Abstract
Despite remarkable progress in the identification of mutations that drive genetic disorders, progress in understanding the effect of genetic background on the penetrance and expressivity of causal alleles has been modest, in part because of the methodological challenges in identifying genetic modifiers. Nonetheless, the progressive discovery of modifier alleles has improved both our interpretative ability and our analytical tools to dissect such phenomena. In this review, we analyze the genetic properties and behaviors of modifiers as derived from studies in patient populations and model organisms and we highlight conceptual and technological tools used to overcome some of the challenges inherent in modifier mapping and cloning. Finally, we discuss how the identification of these modifiers has facilitated the elucidation of biological pathways and holds the potential to improve the clinical predictive value of primary causal mutations and to develop novel drug targets.
The clinical phenotype of a patient with a known driver mutation can be affected by the presence of protective or disease-promoting alleles. The field of retinal disorders has led efforts to understand such modifier alleles.
Since the mapping of the first locus for autosomal dominant retinitis pigmentosa (RP) on the long arm of chromosome 3 (McWilliam et al. 1989) and the subsequent identification of rhodopsin (RHO) as the causal transcript (Dryja et al. 1990), the genetic dissection of disorders in which retinal degeneration comprises a core phenotypic component has accelerated rapidly, leading to the report of more than 250 loci and genes cataloged in RetNet (as of May 2014; see https://sph.uth.edu/retnet/disease.htm). These advances have had a clear contribution to both our understanding of photoreceptor biology and in providing genetic diagnoses to families. At the same time, however, post hoc knowledge of disease-causing genes and mutations had a modest impact on the predictive ability of prognosis with regard to key aspects of disease expressivity, such as age-of-onset, rate of progression, severity, and manifestation of other comorbidities. Numerous reports exist in the literature in which patients with the same primary mutation show stark differences in phenotypic expressivity, with the more striking examples coming from nonidentical twin studies in which patients that grow under a shared environment and carry the same primary mutation display distinct phenotypes, most likely because of genetic determinants that act as modifiers (Walia et al. 2008). The example above is not an isolated oddity. Accumulating clinical and genetic evidence now suggests that, even for classically monogenic disorders, the view that the expressivity of the phenotype is exclusively the property of dominant or recessive mutations at a single locus might be an oversimplification of the biological phenomena (Dipple and McCabe 2000; Badano and Katsanis 2002; Schäffer 2013).
The concept of modifier genes and alleles is not new (Haldane 1941). Nonetheless, the field of retinal degeneration disorders has been in a unique position to lead the efforts to understand genetic architecture, causality, and variable expressivity. This is in part because of (1) large and well-phenotyped multiethnic cohorts; (2) a substantial (and growing) mutational target of almost 300 genes, each known to be necessary for retinal biogenesis, homeostasis, and/or function; and (3) the availability of in vitro and in vivo tools to model genetic interaction phenomena. Here we will discuss the concepts of modifier genes and alleles, including their evolving terminology; we will highlight examples that illustrate the still episodic, yet significant, advances in dissecting the genetic basis of modification of penetrance and expressivity; and we will discuss possible avenues toward improving our interpretative ability of such events.
PROPERTIES OF GENETIC MODIFIERS
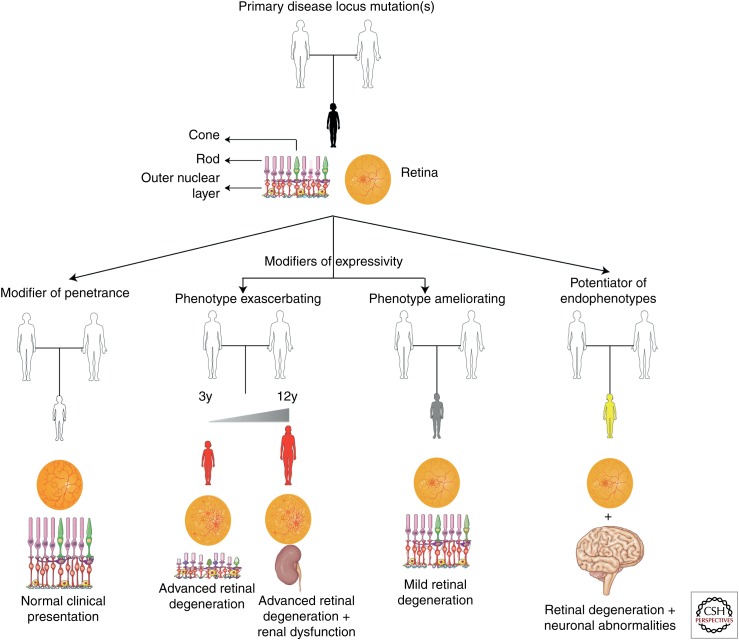
A critical step in understanding genetic modification phenomena consists of providing a clear definition for the term itself. Phenomena described by terms such as epistasis, oligogenic inheritance, or genetic interaction/modification, are all effective synonyms of the same process, namely, the effect of one gene/allele on the phenotypic outcome of a second gene/locus. A distinction that can aid better defining these genetic phenomena consists in establishing whether the primary locus is both necessary and sufficient to cause disease. If yes, then the presence of allele(s) at a second locus/gene exerts purely a modifying role on the severity of the phenotype as defined by the rate of disease progression; the severity of the phenotype with regard to its pleiotropy; or in the manifestation of endophenotypes that necessitate genetic interactions; if not, then the requirement of an allele in a second gene to manifest pathology defines a case of digenic or oligogenic inheritance (Fig. 1). In general, in the context of dissecting modifier phenomena, four major parameters play a prominent role:
Allelic heterogeneity. Phenotypic variability can stem from mutations in different genes that contribute to disease endophenotypes. However, simpler models of genetic heterogeneity and their influence on phenotypic expressivity should first be considered. Locus and allelic heterogeneity are highlighted in Leber congenital amaurosis (LCA), a disorder for which at least 11 causative genes have been reported under a recessive paradigm (reviewed in Davis and Katsanis 2012). In addition to genetic heterogeneity, LCA is also an example of allelic heterogeneity. For example, nonsense and frameshift, as well as missense mutations in CEP290 drive disorders ranging between nonsyndromic LCA to Joubert syndrome (JBTS) to the more severe Meckel Gruber syndrome (MKS) phenotype. These observations have reasonably suggested that modifier genetic factors account for this marked variance in clinical presentation (den Hollander et al. 2006, 2008; Sayer et al. 2006; Valente et al. 2006; Baala et al. 2007; Frank et al. 2007; Perrault et al. 2007). However, a discrete intronic mutation (c.2991+ 1655A>G) in CEP290 that creates a strong splice-donor site and inserts a cryptic exon in the CEP290 mRNA resulting in an aberrantly spliced transcript, appears to be sufficient to cause LCA (den Hollander et al. 2006, 2008; Sayer et al. 2006; Valente et al. 2006; Baala et al. 2007; Frank et al. 2007; Perrault et al. 2007). These data argue that, at least in some instances, this mutational event alone is sufficient to explain the disease without necessarily evoking the action of additional alleles. In this example, recurrence of the specific mutation has facilitated that argument. However, typically bereft of such data, dissecting phenotype–genotype correlations within a primary causal locus from modifier effects can be challenging, especially because most such correlations are made on the basis of a few mutations.
Multiple interactions and gene/allele-specific interactions. The effect of genetic modifiers on penetrance and/or expressivity can vary from a “monogenic” model of modification, in which one modifier can account for most of the observed variability, to a more complex model in which more than one genetic determinant interacts with the primary disease locus. Although identification of major or “panmodifiers” is feasible, the dissection of more than one gene modulating the phenotype caused by the primary locus is challenging, due primarily to lack of genetic power. An example illustrating the mapping of a “panmodifier” was described in a Bardet-Biedl syndrome (BBS) cohort of 226 patients not preselected for mutations at any specific locus. Within this group, an allele in CCDC28B was associated with a more severe BBS phenotype, a discovery based on association analysis and transmission disequilibrium testing (TDT) and dissected further by biochemical studies (Badano et al. 2006). Had the modifying effect of CCDC28B been specific to a particular BBS locus, unmasking of such a genetic interaction would have been difficult. In contrast to “panmodifiers,” the interplay of multiple modifiers, each of which has a variable strength of effect on the final disease expression is probably common, but the most challenging to unravel. A good example is cystic fibrosis, a life-shortening recessive disorder in which 90% of the affected patients die because of obstructive pulmonary disease (Cutting 2010). Despite the majority of patients carrying identical genotypes (homozygosity for CFTR F508del), striking differences in lung function have been described (Kerem et al. 1990). Candidate modifier studies reported that functional variants in MBL2 and alleles in the promoter and first exon of TGFb1 were associated with lower lung function measures (Garred et al. 1999; Drumm et al. 2005; Darrah et al. 2010). More recently, genome-wide association studies revealed additional modifiers of lung function; IL-8, IFRD1 and ENDRA associated with neutrophil function (Vanscoy et al. 2007; Hillian et al. 2008). Taken together, the aforementioned genetic modifiers are believed to alter lung disease severity by affecting the ability of the host to tolerate infection, which can be further modified by nongenetic determinants such as air pollutants, secondhand smoking, or even access to medical care and treatment (Rubin 1990; Goss et al. 2004). Under such a model, each modifier explains only a small portion of the variability, similar to the genetic risk factors involved in complex traits.
Modifiers in the same locus/region with the disease driver. Genetic modifiers in cis with the disease driver gene might be considered to be variants tagging and being transmitted in linkage disequilibrium with the primary mutation. An example of a modifier in cis comes from several studies showing that variants within FTO were reported to confer susceptibility to developing obesity (Dina et al. 2007; Frayling et al. 2007; Scuteri et al. 2007; Church et al. 2010). Although FTO was shown to regulate body mass, the variants identified appeared to have no effect on the expression and stability of the protein (Klöting et al. 2008; Grunnet et al. 2009). Instead, the variants within the FTO region are regulating the expression of a transcription factor located in cis, IRX3 that is now thought to potentially act as the primary susceptibility gene for developing obesity (Smemo et al. 2014). An example of trans modification within the same locus was described in a pedigree diagnosed with erythropoietic protoporphyria (EPP), in which the patients carry a splice donor site deletion resulting in skipping of exon 10 (Δex10) of ferrochelatase (FECH). When this change is accompanied by an in trans intragenic variant (IVS3-48C), the phenotype is aggravated with the levels of ferrochelatase in those patients being significantly lower than in asymptomatic carriers (Gouya et al. 1996, 2006). The mechanism underlying this phenomenon consists in IVS3-48C modulating the use of a constitutive aberrant acceptor splice site on FECH resulting in an aberrantly spliced mRNA that is subject to nonsense-mediated decay and therefore leading to FECH deficiency (Gouya et al. 2002).
