Abstract
Recombination is a prominent feature of meiosis in which it plays an important role in increasing genetic diversity during inheritance. Additionally, in most organisms, recombination also plays mechanical roles in chromosomal processes, most notably to mediate pairing of homologous chromosomes during prophase and, ultimately, to ensure regular segregation of homologous chromosomes when they separate at the first meiotic division. Recombinational interactions are also subject to important spatial patterning at both early and late stages. Recombination-mediated processes occur in physical and functional linkage with meiotic axial chromosome structure, with interplay in both directions, before, during, and after formation and dissolution of the synaptonemal complex (SC), a highly conserved meiosis-specific structure that links homolog axes along their lengths. These diverse processes also are integrated with recombination-independent interactions between homologous chromosomes, nonhomology-based chromosome couplings/clusterings, and diverse types of chromosome movement. This review provides an overview of these diverse processes and their interrelationships.
Meiotic recombination increases genetic diversity, but it is also essential for the pairing and segregation of homologous chromosomes. Structures such as the synaptonemal complex play key roles.
The role of the meiotic program is to generate gametes having half the chromosome complement of the original progenitor cell. This task is accomplished by occurrence of a single round of DNA replication followed by two successive rounds of chromosome segregation. Homologs segregate to opposite poles at meiosis I, then sisters separate to opposite poles in meiosis II, analogously to mitosis (Fig. 1A).
Figure 1.
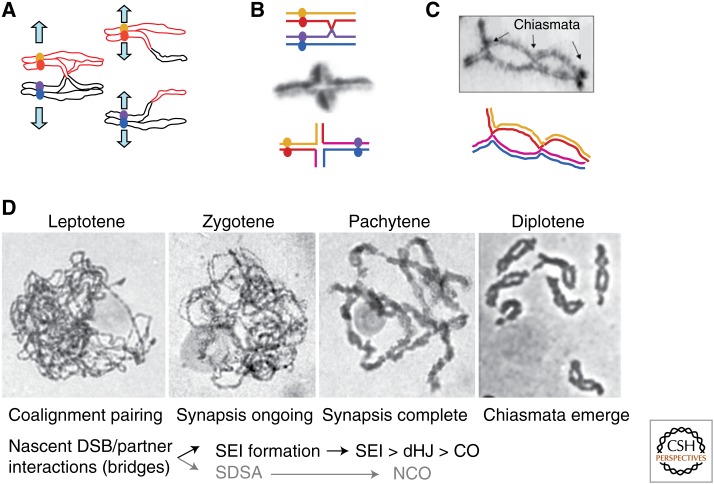
General features of meiosis. (A) At meiosis I, homologs segregate; at meiosis II, sisters segregate. At metaphase I (left), maternal (red) and paternal (black) chromosomes are held together by a chiasma comprising a reciprocal crossover (CO) plus connections along sister arms, which are released during segregation. (B) Monochiasmate bivalent of Locusta after bromodeoxyuridine (BrdU) incorporation. Differential staining of the sister chromatids confirms that exchange has occurred, for example, between red and purple chromatids in corresponding drawings. (From Jones 1987; reprinted, with permission, from Academic Press © 1987.) (C) Diplotene bivalent of grasshopper with three chiasmata (arrows) and corresponding drawing. (From Jones and Franklin 2006; reprinted, with permission, from Elsevier © 2006.) (D) Top: Meiotic prophase in rye microsporocytes; chromosomes are stained by hematoxylin (pictures by D.Z.). Bottom: corresponding timing of the recombination steps from double-strand breaks (DSBs) to COs; timing of intermediates as in budding yeast (Hunter 2007). SEI, Single-end invasion; dHJ, double Holliday junction; SDSA, synthesis-dependent strand annealing; NCO, noncrossover.
During meiosis, a central role of recombination is to increase genetic diversity. However, recombination is also essential for two fundamental features unique to meiotic chromosome mechanics: pairing and segregation of homologous chromosomes (“homologs”). Pairing is mediated by the totality of programmed interhomolog recombinational interactions in association with chromosome structural axes (see below). Segregation is mediated specifically by the carefully chosen subset of those interactions that mature into crossover (CO) products. During segregation of homologs, just as for segregation of sister chromatids, the separating entities must be connected to one another such that regular bipolar alignment on the spindle results in tension on centromere/kinetochore complexes. When all segregating pairs are properly aligned and under tension, anaphase is triggered. Segregation of sisters is ensured by connections between sister centromere/kinetochore regions. Segregation of homologs is ensured by connections along chromosome arms that are provided by the combined effects of an interhomolog CO plus links between sisters (Fig. 1A). These connections can be seen cytologically as chiasmata (Fig. 1B,C). In organisms in which meiosis occurs without recombination, other features have evolved that hold homologs together to ensure regular segregation (Zickler and Kleckner 1998, 1999; reviewed in Stewart and Dawson 2008; Tsai and McKee 2011; Lake and Hawley 2012; Obeso et al. 2014).
THE CANONICAL MEIOTIC PROGRAM
In most organisms (e.g., plants, mammals, budding yeast, and filamentous fungi), recombination mediates an ordered program of interactions between homologs that plays out during an extended postreplicative prophase period. Recombination initiates by programmed double-strand breaks (DSBs). A subset of these breaks proceeds to CO products, which emerge at the end of the “pachytene” stage (reviewed in Hunter 2007; de Massy 2013). All of these events are integrated with the broader program of meiotic chromosome dynamics. At the whole chromosome level, homologs become intimately associated by what appears to be a smooth progressive process (Fig. 1D). However, three distinct aspects can be distinguished.
Programmed DSBs occur during G2/leptotene (Padmore et al. 1991) and result in linkage of the two individual interacting DNA segments. However, these interactions also mediate the spatial coalignment of whole homologous chromosomes. This process, which we will refer to as “pairing,” is concomitant with development of individualized, organized chromosomes.
Following coalignment, homologs become much more closely associated. This association, referred to as “synapsis,” corresponds to installation of a robust structure, the synaptonemal complex (SC), between the homolog axes all along their lengths. The period when SC is forming defines “zygotene”; presence of complete SC defines “pachytene.” SC formation is usually nucleated at the sites of recombinational pairing interactions (see below). The latter steps of recombination occur in the context of the SC. After COs appear at the end of pachytene, the SC disassembles and homologs become more compact and separate along their lengths, except at the sites of COs (chiasmata), thus defining the stage of “diplotene.”
As a prelude and/or complement to coalignment/pairing and synapsis, other types of interchromosomal interactions (e.g., clustering of centromeres and/or telomeres) occur. These effects include both homology-dependent and homology-independent interactions.
ALTERNATIVE PROGRAMS
In some organisms, the program of prophase events is somewhat different. In fission yeast (Schizosaccharomyces pombe) and filamentous fungus Aspergillus nidulans, recombination-independent and recombination-mediated pairing occurs but SC is absent (Egel-Mitani et al. 1982; Bähler et al. 1993). In female Drosophila and Caenorhabditis elegans, pairing and synapsis occur independently of recombination, which then occurs later in the context of the SC (reviewed in Lake and Hawley 2012; Liu and Colaiácovo 2013; Rog and Dernburg 2013). Finally, in male Drosophila, the entire meiotic program occurs in the absence of recombination, with recombination-independent homologous pairing and specialized connections substituting for chiasmata to ensure homolog disjunction (reviewed in Tsai and McKee 2011).
In C. elegans, recombination-independent pairing occurs via specific regions near chromosome ends (“pairing centers” or PCs) (reviewed in Tsai and McKee 2011; Rog and Dernburg 2013). In Drosophila, in both male and female meiosis, chromosome pairing occurs by significantly modulated versions of the somatic pairing characteristic of that organism (reviewed in McKee 2004; 2009; Tsai and McKee 2011; Cahoon and Hawley 2013; see below). Interestingly, however, in a certain mutant of C. elegans, SC formation is now dependent on recombination (Smolikov et al. 2008); and in Drosophila, some SC initiation sites colocalize with recombination sites (Tanneti et al. 2011). Moreover, conversely, in the canonical program, recombination-independent homologous interactions likely make a significant contribution to pairing. Thus, the canonical and alternative programs overlap to some degree.
RECOMBINATION-MEDIATED HOMOLOG PAIRING
In accord with the central role of recombination in the canonical homolog pairing and synapsis program, DSBs are essential for both processes (reviewed in Kleckner et al. 2012; Baudat et al. 2013). Correspondingly, in Sordaria, budding yeast and mouse, analysis of mutants showing that varying numbers of DSBs reveal direct relationships between the number of DSBs and the extents of presynaptic coalignment and/or SC formation (Tessé et al. 2003; Henderson and Keeney 2004; Kauppi et al. 2013; Rockmill et al. 2013).
Spo11-Mediated DSBs
In all organisms studied so far, meiotic DSBs are catalyzed by the topoisomerase-like protein Spo11. The Spo11 protein per se is not strictly required. Irradiation-induced DSBs can rescue pairing and SC formation in the absence of Spo11. However, both processes are less robust than in normal meiosis, at least in part because some nuclei get too few breaks (e.g., Dernburg et al. 1998; Storlazzi et al. 2003; Tessé et al. 2003; Yokoo et al. 2012).
The number of Spo11-induced DSBs per genome is regulated on a species basis (reviewed in de Massy 2013). As a general rule, in organisms with the canonical program, DSB numbers tend to be higher in organisms with longer chromosomes, in accord with their role for pairing. Moreover, DSBs are less frequent in organisms where they are not required for pairing. Thus, DSBs per meiotic nucleus are ∼200–300 per cell in mouse and about 12 and 14 in Drosophila and C. elegans, respectively (reviewed in Lichten and de Massy 2011; de Massy 2013). In contrast, in nearly all organisms, the number of COs/chiasmata is relatively few, with one to several per homolog pair, even in cases with large genome sizes and long chromosomes. Correspondingly, only one among 30 DSBs mature into a CO in Arabidopsis, whereas half are ultimately matured to COs in C. elegans (Serrentino and Borde 2012). Most DSBs that do not mature into COs are matured into interhomolog events not accompanied by crossing over (i.e., noncrossovers [NCOs]) plus some intersister recombination (Hunter 2007).
Recombination Occurs in the Context of Chromosome Structural Axes (and the SC)
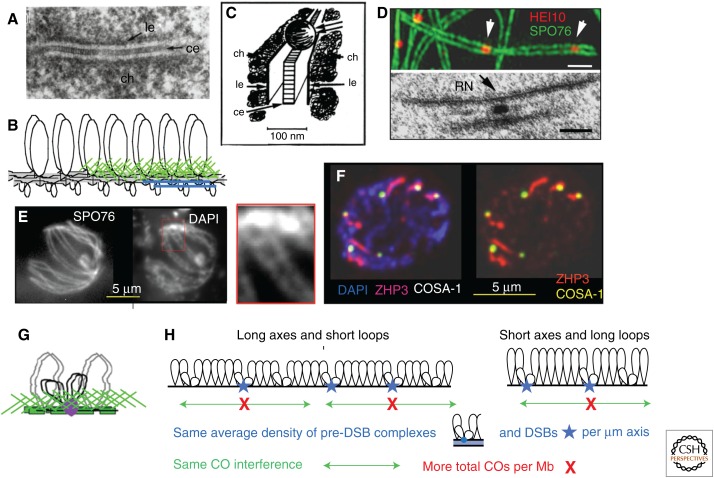
Meiotic chromosomes are highly organized. Electron microscope (EM) analysis of spread preparations and whole sectioned nuclei (Zickler and Kleckner 1999) along with immuno-EM and fluorescence imaging indicate that synapsed pachytene bivalents are organized in the following way (Fig. 2A,B). (1) Each chromatid is organized into a linear array of loops, the bases of which comprise a structural axis, delineated in EM by the “axial element” (AE). (2) Homolog axes are linked along their entire lengths via transverse filaments, which link AEs to form the SC (e.g., Schmekel and Daneholt 1995; Page and Hawley 2004). At this stage, the AEs are now called the lateral elements (LEs) of the SC. (3) Sister-chromatid axes are tightly conjoined and their loops emanate outward from the SC.
