Abstract
To effectively monitor biodegrading populations, a comprehensive 50-mer-based oligonucleotide microarray was developed based on most of the 2,402 known genes and pathways involved in biodegradation and metal resistance. This array contained 1,662 unique and group-specific probes with <85% similarity to their nontarget sequences. Based on artificial probes, our results showed that under hybridization conditions of 50°C and 50% formamide, the 50-mer microarray hybridization can differentiate sequences having <88% similarity. Specificity tests with representative pure cultures indicated that the designed probes on the arrays appeared to be specific to their corresponding target genes. The detection limit was ∼5 to 10 ng of genomic DNA in the absence of background DNA and 50 to 100 ng of pure-culture genomic DNA in the presence of background DNA or 1.3 × 107 cells in the presence of background RNA. Strong linear relationships between the signal intensity and the target DNA and RNA were observed (r2 = 0.95 to 0.99). Application of this type of microarray to analyze naphthalene-amended enrichment and soil microcosms demonstrated that microflora changed differently depending on the incubation conditions. While the naphthalene-degrading genes from Rhodococcus-type microorganisms were dominant in naphthalene-degrading enrichments, the genes involved in naphthalene (and polyaromatic hydrocarbon and nitrotoluene) degradation from gram-negative microorganisms, such as Ralstonia, Comamonas, and Burkholderia, were most abundant in the soil microcosms. In contrast to general conceptions, naphthalene-degrading genes from Pseudomonas were not detected, although Pseudomonas is widely known as a model microorganism for studying naphthalene degradation. The real-time PCR analysis with four representative genes showed that the microarray-based quantification was very consistent with real-time PCR (r2 = 0.74). In addition, application of the arrays to both polyaromatic-hydrocarbon- and benzene-toluene-ethylbenzene-xylene-contaminated and uncontaminated soils indicated that the developed microarrays appeared to be useful for profiling differences in microbial community structures. Our results indicate that this technology has potential as a specific, sensitive, and quantitative tool in revealing a comprehensive picture of the compositions of biodegradation genes and the microbial community in contaminated environments, although more work is needed to improve detection sensitivity.
The transformation of environmental contaminants is a complex process that is influenced by the nature and amount of the contaminant present, the structure and dynamics of the indigenous microbial community, and the interplay of geochemical and biological factors at contaminated sites (1, 14, 26, 36). A better understanding of the processes inherent in natural bioremediation requires, in part, a better understanding of microbial ecology. However, conventional molecular methods (PCR-based technologies, such as gene cloning, terminal-restriction fragment length polymorphism, denaturing gradient gel electrophoresis, and in situ hybridization) for assessing microbial community structure and activities are labor-intensive. Rapid, quantitative, and cost-effective tools that can be operated in field scale heterogeneous environments are needed for measuring and evaluating bioremediation strategies and endpoints.
The microarray is a powerful genomic technology that is widely used to study biological processes. Although microarray technology has been used successfully to analyze global gene expression in pure cultures (8, 30, 32, 39, 41, 45, 52, 55), adapting microarray hybridization for use in environmental studies presents great challenges in terms of specificity, sensitivity, and quantitation (56, 58). Although microarray-based genomic technology has attracted tremendous interest among microbial ecologists, it has only recently been extended to study microbial communities in the environment (6, 17, 33, 42, 46, 47, 48, 53).
Based on the types of probes arrayed, microarrays used in environmental studies can be divided into three major classes (56, 58): functional gene arrays (FGAs), community genome arrays (CGAs), and phylogenetic oligonucleotide arrays. FGAs contain probes corresponding to genes encoding key enzymes involved in various ecological and environmental processes, such as carbon fixation, nitrification, denitrification, sulfate reduction, and contaminant degradation. Both PCR-amplified DNA fragments (53) and oligonucleotides derived from functional genes can be used to fabricate FGAs. To avoid confusion, the former are referred to as PCR product-based FGAs, whereas the latter are referred to as oligonucleotide-based FGAs. These types of arrays are useful in studying the physiological status and functional activities of microbial communities in natural environments (58). CGAs are constructed using whole genomic DNA isolated from pure-culture microorganisms and can be used to describe a microbial community in terms of its cultivable component (56). Phylogenetic oligonucleotide arrays are constructed with short synthetic oligonucleotides from rRNA genes and can be used for phylogenetic analyses of microbial-community composition and structure in environmental samples.
To construct FGAs containing large DNA fragments, the probes used for microarray fabrication are generally amplified by PCR from environmental clones or from pure genomic DNA. However, obtaining all the diverse environmental clones and bacterial strains from various sources as templates for amplification is a big challenge. Constructing comprehensive microarrays is also very difficult. Due to their high specificity and ease of construction, oligonucleotide-based microarrays are an important array-based approach for microbial ecology. However, although it holds great promise for environmental studies, this type of microarray has not been rigorously tested and validated within the context of environmental applications. In this study, for monitoring biodegradation potential and activity, we developed comprehensive 50-mer oligonucleotide microarrays containing 1,657 probes from all 2,402 known genes involved in biodegradation and metal resistance. Our results demonstrated that the 50-mer microarrays developed offer a new rapid, powerful, high-throughput tool for monitoring bioremediation potential and functional activity.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and soil samples.
Pseudomonas putida Gpo1 (γ-proteobacterium; aerobic alkane degrader) and Thauera aromatica K-172 (β-proteobacterium; anaerobic toluene, phenol, and benzoate degrader) used in this study were obtained from the American Type Culture Collection and the German Collection of Microorganisms and Cell Cultures, respectively. P. putida PpG7 (γ-proteobacterium; aerobic naphthalene degrader) was a gift of C. O. Jeon of Cornell University. These strains were grown in M1 minimal medium (hereafter referred to as M1) (35) supplemented with the appropriate carbon source (e.g., 10 mM naphthalene), which was added using 1 M diethyl ether. After evaporation of the diethyl ether for 2 h in a fume hood, the cells were seeded and incubated under the appropriate conditions. Shewanella oneidensis MR-1 (γ-proteobacterium; metal-reducing bacterium) was available in our laboratory (45) and was grown in Luria-Bertani broth. In all experiments, both P. putida and S. oneidensis were grown aerobically at 25°C with shaking at 200 rpm. T. aromatica K-172 was grown under denitrifying conditions using 10 mM nitrate as described by Breese et al. (4). Cells in the exponential phase were quickly harvested and frozen at −80°C for the extraction of DNA and RNA. Genomic DNA of Rhodopseudomonas palustris (α-proteobacterium; anaerobic toluene, phenol, and benzoate degrader) was a gift of C. S. Harwood (University of Iowa).
Four soils, three of which were contaminated, were used to evaluate the performance of the developed oligonucleotide microarray. The TFD site was located in Mercer County, N.J., where a steel and fiber drum-reconditioning facility operated from 1965 to 1985 (21). The soil sample contained 27.5 mg of background polyaromaric hydrocarbons (PAHs) and various kinds of heavy metals/kg of soil. The PCT18 site was officially established in 1980 in Morris County, N.J., and currently serves as a munitions and weapons research and development installation. PCT18 soil contained 3.3 mg of PAHs/kg of soil. The IDT site was owned by the Indiana Department of Transportation, and ∼20 years ago, paints and various kinds of solvents were dumped at the site. The soil sample from IDT was from the subsurface (1 m below the surface) and contained 3.0 mg of benzene, toluene, ethylbenzene, and xylene (BTEX), 17 mg of Pb, and 6.3 mg of Cr/kg of soil. This soil sample was kindly provided by A. Konopka (Purdue University). A forest soil (OR) sample (<1 mg of PAHs/kg of soil) was collected from the free-air carbon dioxide enrichment site at Oak Ridge National Laboratory, Oak Ridge, Tenn., and was used as an uncontaminated control. Three soil samples were collected from each site and mixed to make composite samples. All soil samples collected were stored at −70°C prior to use.
Establishment of enrichment cultures and microcosms.
One gram of TFD soil was added to 50 ml of M1 supplemented with 5 mM naphthalene in sealed shake flasks. After 2 weeks of incubation at 30°C, 1% (vol/vol) of each culture was transferred to fresh M1 supplemented with 5 mM naphthalene and incubated under the same conditions. Similarly, 1 g of TFD soil was added to 50 ml of M1 supplemented with 10 mM pyruvate to establish a control enrichment. The cells were harvested by centrifugation (4,000 × g for 10 min at 4°C) in the middle of the growth phase, frozen quickly, and stored at −70°C before simultaneous extraction of RNA and DNA.
Three different soil microcosms were established using 10 g of TFD soil in a 150-ml capped serum bottle with three replicates for each microcosm. The control contained no naphthalene vapor, one microcosm contained naphthalene vapor, and one microcosm contained naphthalene but used sterilized TFD soil. To provide naphthalene vapor, naphthalene particles (0.1 g) were spiked before incubation. The serum bottle was incubated in a static condition at 30°C. After 2 weeks of incubation, naphthalene consumption and bacterial numbers in the microcosm were measured (see below), and 5 g of microcosm soil was collected for DNA extraction.
To measure naphthalene concentrations, 10 ml of hexane-butanol (9:1) was added to each microcosm and shaken overnight (52). After the soil settled, 5 μl of the organic phase was analyzed using a Hewlett-Packard model 5890 series II gas chromatograph equipped with a 5% phenyl methyl silicone fused silica column (H-P5). The injector and detector temperatures were maintained at 250 and 300°C, respectively. The temperature profile for gas chromatography analysis was 100°C for 1 min, followed by an increase of 10°C/min to 220°C. The total number of bacteria was determined using acridine orange fluorescent direct counting as described by Kirchman et al. (24). One gram of soil was shaken in 10 ml of saline solution (0.85% NaCl) for 2 h. After the soil settled, the supernatant was filtered using a 1.2-μm-pore-size filter (Acrodisc; German Laboratory) to remove soil particles. The filtrate was stained using 0.05% acridine orange (final concentration, 0.005%) for 5 min and filtered using a Nuclepore polycarbonate filter (0.2-μm pore size). The stained cells on the filter were examined by microscopy.
Nucleic acid extraction and quantification.
Genomic DNA was extracted using a Wizard Genomic DNA Purification kit (Promega, Madison, Wis.). Total RNA was extracted using TRIzol LS reagent (Life Technology, Gaithersburg, Md.) and purified with an RNeasy kit (Qiagen, Valencia, Calif.).
DNA and RNA from the naphthalene-degrading enrichment culture were extracted and purified simultaneously as previously described (22). RNA was separated from DNA and purified using a Qiagen RNA/DNA minikit. The bulk community DNA from the soil microcosms was extracted by using an UltraClean Soil DNA Mega Prep kit (Mo Bio Laboratories, Solana Beach, Calif.) with the following modification. Frozen soil samples (5 g) were ground with a mortar and pestle in the presence of sterile sand and liquid nitrogen (22, 57). The ground soil was added to the bead solution tube and processed for DNA extraction according to the manufacturer's protocol. The extracted DNA was concentrated and further purified using a Wizard Genomic DNA Purification kit following the manufacturer's protocol with the exception that the soil DNA was mixed with resin and allowed to sit for >30 min instead of 1 min.