Frequency of the modifying variants in the general population. Assuming that a modifier allele alone is not sufficient to induce pathology, it might be relatively common in the general population, thus complicating the dissection of phenotypically relevant versus benign variants. In the study that identified an allele that confers susceptibility to Hirschsprung disease (HSCR), the causal variant had a minor allele frequency (MAF) of 0.45 in Asia, 0.25 in Europe and was virtually absent (0.01) in the African population (Emison et al. 2005). Unmasking such common variants may require the use of multiple methods; in this case the combinatorial use of careful phenotyping combined with human genetic, comparative genomic, functional, and population genetic analyses (Emison et al. 2005). At a second tier, there are alleles of intermediate frequency in the general population. An example of this is the p.A229T allele in RPGRIP1L which aggravates the retinal phenotype in ciliopathy patients, present in 3% of Northern Europeans (Khanna et al. 2009). Finally, rare variants that are enriched in patients compared with controls can have a modifying effect on disease expressivity and penetrance. In age-related macular degeneration (AMD), in which most primary driver variants are common, rare alleles (MAF<1%) within the complement pathway can alter the disease phenotype. The first example described the rare p.R1210C mutation in CFH as a penetrant allele that confers high risk to AMD and was followed by the discovery of an additional five risk and 15 protective variants through a study that evaluated components of the complement pathway (Raychaudhuri et al. 2011; Seddon et al. 2013). Although it is easier to establish the relevance of a rare variant to disease causality or predisposition, the same is not true for common variants that are overrepresented in the control population. In the context of genetic modification the true debate is not how common or rare a variant is, but rather how those variants work together and what is their cumulative effect (Gibson 2012).
Figure 1.
Schematic of the effects of modifiers on causal mutations. A genetic modifier can exert its effect on the phenotype established by mutation(s) at the primary locus in a number of ways. Depending on the nature of the modifier, this interaction can result in reduced penetrance of specific endophenotype(s) and/or disorders; in variable expressivity with the modifying locus resulting in either exacerbated phenotypes (i.e., earlier age of onset and faster progression of the disease); in a milder clinical presentation (i.e., later age of onset, attenuated disease progression, and absence/protection from specific endophenotypes); and modifiers that potentiate the manifestation of specific endophenotypes resulting in what is described as novel forms of disease. This figure was prepared using Servier Medical Art (http://www.servier.com/Powerpoint-image-bank).
CHALLENGES IN IDENTIFYING MODIFIER GENES/ALLELES
In contrast to the remarkable progress in identification of disease causing genes, the dissection of oligogenic and epistatic phenomena is lagging, in part because of the reduced power of traditional genetic analyses to uncover such phenomena. Beyond methodological limitations, there are additional challenges to consider:
Availability of accurate clinical evaluation and longitudinal studies. The identification of genetic modifying phenomena relies heavily on the detailed characterization of the aspect(s) of the phenotype under modification. It is thus imperative to have detailed clinical longitudinal information. Historically, cohorts have been characterized qualitatively for the presence/absence of a trait (i.e., RP or macular degeneration), with since-point proxies used as an imperfect means of expressing disease progression; these have included age of onset of a specific phenotype (Badano et al. 2006), thickness of the outer nuclear layer (ONL) (Danciger et al. 2008), or number of photoreceptors (Haider et al. 2008). The literature however, is populated by incomplete phenotypic information on the cases presented, often focusing on specific aspects of disease. In BBS, a multisystemic disorder characterized by rod–cone dystrophy, central obesity, polydactyly, and retinal degeneration, a cardinal symptom of the disorder, renal dysfunction had been overlooked (Brattgard 1949; Landau et al. 1949). A reason for this could have been that, although renal structural abnormalities might be present, only a small fraction of the patients develop symptoms consistent with kidney functional impairment that would prompt clinical evaluation (Magro and Peres 1970; Labrune et al. 1974; Gershoni-Baruch et al. 1992; Beales et al. 1999). In the case of RPGRIP1L, thorough description of the retinal phenotype across a cohort of patients with syndromic disorders falling under the umbrella of the ciliopathies, led to the dissection of the p.A229T allele as a variant associated with increased loss of photoreceptors, whereas absence of this change was proposed to have a potential protective effect from RP (Khanna et al. 2009).
Nongenetic factors. The effect of nongenetic factors poses a major confounder. In humans, one way to determine whether the phenotypic variability observed is caused by genetic modifiers or environmental factors is to compare inter- and intrafamilial variability. A second approach is that of comparing the phenotype in monozygotic (MZ) and dizygotic (DZ) twins that share the same environmental influence. One of the best characterized examples of nongenetic determinants leading to disease manifestation comes from studies linking pregnancies complicated by fetal neural tube defects (NTDs) with folate insufficiency, and the evidence that periconceptional folate supplementation can reduce the risk of NTDs and other congenital abnormalities (Laurence et al. 1980; Tolarova 1982; Czeizel and Dudás 1992; Wald et al. 2001).
STRATEGIES FOR CLONING GENETIC MODIFIERS
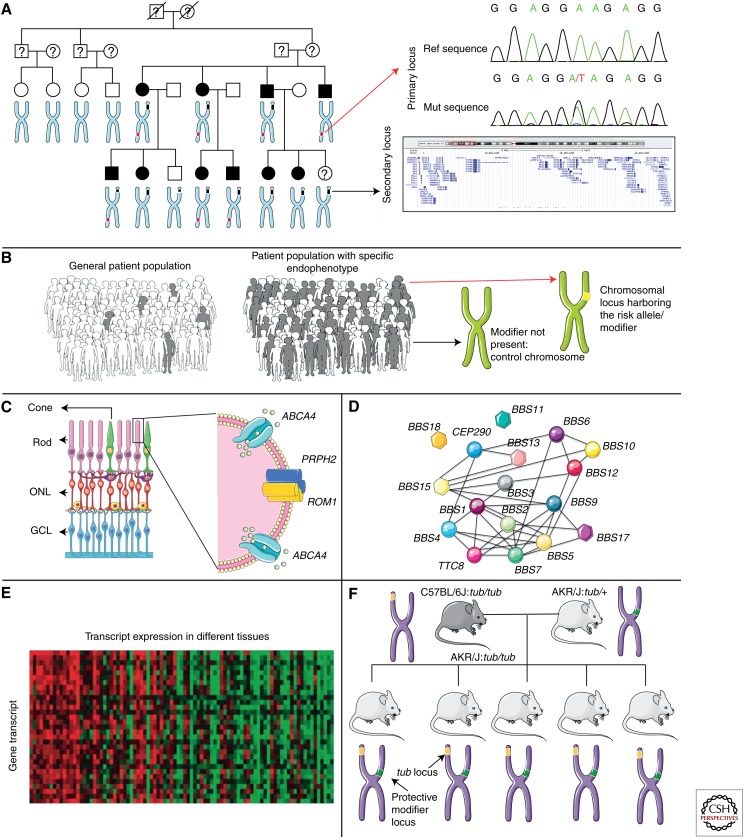
The examples of disease modifier identification existing in the literature have used a plethora of techniques and approaches (Fig. 2), typically combining several lines of investigation:
Linkage analyses. If nuclear families with individuals carrying the same primary locus mutation and displaying discordant phenotypes are available, linkage analyses to identify alleles identical by descent (IBD) in individuals sharing a specific trait or endophenotype can be performed. The same can be applied in sib pairs concordant or discordant for a given phenotype. Knowing the primary disease causing mutation, linkage analysis for the detection of modifier loci can be performed by evaluating the sample set for qualitative phenotypes (mutation carriers with the phenotype, mutation carriers without the phenotype, and mutation carriers with more severe disease form), or for quantitative phenotypes (i.e., age of onset). One of the reasons that could account for the inability to map modifiers is that, especially in human studies, large pedigrees are often necessary to map the regions harboring the genes of interest. This is exemplified in Fuchs corneal dystrophy (FCD), in which genome-wide linkage in a large multigenerational pedigree carrying the TCF8 p.Q840P allele that drives disease, revealed a second locus on 9p that, when combined with TCF8 p.Q840P, results in a more severe form of disease (Riazuddin et al. 2010). Even when large pedigrees are available, variable intrafamilial phenotypic expression, in association with the difficulty to identify families with uniform phenotypes, can complicate mapping of modifiers. To address this, screening pedigrees from genetic isolates can prove beneficial. Such a study was performed on the Menonite genetic isolate to identify genes underlying HSCR (Carrasquillo et al. 2002). Using a genome-wide association study coupled with multipoint-linkage disequilibrium analysis, which searches for association arising from common mutation ancestry, linkage over known HSCR loci was detected. However, this signal was insufficient to explain disease manifestation (Carrasquillo et al. 2002). Instead, it was an epistatic interaction between the receptor tyrosine kinase (RET) and the endothelin receptor type B (EDNRB) that was necessary for disease manifestation (Carrasquillo et al. 2002).
Candidate locus association studies. The advantage of this approach consists mainly in that data from singletons can be compared with other unrelated patients with similar phenotypes. A limitation of this approach is that once association is established, it is difficult to discern whether it is truly owing to the polymorphism/marker identified, to a variant that is in linkage disequilibrium (LD) with the marker used, or to an unknown confounding factor that might not even be genetic (Génin et al. 2008). An alternative is family-based association and TDT, in which the preferential transmission of alleles from parent to affected offspring is evaluated (Speilman et al. 1993). In HSCR, linkage of modifiers has been shown consistently to the RET locus, harboring the main HSCR driver, RET, although in most of these studies no coding mutations could be identified (Bolk et al. 2000; Borrego et al. 2000; Gabriel et al. 2002; Garcia-Barceló et al. 2003; Sancandi et al. 2003). In these studies, it was hypothesized that the genetic determinant conferring increased susceptibility to HSCR when combined with mutations in other loci, would lie within the RET locus (Emison et al. 2005). As such, a combination of human genetic, comparative genomic, functional and population genetic analyses were undertaken to show that the variant sought was an intronic change within a conserved enhancer sequence in intron 1 of RET (Emison et al. 2005). A similar approach was used to show the overtransmission of the CCDC28B modifier in BBS (Badano et al. 2006).
Candidate gene approach. One strategy to identify genetic modifiers is by applying a hypothesis-based candidate gene approach through the screening of genes that operate within the same biological pathways/processes as the primary disease driver gene(s). Such hypotheses can be formulated either through current experimental or in silico knowledge of gene networks or from evidence from model organisms. Using this approach, Poloschek and colleagues screened 15 family members for mutations in genes known to cause retinal degeneration. They identified heterozygous mutations in PRPH2, ABCA4, and ROM1; family members carrying the p.R172W mutation in PRPH2 were severely affected, patients carrying heterozygous alterations in only one of those genes displayed a milder phenotype and patients with heterozygous mutations in all three genes evaluated, show an even more severe form of disease (Poloschek et al. 2010). Another example based on the candidate gene approach involved the identification of the genetic interaction between NPHP1 and AHI1, with NPHP1 accounting for the nephronopthisis phenotype and the AHI1 p.R830W allele being the major determinant driving the retinal phenotype in the NPHP cohort (Louie et al. 2010).
