 | |
|---|---|---|
| K0 | 0.48 ± 0.03 | 0.5 ± 0.01 |
| ΔVA (cm3/mol) | 10.4 ± 1.7 | 0.81 ± 0.03 |
| ΔH (kJ/mol) | 39.7 ± 7.9 | 13.2 ± 2.0 |
| ΔS (kJ) | 0.15 ± 0.02 | 0.05 ± 0.01 |
As shown in Table 1, NEMO conformational change is exothermic, with a positive activation volume for both ANS and Trp6 emission. The extracted ΔH values for ANS and Trp6 emission are significantly smaller than that for the unfolding of NEMO monitored via CD (Fig. 2A, inset): 39.7 ± 7.9 and 13.2 ± 2.0 kJ/mol versus 114.8 ± 18.48 kJ/mol, respectively. These data are then consistent with our assertion that the pressure and temperature range studied is non-denaturing, instead reflecting the transition between natively accessible conformational states. That the extracted ΔH values are different for Trp6 and ANS emission is not surprising because these fluorophores report on different aspects of NEMO structural change. ANS emission is a global metric of NEMO conformational change, and Trp6 emission is very local and specific to the N-terminal environment. These data are therefore evidence that both ANS emission and Trp6 emission are excellent reporters of NEMO conformational change and are able to quantitatively detect variation in the equilibrium of NEMO conformational states.
Pressure acts to perturb a pre-existing equilibrium of conformational states. Combined with our CD data, which show a very significant fraction of structural disorder, these data suggest that, in the native state, NEMO exists in an equilibrium of significantly different conformational states. We observed the significant structural disorder and equilibrium of states at low temperatures. In the context of a free energy landscape interpretation of protein structure, our data therefore suggest a relatively rugged free energy landscape, i.e. NEMO adopts a range of equilibrated conformational states, separated by relatively low energetic barriers.
Ligand Binding Is Coupled to NEMO Conformational Change
NEMO has a huge number of potential interaction partners (33). We hypothesized that the high degree of NEMO structural disorder and the ability to access a range of equilibrium conformational states may allow NEMO to adopt discrete ligand-bound conformational states. To explore this hypothesis, we opted to focus on three key interaction partners of NEMO: IKKβ, IκBα, and free polyubiquitin chains. We monitored conformational change using the fluorescence reporters established above (ANS and Trp6 emission).
We probed the effect of Met1-linked polyubiquitin chain length on the signal arising from both ANS and Trp6 emission (Fig. 3). Ubiquitin does not possess any native tryptophan residues, meaning that the Trp6 signal arising from the NEMO-polyubiquitin complex is attributable to NEMO conformational change alone. We found that polyubiquitin binding to NEMO gave rise to measurable conformational change as assessed from both fluorescence reporter signals (Fig. 5A). However, the magnitude and sign of the signal change were strongly chain length-dependent (oligomers termed Ub2–10). Specifically, there was a pronounced decrease in the relative ANS emission up to Ub6 (12.5% decrease), but a pronounced increase in relative ANS emission for longer chain lengths (Ub10, 16% increase). For Trp6 emission, the signal showed a monotonic decrease only at longer chain lengths (Ub10, 15% decrease). We therefore found two separate conformational “regimes” with increasing Met1-linked polyubiquitin chain length. At low chain lengths, we found a “compact” state with buried hydrophobic residues. At high chain lengths, we found an “expanded” state with solvent-exposed hydrophobic and Trp6 residues.
FIGURE 5.
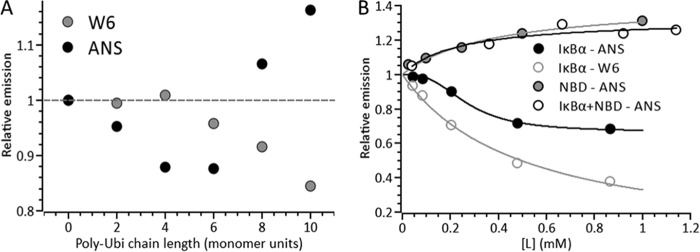
Ligand binding is coupled to NEMO conformational change. Shown is the concentration dependence of the ligand on either ANS or Trp6 emission for polyubiquitin (Poly-Ubi; A) or the NBD and IκBα peptides (B). A signal value of 1 represents the background-subtracted NEMO emission. Solid lines are fits to a weak binding isotherm (Em = (Emax[L])/(K + L), where L is ligand) or the Hill equation (Em = (Emax[L]n)/(Kn + Ln)) (B, black lines). Conditions were 50 mm Tris-Cl (pH 8), 50 mm NaCl, 5 mm DTT, 25 μg/ml polyubiquitin, 2 mm IκBα, and 6 μm NEMO at 10 °C. W6, Trp6.
