Abstract
Genomic DNA extracts from four sites at Kilauea Volcano were used as templates for PCR amplification of the large subunit (coxL) of aerobic carbon monoxide dehydrogenase. The sites included a 42-year-old tephra deposit, a 108-year-old lava flow, a 212-year-old partially vegetated ash-and-tephra deposit, and an approximately 300-year-old forest. PCR primers amplified coxL sequences from the OMP clade of CO oxidizers, which includes isolates such as Oligotropha carboxidovorans, Mycobacterium tuberculosis, and Pseudomonas thermocarboxydovorans. PCR products were used to create clone libraries that provide the first insights into the diversity and phylogenetic affiliations of CO oxidizers in situ. On the basis of phylogenetic and statistical analyses, clone libraries for each site were distinct. Although some clone sequences were similar to coxL sequences from known organisms, many sequences appeared to represent phylogenetic lineages not previously known to harbor CO oxidizers. On the basis of average nucleotide diversity and average pairwise difference, a forested site supported the most diverse CO-oxidizing populations, while an 1894 lava flow supported the least diverse populations. Neither parameter correlated with previous estimates of atmospheric CO uptake rates, but both parameters correlated positively with estimates of microbial biomass and respiration. Collectively, the results indicate that the CO oxidizer functional group associated with recent volcanic deposits of the remote Hawaiian Islands contains substantial and previously unsuspected diversity.
Aerobic carbon monoxide oxidation occurs ubiquitously, with significant activity reported for a wide range of aquatic and terrestrial systems, plant roots, and macroalgal tissues (10, 14, 20, 21, 26, 27). Nonetheless, past efforts to enrich CO oxidizers have resulted in only 12 species in 11 genera (12, 20, 21). More-recent efforts, including genome studies, have identified at least 25 CO-oxidizing species in 10 genera (12, 13). Although these results suggest that CO oxidizers may be as diverse as the habitats in which they are found, essentially nothing is known about in situ diversity.
Analyses of CO oxidizer diversity have been constrained by several factors. High CO concentrations traditionally used for enrichments inhibit growth of many newly recognized strains (12). Thus, simple enrichment strategies based on high CO concentrations may not be suitable for culture-based diversity surveys. The diversity of known CO oxidizers has also precluded development of signature 16S rRNA gene oligonucleotides analogous to those used for phylogenetically coherent groups, such as methanotrophs and ammonia oxidizers (3, 5, 7, 15, 31, 34).
However, primers developed for amplification of a portion of coxL, the large subunit of carbon monoxide dehydrogenase (CODH), have provided a new tool for assessing CO oxidizer diversity (12). coxL PCR primers amplify a 1,260- to 1290-bp fragment containing the CODH active site and sites for binding the molybdopterin cytosine dinucleotide (MCD) cofactor (9, 28). Two sets of primers have been developed and evaluated, one for each of two CO-oxidizing clades (12).
One clade, designated OMP, contains coxL sequences derived from known CO oxidizers (e.g., Oligotropha carboxidovorans, Mycobacterium tuberculosis, and Pseudomonas thermocarboxydovorans) and newly recognized strains (e.g., Mycobacterium smegmatis, Silicibacter pomeroyi, and Stenotrophomonas strain LUP) (12). The second clade, designated BMS, contains putative CODH sequences derived primarily from newly recognized CO oxidizers (e.g., Burkholderia strain LUP, Mesorhizobium loti, and Sinorhizobium meliloti) and from taxa that possess both BMS and OMP coxL genes (Bradyrhizobium japonicum, Burkholderia fungorum LB400, and Stappia stellulata [12]).
We report here analyses of OMP coxL diversity based on PCR amplification of DNA extracts from volcanic deposits at four varied sites in and near the Kilauea Volcano caldera. Deposits at all sites consume CO actively, and CO contributes significantly to respiratory reducing equivalent flow at three of the four sites. Phylogenetic and statistical analyses of sequences from clone libraries indicate that distinct and complex CO-oxidizing populations occur at each site. Although some clone sequences could be associated with known CO oxidizers in proteobacterial and Actinobacteria and Firmicutes lineages, most of the sequences appear to have been derived from novel organisms with uncertain phylogenetic affiliations. These results represent the first molecular ecological analyses of CO oxidizers and reveal a substantial and previously unsuspected diversity.
MATERIALS AND METHODS
Site description and sampling.