System-based candidates. This approach requires a priori knowledge of the specific genes driving the pathogenesis of the trait of interest. In the human disease network hypothesis, genes associated with similar disorders show higher likelihood of physical interactions and higher expression profiling similarity between their products (Goh et al. 2007). Such an interaction network was constructed for 54 proteins involved in 23 distinct types of human inherited ataxias (Lim et al. 2006). Within this network, different ataxia-causing proteins showed physical interactions and revealed common cellular pathways potentially relevant to disease (Lim et al. 2006). To test the robustness of this approach, the ataxia network was evaluated for its ability to unravel both modifiers and disease drivers. Two major modifiers of spinocerebellar ataxia type 1 (SCA1), ataxin-2 (ATXN2) and RNA binding protein with multiple splicing (RBPMS) were present on the “ataxia-genome” map as nodes that shared strong physical interactions with ataxin-1 (ATXN1) (Lim et al. 2006). BBS represents another example of this approach, with more than 20 causative genes participating in the function of the cilium and thus showing that they are related to each other at a higher level of cellular and organismal organization (Davis and Katsanis 2012). As such, other proteins localizing and operating within the cilium emerge as candidates to either drive disease pathology or modulate phenotypic expressivity. Starting with the bona fide genes that can cause BBS under a recessive paradigm in some families, and thus representing de facto contributors to the molecular etiology of BBS, an excess of pathogenic heterozygous BBS alleles were found in families with recessive mutations in other BBS loci (Katsanis et al. 2001; Badano et al. 2003; Beales et al. 2003; Stoetzel et al. 2006, 2007; Leitch et al. 2008). These phenomena were not restricted to genes mutated in BBS under a recessive paradigm; the BBS modifier CCDC28B was screened based on its interaction and colocalization with multiple BBS proteins, in addition to the fact that it was belonging to the ciliary proteome and hence operating in the same system as the other BBS proteins (Li et al. 2004; Badano et al. 2006; Gherman et al. 2006).
Transcriptome analyses. Methodologies that evaluate the expression of proteins such as microarrays and RNA-seq present powerful tools for studying the transcriptional landscape of a given cell or tissue. Transcriptome analyses compare the expression levels of proteins between different phenotypic groups, such as control versus disease organism. The comparison can be applied to the whole transcriptome of the tissue of interest, or it can be combined with other methodologies such as linkage analyses, to identify only the genes that have altered expression within the locus of interest. Whole transcriptome comparison was performed between four siblings (two of which were asymptomatic) and two unrelated patients affected by spinal muscular atrophy (SMA), all of which carried identical SMN1/SMN2 genotypes (Oprea et al. 2008). One transcript showing statistically significant differential expression between the individuals with discordant phenotypes was plastin 3 (PLS3), a protein that elevates the F actin levels and promotes axonogenesis, comprising the first protective modifier in SMA reported (Oprea et al. 2008). The protective effect of PLS3 was illustrated further in a conditional mouse model overexpressing PLS3 into an SMA background (SMAPLS3), with the motor neurons of the SMAPLS3 mice showing restored actin dynamics, increased size of the somata, increased quantal content of the neuromuscular junctions, axon stabilization, increase in the size of innervated muscle fibers, and overall prolonged survival (Ackermann et al. 2013; Lyon et al. 2014).
Considering whole-exome data in toto. A more unbiased approach to screen for genetic modifiers is through interrogation of the whole exome. Advances in massively parallel sequencing over the past years have made such analyses feasible. The caveat in using such large-scale screens lies in the small sample size available to screen for modifiers, the failure to correct for multiple hypotheses testing when several variants and genetic markers are examined, as well as the effects of population stratification (Gusella and MacDonald 2009). Nonetheless, the advantage of using these technologies consists mainly in the fact that they provide an unbiased approach. This is illustrated by recent studies in Gordon Holmes syndrome (GHS), a rare form of cerebellar ataxia associated with hypogonadotropic hypogonadism. Analysis of whole-exome sequencing (WES) identified rare alleles in a large consanguineous pedigree and when the genetic findings were coupled to functional studies concluded that, in that family GHS was hypomorphic caused by combinatorial mutations in two independent genes (RNF216 and OTUD4), both of which participate in the ubiquitin proteasome pathway (Margolin et al. 2013). There are several conclusions that can be drawn by this study: (1) if significant linkage was obtained over the locus of one or both of the genes identified, it would have most probably led to the mapping of the one of the genes of this interaction, leaving the second locus unexplored; (2) if the epistatic interaction in GHS had not been underlay by homozygous hits in both genes, but rather was mediated by a homozygous hit in the primary locus and a heterozygous hit in the second or epistatic locus, it would have likely been missed by “typical” WES allele filtering; (3) without the use of the in vivo functional model uncovering the functional synergy of the two genes, RNF216 would have been proposed as the gene underlying GHS because of identification of mutations in this gene in unrelated patients, leading to the false conclusion that OTUD4 is not relevant to disease.
Modifiers in inbred and mutation-induced mouse strains. The most successful approach to map genetic modifiers are mouse models that show phenotypic divergence caused by their different genetic background. A study illustrating the power of this approach to identify genetic modifiers consisted of crossing C57BL/6J-tub/tub mice that display retinal and cochlear degeneration (Heckenlively et al. 1995; Ohlemiller et al. 1997) with AKR/J−/+ animals. In the F2 generation, the C57BL/6J:AKR/J-tub/tub mice were protected from retinal degeneration, having an almost normal ONL thickness and number of photoreceptors when compared with the C57BL/6J-tub/tub littermates (Ikeda et al. 2002a). Subsequent analysis identified significant linkage over the motr1 locus on chromosome 11 and yielded suggestive evidence for linkage at two loci on chromosomes 2 and 8, respectively (Ikeda et al. 2002a).
Figure 2.
Examples of strategies for cloning modifiers. A. Linkage analysis, with the affected family members manifesting specific endophenotypes or differential disease severity. By evaluating the sample set for the endophenotype or trait of interest, the candidate modifier loci can be mapped. The snapshot of chromosome 9 in panel A is from the UCSC browser (http://genome.ucsc.edu/). B. Association studies compare patient cohorts with (gray figures) or without (white figures) a modifier of disease severity/expressivity, to obtain a signal for the locus/position that harbors the modifying allele(s). C. The candidate gene approach evaluates whether genes whose products operate within the same pathway can exert a modifying effect on each other. The example illustrated comes from the observation that mutations in PRPH2 and ABCA4 can aggravate disease severity when combined with primary disease mutations in ROM1. D. The system-based hypothesis evaluates the likelihood of specific genes to physically interact or manifest similar expression profile changes in control versus affected individuals to build genetic networks that drive disease pathogenesis. Such network interactions among genes that cause the same or similar disorders is exemplified by BBS, in which the responsible genes are shown to operate and interact together. E. In transcriptome analyses, heatmaps compare changes in expression levels of genes, between healthy and affected individuals. F. The use of mouse congenic strains to map modifiers in a more simplified genetic background. This figure was prepared using Servier Medical Art (http://www.servier.com/Powerpoint-image-bank).
GENETIC MODIFIERS OF RETINAL DEGENERATION IN HUMANS
The report of a nuclear family with a 3 bp deletion in peripherin (RDS) presenting with three distinct clinical phenotypes (RP, pattern macular dystrophy, and fundus flavimaculatus; Table 1), provided the first example of complex inheritance in retinal degeneration (Weleber et al. 1993). The identification of additional families with RDS mutations and significant inter- and intrafamilial variability ensued (Apfelstedt-Sylla et al. 1995). Kajiwara and colleagues then chose three clinically variable families carrying the heterozygous RDS p.L185P allele; the investigators undertook a candidate gene approach and screened for mutations in the gene encoding the retinal outer segment membrane protein 1 (ROM1), whose homodimer protein products complex with those encoded by RDS (Goldberg and Molday 1996). This study revealed that only family members that were double heterozygotes for the RDS p.L185P alleles and the ROM1 p.114fsX developed RP, whereas individuals that were heterozygous for either one of these mutations did not (Kajiwara et al. 1994; Rivolta et al. 2002). The mechanism through which such a genetic interaction can affect the clinical phenotype has also been elucidated, revealing that RDS mutant subunits fail to homodimerize and that the formation of functional complexes is further decreased by the frameshifting mutation in ROM1, thus leading to the ultimate degeneration of photoreceptors in the retina (Goldberg and Molday 1996; Loewen et al. 2001). The effect of ROM1 as a modifier of phenotype was highlighted further in a study in which cases with heterozygous ROM1 mutations (p.R172W or p.R229H) combined with mutations in RDS result in a more severe phenotype, with pronounced loss in rod but not cone function (Poloschek et al. 2010). In the same study, a subset of patients with mutations in yet a third gene (ABCA4: p.R172W, p.R229H or p.V2050L) were present together with ROM1 and RDS mutations; the phenotype was even more severe, with advanced macular degeneration, nyctalopia, and severe rod and cone function decline (Poloschek et al. 2010).
Table 1.