To probe the effect of IKKβ and IκBα binding, we used established peptide mimics of these proteins. Specifically, we used a peptide derived from the NEMO-binding domain (NBD) of IKKβ (TALDWSWLQTE), representing the immediate C terminus of IKKβ, and a peptide derived from the phosphorylated region of IκBα (DDRHDSGLDSMKD), incorporating the Ser residues (positions 32 and 36) that are phosphorylated by IKKβ. Together, these peptides act as convenient probes of the parent ligands and, in both cases, have been shown to bind to NEMO and behave as good mimics for the full-length proteins. Due to the two Trp residues present in the NBD peptide, we restricted analysis of Trp6 to the IκBα peptide.
Fig. 5B shows the concentration dependence of the NBD and IκBα peptides on the relative ANS and Trp emission in the presence of NEMO. The data extracted from fitting are given in Table 2. For the NBD peptide, the data show saturating behavior with a large increase in ANS emission with increasing NBD concentration. Fitting these data to a weak binding isotherm (Fig. 5B) gives an emission maximum (Emax) of 1.35 ± 0.02 and an equilibrium constant for NEMO conformational change (K) of 0.14 ± 0.03 mm. In contrast, the IκBα peptide shows a significant decrease in ANS emission and fits with a sigmoidal relationship (Hill equation) (Fig. 2B), giving Emax = 0.67 ± 0.02, K = 0.27 ± 0.02 mm, and n = 2.8 ± 0.5. The IκBα peptide shows simple saturation behavior for Trp6 emission decreasing to essentially zero (Fig. 5B), giving Emax = 0.0 ± 0.08 and K = 0.5 ± 0.08 mm. A peptide that has no known affinity for NEMO but a similar overall hydrophobicity for the NBD peptide (KDVQEFRGVTAVIRCKGK, derived from aggrecan) did not give rise to a change in NEMO ANS emission (Fig. 6). As such, the reported conformational changes did not arise from nonspecific interactions of the peptides with NEMO.
TABLE 2.
Extracted parameters from fitting data in Fig. 2
| Emax | K | n | |
|---|---|---|---|
| mm | |||
| IκBα (Trp6) | 0.0 ± 0.08 | 0.5 ± 0.08 | |
| IκBα (ANS) | 0.67 ± 0.02 | 0.27 ± 0.02 | 2.8 ± 0.5 |
| NBD (ANS) | 1.35 ± 0.02 | 0.14 ± 0.03 | |
| IκBα + NBD (ANS) | 1.32 ± 0.06a | 0.23 ± 0.1 |
a Magnitude of change relative to NEMO ANS emission alone.
FIGURE 6.
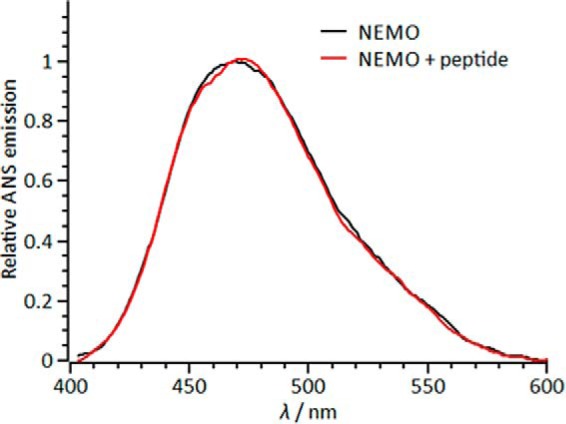
Deconvolved ANS emission spectra in the presence of NEMO (black line) and NEMO with a peptide derived from collagen (red line). The spectra do not show any significant numerical difference outside of standard error.
We explored NEMO conformational change in the ternary NEMO-IKKβ-IκBα complex. We monitored the ANS emission of the NEMO-IκBα complex with increasing concentrations of the NBD, as shown in Fig. 5B. Incubation of NEMO with IκBα induced a significant decrease in ANS emission as discussed above. Subsequent titration of the NBD peptide gave rise to an increase in ANS emission. We found that these data fit to a simple weak binding isotherm as observed for NBD binding to NEMO alone. The results of the fitting are given in Table 2. Relative to the NEMO ANS emission alone, the ANS emission increased with a similar magnitude and equilibrium constant as observed for NBD binding to NEMO alone. These data suggest that binding of IκBα to NEMO does not affect the interaction with IKKβ, suggesting that the NEMO-binding sites for these ligands are independent.