Surface deposits (0 to 2 cm depth of four sites in or near Kilauea Volcano caldera were sampled in April 2002. The sites range in age, water availability, plant community development, and organic contents, among other parameters (11). The Halema'uma'u and Caldera Rim sites occur in a region of the caldera with relatively limited water availability, based on water contents (11). Both sites consist of ash and tephra overlying weathered lava flows and support limited patchy growth of pioneering ferns, shrubs, and the Ohia tree, Metrosideros polymorpha (11). The youngest site, Pu'u Puai, consists of coarse tephra and cinders deposited in 1959 and supports patches of Ohia and an invasive tree, Myrica faya (32). The oldest site, Forest, consists of ash deposits overlying lava that is about 300 years old and contains mixed stands of Ohia and M. faya trees, with a limited litter layer (4).
DNA extraction and coxL PCR amplification.
Surface samples from each site were obtained with a sterile trowel and transferred to sterile Whirlpak sample bags. Within 1 to 2 h, samples were frozen and held at −20°C until transport on dry ice (−80°C) to Maine. Samples were held at −20°C until DNA extraction. DNA from each site was extracted from triplicate samples 10 g [fresh weight] by a bead-beating method with an Ultraclean Mega Soil DNA extraction kit (MoBio Labs, Inc., Carlsbad, Calif.). DNA quality and size distribution were determined by electrophoresis in 0.7% agarose.
Bacillus schlegelii and Pseudomonas carboxydohydrogena were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (German Collection of Microorganisms and Cell Cultures) and grown as recommended. coxL sequences were obtained by extraction of DNA from pure cultures by a bead-beating technique (MoBio Labs, Inc.) and PCR as described below. The nucleotide sequences of the PCR products were determined by the University of Maine Sequencing Facility with primers OMPf and O/Br and an ABI model 377 sequencer (Applied Biosystems, Foster City, Calif.).
A 1,260- to 1,290-bp fragment of coxL was amplified with primers OMPf (5′-GGCGGCTT[C/T]GG[C/G]AA[C/G]AAGGT-3′) and O/Br (5′-[C/T]TCGA[T/C]GATCAT CGG[A/G]TTGA-3′) (13). CO oxidizers that yield an OMP coxL product include classic carboxydotrophs, such as Oligotropha carboxydovorans and Hydrogenophaga pseudoflava, and newly recognized CO oxidizers, such as Bradyrhizobium japonicum, Mycobacterium smegmatis, Stappia aggregata, and Silicibacter pomeroyi (12).
PCR mixtures (50 μl) contained 5 μl of 10× PCR buffer, 10 μl of Eppendorf Taqmaster buffer, 100 μM concentrations of each deoxynucleoside triphosphate, 3.5 mM magnesium ion, 0.1 μM concentrations of primers, and 1.25 U of MasterTaq DNA polymerase (Brinkmann, Inc., Westbury, N.Y.). PCR was carried out in an Eppendorf Mastercycler thermocycler (Brinkmann, Inc.), with an initial denaturation step of 3 min at 94°C and a Taq DNA polymerase “hot start” addition at 80°C. The reaction continued with 30 cycles of 45 s at 94°C, 60 s at 58°C, and 90 s at 72°C, with a final extension for 20 min at 72°C. Approximately 1 to 2 ng of culture DNA extracts was used as PCR templates. Concentrations of volcanic extract DNA were varied to optimize yields for each of the sites. The presence and size of PCR products were determined by electrophoresis in 1% agarose and ethidium bromide staining. PCR products were stored at −20°C overnight.
Construction of coxL clone libraries and sequence analysis.
Triplicate PCR mixtures for each site were combined and cloned into E. coli with the use of a TOPO TA cloning kit for sequencing (Invitrogen Life Technologies, Carlsbad, Calif.) according to the manufacturer's instructions. Arbitrarily selected transformed colonies (n = 96) from each library were grown overnight in Luria-Bertani broth. Plasmids were extracted with a PerfectPrep Plasmid 96 Spin Direct kit (Brinkmann) according to manufacturer's instructions. Plasmids were screened for the appropriate insert size (about 1,260 bp) by comparison to a super-coiled DNA marker (2 to 10 kb; Promega Corp., Madison, Wis.) by agarose (1%) gel electrophoresis and ethidium bromide staining. Cloned PCR products were sequenced by the University of Maine Sequencing Facility with vector primer T7 and an ABI model 377 sequencer (Applied Biosystems).