Genetic modifiers in retinal degeneration from studies in humans
| Disorder | Primary disease driver | Second-site modifier | Clinical aspect affected by modifier | Reference(s) |
|---|---|---|---|---|
| BBS | BBS2 (p.Y24X, p.Q59X) | BBS6 (p.Q147X) | Disease penetrance; digenic inheritance | Katsanis et al. 2001 |
| BBS | MGC1203 (p.C430T) | Earlier onset of RP; accelerated disease progression | Badano et al. 2006 | |
| BBS | BBS1 (p.M390R) | MGC1203 (p.C430T) | Disease penetrance; digenic inheritance | Badano et al. 2006 |
| Ciliopathies | RPGRIP1L (p.A229T) | Retinal degeneration | Khanna et al. 2009 | |
| Dominant cone or cone–rod dystrophy (COD3) | GUCA1A (p.P50 L) | Unknown | Disease severity | Downes et al. 2001 |
| LCA | CEP290 | AHI1 (p.N811K and p.H758P) | Neurological phenotype; disease severity | Coppieters et al. 2010 |
| LCA | AIPL1 (p.Q163X) | CRX (p.T273L) | Earlier disease onset; disease severity | Zernant et al. 2005 |
| LCA | AIPL1 (p.R302 L) | CRB1 (p.R1331H) | Accelerated disease progression | Zernant et al. 2005 |
| LCA | RPE65 (p.E102X) | GUCY2D (p.I573V) | Disease severity | Zernant et al. 2005 |
| LCA | GUCY2D (p.S448X) | CRX (p.A158T) | Development of macular coloboma in both eyes | Zernant et al. 2005 |
| Macular dystrophy | PRPH2 | ROM1 (p.R172 W or p.R229H) | Disease severity; loss of rod function | Poloschek et al. 2010 |
| Macular dystrophy | PRPH2 | ABCA4 (p.R172 W, p.R229H or p.V2050L) | Aggravated disease course; loss of cone and rod function | Poloschek et al. 2010 |
| Nephronopthisis | NPHP1 | AHI1 (p.R830 W) | Retinal degeneration | Louie et al. 2010 |
| Oguchi’s disease/ autosomal recessive RP | SAG (p.383fsX) | Unknown | Development of macular degeneration and golden-yellow fundus reflex | Nakazawa et al. 1998 |
| Retinoschisis (X-linked) | XLRS1 (p.125fsX) | Unknown | Disease severity and progression | Tantri et al. 2003 |
| RP, pattern macular dystrophy, fundus flavimaculatus | RDS (p.L185P) | ROM1 (p.114fsx) | Development of RP | Kajiwara et al. 1993; Weleber et al. 1993 |
| RP (X-linked) | RPGR | RPGRIP1L (p.R744Q) | Disease severity | Fahim et al. 2011 |
| RP (X-linked) | RPGR | IQCB1 (p.I393N) | Disease severity | Fahim et al. 2011 |
| RP (autosomal dominant) | PRPF31 | CNOT3 | Disease penetrance through transcriptional repression | Venturini et al. 2012 |
| RP (autosomal dominant) | PRPF31 | Unknown (linkage on 14q21-23) | Disease penetrance | Frio et al. 2008 |
| RP1 | RP1 (p.R677X) | Unknown | Disease severity | Jacobson et al. 2000 |
| RP9 | PAP-1 (p.H137 L) | Unknown | Disease severity | Keen et al. 2002; Kim et al. 1995 |
| RP48 | GUCA1B (p.G157R) | Unknown | Disease severity | Sato et al. 2005 |
| Stargardt type 3 | ELOVL4 (5 bp deletion) | ABCA4 | Disease severity ranging from Stargardt-like macular dystrophy to pattern dystrophy | Zhang et al. 1999; Bernstein et al. 2001 |
| Sorby fundus dystrophy (SFD) | TIMP3 (S181C) | Unknown | Pathological findings ranging from absence of fundus spots to yellow deposits associated with macular degeneration | Hamilton et al. 1989; Weber et al. 1994; Wijesuriya et al. 1996; Felbor et al. 1997 |
| Usher syndrome | USH2A (p. C1447QfsX29) | PDZD7 (p.R56PfsX24) | Earlier disease onset; aggravation of RP severity | Ebermann et al. 2010 |
| Usher syndrome | USH2C (GPR98 p.A5713LfsX3) | PDZD7 | Disease penetrance; digenic inheritance | Ebermann et al. 2010 |
| Usher syndrome | USH1D (p.R1746Q) | Unknown | Severity and progression of RP | Bolz et al. 2001; Astuto et al. 2002 |
| Usher syndrome | USH3A (p.N48 K) | Unknown | Progression of RP | Ness et al. 2003 |
| Vitelliform macular dystrophy type 2 | VMD2 | Unknown | Disease severity and penetrance; development of macular degeneration | Nordström and Thorburn 1980 |
A second modifier of retinal degeneration identified through the candidate gene approach was AHI1 in a study evaluating patients with nephronophthisis (NPHP), a hereditary kidney disease (Louie et al. 2010; Table 1). More specifically, within a pedigree with three NPHP diagnosed patients, one brother was also carrying the heterozygous AHI1 p.R830W mutation (Louie et al. 2010). This variant was shown to be associated with retinal degeneration both in this familial case but also among sporadic NPHP patients (Louie et al. 2010), thus attributing a retinal degeneration modifying effect role to AHI1. In addition to the NPHP cohort, AHI1 was also screened for mutations and reported as a genetic modifier in a Belgian cohort of eight CEP290 mutation positive patients affected by LCA and mental retardation (Coppieters et al. 2010). The patients most severely affected were carriers of AHI1 p.N811K and p.H758P, leading to the conclusion that CEP290 and AHI1 are likely to be operating in the same pathway and that within this cohort AHI1 serves as a modifier of the neurological phenotype (Coppieters et al. 2010).
A third example came from the screening of RPGRIP1L, a gene known to cause MKS and JBTS (Arts et al. 2007; Delous et al. 2007), for a cohort of patients with various ciliary disorder diagnoses (Khanna et al. 2009). Among the 10 novel nonsynonymous changes identified in RPGRIP1L, one allele (p.A229T) was enriched significantly in patients with ciliary disease coupled by retinal degeneration (Khanna et al. 2009; Table 1). The effect of the modifier allele was established in a zebrafish model in which it scored as pathogenic, and in a yeast two-hybrid screen in which the mutant RPGRIP1L was exerting a significantly lower affinity to form complexes with the retinitis pigmentosa GTPase regulator protein (RPGR), with which it interacts (Khanna et al. 2009). It is thought that the observed photoreceptor degeneration in patients with the modifying RPGRIP1L p.A229T allele are because of the attenuation of the RPGRIP1L–RPGR interaction (Khanna et al. 2009). The observation that RPGRIP1L contributes modifying alleles was shown further in X-linked RP caused by mutations in RPGR (Fahim et al. 2011). Affected males from 56 families with RPGR mutations were genotyped for common alleles in each of RPGRIP1, RPGRIP1L, CEP290, and IQCB1 based on the fact that they all interact with RPGR as determined by coimmunoprecipitation experiments (Boylan and Wright 2000; Otto et al. 2005; Chang et al. 2006; Khanna et al. 2009). Two alleles, the IQCB1 p.I393N and the common RPGRIP1L p.R744Q, showed significant association with disease severity, with the minor allele in IQCB1 and the common allele in RPGRIP1L being associated with more severe disease (Fahim et al. 2011).
In Usher syndrome, a genetically heterogeneous recessive disease characterized by hearing loss and RP, a genetic modifier of the retinal phenotype was identified by blasting for homologous proteins to harmonin causing Usher syndrome subtype 1C (USH1C) and whirlin underlying USH2D that cross-link other Usher proteins to form a multimolecular complex in the photoreceptors and hearing cells (Kremer et al. 2006; Ebermann et al. 2007). PDZD7 was identified and screened for mutations in patients with an Usher syndrome diagnosis (Ebermann et al. 2010; Table 1). Although no changes in PDZD7 were identified in Usher patients with any molecular diagnosis, in familial cases with mutations in known Usher genes, PDZD7 seemed to be contributing to a digenic mode of inheritance and to also be contributing modifying alleles (Ebermann et al. 2010). For example, in a family with two USH2A (p. C1447QfsX29) affected sisters, the one that also carried a heterozygous frameshift mutation in PDZD7 (p.R56PfsX24) displayed an earlier age of onset and more severe progression of RP, whereas in a distinct case of a family with two affected offspring, only the individual with a heterozygous USH2C (GPR98 p.A5713LfsX3) mutation and a second heterozygous hit in PDZD7 is affected, suggesting that GPR98 and PDZD7 seem to be participating in a digenic mode of inheritance (Ebermann et al. 2010). Similar to the study identifying RPGRIP1L as a genetic modifier, the direction of effect of the PDZD7 alleles as pathogenic and the digenic inheritance of GPR98 and PDZD7 were confirmed in a zebrafish in vivo model for RP (Ebermann et al. 2010).
BBS has contributed substantially to understanding the contribution of genetic modifiers to disease (Katsanis et al. 2001; Badano et al. 2006; Table 1). The disorder is genetically and clinically heterogeneous, with well-documented examples of inter- and intrafamilial phenotypic variability (Carmi et al. 1995; Riise et al. 1997). Although initially reported to follow an autosomal recessive mode of inheritance, BBS was the first disorder to follow a “triallelic” model of disease transmission with three alleles at two independent loci being required for disease manifestation (Katsanis et al. 2001). More specifically, in a nuclear family with one affected and two unaffected siblings, compound heterozygosity in BBS2 for the nonsense alleles p.Y24X and p.Q59X was not sufficient to cause the phenotype, but required the presence of a third allele at a second BBS locus, BBS6 p.Q147X, which acts essentially as a modifier of penetrance (Katsanis et al. 2001). A second example of genetic modification coming from the study of BBS samples is the identification of CCDC28B p.C430T as a modifier affecting RP; the patients that carried p.C430T displayed an earlier onset of RP and accelerated disease progression (Badano et al. 2006). In addition to contributing modifying alleles, CCDC28B has also been postulated to represent an example of complex inheritance because it seems to be required for disease manifestation in a family with BBS1 p.M390R homozygous siblings and also asymptomatic father. It is therefore hypothesized that the third allele (p.C430T) located in CCDC28B might be required for disease manifestation or be acting as a modifier of penetrance similar to the BBS6 and BBS2 epistatic interaction (Beales et al. 2003; Badano et al. 2006).
GENETIC MODIFIERS OF RETINAL DEGENERATION IN MICE
The cloning of modifiers in human genetic disorders is confounded by genetic and allelic heterogeneity. Some of these challenges can be overcome in model organisms. Laboratory mice in particular, have been successfully used to map modifier genes and alleles in a wide range of genetic disorders (Table 2). Although in most successful studies the modification of the primary phenotype is because of the effects of a single locus, independent from the primary locus (Danciger et al. 2000; Ikeda et al. 2002a), mapping of modifiers can be complicated when the modification results from the combined effects of several genes at discrete loci (Table 2) (Ikeda et al. 2002a).
Table 2.