Combined, these data are evidence that NBD binding is coupled to the exposure of hydrophobic residues and that binding of the IκBα peptide is coupled to the burying of hydrophobic residues at distinct interaction sites. These different modes of action are also reflected in the observation of significant cooperativity with the IκBα peptide (n = 2.8 ± 0.5), but not the NBD peptide. Furthermore, the significant decrease in Trp6 emission on IκBα peptide binding suggests that Trp6, and by inference the IKKβ-binding region, becomes significantly more solvent-exposed on binding IκBα. The observation of cooperativity on IκBα peptide binding to NEMO (monitored by ANS emission) may be indicative of the complexity of this binding interaction and the coupling to conformational change, particularly because ANS emission is a global metric of NEMO conformational change. For example, the data may reflect several different conformational changes induced on IκBα binding. The key finding therefore is that different ligands induce radically different types and magnitudes of NEMO conformational change, i.e. NEMO does not use a simple binary off and on conformational switch, but is able to adopt a broad range of conformational states that are ligand-specific. This finding therefore provides very strong evidence for our hypothesis that NEMO is able to adopt discrete unique conformational states in response to binding of specific ligands.
Polyubiquitin “Functionalizes” NEMO
In light of our findings that polyubiquitin induces NEMO conformational change (Fig. 5A), we wished to explore whether the NEMO-Ub complex affects the molecular interactions with IKKβ and IκBα and the relationship to ligand-induced conformational change. To achieve this, we monitored the kinetics of ligand (NBD and IκBα peptide)-induced NEMO conformational change using rapid-mixing stopped-flow fluorescence spectroscopy following the change in ANS emission on ligand binding. Example transients are shown in Fig. 7A. In both cases, ligand-induced conformational change was fast, occurring across a broad range of timescales, but with the majority of amplitude occurring in <10 s. In both cases, the data fit to a multiexponential relationship (Equation 4),
where A is the amplitude, k is the observed rate constant (kobs) for the ith exponential component (up to three), and ΔA is the total amplitude change. We suggest that the observation of multiple phases in our stopped-flow transients is indicative of a range of equilibrium NEMO conformational states, as is consistent with our p/T studies above.
FIGURE 7.
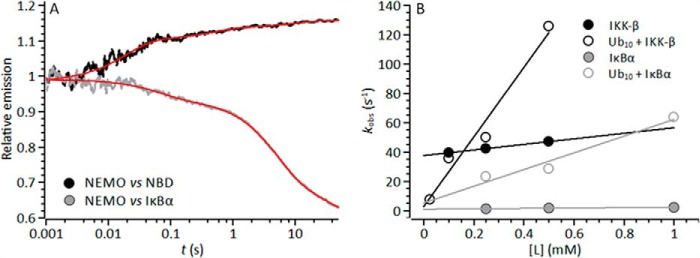
Kinetics of ligand-induced NEMO conformational change. A, example stopped-flow transient showing the multiphasic nature of the emission change detected upon rapid mixing of NEMO versus 0.5 mm NBD (black line) or IκBα (gray line). The solid red line is the fit to Equation 4. B, concentration dependence of NBD/IκBα on the extracted observed rate constants in the absence (closed circles) and presence (open circles) of Ub10. Solid lines are the fit to Equation 6. Conditions were 50 mm Tris-Cl (pH 8), 50 mm NaCl, 5 mm DTT, and 5 μm NEMO at 10 °C.
