Abstract
The past two decades have witnessed incredible progress toward understanding the genetic and cellular mechanisms of organogenesis. Among the organs that have provided key insight into how patterning information is integrated to specify and build functional body parts is the Drosophila salivary gland, a relatively simple epithelial organ specialized for the synthesis and secretion of high levels of protein. Here, we discuss what the past couple of decades of research have revealed about organ specification, development, specialization and death, and what general principles emerge from these studies.
Introduction
Drosophila has proven to be an ideal model system for revealing the molecular and cellular underpinnings of organ development. In particular, the salivary gland (SG) has been crucial for studying how polarized epithelial organs form and specialize. The SG starts out as two plates of approximately one hundred fifty cells each on the ventral surface of the embryo (Figure 1). Following internalization and initial tube formation, the fully polarized SG elongates and actively migrates to its final position in the embryo. Each of the secretory glands connects to an individual duct. Individual ducts connect to one another in a central common duct, which attaches to the mouthparts. The SG that forms during embryonic development persists through larval life to the beginning of pupation, when it produces a final burst of secretion before being destroyed during metamorphosis. Subsequently, the imaginal ring cells, which are found between the SG duct and secretory cells and that proliferate during larval and pupal stages, form the adult gland.
Figure 1.
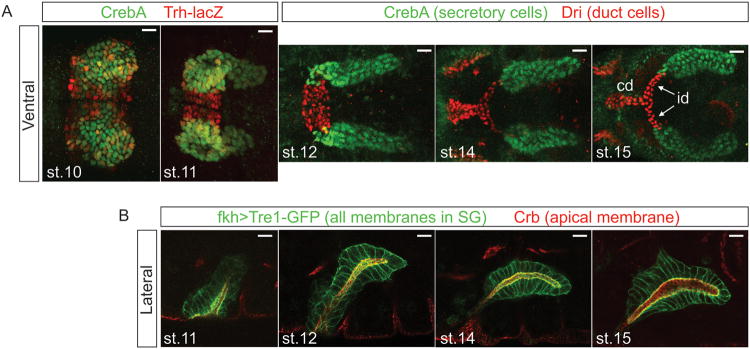
Confocal images of the embryonic SG. (A) Ventral views of the SG stained with nuclear markers. The secretory portion of the SG forms from two placodes of cells on the surface of the embryo, with the duct precursors located between the two secretory placodes (st 10). At this stage, the expression of duct (red) versus secretory (green) markers is not so clear. During st 11, the gland invaginates into the embryo. At this stage, the distinction between duct (red) and secretory markers (green) is more evident. As development progresses (st 12-15), the secretory tubes elongate, and the individual duct (id) and common ducts (cd) form. (B) Lateral views of SG during elongation and migration. All membranes are marked in green, with the apical membrane specifically marked in red. Following invagination, the SG moves dorsally (st 11) and then turns (st 12) and migrates posteriorly. Posterior migration continues (st 14-16) until the SG reaches its final resting place. Throughout this dynamic process, the SG migrates as an intact fully polarized tissue.
As a model system, the SG has been a valuable platform for addressing questions of cell fate specification and maintenance, and for learning how cells coordinate their activities to build an organ of the right size, shape and position in the animal. The SG has been excellent in revealing how the sequential deployment of gene expression programs controls all aspects of epithelial tube form and function. Morphogenetic processes, including invagination, tube elongation and migration, continue to be studied in the SG, and new molecules driving these events are being uncovered. The SG has also been used extensively to study programmed cell death in the context of a living organism. In the following review, we discuss our current understanding of SG formation, maintenance and death, and how findings from these studies have provided insight into more general aspects of organ development and gene function.
Specification of Salivary Glands
SG specification and the distinction between secretory, duct and imaginal ring (pre-adult) cell populations occur through the integration of anterior-posterior patterning information, largely mediated by HOX genes, and the dorsal-ventral patterning system, specifically Dpp-signaling in dorsal cells and EGF-signaling along the ventral midline. Since SG precursors stop dividing once specified, the process of specification determines both where the primordia will arise and the final number of cells in the fully formed tissue.
Anterior-posterior patterning genes and SG specification
Scr, Exd and Hth: positive determinants of SG fates
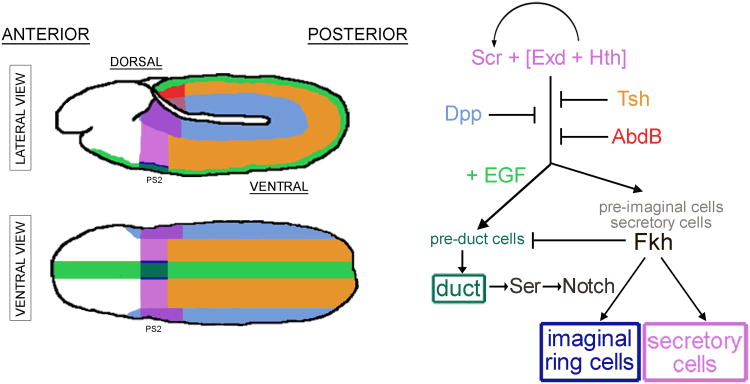
SG specification requires positive input from three transcription factors: Sex combs reduced (Scr), Extradenticle (Exd) and Homothorax (Hth) (Figure 2)1-3. SGs form from the ventral ectodermal cells of parasegment 2 (PS2), which express Scr, the only spatially limited component required to activate SG formation. Exd, a TALE (three amino acid loop extension) homeodomain protein that is expressed throughout the embryo3, is also required for SG formation. Scr and Exd bind each other and bind DNA. The physical association of Exd with Scr distorts the N-terminus of the Scr homeodomain, allowing it to fit into the relatively narrow minor groove of its target DNA sites4. Exd activity is limited by regulated nuclear entry, mediated through binding to its essential cofactor Hth5. Hth, another broadly-expressed TALE protein with highly conserved mammalian orthologues (MEIS1,2,3 and PREP1,2 proteins) is also absolutely required for SG formation2. Whether Hth is directly involved in the binding of the Scr-Exd complex to target sequences is not clear, but the homeodomain of Hth, which is present in only one of two alternative splice forms, is not required for Scr-Exd dependent activation of the single SG target reporter that has been directly tested6. Early expression of Scr in SG precursors requires both Exd and Hth, revealing that TALE proteins function at multiple levels in SG specification2. Most other Hox proteins also require Exd and Hth for target gene regulation3,7. One exception is Abdominal-B (Abd-B), which instead represses Exd and Hth expression to mediate specification of another embryonic organ in Drosophila, the posterior spiracle8,9.
Figure 2.
SGs are specified by the integration of patterning information along both major body axes. Scr (purple in cartoon), a Hox protein expressed in PS2 and dorsal cells of PS3, is the only spatially-regulated activator of SG cell fates. The more globally expressed Exd and Hth Tale-homeodomain proteins are also required for SG formation and these proteins function at multiple levels. Tsh (brown in cartoon), expressed in PS3-13, and AbdB (red in cartoon), expressed in PS14, block SG activation in the trunk and abdomen. SG formation is limited to the ventral cells of PS2 by Dpp signaling (blue in cartoon) in the dorsal cells. EGF-signaling (green in cartoon) along the ventral midline specifies the duct cell fate by blocking expression of Fkh. In turn, Fkh plays a major role in maintaining the secretory cell fate by regulating itself as well as multiple other secretory-specific genes and by blocking expression of duct-specific genes. Ser, which is expressed in duct cells, signals adjacent Notch expressing cells in the common secretory/imaginal ring cell primordia to become imaginal ring cells (dark blue in diagram) – the precursors to the adult SG.
Ectopic expression of Scr driven by heat shock or by ubiquitous Gal4 causes ectopic expression of many SG markers in all segments at early stages1,4,10,11; expression of SG markers persists, however, only in PS2 and the two parasegments more anterior, PS0 and PS1. Expression of SG markers in posterior segments is transient, disappearing completely by mid-embryogenesis11. Moreover, SG marker-expressing cells in posterior regions do not invaginate to form SGs – which does occur in the more anterior regions. Persistent expression of SG markers induced by global expression of Scr is only observed in PS3-13 in the complete absence of teashirt (tsh) function (see below)10.
Posterior Hox genes and Tsh: Negative regulators of SG fates
SG formation is further defined by negative regulation from two transcription factors. The major block to SG activation in PS3-13 is Teashirt, a zinc finger containing transcription factor (Figure 2)10,12. In the absence of Tsh, Scr early expression expands to PS3 and SGs form in PS3. A role for Tsh in blocking Scr-induced SG formation fits well with the previously described role for Tsh in distinguishing trunk from head13,14. Tsh directly regulates at least one SG target gene 12 and Tsh has been shown to physically contact Scr 15; the exact mechanisms where by Tsh prevents Scr's SG-inducing activity, however, remain unclear. Nonetheless, Tsh appears to function at two levels: repression of early Scr expression in the ventral cells of PS3 and repression of Scr's SG-inducing activities when that transcriptional regulation is overridden 10.