Genetic modifiers in retinal degeneration from studies in mice
| Disorder | Primary disease driver/mouse model | Second-site modifier | Phenotypic aspect affected by modifier | Strains involved | Reference(s) |
|---|---|---|---|---|---|
| Fibrous retrolental tissue | p53 | Not identified | Disease penetrance | C57BL/6J, 129/SvJ | Ikeda et al. 1999 |
| Glaucoma | BMP4 | Not identified | Disease severity | C57BL/6J, BliA, CAST/Ei, C3H, AKR/J, BALB/C, 129/SvEvTac | Hong et al. 1999 |
| Iris atrophy/Glaucoma | Ipd | Isa (Tyrp1) | Disease manifestation | C57BL/6J, DBA/2J | Chang et al. 1999 |
| Iris digment dispersion | Ipd | Not identified | Disease penetrance | AKXD-28/Ty | Anderson et al. 2001 |
| Iris stromal atrophy | Isa | Not identified | Disease severity; increased cell death | AKXD-28/Ty | Anderson et al. 2001 |
| LCA | Rd16 | Bbs6 | Variability in photoreceptor degeneration | C57BL/6J, 129/Sv, C57BL/6 | Rachel et al. 2012 |
| Light-induced retinal degeneration | Rpe65 | Rpe65 p.Leu450, Rpe65 p.Met450 | Disease penetrance | BALB/c2J | Danciger et al. 2000; Wenzel et al. 2003 |
| Ocular retardation | Chx10 | Not identified | Disease penetrance | CASA/Rk | Bone-Larson et al. 2000 |
| Oguchi disease (stationary night blindness) | Arrestin−/− | ArrestinP365/P365 | Light-damage susceptibility | C57BL/6, 129/SvJ | Chen et al. 1999 |
| Light-induced RP | Rho−/− | Rho p.V20G, Rho p.P23H, Rho p.P27L | Disease penetrance | C57BL/6, FVB | Naash et al. 1996; Wang et al. 1997 |
| Retinoschisis | Rs1 | Tyr (Mor1) | Disease penetrance | C57BL/6J, AKR/J | Johnson et al. 2010 |
| RP (recessive) | Pde6a | Not identified | Disease severity; photoreceptor loss rate | G3 A.B6-Tyr+/J | Sakamoto et al. 2009 |
| RP1 | Rp1h | Not identified | Disease severity | (N6) B6.129S, DBA.129S(B6), A.129S(B6) | Liu et al. 2009 |
| RP | Tub or Rd5 | Mtap1a (motr1) | RP penetrance; disease progression | B6.Cg-rd5/rd5, AKR/J | Ikeda et al. 2002b; Maddox et al. 2012 |
| Retinal degeneration | Rd3 | Not identified | Onset and progression of photoreceptor degeneration | RBF/Dn, Meta-In(1)Rk, Rb(11.13)4Bnr, In-30 | Heckenlively et al. 1993 |
| Retinal degeneration | Rd7 | Chr 8 and 19 (CAST/EiJ), Chr 7 and 11 (AKR/J) | Suppression of retinal spotting | B6.Cg-rd7/rd7, CAST/EiJ, AKR/J, NOD.NON-H2nb1 | Akhmedov et al. 2000; Haider et al. 2008 |
| Retinal degeneration | Rho−/− | Not identified | Variability in photoreceptor degeneration | 129/SV, C57BL/6 | Humphries et al. 2001 |
| Retinal degeneration | Rd16 | Bbs4 | Variability in photoreceptor degeneration | 129/SV, BXD24 | Zhang et al. 2014 |
The first report of a genetic modifier of retinal degeneration in mice was the identification of protective alleles in the rd5 or tubby mouse for both the hearing and retinal degeneration phenotypes (Danciger et al. 2000; Ikeda et al. 2002a,b). The tubby mouse has been proposed as animal model for Usher syndrome 1, BBS and Alström syndrome because of the phenotypic convergence of the clinical phenotypes of retinal and cochlear degeneration, and maturity onset insulin-resistant obesity (Coleman and Eicher 1990; Heckenlively et al. 1995). Originally, the profound hearing loss in B6-tub/tub mice was fully rescued in F2 tub/tub homozygotes crossed with AKR/J, 129P2/OlaHsd and CAST/Ei mice, suggesting the presence of a protective allele (Ikeda et al. 2002a). Linkage analysis and subsequent sequencing identified 10 amino acid substitutions and an Ala-Pro repeat length difference in the microtubule-associated protein 1A (Mtap1a) residing within the modifier of tubby hearing 1 (moth1) locus on chromosome 2, between the B6-tub/tub and the tub/tub homozygotes in protective strains (Ikeda et al. 1999, 2002a). Mtap1a was suggested to play a role in vesicular trafficking of proteins to the neuronal synaptic junctions and further work demonstrating differential binding affinities of Mtap1a with the postsynaptic marker PSD95 support a role of the Mtap1a modifier gene in synaptic maintenance and architecture (Brenman et al. 1998; Halpain and Dehmelt 2006). The second modifier hunt in the tubby strain consisted in the identification of the protective allele for the retinal degeneration phenotype of this model (Ikeda et al. 2002a). The degree of retinal degeneration was quantitatively assessed by measuring the thickness of the ONL and the number of photoreceptors in a defined area of the ONL between C57BL/6J-tub/tub mice and tub/tub homozygotes in the protective AKR/J strain (Ikeda et al. 2002a). Linkage analysis comparing the two strains identified a significant signal on chromosome 11, a locus termed modifier of tubby retinal degeneration 1 (motr1) whereby a recessive mode of inheritance was assumed for the mapping of the protective allele and two suggestive signals on chromosomes 2 and 8, where dominant protective alleles were likely to segregate (Ikeda et al. 2002a). Interestingly, the locus on chromosome 2 identified in the study for the retinal degeneration modifier, maps to the same locus as moth1 harboring the Mtap1a hearing modifier (Ikeda et al. 2002a,b). Subsequent screening of Mtap1a revealed nonsynonymous sequence alterations between the C57BL/6J-tub/tub and AKR/J-tub/tub mice, suggesting that the same gene contributes protective alleles that account for the amelioration of both the hearing and retinal degeneration phenotypes of the tubby mouse (Maddox et al. 2012).
The use of ONL thickness as a quantitative trait that reflects retinal damage to perform linkage analysis led to the successful identification of a protective modifier in the Rpe65 mouse model for light-induced photoreceptor damage, corresponding to human LCA (Akhmedov et al. 2000). The two strains compared were C57BL/6J-c(2J) (c2J) albino mice that showed less damage to their photoreceptors after exposure to prolonged light than BALB/c mice and other albino strains, suggesting that the c2J strain was contributing protective allele(s) (Danciger et al. 2000). The strongest linkage signal was on chromosome 3; subsequent analyses let to the mapping of Rpe65, in which p.Leu450 was present on the BALB/c strain and was associated with light-induced photoreceptor damage, although the c2J strain carried the p.Met450 that appeared to be driving the protection against light-induced damage (Danciger et al. 2000). The protective effect of Rpe65 p.Met450 was also confirmed in other forms of retinal degeneration (Samardzija et al. 2006) and was supported further by introducing the allele in a c-Fos−/− mouse (Wenzel et al. 2003). C-Fos is essential for light-induced photoreceptor apoptosis because of a sustained increase in the DNA-binding activity of AP-1 (Hafezi et al. 1997; Wenzel et al. 2000) and as a result c-Fos−/− mice appear resistant to light-induced damage (Wenzel et al. 2000). When the endogenous Rpe65 gene (carrying p.Met450) in c-Fos−/− mice is substituted by Rpe65 p.Leu450, Rpe65 p.Leu450 resulted in an increase in the levels of rhodopsin and therefore led to light-induced photoreceptor loss despite the protective role of c-Fos deficiency (Wenzel et al. 2003).
A third successful application of linkage analysis for the mapping of a quantitative trait locus (QTL) was performed in Rs1 mouse model for X-linked retinoschisis (XLRS) (George et al. 1995; Johnson et al. 2010). XLRS is a form of juvenile-onset macular degeneration, whereby there is splitting of the retina (schisis) and loss of synaptic transmission (George et al. 1995; Johnson et al. 2010). When C57BL/6J Rs1tmgc1 mice were backcrossed into the AKR/J background, a recessive modifier seemed to be protecting Rs1tmgc1 mice from retinoschisis (George et al. 1995; Johnson et al. 2010). Linkage analysis identified the modifier for Rs1 (Mor1) locus on chromosome 7 (Johnson et al. 2010). Within the critical interval mapped in AKR/J Rs1tmgc1 mice, a recessive mutation in tyrosinase (Tyrc-2J) was identified as the Mor1 gene responsible for protection from the development of the schisis phenotype (George et al. 1995; Johnson et al. 2010).
CONCLUSIONS AND FUTURE PERSPECTIVES
Knowledge of the primary disease-causing mutation is rarely coupled by accurate prognosis. In many instances, the clinical phenotype of a patient with an identified driver mutation can either be ameliorated on the presence of protective alleles or exacerbated when disease-promoting alleles are present in the genome. Despite the numerous reports supporting the presence and importance of genetic modifiers, this field has not progressed as rapidly compared with the identification of primary disease drivers. One of the reasons for this difference, is our inability to recognize interactions among alleles and variants (Eichler et al. 2010). This is true not only for disorders considered to be monogenic but also for complex traits, in which “missing heritability,” the fraction of the genetic contribution not explained/explainable by single-locus analysis for rare or common alleles, has been attributed, in part, to the presence of locus interactions and modifiers (Marchini et al. 2005; Zuk et al. 2012). It is possible to consider epistasis and genetic modification to uncover additional genetic contributions; in a study of ankylosing spondylitis, a common form of inflammatory arthritis, nonancestral ERAP1 alleles have been shown to interact with disease predisposition alleles in HLA-B27 to confer protection from disease manifestation (Evans et al. 2011). The protective role of ERAP1 was validated further in a second multifactorial disorder, psoriasis, uncovering the interaction of this gene with HLA-Cw6, one of the major psoriasis susceptibility genes (Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2, 2010). Despite the advances in the sequencing technologies, the number of successful genetic modifier identification studies remains small to date owing to many reasons such as the limited disease population sizes, genetic heterogeneity, the low frequency at which any particular combination of alleles is present in the population, and incomplete clinical descriptions (Flint and Mackay 2009). Animal models have provided a powerful tool for the identification of genetic modifiers. Congenic mouse strains have facilitated the mapping of protective and disease-aggravating alleles in a number of cases, some of which have been illustrated here. Zebrafish in vivo assays have aided in the functional annotation and direction of effect of candidate alleles arising by such screens.
The most efficient method to unravel phenomena of genetic modification and oligogenic or complex inheritance, is to interrogate the identified candidate alleles starting from a discrete number of likely interacting loci. Higher throughput technologies provide a unique opportunity toward this goal; a complete genetic profile of cohorts or even specific cell types such as photoreceptors in retinal disorders can be generated through whole-exome or whole-genome sequencing providing the opportunity to mind the same data set multiple times to dissect specific and discrete endophenotypes associated with a genetic condition and its progression, and the development of induced pluripotent stem (iPS) cell technology offers the promise to provide differentiated products from individual human subjects. Although each of these methodologies appear promising, the majority of successful genetic modifier identification reports have relied on the combinatorial use of such tools. Shedding light into the field of genetic modifiers will lead to a more insightful comprehension of the affected pathways and cellular processes. Understanding the basis of disease is the only way toward designing new and more efficient therapeutic paradigms, improve prognosis and even prevent disease manifestation by attempting to mimic the effect of modifiers that affect disease penetrance.