Fig. 7B shows the change in the observed rate of NEMO conformational change with increasing NBD and IκBα peptide concentrations. For NBD binding, the observed rates were extracted by fitting the major exponential component (>75% of the total amplitude). For IκBα binding, there was no clear major exponential phase, and the amplitudes of the extracted exponential phases show a complex relationship with respect to ligand concentration. In this case, primarily to reduce the very significant complexity of these transients, we extracted a single rate constant from the transients by weighting the rate constant from each exponential phase with the corresponding amplitude (Equation 5) as described previously (34).
 |
Within the accessible concentration range, defined by ligand solubility and the detection limit of the instrument, we found that the data for both ligands are best fit by a linear function. In the simplest mechanistic scheme, an initial ligand binding step is followed by NEMO conformational change, and a plot of kobs versus ligand concentration should show significant curvature (35). However, our observation of a linear relationship is consistent with an apparent second-order process, where NEMO conformational change is concomitant with ligand binding. Within this model, the fitted linear function shown in Fig. 3 takes the form shown in Equation 6,
where kb and k−b reflect the on- and off-rates for NBD/IκBα binding, respectively. The equilibrium constant extracted from these values then represents an apparent dissociation constant for ligand binding, kb/k−b = Kd(app). These data are given in Table 3. For both the NBD and IκBα, kb is very similar to k−b, giving Kd(app) = 2.0 ± 0.1 and 0.6 ± 0.2 mm, respectively. These data therefore provide a kinetic rationale for the high-micromolar equilibrium constants extracted from the steady-state fluorescence experiments shown in Fig. 5B and given in Table 2, i.e. the off-rate is very similar to the on-rate for ligand binding.
TABLE 3.
Kinetic parameters resulting from fitting stopped-flow concentration dependence studies
| kobsa | kb | k−b | Kd(app) | |
|---|---|---|---|---|
| s−1 | mm−1 s−1 | s−1 | mm | |
| NEMO + NBD | 47.1 ± 0.7 | 19.0 ± 0.9 | 37.5 ± 0.3 | 2.0 ± 0.1 |
| NEMO-Ub10 + NBD | 125.7 ± 6.3 | 237.3 ± 30.9 | 2.7 ± 8.8 | 0.01 ± 0.04 |
| NEMO + IκBα | 2.2 ± 0.45 | 1.4 ± 0.4 | 0.9 ± 0.2 | 0.6 ± 0.2 |
| NEMO-Ub10 + IκBα | 63.9 ± 6.1 | 56.5 ± 12.4 | 5.2 ± 8.2 | 0.09 ± 0.14 |
a Data extracted from transients at either 0.5 mm NBD or 1 mm IκBα peptide.
Incubating NEMO with Ub10 resulted in a significant change in the extracted kinetic parameters for ligand-induced conformational change (Fig. 7B and Table 3). The observed rate increased for both the NBD and IκBα (Table 3). The on-rate becomes much larger in the presence of Ub10, and in the case of the NBD, the off-rate becomes smaller, giving rise to a significant decrease in Kd(app) in the presence of Ub10 (Table 3): Kd(app) = 0.01 ± 0.04 and 0.09 ± 0.14 mm for the NBD and IκBα, respectively. The affinity of both the NBD and IκBα for NEMO is enhanced for the NEMO-Ub10 complex compared with NEMO alone. We note that this increase in affinity brings Kd(app) more in line with expected intracellular protein concentrations, but does not give rise to overly “tight” binding. High-specificity low-affinity interactions are a hallmark of disorder-mediated interactions involved in signaling processes (14) and potentially reflect that, for signaling processes, turning a signal off is as important as turning a signal on. Tight binding (in the nanomolar and picomolar range) means that molecular interactions are potentiated in the long term, and this would seem incompatible with the diverse and transient signaling interactions of NEMO.
NEMO binding to Ub10 gave rise to an increase in the solvent exposure of both NEMO hydrophobic residues (increase in ANS emission) and Trp6 (decrease in Trp6 emission) as discussed above (Fig. 5A). These findings provide a rationale for the enhanced ligand affinity, i.e. the exposed hydrophobic residues promote protein-protein interactions, and the altered environment at the NEMO N terminus (characterized by the changing Trp6 emission) reflects improved binding of the NBD. On the basis of our findings, we hypothesized that non-covalent binding of long chains of Met1-linked polyubiquitin functionalizes NEMO and serves to regulate the interaction of NEMO with the key species governing the canonical NF-κB transcriptional regulation pathway. The polyubiquitin-binding site is located at the C terminus of NEMO, whereas the IKKβ-binding site is located at the N terminus (Fig. 1). There is no evidence that the peptide mimics bind to or interact with polyubiquitin. As such, it would appear that polyubiquitin achieves the observed enhancement of ligand binding by inducing NEMO conformational change, potentially in an allosteric fashion.