In PS14, Scr-induced SG gene activation is blocked by another Hox gene, Abdominal B (Abd-B) (Figure 2)10. The block by Abd-B is not absolute and may be due to “posterior prevalence”, a phenomenon wherein more posteriorly expressed Hox proteins block the activities of more anteriorly expressed Hox proteins16-21. Recent work suggests that posterior prevalence occurs because posteriorly expressed Hox proteins compete more successfully for the shared essential cofactor Exd6. Alternatively, given recent findings that Abd-B shuts off expression of Exd and Hth, the block by Abd-B could be due to the absence of these essential SG-inducing cofactors in PS148.
Dorsal-ventral patterning genes further refine SG coordinates
Although Scr, Exd and Hth drive SG formation, not all cells that express these transcription factors become SGs; SGs form only from ventral ectodermal cells. Thus, dorsal-ventral patterning information also feeds into the system. Loss of dorsal (dl), a major early determinant of ventral cell fates, results in a complete loss of SG marker expression1. Dl promotes SG development by blocking ventral expression of the gene encoding the BMP ligand Decapentaplegic (Dpp), which is normally expressed in only dorsal cells. In turn, Dpp signaling blocks SG formation (Figure 2); loss of any one component in the Dpp signaling pathway results in the dorsal expansion of all tested SG markers1,22,23.
Maintaining the SG fate
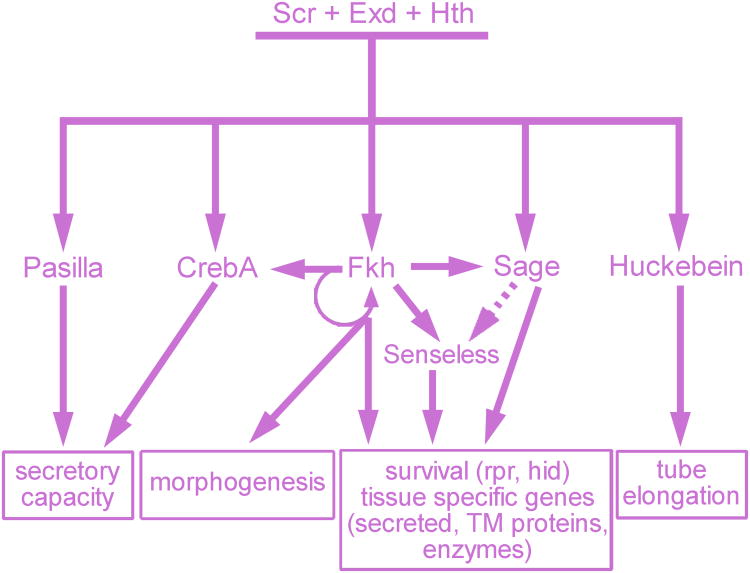
Although Scr, Exd and Hth are absolutely required for SG formation, their expression/nuclear localization disappears from the SG shortly after the onset of morphogenesis2. So, how is cell fate maintained in this tissue? Among the genes activated by Scr, Exd and Hth is the winged helix transcription factor Fork head (Fkh), a key player in maintaining SG fates (Figure 3). Fkh, the single FoxA family protein in flies, maintains its own expression24 and the expression of two other transcription factor genes that are highly expressed in the secretory cells – CrebA, which encodes a bZip transcription factor related to the Creb3/Creb3L family of mammalian proteins, and Sage, which encodes a less well conserved bHLH transcription factor distantly related to several mammalian proteins25-27. fkh affects the expression, either directly or indirectly, of about 60% of all SG genes, including those it represses in the SG duct (see below), as well as downstream targets of CrebA and Sage11,28. As will be discussed later, sustained expression of Fkh, Sage and CrebA is critical for SG function26-29. Fkh, Sage and another downstream transcription factor Senseless (Sens) are also required for secretory cell survival; embryos mutant for any one of these three genes undergo extensive apoptotic SG cell death following elevated expression of the apoptotic activator genes, reaper (rpr) and head involution defective (hid)25,28,30. Moreover, Fkh is essential for SG morphogenesis25.
Figure 3.
The secretory specific genes directly activated by Scr, Exd and Hth include several transcription factors – CrebA, Fork head (Fkh), Sage and Huckebein – as well as a splicing factor – Pasilla. The early expressed genes both maintain and implement the secretory cell fate decision. Pasilla and CrebA function to increase secretory capacity in the professional secretory cells of the SG. Fkh maintains its own expression as well as expression of CrebA and Sage. Fkh also controls morphogenesis and in collaboration with Sage and their downstream target Senseless, activates expression of SG-specific genes and represses the apoptotic genes reaper and hid, keeping the SGs alive. Huckebein mediates tube elongation.
Duct versus secretory fate specification
Ducts form from approximately 25-30 cells on each side of PS2 between the ventral midline and the more laterally positioned SG placodes. Once the SG has internalized, duct and secretory cells can be distinguished at the molecular level since expression of several genes is restricted to only one of these domains (Figure 1). For example, fkh and its many transcriptional targets, are expressed to high levels in the gland cells and levels are either reduced or absent in duct cells. In contrast, expression of several other genes, including Serrate (Ser)31,32, breathless (btl)33 and dead ringer (dri)34, is duct-specific. The secretory versus duct cell distinction is not so clear prior to invagination. Although levels of secretory gene expression in the cells flanking the midline are generally lower, there is considerable cell-to-cell variation, variation that disappears as morphogenesis proceeds (Figure 1). The changes in secretory versus duct cell marker expression may reflect the graded distribution of the signal(s) that induce(s) specific cell fates (e.g. EGF – see below) followed by boundary sharpening through repression and/or activation of gene expression by the transcription factors expressed to higher levels in one versus the other cell type.
Dorsal specifies duct cell fates through activation of EGF signaling
Dl plays an instrumental role in distinguishing SG duct versus secretory cell fates (Figure 4). Dl (through both direct and indirect mechanisms) activates expression of the bHLH transcription factor Single minded (Sim) along the ventral midline35. In turn, Sim activates expression of rhomboid (rho)36, which encodes the spatially limited component required for Epidermal Growth Factor (EGF) signaling37. Sim, Rho and the EGF ligand Spitz are all required for duct specification; loss-of-function mutations in any one of these genes results in a ventral expansion of secretory cell markers at the expense of duct cell markers1,38.
Figure 4.
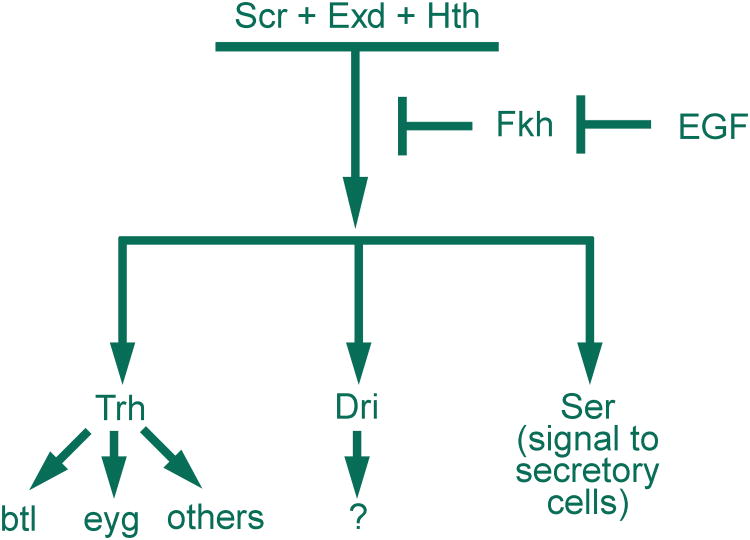
The duct is specified by Scr, Exd and Hth in combination with EGF signaling. EGF signaling blocks expression of Fkh in the most ventral cells of PS2, allowing the expression of duct-specific genes such as Trh and its downstream targets, btl, eyg and, presumably, other genes. The absence of Fkh expression in the ventral cells also allows expression of two other duct-specific factors: Dri and Ser. Ser in duct cells is important for establishing the imaginal ring cell fate.
EGF signaling specifies duct cell fates by repression of fkh
The boundary between gland and duct cells is determined by the combination of two negatively regulated steps; the EGF signaling pathway represses fkh expression in the duct cells and Fkh represses expression of duct-specific genes in the gland cells38,39. Trachealess (Trh), a basic helix loop helix (bHLH)-PAS transcription factor, is initially expressed throughout the entire SG primordia22. As with other duct genes, trh expression is repressed by Fkh, remaining on in only duct cells22,38. Early experiments suggested that loss of trh resulted in a loss of expression of all other tested duct markers38; Trh appears, however, to affect expression of only a subset of duct genes, in many cases simply boosting their expression levels39. Thus, EGF-dependent duct cell specification appears to be mediated largely by repression of fkh expression in future duct cells (Figure 4).
Notch signaling specifies the adult SG primordia
Notch signaling through Ser, one of two Drosophila Notch ligands, specifies the imaginal ring cells (Figure 2)39. Ser expression is duct specific31,32, whereas Notch is transiently upregulated in secretory cells40. Ser loss results in the absence of imaginal ring cells39. Correspondingly, a mutant allele of Notch that specifically affects its response to Ser, but not to Delta, the other Notch ligand in flies, also results in a loss of the imaginal ring cells41. Although it is clear that imaginal ring cells are specified during embryogenesis39, whether this happens prior to or during tube morphogenesis is unknown. Roles for Notch signaling in specifying distinct cell types within developing organs has also been documented in the vertebrate vasculature42, pituitary gland43, pancreas44 and central nervous system45.