DNA extracted from the microbial community was quantified by fluorometry using a SPECTRAmax fluorometer (Molecular Devices, Sunnyvale, Calif.). The fluorometer was calibrated with pure herring sperm DNA (Invitrogen, Carlsbad, Calif.). The DNA concentrations were determined in triplicate. The quality of RNA extracted from the enrichment culture was checked using agarose gel electrophoresis. Intactness of 16S and 28S rRNA bands on the gel was used as the indicator of RNA quality. DNA and RNA purified from pure culture were quantified using a spectrophotometer (Nanodrop Technologies, Rockland, Del.).
Oligonucleotide probe (50-mer) design.
The oligonucleotide-based microarray for detecting bioremediation activity contained probes from various groups of genes involved in organic contaminant degradation and metal resistance. The University of Minnesota Biocatalysis/Biodegradation Database (http://umbbd.ahc.umn.edu/) was used to identify the degradation pathways of known pollutants and their metabolites. The names of the pollutants known to be in the soil and their metabolites were used as keywords for identifying appropriate genes in the GenBank database (September 2002) through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). The gene identification (gi) numbers for the genes of interest were retrieved and used for automatically downloading gene sequences. The gene sequences of toluene monoxygenases from various toluene degraders in our laboratory were also included. Altogether, 2,402 sequences from eight functional groups involved in organic-contaminant degradation and metal resistance were collected (Table 1).
TABLE 1.
Summary of probes designed
| Chemical class | Biodegradation type | Target chemicala | No. of genes | Total no. of probes synthesized | Representative genes for overall diversity evaluation | No. of genes for similarity calculation | Avg (range) of similarity (%) |
|---|---|---|---|---|---|---|---|
| Organic | Aerobic degradation | Monoaromatic compoundsb | 697 | 542 | Phenol hydroxylase | 58 | 70 (30-85) |
| Polyaromatic compoundsb | 285 | 212 | Naphthalene dioxygenase | 17 | 51 (31-94) | ||
| Biphenylb | 125 | 76 | Biphenyl dioxygenase | 49 | 34 (21-93) | ||
| Catechol and its derivatives | 669 | 417 | Catechol dioxygenase | 71 | 45 (25-94) | ||
| Aliphatic compounds | 218 | 131 | Alkane monoxygenase | 18 | 34 (21-83) | ||
| Anaerobic degradation | Monoaromatic compounds | 120 | 99 | NAc | |||
| Metal | Heavy metal reduction | 153 | 120 | Arsenate reductase | 32 | 31 (18-86) | |
| Heavy metal export | 135 | 73 | Cadmium-transporting ATPase | 18 | 32 (22-74) | ||
| Total | 2,402 | 1,662 | NA |
Monoaromatic compounds: benzene, toluene, xylene, phenol, halophenol, phthalate, cymene, halobenzoate, 2,4-D,2,4,5-T, hydroxyphenolpropionate, cyclohexanol, hydroxybenzoate, nitrotoluene, hydroxyacetophenone, cyclohexane, alkylbenzene, aniline, anthranilate, styrene, gentisate, salicylate, and nitrobenzene. Polyaromatic compounds: naphthalene, phenanthrene, pyrene, dibenzothiophene, and dioxin carbazole. Biphenyl: polychlorinated biphenyl, biphenyl, aminobiphenyl, and trihydroxybiphenyl. Aliphatic compounds: alkane, haloalkane, alkene, haloethylene, methane acetone, nitrile, chloroacrylic acid, hanomethane, haloacid, and parathion.
Genes involved only in the transformation of aromatic compounds to catechol and its derivatives.
NA, not available.
The 50-mer oligonucleotide probes were designed using the software PRIMEGENS (54) with modified parameters. To design 50-mer oligonucleotide probes, each individual gene sequence was compared against the entire downloaded sequence database using pairwise BLAST and aligned with the other sequences using dynamic programming. Based on these global optimal alignments, segments of 50-mer with <85% nucleotide identity to the corresponding aligned regions of any of BLAST hit sequences were selected as potential probes. In this process, we considered stretches of matches between aligned regions. Probes with >15-bp matching stretches were removed from the potential probes (23). Among the identified potential probes, one was finally selected by considering melting temperature and self-complementarity. About 10% of the probes selected were searched against the GenBank database and proved to be unique in the database.
In summary, a total of 1,662 oligonucleotide probes were designed. The information on the probe sequences, control probes, melting temperatures, organisms of origin, and gene functions can be found at our website (http://www.esd.ornl.gov/facilities/genomics/index.html).
Microarray construction and postprocessing.
The designed oligonucleotide probes were synthesized without modification by MWG Biotech, Inc. (High Point, N.C.), in 96-well plate format. The oligonucleotides were diluted to a final concentration of 50 pmol μl−1 in 50% dimethyl sulfoxide (Sigma, St. Louis, Mo.). Ten microliters of each probe was transferred to a 384-well microplate for printing. The probes were arrayed with eight pins (Chipmaker 3; Telechem International, Inc., Sunnyvale, Calif.) at a spacing of 250 μm onto 25- by 75-mm Superamine glass slides (Telechem International, Inc.) using a PixSys 5500 Printer (Cartesian Technologies, Inc., Irvine, Calif.) at 55 to 58% relative humidity. Each probe set was printed in duplicate on a different part of the slide. The slides were cross-linked by using a UV Stratalinker 1800 (Stratagene, La Jolla, Calif.). The slides were exposed to 80 mJ of UV irradiation and washed at room temperature with 0.1% sodium dodecyl sulfate (SDS) for 4 min, followed by washing with water for 2 min. The slides were dried by centrifugation at 500 × g for 5 min and stored in a clean slide box at room temperature.
Fluorescent labeling of target DNA and RNA.
Two methods were used to fluorescently label DNA. For genomic DNA labeling, we used the BioPrime DNA Labeling kit (Invitrogen). Genomic DNA (500 ng) was mixed with 15 μg of random octamer, denatured by being boiled for 2 min, and immediately chilled on ice. The denatured genomic-DNA solution was then mixed with 15 μl of a labeling reaction solution containing 5 mM (each) dATP, dTTP, and dGTP; 2.5 mM dCTP (New England Biolabs, Beverly, Mass.); 1 mM Cy3 dUTP (Amersham Pharmacia Biotech, Piscataway, N.J.); and 40 U of Klenow fragment (Invitrogen). The reaction mixture was incubated at 37°C for 3 h. The labeled target DNA was purified using a QIAquick PCR purification column (Qiagen), concentrated in a Speedvac at 40°C for 1 h, and resuspended in an appropriate volume of distilled water. To quantify the target gene, human gene PCR products of known concentrations were mixed with the target DNA prior to labeling it (46, 47).
For PCR product labeling, each gene was amplified using gene-specific primers. The PCR amplification conditions were as follows: 95°C for 2 min for 1 cycle; 94°C for 1 min, 53°C for 30 s, and 72°C for 30 s for 30 cycles; and 72°C for 5 min for 1 cycle. After purification with a QIAquick PCR purification column (Qiagen), 1 ng of PCR product was labeled using random priming.
Labeled DNA template was produced from RNA with Cy3 or Cy5 using reverse transcription. First, 5 μg of total RNA was mixed with 10 μg of random primer in a 16.5-μl volume, incubated at 70°C for 5 min, and chilled on ice. RNA was labeled by mixing it with 13.5 μl of labeling solution containing 10 mM (each) dATP, dGTP, and dCTP; 0.5 mM dTTP; 3 μl of 0.1 M dithiothreitol; 40 U of RNase inhibitor (Gibco BRL and Invitrogen); 1 mM Cy3-dUTP or Cy5 dUTP (Perkin-Elmer/NEN Life Science Products, Boston, Mass.); and 200 U of Superscript RNase H− reverse transcriptase in 1× First Strand buffer. Two different dyes (Cy3-dUTP and Cy5-dUTP) were used for labeling RNAs from treatment and control samples, respectively. To quantify the target RNA, mRNA (AF159801) of Arabidopsis thaliana (0.1 ng; Stratagene) was mixed with total RNA prior to RNA labeling.
Hybridization.
All hybridizations were carried out in triplicate. The probe was completely dried under vacuum and mixed with hybridization solution. The hybridization solution contained 8 μl of formamide, 3× SSC (1× SSC is 150 mM NaCl and 15 mM trisodium citrate), 1 μg of unlabeled herring sperm DNA (Promega), and 0.31% SDS in a total volume of 17.5 μl. The hybridization solution was denatured at 95°C for 5 min. After heat denaturation, the hybridization solution was kept at >50°C until it was washed to prevent cross-hybridization. The hybridization mixture was deposited directly onto slides, which were prewarmed to 50°C, and covered with a coverslip. The microarray was placed into a self-contained flow cell (Telechem International) and plunged into the 50°C water bath immediately for overnight hybridization. After hybridization, the time that the slide remained at room temperature was minimized in order to prevent cross-hybridization. Each microarray slide was taken out, and the coverslip was immediately removed in wash solution 1 (1× SSC and 0.2% SDS). The slides were washed using wash solution 1, wash solution 2 (0.1× SSC and 0.2% SDS), and wash solution 3 (0.1× SSC) for 5 min each at ambient temperature prior to being dried. The slides were dried using centrifugation as described above.
Image processing and data analysis.
The microarrays were scanned with a ScanArray 5000 Microarray Analysis system (Perkin-Elmer, Wellesley, Mass.) at a resolution of 10 μm. For detection sensitivity experiments, the laser power and photomultiplier tube (PMT) gain were both 100%. For all other experiments, the laser power and PMT gain were adjusted to avoid saturation of spots.
The scanned images were saved as 16-bit TIFF files, and each spot was quantified using ImaGene version 4.0 software (Biodiscovery Inc., Los Angeles, Calif.). A grid of individual circles defining the location of each DNA spot on the array was superimposed on the image to designate each fluorescent spot to be quantified. The mean signal intensity was determined for each spot. The local background signal was subtracted automatically from the hybridization signal of each separate spot. Fluorescence intensity values for all replicates of the human genes (negative controls) were averaged and then subtracted from the background-corrected intensity values for each hybridization signal. Poor-quality spots were automatically flagged by the software. The signal-to-noise ratio (SNR) was also calculated based on the following formula (49): SNR = (signal intensity − background)/standard deviation of background, in which the background measurement refers to the local spot background intensity and the standard deviation of background was calculated across all pixels measured by the ImaGene software. The SNRs from six replicate data sets were then averaged to represent the SNR for a particular probe. Spots that appeared to be lower than the threshold value were also removed from the data set for further analysis. A commonly accepted criterion for the minimum signal (threshold) that can be accurately quantified is an SNR of ≥3 (49).
For the hybridization experiments in which two fluorescent dyes (Cy3 and Cy5) were used simultaneously in hybridization, the signal intensity for each spot was normalized based on the signal intensity of the human control genes (0.1 ng) spiked equally prior to being labeled. The signal ratios of Cy5 to Cy3 for the three human genes were calculated and averaged. The signal intensity for each spot from the Cy3 channel was then multiplied by the averaged Cy5/Cy3 ratio for human genes to obtain corrected Cy3 signals for individual hybridization spots. Outlier detection and removal were carried out using ArrayStat (Imaging Research, Inc., St. Catharine, Ontario, Canada). The signal intensity ratio for each spot between the treatment and control samples was then calculated and used for further analysis. Statistical analysis was performed using SigmaPlot version 5.0 (Jandel Scientific, San Rafael, Calif.). The relationships of the genes detected in various soil samples were determined using hierarchical cluster analysis (CLUSTER) and visualized with TREEVIEW (12).