We wished to consider the alternative possibility that exposure of NEMO hydrophobic residues serves to localize NEMO to the membrane, e.g. mediated by the exposure of a hydrophobic membrane insertion loop (36). Whole cell experiments have provided evidence that polyubiquitin may have a role in localizing NEMO to the plasma membrane (37–39), without the involvement of microtubules or actin (39). To investigate the potential for NEMO binding to the membrane environment, we performed a simple liposome binding assay. We incubated the binary (NEMO with peptide) and ternary (NEMO, peptide, and Ub10) complexes in the presence of artificial liposomes and examined the bound protein fraction from the centrifuged pellet (see “Experimental Procedures” for details). The resulting SDS-PAGE image is presented in Fig. 8, showing strong banding for those conditions under which NEMO was incubated with Ub10. We therefore found that a much more significant amount of NEMO was localized with the membranes when Ub10 was present, suggesting that Ub10 binding to NEMO promotes liposome association. We note that there was some variation in the concentrations of NEMO used (between 0.45 and 0.75 μm), and this would appear to account for the differing intensities of the observed bands. In summary, NEMO alone or NEMO bound to the NBD or IκBα may have some propensity to bind to membrane environments, but this localization is enhanced by the presence of non-covalently bound Ub10.
FIGURE 8.
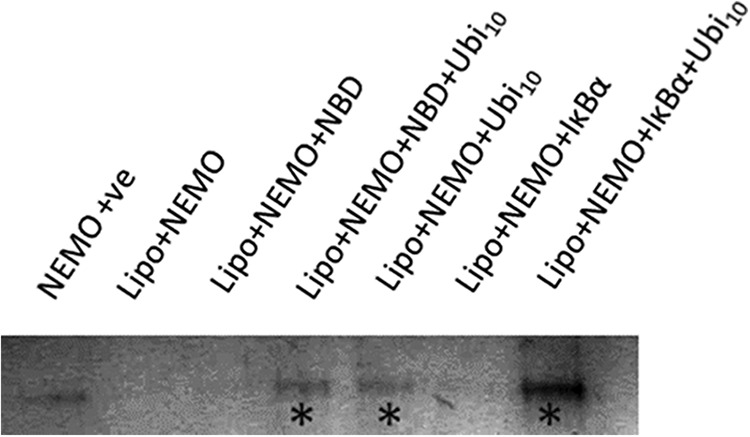
SDS-PAGE showing the results of the liposome binding assay. The protein bands shown ran at ∼50 kDa and are indicative of full-length NEMO. The contrast has been enhanced evenly across the image to better visualize the bands. The asterisks mark those samples in which polyubiquitin (Ubi10) was present and are coincident with the occurrence of significantly observable protein bands. The polyubiquitin concentration was 0.5 μm, and the NBD and IκBα concentrations were 1 mm. Lipo, liposome; +ve, positive control.
Molecular Mechanistic Model for NEMO Activity
We found spectroscopic evidence that NEMO is a native molten globule that can undergo ligand-induced conformational change and that polyubiquitin binding, at least for the chain length and linkage type we studied, functionalizes NEMO. On the basis of these findings, we developed a molecular mechanistic model for the activity of NEMO that specifically incorporates NEMO conformational change, shown as a mechanistic scheme in Fig. 9A and as a diagrammatic scheme in Fig. 9B.
FIGURE 9.