Clone sequences were subjected to analysis by GenBank's BLAST utility to determine similarity to known coxL sequences. Sequences that were most similar to CODHs were submitted to ExPASy (http://us.expasy.org/tools) to obtain inferred amino acid sequences. The correct reading frames were determined from the presence of diagnostic amino acid motifs, including the active site (CSFR) and binding sites (HETT and SRS) (27) for MCD cofactor. Inferred amino acid sequences containing the proper motifs were aligned with corresponding coxL sequences from known CO-oxidizing bacteria by using ClustalX software version 1.8, with manual adjustments as necessary. A 163-residue region containing the active site and two MCD-binding sites was chosen for further analyses.
Each of the clone sequences was unique and was defined operationally as an operational taxonomic unit (OTU). Phylogenetic analysis of the clone libraries was performed by using a neighbor-joining algorithm with 1,000 bootstrap replicates implemented with PAUP 4.0b (Sinauer Associates, Inc., Sunderland, Mass.). A putative coxL sequence from the archeal species Pyrobacalum aerophilum was used to root the neighbor-joining tree. Statistical comparisons of the clone sequence libraries for each of the sites were accomplished with LIBSHUFF (30) and Arlequin version 1.1 (29).
LIBSHUFF estimated homologous and heterologous coverages of clone libraries as a function of evolutionary distance for pairwise reciprocal comparisons of libraries. A bootstrap procedure was used to estimate the significance of differences in coverage versus evolutionary distance between libraries. Differences in coverage as a function of evolutionary distance (Δc) were considered significant for P values of ≤0.05.
Arlequin estimated the significance of differences in population pairwise fixation index (FST) values and average pairwise difference among libraries by using analysis of molecular variance (AMOVA), and the software calculated nucleotide diversity, average pairwise differences, and mismatch distributions for each library. FST values were derived from the genetic diversity within a library and the total diversity of a pair of pooled libraries (23, 29, 35). Nucleotide diversity was estimated from the number of variable positions for aligned sequences in a given library; average pairwise differences were estimated from comparisons within a library of the number of sequence differences between a given clone and all other clones; and mismatch distributions were derived for each library from the frequency of pairwise differences.
A P-test was used to determine the significance of covariation between the distribution of unique sequences within libraries and phylogeny (18, 22). Briefly, PAUP 4.0b was used to generate 1,000 random trees from the combined clone libraries. The lengths of maximum parsimony trees for the clones were compared by P-test to the 95% lower confidence limit of the random tree lengths obtained by maximum parsimony with a heuristic search algorithm.
Nucleotide sequence accession numbers.
The sequence of the volcanic deposits identified in this study have been deposited in GenBank under accession numbers AY463248 to AY463356.
RESULTS
Phylogeny of clone libraries.
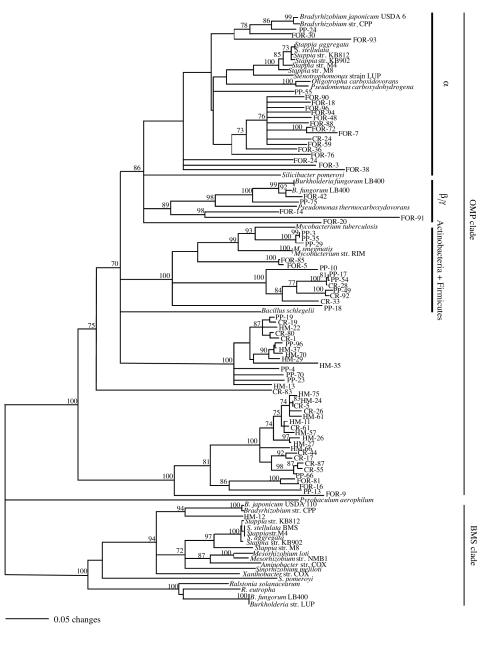
PCR products of the appropriate length (approximately 1,260 bp) were successfully amplified from volcanic deposit DNA. A total of 109 coxL clones were sequenced. BLAST analysis indicated that 108 of the clone sequences were most similar to OMP coxL genes, while one, HM-12, was most similar to a BMS putative coxL gene. A majority of Forest and Pu'u Puai clone sequences clustered with sequences from known CO oxidizers (Fig. 1). Sixty-two percent of Forest clones clustered with known α-Proteobacteria sequences (e.g., Bradyrhizobium japonicum and Oligotropha carboxydovorans) and 15% clustered with known β/γ-Proteobacteria (e.g., Burkholderia fungorum and Pseudomonas thermocarboxydovorans) (Table 1). Fifty-six percent of Pu'u Puai clones clustered with known Actinobacteria and Firmicutes (e.g., Mycobacterium and Bacillus schlegelii). In contrast, a majority of Caldera Rim (81%) and Halema'uma'u (97%) clone sequences did not cluster with any known CO oxidizers. Few of the Pu'u Puai, Caldera Rim, and Halema'uma'u clones clustered with Proteobacteria (12, 4 and 0%, respectively), while few Caldera Rim, Forest, and Halema'uma'u clones clustered with Actinobacteria and Firmicutes (15, 11, and 0%, respectively).