Figure 2.
Chromosome axes and synaptonemal complex. (A) Electronic microscope (EM) longitudinal section of Blaps cribrosa synaptonemal complex (SC) with distinct central region transverse filaments. (From Schmekel et al. 1993; reprinted, with permission, from Springer © 1993.) (B) Co-oriented sister linear loop arrays cojoined by meshwork of structural proteins (green; see text). (C) SC with recombination nodule; reconstruction from serial section through Drosophila female SC. (From Carpenter 1975; with permission from the National Academy of Sciences © 1975.) (D) Sordaria bivalent at pachytene by immunofluorescence microscopy (top) and by EM (bottom). Top: Crossover (CO) sites marked by the E3 ligase Hei10-mCherry and chromosome axes visualized by cohesin-associated Spo76-GFP (arrows point to Hei10 foci). Bottom: Sordaria SC with late recombination nodule (RN) (both from D.Z.). Scale bars, 2 μm (top); 100 nm (bottom). (E) Sordaria bouquet stage stained by axis component Spo76/Pds5-GFP (left) and by DAPI (right). Note that axis width comprises a significant fraction of the total DAPI width, suggesting that, albeit with limitations of imaging resolution, the axis meshwork may include a significant fraction of the DNA. DAPI staining indicates that chromatin bridges join the two homologs (magnification right) (from D.Z.). (F) COSA-1 foci mark sites of COs in C. elegans. SC marked by ZHP3 staining. (From Yokoo et al. 2012; reprinted, courtesy of a PMC Open Access license.) (G) Recombination complexes are indirectly tethered to underlying chromosome axes (see discussion in Blat et al. 2002). (H) Coordinate variation in axis/SC length and CO frequency can be explained by development of axis-associated pre-double-strand break (DSB) recombination complexes at constant spacing along axes followed by identical probabilities of DSB formation per complex and identical CO-designation/interference (Adapted from Kleckner et al. 2003). ch, Chromatin; le, lateral element; ce, central element.
Interestingly, the spacing of loops along pachytene axes is evolutionarily conserved (∼20 per micrometer) (Kleckner 2006). Conserved loop spacing can be manifested in mutants, in which axis length and loop size show opposite and compensatory changes. For example, in mutants altered for the mammalian meiotic-specific cohesin Smc1β and the SC lateral component Sycp3, longer and shorter chromosome axes are accompanied by shorter and longer loops (Revenkova and Jessberger 2006; Novak et al. 2008; Kauppi et al. 2011). Also, different organisms show different genome sizes, and these differences are accommodated by variations in loop size and axis length rather than loop spacing (reviewed in Zickler and Kleckner 1999; Page and Hawley 2004; Kleckner 2006).
Chromosome structural axes develop during prophase, concomitant with developing organization (Zickler and Kleckner 1999). Axes comprise a complex meshwork of protein/DNA interactions and likely include a significant amount of DNA (Fig. 2E) (Kleckner 2006; Zhang et al. 2014c). How these axes develop is not known. Interestingly, however, while preferred DNA sequence regions may be involved in axis formation (e.g., a queue of locally AT-rich regions in budding yeast) (Blat et al. 2002), when a chromosome from one organism is introduced into another organism, it assumes the axis/loop organization of its host (Loidl et al. 1995) showing that sequence features are important but are not, per se, determinative of loop size. Mitotic prophase chromosome organization may turn out to be the same as that of meiotic prophase chromosomes (Kleckner et al. 2012).
Molecular studies of pachytene chromosomes have identified several types of axis components: mitotic structural proteins like topoisomerase II, condensins, cohesins, and cohesin-associated proteins, including meiosis-specific versions of some of these proteins; meiosis-specific axis components; and structural components of the SC, which are also unique to meiosis (Moens and Earnshaw 1989; Page and Hawley 2004; Revenkova and Jessberger 2006; Wojtasz et al. 2009; Wood et al. 2010, and references therein; Liu and Colaiácovo 2013). In many organisms (including budding and fission yeast, mammals, Arabidopsis, rice, Drosophila, and C. elegans), prominent meiotic axis components include one or more HORMA-domain proteins. These molecules and associated partner proteins occur at the nexus of axis structure and recombination and are also intimately involved in regulatory surveillance responses to defects in synapsis and/or recombination (e.g., Hollingsworth and Byers 1989; Molnar et al. 2003; Couteau and Zetka 2005; Nonomura et al. 2006; Carballo et al. 2008; Sanchez-Moran et al. 2008; Wojtasz et al. 2009, 2012; Roig et al. 2010; Daniel et al. 2011; MacQueen and Hochwagen 2011; reviewed in Liu and Colaiácovo 2013).
The biochemical complexes that carry out recombination are intimately associated with chromosome structural axes, both physically and functionally, at all stages. This association was first revealed by the discovery of CO-correlated “nodules” along the SC (Fig. 2C,D,F) (Carpenter 1975, 1987; von Wettstein et al. 1984). This association is set up during development of pre-DSB recombination complexes (e.g., Blat et al. 2002; Peoples et al. 2002; Storlazzi et al. 2010; Panizza et al. 2011). Molecular studies show that recombination complexes form in sequences that are not, per se, axis ssociated, implying indirect tethering to axes in “tethered-loop axis complexes” (Fig. 2G) (Blat et al. 2002; Miyoshi et al. 2013). Recombinosome/axis-SC association persists through the pairing process and, for CO-fated recombination complexes, until the end of the pachytene and sometimes beyond. NCO-fated complexes appear to be released from axes at earlier stages (see discussion in Terasawa et al. 2007). Axis-association of recombination complexes is one of the central hallmarks of meiosis. It is present in noncanonical prophase programs as well as the canonical program (e.g., Martinez-Perez et al. 2005; Mehrotra and McKim 2006; Mets and Meyer 2009; Libuda et al. 2013; Miyoshi et al. 2013) and likely has many roles (e.g., Blat et al. 2002; Panizza et al. 2011; see below).
DSB/Partner Interactions Mediate Coalignment via Interaxis Bridges
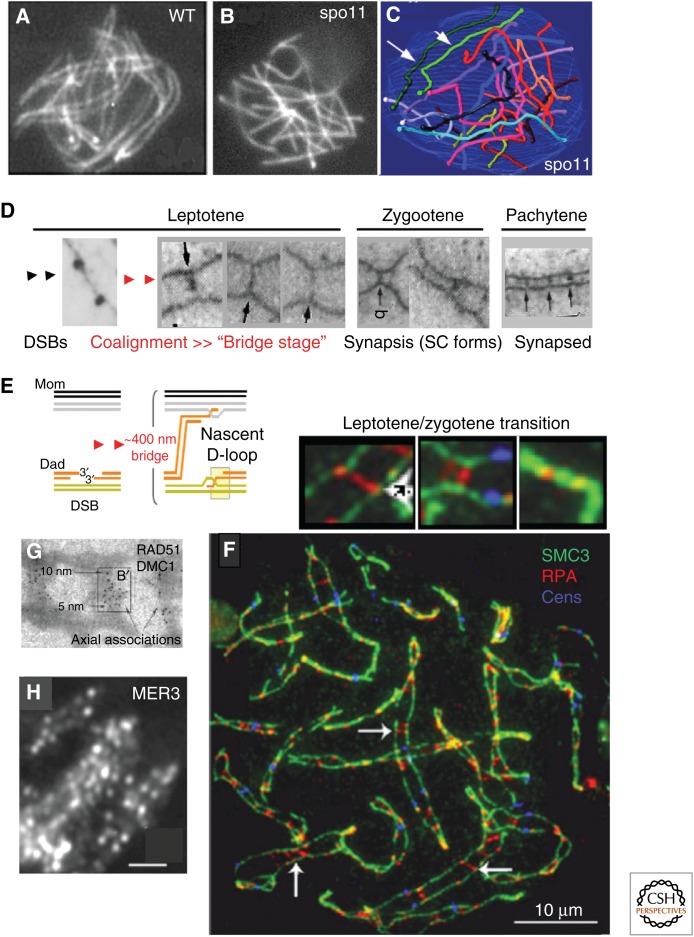
DSBs mediate homolog pairing. This process culminates in a discrete state at which homologs are coaligned at a distance of ∼400 nm (vs. the ∼100 nm of the SC) (Figs. 2E, 3A,D,F,G). Notably, axial chromosome organization, with the chromatin of the two sister chromatids emanating from the same side of the axis (see above), makes possible close juxtaposition of homolog axes in a way that would not be possible if chromatin surrounded each axis.
Figure 3.
Interhomolog recombination-dependent interactions. (A–C) Sordaria leptotene. (A) In wild type (WT), homologs align at a 300- to 400-nm distance all along their lengths. (B,C) In the absence of Spo11, axes do not align (asynapsis; B) except C in a few meiocytes when one chromosome pair (light and dark green; indicated by white arrows) is seen aligned. Chromosomes are stained by Spo76-GFP in A and B. (A from Storlazzi et al. 2010; reprinted, with permission, from the authors; B and C from Storlazzi et al. 2003; reprinted, with permission, from the authors.) C is a reconstruction from serial sections of a spo11 mutant. (D) Axis association of early recombination nodules and interhomolog bridges from early leptotene (left) through pachytene (right). (From Albini and Jones 1987; reprinted, with permission, from Springer © 1987, except for the image on the left, which is from Stack and Anderson 1986; reprinted, with permission, from JSTOR, Early Journal Content © 1986.) (E) Presumptive recombination intermediate at the interaxis coalignment bridge stage. Double-strand break (DSB) engages a homolog partner chromatid and directs juxtaposition of associated donor and partner chromosome axes to a distance of ∼400 nm (as in panels D and F–H). (F) Replication protein A (RPA) staining in human cells at leptotene/zygotene identifies interaxis bridges and configurations showing approaching and completed synapsis (insets, compare with panel D). (From Oliver-Bonet et al. 2007; reprinted, with permission, from Oxford University Press © 2007.) (G) RecA homologs Rad51 and Dmc1 decorate interaxis bridges in mouse. (From Tarsounas et al. 1999; reprinted, with permission, from Rockefeller University Press under a Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported license.) (H) Mer3 foci face each other on coaligned late leptotene homologs. Scale bar, 2 μm. (From Storlazzi et al. 2010; reprinted, with permission, from the authors.) SC, Synaptonemal complex.
Cytologically, coalignment can be seen to comprise the linkage of homolog axes via a series of “bridges” (Fig. 3D) (e.g., Albini and Jones 1987). Each bridge corresponds to the site of a DSB-mediated interhomolog interaction. Correspondingly, coalignment is absent in mutants lacking DSB transesterase Spo11 or the relevant RecA homolog strand exchange protein (compare Fig. 3B and 3A) (Storlazzi et al. 2003) and is reduced in relation to lowered DSB levels (e.g., Tessé et al. 2003). Moreover, these bridges comprise recombinational interactions as marked by RecA homologs (e.g., in mouse) (Fig. 3G) (Tarsounas et al. 1999), RPA in human spermatocytes (Fig. 3F) (Oliver-Bonet et al. 2007), and matched pairs of Mer3 helicase complexes in Sordaria (Fig. 3H) (Storlazzi et al. 2010).