Construction of the Secretory Tubes
Once specified, the SG primordial cells undergo morphological changes and internalize to form epithelial tubes. Since no further cell division or cell death occurs during SG differentiation, all subsequent changes take place within and between pre-existing cells. Polarity is maintained throughout the entire process of tube morphogenesis and, in the fully internalized SG tubes, the apical domain forms the luminal surface and the basal domain contacts surrounding tissues (Figure 1).
Regulated sequential secretory cell internalization
The first SG cells to internalize are located in the dorsal-posterior regions of the SG placodes, plate-like structures of columnar epithelial cells that form shortly after Scr expression is first observed in the primordia. The dorsal-posterior cells undergo apical constriction, a process whereby nuclei move to the basal domain and the apical domain constricts to create the pyramidal shaped cells thought to drive tube internalization46. Subsequently, neighboring cells invaginate to create nascent epithelial tubes, with their newly formed lumens contiguous with the apical surface of the SG cells still on the surface. Through a series of less well-characterized shape changes and cell rearrangement, the remaining placode cells internalize to form elongated fully internalized epithelial tubes.
Fkh is required for secretory cell internalization
Fkh plays a major role in SG morphogenesis; the SG primordia in fkh mutant embryos rescued from cell death do not undergo apical constriction and completely fail to internalize. Basal movement of nuclei, however, is unaffected by fkh loss, indicating that the two processes – apical constriction and basal nuclear movement – are separable25. Fkh, as a transcription factor, likely controls SG invagination (and other processes) indirectly, through its downstream targets; the Fkh targets that mediate internalization, however, remain to be identified and/or characterized.
Rho-GTPases and the actinomyosin cytoskeleton mediate secretory gland internalization
Several molecules that directly affect SG internalization have been discovered, many of which are known to be more generally involved in cell shape changes (Table 1). Mutations in folded gastrulation (fog), a secreted ligand for the G protein-coupled receptor Mist47, and RhoGEF2, a Rho GTPase exchange factor required for invagination of the ventral furrow48,49, result in partial failure of SG internalization49,50. RhoGEF2 affects apical constriction by regulating apical accumulation of Spaghetti squash (Sqh), the Myosin regulatory light chain49. Embryos mutant for 18 wheeler (18W), a Toll-like receptor protein, and two Rho-GAPs, RhoGAP5A and RhoGAP88C/Crossveinless-c (CV-C), also show delays and/or partial failure of SG internalization, further supporting the idea that Rho signaling is critical50. Indeed, both Rho1 mutants and embryos with SG-specific expression of a dominant-negative Rho1 construct showed partial defects in SG invagination51. Rho1 regulates SG internalization by two mechanisms: 1) upregulation and apical localization of transcripts for crumbs (crb), which encodes an apical membrane protein necessary for the establishment and maintenance of apical-basal polarity and for apical membrane expansion52-55, and 2) induced apical constriction and cell shape changes mediated by Rho-kinase (Rok)51.
Table 1. Genes Implicated in Salivary Gland (SG) Morphogenesis with Their Currently Understood Roles and Interactions.
| Process: Secretory cell invagination | ||
|---|---|---|
| MOLECULE | ROLE | |
| Fkh | Transcription factor Regulates expression of ≈60% of SG genes Maintains SG viability (with Sage and Sens) Mediates invagination |
|
| Fog | Ligand for GPCR Mist in mesoderm Coordinates invagination |
|
| Rho1 | GTPase Increases Crb levels Maintains Crb, aPKC, and Stardust apical localization |
|
| Crb | Apical membrane protein Establishes and maintains apical/basal polarity Along with aPKC, prevents Rok accumulation and Myoll cable formation in SG |
|
| aPKC | Along with Crb, negatively regulates Rok and Myoll | |
| Rok | Rho kinase Induces cell-shape changes and apical constriction Positive regulator of Myoll |
|
| RhoGEF2 | Rho guanine nucleotide exchange Factor Regulates apical constriction via Sqh localization |
|
| Sqh | Myosin regulatory light chain Creates intracellular myosin network |
|
| RhoGAP88C | Basolaterally localized GTPase activating protein Coordinates invagination |
|
| 18-Wheeler | Toll-like receptor Component of Rho pathway Coordinates invagination |
|
| Tec29/Btk29A | Non-receptor tyrosine kinase Affects F- and G- actin dynamics Coordinates invagination and migration |
|
| Gyc76C | Guanylyl cyclase Upstream of DG1 Regulates Talin and laminin localization |
|
| DG1 | cGMP-dependent kinase 1 Downstream of Gyc76C Regulates Talin and laminin localization |
|
| Process: Tube elongation via cell elongation | ||
| MOLECULE | ROLE | |
| Hkb | Transcription factor Upstream of Klar, Crb Mediates apical expansion |
|
| Klar | Mediates apical membrane delivery via vesicle transport | |
| Crb | Transmembrane protein Mediates apical membrane expansion |
|
| Rib | Transcription factor Upstream of Crb Regulates moesin activity (indirect) |
|
| Lolal | Mediates nuclear localization of Rib in the SG (and other ectoderm) | |
| Moesin | ERM family member; actin linker Increases apical stiffness when phosphorylated |
|
| Cdc42 | Rho GTPase; activates Pakl | |
| Pakl | Serine-threonine kinase Regulates lateral E-Cad endocytosis through Merlin, Dynamin, and Rab5 |
|
| E-Cadherin | Cell-cell adhesion Mediates cell shape changes in the SG [e.g. expansion along the apical P/D axis] |
|
| Process: Tube elongation via cell rearrangement | ||
| MOLECULE | ROLE | |
| Racl | Rho GTPase; regulates junctional E-Cad turnover | |
| E-Cadherin | Regulates cell-cell adhesion [too little, cells disperse; too much, cells are unable to rearrange properly] | |
| Gonl | Apically secreted metalloprotease Influences apical membrane release from ECM and consequent cell rearrangements Genetically interacts with Cad99C |
|
| Cad99C | Apically secreted protocadherin Influences apical membrane release from ECM and consequent cell rearrangements |
|
As with other morphogenetic processes, cytoskeletal events associated with cell shape changes are key for SG tube formation. Indeed, a prominent multi-cellular Myosin II cable forms around the SG placode prior to invagination. This cable is maintained throughout the process of SG internalization and cinches up as more cells are internalized, suggesting that tension created by this cable provides a motive force driving internalization56. The myosin cable forms at the interface between peripheral SG cells and their immediate non-SG neighbors through downregulation of Rok. The high levels of Crb and atypical protein kinase C (aPKC) found within SG cells (and not in neighboring non-SG cells) negatively regulate Rok accumulation, preventing the formation of the Myosin II cable within the placodes56. Actin reorganization is also essential for timely invagination of the SG. Null mutations in Tec29/Btk29A, a member of the Tec family of non-receptor tyrosine kinases, cause a delay in SG invagination due, in part, to a shift in the equilibrium between F- and G-actin57. Although most mutants with invagination defects appear to affect the apical surface, a recent study has revealed that Guanylyl cyclase at 76C (Gyc76C) and its downstream cGMP-dependent kinase 1 (DG1) affect invagination, collective migration and SG lumen shape partly by regulating localization of Talin and the laminin matrix surrounding the basal surface of the SG58.
Elongation of the Secretory Tubes
As the SG primordia internalize and the resulting tube moves to its final correct position in the embryo, the tube elongates through both cell shape change and cell rearrangement.
Tube elongation by cell elongation
Hkb regulates polarized growth and delivery of apical membrane
Polarized growth and delivery of apical membranes is critical for tube elongation59. In embryos mutant for huckebein (hkb), which encodes a zinc finger transcription factor60, the SG cells internalize but almost completely fail to elongate, resulting in small ‘puck-shaped’ SGs with very little apical surface area46,59. Hkb controls SG apical expansion through increased translation and/or stabilization of Crb52-55, and increased transcription of klarsicht (klar), which encodes a putative regulator of the dynein ATPase that mediates microtubule-dependent vesicle transport to the apical surface61,62. Thus, Hkb facilities tube elongation through both Crb-mediated apical membrane expansion and Klar-driven apical targeting of membrane vesicles (Table 1).
Ribbon reduces apical stiffness to facilitate membrane expansion
Ribbon (Rib), a BTB-containing transcription factor, modulates apical membrane expansion to elongate the SG, and two other tubes, the trachea and Malpighian tubules63-65. rib mutant SGs achieve only 60% of the WT lumen length and live imaging studies indicate that rib mutant SGs elongate more slowly than WT66. Rib interacts with Lola like, another BTB-domain containing protein required for robust nuclear localization of Rib, to upregulate crb transcription and to downregulate the activity of Moesin (Moe), a protein that cross-links the apical membrane to the apical cytoskeleton65,67,68. Genetic and mechanical analyses suggest that the increased apical stiffness caused by increased Moe activity (increased phosphorylated Moe) is a major contributor to the rib mutant phenotype65,66. Thus, tube elongation requires not only the generation of sufficient apical membrane for expansion but also modulation of the mechanical properties of the apical domain (Table 1).