The free-energy change (ΔG°) of probe-target hybrids was calculated at a temperature of 65°C (ΔG°65) for prediction of DNA hybridization, using mfold version 3.1 together with SantaLucia free-energy parameters (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form6.cgi) (59).
Real-time PCR quantification.
In order to validate the microarray hybridization results, four genes detected in the TFD sample and associated enrichments and microcosms were selected for further analysis by real-time PCR using Sybrgreen (Molecular Probes, Eugene, Oreg.). A specific primer set was designed for the detection of each of the following genes (∼200-bp amplicon): naphthalene dioxygenase large subunit (gi4704462), naphthalene dioxygenase small subunit (gi4704463), cis-naphthalene dihydrodiol dehydrogenase (gi4827073), salicylate-5-hydroxylase large oxygenase component (gi2828015), salicylate-5-hydroxylase small oxygenase component (gi2828016), and naphthalene 1,2-dioxygenase large oxygenase component (gi2828018). Prior to real-time PCR, conventional PCR amplification was performed to test the specificities of the designed primers and the purity of community DNA templates with the conditions of denaturation of DNA (15 s at 94°C), annealing of primers (30 s at 60°C), and elongation (30 s at 72°C) in 45 cycles with different series of dilutions of community DNA templates. The lowest dilution (10−2) of community DNA which did not inhibit PCR amplification was used. Only the primers that generated single PCR products were used for further real-time analysis.
Real-time PCR (50 μl) was performed using Thermo-fast 96 PCR plates (Bio-Rad Laboratories, Hercules, Calif.), which were sealed with iCycler IQ optical-quality tapes (Bio-Rad Laboratories) on an iCycler IQ thermocycler (Bio-Rad Laboratories). Each measurement was performed in three replicates. A dilution series of positive control DNA was used in the same plates as the calibration standards. Positive control DNA was generated by the amplification of the gene from samples. The amplicon was purified, cloned into TOPO TA cloning vector (Invitrogen), and reamplified. After purification, the concentrations of PCR products were determined using a fluorometer as described above and were serially diluted to generate calibration standards. Data analysis was carried out with iCycler software (Bio-Rad Laboratories). Based on the standard curve, a threshold cycle measured in a sample was converted to the copy number of the gene in a sample.
RESULTS
Probe design.
Altogether, 1,662 probes were designed based on 2,402 sequences downloaded from databases (available athttp://www.esd.ornl.gov/facilities/genomics/index.html). A probe was designed for any sequence with <85% nucleic acid sequence identity to any downloaded sequence. When a group of sequences had >85% identity, one probe was designed to represent that group. The target genes of these probes are involved in degradation-transformation processes of a variety of chemical compounds, such as monoaromatic-compound oxidation (546 probes), PAH oxidation (212 probes), biphenyl oxidation (76 probes), ring cleavage reaction (417 probes), aliphatic-compound transformation (131 probes), anaerobic degradation (100 probes), and heavy-metal reduction (120 probes) and export (73 probes) (Table 1). Almost all of the genes are from bacteria. While the majority of the probes (68%) were designed to be specific for individual genes, ∼30% of the probes were specific for groups of genes.
Specificity and resolution determined with artificial probes.
A previous study demonstrated that genes having <80 to 85% sequence identity could be differentiated at high stringency using PCR-amplified DNA fragments (400 to 800 bp) as probes (53). Based on this finding, all of the oligonucleotide probes that were selected for microarray fabrication in this study had sequence identities of <85%.
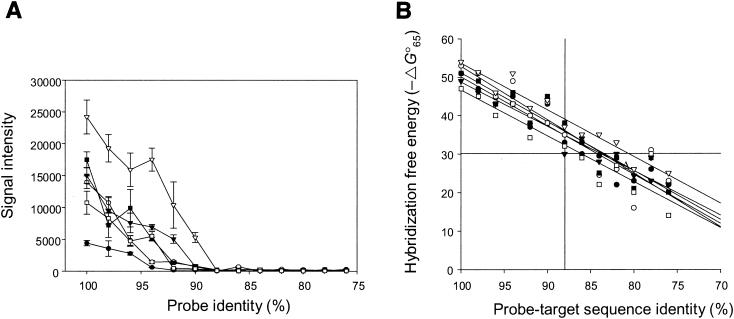
Since much shorter sequences were used as probes, the 50-mer FGAs should have had higher resolution than the PCR product-based FGAs. To experimentally determine the resolution power of the hybridization with the 50-mer FGAs, the effects of probe sequence similarity on the hybridization signal intensities of six target genes (Fig. 1A) from P. putida Gpo1 and PpG7 were evaluated. The probes were artificially designed to differ by 2% (i.e., 1-bp length) (for artificial-probe sequences, see http://www.esd.ornl.gov/facilities/genomics/index.html). All of the artificial probes had the same length; mismatches were incorporated in random positions, and the numbers of mismatches were increased from 1 to 12, corresponding to 2 to 24% differences from target sequences. PCR fragments (400 bp long) containing the probe region were amplified from genomic DNA, labeled, and hybridized with the arrays. The highest signal intensity was observed from oligonucleotide probes having 100% similarity to their target genes. As the similarity decreased, the signal intensities of all probes also decreased. Little hybridization was observed for probes showing 76 to 88% identity to the target DNA for all six genes, whereas the signal intensity increased substantially for probes showing >96% similarity to the target DNA (Fig. 1A). In addition, the effect of sequence identity on the signal intensity appeared to be gene dependent (Fig. 1A). For example, very little hybridization was observed for the naphthalene dioxygenase gene (gi151388) when the probe was 94% similar to the target gene, whereas very strong hybridization was observed for the alkane 1-monooxygenase gene (gi5824143) when the probe was 94% similar.
FIG. 1.
Effect of probe-target sequence identities on hybridization signal intensity. The symbols correspond to the genes from P. putida PpG7 (•, gi151388 [naphthalene dioxygenase, large subunit]; ○, gi151385 [naphthalene dioxygenase reductase]; ▾, gi5070351 [dihydroxynaphthalene dehydrogenase]) and Gpo1 (▿, gi5824143 [alkane 1-monoxygenase]; ▪, gi5824146 [aldehyde dehydrogenase]; □, gi5824147 [alcohol dehydrogenase]). The error bars indicate the standard deviations from four replicates. (A) Artificial probes with 76 to 100% sequence identity to the target genes were hybridized with PCR-amplified target genes in triplicate. (B) Change of hybridization free energy between probes and target genes. The intersecting vertical line indicates the threshold value of cross-hybridization signal at 88% sequence homology. The intersecting horizontal line indicates the suggested −ΔG°65 value (30 in this case) under which little cross-hybridization occurred.
The signal intensities for the probes that were <88% similar to the target sequences were comparable to background signal noise under the hybridization conditions of 50°C with 50% formamide. To determine whether the hybridization signal intensities were true signals from target sequences or signal noise from background hybridization, the SNR was calculated (Table 2). The SNRs varied from 0 to 0.5 for all of the genes when the probe sequence similarities were <88%, which was much smaller than the generally accepted threshold value (SNR, 3.0). When the probes were 90% similar to the target genes, all of the SNRs were <3, except for one gene (gi5824143), which had an SNR equal to 7.5. For gene gi151388, the SNR ratio was <3 even at a sequence similarity of 94%. These results indicate that, under the hybridization conditions of 50°C plus 50% formamide, oligonucleotide microarray hybridization can differentiate sequences having <88% similarity, and sequences having higher similarity can also be differentiated for some genes. These results also suggest that all probes (>85% sequence identity) on the designed arrays could be specific for their corresponding target genes.
TABLE 2.
SNRs of microarray hybridization to probes with different sequence similarities
| Probe sequence identity (%) | NRa
|
|||||
|---|---|---|---|---|---|---|
| gi151388b | gi151385 | gi5070351 | gi5824143 | gi5824146 | gi5824147 | |
| 100 | 8.5 ± 2.5 | 19.2 ± 5.2 | 20.6 ± 6.2 | 31.2 ± 12.3 | 25.5 ± 8.5 | 10.5 ± 3.8 |
| 98 | 3.7 ± 1.1 | 18.3 ± 3.6 | 10.2 ± 3.5 | 25.6 ± 8.5 | 16.2 ± 7.6 | 12.6 ± 4.2 |
| 96 | 4.9 ± 1.3 | 12.7 ± 1.3 | 11.8 ± 3.6 | 20.4 ± 9.3 | 11.3 ± 4.2 | 6.4 ± 3.2 |
| 94 | 0.9 ± 0.1d | 12.6 ± 2.3 | 10.4 ± 2.8 | 22.2 ± 8.9 | 12.1 ± 0.8 | 7.2 ± 2.1 |
| 92 | 0.2 ± 0.1 | 7.5 ± 3.2 | 6.3 ± 3.2 | 11.5 ± 6.2 | 8.2 ± 0.2 | 0.5 ± 0.2 |
| 90 | 0.2 ± 0.0 | 0.8 ± 0.3 | 1.2 ± 0.5 | 7.5 ± 2.5 | 1.5 ± 0.4 | 0.3 ± 0.1 |
| 88 | 0.1 ± 0.1 | 0.0 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.0 | 0.3 ± 0.2 | 0.0 ± 0.0 |
| 86c | 0.1 ± 0.0 | 0.9 ± 0.3 | 0.3 ± 0.2 | 0.5 ± 0.3 | 0.1 ± 0.0 | 0.3 ± 0.1 |
The data represent mean values and standard deviations derived from three independent microarray slides, with two replicates on each slide.
gi151388, naphthalene dioxygenase large subunit; gi151385, naphthalene dioxygenase (reductase subunit); gi5070351, cis-1,2-dihydro-1,2-dihydroxynaphthalene dehydrogenase; gi5824143, alkane 1-monooxygenase; gi5824146, aldehyde dehydrogenase; gi5824147, alcohol dehydrogenase.
SNRs for probes of identities from 76 to 84% are all <0.3.
Numbers for threshold points above which SNR is >3 are in boldface.
To facilitate probe design, the relationships between the theoretical values of hybridization free-energy changes and sequence identities were examined (Fig. 1B). The mfold program (see Materials and Methods) was used for the calculation of hybridization free energy for mismatched probe-target hybrids (ΔG°65). As discussed above, no hybridization was observed when a probe was <88% similar to the target, which corresponds to −30 (Fig. 1B). Thus, an approximate ΔG°65 value of >−30 (∼60% of the total free energy of the perfect-match hybrid) could be used for the design of probes under which little cross-hybridization would be expected.
Specificity evaluation with genomic DNAs from pure cultures.