To a first approximation, the recombinational interactions responsible for coalignment bridges represent the totality of DSB-mediated interhomolog contacts. Given the stage at which they occur, each bridge can be inferred to represent a DSB-mediated interaction in which a “leading” DSB end has made a nascent D-loop with a homolog partner chromatid, whereas the “lagging” DSB end remains associated with the original donor chromosome (Fig. 3E). Thus, meiotic DSBs and their ensuing identification of homolog partner chromatids in DNA/DNA interactions provide the informational basis for recognition of homologs. However, juxtaposition of local chromatin/DNA segments will not per se propagate over significant distances as required to affect homolog pairing at the level of whole chromosomes. Association of recombination complexes with chromosome axes is therefore critical, because it permits local DNA interactions to mediate overall chromosome juxtaposition.
The inferences that (1) DSB complexes occur in tethered loop/axis complexes, and (2) initial DSB/partner interactions involve only one DSB end, lead to the hypothesis that one end of a DSB is released from its axis to create a genome-scaled “tentacle” that can search chromosomal space (Kim et al. 2010; Panizza et al. 2011). This situation would seem to contrast with the case for sister-directed mitotic DSB repair where the two DSB ends appear to remain together, at least for much of the process (reviewed in Jasin and Rothstein 2013). In the context of the tentacle hypothesis, a homology-searching DSB might usually identify its partner sequence in a chromatin loop. This new interaction might then become associated to its underlying axis either actively (by “reeling-in”) or passively (by fortuitous collision stabilized by protein/protein interactions), thus creating a bridge (see cartoon in Kim et al. 2010; Storlazzi et al. 2010).
Homolog Bias
A basic feature of meiosis is that DSB-mediated interactions/repair occurs differentially between homologous nonsister chromatids, rather than between sisters as in mitotic DSB repair. This “homolog bias” is critical for all of the functions of recombination for meiosis: recombination-mediated homolog pairing; the appropriate genetic outcome (an interhomolog CO); and the corresponding interhomolog connections (chiasmata) that ensure regular homolog disjunction. Accordingly, homolog bias is established early in the recombination reaction, as a DSB first identifies its partner. However, this bias must also be actively maintained at a later stage, as the lagging DSB end becomes engaged in the reaction to form a double Holliday junction intermediate for CO recombination (Kim et al. 2010).
In budding yeast, meiotic and mitotic RecA homologs (Dmc1 and Rad51) and the meiotic axis complex Red1/Hop1/Mek1 all play active roles in this process, reflecting the central role of recombinosome/axis association for recombination and pairing (Callender and Hollingsworth 2010; Hong et al. 2013; Lao et al. 2013). A central role of the axis complex is to switch recombination from a mitotic-like mode (in which Rad51 carries out strand exchange and meiotic components are not involved) to the meiotic mode. Recent findings further suggest that partner choice shows an unexpected basic logic (Hong et al. 2013). In the absence of all meiotic functions, and of meiotic cohesin Rec8, homolog bias is still established. Restoration of cohesin alone then results in intersister bias. And restoration of both cohesin and meiotic functions then fully restores homolog bias. Thus, homolog bias could be the intrinsic default option for mitotic recombination, even in the S/G2 stages of the cell cycle, perhaps because a genome-wide homology search is required for DSB repair in G0/G1. When a sister is present, cohesin would channel the process so that the sister is used, in meiosis and mitotic G2. And for meiosis, where recombination occurs after replication, meiotic functions would then act to overcome or eliminate this channeling effect of cohesin to restore genome-wide searching and homolog bias.
RECOMBINATION-INDEPENDENT HOMOLOGOUS PAIRING
A prominent, but still mysterious, feature of chromosome biology is the ability of homologous chromosomes, or chromosomal regions, to specifically recognize and pair with one another in the apparent absence of DNA lesions (DSBs) or recombination. The paradigmatic example is Drosophila somatic pairing (e.g., Joyce et al. 2013). Genome-wide somatic pairing also occurs in budding yeast (Weiner and Kleckner 1994; Keeney and Kleckner 1996; Burgess et al. 1999; Burgess and Kleckner 1999; Cha et al. 2000; Dekker et al. 2002), and in stabilized S. pombe diploids (Scherthan et al. 1994), including telomere pairing in both organisms (Klein et al. 1992; Molnar and Kleckner 2008). Pairing in somatic cells also occurs locally, for example, in cases of monoallelic expression in mammalian cells (e.g., X-chromosome inactivation, V(D)J recombination and imprinting; references in Joyce et al. 2013; Barakat et al. 2014).
Recombination-independent pairing also plays prominent roles for premeiotic and meiotic programs, where it is defined as pairing that occurs before and/or without Spo11-mediated DSBs.
Examples
RIP/MIP
One prominent example is provided by RIP and MIP in the filamentous fungi Neurospora crassa and Ascobolus immersus, respectively (reviewed in Selker 1990). In haploid nuclei preparing for karyogamy and onset of meiotic prophase, all repeated sequences (except the rDNA) undergo methylation (MIP) or methylase-directed point mutation (RIP), independent of Spo11 or Rad51 (Gladyshev and Kleckner 2014).
Drosophila Meiosis
In Drosophila female meiosis, pairing and SC formation precede and are independent of recombination with DSBs then occurring in the context of the SC (Lake and Hawley 2012). It was long thought that meiotic DSB-independent pairing was a simply continuation and enhancement of somatic pairing. However, recent studies show that germline stem cells do not have somatic pairing, genome wide or specifically in centromere regions. Pairing reestablished five mitotic divisions before the onset of meiosis (Cahoon and Hawley 2013; Christophorou et al. 2013; Joyce et al. 2013). Interestingly, homologous heterochromatic regions remain paired after prophase, permitting regular segregation of homologs that have failed to acquire a CO/chiasma (Dernburg et al. 1996).
In Drosophila male meiosis, DSB-independent pairing occurs genome wide and permits formation of persistent interhomolog links that take the place of chiasmata to ensure segregation (McKee 2009; McKee et al. 2012). Additionally, a specific pairing site in the rDNA effects pairing of the X and Y chromosomes (reviewed in McKee 2009; Tsai and McKee 2011; McKee et al. 2012).
Budding Yeast
Somatic pairing in yeast persists through the G1/G0 period that immediately precedes meiotic S phase, is diminished during S phase as in cycling cells, and is then restored, dependent on Spo11 protein but independent of its role in DSBs (Weiner and Kleckner 1994; Cha et al. 2000). DSB-independent pairing interactions, before and during meiosis, may underlie observed trans effects of the homolog on DSB formation (Xu and Kleckner 1995; Rocco and Nicolas 1996; Zhang et al. 2011).
Sordaria
Spo11-independent homolog pairing can occur in this filamentous fungus, as shown by parallel axis coalignment in spo11Δ (Fig. 3C) (Storlazzi et al. 2003).
Mouse
Global DSB-independent pairing is seen very early in mouse meiosis (Boateng et al. 2013; Ishiguro et al. 2014). As for yeast, this pairing is dependent on the Spo11 protein but independent of its role in DSBs (Boateng et al. 2013). Pairing is also dependent on meiotic cohesin Rec8 (Ishiguro et al. 2014).
C. elegans
Homology-dependent recombination-independent interactions occur between specific PCs located near one end of each C. elegans chromosome (reviewed in Tsai and McKee 2011; Liu and Colaiácovo 2013; Rog and Dernburg 2013). PCs comprise complex arrays of DNA repeat sequences with interspersed binding sites for zinc-finger (ZnF) proteins. ZnF proteins are necessary for pairing but not sufficient for homology discrimination, implying roles for sequences within PCs or in adjacent regions. PCs have diverse additional roles including stabilization of homologous interactions and nucleation of SC formation. Binding sites for ZnF proteins also occur along chromosome arms (Phillips et al. 2009) and DSB-independent interactions can likely occur along the lengths of chromosomes as seen when Spo11 and the SC are both absent (Smolikov et al. 2008).
S. pombe
Recent S. pombe studies reveal robust locus-specific Spo11-independent pairing that is dependent on both a locus-specific binding protein and a long noncoding RNA (Ding et al. 2012). Elimination of this locus has no discernible effect on overall chromosome pairing leading to the speculation that analogous RNA-mediated pairing might be occurring, undetected, genome wide (Ding et al. 2012, 2013).
DSB-Independent Pairing at Centromeres and Telomeres
Many organisms show local homologous pairing, early in meiosis, between centromeres (reviewed in Christophorou et al. 2013; Zhang et al. 2013b) or telomeres (Armstrong et al. 2001; Armstrong and Jones 2003; Boateng et al. 2013). In many, possibly all, of these cases, pairing is not directly mediated by DSBs but rather by direct local homology-dependent associations.
Mechanism(s)
The informational basis by which homologous chromosomes recognize one another in the absence of recombination is unclear. The most straightforward possibility is recognition of homology at the DNA level, by direct DNA/DNA interactions. This is favorable a priori for RIP and MIP because they involve repeat-induced DNA modifications that can be triggered by any repeat, regardless of origin, sequence or genetic activity (Kleckner and Weiner 1993). This possibility has recently been supported directly by the demonstration that RIP involves direct interaction of coaligned, intact DNA duplexes via triplet contacts (Gladyshev and Kleckner 2014). More generally, direct homology-dependent pairing between intact B-DNA duplexes, in the absence of supercoiling, has been observed experimentally in vitro in the absence of aggregation-promoting divalent metal ions, proteins and crowding agents (Danilowicz et al. 2009). However, other models for DSB-independent homology recognition include protein/protein interactions guided by underlying DNA/protein binding sequences and simple sequence “bar codes.” In cases in which RNA is required for pairing, notably X-chromosome inactivation and recently identified pairing in S. pombe, the involved RNA may be the direct mediator via RNA/RNA/DNA pairing and/or could play an indirect role as a modulator of chromatin structure (e.g., Ding et al. 2013).
Roles of Early Homologous Pairing and Nonhomologous Clustering
DSB-independent pairing, either global or local, at centromeres or telomeres, as well as homology-independent clustering, will promote DSB-mediated pairing in two ways. First, a tendency for colocalization will increase the proximity of a DSB and its cognate partner sequence while concomitantly reducing the complexity of irrelevant sequences that must be scanned and rejected (e.g., Goldman and Lichten 2000). Second, such pairing will tend to place homologous chromosomes in topologically acceptable joint domains, thus reducing the likelihood that creation of many DSB-mediated connections between different homologs will create a tangled mess (Kleckner and Weiner 1993; Klutstein and Cooper 2014).
DYNAMICS OF DSB-MEDIATED HOMOLOG PAIRING
Interplay between homologous chromosomes during meiosis is, by its nature, a dynamic process. The nature of the forces behind these dynamics, which are likely of diverse origins, are only partially understood.
DSB-Mediated Partner Identification
DNA/chromatin/chromosome movement is required for a DSB to identify the homologous sequence on the partner chromosome. In vitro, a RecA filament formed on a short oligonucleotide can find an appropriate partner extremely rapidly, suggesting that molecular scanning for homology, per se, is not time limiting for the pairing process (Yancey-Wrona and Camerini-Otero 1995). For a meiotic DSB, however, the challenge is greater because the searching region is part of a whole chromosome. In principle, homologous regions might find one another because of “stirring forces” and/or because a DSB end (e.g., as an elongated “tentacle”; see above) can search through chromosome space irrespective of whole chromosome movement. Stirring forces might be provided by thermal motion, chromatin remodeling, DNA/RNA metabolism, assembly of prophase chromosome structure, and/or the ongoing process of DSB-mediated homolog recognition and juxtaposition itself. Yeast studies identify rapid movements of a fluorescently tagged locus that appear to be involved in primary DSB/partner recognition (Lee et al. 2012). These movements require association of telomeres with the nuclear periphery and begin around the time of DSB formation. Also, in vegetatively growing yeast cells, a DSB triggers increased global chromatin mobility, presumably allowing the DSB to explore a larger fraction of the genome (Miné-Hattab and Rothstein 2013); perhaps meiosis has an analogous response.