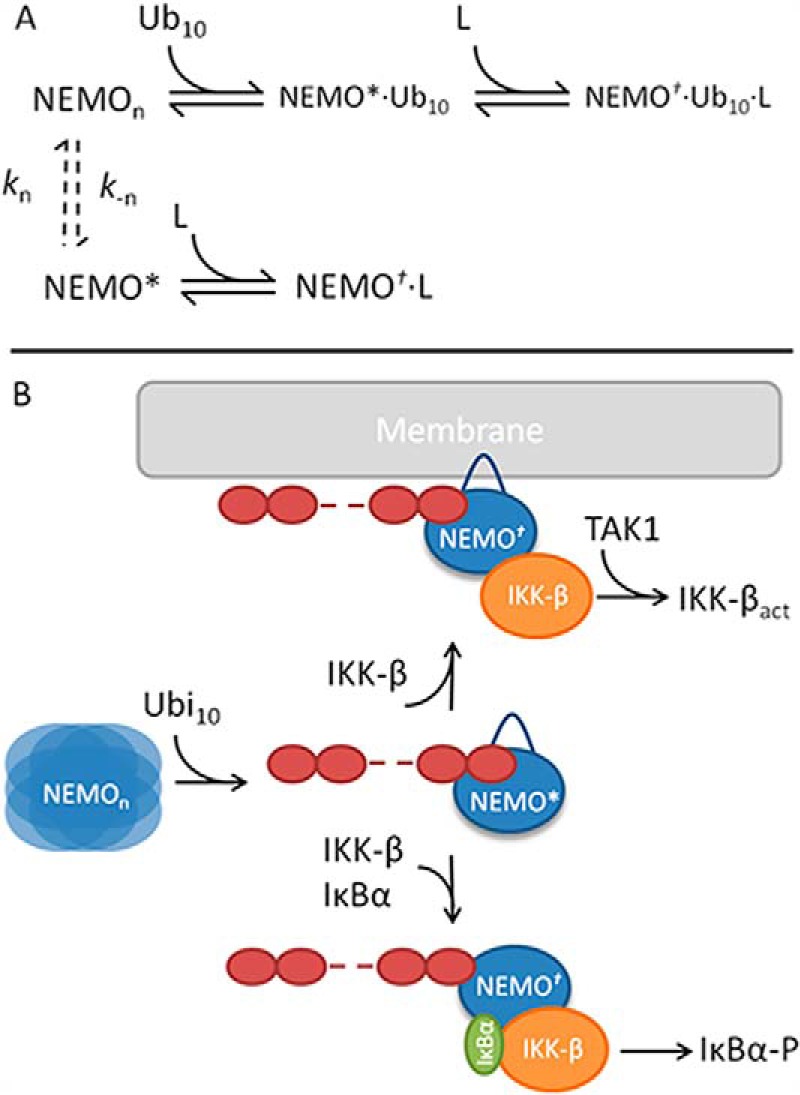
Molecular mechanistic model for the activity of NEMO, incorporating ligand-induced conformational change. A, mechanistic scheme outlining the steps associated with ligand binding and subsequent conformational change. B, molecular model for the role of free polyubiquitin chains in mediating the recruitment of both IKKβ and IκBα but also recruitment of the ternary complex to the membrane.
Evidence from far-UV CD suggests that NEMO is composed of a large fraction of structurally disordered and helical content (Fig. 2). In this state, NEMO can undergo conformational change without a significant variation in secondary structure composition (Fig. 2B) and is able to bind to ligands (Figs. 5 and 7). Furthermore, we found that the structure can be perturbed by high pressure, which acts to perturb pre-existing equilibria. Together, these findings suggest that NEMO exists in a broad equilibrium of interconverting conformational substates (NEMOn) (Fig. 9A, dashed arrows), potentially indicative of a native molten globule-like protein. As such, the conformational states that are competent for ligand binding (high-affinity state, NEMO*, Fig. 9) may only be a fraction of the total population, giving rise to a slow on-rate and so weak binding of ligands (Tables 2 and 3).
Polyubiquitin (at least Met1-linked Ub10) binding selects for or induces a conformational state that is structurally similar to NEMO*, giving rise to an enhanced affinity for ligands (faster binding and smaller equilibrium constant) (Table 3) and promoting membrane association (Fig. 8). Met1-linked Ub10 and IKKβ binding occurs at distinct domains (Fig. 1), and we have provided evidence the IκBα- and IKKβ-binding sites are independent (Fig. 5B). There is no evidence that the peptide mimics we used interact with Met1-linked Ub10. As such, taken together, our kinetic and equilibrium data support a mechanism in which this polyubiquitin-mediated functionalization of NEMO arises from allosteric conformational change.
Several theoretical models exist within which one can interpret ligand-induced conformational change. In the simplest cases, either ligand binding induces the conformational change on binding (induced fit), or the appropriate conformational state is selected from a broad equilibrium of pre-existing states (population shift) (40). We favor a population shift model rather than induced fit for this interaction primarily because of the evidence we provided (see above) showing that native NEMO exists in an equilibrium of significantly different conformational states. However, we note that developing theory suggests that the two models are not mutually exclusive, but act together (40), and we expect this to be the case with the NEMO-polyubiquitin interaction. We stress that our findings were obtained with free polyubiquitin chains. The binding determinants of free chains to NEMO are different from covalently bound ubiquitin, and our findings are therefore very specific to the non-covalent molecular interaction.