Tube elongation by cell rearrangement
Rho1 controls tube length by cell elongation and rearrangement
To achieve fully elongated tubes, the SG also undergoes cell rearrangement. This process simultaneously reduces the number of cells in circumference and increases the number of cells along the length of the tube. Some molecules, such as the Rho1 GTPase, control SG lumen size by regulating both cell rearrangement and cell shape. Proximal SG cells of Rho1 mutant embryos fail to rearrange and the apical domains do not elongate fully69. SG-specific knockdown of Rok by RNAi causes similar defects as observed in Rho1 mutants, suggesting that Rho1 affects tube architecture, at least in part, through Rok regulation of myosin mechanics69. Rho1 also appears to work with Rib to limit apical phosphorylated Moe, suggesting more direct effects on the apical actin cytoskeleton, an idea supported by the observed changes in F-actin distribution69. Interestingly, cell rearrangements mediated by Rho, Rock and Myosin II also drive tube elongation in the vertebrate gut70, suggesting a conserved role for Rho1 in cell rearrangement.
Reduced cell-cell adhesion facilitates cell rearrangement
Allowing cells to rearrange while maintaining polarity requires tight regulation of the junctional complexes that hold epithelial cells together. Since septate junctions (structures equivalent to vertebrate tight junctions) do not fully mature until late embryogenesis71, the major junction requiring modulation during SG cell rearrangement is the adherens junction (AJ). Regulation of cell rearrangement in the SG appears to be through Rac1 modulation of the AJ protein E-Cadherin (E-Cad)72. Too little Rac1 – through either mutations in multiple Rac1 genes [Rac1, Rac2 and Mig 2-like] or through SG expression of a dominant-negative Rac1 construct – results in increased E-Cad (and another AJ protein – βcatenin) at the AJs. The increased pools of AJ-localized E-Cad blocks cell rearrangement. Consequently, the distal gland, which likely forms without significant cell rearrangement, forms relatively normally, but many of the proximal SG cells, which must rearrange during internalization, remain on the embryo surface. As the distal cells continue to migrate posteriorly in the rac1 mutants, the gland often breaks, resulting in multiple small glands surrounding separate lumens. Correspondingly, excessive Rac activity results in reduced E-Cad at the AJs and a corresponding loss of adhesion – SG cells disperse and eventually die. The dispersion phenotype driven by SG expression of constitutively-active Rac can be rescued simply by overexpressing E-Cad, supporting the idea that Rac affects cell rearrangement largely through the localization of E-Cad. Rac1 appears to modulate E-Cad pools in the different membrane domains by regulated endocytosis, since altering the endocytic pathway also alters the Rac1 phenotypic outcomes72.
Not surprisingly, E-Cad turnover is also key to the proximal-distal elongation of individual SG cells. In this case, endocytic turnover of E-Cad is differentially regulated along the axis of polarity by p21-activated kinase 1 (Pak1). Pak1 belongs to a serine-threonine kinase family that in other systems binds and is activated by Cdc42 and/or Rac to regulate diverse biological processes73,74. SG expression of dominant-negative Cdc42 and/or loss of Pak1 results in a complete loss of lateral membrane pools of E-Cad and a widening of cells along the dorsal-ventral axis. Correspondingly, high-level expression of activated Pak1 depletes the apical pools of E-Cad, resulting in the loss of the single shared apical lumen and the appearance of multiple intercellular lumena. The intercellular lumena arise between cells in the lateral domains from endocytosed E-Cad containing vesicles75. Pak1 activity in the SG depends on Rab5, Dynamin and the ERM protein Merlin, a substrate of Pak175,76.
Just as cells have to release their attachment to their neighbors to rearrange, they also have to release their attachment to the matrix77. SGs mutant for AdamTS-A, which encodes an apically-targeted and secreted zinc metalloproteinase, have highly irregular luminal surfaces due to a failure of the cells to easily rearrange during tube elongation and posterior migration. AdamTS-A null mutants also have over-stretched apical domains in the distal-most SG cells and show increased accumulation of apical actin, suggesting that these cells are under increased tension. Further support for a role of AdamTS-A in releasing the apical cell surface from the apical matrix emerges from the finding that null mutations in Cadherin99C (Cad99C) rescue the apical irregularities associated with AdamTS-A loss77. Cad99C, an atypical cadherin with a large extracellular domain, localizes to the apical surface of SG cells and other epithelia78,79,80. Through its attachment to (currently unidentified) apically secreted proteins, Cad99C is proposed to resist and balance the forces driving tube elongation80. Thus, associations between the apical surface and apical matrix are important in fine-tuning overall SG tube shape.
Positioning the Salivary Glands
To attain its final correct position in the embryo, the SG actively migrates in direct contact with several other tissues. Whereas some of the contacting tissues may serve only as tracks or barriers to migration, others also provide guidance cues to either attract or repel the gland.
Surrounding tissues promote SG movement
The visceral mesoderm provides a track forSG migration
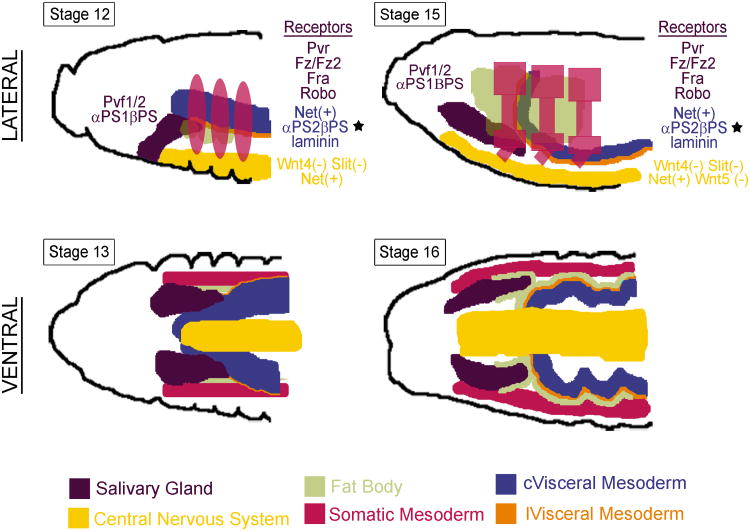
The internalization process positions the secretory tubes in an approximate dorsal-posterior orientation, with the distal most cells directly contacting the dorsally-positioned visceral mesoderm (VM) (Figure 5). As the secretory cells sequentially contact this tissue, they turn and migrate along it to eventually arrive at their final position, with the long axis of the tube aligned with the long axis of the embryo. In mutants where the VM is discontinuous, the SG will often continue to move dorsally instead of turning posteriorly, although the gland still appears to maximize contact with whatever VM tissues remain in these mutants81. Posterior migration of the SG absolutely requires integrin expression both in the SG, which expresses αPS1βPS, and in the VM, which expresses not only αPS2βPS, and also an essential laminin that is bound by both integrins77,81. With loss of any of these molecules, the SG completely fails to migrate; SG tubes still elongate, however, resulting in buckled, U-shaped tubes. These findings suggest that the VM provides a suitable substrate for SG migration. At later stages, contact between the SG and VM is abrogated by the ingression of caudal mesoderm cells between the gland and VM. Failure of this population to migrate results in the continued attachment of the SG to the VM and, during the process of head involution, this attachment stretches the glands to about twice their normal length82.
Figure 5.
The SG contacts or comes close to several tissues as it migrates to its correct final position in the embryo, including the circular (c) visceral mesoderm (cVM), the longitudinal (l) visceral mesoderm (lVM), the fat body, the somatic musculature and the central nervous system. The cVM provides a suitable substrate for posterior SG migration through the expression of αPS2βPS integrin that binds a secreted laminin also expressed in the cVM. The SG expresses αPS1βPS integrin, which also binds the secreted laminin. Both the integrins and laminin are essential for posterior migration (starred). The lVM migrates between the SG and the cVM to detach these two cell types. The SG also expresses several receptor genes, which allow it to properly navigate to its final correct position in response to local sources of the corresponding ligands. In turn, the SG is likely to also provide cues for the migration of other cell types in the embryo. For example, the fat body migrates over specific parts of the SG at late embryonic stages.
Other tissues also provide SG migration cues
The SG either directly contacts or comes near several other tissues during migration, including the somatic muscle, the fat body, the central nervous system (CNS) and the gastric caeca, long tubular extensions of the midgut that emerge during later embryonic stages (Figure 5)82. Studies suggest that most, perhaps all, of these tissues provide cues that direct SG movement. Several guidance cues have been discovered that seem to function either to attract or repel the migrating SG.