Other unique features of meiosis could be important for the dynamics of homology searching in as-yet mysterious ways. A dramatic increase in nuclear volume is observed almost universally at premeiotic G1 (e.g., Zickler 1977), perhaps to facilitate chromosome movement? Also, DSBs occur in the context of local chromosome axis ensembles, implying that searching may involve partially organized, compact chromosomes or chromosomal regions. A prediction of this condition is that local pairing at one position will automatically be propagated for a significant distance along the chromosomes.
Preventing Chromosome Entanglements from Arising
If DSB-mediated pairing interactions occurred randomly and simultaneously throughout the genome, without any prior relationship between homologs, the result would be massive entanglement. Pre-DSB disposition of homologs via global pairing or local pairing, coupling or clustering (see above) will clearly help to minimize irregular relationships. However, the existence of an additional effect is suggested in Sordaria by identification of a mutant, which shows both dramatically interwoven chromosomes and delayed homolog coalignment (Fig. 4D) (Storlazzi et al. 2010). It was proposed that, during normal coalignment, rapid DSB-triggered juxtaposition at one position would draw adjacent regions into the same space, thereby increasing the probability that subsequent DSBs at nearby positions will find a partner efficiently and without creating an entanglement. If juxtaposition were delayed or inefficient, this simplifying process would be absent and interwoven chromosomes would result.
Figure 4.
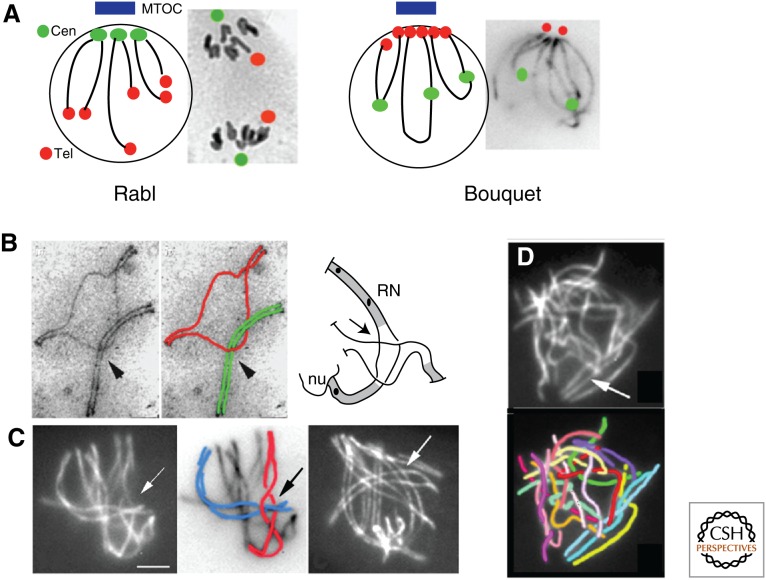
Rabl/Bouquet and Interlockings. (A) Left: Cartoon of anaphase of mitotic division; chromosomes segregate with their centromeres facing the spindle pole (and corresponding microtubule organizing center [MTOC], in blue) plus telomeres at arm-size latitude and remain in this “Rabl” disposition. Right: During meiotic prophase, telomeres turn position and now cluster facing the MTOC with centromeres more or less dispersed in the nucleus (Sordaria image from D.Z.). (B) Interlockings. Left: Interlocking of one bivalent (green) in a half-synapsed second bivalent (red) from spread zygotene chromosomes of the silkworm. (From Rasmussen 1986; reprinted, with permission, from Elsevier © 1982.) Right: Interlocking of Sordaria in which the lower bivalent shows one open end (no synaptonemal complex [SC], in gray), thus allowing easy resolution by sliding of the entrapped bivalent (from D.Z.). (C) Interlocking (arrow) during the bouquet stage (left) and corresponding drawing; right entanglements (arrow) start at late leptotene during coalignment. Scale bar, 2 μm. (From Storlazzi et al. 2010; reprinted, with permission, from the authors.) (D) An example of interwoven chromosomes in Sordaria mer3 null mutant with corresponding drawing. (From Storlazzi et al. 2010; with permission, from the authors.) RN, Recombination nodule; nu, nucleus.
Dynamic Telomere-Led Chromosome Movements, the “Bouquet” Configuration and Entanglement Resolution
One prominent feature of meiotic prophase is chromosome spatial organization at the “bouquet stage.” Telomeres associate with the nuclear envelope during leptotene coalignment and then tend to cluster, more or less tightly, in a restricted area of the nuclear envelope (Fig. 4A, right). Usually, this cluster faces the microtubule organizing center (MTOC; centrosome or spindle pole body); however, the bouquet also forms in plants where no defined MTOC is present (reviewed in Bähler et al. 1993; Zickler and Kleckner 1998; Scherthan 2001; Ding et al. 2004; Zickler 2006). Importantly, the bouquet is not a static configuration. Instead, it occurs as part of a program of dramatic back-and-forth chromosome movements mediated by interaction with telomere regions, through the nuclear envelope, to proximal cytoskeletal components. These motions were first discovered in S. pombe (reviewed in Ding et al. 2004; Chikashige et al. 2006; Hiraoka and Dernburg 2009) and are also seen in budding yeast (Scherthan et al. 2007; Conrad et al. 2008; Koszul et al. 2008). Also, mouse nuclei show both coordinate rotational motions of chromosomes and individual movements of telomeres (Parvinen and Soderstrom 1976; Shibuya et al. 2013).
Given the conservation of the bouquet stage and the involved proteins among plants, mammals and fungi, this feature of meiosis is quite likely universal among organisms that carry out the “canonical” program. Also, in C. elegans, in which pairing and SC formation precede initiation of recombination, chromosomes show analogous movements, led by their chromosome-terminal PCs (reviewed in Rog and Dernburg 2013).
For organisms that carry out the canonical meiotic program, long-standing dogma has held that the role of the bouquet is to bring homologs together in a simplified spatial arrangement that facilitates their efficient and topologically regular pairing (e.g., Scherthan 2001). However, recent observations suggest that initial homolog juxtaposition may not be the major role of this configuration. Homologs are brought together much earlier via global pairing and/or local pairing, coupling and general clustering processes (see above), after which DSB-mediated coalignment occurs, during leptotene. Only then do the “bouquet stage” and associated movements occur, during zygotene, contemporaneous with SC formation. Thus, in plants and filamentous fungi like Sordaria and Neurospora, coalignment is almost complete before telomere clustering into the bouquet (reviewed in Zickler 2006); in human male meiosis, synapsis initiates before any bouquet is present (Tankimanova et al. 2004); and in budding yeast, both the bouquet and the associated dynamic chromosome movements occur only at zygotene and pachytene, and thus after the main job of pairing has been substantially accomplished (Scherthan et al. 2007; Koszul et al. 2008). Correspondingly, earlier “rapid prophase movements” that promote pairing in yeast (see above) are not only earlier than but are not functionally related to occurrence of the bouquet (Lee et al. 2012).
What, then, is the role of the bouquet configuration and, more generally, the mid-prophase telomere movements that, in the classical program, create this state? Once the pairing process is underway, telomere-led motion could help to move chromosomes out of the way of one another to permit full-length pairing. Another attractive possibility is that these motions promote resolution of entanglements (Zickler 2006; Koszul et al. 2008; Storlazzi et al. 2010; Kleckner et al. 2012; Klutstein and Cooper 2014). Whole-chromosome entanglements, known as “interlocks,” occur during the leptotene pairing process and during synapsis but are absent (and thus must be actively resolved) by the end of pachytene (reviewed in von Wettstein et al. 1984; Zickler and Kleckner 1999; Storlazzi et al. 2010). In interlock configurations, either one chromosome or a pair of chromosomes is entrapped between two aligned homologs held in place by SC formation to either side (Fig. 4B,C). One model proposes resolution by DNA topoisomerase II-mediated passage of the trapped chromosome through the encircling one (von Wettstein et al. 1984). Another model proposes telomere-led motion promoting the movement of an interlocked chromosome out the ends of the entrapping pair of homologs (Kleckner and Weiner 1993; Storlazzi et al. 2010).
Topological resolution of chromosome interlockings will also require resolution of constraining DNA connections resulting from recombination intermediates already formed in the synapsed regions flanking the entrapped chromosome or bivalent. Recombination protein Mlh1, which has been found to be required for interlocking resolution in Sordaria, could play such a role (Storlazzi et al. 2010). The recombination complex might sense the presence of constraining DNA connections and trigger their Mlh1-mediated dissolution. This role for Mlh1 might be related to its general ability to trigger the disassembly of recombination complexes whose interacting duplexes contain DNA mismatches (e.g., Hunter 2007). Active chromosome movement should similarly facilitate elimination of unwanted DNA links of other types, for example, ectopic recombinational interactions between homologous regions located on different chromosomes (Goldman and Lichten 2000; reviewed in Ding et al. 2004; Davis and Smith 2006; Conrad et al. 2008; Koszul and Kleckner 2009; further discussion in Klutstein and Cooper 2014).
This view of the canonical program is supported by findings in other programs. In C. elegans, PC-directed movement not only helps to bring homologous chromosomes into contact, but acts as a stringency factor to eliminate unwanted contacts, for example, between PCs on nonhomologous chromosomes that share common ZnF proteins (reviewed in Liu and Colaiácovo 2013; Rog and Dernburg 2013). Similarly, in S. pombe, movement disfavors ectopic interactions as well as promoting homologous interactions (Ding et al. 2004; Davis and Smith 2006).
THE SYNAPTONEMAL COMPLEX
Structure
The structure of the SC is as evolutionarily conserved as meiosis itself. Correspondingly, its components share a common underlying structural organization but the level of their primary amino acid sequence homology is very low, reflecting the fact that these molecules play primarily structural roles, rather than catalytic roles (Page and Hawley 2004; Mercier and Grelon 2008; Wojtasz et al. 2009; Yang and Wang 2009; Schild-Prüfert et al. 2011; Fraune et al. 2012). Also, as this structural role depends heavily on contacts with other molecules, within a given organism, evolution of these molecules must be constrained by their need to interact with one another, rather than by the need to conserve specific catalytic elements of the molecules individually. For example, dimers of the large coiled-coil protein Zip1/SCP1/SYCP1/C(3)G/SYP-1/ZEP1/ZIP1 form the transverse filaments of the SC central region in all organisms (e.g., Higgins et al. 2005; Hawley 2011; Miao et al. 2013). They were previously thought to be the sole SC central element components, but they are in fact associated with additional proteins different in number, localization, and perhaps function from one organism to the other: SYCE1, SYCE2, SYCE3, and Tex12 in mammals; Cona in Drosophila, SYP-2, SYP-3, SYP-4 in worm; and Ecm11, Gmc2 in budding yeast (reviewed in Bolcun-Filas et al. 2007; Hawley 2011; Schild-Prüfert et al. 2011; Davies et al. 2012; Fraune et al. 2012; Gómez et al. 2013; Humphryes et al. 2013). However, despite the identification of several new AE and central region components in recent years, we are only beginning to understand how they interact precisely to form the SC (Davies et al. 2012), how they are regulated, and, more importantly, what is/are the function(s) of the SC.