The detection specificity of the microarray was tested using genomic DNAs of three reference strains (Table 3). Probes corresponding to all known genes reported in the reference strains had strong hybridization signals to their corresponding genomic DNAs (Table 3). Several additional genes not reported previously in P. putida strain PpG7 were also detected. Many of these additional genes have been reported in other Pseudomonas strains and are known to be involved in the degradation of metabolites of naphthalene degradation pathways (Table 3). To confirm the presence of these additional genes in strain PpG7, PCR primer sets were designed for four of the genes (gi294344, gi1008926, gi22000712, and gi22000714). Single PCR amplification products with the expected sizes were observed for all of these genes. DNA sequencing analysis of the amplified PCR products showed 92 to 99% nucleotide sequence identity to their corresponding genes. In addition, all of probes from T. aromatica K-172 and R. palustris had SNRs of >3. In contrast to the data for P. putida PpG7, no additional unknown probes were hybridized to the genomic DNAs from these two strains (Table 3). Finally, as expected, no probes showed cross-hybridization (SNR, <1) with the genomic DNA of S. oneidensis strain MR-1, which was used as a negative control. These results indicated that the designed probes were specific for their corresponding target genes from the genomes examined.
TABLE 3.
Detection by microarrays of biodegradation genes in genomes of reference microorganisms used in this study
| Strain | Chemical(s) | gi no. | Gene function | SNR | References |
|---|---|---|---|---|---|
| P. putida PpG7 | PAH: naphthalene | 45703 | Catechol 2,3-dioxygenase | 13.5 ± 1.5 | 10, 16, 18, 19, 40 |
| 151379 | 1,2-Dihydroxynaphthalene dioxygenase | 3.2 ± 0.3 | |||
| 151381 | Salicylate hydroxylase | 5.4 ± 0.8 | |||
| 151385 | Naphthalene dioxygenase reductase (Fd) | 13.4 ± 1.2 | |||
| 151386 | Naphthalene dioxygenase (Fd) | 16.5 ± 2.3 | |||
| 151387 | Naphthalene dioxygenase (large subunit) | 22.7 ± 2.1 | |||
| 151388 | Naphthalene dioxygenase (small subunit) | 6.10 ± 0.5 | |||
| 551908 | Salicylate hydroxylase | 5.6 ± 0.6 | |||
| 4235477 | 4-Oxalocrotonate decarboxylase | 19.0 ± 3.1 | |||
| 5070351 | Dihydroxynaphthalene dehydrogenase | 7.2 ± 0.9 | |||
| 483793 | 2-Hydroxylchromene2-carboxylate isomerase | 12.0 ± 2.0 | |||
| Genes not previously reported in strain PpG7a | 294344 | Protocatechuate 3,4-dioxygenase | 10.9 ± 1.3 | ||
| 294357 | Trans-o-hydroxybenzylidenepyruvate hydratase-aldolase | 7.7 ± 0.9 | |||
| 595675 | 4-Oxalocrotonate decarboxylase | 10.3 ± 1.6 | |||
| 1008926 | Catechol 1,2-dioxygenase | 3.9 ± 0.2 | |||
| 1255676 | Isomerase | 10.8 ± 1.7 | |||
| 2252625 | Salicylaldehyde dehydrogenase | 20.8 ± 1.9 | |||
| 3641341 | Lactone-specific esterase | 3.9 ± 0.5 | |||
| 4104766 | Catechol 2,3-dioxygenase | 4.9 ± 0.6 | |||
| 4235477 | 4-Oxalocrotonate decarboxylase | 19.0 ± 3.2 | |||
| 11546053 | Benzoate dioxygenase | 17.8 ± 2.7 | |||
| 22000712 | 2-Hydroxymuconic semialdehyde dehydrogenase | 5.9 ± 0.5 | |||
| 22000714 | Acetaldehyde dehydrogenase | 4.9 ± 0.6 | |||
| 119911694 | 4-Chlorobenzoate CoA ligase | 5.6 ± 0.4 | |||
| R. palustris | Monoaromatics: benzoate, toluene | 1730295 | 4-Hydroxybenzoyl-CoA reductase HbaB subunit | 13 ± 1.1 | 11, 15 |
| 1730296 | 4-Hydroxybenzoyl-CoA reductase HbaC subunit | 9.5 ± 0.9 | |||
| 1730297 | 4-Hydroxybenzoyl-CoA reductase HbaD subunit | 14.3 ± 2.0 | |||
| 2190573 | Cyclohex-1-ene-1-carboxylate CoA ligase | 24.1 ± 2.1 | |||
| 2190574 | Cyclohexanecarboxyl-CoA dehydrogenase | 7.2 ± 0.6 | |||
| 2190576 | 2-Hydroxycyclohexanecarboxyl-CoA dehyrogenase | 15.8 ± 1.8 | |||
| 2190579 | Benzoyl-CoA reductase subunit | 18.0 ± 2.2 | |||
| 3165396 | Benzoyl-CoA reductase subunit | 9.6 ± 0.9 | |||
| 3243085 | 2-Ketocyclohexanecarboxyl-CoA hydrolase | 8.7 ± 0.9 | |||
| T. aromatica K172 | Monoaromatics: benzoate, phenol, toluene | 3184130 | Benzylsuccinate synthase gamma subunit | 11.3 ± 1.0 | 4, 5, 25, 28, 29 |
| 3184131 | Benzylsuccinate synthase alpha subunit | 4.0 ± 0.5 | |||
| 3184132 | Benzylsuccinate synthase beta subunit | 11.4 ± 1.4 | |||
| 3724166 | 6-Oxocyclohex-1-ene-1-carbonyl-CoA hydratase | 13.4 ± 1.1 | |||
| 3724167 | Cyclohexa-1,5-diene-1-carbonyl-CoA hydratase | 10.7 ± 0.8 | |||
| 3724168 | C subunit of benzoyl-CoA reductase | 8.9 ± 0.9 | |||
| 3724169 | B subunit of benzoyl-CoA reductase | 4.1 ± 0.5 | |||
| 3724170 | A subunit of benzoyl-CoA reductase | 9.7 ± 1.3 | |||
| 3724171 | D subunit of benzoyl-CoA reductase | 10.3 ± 0.9 | |||
| 3724172 | Ferredoxin | 11.3 ± 1.2 | |||
| 9622531 | BbsA; toluene metabolism | 3.9 ± 0.5 | |||
| 9622533 | BbsC; toluene metabolism | 7.0 ± 0.6 | |||
| 9622534 | BbsD; toluene metabolism | 14.4 ± 1.7 | |||
| 9622535 | BbsE; toluene metabolism | 9.6 ± 0.8 | |||
| 9622536 | BbsF; toluene metabolism | 6.6 ± 0.7 | |||
| 9622538 | E-phenylitaconyl-CoA hydratase; toluene metabolism | 15.7 ± 1.3 | |||
| 9622539 | BbsI; toluene metabolism | 14.2 ± 1.3 | |||
| 10697118 | Hypothetical protein; phenol metabolism | 12.4 ± 1.4 | |||
| 10697119 | Hypothetical protein; phenol metabolism | 16.8 ± 1.5 | |||
| 10697121 | Hypothetical protein; phenol metabolism | 15.9 ± 1.3 | |||
| 10697123 | Hypothetical protein; phenol metabolism | 7.9 ± 0.6 | |||
| 10697124 | Hypothetical protein; phenol metabolism | 7.1 ± 0.8 | |||
| 10697125 | Hypothetical protein; phenol metabolism | 17.9 ± 1.9 | |||
| 19571178 | 2-Oxyglutarate ferredoxin-oxidoreductase beta subunit | 33.8 ± 2.8 | |||
| 19571179 | 2-Oxoglutarate ferredoxin-oxidoreductase alpha subunit | 7.2 ± 0.6 | |||
| 19571180 | 6-Hydroxycylohex-1-ene-1-carbonyl-CoA dehydrogenase | 4.9 ± 0.4 | |||
| 12667039 | Putative alcohol dehydrogenase | 4.2 ± 0.5 | |||
| 12667041 | Putative alcohol dehydrogenase | 5.9 ± 0.8 | |||
| 12667042 | Putative acyl-CoA dehydrogenase | 15.9 ± 1.4 | |||
| 12667043 | 3-Hydroxybenzoate CoA ligase | 12.9 ± 1.6 | |||
| 12667044 | Putative hydrolase | 21.0 ± 2.3 |
Additional genes not reported but detected in strain PpG7.
Detection of gene expression in strain PpG7 induced by naphthalene.
To determine whether the 50-mer-based FGAs can be used to monitor the functional activities of the genes of interest, mRNA-based gene expression was examined with P. putida PpG7. Strain PpG7 was incubated in the presence of naphthalene as a sole carbon source to induce expression of naphthalene degradation pathways. All known genes involved in naphthalene degradation were highly expressed for the cells grown with naphthalene (not shown) but not for the cells grown with pyruvate (not shown). As expected, strong hybridization signals were obtained for the 16S rRNA gene probe under both growth conditions. In addition, several additional genes (gi294357, gi595675, gi1255676, gi22000712, and gi22000714) (Table 3) which were not previously reported in this strain were also highly expressed (average SNR, 13) under naphthalene growth conditions but not under pyruvate growth conditions. However, some of the additional genes detected (Table 3) were not expressed (average SNR, 0.1) under both conditions, suggesting that the genes could not be induced by naphthalene. These results indicated that the developed 50-mer FGAs could be used to monitor mRNA-based functional activity.
Determination of detection sensitivity of 50-mer FGA-based hybridization.
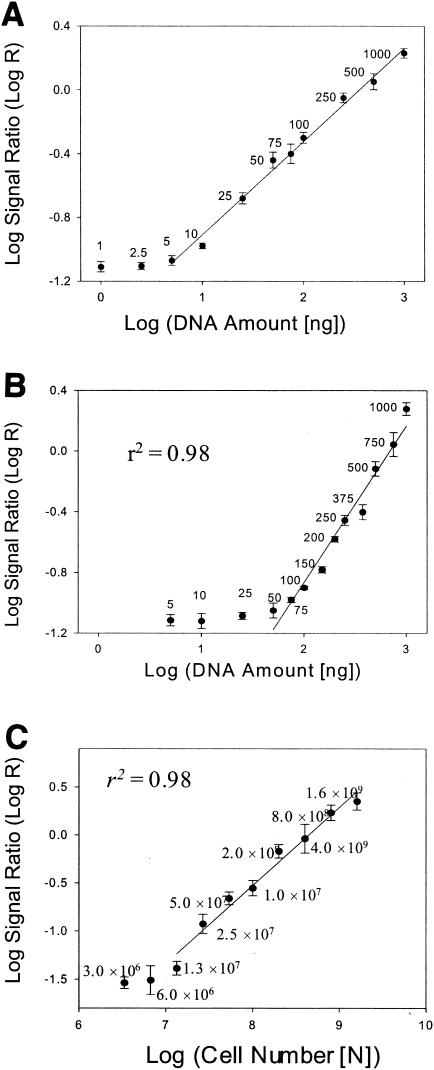
The detection sensitivity of the 50-mer FGA-based hybridization was determined using genomic DNA extracted from a pure culture of T. aromatica K172. Genomic DNA (1 to 1,000 ng) from T. aromatica K172 was randomly labeled with Cy5. At a hybridization temperature of 50°C in the presence of 50% formamide, the strongest hybridization signals were observed for the 2-oxoglutarate ferredoxin-oxidoreductase beta subunit gene (gi19571178) with 5 ng of T. aromatica K172 DNA (average SNR, 13) (not shown). The signal intensity of this gene was comparable to the background level when the amount of genomic DNA used for labeling was 1 to 2.5 ng (see Fig. 3A). However, it appeared that detection sensitivity was gene dependent, ranging from 5.0 to 10.0 ng. Therefore, the maximum detection limit with randomly labeled pure genomic DNA under these hybridization conditions was estimated to be ∼5 ng.