Ligand binding to the NEMO-polyubiquitin complex selects for or induces NEMO conformational change to produce the functionally active complex (Fig. 9; NEMO†). In the context of IKKβ and IκBα binding, we suggest that polyubiquitin drives the recruitment of the substrate (IκBα) to IKKβ, mediated via the enhanced affinity for both ligands. Recruitment of IκBα putatively promotes the phosphorylation of IκBα, releasing the NF-κB transcriptional regulation machinery. The functional role of NEMO localization to the plasma membrane may be 2-fold. First, NEMO promotes TAK1-catalyzed phosphorylation of IKKβ, which is required for IKKβ to phosphorylate exogenous substrates such as IκBα (37). TAK1 is located near the membrane associated with TAB1. Second, IKKβ phosphorylates exogenous substrates that are localized to the membrane, e.g. promoting the phosphorylation and subsequent degradation of IRS-1 (insulin receptor substrate 1) (38).
In conclusion, we have developed a detailed molecular mechanistic model for NEMO activity based on quantitative evidence that, for the first time, incorporates the ability of NEMO to undergo ligand-induced conformational change. Our study further demonstrates that the functional conformational states accessible to NEMO are regulated by free polyubiquitin and that this regulation appears to be allosteric in nature. This model therefore provides a mechanistic rationale for many of the competing hypotheses relating to the functional activity and regulation of NEMO.
Footnotes
- IKKβ
- IκB kinase-β
- NEMO
- NF-κB essential modulator
- ANS
- 8-anilinonaphthalene-1-sulfonic acid
- Ub
- ubiquitin
- NBD
- NEMO-binding domain.
References
- 1. Gilmore T. D. (2006) Introduction to NF-κB: players, pathways, perspectives. Oncogene 25, 6680–6684 [DOI] [PubMed] [Google Scholar]
- 2. Fontan E., Traincard F., Levy S. G., Yamaoka S., Véron M., Agou F. (2007) NEMO oligomerization in the dynamic assembly of the IκB kinase core complex. FEBS J. 274, 2540–2551 [DOI] [PubMed] [Google Scholar]
- 3. Solt L. A., Madge L. A., May M. J. (2009) NEMO-binding domains of both IKKα and IKKβ regulate IκB kinase complex assembly and classical NF-κB activation. J. Biol. Chem. 284, 27596–27608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Courtois G., Israël A. (2011) IKK regulation and human genetics. Curr. Top. Microbiol. Immunol. 349, 73–95 [DOI] [PubMed] [Google Scholar]
- 5. Schröfelbauer B., Polley S., Behar M., Ghosh G., Hoffmann A. (2012) NEMO ensures signaling specificity of the pleiotropic IKKβ by directing its kinase activity toward IκBα. Mol. Cell 47, 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu G., Lo Y.-C., Li Q., Napolitano G., Wu X., Jiang X., Dreano M., Karin M., Wu H. (2011) Crystal structure of inhibitor of κB kinase β. Nature 472, 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emmerich C. H., Ordureau A., Strickson S., Arthur J. S., Pedrioli P. G., Komander D., Cohen P. (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. U.S.A. 110, 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xia Z.-P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., Chen Z. J. (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ivins F. J., Montgomery M. G., Smith S. J., Morris-Davies A. C., Taylor I. A., Rittinger K. (2009) NEMO oligomerization and its ubiquitin-binding properties. Biochem. J. 421, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kensche T., Tokunaga F., Ikeda F., Goto E., Iwai K., Dikic I. (2012) Analysis of nuclear factor-κB (NF-κB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J. Biol. Chem. 287, 23626–23634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 12. Ea C.-K., Deng L., Xia Z.-P., Pineda G., Chen Z. J. (2006) Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 [DOI] [PubMed] [Google Scholar]
- 13. Wu C.-J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. (2006) Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat. Cell Biol. 8, 398–406 [DOI] [PubMed] [Google Scholar]
- 14. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 15. Uversky V. N. (2013) A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 22, 693–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferreon A. C., Ferreon J. C., Wright P. E., Deniz A. A. (2013) Modulation of allostery by protein intrinsic disorder. Nature 498, 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schulenburg C., Hilvert D. (2013) Protein conformational disorder and enzyme catalysis. Top. Curr. Chem. 337, 41–67 [DOI] [PubMed] [Google Scholar]