Several lines of evidence suggest that the small ubiquitin-like modifier (SUMO) is involved in the regulation of SC formation. First, sumoylation of budding yeast E2, Ubc9 enzyme is required for SC formation (Klug et al. 2013; reviewed in Watts and Hoffmann 2011). Mammalian UBC9, the SUMO E2-conjugating enzyme, associates with the SC and interacts with several SC proteins (Kovalenko et al. 1996; Tarsounas et al. 1997). Also, SC central-component Zip1 colocalizes with SUMO both along the SC and in polycomplexes (Cheng et al. 2006; Hooker and Roeder 2006; Voelkel-Meiman et al. 2013). Second, SUMO-1, one of the four members of SUMO family, colocalizes with the SC only at synapsed regions; moreover, both lateral and central components are SUMOylated in human cells (Brown et al. 2008). Third, Ecm11, a member of the SC central region of budding yeast, is SUMOylated, and this modification is important for the assembly of the Zip1 traverse filaments along homologs but is dispensable for their assembly into polycomplexes, suggesting a specific role in promoting chromosome-associated SC polymerization (Zavec et al. 2008; Humphryes et al. 2013). However, the precise role of SUMO for SC formation remains to be defined (see discussion in Watts and Hoffmann 2011).
Interestingly, the complexes that mediate recombination are physically first associated with chromosome axes and, after SC nucleation, with the SC central components (e.g., Moens et al. 2002; Storlazzi et al. 2010). Such association was revealed initially from EM studies identifying CO-correlated “nodules” localized in the SC central region (Carpenter 1975, 1987) and confirmed since by immunolocalization of several recombination proteins (e.g., Moens et al. 2002; Higgins et al. 2004; Anderson and Stack 2005; de Boer et al. 2006; Oliver-Bonet et al. 2007). The SC central components are required for reorganization of the recombination complexes (Rad51, Mer3, and Msh4) from an on-axis position to a between-axis (thus, on the SC central region) position concomitant with SC installation (e.g., Espagne et al. 2011). Thus, whereas in most organisms, DSB-initiated recombinational interactions directly mediate both homology searching and presynaptic homolog coalignment, the SC is required, through its central components, for the maintenance and/or turnover of the recombination proteins required for maturation of the DSBs into crossovers (e.g., Börner et al. 2004; Storlazzi et al. 2010; Espagne et al. 2011; Qiao et al. 2012; Yokoo et al. 2012; Reynolds et al. 2013; De Muyt et al. 2014).
Installation: Nucleation and Limited Extension Followed by Nonhomologous Synapsis
In wild-type meiosis, SC installation is specifically nucleated at particular sites, rather than initiating randomly along the chromosomes. In the canonical program, SC nucleations occur at multiple sites throughout the genome (e.g., Zickler 1977; von Wettstein et al. 1984; Albini and Jones 1987; Zickler and Kleckner 1999; Tankimanova et al. 2004; Henderson and Keeney 2005; Tsubouchi and Roeder 2005). These nucleations occur at sites of DSB-mediated coaligment pairing contacts as seen cytologically (Albini and Jones 1987; Oliver-Bonet et al. 2007) and as inferred from association of SC nucleation sites with recombination ensembles (Zickler et al. 1992; Fung et al. 2004; N Hunter, pers. comm.).
Following coalignment, the totality of recombination-mediated interhomolog interactions ultimately gives rise to a much smaller number of COs/chiasmata, which are spatially patterned in the phenomenon of “CO interference” (see further discussion below). In budding yeast, Sordaria, and human, CO interference is imposed during the leptotene/zygotene transition as shown by appearance of CO nodules or CO-correlated protein foci at that stage (Bojko 1985; Zickler et al. 1992; Fung et al. 2004; Zhang et al. 2014b). In budding yeast, SC nucleation occurs specifically at sites of CO-fated interactions, apparently with a 1:1 relationship between the two (Fung et al. 2004). In Sordaria, SC nucleations occur at CO-fated sites and a subset of other recombination interaction precursor sites. Observed patterns point to a single interference-mediated process that gives evenly spaced SC nucleations within, which are embedded the CO-fated sites that show classical CO interference (Zhang et al. 2014b). In other organisms, precise timing of CO interference is not established, but the number of SC nucleations is always greater than the number of COs, dramatically so in higher plants with very long chromosomes and very few COs. However, CO recombination complexes presumably occur in a special structural context, as compared with complexes that mature to other fates. Thus, it is attractive to believe that, in many organisms, as in yeast and Sordaria, SC is preferentially nucleated at CO-designated sites.
SC formation is also specifically nucleated in organisms in which SC forms before and independent of DSBs. In C. elegans, SC nucleates preferentially at the PCs located near chromosome ends (Rog and Dernburg 2013). In Drosophila, SC first appears at centromeres and then initiates internally (Takeo et al. 2011; Tanneti et al. 2011).
Interestingly, in hypomorphic spo11 and ski8 mutants of Sordaria and budding yeast, which show reduced numbers of DSBs, only partial SC formation is observed (Tessé et al. 2003; Henderson and Keeney 2004; Rockmill et al. 2013). Thus, nucleation of SC formation at a single or few sites is not sufficient to permit spreading of SC all along the length of a chromosome. What is preventing the SC from polymerizing indefinitely once it has been nucleated? A link to spreading “interference” signals has been suggested (Börner et al. 2004; Zhang et al. 2014b). Interestingly, also, a minimum DSB number is required for normal timing of SC initiation and for efficient synapsis of the smaller chromosomes in mouse (Kauppi et al. 2013) and yeast (Lee et al. 2012).
SC normally forms specifically between coaligned homologs. However, when a chromosome or chromosome region lacks a homolog partner, SC can form between nonhomologous chromosomes, or between adjacent regions along a single chromosome in a “hairpin” structure (e.g., in haploid meiosis) (reviewed in Zickler and Kleckner 1999; Gong et al. 2011). Nonhomologous synapsis usually occurs late in the pachytene stage (reviewed in von Wettstein et al. 1984; Zickler and Kleckner 1999), highlighting the fact homologous synapsis is specifically dependent on nucleation.
In certain unusual situations, SCs can form between two “single” AEs, provided either by sisters (e.g., in the cohesin rec8 mutant) or by unreplicated homologs (Pukkila et al. 1995; Xu et al. 2005). Specific features must preclude such events in normal meiosis. SC can also assemble along chromosomes in the absence of key axis components (Pelttari et al. 2001). Moreover, SC components have a tendency to self-assemble outside of chromosomes leading to the formation of aggregates called polycomplexes, particularly in situations in which normal synapsis is perturbed (Sym and Roeder 1995; Zickler and Kleckner 1999; Ollinger et al. 2005), reflecting a balance between normal installation and aggregation.
Roles: Recombination and Beyond
The SC likely has both global roles for chromosomes and local roles at sites of recombination (see also, e.g., de Boer and Heyting 2006; Yang and Wang 2009; Fraune et al. 2012; Lake and Hawley 2012; Qiao et al. 2012; Liu and Colaiácovo 2013).
The most obvious global role of the SC is to provide order within the nucleus during late prophase. At early stages, homologs are linked along their lengths by the large number of coalignment linkages that represent total DSB-mediated recombinational interactions. However, at ensuing stages, after CO/NCO differentiation, NCO-fated DSBs lose their link to the homolog partner at the DNA level (Hunter 2007) and also are likely released from the SC (see discussion in Terasawa et al. 2007). After this point, because COs are very few, the SC is essential for holding homologs together during CO maturation and until onset of diplotene (Zickler and Kleckner 1999; Qiao et al. 2012).
Diverse local SC roles have been identified or proposed:
The SC may stabilize chromosome structure around sites of COs. CO-designation can lead to remodeling of chromosome axes (e.g., Martinez-Perez et al. 2008; Storlazzi et al. 2008). In addition, crossing-over at the DNA level requires some accompanying structural axis modification (“axis exchange”) (Blat et al. 2002; Storlazzi et al. 2008). Thus, a role of the SC could be to stabilize chromosome structure against this local turbulence. Indeed, in mouse, retention of SC coincides with axis exchange, consistent with a role for the SC in guiding that process while diplotene chiasmata are forming (Qiao et al. 2012). SC may also prevent aberrant axis remodeling as shown by occurrence of telomeric fusions in mouse sycp1 mutants (Qiao et al. 2012).
SC plays direct positive roles in recombination. Mutations in SC components universally confer recombination defects, particularly for CO recombination. In yeast, SC component Zip1 is required specifically for progression of CO-fated events just after the point of CO/NCO differentiation, concomitant with SC nucleation, thus reflecting local roles of SC components (e.g., Börner et al. 2004; Shinohara et al. 2008). Subsequently, fully formed SC may help constrain the resolution of double Holliday junctions, specifically into CO products versus NCO products (De Muyt et al. 2012; Zakharyevich et al. 2012), among other possible roles.
COs show interesting spatial patterning along chromosomes (CO interference; see below). Neither the SC nor SC components nor continuous SC is required for this process in budding yeast (Fung et al. 2004; Shinohara et al. 2008; Zhang et al. 2014c). In mouse, and also in Drosophila, in which interference is imposed after SC formation, continuous SC is not required (Page and Hawley 2001; de Boer et al. 2007). However, in C. elegans, in which interference also occurs after SC formation, partial depletion of an SC central region component confers defects in the CO-designation/interference process (Libuda et al. 2013) and complete SC seems to be required (reviewed in Liu and Colaiácovo 2013). Presumably, CO interference requires axial chromosome structure, either homolog axes alone or axes linked by SC, according to the details of the particular chromosomal program involved.
The SC might be used for monitoring of interhomolog interactions, with its formation signaling that homologs are properly interacting along their lengths. This could be part of the mechanism used to sense the presence of aberrantly entangled chromosomes (Storlazzi et al. 2010). Analysis of mutants in the mice HORMAD genes (Wojtasz et al. 2009; Daniel et al. 2011) and of Arabidopsis mutants lacking SC central region (Higgins et al. 2005) suggests that SC could be centrally important in regulatory surveillance of recombination.
Recent studies also suggest that SC formation or some related/preceding event is important for shutting off recombination initiation in cis. In mouse spo11 hypomorphic mutants, chromosome axes that fail to synapse show continued DSB formation. The investigators suggest that these extra DSBs, specifically targeted to asynapsed regions, reflect a “feedback process” that helps ensure that the smallest chromosomes find their homologs (Kauppi et al. 2013). Inhibitory feedback between homolog engagement and DSB formation also occurs in budding yeast (Lee et al. 2012; Lao et al. 2013; Thacker et al. 2014).
The SC sometimes plays a direct role in meiosis I homolog segregation per se. In some organisms that do not show recombination and thus do not form chiasmata, but do nevertheless build SCs, modified SC structures remain between bivalents, providing the connection that ensures homolog segregation (e.g., female silkworm) (Rasmussen 1977; for other examples, see Zickler and Kleckner 1999). Similarly, in yeast, SC at centromeres promotes segregation of occasional achiasmate chromosomes (Newnham et al. 2010; reviewed in Obeso et al. 2014). In budding yeast and mouse, as a regular feature of meiosis, transient retention of SCs at centromeres after pachytene is proposed to promote biorientation of sister kinetochores (Bisig et al. 2012; Qiao et al. 2012; Obeso et al. 2014).
SC components can be important for centromere couplings and/or clusterings early in, or even before, meiosis, for example, in yeast, Drosophila, and maize (Tsubouchi and Roeder 2005; Takeo et al. 2011; Tanneti et al. 2011; Lake and Hawley 2012; Zhang et al. 2013a,b; reviewed in Obeso et al. 2014).