FIG. 3.
Evaluation of quantitative potential of 50-mer FGA-based hybridization. The log ratios of hybridization signals between the target gene and the positive control human gene HSPC120 (AF161469) were calculated and plotted against the log of the concentration of genomic DNA. (A) Quantitative relationship of 50-mer FGA-based hybridization with pure genomic DNA in the absence of heterogeneous background DNAs. Genomic DNA of strain K-172 was serially diluted in 1× Tris-EDTA buffer, and the diluted genomic DNAs at concentrations of 1 to 1,000 ng were labeled with Cy5 using a random-primer labeling method. The labeled DNAs were hybridized with the microarrays. The quantitative relationship shown is that of the hybridization signals from the gene for the 2-oxoglutarate ferredoxin-oxidoreductase beta subunit, gi19571178. The quantitative relationships of other genes are not shown. (B) Quantitative relationship of 50-mer FGA-based hybridization with pure genomic DNA in the presence of heterogeneous background DNAs. See the legend to Fig. 2A for details. The quantitative relationship shown is that of the hybridization signals from the gene for the 2-oxoglutarate ferredoxin-oxidoreductase beta subunit, gi19571178. The quantitative relationships of other genes are not shown. (C) Quantitative relationship of FGA-based hybridization with total RNAs from known numbers of cells. Cell preparation and array analysis are shown in Fig. 2B. The numbers of P. putida cells mixed with S. oneidensis MR-1 cells (1.9 × 109) are indicated. The quantitative relationship shown is that of the hybridization signals from the gene for the 2-oxoglutarate ferredoxin-oxidoreductase beta subunit, gi19571178. The quantitative relationships of the other genes are not shown. The error bars represent standard deviations.
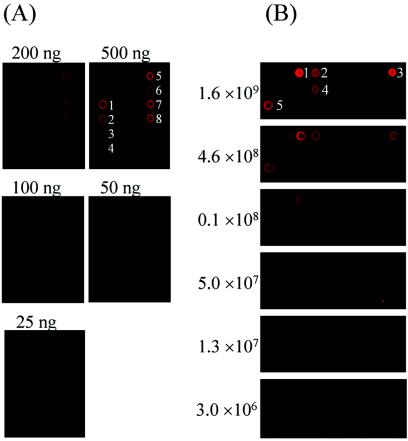
In environmental samples, the microorganisms of interest are present together with other diverse microorganisms. The existence of other nontarget DNAs may affect hybridization with target DNA and hence decrease the detection sensitivity. To evaluate the detection sensitivity in the presence of heterogeneous nontarget DNAs, genomic DNA (10 to 1,000 ng) from T. aromatica K172 was mixed with 1 μg of S. oneidensis MR-1 DNA and randomly labeled with Cy5. The strongest hybridization signals were observed with 50 ng of T. aromatica K172 DNA for the target 2-oxoglutarate ferredoxin-oxidoreductase beta subunit gene (gi19571178) (Fig. 2A). Hybridization signals using 25 ng of genomic DNA, however, were barely detectable. Similarly, the detection sensitivity was also gene dependent, ranging from 50 to 100 ng for the genes examined. In addition, the detection sensitivity in the presence of heterogeneous DNA was also evaluated with various amounts of PCR-amplified products (∼400 bp) from genes for naphthalene degradation and alkane degradation: the Fe-S protein of naphthalene dioxygenase (gi151388), the reductase of naphthalene dioxygenase (gi151385), cis-1,2-dihydro-1,2-dihydroxynaphthalene dehydrogenase (gi5070351), alkane 1-monooxygenase (gi5824143), aldehyde dehydrogenase (gi5824146), and alcohol dehydrogenase (gi5824147).
FIG. 2.
Detection sensitivity of 50-mer FGA-based hybridization. (A) Fluorescence images showing DNA detection sensitivity with pure genomic DNA. Genomic DNA of T. aromatica K-172 was serially diluted in 1× Tris-EDTA buffer. The diluted genomic DNAs (5 to 1,000 ng) were mixed with 1 μg of the negative control genomic DNA from S. oneidensis MR-1, labeled with Cy5 in a total hybridization solution volume of 20 μl, and hybridized with the microarrays. The arrays were scanned with 100% laser power and 100% PMT gain. A portion of the image, which contained probes from the following genes, is presented: succinyl-CoA-benzylsuccinate CoA-transferase (1, gi9622538; 2, gi9622535; 3, gi9622533; 4, gi9622531), benzoyl-CoA reductase (5, gi3724172; 6, gi3724140; 7, gi3724168), and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydratase (8, gi3724166). (B) Fluorescence images showing RNA detection sensitivities with total RNAs from known numbers of cells (on the left). Cells of strain PpG7 were serially diluted with cells of strain MR-1 (1.9 × 109) grown on M4 medium before extraction of total RNA. Total RNAs from the cell mixture were randomly labeled with Cy5 using reverse transcriptase and hybridized with the microarrays. A portion of the image is presented to show the RNA hybridization sensitivity. 1, gi2246756 (1,2-dihydroxy-1,2-dihydronaphthalene dehydrogenase); 2, gi22000712 (hydroxymuconic semialdehyde hydrolase); 3, gi17863959 (naphthalene dioxygenase); 4, gi551908 (salicylate hydroxylase); 5, gi45703 (catechol 2,3-dioxygenase).
Various amounts of PCR products (1 to 1,000 pg) were mixed with 1 μg of S. oneidensis MR-1 DNA and labeled with Cy5. The hybridization signals were significantly higher than background noise when the PCR product was >10 pg (not shown). If we assume that the genome size is ∼4 Mb, the detection limit with PCR products will be equivalent to 100 ng of genomic DNA for the gene in the presence of nontarget DNAs, which is consistent with the results using the labeled genomic DNA. These results suggested that the detection limit of genomic DNA of the 50-mer FGA-based hybridization in the presence of nontarget genomic DNAs is ∼50 to 100 ng.
The RNA detection sensitivity was also determined with P. putida PpG7 cells grown with naphthalene as a sole carbon source. Concentrations of 3.0 × 106 to 1.6 × 109 cells of strain PpG7 were mixed with 1.9 × 109 cells of S. oneidensis MR-1. Total RNAs were extracted from the mixture, used for production of labeled DNA with Cy5 using reverse transcriptase, and hybridized with the 50-mer FGAs. The hybridization signal intensity of the gene (gi17863959) was significantly higher than the background signal when the cell number was >1.3 × 107 (Fig. 3C). However, the signal intensity was not significantly different from the background level when the cell numbers ranged from 3.0 × 106 to 6.0 × 106. Similarly, the detection sensitivity was also gene dependent, ranging from 1.3 × 107 to 5.0 × 107 cells for the genes examined. Thus, the detection limit of mRNA is ∼1.3 × 107 to 5.0 × 107 cells, depending on the genes and the experimental conditions.
Quantification of the 50-mer FGA-based hybridization.
The assessment of bioremediation potential requires the quantification of individual target genes and microorganisms. The capacity of the 50-mer FGA-based hybridization to serve as a quantitative tool was explored by examining the relationship between the concentration of target DNA and the hybridization signal intensity. Genomic DNA from strain K-172 (1 to 1,000 ng) was fluorescently labeled with Cy5 as described above and hybridized in triplicate with the microarray. To avoid signal saturation, the slides were scanned using different combinations of laser power and PMT. The log ratios of the hybridization signals between target genes and the positive control human gene HSPC120 (AF161469) were calculated and plotted against the log of the concentrations of genomic DNA. For the gene gi19571178, strong linear relationships were observed between the signal intensity and target DNA concentrations from 5 to 1,000 ng (r2 = 0.98; P < 0.01) (Fig. 3A). Significant correlations between the signal intensity and DNA concentrations from 10 to 1,000 ng were also observed for all other genes (r2 = 0.97 to 0.99). These results indicated that the 50-mer FGA-based hybridization is quantitative for pure bacterial cultures within a wide range of DNA concentrations.
To determine whether the 50-mer FGA-based hybridization is quantitative for targeted organisms in the presence of other nontarget DNAs, the quantitative relationships between the signal intensity and the DNA concentration were further examined. As described above, various amounts of genomic DNA (5 to 1,000 ng) from T. aromatica K172 were mixed with 1 μg of S. oneidensis MR-1 DNA and randomly labeled with Cy5. A significant linear relationship (r2 = 0.97; P < 0.01) was observed for the gene gi19571178 between the signal intensity and the target DNA concentration within 75 and 1,000 ng (Fig. 3B). Significant correlations between the signal intensity and DNA concentrations ranging from 75 to 1,000 ng were also observed for all other genes (r2 = 0.95 to 0.99). These results suggested that the 50-mer FGA-based hybridization could also be quantitative in the presence of nontarget DNAs.
To determine whether the 50-mer FGA-based hybridization is quantitative for detecting functional activity, the quantitative capability of mRNA-based hybridization was also evaluated. Concentrations of 3.0 × 106 to 1.6 × 109 cells of P. putida PpG7 were mixed with 1.9 × 109 S. oneidensis MR-1 cells as described above. Total RNAs were extracted, randomly labeled with Cy5, and hybridized with the 50-mer FGAs. A significant linear relationship (r2 = 0.98; P < 0.01) was observed for the naphthalene dioxygenase gene (gi17863959) between the signal intensity and target cell numbers from 1.3 × 107 to 1.6 × 109 cells (Fig. 3C). Significant correlations between the signal intensity and cell numbers from 5.0 × 107 to 1.6 × 109 were also observed for all other genes (r2 = 0.96 to 0.99). These results suggested that the 50-mer FGA-based hybridization could be quantitative for target RNA determination in the presence of nontarget RNAs.
Detection of naphthalene-degrading genes and their expression in enrichment cultures.
To determine the potential applicability of the developed FGAs for detecting microorganisms in samples of mixed populations, bacterial enrichments were established under aerobic conditions from a naphthalene-contaminated TFD soil sample using naphthalene or pyruvate as a carbon source. The cells were harvested in the middle of the growth phase, and DNAs were extracted. The purified DNAs from both naphthalene and pyruvate enrichments were labeled with Cy5 and Cy3 and hybridized with the microarrays. Thirteen genes obtained from the naphthalene enrichment were highly different from those obtained from the pyruvate enrichment (Table 4). Among them, four genes were involved in the naphthalene degradation pathway. Interestingly, all genes except one were from gram-positive soil bacteria (Rhodococcus), suggesting that this group was specifically enriched.
TABLE 4.