FIG. 1.
Neighbor-joining analysis (1,000 bootstrap replicates) of inferred amino acid sequences for nonredundant coxL clone sequences and known CO-oxidizing isolates. A total of 109 clone sequences (163 amino acid residues each) from the Forest (FOR), Pu'u Puai (PP), Halema'uma'u (HM), and the Caldera Rim (CR) libraries were analyzed. OMP and BMS clades are indicated based on known coxL gene sequences (13). Phylogenetic lineages are designated based on affiliations with known CO oxidizers. Bootstrap values of <70% are not shown. The Pyrobaculum aerophilum putative coxL sequence was used as an outgroup. The GenBank accession numbers for the following strains are given, with the clade indicated in parentheses. Aminobacter strain (str.) COX, AY307908 (BMS); Bradyrhizobium japonicum USDA 6, AY307921 (BMS); Bradyrhizobium strain CPP, AY307900 (BMS) and AY307913 (OMP); Burkholderia fungorum LB400, AY307901 (BMS), AY307916 (OMP); Burkholderia strain LUP, AY307907 (BMS); Mesorhizobium strain NMB1, AY307906 (BMS); Mycobacterium smegmatis, AY307916 (OMP); Oligotropha carboxydovorans, X82447(OMP); Pseudomonas thermocarboxydovorans, Y77931 (OMP); Stappia aggregata, AY307904 (BMS) and AY307918 (OMP); Stappia strain KB812, AY307898 (BMS) and AY307193 (OMP); Stappia strain KB902, AY307899 (BMS), AY307914 (OMP); Stappia strain M4, AY307902 (BMS) and AY307916 (OMP); Stappia strain M8, AY307903 (BMS) and AY307917 (OMP); Stappia stellulata, AY307905 (BMS) and AY307919 (OMP); Stenotrophomonas strain LUP, AY307920 (OMP); and Xanthobacter strain COX, AY307911 (BMS).
TABLE 1.
Phylogenetic distribution of coxL clone sequences based on relationships with known CO-oxidizing isolates
| Site of sample | Relative occurrence (%) of:
|
||||
|---|---|---|---|---|---|
| α-Proteo- bacteria | β/γ-Proteo- bacteria | Actinobacteria + Firmicutes | Unknown | BMS | |
| Halema'uma'u | 0 | 0 | 0 | 97 | 3 |
| Caldera rim | 4 | 0 | 15 | 81 | 0 |
| Pu'u Puai | 8 | 4 | 56 | 32 | 0 |
| Forest | 62 | 15 | 11 | 11 | 0 |
Phylogenetic analysis suggested that each of the libraries was unique, and statistical analyses supported this notion. Pairwise reciprocal comparisons of libraries by using LIBSHUFF revealed significant differences among libraries (P ≤ 0.04) (Table 2). Homologous coverage, a measure of the extent of similarity within a given library, ranged from 0% for the Forest library to 25.8% for the Halema'uma'u library, with those for Pu'u Puai (16%) and Caldera Rim (22.2%) between these percentages. Heterologous coverage, a measure of the representation of a given library in another, and vice versa, ranged from 0 to 6.65%. In addition, a P-test indicated that lengths of maximum parsimony trees for the clone libraries were significantly less than the lengths of randomly generated trees (P < 0.05).
TABLE 2.
LIBSHUFF comparisons of coxL clone librariesa
| Comparison | Covhom | Covhet | P |
|---|---|---|---|
| Caldera Rim (A) and Pu'u Puai (B) | 22.0 | 0.0 (A vs B) | 0.001 |
| 16.0 | 0.0 (B vs A) | 0.001 | |
| Caldera Rim (A) and Halema'uma'u (B) | 3.7 (A vs B) | 0.001 | |
| 25.8 | 6.5 (B vs A) | 0.002 | |
| Caldera Rim (A) and Forest (B) | 0.0 (A vs B) | 0.001 | |
| 0.0 | 0.0 (B vs A) | 0.021 | |
| Pu'u Puai (A) and Halema'uma'u (B) | 0.0 (A vs B) | 0.001 | |
| 0.0 (B vs A) | 0.001 | ||
| Pu'u Puai (A) and Forest (B) | 0.0 (A vs B) | 0.001 | |
| 0.0 (B vs A) | 0.017 | ||
| Forest (A) and Halema'uma'u (B) | 0.0 (A vs B) | 0.001 | |
| 0.0 (B vs A) | 0.001 |
Homologous coverage (Covhom) and heterologous coverage (Covhet) of libraries (as a percentage) for the comparisons indicated. Probability values (P) are given for the significance of differences between homologous and heterologous coverages in a reciprocal comparison as a function of evolutionary distance. See reference 30 for details.