CO Frequency and SC Length Are Correlated Independently of Genome Size
In a number of organisms, the same (genetic) chromosome can, in different situations, show higher versus lower CO frequencies, which are correlated with longer versus shorter axis/SC lengths. These relationships are seen in female versus male in mouse and human (Bojko 1985; Lynn et al. 2002; Tease and Hultén 2004; Hou et al. 2013) and similarly in Arabidopsis (Drouaud et al. 2007; Giraut et al. 2011). Such effects are also seen in budding yeast, between different strain backgrounds and in a condensin mutant (Zhang et al. 2014a,c). These relationships have sometimes been attributed to variations in CO interference. However, this is not correct. The metric of CO interference is physical distance along the chromosome (see below). By this criterion, the strength of interference is the same along a particular chromosome regardless of total SC length. Instead, these cases can all be explained as reflecting only one variable; the way chromosome axes and chromatin loops arise during leptotene (Kleckner et al. 2003) as follows.
As discussed above, meiotic chromosomes are organized in loops anchored along a proteinaceous axis, with variations in axis length correlating with variations in chromatin loop size. Because pre-DSB recombination complexes form in chromatin loop sequences, but in association with chromosome axes, longer axes with shorter loops would imply more axis-associated complexes and thus more DSBs, but with the same probability of DSBs per axis length in all cases. Further, the metric for CO interference is physical distance along the chromosome (Drouaud et al. 2007; Petkov et al. 2007; Hou et al. 2013; Zhang et al. 2014a). If the same genome complement is organized into longer versus shorter chromosomes with a constant density of interaxis interactions per unit length, and CO interference is then imposed, the result will be a higher versus lower frequency of COs with no alteration in interference distances (Fig. 2H) (Kleckner et al. 2003).
In accord with this suggestion: (1) analysis of mice hypomorphic spo11 mutants suggests that DSBs are distributed among chromosomes proportionately to axis length (Kauppi et al. 2013); (2) evidence of a direct, mechanistic relationship between axis length per se and DSB number and distribution is provided by analysis of mutants of C. elegans condensin I complex; its absence leads to increased axis length, which is correlated with both increased DSB numbers and altered localization (Mets and Meyer 2009); (3) links between loop sizes and DSB formation are evidenced in the mice PAR regions of the X and Y sex chromosomes; per DNA kilobase, DSBs occur at a 10- to 20-fold higher rate than on autosomes, chromatin loops are fivefold shorter in the PAR, and the axis length relative to DNA content is 10-fold longer in the PAR relative to the autosomes (Kauppi et al. 2011); and (4) most strikingly, male and female human chromosomes show all of the predicted differences: longer loops, shorter axes, fewer DSBs (Rad51 foci) and fewer COs in male versus shorter loops, longer axes, more Rad51 foci and more COs in female (Gruhn et al. 2013).
SPATIAL PATTERNING DURING RECOMBINATION
One of the most interesting features of meiotic recombination is the fact that both DSBs and COs tend to be evenly spaced along the chromosomes by mechanisms that remain to be discovered.
CO Interference
COs occur stochastically, at different positions along the chromosomes in different meiotic nuclei. Nonetheless, along any given chromosome in any given nucleus, COs tend to be evenly spaced. This feature was discovered during the genetic elucidation of recombination in Drosophila as the classical phenomenon of “CO interference” (Sturtevant 1915; Muller 1916; reviewed in Berchowitz and Copenhaver 2010). If a CO has occurred at one position along a chromosome, there is a reduced probability that another CO will occur nearby; moreover, the strength of this interference decreases with increasing distance between the two positions. This patterning process is interesting because it implies the existence of communication along the chromosomes, the basis for which is unknown.
Models for CO Interference
Models for CO patterning can be considered either on the basis of their mechanism or their underlying logic. In all models, a CO-designation process operates on an array of “precursor” interactions that correspond to DSB-mediated interhomolog interactions (e.g., bridges, see above). In the first detailed model by King and Mortimer (1990), a CO designation at a given precursor site triggered a polymerization signal that then spread outward until it ran into another polymerization signal triggered by a CO designation occurring elsewhere. In this model, the final outcome is determined by the kinetics of CO designation and signal spreading. A second model by Stahl and colleagues (Lande and Stahl 1993), proposed that a mechanism began at one end of a chromosome and “counted” precursor interactions, with CO-designation occurring after a specific (nearly) fixed number of precursors. Arguing against this model is the finding that the length of CO interference does not change if the density of precursors is decreased (Martini et al. 2006). A third model emerged from the idea that communication for CO interference might occur via redistribution of mechanical stress (the “beam-film model”) (Kleckner et al. 2004; Zhang et al. 2014a). However, this model also implies a general logic for the process. In essence, CO-designations occur sequentially along a chromosome, with each designation event triggering an interference signal that decreases exponentially with distance away from the nucleating site. The latter feature contrasts with the logic of the King and Mortimer model or the counting model. In a mechanical mechanism, all precursors come under mechanical stress; CO-designation is promoted when that stress reaches a critical level and, consequently, results in local relief of stress; finally, interference results from redistribution of that stress relief outward from the nucleation site. Importantly, however, the basic logic of the beam-film model, and a specific mathematical formulation developed in a mechanical context, can apply to any type of mechanism for communication including a dissipating molecular signal without any role for a mechanical effect or a reaction, diffusion mechanism (e.g., Vecchiarelli et al. 2014). Application of beam-film analysis suggests that this logic can very accurately explain CO patterning in a wide variety of organisms and mutant situations (Zhang et al. 2014a,c). Another way of modeling CO patterning is to ask whether observed patterns are explained by any particular mathematical distribution pattern. Considerable attention has been given to modeling by the γ distribution, (often used to analyze CO patterns) of which the “counting model” is a special case (e.g., McPeek and Speed 1995; Falque et al. 2007). This approach only describes the final outcome of the process, without regard to any other features. Finally, the possibility has been raised that the final CO interference pattern might arise in two stages (de Boer et al. 2006; Yokoo et al. 2012). Such a model appears unlikely in Sordaria and some other organisms (De Muyt et al. 2014; Zhang et al. 2014b) but remains to be further assessed (further discussion below).
The Obligatory CO and the Precursor Distribution
To a first approximation, regular segregation of a pair of homologs requires at least one CO (chiasma). This biological requirement is manifested in the fact that, in most systems, the frequency of zero-CO chromosomes is extremely low. Indeed, in a number of cases, a particular pair of homologs (or, in C. elegans, all pairs of homologs), always acquires one and only one CO. Given the biological imperative, this is sometimes said to imply the existence of a first “obligatory CO” (Jones and Franklin 2006). In the King and Mortimer model, precursors were assumed to be distributed randomly along and among chromosomes. As a result, some chromosomes would fail to acquire even one precursor and thus would also fail to acquire even one CO. To address this possibility, the model envisioned that the interference signal triggered release and recycling of precursors to regions in which interference had not yet had an effect, thus ensuring that eventually all chromosomes would get at least one CO. The γ model (and the counting model) also both assume a random distribution of precursors. However, experimental evidence suggests that this assumption is wrong. This is particularly obvious in Sordaria, in which DSB-mediated interactions are very evenly spaced along the chromosomes, likely also with similar numbers per chromosome among all nuclei (Storlazzi et al. 2010; D Zickler and N Kleckner, unpubl.). These same tendencies are likely present in other organisms (e.g., Zhang et al. 2014a). Furthermore, the implication of the King and Mortimer model is that CO interference would be required for ensuring a low level of zero-CO chromosomes (the obligatory CO). In contrast, in the logic of the beam-film model, interference is irrelevant to the obligatory CO, which is ensured instead by an appropriate constellation of effects including precursor distribution and the “strength” of the CO-designation process (Zhang et al. 2014a). In accord with such a model, in budding yeast, a significant reduction in interference can occur without any discernible effect on the frequency of zero-CO chromosomes (Zhang et al. 2014c).
CO Homeostasis
When the density of precursors (i.e., DSBs) is decreased, there is a less than proportionate decrease in the number of COs (Martini et al. 2006). This “homeostatic” effect results from the fact that, for a given precursor, its probability of being subjected to interference (and thus its probability of giving a CO) will decrease if there are fewer precursors (Zhang et al. 2014a,c). Analysis of several mutants showed that TopoII is required for both interference and homeostasis, suggesting a common underlying process, supporting the direct relationship originally proposed (Martini et al. 2006). The beam-film model can quantitatively explain the link between CO and homeostasis in budding yeast and Drosophila (Zhang et al. 2014a,c).
Mechanism of CO Interference
There is relatively little information about the mechanism of CO patterning. However, it is clear that chromosome continuity is required for transmission of interference in C. elegans (Hillers and Villeneuve 2003) and that the “metric” for interference is physical distance along the chromosomes (micrometers) rather than “genomic distance” (Mb) (see above). It is also clear that the structural axes of the chromosomes are required, in accord with the fact that all aspects of recombination occur in association with these axes. In most organisms, CO interference does not require any, or continuous SC. However, the SC is required in C. elegans, presumably because, in this organism, it is an integral component of the structural axes at the relevant time (see above). A number of mutants defective in CO patterning have been described and are, operationally, defective in interference as defined by classical genetic tests (e.g., Crismani et al. 2012). It is often difficult to know whether the mutant defect actually reflects an alteration in the patterning process per se or some more general aberration in the recombination process. One recent study has identified a pathway of events that appears to be directly important for the interference process in budding yeast. This pathway involves the catalytic activity of Topoisomerase II plus Ubc9-mediated SUMOylation and Slx5/8-mediated STUbL (SUMO-targeted ubiquitin ligase) activity (Zhang et al. 2014c).
DSB Patterning
The nonrandom distribution of CO precursors along and among chromosomes (see above) implies the existence of process(es) that act at an early stage to determine the patterning of DSBs and/or DSB-initiated interhomolog interactions. Correspondingly, in yeast, occurrence of a DSB at one position is accompanied by a decreased probability that another DSB will occur nearby (Wu and Lichten 1994). The nature of this communication process, which could potentially be linked to concomitantly occurring chromosome axis development, remains to be determined.
SUMMARY
During meiosis, DNA events of recombination are directly integrated with structural features of chromosomes and complex global whole chromosome behaviors at all stages of the program, with functional interplay in both directions. Interestingly, these processes follow a general principle of progressive stability. During the meiotic interhomolog interaction process, initial interactions are unstable, thus permitting rapid reversible sampling subject to correction, but become more molecularly robust as the process proceeds. DSB-independent pairing, DSB-mediated homology searching, nascent DSB/partner interactions and interaxis bridges, predouble Holliday junction intermediates, and double Holliday junctions in the context of the SC occur in succession, each involving more stable interactions than the stages before, until finally a molecularly irreversible CO product arises (Weiner and Kleckner 1994; Boateng et al. 2013).
These fascinating events, which underlie the fundamental process of sexual reproduction, provide fertile ground for future explorations.