- 18. Tantos A., Han K.-H., Tompa P. (2012) Intrinsic disorder in cell signaling and gene transcription. Mol. Cell. Endocrinol. 348, 457–465 [DOI] [PubMed] [Google Scholar]
- 19. Uversky V. N., Oldfield C. J., Dunker A. K. (2005) Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 18, 343–384 [DOI] [PubMed] [Google Scholar]
- 20. Smock R. G., Gierasch L. M. (2009) Sending signals dynamically. Science 324, 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsai C.-J., Ma B., Sham Y. Y., Kumar S., Nussinov R. (2001) Structured disorder and conformational selection. Proteins 44, 418–427 [DOI] [PubMed] [Google Scholar]
- 22. Motlagh H. N., Wrabl J. O., Li J., Hilser V. J. (2014) The ensemble nature of allostery. Nature 508, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uversky V. N. (2009) Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counterions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 28, 305–325 [DOI] [PubMed] [Google Scholar]
- 24. Stryer L. (1965) The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. J. Mol. Biol. 13, 482–495 [DOI] [PubMed] [Google Scholar]
- 25. Semisotnov G. V., Rodionova N. A., Razgulyaev O. I., Uversky V. N., Gripas' A. F., Gilmanshin R. I. (1991) Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 31, 119–128 [DOI] [PubMed] [Google Scholar]
- 26. Agashe V. R., Shastry M. C., Udgaonkar J. B. (1995) Initial hydrophobic collapse in the folding of barstar. Nature 377, 754–757 [DOI] [PubMed] [Google Scholar]
- 27. Engelhard M., Evans P. A. (1995) Kinetics of interaction of partially folded proteins with a hydrophobic dye: evidence that molten globule character is maximal in early folding intermediates. Protein Sci. 4, 1553–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pan C.-P., Barkley M. D. (2004) Conformational effects on tryptophan fluorescence in cyclic hexapeptides. Biophys. J. 86, 3828–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akasaka K. (2006) Probing conformational fluctuation of proteins by pressure perturbation. Chem. Rev. 106, 1814–1835 [DOI] [PubMed] [Google Scholar]
- 30. Wiedersich J., Köhler S., Skerra A., Friedrich J. (2008) Temperature and pressure dependence of protein stability: the engineered fluorescein-binding lipocalin FluA shows an elliptic phase diagram. Proc. Natl. Acad. Sci. U.S.A. 105, 5756–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tognotti D., Gabellieri E., Morelli E., Cioni P. (2013) Temperature and pressure dependence of azurin stability as monitored by tryptophan fluorescence and phosphorescence. The case of F29A mutant. Biophys. Chem. 182, 44–50 [DOI] [PubMed] [Google Scholar]
- 32. Ravindra R., Winter R. (2003) On the temperature-pressure free-energy landscape of proteins. ChemPhysChem 4, 359–365 [DOI] [PubMed] [Google Scholar]
- 33. Fenner B. J., Scannell M., Prehn J. H. (2010) Expanding the substantial interactome of NEMO using protein microarrays. PLoS ONE 5, e8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pudney C. R., Hay S., Pang J., Costello C., Leys D., Sutcliffe M. J., Scrutton N. S. (2007) Mutagenesis of morphinone reductase induces multiple reactive configurations and identifies potential ambiguity in kinetic analysis of enzyme tunneling mechanisms. J. Am. Chem. Soc. 129, 13949–13956 [DOI] [PubMed] [Google Scholar]
- 35. Hammes G. G., Chang Y.-C., Oas T. G. (2009) Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brunecky R., Lee S., Rzepecki P. W., Overduin M., Prestwich G. D., Kutateladze A. G., Kutateladze T. G. (2005) Investigation of the binding geometry of a peripheral membrane protein. Biochemistry 44, 16064–16071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang J., Clark K., Lawrence T., Peggie M. W., Cohen P. (2014) An unexpected twist to the activation of IKKβ: TAK1 primes IKKβ for activation by autophosphorylation. Biochem. J. 461, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakamori Y., Emoto M., Fukuda N., Taguchi A., Okuya S., Tajiri M., Miyagishi M., Taira K., Wada Y., Tanizawa Y. (2006) Myosin motor Myo1c and its receptor NEMO/IKK-γ promote TNF-α-induced serine 307 phosphorylation of IRS-1. J. Cell Biol. 173, 665–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tarantino N., Tinevez J.-Y., Crowell E. F., Boisson B., Henriques R., Mhlanga M., Agou F., Israël A., Laplantine E. (2014) TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. J. Cell Biol. 204, 231–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nussinov R., Ma B., Tsai C.-J. (2014) Multiple conformational selection and induced fit events take place in allosteric propagation. Biophys. Chem. 186, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]