Footnotes
Editors: Eric A. Pierce, Richard H. Masland, and Joan W. Miller
Additional Perspectives on Retinal Disorders: Genetic Approaches to Diagnosis and Treatment available at www.perspectivesinmedicine.org
REFERENCES
- Ackermann B, Kröber S, Torres-Benito L, Borgmann A, Peters M, Hosseini Barkooie SM, Tejero R, Jakubik M, Schreml J, Milbradt J, et al. 2013. Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum Mol Genet 22: 1328–1347. [DOI] [PubMed] [Google Scholar]
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, et al. 2000. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci 97: 5551–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M, Smith R, Savinova O, Hawes N, Chang B, Zabaleta A, Wilpan R, Heckenlively J, Davisson M, John S. 2001. Genetic modification of glaucoma associated phenotypes between AKXD-28/Ty and DBA/2J mice. BMC Genet 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfelstedt-Sylla E, Theischen M, Rüther K, Wedemann H, Gal A, Zrenner E. 1995. Extensive intrafamilial and interfamilial phenotypic variation among patients with autosomal dominant retinal dystrophy and mutations in the human RDS/peripherin gene. Br J Ophthalmol 79: 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts HH, Doherty D, van Beersum SEC, Parisi MA, Letteboer SJF, Gorden NT, Peters TA, Marker T, Voesenek K, Kartono A, et al. 2007. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet 39: 882–888. [DOI] [PubMed] [Google Scholar]
- Astuto LM, Bork JM, Weston MD, Askew JW, Fields RR, Orten DJ, Ohliger SJ, Riazuddin S, Morell RJ, Khan S, et al. 2002. CDH23 mutation and phenotype heterogeneity: A profile of 107 diverse families with Usher Syndrome and nonsyndromic deafness. Am J Hum Genet 71: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L, Audollent S, Martinovic J, Ozilou C, Babron M-C, Sivanandamoorthy S, Saunier S, Salomon R, Gonzales M, Rattenberry E, et al. 2007. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet 81: 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Katsanis N. 2002. Beyond Mendel: An evolving view of human genetic disease transmission. Nat Rev Genet 3: 779–789. [DOI] [PubMed] [Google Scholar]
- Badano JL, Kim JC, Hoskins BE, Lewis RA, Ansley SJ, Cutler DJ, Castellan C, Beales PL, Leroux MR, Katsanis N. 2003. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet–Biedl patients with two mutations at a second BBS locus. Hum Mol Genet 12: 1651–1659. [DOI] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. 2006. The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 7: 125–148. [DOI] [PubMed] [Google Scholar]
- Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. 1999. New criteria for improved diagnosis of Bardet-Biedl syndrome: Results of a population survey. J Med Genet 36: 437–446. [PMC free article] [PubMed] [Google Scholar]
- Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, Mein CA, Froguel P, Scambler PJ, Lewis RA, et al. 2003. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet 72: 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein PS, Tammur J, Singh N, Hutchinson A, Dixon M, Pappas CM, Zabriskie NA, Zhang K, Petrukhin K, Leppert M, et al. 2001. Diverse macular dystrophy phenotype caused by a novel complex mutation in the ELOVL4 gene. Invest Ophthalmol Vis Sci 42: 3331–3336. [PubMed] [Google Scholar]
- Bolk S, Pelet A, Hofstra RMW, Angrist M, Salomon R, Croaker D, Buys CHCM, Lyonnet S, Chakravarti A. 2000. A human model for multigenic inheritance: Phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci 97: 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, Cabrera MdCS, Vila MC, Molina OP, et al. 2001. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet 27: 108–112. [DOI] [PubMed] [Google Scholar]
- Bone-Larson C, Basu S, Radel JD, Liang M, Perozek T, Kapousta-Bruneau N, Green DG, Burmeister M, Hankin MH. 2000. Partial rescue of the ocular retardation phenotype by genetic modifiers. J Neurobiol 42: 232–247. [DOI] [PubMed] [Google Scholar]
- Borrego S, Ruiz A, Saez ME, Gimm O, Gao X, López-Alonso M, Hernández A, Wright FA, Antiñolo G, Eng C. 2000. RET genotypes comprising specific haplotypes of polymorphic variants predispose to isolated Hirschsprung disease. J Med Genet 37: 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JP, Wright AF. 2000. Identification of a novel protein interacting with RPGR. Hum Mol Genet 9: 2085–2093. [DOI] [PubMed] [Google Scholar]
- Brattgard S. 1949. The pathology of the Laurence-Moon-Biedl syndrome. Acta Path Microbiol Scand 26: 525–537. [PubMed] [Google Scholar]
- Brenman JE, Topinka JR, Cooper EC, McGee AW, Rosen J, Milroy T, Ralston HJ, Bredt DS. 1998. Localization of postsynaptic density-93 to dendritic microtubules and interaction with microtubule-associated protein 1A. J Neurosci 18: 8805–8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi R, Elbedour K, Stone EM, Sheffield VC. 1995. Phenotypic differences among patients with Bardet-Biedl syndrome linked to three different chromosome loci. Am J Med Genet 59: 199–203. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, McCallion AS, Puffenberger EG, Kashuk CS, Nouri N, Chakravarti A. 2002. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet 32: 237–244. [DOI] [PubMed] [Google Scholar]
- Chang B, Smith RS, Hawes NL, Anderson MG, Zabaleta A, Savinova O, Roderick TH, Heckenlively JR, Davisson MT, John SWM. 1999. Interacting loci cause severe iris atrophy and glaucoma in DBA/2J mice. Nat Genet 21: 405–409. [DOI] [PubMed] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, et al. 2006. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet 15: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Simon MI, Matthes MT, Yasumura D, LaVail MM. 1999. Increased susceptibility to light damage in an arrestin knockout mouse model of Oguchi disease (stationary night blindness). Invest Ophthalmol Vis Sci 40: 2978–2982. [PubMed] [Google Scholar]
- Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Bruning JC, Nolan PM, Ashcroft FM, et al. 2010. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet 42: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DL, Eicher EM. 1990. Fat (fat) and tubby (tub): Two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered 81: 424–427. [DOI] [PubMed] [Google Scholar]
- Coppieters F, Casteels I, Meire F, De Jaegere S, Hooghe S, van Regemorter N, Van Esch H, Matulevičienė A, Nunes L, Meersschaut V, et al. 2010. Genetic screening of LCA in Belgium: Predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum Mutat 31: E1709–E1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting GR. 2010. Modifier genes in Mendelian disorders: The example of cystic fibrosis. Ann NY Acad Sci 1214: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeizel AE, Dudás I. 1992. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N Engl J Med 327: 1832–1835. [DOI] [PubMed] [Google Scholar]
- Danciger M, Matthes MT, Yasamura D, Akhmedov NB, Rickabaugh T, Gentleman S, Redmond TM, La Vail MM, Farber DB. 2000. A QTL on distal Chromosome 3 that influences the severity of light-induced damage to mouse photoreceptors. Mamm Genome 11: 422–427. [DOI] [PubMed] [Google Scholar]
- Danciger M, Ogando D, Yang H, Matthes MT, Yu N, Ahern K, Yasumura D, Williams RW, LaVail MM. 2008. Genetic modifiers of retinal degeneration in the rd3 mouse. Invest Ophthalmol Vis Sci 49: 2863–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah R, McKone E, O’Connor C, Rodgers C, Genatossio A, McNamara S, Gibson R, Stuart Elborn J, Ennis M, et al. 2010. EDNRA variants associate with smooth muscle mRNA levels, cell proliferation rates, and cystic fibrosis pulmonary disease severity. Physiol Genomics 41: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EE, Katsanis N. 2012. The ciliopathies: A transitional model into systems biology of human genetic disease. Curr Opin Genet Dev 22: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M, Baala L, Salomon R, Laclef C, Vierkotten J, Tory K, Golzio C, Lacoste T, Besse L, Ozilou C, et al. 2007. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet 39: 875–881. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KEJ, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, et al. 2006. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 79: 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Roepman R, Koenekoop RK, Cremers FPM. 2008. Leber congenital amaurosis: Genes, proteins and disease mechanisms. Prog Retin Eye Res 27: 391–419. [DOI] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C, et al. 2007. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 39: 724–726. [DOI] [PubMed] [Google Scholar]
- Dipple KM, McCabe ERB. 2000. Phenotypes of patients with “simple” Mendelian disorders are complex traits: Thresholds, modifiers, and systems dynamics. Am J Hum Genet 66: 1729–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes SM, Holder GE, Fitzke FW, et al. 2001. Autosomal dominant cone and cone–rod dystrophy with mutations in the guanylate cyclase activator 1a gene-encoding guanylate cyclase activating protein-1. Arch Ophthalmol 119: 96–105. [PubMed] [Google Scholar]
- Drumm ML, Konstan MW, Schluchter MD, Handler A, Pace R, Zou F, Zariwala M, Fargo D, Xu A, Dunn JM, et al. 2005. Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med 353: 1443–1453. [DOI] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Hahn LB, Cowley GS, Olsson JE, Reichel E, Sandberg MA, Berson EL. 1990. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med 323: 1302–1307. [DOI] [PubMed] [Google Scholar]
- Ebermann I, Scholl HN, Charbel Issa P, Becirovic E, Lamprecht J, Jurklies B, Millán J, Aller E, Mitter D, Bolz H. 2007. A novel gene for Usher syndrome type 2: Mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet 121: 203–211. [DOI] [PubMed] [Google Scholar]
- Ebermann I, Phillips JB, Liebau MC, Koenekoop RK, Schermer B, Lopez I, Schaefer E, Roux A-F, et al. 2010. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J Clin Invest 120: 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11: 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. 2005. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434: 857–863. [DOI] [PubMed] [Google Scholar]
- Evans DM, Spencer CCA, Pointon JJ, Su Z, Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, et al. 2011. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet 43: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahim AT, Bowne SJ, Sullivan LS, Webb KD, Williams JT, Wheaton DK, Birch DG, Daiger SP. 2011. Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS ONE 6: e23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felbor U, Benkwitz C, Klein ML, Greenberg J, Gregory CY, Weber BF. 1997. Sorsby fundus dystrophy: Reevaluation of variable expressivity in patients carrying a TIMP3 founder mutation. Arch Ophthalmol 115: 1569–1571. [DOI] [PubMed] [Google Scholar]