ACKNOWLEDGMENTS
We apologize to the many colleagues whose research is cited only in reviews because of the need to cover a range of topics in a relatively short space. The authors’ research is supported by grants to N.K. (National Institutes of Health Grant GM-044794) and D.Z. (Centre National de la Recherche Scientifique; Unité Mixte de Recherche 8621). We thank Jim Henle, Liangran Zhang, and Beth Weiner for help with manuscript preparation.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
- Albini SM, Jones GH. 1987. Synaptonemal complex spreading in Allium cepa and A. fistulosum. I: The initiation and sequence of pairing. Chromosoma 95: 324–338. [Google Scholar]
- Anderson LK, Stack SM. 2005. Recombination nodules in plants. Cytogenet Genome Res 109: 198–204. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Jones GH. 2003. Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana. J Exp Bot 54: 1–10. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Franklin FC, Jones GH. 2001. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci 114: 4207–4217. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wyler T, Loidl J, Kohli J. 1993. Unusual nuclear structures in meiotic prophase of fission yeast: A cytological analysis. J Cell Biol 121: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat TS, Loos F, van Staveren S, Myronova E, Ghazvini M, Grootegoed JA, Gribnau J. 2014. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell 53: 965–978. [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. 2013. Meiotic recombination in mammals: Localization and regulation. Nat Genet 14: 794–806. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP. 2010. Genetic interference: Don’t stand so close to me. Curr Genomics 11: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisig CG, Guiraldelli MF, Kouznetsova A, Scherthan H, Höög C, Dawson DS, Pezza RJ. 2012. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet 8: e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. 2013. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell 24: 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojko M. 1985. Human meiosis IX: Crossing over and chiasma formation in oocytes. Carlsberg Res Commun 50: 43–72. [Google Scholar]
- Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R, De Rooij DG, Cooke HJ. 2007. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol 176: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Brown PW, Hwang K, Schlegel PN, Morris PL. 2008. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum Reprod 23: 2850–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Kleckner N. 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev 13: 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Kleckner N, Weiner BM. 1999. Somatic pairing of homologs in budding yeast: Existence and modulation. Genes Dev 13: 1627–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon CK, Hawley RS. 2013. Flies get a head start on meiosis. PLoS Genet 9: e1004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender TL, Hollingsworth N. 2010. Mek1 suppression of meiotic double-strand break repair is specific to sister chromatids, chromosome autonomous and independent of Rec8 cohesin complexes. Genetics 185: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG, Cha RS. 2008. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132: 758–770. [DOI] [PubMed] [Google Scholar]
- Carpenter ATC. 1975. Electron microscopy of meiosis in Drosophila melanogaster females. II: The recombination nodule—A recombination-associated structure at pachytene? Proc Natl Acad Sci 72: 3186–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AT. 1987. Gene conversion, recombination nodules, and the initiation of meiotic synapsis. Bioessays 6: 232–236. [DOI] [PubMed] [Google Scholar]
- Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. 2000. Progression of meiotic DNA replication is regulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev 14: 493–503. [PMC free article] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev 20: 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69. [DOI] [PubMed] [Google Scholar]
- Christophorou N, Rubin T, Huynh J-R. 2013. Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet 9: e1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133: 1175–1187. [DOI] [PubMed] [Google Scholar]
- Couteau F, Zetka M. 2005. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev 19: 2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. 2012. FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M, et al. 2011. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nature Cell Biol 13: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilowicz C, Lee CH, Kim K, Hatch K, Coljee VW, Kleckner N, Prentiss M. 2009. Single molecule detection of direct, homologous, DNA/DNA pairing. Proc Natl Acad Sci 106: 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies OR, Maman JD, Pellegrini L. 2012. Structural analysis of the human SYCE2-TEX12 complex provides molecular insights into synaptonemal complex assembly. Open Biol 2: 120099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Smith GR. 2006. The meiotic bouquet promotes homolog interactions and restricts ectopic recombination in Schizosaccharomyces pombe. Genetics 174: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Heyting C. 2006. The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115: 220–234. [DOI] [PubMed] [Google Scholar]
- de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. 2006. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci 103: 9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Dietrich AJ, Höög C, Stam P, Heyting C. 2007. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J Cell Sci 120: 731–736. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- De Massy B. 2013. Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu Rev Genet 47: 563–599. [DOI] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D. 2014. E3 ligase Hei10: A multi-faceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev 28: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JF, Hawley RS. 1996. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86: 135–146. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Yamamoto A, Haraguchi, Hiraoka Y. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell 6: 329–341. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y. 2012. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science 336: 732–736. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Haraguchi T, Hiraoka Y. 2013. The role of chromosomal retention of noncoding RNA in meiosis. Chromosome Res 21: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M, Martin O, Zanni V, Brunel D, Mézard C. 2007. Sex-specific crossover distributions and variations at the interference level along Arabidopsis thaliana chromosome 4. PLoS Genet 3: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel-Mitani M, Olson LW, Egel R. 1982. Meiosis in Aspergillus nidulans: Another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas 97: 179–187. [DOI] [PubMed] [Google Scholar]
- Espagne E, Vasnier C, Storlazzi A, Kleckner NE, Silar P, Zickler D, Malagnac F. 2011. Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc Natl Acad Sci 108: 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falque M, Mercier R, Mézard C, de Vienne D, Martin OC. 2007. Patterns of recombination and MLH1 foci density along mouse chromosomes: Modeling effects of interference and obligate chiasma. Genetics 176: 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune J, Schramm S, Alsheimer M, Benavente R. 2012. The mammalian synaptonemal complex: Protein components assembly and role in meiotic recombination. Exp Cell Res 318: 1340–1346. [DOI] [PubMed] [Google Scholar]
- Fung JC, Rockmill B, Odell M, Roeder GS. 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C. 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7: e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev E, Kleckner N. 2014. Direct recognition of homology between double helices of DNA in Neurospora crassa. Nat Commun 5: 3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AS, Lichten M. 2000. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci 97: 9537–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez R, Jordan PW, Viera A, Alsheimer M, Fukuda T, Jessberger R, Liano E, Pendás AM, Handel MA, Suja JA. 2013. Dynamic localization of SMC5/6 complex proteins during mammalian meiosis and mitosis suggests functions in distinct chromosome processes. J Cell Sci 126: 4239–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Liu X, Tang D, Yu H, Yi C, Cheng Z, Gu M. 2011. Non-homologous chromosome pairing and crossover formation in haploid rice meiosis. Chromosoma 120: 47–60. [DOI] [PubMed] [Google Scholar]
- Gruhn JR, Rubio C, Broman KW, Hunt PA, Hassold T. 2013. Cytological studies of human meiosis: Sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS ONE 8: e85075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RS. 2011. Solving a meiotic LEGO puzzle: Transverse filaments and the assembly of the synaptonemal complex in Caenorhabditis elegans. Genetics 189: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Keeney S. 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci 101: 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Keeney S. 2005. Synaptonemal complex formation: Where does it start? Bioessays 27: 995–998. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FC, Jones GH. 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev 18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. 2005. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers KJ, Villeneuve AM. 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol 13: 1641–1647. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. 2009. The SUN rises on meiotic chromosome dynamics. Dev Cell 17: 598–605. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Byers B. 1989. HOP1: A yeast meiotic pairing gene. Genetics 121: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Sung Y, Yu M, Lee M, Kleckner N, Kim KP. 2013. The logic and mechanism of homologous recombination partner choice. Mol Cell B51: 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker GW, Roeder GS. 2006. A role for SUMO in meiotic chromosome synapsis. Curr Biol 16: 1238–1243. [DOI] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, et al. 2013. Genome analyses of single human oocytes. Cell 155: 1492–1506. [DOI] [PubMed] [Google Scholar]
- Humphryes N, Leung WK, Argunhan B, Terentyev Y, Dvorackova M, Tsubouchi H. 2013. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet 9: e1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2007. Molecular recombination. In Molecular genetics of recombination (ed. Aguilera A, Rothstein R), pp. 381–442. Springer, New York. [Google Scholar]
- Ishiguro K, Kim J, Shibuya H, Hernández-Hernández A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, et al. 2014. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev 28: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GH. 1987. Chiasmata. In Meiosis (ed. Moens PB). Academic, New York. [Google Scholar]
- Jones GH, Franklin FC. 2006. Meiotic crossing-over: Obligation and interference. Cell 126: 246–248. [DOI] [PubMed] [Google Scholar]
- Joyce EF, Apotolopoulos N, Beliveau BJ, Wu C-T. 2013. Germline progenitors escape the widespread phenomenon of homolog pairing during Drosophila development. PLoS Genet 9: e1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M. 2011. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Kleckner N. 1996. Communication between homologous chromosomes: Genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells 1: 475–489. [DOI] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. 2010. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143: 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JS, Mortimer RK. 1990. A polymerization model of chiasma interference and corresponding computer simulation. Genetics 126: 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. 2006. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Weiner BM. 1993. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp Quant Biol 58: 553–565. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Storlazzi A, Zickler D. 2003. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet 19: 623–628. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, Hutchinson J. 2004. A mechanical basis for chromosome function. Proc Natl Acad Sci 101: 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Zhang L, Weiner B, Zickler D. 2012. Meiotic chromosome dynamics. In Genome organization and function in the mammalian cell nucleus (ed. Rippe K). Wiley-VCH, Weinheim, Germany. [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM. 1992. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol 117: 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug H, Xaver M, Chaugule VK, Koidl S, Mittler G, Klein F, Pichler A. 2013. Ubc9 sumoylation controls SUMO chain formation and meiotic synapsis in Saccharomyces cerevisiae. Mol Cell 50: 625–636. [DOI] [PubMed] [Google Scholar]
- Klutstein M, Cooper JP. 2014. The chromosomal courtship dance—Homolog pairing in early meiosis. Curr Opin Cell Bio 26: 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kleckner N. 2009. Dynamic chromosome movements during meiosis: A way to eliminate unwanted connections? Trends Cell Biol 19: 716–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133: 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko OV, Plug AW, Haaf T, Gonda DK, Ashley T, Ward DC, Radding CM, Golub EI. 1996. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc Natl Acad Sci 93: 2958–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake CM, Hawley RS. 2012. The molecular control of meiotic chromosomal behavior: Events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol 74: 425–451. [DOI] [PubMed] [Google Scholar]
- Lande R, Stahl FW. 1993. Chiasma interference and the distribution of exchanges in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol 58: 543–552. [DOI] [PubMed] [Google Scholar]
- Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, Lee CY, Dresser ME, Hunter N, Bishop DK. 2013. Meiotic crossover control by concerted action of rad51-dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet 9: e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Conrad MN, Dresser ME. 2012. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet 8: e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, de Massy B. 2011. The impressionistic landscape of meiotic recombination. Cell 147: 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Scherthan H, Den Dunnen JT, Klein F. 1995. Morphology of a human-derived YAC in yeast meiosis. Chromosoma 104: 183–188. [DOI] [PubMed] [Google Scholar]