Molecular guidance
Like most migrating tissues, the SG responds to a range of guidance cues during its posterior migration. The Netrin/Frazzled (Fra), Slit/Robo, and Wnt/Derailed (Drl)/Frizzled (Fz) pathways have all been shown to influence SG migration83,84 (Figure 5). The Netrin ligands are expressed in both the VM and CNS, whereas their receptor Fra is expressed in the SG. Loss-of-function mutations in the netrin genes or in fra cause a mild migration defect of the glands curving away from the midline84. Overexpression of NetrinB (NetB) causes more pronounced mis-migration of the SG towards the source of expression, suggesting that NetB acts as an attractant for the fra-expressing SGs. Conversely, Slit acts as a strong SG repellent. Loss of Robo1 and Robo2, receptors for Slit, which is expressed in midline cells, cause the SG to mis-migrate towards the midline84. Wnt signaling also acts to repel the migrating SG83. Wnt4 and Wnt5 are expressed in the CNS; their respective receptors Fz/Fz2 and Drl are expressed in the SG. Loss of any of these factors causes the SG to curve towards the CNS. The PDGF/VEGF pathway has been implicated in SG migration: mutations in both the receptor and two of the ligands result in ventrally curved SGs83. It remains unclear, however, whether this pathway affects migration or some other aspect of SG morphology. Additional cues, provided by other cell types, will likely be discovered to influence final SG placement; disrupting these pathways, however, may lead to relatively subtle defects in SG placement given the number of tissues providing guidance cues. How an intact epithelial tissue, such as the SG, integrates signals from multiple different sources to control its movement remains to be discovered.
Formation of the Salivary Duct
Duct cells internalize by wrapping and convergent extension
Duct cells internalize immediately following the secretory cells, beginning with the cells that will form the individual ducts (Figure 1)85. These cells form tubes by a wrapping type mechanism, whereby the cells become wedge-shaped – wider on the basal side, narrower on the apical side - as they sink below the embryo surface86. The internalizing individual duct cells eventually meet and close, beginning distally and extending proximally until they meet up with the common duct primordia at the ventral midline. Once the individual ducts tubes have formed, the common duct tube forms from the more anterior ventral primordia using a similar mechanism. The duct tubes also continue to elongate, eventually forming tubes that are about two cells in diameter for the individual ducts and 3-4 cells for the common duct. Thus, duct elongation occurs by convergent extension85, a common developmental process that narrows and elongates tissue by cell intercalation87 and that has been implicated in tube elongation in several Drosophila tissues88-91 as well as in the vertebrate neural tube92. Posterior migration of the gland coupled with the attachment of the duct to the mouthparts may provide the tensile forces required for duct elongation by cell intercalation.
Trh and Eyg are required for duct cell invagination
Trh plays a critical role in internalizing both individual and common ducts since the entire duct fails to invaginate and remains on the ventral surface in trh mutant embryos22,38. eye gone (eyg), which encodes a Pax transcription factor positively regulated by Trh, is required to distinguish individual from common duct domains85. Eyg is also necessary for morphogenesis of the individual duct tubes85; in eyg mutants, the individual ducts often fail to elongate to connect the secretory cells to the common duct, resulting in closed internalized secretory tubes that are disconnected from the rest of the digestive tract. We expect that many more genes that function in duct morphogenesis await discovery.
Salivary Gland Function: Secretion and Production of Tissue-Specific Gene Products
As the SG undergoes the process of morphogenesis, it simultaneously begins to specialize into a secretory organ. Several transcription factor genes that are induced in the earliest stages of SG formation – fkh, sage and CrebA - continue to be expressed throughout the life of this organ. These same genes are also expressed in the adult gland, suggesting that they may play similar roles in both the larval and adult tissues. As will be discussed, Fkh, Sage and CrebA play key roles in the main function of the SG – the synthesis and secretion of high levels of protein (Figure 3).
Fkh, Sage (and Sens) regulate SG-specific gene products
fkh is required for expression of most SG genes
As mentioned previously, Fkh controls many aspects of SG development, from specification to internalization to gland maintenance. How can Fkh have so many distinct functions in the SG when it is also expressed and required in multiple other tissues93? Whereas there may be some commonality in Fkh's role in some tissues, Fkh also has distinct tissue-specific functions. In the SG, tissue-specific Fkh function is largely mediated by Sage28, a SG-specific bHLH transcription factor.
Sage – a SG-specific bHLH transcription factor – provides specificity to Fkh function
In embryos, Sage (salivary gland E-box binding protein) is expressed only in the SG94, making it a prime candidate for controlling SG identity and specialization. sage null mutant SG cells internalize and form elongated secretory tubes, but undergo massive apoptotic cell death once fully internalized. This finding suggests that even after gland formation and internalization, the SG must be actively kept alive throughout development. sage mutant glands can be rescued from death by co-expression of the anti-apoptotic P35 protein. Although overall gland morphology and polarity is normal, the rescued glands have thin, irregular lumens28.
Microarray studies reveal that Sage regulates SG-specific secreted or transmembrane proteins and their modifiers. Interestingly, expression of Sage targets entirely depends on Fkh; loss of fkh resulted in a complete loss of SG expression of Sage targets. Moreover, experiments expressing either fkh or sage alone or co-expressing both genes reveal that the combined activity of Fkh and Sage is required for inducing Sage target gene expression in multiple distinct cell types27,28. Moreover, Sage, Fkh and another downstream SG transcription factor – Sens – localize to largely overlapping sites on SG polytene chromosomes28. Thus, Fkh and Sage (+/- Sens) clearly work together to directly regulate expression of SG-specific genes; the exact mechanism by which these proteins collaborate, however, remains to be elucidated. Similar collaborations between FoxA and Sage-related bHLH proteins are likely to underlie tissue specific gene expression in mammalian dopaminerginic neurons95,96, pancreas97-99, as well as the secretory cells of the C. elegans pharynx100.
CrebA upregulates secretory capacity
CrebA and expression of secretory machinery
Early experiments designed to learn how core components of the secretory machinery are regulated in professional secretory cells, such as the SG, revealed that regulation occurs at the transcriptional level. Thirty-four genes encoding protein components of the machinery required at all early steps of the secretory pathway are expressed to significantly higher levels in the SG secretory cells than in surrounding tissues26. A combination of in vitro and in vivo DNA binding experiments, as well as in vivo expression studies, established that CrebA directly regulates secretory pathway component gene (SPCG) expression through a consensus sequence identified by computational analysis of the enhancers for all 34 SPCGs29. Genome-wide microarray studies revealed that CrebA is largely dedicated to the regulation of secretory capacity – well over 200 genes encoding core secretory machinery proteins, as well as secreted cargo, require CrebA for their full expression29. Moreover, expression of every SPCG that has been tested can be activated in additional cells simply by overexpressing CrebA – or the activated form of any of its five human orthologues, the Creb3/Creb3L family of bZip transcription factors - in those cells29,101. Thus, CrebA is both necessary and sufficient for elevated secretory capacity and Creb3/Creb3L have similar activities. CrebA and its human orthologues also appear to boost expression of secreted cargo genes. Whereas direct regulation of cargo genes by the mammalian proteins has been observed102, studies in flies suggest CrebA may work indirectly by up-regulating expression of Sage (see above); sage transcript levels decrease about twofold in CrebA mutants29.
Pasilla encodes a splicing factor also required for high-level SG secretion
Among the early expressed SG genes that come on and stay on in the SG, is pasilla (ps), which encodes a KH domain-containing nuclear splicing factor related to two mammalian proteins, Nova1 and Nova2103. Loss of ps in the SG results in late-stage apical lumen irregularities that are linked to a significant reduction in material secreted into the lumen and a corresponding reduction in the size and number of secretory vesicles. Although more than 400 genes have been identified as PS splicing targets in S2 cells, the link to SG secretion remains to be discovered104.
Diaphanous targets apical secretion
Apical targeting of secretory vesicles in multiple epithelial tubular organs in Drosophila, including the SG, is mediated by Diaphanous (Dia), an actin-nucleation factor that localizes tightly to the apical surface, and by Myosin V (MyoV)105. Loss of Dia, which is expressed to high levels in the SG and other secretory organs, or of MyoV has no effect on overall apical-basal polarity but significantly compromises apical secretion. Apical localization of Dia is mediated by its interactions with PIP2 and Rho1, both of which are enriched in the apical membrane106. Dia is proposed to nucleate apically directed actin filaments, towards which secretory vesicles are targeted via MyoV-based transport. Mammalian Dia plays a similar role in other secretory organs, including the pancreas and submandibular gland107.