Genes detected in enrichment culture from TFD soil using naphthalene as sole carbon source
| gi no. | Gene function | Microorganism | Log mean signal ratioa | SNRb |
|---|---|---|---|---|
| 4704462 | Naphthalene dioxygenase large subunitc | Rhodococcus sp. strain NCIMB12038 | 1.79 ± 0.62 | 45.12 ± 3.24 |
| 4704463 | Naphthalene dioxygenase small subunitc | Rhodococcus sp. strain NCIMB12038 | 1.46 ± 0.56 | 22.54 ± 3.27 |
| 1771524 | Muc7onate cycloisomerasec | R. opacus | 1.43 ± 0.56 | 17.62 ± 2.83 |
| 1771525 | Muconolactone isomerasec | R. opacus | 1.36 ± 0.54 | 15.86 ± 1.29 |
| 8978311 | 2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase | Rhodococcus sp. strain RHA1 | 1.35 ± 0.55 | 15.69 ± 1.45 |
| 10933936 | Putative alkane hydroxylase | Rhodococcus sp. strain 1BN | 1.31 ± 0.54 | 18.83 ± 2.96 |
| 8926387 | 3-(2,3-Dihydroxylphenyl) propionic acid dioxygenase | Rhodococcus sp. | 1.12 ± 0.51 | 10.56 ± 0.73 |
| 6652685 | Biphenyl dioxygenase | Pseudomonas pseudoalcaligens | 1.01 ± 0.49 | 10.44 ± 1.72 |
| 14289339 | Benzoate dioxygenase | Rhodococcus sp. strain 19070 | 0.86 ± 0.47 | 9.25 ± 0.43 |
| 9711314 | 2,3-Dihydroxybiphenyl 1,2-dioxygenase | Rhodococcus sp. strain RHA1 | 0.85 ± 0.47 | 5.98 ± 0.83 |
| 2935027 | 3-Oxoadipate enol-lactone hydrolase | Rhodococcus sp. | 0.80 ± 0.47 | 10.44 ± 0.75 |
| 8978307 | 2,3-Dihydroxybiphenyl 1,2-dioxygenase | Rhodococcus sp. strain RHA1 | 0.77 ± 0.46 | 14.02 ± 1.19 |
| 2398775 | Putative 1,2-catechol dioxygenase | R. opacus | 0.74 ± 0.46 | 10.21 ± 0.98 |
Hybridization signal ratios (log mean ± standard deviation from six replicates) of the naphthalene-enriched consortium to the pyruvate-enriched consortium.
Mean ± standard deviation from six replicates.
Gene possibly involved in naphthalene degradation pathway.
To evaluate whether microarrays can detect differences in gene expression, the naphthalene-enriched cells were divided into two aliquots: one aliquot was grown on pyruvate for 3 h, and the other was grown on naphthalene for 3 h. It was expected that the expression of the genes involved in the naphthalene degradation pathway would be repressed in the presence of pyruvate. Total RNAs were extracted from these two samples, labeled with Cy3 and Cy5 separately, and hybridized with the microarrays. Three genes (naphthalene dioxygenase large and small subunits, gi4704462 and gi4704463, and cis-naphthalene dihydrodiol dehydrogenase, gi4827073) were highly expressed (40- to 100-fold) under growth conditions with naphthalene. These three genes were known to be involved in the first and second steps of the naphthalene degradation pathway and were reported to originate from a plasmid of Rhodococcus sp. strain NCIMB12038. These results indicated that Rhodococcus sp.-type microorganisms were among the major naphthalene degraders in the enrichment, which was consistent with the DNA-based microarray hybridization described above. However, the genes controlling catechol oxidation (potential naphthalene degradation genes) from Rhodococcus opacus were not highly expressed in the enrichment, although they were abundant, as indicated by DNA-based microarray hybridization.
Detection of naphthalene-degrading genes in soil microcosms.
To determine whether the developed microarrays could be used to detect microorganisms within the context of environmental applications, two microcosms were established with soil samples from TFD, one with naphthalene vapor, and the other without naphthalene vapor as a control. To accurately estimate naphthalene degradation, an additional microcosm was established containing naphthalene and sterilized TFD soil. Gas chromatography analysis revealed that the amount of naphthalene in the microcosm decreased by 0.61 mg · g of soil−1 after 2 weeks of incubation, whereas little change in the concentration of naphthalene in the vapor phase was observed in the microcosm with sterilized soil, indicating that the decrease in the naphthalene concentration was mostly due to biological degradation (not shown). Direct cell counting also showed that cell numbers increased threefold (from 0.7 × 1010 to 2.3 × 1010) in the treatment microcosm containing naphthalene, whereas no significant change in cell numbers was observed in the control microcosm without naphthalene, indicating that microbial populations were stimulated significantly by the presence of naphthalene (data not shown).
The bulk community DNAs were extracted from the treatment and control microcosm soil samples after 2 weeks of incubation, purified and labeled with Cy5, mixed with the Cy3-labeled reference DNA (OR), and hybridized with the developed microarrays. The ratios of the hybridization signals from the treatment samples to those from the control samples, (Cy5-treatment/Cy3-ref)/(Cy5-control/Cy3-ref), were determinedand used for relative comparison. The hybridization signals of 40 genes were significantly different (P = 0.05). Among them, 22 genes were highly different, with >3-fold difference (Table 5). Interestingly, seven of these genes were from a plasmid-borne naphthalene degradation pathway observed in Ralstonia sp. strain U2. Four of the genes, encoding nitrobenzene dioxygenase (gi18643025) and (D)NT dioxygenase (gi1477920, gi1477923, and gi17942397), were similar to the naphthalene dioxygenase gene and have catalytic activity on naphthalene (27, 43). All of the other genes except one (gi3894249; phenol hydroxylase) were involved in PAH degradation. It appears that microorganisms containing naphthalene degradation-related genes were specifically stimulated in the microcosm. Among them, Ralstonia sp. strain U2-type microorganisms containing the plasmid might be one of the major constituents for degrading naphthalene in the microcosm. However, in contrast to the enrichment experiment, the plasmid-borne genes in Rhodococcus sp. strain NCIMB12038 were not detected with the microarray, indicating that the enrichment process could cause severe bias of the population compositions.
TABLE 5.
Genes detected in soil microcosm incubated with naphthalene
| Gene (gi no.) | Gene function | Microorganism | Log mean signal ratioa | SNRb |
|---|---|---|---|---|
| 1477920 | 2,4-DNT dioxygenase ferredoxin oxidoreductase | Burkholderia sp. strain RASC | 1.64 ± 0.18 | 3.23 ± 0.45 |
| 1477923 | 2,4-DNT dioxygenase large subunit | Burkholderia sp. strain RASC | 2.02 ± 0.23 | 3.42 ± 0.62 |
| 1773277 | 2-NT oxygenase large subunit | Pseudomonas sp. | 1.98 ± 0.23 | 6.32 ± 0.43 |
| 2828015 | Salicylate-5-hydroxylase large subunit | Ralstonia sp. strain U2 | 1.26 ± 0.13 | 13.94 ± 1.03 |
| 2828016 | Salicylate-5-hydroxylase small subunit | Ralstonia sp. strain U2 | 1.94 ± 0.22 | 15.02 ± 0.75 |
| 2828018 | Naphthalene 1,2-dioxygenase large subunit | Ralstonia sp. strain U2 | 1.34 ± 0.14 | 3.77 ± 0.27 |
| 3337416 | Salicylaldehyde dehydrogenase | Ralstonia sp. strain U2 | 1.49 ± 0.16 | 4.33 ± 0.24 |
| 3894249 | Phenol hydroxylase | Unidentified bacterium rN5 | 1.32 ± 0.14 | 6.85 ± 0.45 |
| 4220429 | Putative aldolase | Ralstonia sp. strain U2 | 1.74 ± 0.19 | 3.99 ± 0.12 |
| 4220432 | Glutathione S-transferase protein | Ralstonia sp. strain U2 | 1.65 ± 0.18 | 6.62 ± 0.67 |
| 4220433 | Gentisate 1,2-dioxygenase | Ralstonia sp. strain U2 | 2.12 ± 0.26 | 10.63 ± 0.45 |
| 4220434 | Fumarylpyruvate hydrolase | Ralstonia sp. strain U2 | 2.34 ± 0.28 | 8.14 ± 0.27 |
| 4220435 | Maleylpyruvate isomerase | Ralstonia sp. strain U2 | 2.27 ± 0.27 | 7.74 ± 0.56 |
| 4234963 | Naphthalene dioxygenase | Uncultured bacterium D1b | 1.95 ± 0.22 | 9.58 ± 0.88 |
| 4234998 | Naphthalene dioxygenase | Uncultured bacterium U3b | 1.64 ± 0.18 | 5.62 ± 0.59 |
| 8118285 | Polyaromatic hydrocarbon dioxygenase large subunit | Comamonas testosteroni | 1.22 ± 0.13 | 3.94 ± 0.45 |
| 8118287 | cis-Naphthalene dihydrodiol dehydrogenase | C. testosteroni | 1.63 ± 0.18 | 13.96 ± 1.22 |
| 16943679 | Cyclopentanone 1,2-monooxygenase | C. testosteroni | 0.77 ± 0.08 | 13.92 ± 1.73 |
| 17863951 | Naphthalene dioxygenase | Burkholderia sp. strain S1-17 | 1.72 ± 0.19 | 5.13 ± 0.76 |
| 17942397 | 2,4-DNT oxygenase Fe-S protein | Burkholderia cepacia | 2.04 ± 0.24 | 11.93 ± 0.35 |
| 18307552 | Naphthalene dioxygenase Fe-S protein | Ralstonia sp. strain NI1 | 2.32 ± 0.28 | 7.88 ± 0.67 |
| 18643025 | Nitrobenzene dioxygenase alpha subunit | Comamonas sp. strain JS765 | 1.12 ± 0.12 | 8.53 ± 0.34 |
Hybridization signal ratios (log mean ± standard deviation from six replicates) of the naphthalene-spiked TFD soil to the control TFD soil. The hybridization signals of 40 genes were significantly different (P = 0.05), but only the genes with >3-fold difference are listed.
Mean ± standard deviation of six replicate Cy5 signals from treated samples.
Application of the 50-mer FGA-based hybridization for profiling microbial communities in environmental samples.
The developed microarrays could potentially be used as a generic profiling tool that would reveal differences among various microbial communities. To evaluate such potential, bulk community DNAs were isolated from 5 g of contaminated soil samples. For comparison, DNA from an uncontaminated soil sample was used as a reference. Five micrograms of the purified bulk community DNA from the contaminated soil samples was directly labeled with Cy5 using random-primer labeling. The DNA from the reference forest sample was labeled with Cy3. Cy3- and Cy5-labeled DNAs were mixed together and hybridized with the microarrays in triplicate. All spots with SNRs of >3 were considered positive signals. Overall, the numbers of arrayed probes with statistically significant positive signals were 30 in TFD soil, 37 in TFD-NP soil, 36 in PCT18 soil, and 24 in IDT soil. The average variation in signal intensity among all three replicates in these samples was 25.0%, with a standard deviation of 7.8%.
Clustering analysis revealed five main groups (Fig. 4). Many genes related to naphthalene degradation were clustered together, and they were abundant in naphthalene-amended soil. However, they appeared to be present at very low levels in all three soils. The genes involved in anaerobic benzoate degradation (4-hydroxylbenzoyl-coenzyme A [CoA] reductase, 3-hydroxylbenzoate CoA reductase, benzoyl-CoA ligase, benzoyl-CoA, and cyclohex-1-ene-1-carboxylate CoA ligase) were grouped together and were observed only in TFD soil. In addition, 4-chlorobenzate CoA ligase, alkane-1 monooxygenase, benzoyl CoA ligase, and cis-naphthalene dihydrodiol dehydrogenase genes were relatively abundant among all the soil samples (Fig. 4). Overall, PCT18 soil was more closely clustered with TFD soil than IDT soil, suggesting that the community structure in terms of the biodegrading components in PCT18 soil was more similar to that of TFD soil than to that of IDT soil. This was expected, because both TFD and PCT18 soils were contaminated with PAHs, whereas IDT soil contained BTEX.