Statistical comparisons of clone sequences with Arlequin revealed that the Forest library was most diverse, followed by those of the Caldera Rim and Pu'u Puai; the Halema'uma'u library was least diverse (Table 3). AMOVA comparisons revealed significant differences (P < 0.05) among libraries for pairwise fixation indices and average pairwise differences (Table 4). Both values were lowest when Caldera Rim and Halema'uma'u libraries were compared, followed by the Pu'u Puai and Forest comparison. The highest FST values and average pairwise differences were observed for comparison of the Halema'uma'u library with that of either Pu'u Puai or Forest.
TABLE 3.
Characteristics of coxL clone librariesa
| Site | No. of clones | No. of variable positions | Nucleotide diversity | θ(π) |
|---|---|---|---|---|
| Halema'uma'u | 31 | 334 | 0.28 (0.14)a | 128.8 (63.2)a |
| Caldera Rim | 27 | 339 | 0.37 (0.18)b | 177.9 (87.6)b |
| Pu'u Puai | 25 | 355 | 0.38 (0.19)b | 179.7 (88.7)b |
| Forest | 26 | 429 | 0.46 (0.23)c | 221.5 (109.1)c |
Variable positions indicate the number of variable nucleotide sites (total n = 488) within each library. Values with different letters (superscripts) are significantly different (comparisons with Halema'uma'u [P ≤ 0.05] and Forest [0.05 ≤ P ≤ 0.1] by a one-tailed t-test).
TABLE 4.
Corrected average pairwise differences (θ[π], above diagonal) and pairwise fixation indices (FST, below diagonal)
| Site | Result for sample from:
|
|||
|---|---|---|---|---|
| Halema'uma'u | Caldera Rim | Pu'u Puai | Forest | |
| Halema'uma'u | 11.2 | 94.1 | 98.9 | |
| Caldera Rim | 0.067 | 57.2 | 72.3 | |
| Pu'u Puai | 0.371 | 0.236 | 37.6 | |
| Forest | 0.357 | 0.261 | 0.153 | |
All values are significant (P ≤ 0.05).
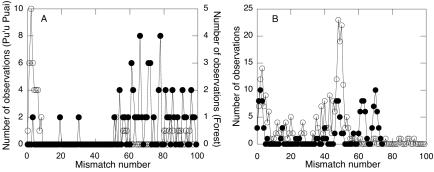
Frequency distributions of sequence mismatches for Forest and Caldera Rim libraries were multimodal, or ragged (Fig. 2). In contrast, the mismatch distribution for Pu'u Puai was largely unimodal, with the greatest frequency of mismatches occurring between 0 and 6 mismatches (Fig. 2B). The Halema'uma'u library distribution was approximately bimodal, with a prominent peak at approximately 50 mismatches and a smaller secondary peak between 0 and 4 mismatches (Fig. 2B).
FIG. 2.
Frequency distribution of pairwise mismatches within coxL clone libraries. (A) Caldera Rim (•) and Halema'uma'u (○); (B) Forest (•) and Pu'u Puai (○).
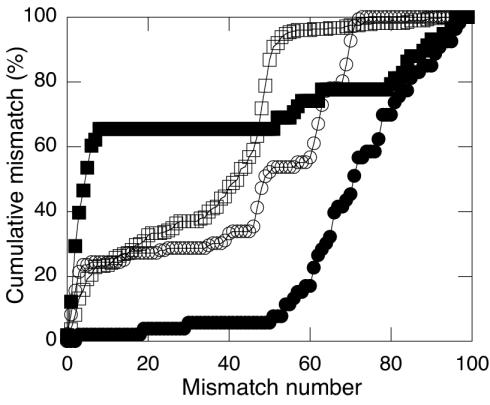
Plots of cumulative percentage of the total mismatch for each library revealed unique patterns, the most unique of which occurred for Pu'u Puai, where a large fraction of closely related clone sequences (<10 mismatches) accounted for nearly 70% of total cumulative mismatch (Fig. 3). The cumulative percentage of total mismatch increased approximately exponentially for the Forest library, with the majority of sequences having greater than 70 mismatches. The cumulative percent of total mismatch increased approximately sigmoidally with mismatch level in the Caldera Rim and Halema'uma'u libraries, with the majority of sequences showing greater than 50 mismatches.