The Larval-Pupal Salivary Gland
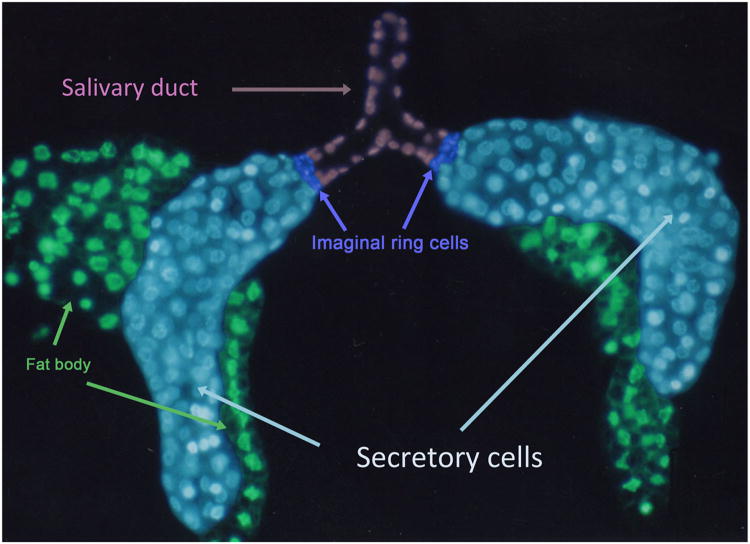
The larval SG of Drosophila has been used to study several basic cellular functions, including secretion, hormone responsiveness and the cell death pathways. Morphologically, the larval SG looks quite similar to the late embryonic SG, with the salivary duct, imaginal ring cells and secretory gland arranged from proximal to distal (Figure 6). In the larva, the proximal region of the secretory gland is termed the transition zone and the cells in the most distal portion of the secretory tube are called the corpus cells. The SG is among the last larval tissues to be destroyed prior to pupation, and its contents are vital to pupae formation as the fly transitions to adulthood.
Figure 6.
Confocal image of the larval salivary gland with the different cell types artificially colorized. The larval salivary gland includes the large polytenized secretory cells (light blue), the medium sized duct cells (light purple) and the small imaginal ring cells (blue). The fat body (green) attaches to the secretory cells at several places.
Ecdysone signaling in the SG
One of the most notable characteristics of the larval SG is its responsiveness to the hormone 20-hydroxy-ecdysone (20E or ecdysone). During the third instar stage, the final stage of larval development, there are three relatively small pulses of ecdysone, followed by a large pulse that signifies the transition from larva to prepupa108 (Figure 7). This final pulse of ecdysone is approximately five times larger than the earlier pulses109. An early observation of insect SGs was that the presence of ecdysone led to the formation of puffs along the polytenized chromosomes110. Successful in vitro culture of the Drosophila larval SGs allowed for a more complete study of this phenomenon as well as the mapping of the ecdysone-induced puffs to specific chromosomal regions111,112. The small pulses of ecdysone induce the intermolt puffs in late larvae that regress during the large ecdysone pulse at the end of the third instar stage113. The next set of ecdysone-sensitive puffs has been separated into three distinct groups based on how rapidly they are induced by the large ecdysone pulse: early genes, early-late genes, and late genes. Many early genes encode transcription factors, such as the Broad complex (Br-C), E74, and E75114-119. The early genes, in turn, activate expression of the late genes and repress their own expression120. There are many more late genes than early genes, indicating a transcriptional hierarchy in the larval SG not unlike that seen the embryonic gland.
Figure 7.
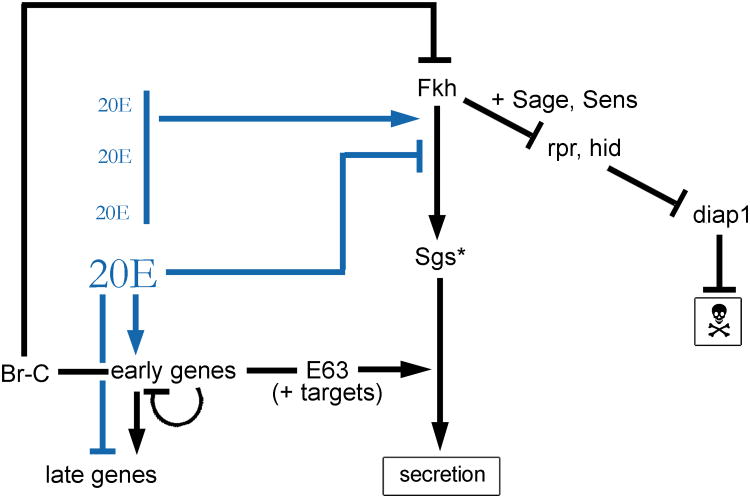
Fkh (likely in collaboration with Sage and Sens) keeps the SG alive until the prepupal stage by preventing expression of the apoptosis inducers reaper (rpr) and head involution defective (hid). Fkh (and Sage +/- Sens) in combination with low level ecdysone signaling activate transcription of the Salivary glue secretion (Sgs) genes in late larvae. High-level ecdysone signaling just prior to pupation activates expression of the early ecdysone-responsive genes, which encode transcription factors. A subset of these transcription factors repress fkh and Sgs transcription and activate expression of genes required for glue secretion. Thus, the glue is secreted when Fkh begins to disappear. In turn, the disappearance of Fkh results in rpr and hid expression, which overcome DIAP and activate the cell death pathway. Thus, the SG dies shortly after it completes its final task of glue secretion.
The identification of the receptor for 20-hydroxyecdysone was made in the early 1990s121. The ecdysone receptor (EcR) was cloned and shown to be a zinc-finger containing nuclear hormone receptor. The expression of three Ec-R isoforms was shown to be upregulated prior to the larval ecdysone pulses122. Of these, EcR-B1 is most highly expressed in the SG123. EcR function is critical for continued SG development. Loss of EcR results in SGs that die before secretion occurs124. Although EcR and another zinc-finger nuclear hormone receptor, Ultraspiracle (Usp), were subsequently shown to function as a heterodimeric receptor for ecdysone125, more recent findings suggest that the EcR-Usp heterodimer may not be the functional ecdysone receptor, at least for the SG. Experiments using RNAi against all three EcR isoforms revealed that larvae without any EcR do not secrete at all, and that expression of any one EcR isoform can rescue this phenotype126. Surprisingly, RNAi of Usp did not block secretion, but its overexpression did. This finding suggested that EcR either forms functional homodimers and/or that EcR forms a heterodimer with a receptor other than Usp. Further studies identified DHR96, a broadly expressed orphan nuclear receptor, as apotential EcR binding partner. Knocking down DHR96 with RNAi results in a block in gland secretion that cannot be rescued by over-expressing any of the EcR isoforms127.
SG glue secretion
The first genes to be induced by the small pulses of ecdysone are those in the intermolt puffs, many of which contain Salivary glue secretion (Sgs) genes. Expression of at least three Sgs genes (Sgs 1, 3, and 4) is controlled by Fkh128,129 and several Sgs genes were identified as potential targets of Sage 130, suggesting that the same transcription factors that cooperate to regulate SG specific expression in embryos also work together to activate larval SG-specific genes. Sgs genes encode glue proteins, several of which are highly glycosylated. Once secreted, glue proteins adhere the pupa to a solid surface during metamorphosis129,131,132. The Sgs proteins are synthesized in both the transition and corpus cells of the SG124, where they accumulate in approximately eleven thousand secretory granules per cell, which coalesce into fewer larger granules prior to their secretion133. The final large pulse of ecdysone has two effects on the Sgs genes: transcriptional repression and glue protein secretion134. Secretion of the glue granules is dependent on Clathrin, the AP adaptor proteins AP-1, AP47, and EpsinR135. Overexpression of E63-1, an early gene that encodes a calcium binding protein, induces premature secretion of glue granules124. Moreover, loss of E63-1 and calmodulin impairs secretion136. Together, these data suggest a coordinated sequence of events that starts with accumulation of Sgs transcripts and glue proteins, followed by a halt in Sgs transcription due to increased ecdysone, which also signals the cells to secrete their contents. Without glue protein secretion, the pupa cannot adhere to solid substrates and further development is arrested. In signaling pupa formation, the large ecdysone pulse also signals the larva to destruct (discussed below).
SG death
In addition to spurring secretion of SG contents, the final large pulse of ecdysone also signals gland death. By this time, many other larval tissues are already partially destroyed. How does the SG remain intact for longer than the surrounding tissues? Two molecules keep the glands alive until after all of their glue is expelled: Fkh, which has kept SG cells alive since the gland was first specified25, and Drosophila inhibitor of apoptosis 1 (Diap1)137. The late pulse of ecdysone induces Br-C transcription, which, in turn, represses fkh transcription and induces the apoptotic cell death cascades. Sustained expression of fkh in the SGs delays death, whereas RNAi knockdown of fkh at earlier stages induces premature cell death137. Although expression of both rpr and hid RNA are repressed by Fkh, hid seems to have the most impact on SG death; overexpression of hid alone can induce death and hid loss results in gland persistence138. Overexpression of other known pro-apoptotic genes - rpr, dronc, sickle or grim -does not lead to cell death, at least not on their own, and loss of rpr alone does not affect gland persistence76. The upregulation of hid (and rpr) that occurs when Fkh is shut off overcomes the action of Diap1, allowing apoptosis to proceed. Overexpression of p53 alone induces some SG death through apoptotic mechanisms, but is insufficient by itself to cause full gland death139. Indeed, SGs also show many signs of autophagic cell death140. Overexpression of the autophagy gene atg1 results in premature cell death139, which can be rescued by the simultaneous knock down of atg12. Moreover, atg8a and atg18 mutant SGS show cleavage of a caspase substrate as well as positive TUNEL staining, suggesting that the apoptotic death pathways remain active when autophagy is blocked. These data reinforce the idea that both autophagy and apoptosis collaborate to ensure the timely death of the SG during metamorphosis.