FIG. 4.
Hierarchical cluster analysis of community relationships based on hybridization signal intensity ratios for genes showing SNRs of >3. The figure was generated using hierarchical cluster analysis (CLUSTER) and visualized with TREEVIEW (12). The hybridization signals of genomic DNAs from each of the four contaminated soils were divided by the hybridization signals from genomic DNA of OR. Microarray hybridization patterns with the labeled genomic DNAs from the soils are shown in each column. Each row represents the hybridization signal observed for each gene when the genomic DNA from the contaminated soil indicated above the column was used for hybridization. Black represents no detectable difference in the hybridization signal, while red represents a significant hybridization signal. The columns correspond to the hybridization patterns obtained with Cy5-labeled genomic DNAs from the following contaminated soils: TFD-NP, IDI, TFD, and PCT18 soil incubated under naphthalene vapor.
Verification of microarray hybridization data using real-time PCR.
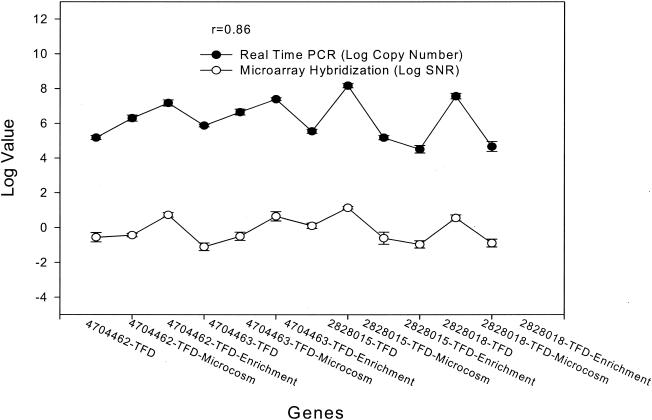
To assess the reliability of microarray hybridization data independently, real-time PCR was used to measure the copy numbers of several key genes in TFD soil, the microcosm, and the enrichment. Six plasmid-borne genes in Rhodococcus sp. strain NCIMB12038 and Ralstonia sp. strain U2 were selected, PCR primers were designed, and PCR amplifications were carried out with the genomic DNAs. Single amplicons with an expected size were generated with four primer sets, which were further used for real-time PCR analysis. All of the genes were present in low copy numbers (104 to 105) in the original TFD soil (Fig. 5), but the microarray hybridization signals of these genes in the TFD sample were negligible (SNR, <3). Ralstonia sp. strain U2-type microorganisms had low abundance in TFD soil but were specifically stimulated in the soil microcosm, as detected by real-time PCR and microarray hybridization (Fig. 5). Real-time PCR data indicated that the abundance of the genes in Rhodococcus-type microorganisms (copy numbers, 5.2 × 105 to 9.8 × 105) was higher than that of Ralstonia sp. strain U2 genes (copy numbers, 2.4 × 104 to 4.0 × 104) in the TFD soil, but they could not be reliably detected by microarray hybridization due to microarray detection sensitivity. Overall, significant correlations between the microarray hybridization signals and the gene copy numbers determined by real-time PCR were obtained (r2 = 0.74 for all genes and r2 = 0.96 for the genes with SNRs of >3) (Fig. 5). These results suggested that microarray hybridization data were consistent with real-time PCR data, and hence, the microarray hybridization signal appeared to be reliable.
FIG. 5.
Comparison between real-time PCR and microarray hybridization for detection of naphthalene degradation genes in the microbial community. Two genes, naphthalene dioxygenase large subunit (gi4704462) and naphthalene dioxygenase small subunit (gi4704463), from Rhodococcus sp. and two genes, salicylate 5-hydroxylase large subunit (gi2828015) and naphthalene dioxygenase large subunit (gi2828018), from Ralstonia sp. strain U2, were used for comparison. Three replicates of real-time PCR were performed for each gene with each sample.
DISCUSSION
Specificity is one of the critical issues in microarray assays, especially for environmental studies. Several pieces of evidence suggested that the developed 50-mer FGAs were specific to their corresponding genes or group of genes, although we could not experimentally examine the specificities for all individual probes. First, all probes on the arrays were <85% similar, which is below the threshold value of 88% determined in this study using artificial probes. Second, the determined threshold value of sequence identity (<88%) with the 50-mer FGAs was consistent with previous results (<80 to 85% sequence identity) with the PCR product-based FGA (53). As expected, higher hybridization specificity was achieved with the 50-mer FGAs than with the PCR product-based FGAs. Our results showed that the developed 50-mer FGAs were able to discriminate gene sequences with <88 to 94% identity under the hybridization conditions of 50°C with 50% formamide (Fig. 1). Third, since probes having longer than 15-mer continuous sequence stretches identical to the nontarget sequences may cause cross-hybridization problems (38), such probes were removed after being designed. In addition, based on the artificial probes, our results suggested that the ΔG°65 value of <60% of the total free energy (corresponding to 88% probe sequence identity) could be used as a threshold value for probe design. This is consistent with the results of Taroncher-Oldenburg et al. (44). They recommended 87% probe-to-target identity and 56% ΔG as threshold values for designing 70-mer oligonucleotide probes. Finally, specific detection of the developed 50-mer FGAs was obtained using the genomic DNAs from a limited number of pure cultures. The overall consistency of the hybridization results with real-time PCR and sequencing results and with our knowledge and prediction of the environmental samples examined (enrichments, microcosms, and soils) also suggested that the developed 50-mer arrays were specific. However, we found that nonspecific hybridization could be significant when the microarray slides were not warmed or the hybridization mixture remained at room temperature for several minutes after hybridization and before washing. To minimize potential nonspecific hybridization, the slides should be prewarmed and the hybridization mixture should be kept above the hybridization temperature through all hybridization steps prior to washing.
Because only probes with <85% similarity were used to construct the 50-mer FGAs, it was important to understand whether they could be specific to the corresponding target genes or groups when used for analyzing environmental samples of unknown composition. To address this question, a representative enzyme from each group in which many gene sequences were available was selected for similarity analysis. The average nucleic acid identities ranged from 31 to 70% (Table 1). The maximum nucleic acid sequence identities were <88% for genes encoding phenol hydroxylase, arsenate reductases, and cadmium-transporting ATPase. Although the maximum nucleic acid identities for genes encoding naphthalene, biphenyl, and catechol dioxygenases were >88%, the average identities were relatively low for these three genes (34 to 51%). These results indicated that the nucleic acid sequences of the genes encoding these enzymes are highly diverse. Since our experimental data showed that little hybridization was observed for the probes with <88% sequence identity, the designed microarrays with probes <85% similar could be specific for the corresponding target genes or groups when they were used for analyzing environmental samples of unknown composition, and cross-hybridizations from unknown homologous sequences in environmental samples may not be a severe problem. However, it is always important to use the 50-mer FGAs for relative comparisons. The effects of any potential cross-hybridization can be cancelled out when the hybridization intensity signals from treatment samples are divided by the hybridization intensity signals from the common reference samples under the assumption that the community compositions of the treatment and reference samples are similar.
Sensitivity is another critical parameter that impacts the effectiveness of the microarray-based approach for detecting genes in environmental samples. With the 50-mer oligonucleotide arrays, genes involved in biodegradation could be detected with 5 ng of genomic DNA in the absence of background DNA and with 50 to 100 ng of genomic DNA in the presence of pure-culture genomic DNA. Since 1,000 ng of nontarget genomic DNA was used for hybridization, a detection limit of 50 ng corresponds to 5% of the target organisms. With this sensitivity, we should be able to detect the dominant microorganisms in a microbial community. The determined sensitivity is consistent with real-time PCR results. For example, when the copy number of the gene (gi2828018) was at the level of 107 (∼3.7 × 107 [170 ng of genomic DNA]) in the corresponding microcosm sample, a relatively strong hybridization signal (SNR, 3.6) was obtained for the gene. However, when it was at the level of 104 (∼3.2 × 104) in TFD soil, very little hybridization was obtained (SNR, 0.11). As expected, the sensitivity of the 50-mer-based FGAs was also ∼10- to 100-fold lower than that of PCR product-based FGAs (53) and CGAs (L. Wu, D. K. Thompson, X. Liu, C. E. Bagwell, M. W. Fields, J. M. Tiedje, and J. Zhou, unpublished data). Using the PCR product-based FGAs, nirS genes were detected with ∼1 ng of labeled pure genomic DNA in the absence of background DNA (53). The detection limit of CGAs was estimated to be ∼0.2 ng with pure labeled genomic DNA in the absence of background DNA and ∼5 ng of genomic DNA, or 2.5 × 105 cells, in the presence of background DNA (Wu et al., unpublished). In addition, the detection limit (∼10 pg of PCR products) was relatively comparable with that determined with the arrays containing DNA fragments as probes (500 to 900 bp) in the presence of background DNA (7).
Based on the experimental results with mixtures of known amounts of DNAs, a known number of cells, and real-time PCR, roughly 107 cells are needed to achieve reasonably strong hybridization. If these values are directly applicable to real environmental samples, the level of the 50-mer-based FGA detection sensitivity should be sufficient for detection of at least the more dominant members of a microbial community. However, it is still not sensitive enough to detect less abundant microbial populations. This is supported by our results from soil microcosm experiments in which naphthalene was added to stimulate naphthalene-degrading populations. Very strong hybridization signals were observed for the genes involved in naphthalene degradation with 2 μg of community genomic DNA from these soil samples, whereas these genes were rarely detectable in the soil sample without naphthalene amendment. To detect rare populations in natural environments, other approaches for increasing hybridization sensitivity are needed. We are exploring ways to enhance the level of detection sensitivity.
Our experimental results showed that a small portion of probes had strong hybridization with the bulk community DNAs (5 μg) from the three soil samples without naphthalene amendment. The overall relationships of these three soils based on the detected genes are consistent with general expectations in terms of the contaminant differences of the soils. Thus, despite the fact that there is an urgent need for improving hybridization sensitivity, the 50-mer FGAs are still valuable to some extent for revealing the differences among various microbial communities as a profiling tool. As we demonstrated, the developed arrays will be particularly useful for monitoring the compositions and activities of biodegrading communities in samples with enhanced microbial activity. However, the capability of revealing microbial population differences with this type of microarray will depend on the compositions, structures, and complexities of the microbial communities of interest.
The quantitative capability of microarray-based hybridizations is another central issue for environmental applications. A good linear relationship was observed between the hybridization signal intensity and the target DNA or RNA concentration for a pure culture, mixed DNA templates, and cells (Fig. 3). This is consistent with previous reports with the PCR product-based FGAs (53) and CGAs (Wu et al., unpublished), as well as with the findings of microarray studies of gene expression (2). However, like other molecular approaches, the quantitative accuracy of the 50-mer-based FGA hybridization will depend on probe specificity. To obtain more accurate results, the probe must be highly specific for the target genes. Any cross-hybridization from closely related genes could distort quantification. Since the functional genes involved in contaminant degradation and metal resistance appear to be highly diverse, cross-hybridization to these probes should not be a severe problem.