- Flint J, Mackay TFC. 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res 19: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank V, Ortiz Brüchle N, Mager S, Frints SGM, Bohring A, du Bois G, Debatin I, Seidel H, Senderek J, Besbas N, et al. 2007. Aberrant splicing is a common mutational mechanism in MKS1, a key player in Meckel-Gruber syndrome. Hum Mutat 28: 638–639. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, et al. 2007. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316: 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frio TR, Civic N, Ransijn A, Beckmann JS, Rivolta C. 2008. Two trans-acting eQTLs modulate the penetrance of PRPF31 mutations. Hum Mol Genet 17: 3154–3165. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Salomon R, Pelet A, Angrist M, Amiel J, Fornage M, Attie-Bitach T, Olson JM, Hofstra R, Buys C, et al. 2002. Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet 31: 89–93. [DOI] [PubMed] [Google Scholar]
- Garcia-Barceló M-M, Sham M-H, Lui VC-H, Chen BL-S, Song Y-Q, Lee W-S, Yung S-K, Romeo G, Tam PK-H. 2003. Chinese patients with sporadic Hirschsprung’s disease are predominantly represented by a single RET haplotype. J Med Genet 40: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garred P, Pressler T, Madsen HO, Frederiksen B, Svejgaard A, Fiby N, Schwartz M, Koch C. 1999. Association of mannose-binding lectin gene heterogeneity with severity of lung disease and survival in cystic fibrosis. J Clin Invest 104: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2, Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, et al. 2010. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Génin E, Feingold J, Clerget-Darpoux F. 2008. Identifying modifier genes of monogenic disease: Strategies and difficulties. Hum Genet 124: 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George ND, Yates JR, Moore AT. 1995. X linked retinoschisis. Br J Ophthalmol 79: 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni-Baruch R, Nachlieli T, Leibo R, Degani S, Weissman I. 1992. Cystic kidney dysplasia and polydactyly in 3 sibs with Bardet-Biedl syndrome. Am J Med Genet 44: 269–273. [DOI] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. 2006. The ciliary proteome database: An integrated community resource for the genetic and functional dissection of cilia. Nat Genet 38: 961–962. [DOI] [PubMed] [Google Scholar]
- Gibson G. 2012. Rare and common variants: Twenty arguments. Nat Rev Genet 13: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L. 2007. The human disease network. Proc Natl Acad Sci 104: 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AFX, Molday RS. 1996. Defective subunit assembly underlies a digenic form of retinitis pigmentosa linked to mutations in peripherin/rds and rom-1. Proc Natl Acad Sci 93: 13726–13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss CH, Newsom SA, Schildcrout JS, Sheppard L, Kaufman JD. 2004. Effect of ambient air pollution on pulmonary exacerbations and lung function in cystic fibrosis. Am J Respir Crit Care Med 169: 816–821. [DOI] [PubMed] [Google Scholar]
- Gouya LDJ, Lamoril J, Da Silva V, Beaumont C, Grandchamp B, Nordmann Y. 1996. Modulation of the phenotype in dominant erythropoietic protoporphyria by a low expression of the normal ferrochelatase allele. Am J Hum Genet 58: 292–299. [PMC free article] [PubMed] [Google Scholar]
- Gouya L, Puy H, Robreau A-M, Bourgeois M, Lamoril J, Da Silva V, Grandchamp B, Deybach J-C. 2002. The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH. Nat Genet 30: 27–28. [DOI] [PubMed] [Google Scholar]
- Gouya L, Martin-Schmitt C, Robreau A-M, Austerlitz F, Da Silva V, Brun P, Simonin S, Lyoumi S, Grandchamp B, Beaumont C, et al. 2006. Contribution of a common single-nucleotide polymorphism to the genetic predisposition for erythropoietic protoporphyria. Am J Hum Genet 78: 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunnet LG, Brøns C, Jacobsen S, Nilsson E, Astrup A, Hansen T, Pedersen O, Poulsen P, Quistorff B, Vaag A. 2009. Increased recovery rates of phosphocreatine and inorganic phosphate after isometric contraction in oxidative muscle fibers and elevated hepatic insulin resistance in homozygous carriers of the A-allele of FTO rs9939609. J Clin Endocrinol Metab 94: 596–602. [DOI] [PubMed] [Google Scholar]
- Gusella J, MacDonald M. 2009. Huntington’s disease: The case for genetic modifiers. Genome Med 1: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafezi F, Steinbach JP, Marti A, Munz K, Wang ZQ, Wagner EF, Aguzzi A, Remé CE. 1997. The absence of c-fos prevents light-induced apoptotic cell death of photoreceptors in retinal degeneration in vivo. Nat Med 3: 346–349. [DOI] [PubMed] [Google Scholar]
- Haider N, Zhang W, Hurd R, Ikeda A, Nystuen A, Naggert J, Nishina P. 2008. Mapping of genetic modifiers of Nr2e3rd7/rd7 that suppress retinal degeneration and restore blue cone cells to normal quantity. Mamm Genome 19: 145–154. [DOI] [PubMed] [Google Scholar]
- Haldane J. 1941. The relative importance of principal and modifying genes in determining some human diseases. J Genet 41: 149–157. [Google Scholar]
- Halpain S, Dehmelt L. 2006. The MAP1 family of microtubule-associated proteins. Genome Biol 7: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WK, Ewing CC, Ives EJ, Carruthers JD. 1989. Sorsby’s Fundus Dystrophy. Ophthalmology 96: 1755–1762. [DOI] [PubMed] [Google Scholar]
- Heckenlively JR, Chang B, Peng C, Hawes NL, Roderick TH. 1993. Variable expressivity of rd-3 retinal degeneration dependent on background strain. In Retinal degeneration (ed. Hollyfield JG, Anderson RE, LaVail MM), pp. 273–280. Plenum, New York. [Google Scholar]
- Heckenlively JR, Chang B, Erway LC, Peng C, Hawes NL, Hageman GS, Roderick TH. 1995. Mouse model for Usher syndrome: Linkage mapping suggests homology to Usher type I reported at human chromosome 11p15. Proc Natl Acad Sci 92: 11100–11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillian AD, Londono D, Dunn JM, Goddard KAB, Pace RG, Knowles MR, Drumm ML. 2008. Modulation of cystic fibrosis lung disease by variants in interleukin-8. Genes Immun 9: 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H-K, Lass JH, Chakravarti A. 1999. Pleiotropic skeletal and ocular phenotypes of the mouse mutation congenital hydrocephalus (ch/Mf1) arise from a winged helix/forkhead transcription factor gene. Hum Mol Genet 8: 625–637. [DOI] [PubMed] [Google Scholar]
- Humphries MM, Kiang S, McNally N, Donovan MA, Sieving PA, Bush RA, Machida S, Cotter T, Hobson A, Farrar J, et al. 2001. Comparative structural and functional analysis of photoreceptor neurons of Rho−/− mice reveal increased survival on C57BL/6J in comparison to 129Sv genetic background. Vis Neurosci 18: 437–443. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Yin Zheng Q, Rosenstiel P, Maddatu T, Zuberi AR, Roopenian DC, North MA, Naggert JK, Johnson KR, Nishina PM. 1999. Genetic modification of hearing in tubby mice: Evidence for the existence of a major gene (moth1) which protects tubby mice from hearing loss. Hum Mol Genet 8: 1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Naggert JK, Nishina PM. 2002a. Genetic modification of retinal degeneration in tubby mice. Exp Eye Res 74: 455–461. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Zheng QY, Zuberi AR, Johnson KR, Naggert JK, Nishina PM. 2002b. Microtubule-associated protein 1A is a modifier of tubby hearing (moth1). Nat Genet 30: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Iannaccone A, Weleber RG, Fishman GA, Maguire AM, Affatigato LM, Bennett J, Pierce EA, Danciger M, et al. 2000. Disease expression of RP1 mutations causing autosomal dominant retinitis pigmentosa. Invest Opthalmol Vis Sci 41: 1898–1908. [PubMed] [Google Scholar]
- Johnson BA, Cole BS, Geisert EE, Ikeda S, Ikeda A. 2010. Tyrosinase is the modifier of retinoschisis in mice. Genetics 186: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K, Sandberg MA, Berson EL, Dryja TP. 1993. A null mutation in the human peripherin/RDS gene in a family with autosomal dominant retinitis punctata albescens. Nat Genet 3: 208–212. [DOI] [PubMed] [Google Scholar]
- Kajiwara K, Berson E, Dryja T. 1994. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 264: 1604–1608. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. 2001. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293: 2256–2259. [DOI] [PubMed] [Google Scholar]
- Keen TJ, Himms MM, McKie AB, Moore AT, Doran RM, Mackey DA, Mansfield DC, Mueller RF, Bhattacharya SS, Bird AC, et al. 2002. Mutations in a protein target of the Pim-1 kinase associated with the RP9 form of autosomal dominant retinitis pigmentosa. Eur J Hum Genet 10: 245–249. [DOI] [PubMed] [Google Scholar]
- Kerem BS, Zielenski J, Markiewicz D, Bozon D, Gazit E, Yahav J, Kennedy D, Riordan JR, Collins FS, Rommens JM. 1990. Identification of mutations in regions corresponding to the two putative nucleotide (ATP)-binding folds of the cystic fibrosis gene. Proc Natl Acad Sci 87: 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna H, Davis EE, Murga-Zamalloa CA, Estrada-Cuzcano A, Lopez I, den Hollander AI, Zonneveld MN, Othman MI, Waseem N, Chakarova CF, et al. 2009. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet 41: 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RY, Fitzke FW, Moore AT, Jay M, Inglehearn C, Arden GB, Bhattacharya SS, Bird AC. 1995. Autosomal dominant retinitis pigmentosa mapping to chromosome 7p exhibits variable expression. Br J Ophthalmol 79: 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöting N, Schleinitz D, Ruschke K, Berndt J, Fasshauer M, Tönjes A, Schön MR, Kovacs P, Stumvoll M, Blüher M. 2008. Inverse relationship between obesity and FTO gene expression in visceral adipose tissue in humans. Diabetologia 51: 641–647. [DOI] [PubMed] [Google Scholar]
- Kremer H, van Wijk E, Märker T, Wolfrum U, Roepman R. 2006. Usher syndrome: Molecular links of pathogenesis, proteins and pathways. Hum Mol Genet 15: R262–R270. [DOI] [PubMed] [Google Scholar]
- Labrune M, Gaux JC, Brault B. 1974. The Laurence-Moon-Bardet-Biedl syndrome. Urographic signs. Ann Radiol 17: 385–389. [PubMed] [Google Scholar]
- Landau J, Bromberg Y, Schorr J. 1949. Laurence-Moon-Biedl syndrome with mulitple congenital malformations of the urinary tract. Acta Med Orient 8: 205–210. [Google Scholar]
- Laurence KM, James N, Miller M, Campbell H. 1980. Increased risk of recurrence of pregnancies complicated by fetal neural tube defects in mothers receiving poor diets, and possible benefit of dietary counselling. BMJ 281: 1592–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, Alfadhel M, Lewis RA, Eyaid W, Banin E, et al. 2008. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet 40: 443–448. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552. [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual J-F, Fisk CJ, Li N, Smolyar A, Hill DE, et al. 2006. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125: 801–814. [DOI] [PubMed] [Google Scholar]
- Liu Q, Saveliev A, Pierce EA. 2009. The severity of retinal degeneration in Rp1h gene-targeted mice is dependent on genetic background. Invest Ophthalmol Vis Sci 50: 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJR, Moritz OL, Molday RS. 2001. Molecular characterization of peripherin-2 and rom-1 mutants responsible for digenic retinitis pigmentosa. J Biol Chem 276: 22388–22396. [DOI] [PubMed] [Google Scholar]