Genes separate from the canonical death pathways also play important roles in SG death. The matrix metalloprotease Mmp1 is upregulated in the dying glands, whereas its inhibitor Timp (Tissue inhibitor of metalloprotease) is downregulated141. Small GTPases are also induced, indicative of cell rearrangements and migration142. Mdh2, a mitochondrial malate dehydrogenase, also plays a unique role in cell death143. mdh2 mutant SGs survive longer than wild-type, and although autophagy is initiated in mdh2 mutant glands, it is not completed. Caspase-3 activation is blocked in mdh2 mutants, but expression of rpr, hid and grim occurs normally. mdh2 mutants also have reduced ATP levels relative to wild-type larva. These studies suggest that Mdh2 functions downstream of the previously described death pathways. The findings that Mdh2 and other factors impact SG death suggest that many additional genes function to provide for both the timely death and clean destruction of the larval SG.
Conclusion
Studies of the Drosophila SG have broader implications regarding how epithelial organs are specified, formed, specialized, maintained and eventually destroyed. Importantly, cell fate is determined by integrating patterning information along both major body axes – anterior-posterior and dorsal-ventral. This is true not only of the SG, but of all other Drosophila tissues that have been studied to the same level of detail, such as the trachea and mesodermal derivatives144,145. Although this finding suggests that there may be no true “organ-specifying” genes, clearly there are genes that play major roles in organ development and homeostasis. For the SG, Fkh has this role, affecting all aspects of gland biology. Nonetheless, approximately 40% of SG expressed genes are unaffected by fkh loss and Fkh (even with its SG-specific partner Sage) is incapable of stably converting other cell types to a SG fate11. Importantly, the only additional cells that persistently express SG markers upon ubiquitous expression of Fkh and Sage, are cells that also express Scr, the Hox gene required for SG formation28. To date, Scr itself is the only gene capable of driving ectopic gland formation1,10.
Excellent progress has been made regarding contributions of the small GTPases and cytoskeletal components to SG morphogenesis, particularly with respect to converting a plate of polarized epithelial cells on the embryo surface into an elongated, fully internalized secretory tube (Table 1)49-51,56,69. Understanding the initiation and coordination of these events within the gland primordia is the next challenge. We expect that the identification and characterization of early-expressed Fkh targets will be key to fully understanding this process, since fkh mutant SGs completely fail to internalize25.
The SG also provides an excellent model for collective cell migration. The SG is the ultimate collective, since the gland migrates (and elongates) as a fully polarized epithelium. Several pathways and tissues that guide migration have been discovered, but very few of these pathways completely impede migration, leading to the hypothesis that additional guidance molecules exist. Much remains to be learned about how the SG responds to each of the signals and integrates this information to reproducibly arrive at its appropriate final destination, where carrying out its functions is presumably optimized and where the SG in turn can provide cues for positioning other body parts. Understanding the forces fueling tube elongation will also be key.
The past decade has been exciting with regards to learning how SGs specialize – specifically, how SG cells prepare for their major function – high-level secretion – and how SG cells become programmed to produce the right products. High-level secretion is controlled by a single transcription factor – CrebA, which appears to directly activate expression of the entire battery of proteins that make up the early secretory machinery26,29. Having a single protein (as in flies) or very few proteins (as in humans) with the capacity to coordinately up-regulate the entire secretory pathway provides a simple mechanism for generating sufficient machinery to meet the very different levels of secretory load experienced by various cell types. The beauty of addressing this issue in flies is that with only a single gene with this activity (instead of up to five potentially redundant genes), the consequences of gene loss are much more apparent.
Fkh – like the vertebrate FoxA proteins – is expressed and required in a broad array of embryonic tissues, from cells of the nervous and immune systems to multiple different organ types. Studies of Sage have revealed that Drosophila Fkh achieves SG specificity by partnering up with this tissue-specific bHLH protein28. We predict that Fkh will partner with other tissue-specific proteins (perhaps also other bHLH transcription factors) to regulate completely different sets of targets in the other cells in which it is expressed and required. The next task is to uncover the mechanisms by which Fkh and Sage cooperate to control SG specific gene expression.
Finally, the SG performs vital functions for the animal up to the minutes and hours before its demise. To ensure that these functions can be achieved, Fkh continues to hold death at bay until the final task is completed137. Fkh (and likely Sage) work together with the ecdysone-signaling pathway to ensure that not only are the right proteins made at the right time, but that the SG is quickly and cleanly disposed of once its function is accomplished. A relatively complex and seemingly redundant set of events – including what appears to be death by multiple mechanisms – occur to ensure that even death is done right.
Contributor Information
SeYeon Chung, Department of Cell Biology, Johns Hopkins University School of Medicine.
Caitlin D. Hanlon, Department of Cell Biology, Johns Hopkins University School of Medicine
Deborah J. Andrew, Department of Cell Biology, Johns Hopkins University School of Medicine
References
- 1.Panzer S, Weigel D, Beckendorf SK. Organogenesis in Drosophila melanogaster: embryonic salivary gland determination is controlled by homeotic and dorsoventral patterning genes. Development. 1992;114:49–57. doi: 10.1242/dev.114.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Henderson KD, Andrew DJ. Regulation and function of Scr, exd, and hth in the Drosophila salivary gland. Developmental biology. 2000;217:362–374. doi: 10.1006/dbio.1999.9560. [DOI] [PubMed] [Google Scholar]
- 3.Ryoo HD, Mann RS. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev. 1999;13:1704–1716. doi: 10.1101/gad.13.13.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mann RS, Abu-Shaar M. Nuclear import of the homeodomain protein extradenticle in response to Wg and Dpp signalling. Nature. 1996;383:630–633. doi: 10.1038/383630a0. [DOI] [PubMed] [Google Scholar]
- 6.Noro B, Lelli K, Sun L, Mann RS. Competition for cofactor-dependent DNA binding underlies Hox phenotypic suppression. Genes & development. 2011;25:2327–2332. doi: 10.1101/gad.175539.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryoo HD, Marty T, Casares F, Affolter M, Mann RS. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development. 1999;126:5137–5148. doi: 10.1242/dev.126.22.5137. [DOI] [PubMed] [Google Scholar]
- 8.Rivas ML, Espinosa-Vazquez JM, Sambrani N, Greig S, Merabet S, Graba Y, Hombria JC. Antagonism versus cooperativity with TALE cofactors at the base of the functional diversification of Hox protein function. PLoS genetics. 2013;9:e1003252. doi: 10.1371/journal.pgen.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castelli Gair Hombria J, Rivas ML, Sotillos S. Genetic control of morphogenesis - Hox induced organogenesis of the posterior spiracles. The International journal of developmental biology. 2009;53:1349–1358. doi: 10.1387/ijdb.072421jc. [DOI] [PubMed] [Google Scholar]
- 10.Andrew DJ, Horner MA, Petitt MG, Smolik SM, Scott MP. Setting limits on homeotic gene function: restraint of Sex combs reduced activity by teashirt and other homeotic genes. Embo J. 1994;13:1132–1144. doi: 10.1002/j.1460-2075.1994.tb06362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maruyama R, Grevengoed E, Stempniewicz P, Andrew DJ. Genome-wide analysis reveals a major role in cell fate maintenance and an unexpected role in endoreduplication for the Drosophila FoxA gene Fork head. PloS one. 2011;6:e20901. doi: 10.1371/journal.pone.0020901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandre E, Graba Y, Fasano L, Gallet A, Perrin L, De Zulueta P, Pradel J, Kerridge S, Jacq B. The Drosophila teashirt homeotic protein is a DNA-binding protein and modulo, a HOM-C regulated modifier of variegation, is a likely candidate for being a direct target gene. Mechanisms of development. 1996;59:191–204. doi: 10.1016/0925-4773(96)00594-1. [DOI] [PubMed] [Google Scholar]
- 13.Fasano L, Roder L, Core N, Alexandre E, Vola C, Jacq B, Kerridge S. The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell. 1991;64:63–79. doi: 10.1016/0092-8674(91)90209-h. [DOI] [PubMed] [Google Scholar]
- 14.Roder L, Vola C, Kerridge S. The role of the teashirt gene in trunk segmental identity in Drosophila. Development. 1992;115:1017–1033. doi: 10.1242/dev.115.4.1017. [DOI] [PubMed] [Google Scholar]
- 15.Taghli-Lamallem O, Gallet A, Leroy F, Malapert P, Vola C, Kerridge S, Fasano L. Direct interaction between Teashirt and Sex combs reduced proteins, via Tsh's acidic domain, is essential for specifying the identity of the prothorax in Drosophila. Developmental biology. 2007;307:142–151. doi: 10.1016/j.ydbio.2007.04.028. [DOI] [PubMed] [Google Scholar]
- 16.Struhl G. Role of the esc+ gene product in ensuring the selective expression of segment-specific homeotic genes in Drosophila. Journal of embryology and experimental morphology. 1983;76:297–331. [PubMed] [Google Scholar]