Altogether, the oligonucleotide arrays developed in this study contained 1,662 unique probes to target a variety of genes involved in the biodegradation of various contaminants and in metal resistance (Table 1). To our knowledge, this is the first comprehensive array available for environmental studies. In addition, the probes should well represent the known microbial-gene diversity involved in biodegradation, because the probes were designed based on up-to-date, available sequence information from public databases. The developed microarray should therefore be useful as a general tool for monitoring the compositions, structures, activities, and dynamics of microbial populations involved in biodegradation and metal resistance across different environments. In addition, the arrays contain probes from different steps of the catabolic pathways involved in degrading a variety of chemical compounds. The developed arrays should be useful for assessing biodegrading populations in a variety of environments contaminated with different chemicals and extremely useful for characterizing pure isolates involved in biodegradation. This was demonstrated by our discovery of many new catabolic genes in P. putida PpG7 using microarray hybridization, which was then validated by PCR and sequencing (Table 3). Finally, most of the gene probes were derived from cultivated microorganisms. Only a limited number of genes were cloned directly from environmental samples using PCR amplification with relatively conserved degenerate primers (e.g., genes for benzylsuccinate synthase, phenol oxygenase, naphthalene dioxygenase, and alkane monooxygenase) (3, 31, 34, 50). Thus, the developed microarray should be useful for assessing possible horizontal gene transfer events among different cultivated biodegrading organisms (20). It is also important to note that the knowledge gained in this study could be useful in addressing relevant microbial problems associated with human health, water and food safety, animal and plant health and productivity, biodiversity, bioprocessing of industrial products, and wastewater treatment, because microbial communities are important in each of these areas.
Using the developed 50-mer FGAs, we successfully detected changes in the microbial community structures in enrichments and soil microcosms. In soil microcosms, naphthalene (salicylate) degradation genes from the gentisate pathway originally found in a plasmid of Ralstonia sp. strain U2 (13) were detected. Interestingly, although the gentisate pathway for salicylate degradation is less common than other pathways, our results show that this pathway could be more common in soil ecosystems than suspected. Thus, the Ralstonia-type microorganisms might be actively involved in naphthalene degradation.
Although the same TFD soil was used for establishing the enrichments and soil microcosms with naphthalene as a sole carbon source, different microorganisms involved in naphthalene degradation were detected. The genes, possibly carried in a plasmid of gram-positive Rhodococcus, were detected in the enrichment but not in microcosms. Instead, Ralstonia-, Burkholderia-, and Comamonas-type microorganisms appeared to be dominant in soil microcosms. These results suggested that the enrichment process could cause severe bias of the population compositions, and hence the isolation-based approaches for monitoring community structure and dynamics may not be an appropriate choice. The culture-independent microarray-based approach has advantages over the traditional culture-(in)dependent method because large amounts of data on microbial community dynamics could rapidly be obtained without or with little bias.
Although our results demonstrated that the developed 50-mer FGAs can potentially be used as specific and quantitative tools for monitoring biodegrading populations, their usefulness and power should be further evaluated with diverse samples from a variety of contaminated environments by addressing various environmental and ecological questions. We are using the developed microarrays to address research questions related to bioremediation of groundwater contaminated with mixed wastes of uranium and organic solvents. In addition, the probes on arrays may not represent the indigenous microbial communities, because the majority of microorganisms in natural environments are not cultivated. Further efforts to understand the sequence diversity of genes related to the biodegradation of various compounds are needed.
In summary, this work evaluated the specificity, sensitivity, and quantitation of the 50-mer oligonucleotide-based microarrays. This type of microarray was successfully used for detecting biodegradation genes in soil microcosms and enrichments and for profiling the differences in microbial community structures of soils contaminated with PAH and BTEX. Our results indicated that this technology has potential as a specific, sensitive, and quantitative tool in revealing a comprehensive picture of the compositions of biodegradation genes and the microbial community in contaminated environments, although more work is needed to evaluate the performance of the 50-mer FGAs with diverse contaminated environmental samples. It is our hope that the methods presented here will serve as useful tools to monitor bioremediation processes in situ.
Acknowledgments
We thank Lynn Kszos for editorial assistance.
This research was supported by the U.S. Department of Energy under the Natural and Accelerated Bioremediation Research Program of the Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
REFERENCES
- 1.Atlas, R. M. 1981. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol. Rev. 7:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosiewicz, M., M. Trounstine, D. Barker, R. Johnston, and A. Buckpitt. 2000. Development of a toxicological gene array and quantitative assessment of this technology. Arch. Biochem. Biophys. 376:66-73. [DOI] [PubMed] [Google Scholar]
- 3.Beller, R. B., S. R. Kane, T. C. Legler, and J. J. Alvarez. 2002. A real time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. [DOI] [PubMed] [Google Scholar]
- 4.Breese, K., M. Boll, J. Alt-Morbe, H. Schagger, and G. Fuchs. 1998. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium Thauera aromatica. Eur. J. Biochem. 256:148-154. [DOI] [PubMed] [Google Scholar]
- 5.Breinig, S., E. Schiltz, and G. Fuchs. 2000. Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica. J. Bacteriol. 182:5849-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Call, D., D. Chandler, and F. Brockman. 2001. Fabrication of DNA microarrays using unmodified oligonucleotide probes. BioTechniques 30:368-379. [DOI] [PubMed] [Google Scholar]
- 7.Cho, J. C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 9.Eaton, R. W. 1994. Organization and evolution of naphthalene catabolic pathways: sequence of the DNA encoding 2-hydroxychromene-2-carboxylate isomerase and trans-o-hydroxybenzylidenepyruvate hydratase-aldolase from the NAH7 plasmid. J. Bacteriol. 176:7757-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, R. W., and P. J. Chapman. 1992. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J. Bacteriol. 174:7542-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, D. T., and G. S. Sayler. 1992. Scientific foundations of bioremediation: current status and future needs. American Academy of Microbiology, Washington, D.C.
- 15.Gibson, J., M. Dispensa, and C. S. Harwood. 1997. 4-Hydroxybenzoyl coenzyme A reductase (dehydroxylating) is required for anaerobic degradation of 4-hydroxybenzoate by Rhodopseudomonas palustris and shares features with molybdenum-containing hydroxylases. J. Bacteriol. 179:634-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimm, A. C., and C. S. Harwood. 1999. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 181:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittman, and A. D. Mitzabekov. 1997. Oligonucleotide microarrays as genosensors for determinative environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harayama, S., and M. Rekik. 1989. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J. Biol. Chem. 264:15328-15333. [PubMed] [Google Scholar]
- 19.Harayama, S., M. Rekik, A. Wasserfallen, and A. Bairoch. 1987. Evolutionary relationships between catabolic pathways for aromatics: conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids. Mol. Gen. Genet. 210:241-247. [DOI] [PubMed] [Google Scholar]
- 20.Herrick, J. B., K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen. 1997. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl. Environ. Microbiol. 63:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillegass, J. 2003. Determination of New Jersey-specific soil contaminant bioaccumulation factors for polycyclic aromatic hydrocarbons, polychlorinated biphenyls, and heavy metals using the earthworm, Lumbricus terrestris. M.S. thesis. Rutgers University, New Brunswick, N.J.
- 22.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane, M. D., T. A. Jatkoe, C. R. Stumpt, J. Liu, J. D. Thomas, and S. J. Madore. 2000. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 28:4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchman, D., J. Sigda, R. Kapuscinski, and R. Mitchell. 1982. Statistical analysis of the direct count method for enumerating bacteria. Appl. Environ. Microbiol. 44:376-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laempe, D., M. Jahn, K. Breese, H. Schagger, and G. Fuchs. 2001. Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica. J. Bacteriol. 183:968-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leahy, J. G., and R. R. Colwell. 1990. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leuthner, B., and J. Heider. 2000. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of beta oxidation of the intermediate benzylsuccinate. J. Bacteriol. 182:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leuthner, B., C. Leutwein, H. Schulz, P. Horth, W. Haehnel, E. Schiltz, H. Schagger, and J. Heider. 1998. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol. Microbiol. 28:615-628. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Y., J.-Z. Zhou, M. Omelchenko, A. Beliaev, A. Venkateswaran, J. Stair, L. Wu, D. K. Thompson, D. Xu, I. B. Rogozin, E. K. Gaidamakova, M. Zhai, K. S. Makarova, E. V. Koonin, and M. J. Daly. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 100:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd-Jones, G., A. D. Laurie, D. W. F. Hunter, and R. Fraser. 1999. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol. Ecol. 29:69-79. [Google Scholar]
- 32.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 33.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA-based detections of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margesin, R., D. Labbe, F. Schinner, C. W. Greer, and L. G. Whyte. 2003. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine alpine soils. Appl. Environ. Microbiol. 69:3085-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, C. R., and J. M. Myers. 1992. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174:3429-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Research Council. 1993. In situ bioremediation: when does it work? National Academies Press, Washington, D.C.
- 37.Parales, J. V., A. Kumar, R. E. Parales, and D. T. Gibson. 1996. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181:57-61. [DOI] [PubMed] [Google Scholar]
- 38.Relogio, A., C. Schwager, A. Richter, W. Ansorge, and J. Valcarcel. 2002. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 30:e51. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schell, M. A. 1986. Homology between nucleotide sequences of promoter regions of nah and sal operons of NAH7 plasmid of Pseudomonas putida. Proc. Natl. Acad. Sci. USA 83:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schena, M., D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suen, W. C., B. E. Haigler, and J. C. Spain. 1996. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J. Bacteriol. 178:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, H. Lim, J. Yates III, C. C. Brandt, J. M. Tiedje, and J.-Z. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiquia, S. M., S. C. Chong, M. W. Fields, and J. Zhou. Oligonucleotide-based functional gene arrays for analysis of microbial communities in the environment. In Kowalchuk, G. A., F. J. de Bruijn, I. M. Head, A. D. Lakkennans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Tiquia, S. M., L. Wu, S. C. Chong, S. Passovets, D. Xu, Y. Xu, and J.-Z. Zhou. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664-670, 672, 674-675. [DOI] [PubMed] [Google Scholar]
- 48.Valinsky, L., G. D. Vedova, A. J. Scupham, A. Figueroa, B. Yin, R. J. Hartin, M. Chroback, D. E. Crowley, T. Jiang, and J. Borneman. 2002. Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl. Environ. Microbiol. 68:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdnick, D., S. Handran, and S. Pickett. 2002. Key considerations for accurate microarray scanning and image analysis, p. 83-98. In G. Kamberova (ed.), DNA image analysis: nuts and bolts. DNA Press, Salem, Mass.
- 50.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. 1998. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, M. S., J. B. Herrick, C. O. Jeon, D. E. Hinman, and E. L. Madsen. 2003. Horizontal transfer for phnAc dioxygenase genes within one of two phenotypically and genotypically distinctive naphthalene-degrading guilds from adjacent soil environments. Appl. Environ. Microbiol. 69:2172-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 53.Wu, L. Y., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, D., G. Li, L. Wu, J.-Z. Zhou, and Y. Xu. 2002. PRIMEGENS: a computer program for robust and efficient design of gene-specific targets on microarrays. Bioinformatics 18:1432-1437. [DOI] [PubMed] [Google Scholar]
- 55.Ye, R. W., W. Tao, L. Beddzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, J.-Z., and D. K. Thompson. 2002. Challenges in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 13:202-204. [DOI] [PubMed] [Google Scholar]
- 59.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]