FIG. 3.
Cumulative distribution of mismatch as a function of the number of mismatches. Forest, •; Caldera Rim, ▪; Halema'uma'u, □; Pu'u Puai, ○.
DISCUSSION
Recent enrichment and genomic studies have shown that aerobic CO oxidation occurs among numerous lineages within the α-, β-, and γ-Proteobacteria and Actinobacteria and Firmicutes (12). However, the extent of CO oxidizer diversity in situ remains largely unknown, since the only information available to date has been limited to culture descriptions. Molecular studies have been precluded until recently due to the lack of suitable CODH or 16S rDNA primers. Nonetheless, insights from enrichments and cultures have indicated that CO oxidizer diversity may parallel that of the wide range of systems in which CO oxidation occurs.
Primers based on sequences derived from a variety of representative CO oxidizers (12) have been used to assess the presence and diversity in volcanic deposits of coxL in a clade (OMP) characterized by several well-known carboxydotrophs (e.g., Oligotropha carboxidovorans and Pseudomonas thermocarboxydovorans) and a number of newly recognized CO oxidizers (12). In addition to documenting greater diversity than previously recognized in known lineages, results presented here also document substantial diversity in yet-unidentified bacterial lineages.
Since phylogenetic analyses of inferred amino acid sequences obtained from a variety of isolates have indicated that the coxL phylogeny is congruent with 16S rDNA phylogeny (12), it is possible tentatively to assign clones to major bacterial divisions and, in some cases, subdivisions (Fig. 1; Table 1). Accordingly, about 40% (44 of 109) of the clones overall cluster with Proteobacteria and Actinobacteria and Firmicutes. However, 60% of the clone sequences cannot be associated with bacterial divisions known to harbor CO oxidizers. This finding suggests that aerobic CO oxidation is more widespread among the bacterial domain than previously imagined and that CO oxidizers may occur in common terrestrial divisions such as Acidobacterium and Verrucomicrobium (1, 2, 6, 8, 16, 17, 19, 25), representatives of which have been identified in Hawaiian volcanic deposits by 16S rDNA sequence analysis (24; V. Gomez-Alvarez, G. M. King, and K. Nuesslein, unpublished data).
For the Forest and Pu'u Puai libraries, modest numbers of the clones, 12 and 32%, respectively, clustered with unknown lineages. However, the majority of clones from the Caldera Rim (81%) and Halema'uma'u (96%) could not be assigned to a bacterial phylum. This result may reflect differences in successional state and various biological and abiological parameters that distinguish the Caldera Rim and Halema'uma'u samples from the Forest and Pu'u Puai samples. In particular, the latter sites occur within a well-vegetated, relatively moist area of the caldera, while the former occur in a region with reduced water availability and substantially less plant growth (11).
It should also be noted that one of the Halema'uma'u clones, HM-12, clustered with BMS putative coxL sequences. This occurrence likely results from the similarity between OMP and BMS sequences in the regions targeted for primer development (12). Nonetheless, the rare occurrence of a BMS-like clone indicates that the primers used here are relatively specific for environmental OMP sequences across a broad phylogenetic range.
Patterns for nucleotide diversity and average pairwise difference generally agree with clone phylogeny. For instance, the lowest values for both parameters are obtained for the Halema'uma'u library (Table 3), which also has the most-limited phylogenetic distances among clones, while the Forest site supports the most-divergent sequences and has the highest nucleotide diversity and average pairwise difference. However, it is also evident from results for the Pu'u Puai and Caldera Rim libraries that similar levels of diversity can be obtained from libraries with substantially different phylogenetic compositions (Tables 1 and 3). Therefore, without additional information about population structure, such indices may have limited value.
Paired reciprocal comparisons performed using LIBSHUFF indicate that each of the libraries differs significantly from the others (Table 2), in general agreement with phylogenetic results. This result is true even for sites with similar nucleotide diversity and average pairwise difference values, e.g., Pu'u Puai and the Caldera Rim (Tables 3 and 4). Thus, LIBSHUFF comparisons, which have thus far been used primarily for 16S rDNA sequence data, appear to provide useful insights for functional genes and to complement other analyses.
However, unlike the case for 16S rDNA sequence analyses, it is not yet evident what level of nucleotide similarity should be used for distinguishing coxL OTUs. For the study described herein, only 100% identical sequences were considered equivalent OTUs. This decision reflects the fact that essentially identical coxL sequences have been obtained from taxonomically distinct cultures (e.g., Mycobacterium bovis, M. tuberculosis, and M. microti) (13). Accordingly, LIBSHUFF data may underestimate the true level of library coverage.