- Lui DY, Colaiácovo MP. 2013. Meiotic development in Caenorhabditis elegans. Adv Exp Med Biol 757: 133–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ. 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296: 2222–2225. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Hochwagen A. 2011. Checkpoint mechanisms: The puppet masters of meiotic prophase. Trends Cell Biol 21: 393–400. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM. 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev 19: 2727–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Schvarzstein M, Barroso C, Lightfoot J, Dernburg AF, Villeneuve AM. 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev 22: 2886–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee BD. 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta 1677: 165–180. [DOI] [PubMed] [Google Scholar]
- McKee BD. 2009. Homolog pairing and segregation in Drosophila meiosis. Genome Dyn 5: 56–68. [DOI] [PubMed] [Google Scholar]
- McKee BD, Yan R, Tsai J-H. 2012. Meiosis in male Drosophila. Spermatogenesis 2: 167–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Speed TP. 1995. Modeling interference in genetic recombination. Genetics 139: 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, McKim KS. 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet 2: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Grelon M. 2008. Meiosis in plants: Ten years of gene discovery. Cytogenet Genome Res 120: 281–290. [DOI] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ. 2009. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z. 2013. Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25: 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miné-Hattab J, Rothstein R. 2013. DNA in motion during double-strand break repair. Trends Cell Biol 23: 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito M, Ohta K. 2013. Spatiotemporal regulation of meiotic recombination by Liaisonin. Bioarchitecture 3: 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Earnshaw WC. 1989. Anti-topoisomerase II recognizes meiotic chromosome cores. Chromosoma 98: 317–322. [DOI] [PubMed] [Google Scholar]
- Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA–DNA interactions without reciprocal recombination. J Cell Sci 115: 1611–1622. [DOI] [PubMed] [Google Scholar]
- Molnar M, Kleckner N. 2008. Examination of interchromosomal interactions in vegetatively growing diploid Schizosaccharomyces pombe cells by Cre/loxP site-specific recombination. Genetics 178: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Doll E, Yamamoto A, Hiraoka Y, Kohli J. 2003. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J Cell Sci 116: 1717–1731. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1916. The mechanism of crossing over. Am Nat 50: 193–434. [Google Scholar]
- Newnham L, Jordan P, Rockmill B, Roeder GS, Hoffmann E. 2010. The synaptonemal complex protein, Zip1, promotes the segregation of nonexchange chromosomes at meiosis I. Proc Natl Acad Sci 107: 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N. 2006. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119: 217–225. [DOI] [PubMed] [Google Scholar]
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Höög C. 2008. Cohesin Smc1β determines meiotic chromatin axis loop organization. J Cell Biol 180: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeso D, Pezza RJ, Dawson D. 2014. Couples, pairs, and clusters: Mechanisms and implications of centromere associations in meiosis. Chromosoma 123: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Bonet M, Campillo M, Turek PJ, Ko E, Martin RH. 2007. Analysis of replication protein A (RPA) in human spermatogenesis. Mol Hum Reprod 13: 837–844. [DOI] [PubMed] [Google Scholar]
- Ollinger R, Alsheimer M, Benavente R. 2005. Mammalian protein SCP1 forms synaptonemal complex-like structures in the absence of meiotic chromosomes. Mol Cell Biol 16: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1238–1256. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. 2004. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Biol 20: 525–528. [DOI] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. [DOI] [PubMed] [Google Scholar]
- Parvinen M, Soderstrom K-O. 1976. Chromosome rotation and formation of synapsis. Nature 260: 534–535. [DOI] [PubMed] [Google Scholar]
- Pelttari J, Hoja MR, Yuan L, Liu JG, Brundell E, Moens P, Santucci-Darmanin S, Jessberger R, Barbero JL, Heyting C, et al. 2001. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol Cell Biol 21: 5667–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples TL, Dean E, Gonzalez O, Lambourne L, Burgess SM. 2002. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev 16: 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Broman KW, Szatkiewicz JP, Paigen K. 2007. Crossover interference underlies sex differences in recombination rates. Trends Genet 23: 539–542. [DOI] [PubMed] [Google Scholar]
- Phillips CM, Meng X, Zhang L, Chretien JH, Umov FD, Dernburg AF. 2009. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C.elegans. Nat Cell Biol 11: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila PJ, Shannon KB, Skrzynia C. 1995. Independent synaptic behavior of sister chromatids in Coprinus cinereus. Can J Bot 73: S215–S220. [Google Scholar]
- Qiao H, Chen JK, Reynolds A, Höög C, Paddy M, Hunter N. 2012. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet 8: e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SW. 1977. The transformation of the synaptonemal complex into the “elimination chromatin” in Bombyx mori oocytes. Chromosoma 60: 205–221. [DOI] [PubMed] [Google Scholar]
- Rasmussen SW. 1986. Initiation of synapsis and interlocking of chromosomes during zygotene in Bombyx spermatocytes. Carlsberg Res Commun 51: 401–432. [Google Scholar]
- Revenkova E, Jessberger R. 2006. Shaping meiotic prophase chromosomes: Cohesins and synaptonemal complex proteins. Chromosoma 115: 235–240. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco V, Nicolas A. 1996. Sensing of DNA non-homology lowers the initiation of meiotic recombination in yeast. Genes Cells 1: 645–661. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Lefrançois P, Voelkel-Meiman K, Oke A, Roeder GS, Fung JC. 2013. High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLoS Genet 9: e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Dernburg AF. 2013. Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr Opin Cell Biol 25: 349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, Keeney S. 2010. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet 6: e10010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E, Osman K, Higgins JD, Pradillo M, Cuñado N, Jones GH, Franklin FC. 2008. ASY1 coordinates early events in the plant meiotic recombination pathway. Cytogenet Genome Res 120: 302–312. [DOI] [PubMed] [Google Scholar]
- Scherthan H. 2001. A bouquet makes ends meet. Nat Rev Mol Cell Biol 2: 621–627. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Bähler J, Kohli J. 1994. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol 127: 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. 2007. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci 104: 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Prüfert K, Saito TT, Smolikov S, Gu Y, Hincapie M, Hill DE, Vidal M, McDonald K, Colaiácovo MP. 2011. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics 189: 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmekel K, Daneholt B. 1995. The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol 5: 239–242. [DOI] [PubMed] [Google Scholar]
- Schmekel K, Wahrman J, Skoglund U, Daneholt B. 1993. The central région of the synaptonemal complex in Blaps cribrosa studied by electron microscope tomography. Chromosoma 102: 669–681. [DOI] [PubMed] [Google Scholar]
- Selker EU. 1990. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet 24: 579–613. [DOI] [PubMed] [Google Scholar]
- Serrentino ME, Borde V. 2012. The spatial regulation of meiotic recombination hotspots: Are all DSB hotspots crossover hotspots? Exp Cell Res 318: 1347–1352. [DOI] [PubMed] [Google Scholar]
- Shibuya H, Ishiguro K, Watanabe Y. 2013. The TERF-1 binding protein TERB1 promotes chromosome movement and telomere rigidity. Nat Cell Biol 16: 145–156. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A. 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309. [DOI] [PubMed] [Google Scholar]
- Smolikov S, Schild-Prüfert K, Colaiácovo MP. 2008. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet 4: e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack SM, Anderson LK. 1986. Two dimensional spreads of synaptonemal complexes from solanaceous plants. II: Synapsis in Lycopersicon esculentum (tomato). Am J Bot 73: 264–281. [Google Scholar]
- Stewart MN, Dawson DS. 2008. Changing partners: Moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet 24: 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Tessé S, Gargano S, James F, Kleckner N, Zickler D. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Tesse S, Ruprich-Robert G, Gargano S, Pöggeler S, Kleckner N, Zickler D. 2008. Coupling meiotic chromosome axis integrity to recombination. Genes Dev 22: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. 2010. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141: 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. 1915. The behavior of the chromosomes as studied through linkage. Z Induct Abstammungs Vererbungsl 13: 234–287. [Google Scholar]
- Sym M, Roeder GS. 1995. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J Cell Biol 128: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S, Lake CM, Morais-de-Sa E, Sunkel CE, Hawley RS. 2011. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr Biol 21: 1845–1851. [DOI] [PubMed] [Google Scholar]
- Tankimanova M, Hultén MA, Tease C. 2004. The initiation of homologous chromosome synapsis in mouse fetal oocytes is not directly driven by centromere and telomere clustering in the bouquet. Cytogenet Genome Res 105: 172–181. [DOI] [PubMed] [Google Scholar]
- Tanneti NS, Landy K, Joyce EF, McKim KS. 2011. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol 21: 1852–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Pearlman RE, Gasser PJ, Park MS, Moens PB. 1997. Protein–protein interactions in the synaptonemal complex. Mol Biol Cell 8: 1405–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Morita T, Pearlman RE, Moens PB. 1999. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 147: 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tease C, Hultén MA. 2004. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet Genome Res 107: 208–215. [DOI] [PubMed] [Google Scholar]
- Terasawa M, Ogawa H, Tsukamoto Y, Shinohara M, Shirahige K, Kleckner N, Ogawa T. 2007. Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation. Proc Natl Acad Sci 104: 5965–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci 100: 12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S. 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, McKee BD. 2011. Homologous pairing and the role of pairing centers in meiosis. J Cell Sci 124: 1955–1963. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T, Roeder GS. 2005. A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308: 870–873. [DOI] [PubMed] [Google Scholar]
- Vecchiarelli AG, Neuman KC, Mizuuchi K. 2014. A propagating ATPase gradient drives transport of surface-confined cellular cargo. Proc Natl Acad Sci 111: 4880–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Taylor LF, Mukherjee P, Humphryes N, Tsubouchi H, Macqueen AJ. 2013. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet 9: e1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. 1984. The synaptonemal complex in genetic segregation. Annu Rev Genet 18: 331–413. [DOI] [PubMed] [Google Scholar]
- Watts FZ, Hoffman E. 2011. SUMO meets meiosis: An encounter at the synaptonemal complex: SUMO chains and sumoylated proteins suggest that heterogeneous and complex interactions lie at the centre of the synaptonemal complex. Bioassays 33: 529–537. [DOI] [PubMed] [Google Scholar]
- Weiner BM, Kleckner N. 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77: 977–991. [DOI] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, et al. 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5: e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, Anastassiadis K, Stewart AF, Reményi A, Turner JM, et al. 2012. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev 26: 958–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. 2010. Condensin and cohesin complexity: The expanding repertoire of functions. Nat Rev Genet 11: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TC, Lichten M. 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518. [DOI] [PubMed] [Google Scholar]
- Xu L, Kleckner N. 1995. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J 14: 5115–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. 2005. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 8: 949–961. [DOI] [PubMed] [Google Scholar]
- Yancey-Wrona JE, Camerini-Otero RD. 1995. The search for DNA homology does not limit stable homologous pairing promoted by RecA protein. Curr Biol 5: 1149–1158. [DOI] [PubMed] [Google Scholar]
- Yang F, Wang PJ. 2009. The mammalian synaptonemal complex: A scaffold and beyond. Genome Dyn 5: 69–80. [DOI] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavec AB, Comino A, Lenassi M, Komel R. 2008. Ecm11 protein of yeast Saccharomyces cerevisiae is regulated by sumoylation during meiosis. FEMS Yeast Res 8: 64–70. [DOI] [PubMed] [Google Scholar]
- Zhang L, Kim KP, Kleckner NE, Storlazzi A. 2011. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc Natl Acad Sci 108: 20036–20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Lv Z, Pang J, Liu Y, Guo X, Fu S, Li J, Dong Q, Wu HJ, Gao Z, et al. 2013a. Formation of a functional maize centromere after loss of centromeric sequences and gain of ectopic sequences. Plant Cell 25: 1979–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pawlowski WP, Han F. 2013b. Centromere pairing in early meiotic prophase requires active centromeres and precedes installation of the synaptonemal complex in maize. Plant Cell 25: 3900–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liang Z, Hutchinson J, Kleckner N. 2014a. Crossover patterning by the beam-film model: Analysis and implications. PLoS Genet 10: e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Espagne E, De Muyt A, Zickler D, Kleckner N. 2014b. Meiotic crossover interference is embedded within spatial patterning of synaptonemal complex nucleation: A reactivity threshold model. Proc Natl Acad Sci 111: E5059–E5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. 2014c. Topoisomerase II mediates meiotic crossover interference. Nature 511: 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. 1977. Development of the synaptonemal complex and the “recombination nodules” during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora Auersw. Chromosoma 61: 289–316. [DOI] [PubMed] [Google Scholar]
- Zickler D. 2006. From early homologue recognition to synaptonemal complex formation. Chromosoma 115: 158–174. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 1998. The leptotene–zygotene transition of meiosis. Annu Rev Genet 32: 619–697. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 1999. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]
- Zickler D, Moreau PJ, Huynh AD, Slezec AM. 1992. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics 132: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]