- Louie CM, Caridi G, Lopes VS, Brancati F, Kispert A, Lancaster MA, Schlossman AM, Otto EA, Leitges M, Grone H-J, et al. 2010. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet 42: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon AN, Pineda RH, Hao lT, Kudryashova E, Kudryashov DS, Beattie CE. 2014. Calcium binding is essential for plastin 3 function in Smn-deficient motoneurons. Hum Mol Genet 23: 1990–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox DM, Ikeda S, Ikeda A, Zhang W, Krebs MP, Nishina PM, Naggert JK. 2012. An allele of microtubule-associated protein 1A (Mtap1a) reduces photoreceptor degeneration in Tulp1 and Tub mutant mice. Invest Ophthalmol Vis Sci 53: 1663–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro F, Peres E. 1970. Nefropathia do sindrome do Laurence–Moon–Bardet–Biedl. Revista Iberica de Endocrinologia 17: 243–253. [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. 2005. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet 37: 413–417. [DOI] [PubMed] [Google Scholar]
- Margolin DH, Kousi M, Chan Y-M, Lim ET, Schmahmann JD, Hadjivassiliou M, Hall JE, Adam I, Dwyer A, Plummer L, et al. 2013. Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N Engl J Med 368: 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam P, Farrar GJ, Kenna P, Bradley DG, Humphries MM, Sharp EM, McConnell DJ, Lawler M, Sheils D, Ryan C, et al. 1989. Autosomal dominant retinitis pigmentosa (ADRP): Localization of an ADRP gene to the long arm of chromosome 3. Genomics 5: 619–622. [DOI] [PubMed] [Google Scholar]
- Naash ML, Peachey NS, Li ZY, Gryczan CC, Goto Y, Blanks J, Milam AH, Ripps H. 1996. Light-induced acceleration of photoreceptor degeneration in transgenic mice expressing mutant rhodopsin. Invest Ophthalmol Vis Sci 37: 775–782. [PubMed] [Google Scholar]
- Nakazawa M, Wada Y, Tamai M. 1998. Arrestin gene mutations in autosomal recessive retinitis pigmentosa. Arch Ophthalmol 116: 498–501. [DOI] [PubMed] [Google Scholar]
- Ness SL, Ben-Yosef T, Bar-Lev A, Madeo AC, Brewer CC, Avraham KB, Kornreich R, Desnick RJ, Willner JP, Friedman TB, et al. 2003. Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet 40: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström S, Thorburn W. 1980. Dominantly inherited macular degeneration (Best’s disease) in a homozygous father with 11 children. Clin Genet 18: 211–216. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Hughes RM, Lett JM, Ogilvie JM, Speck JD, Wright JS, Faddis BT. 1997. Progression of cochlear and retinal degeneration in the tubby (rd5) mouse. Audiol Neurootol 2: 175–185. [DOI] [PubMed] [Google Scholar]
- Oprea GE, Kröber S, McWhorter ML, Rossoll W, Müller S, Krawczak M, Bassell GJ, Beattie CE, Wirth B. 2008. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 320: 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, et al. 2005. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet 37: 282–288. [DOI] [PubMed] [Google Scholar]
- Perrault I, Delphin N, Hanein S, Gerber S, Dufier J-L, Roche O, Defoort-Dhellemmes S, Dollfus H, Fazzi E, Munnich A, et al. 2007. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum Mutat 28: 416–416. [DOI] [PubMed] [Google Scholar]
- Poloschek CM, Bach M, Lagrèze WA, Glaus E, Lemke JR, Berger W, Neidhardt J. 2010. ABCA4 and ROM1: Implications for modification of the PRPH2-associated macular dystrophy phenotype. Invest Ophthalmol Vis Sci 51: 4253–4265. [DOI] [PubMed] [Google Scholar]
- Rachel RA, May-Simera HL, Veleri S, Gotoh N, Choi BY, Murga-Zamalloa C, McIntyre JC, Marek J, Lopez I, Hackett AN, et al. 2012. Combining Cep290 and Mkks ciliopathy alleles in mice rescues sensory defects and restores ciliogenesis. J Clin Ivest 122: 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Iartchouk O, Chin K, Tan PL, Tai AK, Ripke S, Gowrisankar S, Vemuri S, Montgomery K, Yu Y, et al. 2011. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet 43: 1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin SA, Shahzadi A, Zeitz C, Ahmed ZM, Ayyagari R, Chavali VRM, Ponferrada VG, Audo I, Michiels C, Lancelot M-E, et al. 2010. A mutation in SLC24A1 implicated in autosomal-recessive congenital stationary night blindness. Am J Hum Genet 87: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riise R, Andréasson S, Borgström MK, Wright AF, Tommerup N, Rosenberg T, Tornqvist K. 1997. Intrafamilial variation of the phenotype in Bardet–Biedl syndrome. Br J Ophthalmol 81: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, Sharon D, DeAngelis MM, Dryja TP. 2002. Retinitis pigmentosa and allied diseases: Numerous diseases, genes, and inheritance patterns. Hum Mol Genet 11: 1219–1227. [DOI] [PubMed] [Google Scholar]
- Rubin BK. 1990. Exposure of children with cystic fibrosis to environmental tobacco smoke. N Engl J Med 323: 782–788. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, McCluskey M, Wensel TG, Naggert JK, Nishina PM. 2009. New mouse models for recessive retinitis pigmentosa caused by mutations in the Pde6a gene. Hum Mol Genet 18: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samardzija M, Wenzel A, Naash M, Remé CE, Grimm C. 2006. Rpe65 as a modifier gene for inherited retinal degeneration. Eur J Neurosci 23: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancandi M, Griseri P, Pesce B, Patrone G, Puppo F, Lerone M, Martucciello G, Romeo G, Ravazzolo R, Devoto M, et al. 2003. Single nucleotide polymorphic alleles in the 5′ region of the RET proto-oncogene define a risk haplotype in Hirschsprung’s disease. J Med Genet 40: 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Nakazawa M, Usui T, Tanimoto N, Abe H, Ohguro H. 2005. Mutations in the gene coding for guanylate cyclase-activating protein 2 (GUCA1B gene) in patients with autosomal dominant retinal dystrophies. Graefes Arch Clin Exp Ophthalmol 243: 235–242. [DOI] [PubMed] [Google Scholar]
- Sayer JA, Otto EA, O’Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, et al. 2006. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet 38: 674–681. [DOI] [PubMed] [Google Scholar]
- Schäffer AA. 2013. Digenic inheritance in medical genetics. J Med Genet 50: 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen W-M, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orrú M, Usala G, et al. 2007. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 3: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Yu Y, Miller EC, Reynolds R, Tan PL, Gowrisankar S, Goldstein JI, Triebwasser M, Anderson HE, Zerbib J, et al. 2013. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet 45: 1366–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. 2014. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speilman RS, McGinnis RE, Ewens WJ. 1993. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52: 506–516. [PMC free article] [PubMed] [Google Scholar]
- Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, Badano JL, Leitch CC, Salem N, Chouery E, Corbani S, et al. 2006. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet 38: 521–524. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Muller J, Laurier V, Davis EE, Zaghloul NA, Vicaire S, Jacquelin C, Plewniak F, Leitch CC, Sarda P, et al. 2007. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet 80: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantri A, Vrabec TR, Cu-Unjieng A, Frost A, Annesley WH Jr, Donoso LA. 2003. X-linked retinoschisis: Report of a family with a rare deletion in the xLRS1 gene. Am J Ophthalmol 136: 547–549. [DOI] [PubMed] [Google Scholar]
- Tolarova M. 1982. Periconceptional supplementation with vitamins and folic acid to prevent recurrence of cleft lip. Lancet 320: 217. [DOI] [PubMed] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, et al. 2006. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet 38: 623–625. [DOI] [PubMed] [Google Scholar]
- Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, Algire M, McWilliams R, Beck S, Hoover-Fong J, et al. 2007. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med 175: 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini G, Rose AM, Shah AZ, Bhattacharya SS, Rivolta C. 2012. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet 8: e1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald NJ, Law MR, Morris JK, Wald DS. 2001. Quantifying the effect of folic acid. Lancet 358: 2069–2073. [DOI] [PubMed] [Google Scholar]
- Walia S, Fishman GA, Swaroop A, Branham KEH, Lindemena M, Othman M, Weleber RG. 2008. Discordant phenotypes in fraternal twins having an identical mutation in exon ORF15 of the RPGR gene. Arch Ophthalmol 126: 379–384. [DOI] [PubMed] [Google Scholar]
- Wang M, Lam TT, Tso MOM, Naash MI. 1997. Expression of a mutant opsin gene increases the susceptibility of the retina to light damage. Vis Neurosci 14: 55–62. [DOI] [PubMed] [Google Scholar]
- Weber BHF, Vogt G, Pruett RC, Stohr H, Felbor U. 1994. Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby’s fundus dystrophy. Nat Genet 8: 352–356. [DOI] [PubMed] [Google Scholar]
- Weleber RG, Carr RE, Murphey WH, Sheffield VC, Stone EM. 1993. Phenotypic variation including retinitis pigmentosa, pattern dystrophy, and fundus flavimaculatus in a single family with a deletion of codon 153 or 154 of the peripherin/RDS gene. Arch Ophthalmol 111: 1531–1542. [DOI] [PubMed] [Google Scholar]
- Wenzel A, Grimm C, Marti A, Kueng-Hitz N, Hafezi F, Niemeyer G, Remé CE. 2000. c-fos controls the “private pathway” of light-induced apoptosis of retinal photoreceptors. J Neurosci 20: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A, Grimm C, Samardzija M, Remé CE. 2003. The genetic modifier Rpe65Leu450: Effect on light damage susceptibility in c-Fos-deficient mice. Invest Ophthalmol Vis Sci 44: 2798–2802. [DOI] [PubMed] [Google Scholar]
- Wijesuriya SD, Evans K, Jay MR, Davison C, Weber BH, Bird AC, Bhattacharya SS, Gregory CY. 1996. Sorsby’s fundus dystrophy in the British Isles: Demonstration of a striking founder effect by microsatellite-generated haplotypes. Genome Res 6: 92–101. [DOI] [PubMed] [Google Scholar]
- Zernant J, Külm M, Dharmaraj S, den Hollander AI, Perrault I, Preising MN, Lorenz B, Kaplan J, Cremers FPM, Maumenee I, et al. 2005. Genotyping microarray (disease chip) for Leber congenital amaurosis: Detection of modifier alleles. Invest Ophthalmolo Vis Sci 46: 3052–3059. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kniazeva M, Hutchinson A, Han M, Dean M, Allikmets R. 1999. The ABCR gene in recessive and dominant Stargardt diseases: A genetic pathway in macular degeneration. Genomics 60: 234–237. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Seo S, Bhattarai S, Bugge K, Searby CC, Zhang Q, Drack AV, Stone EM, Sheffield VC. 2014. BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum Mol Genet 23: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. 2012. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci 109: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]