The unique distribution of taxa among sites is supported by several other analyses. Fixation indices (FST) and corrected average pairwise differences derived from paired library comparisons by AMOVA suggest that the most divergent populations occur in sites with distinct plant and water regimes (e.g., Halema'uma'u versus Pu'u Puai or Forest; Table 4), while the least divergent populations occur in sites with more similar plant and water regimes (e.g., Forest and Pu'u Puai or Halema'uma'u and Caldera Rim; Table 4). These observations agree with results from LIBSHUFF and a P-test (17), which show that sequence distributions within libraries covary significantly with phylogenetic position. Collectively, the various statistical analyses suggest that CO oxidizer population structure responds to environmental (moisture and plant growth) and historical (e.g., eruption and date of deposition) differences among sites located within a radius of less than 2.5 km.
Mismatch distributions also varied among libraries (Fig. 2). For the Forest and Caldera Rim samples, mismatch distributions are multimodal, reflecting a relatively even distribution of closely and distantly related taxa. In contrast, mismatch distributions for Pu'u Puai and Halema'uma'u samples are approximately unimodal and bimodal, respectively. The shape of the mismatch curves for the latter sites may result from recent population expansions or constraints on diversity imposed by limited ecosystem complexity. Curves for the former sites may result from increased diversity accompanying greater deposit age and successional development (11, 33). Similar results have been obtained for analyses of the large subunit gene for ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) (22).
Although distinct CO-oxidizing communities exist at each of the sites, variations among sites in coxL diversity indices (e.g., nucleotide diversity and average pairwise difference) do not correlate (r = 0.23) with variations in atmospheric CO uptake rates (11). Indeed, atmospheric CO uptake rates are lowest for samples from Forest, the site with the highest diversity. Similarly, patterns of rbcL diversity for the same sites do not correlate with CO and hydrogen uptake, even though these gases likely support most, if not all, of the facultative lithotrophic activity (22). Whether similar relationships exist between the diversity of other functional genes and their activities in situ is uncertain. However, the results presented here indicate that diversity and function may be uncoupled in at least some systems.
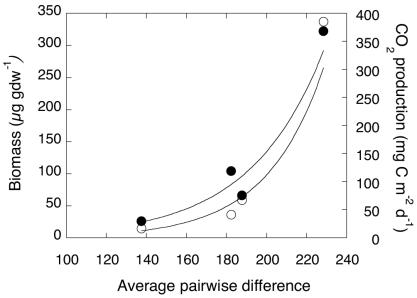
In contrast to the weak correlation between coxL diversity and CO uptake, both microbial biomass and respiration correlate strongly (r = 0.99; Fig. 4) with average pairwise difference (and nucleotide diversity). One explanation for this correlation is that ecosystem level changes that promote increased microbial abundance and activity also increase opportunities for niche differentiation and diversification of CO oxidizers. Changes in the availability and diversity of organic substrates with successional development may be particularly important since almost all CO oxidizers function preferentially as heterotrophs (20, 21).
FIG. 4.
Correlation of microbial biomass (r = 0.99; ○) and respiration (r = 0.99; •) with average pairwise difference within clone libraries (Table 1). Biomass and respiration data from King (11). gdw, grams (dry weight).
Of course, additional analyses, e.g., reverse transcription-PCR, will be necessary to assess the diversity of populations that actively express coxL and that may be involved with atmospheric CO consumption under in situ conditions. However, organic limitation in soils generally and volcanic deposits specifically (11) may select for numerous taxa that use CO as an energy supplement for growth with heterotrophic substrates.
In conclusion, phylogenetic analyses of coxL sequences derived from genomic extracts of recent Hawaiian volcanic deposits have revealed a substantial and previously unrecognized level of CO oxidizer diversity. Some clone sequences cluster with, yet are distinct from, sequences of known CO oxidizers in Proteobacteria, Actinobacteria, and Firmicutes; the majority of sequences occur in unidentified lineages. Phylogenetic and statistical analyses also show that distinct CO-oxidizing communities occur at sites differing in deposit age and ecosystem development, but differences in community structure are not correlated with estimates of atmospheric CO uptake. Remarkably, the level of CO oxidizer diversity observed in a limited regional scale sampling of a remote island appears to exceed reports of diversity for another major group of trace gas-utilizing bacteria, the methanotrophs, which have been assayed more extensively over a much broader range of habitats.
Acknowledgments
This work was supported in part by the National Science Foundation (BIO-0085495).
We thank K. Nanba for technical assistance and W. Gilmartin for use of Hale Mahana.
Footnotes
Contribution no. 393 from the Darling Marine Center.
REFERENCES
- 1.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deForestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruns, M. A., J. R. Stephen, G. A. Kowalchuk, J. I. Prosser, and E. A. Paul. 1999. Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled, and successional soils. Appl. Environ. Microbiol. 65:2994-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews, T. E., H. Farrington, and P. M. Vitousek. 2000. Changes in asymbiotic, heterotrophic nitrogen fixation on leaf litter of Metrosideros polymorpha with long-term ecosystem development in Hawaii. Ecosystems 3:386-395. [Google Scholar]
- 5.Dedysh, S. V., M. Derakshani, and W. Liesack. 2001. Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67:4850-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felske, A., H. Rheims, A. Wolterink, E. Stackebrandt, and A. D. L. Akkermans. 1997. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983-2989 [DOI] [PubMed] [Google Scholar]
- 7.Gulledge, J., A. Ahmed, P. A. Steudler, W. J. Pomerantz, and C. M. Cavanaugh. 2001. Family- and genus-level 16S rRNA-targeted oligonucleotide probes for ecological studies of methanotrophic bacteria. Appl. Environ. Microbiol. 67:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janssen, P. H., P. S. Yates, B. E. Ginton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang, B. S., and Y. M. Kim. 1999. Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava. J. Bacteriol. 181:5581-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King, G. M. 2000. Impacts of land use on atmospheric carbon monoxide consumption by soils. Global Biogeochem. Cycles 14:1161-1172. [Google Scholar]
- 11.King, G. M. 2003. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 69:4067-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, G. M. 2003. Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 69:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King, G. M. 2003. Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl. Environ. Microbiol. 69:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King, G. M., and H. Crosby. 2002. Impacts of plant roots on soil CO cycling and soil-atmosphere CO exchange. Glob. Change Biol. 8:1085-1093. [Google Scholar]
- 15.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. Martin Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β-subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer, O., and H. G. Schlegel. 1983. Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 37:277-310. [DOI] [PubMed] [Google Scholar]
- 21.Morsdorf, G., K. Frunzke, D. Gadkari, and O. Meyer. 1992. Microbial growth on carbon monoxide. Biodegradation 3:61-82. [Google Scholar]
- 22.Nanba, K., G. M. King, and K. Dunfield. 2004. Analysis of facultative lithotrophs distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase. Appl. Environ. Microbiol. 70:2245-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neigel, J. E. 2002. Is FST obsolete? Conserv. Genet. 3:167-173. [Google Scholar]
- 24.Nüsslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Farrell, K. A., and P. H. Janssen. 1999. Detection of verrucomicrobia in a pasture soil by PCR-mediated amplification of 16S rRNA genes. Appl. Environ. Microbiol. 65:4280-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rich, J. J., and G. M. King. 1998. Carbon monoxide oxidation by bacteria associated with the roots of freshwater macrophytes. Appl. Environ. Microbiol. 64:4939-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rich, J. J., and G. M. King. 1999. Aerobic and anaerobic transformations of carbon monoxide in freshwater peats. FEMS Microbiol. Ecol. 28:215-224. [Google Scholar]
- 28.Santiago, B., U. Schubel, C. Egelseer, and O. Meyer. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236:115-124. [DOI] [PubMed] [Google Scholar]
- 29.Schneider, S., D. Roesseli, and L. Excoffier. 2000. Arlequin version 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland.
- 30.Singleton, D. R., M. A. Furlong, S. L. Rathburn, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephen, J. R., G. A. Kowalchuk, M. A. Bruns, A. E. McCaig, C. J. Phillips, T. Martin Embley, and J. I. Prosser. 1998. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitousek, P. M., L. R. Walker, L. D. Whitaker, D. Mueller-Dombois, and P. A. Matson. 1987. Biological invasion by Myrica fava alters ecosystem development in Hawaii. Science 238:802-804. [DOI] [PubMed] [Google Scholar]
- 33.Weckstein, J. D., A. D. Afton, R. M. Zink, and R. T. Alisauskas. 2002. Hybridization and population subdivision within and between Ross's geese and Lesser snow geese: a molecular perspective. Condor 104:432-436. [Google Scholar]
- 34.Wise, M. G., J. V. McArthur, and L. J. Schimkets. 1999. Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 65:4887-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright, S. 1951. The genetic structure of populations. Ann. Eugenics 15:323-354. [DOI] [PubMed] [Google Scholar]