- 17.Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987;325:816–818. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- 18.Gibson G, Schier A, LeMotte P, Gehring WJ. The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell. 1990;62:1087–1103. doi: 10.1016/0092-8674(90)90386-s. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Reyes A, Morata G. Organization of the Drosophila head as revealed by the ectopic expression of the Ultrabithorax product. Development. 1991;113:1459–1471. doi: 10.1242/dev.113.4.1459. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Reyes A, Urquia N, Gehring WJ, Struhl G, Morata G. Are cross-regulatory interactions between homoeotic genes functionally significant? Nature. 1990;344:78–80. doi: 10.1038/344078a0. [DOI] [PubMed] [Google Scholar]
- 21.Mann RS, Hogness DS. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 22.Isaac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes & development. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- 23.Henderson KD, Isaac DD, Andrew DJ. Cell fate specification in the Drosophila salivary gland: the integration of homeotic gene function with the DPP signaling cascade. Developmental biology. 1999;205:10–21. doi: 10.1006/dbio.1998.9113. [DOI] [PubMed] [Google Scholar]
- 24.Zhou B, Bagri A, Beckendorf SK. Salivary gland determination in Drosophila: a salivary-specific, fork head enhancer integrates spatial pattern and allows fork head autoregulation. Developmental biology. 2001;237:54–67. doi: 10.1006/dbio.2001.0367. [DOI] [PubMed] [Google Scholar]
- 25.Myat MM, Andrew DJ. Fork head prevents apoptosis and promotes cell shape change during formation of the Drosophila salivary glands. Development. 2000;127:4217–4226. doi: 10.1242/dev.127.19.4217. [DOI] [PubMed] [Google Scholar]
- 26.Abrams EW, Andrew DJ. CrebA regulates secretory activity in the Drosophila salivary gland and epidermis. Development. 2005;132:2743–2758. doi: 10.1242/dev.01863. [DOI] [PubMed] [Google Scholar]
- 27.Abrams EW, Mihoulides WK, Andrew DJ. Fork head and Sage maintain a uniform and patent salivary gland lumen through regulation of two downstream target genes, PH4alphaSG1 and PH4alphaSG2. Development. 2006;133:3517–3527. doi: 10.1242/dev.02525. [DOI] [PubMed] [Google Scholar]
- 28.Fox RM, Vaishnavi A, Maruyama R, Andrew DJ. Organ-specific gene expression: the bHLH protein Sage provides tissue specificity to Drosophila FoxA. Development. 2013;140:2160–2171. doi: 10.1242/dev.092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox RM, Hanlon CD, Andrew DJ. The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J Cell Biol. 2010;191:479–492. doi: 10.1083/jcb.201004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrasekaran V, Beckendorf SK. senseless is necessary for the survival of embryonic salivary glands in Drosophila. Development. 2003;130:4719–4728. doi: 10.1242/dev.00677. [DOI] [PubMed] [Google Scholar]
- 31.Fleming RJ, Scottgale TN, Diederich RJ, Artavanis-Tsakonas S. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes & development. 1990;4:2188–2201. doi: 10.1101/gad.4.12a.2188. [DOI] [PubMed] [Google Scholar]
- 32.Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development. 1991;111:749–761. doi: 10.1242/dev.111.3.749. [DOI] [PubMed] [Google Scholar]
- 33.Klambt C, Glazer L, Shilo BZ. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes & development. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- 34.Gregory SL, Kortschak RD, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Molecular and cellular biology. 1996;16:792–799. doi: 10.1128/mcb.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasai Y, Stahl S, Crews S. Specification of the Drosophila CNS midline cell lineage: direct control of single-minded transcription by dorsal/ventral patterning genes. Gene expression. 1998;7:171–189. [PMC free article] [PubMed] [Google Scholar]
- 36.Nambu JR, Franks RG, Hu S, Crews ST. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell. 1990;63:63–75. doi: 10.1016/0092-8674(90)90288-p. [DOI] [PubMed] [Google Scholar]
- 37.Klambt C. EGF receptor signalling: roles of star and rhomboid revealed. Current biology: CB. 2002;12:R21–23. doi: 10.1016/s0960-9822(01)00642-x. [DOI] [PubMed] [Google Scholar]
- 38.Kuo YM, Jones N, Zhou B, Panzer S, Larson V, Beckendorf SK. Salivary duct determination in Drosophila: roles of the EGF receptor signalling pathway and the transcription factors fork head and trachealess. Development. 1996;122:1909–1917. doi: 10.1242/dev.122.6.1909. [DOI] [PubMed] [Google Scholar]
- 39.Haberman AS, Isaac DD, Andrew DJ. Specification of cell fates within the salivary gland primordium. Dev Biol. 2003;258:443–453. doi: 10.1016/s0012-1606(03)00140-4. [DOI] [PubMed] [Google Scholar]
- 40.Kidd S, Baylies MK, Gasic GP, Young MW. Structure and distribution of the Notch protein in developing Drosophila. Genes & development. 1989;3:1113–1129. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto S, Charng WL, Rana NA, Kakuda S, Jaiswal M, Bayat V, Xiong B, Zhang K, Sandoval H, David G, Wang H, Haltiwanger RS, Bellen HJ. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science. 2012;338:1229–1232. doi: 10.1126/science.1228745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gridley T. Notch signaling in the vasculature. Current topics in developmental biology. 2010;92:277–309. doi: 10.1016/S0070-2153(10)92009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nantie LB, Himes AD, Getz DR, Raetzman LT. Notch signaling in postnatal pituitary expansion: proliferation, progenitors and cell specification. Molecular endocrinology. 2014 doi: 10.1210/me.2013-1425. me20131425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patten BA, Peyrin JM, Weinmaster G, Corfas G. Sequential signaling through Notch1 and erbB receptors mediates radial glia differentiation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:6132–6140. doi: 10.1523/JNEUROSCI.23-14-06132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myat MM, Andrew DJ. Organ shape in the Drosophila salivary gland is controlled by regulated, sequential internalization of the primordia. Development. 2000;127:679–691. doi: 10.1242/dev.127.4.679. [DOI] [PubMed] [Google Scholar]
- 47.Manning AJ, Peters KA, Peifer M, Rogers SL. Regulation of epithelial morphogenesis by the G protein-coupled receptor mist and its ligand fog. Science signaling. 2013;6:ra98. doi: 10.1126/scisignal.2004427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers SL, Wiedemann U, Hacker U, Turck C, Vale RD. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Current biology: CB. 2004;14:1827–1833. doi: 10.1016/j.cub.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 49.Nikolaidou KK, Barrett K. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Current biology: CB. 2004;14:1822–1826. doi: 10.1016/j.cub.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 50.Kolesnikov T, Beckendorf SK. 18 wheeler regulates apical constriction of salivary gland cells via the Rho-GTPase-signaling pathway. Dev Biol. 2007;307:53–61. doi: 10.1016/j.ydbio.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu N, Keung B, Myat MM. Rho GTPase controls invagination and cohesive migration of the Drosophila salivary gland through Crumbs and Rho-kinase. Developmental biology. 2008;321:88–100. doi: 10.1016/j.ydbio.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Tepass U, Theres C, Knust E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–799. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- 53.Tepass U, Knust E. Crumbs and stardust act in a genetic pathway that controls the organization of epithelia in Drosophila melanogaster. Developmental biology. 1993;159:311–326. doi: 10.1006/dbio.1993.1243. [DOI] [PubMed] [Google Scholar]
- 54.Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 55.Tepass U. Crumbs, a component of the apical membrane, is required for zonula adherens formation in primary epithelia of Drosophila. Dev Biol. 1996;177:217–225. doi: 10.1006/dbio.1996.0157. [DOI] [PubMed] [Google Scholar]
- 56.Roper K. Anisotropy of Crumbs and aPKC drives myosin cable assembly during tube formation. Developmental cell. 2012;23:939–953. doi: 10.1016/j.devcel.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chandrasekaran V, Beckendorf SK. Tec29 controls actin remodeling and endoreplication during invagination of the Drosophila embryonic salivary glands. Development. 2005;132:3515–3524. doi: 10.1242/dev.01926. [DOI] [PubMed] [Google Scholar]
- 58.Patel U, Myat MM. Receptor guanylyl cyclase Gyc76C is required for invagination, collective migration and lumen shape in the Drosophila embryonic salivary gland. Biology open. 2013;2:711–717. doi: 10.1242/bio.20134887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myat MM, Andrew DJ. Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell. 2002;111:879–891. doi: 10.1016/s0092-8674(02)01140-6. [DOI] [PubMed] [Google Scholar]
- 60.Bronner G, Jackle H. Regulation and function of the terminal gap gene huckebein in the Drosophila blastoderm. The International journal of developmental biology. 1996;40:157–165. [PubMed] [Google Scholar]
- 61.Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Current biology: CB. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- 62.Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 1998;92:547–557. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- 63.Bradley PL, Andrew DJ. ribbon encodes a novel BTB/POZ protein required for directed cell migration in Drosophila melanogaster. Development. 2001;128:3001–3015. doi: 10.1242/dev.128.15.3001. [DOI] [PubMed] [Google Scholar]
- 64.Shim K, Blake KJ, Jack J, Krasnow MA. The Drosophila ribbon gene encodes a nuclear BTB domain protein that promotes epithelial migration and morphogenesis. Development. 2001;128:4923–4933. doi: 10.1242/dev.128.23.4923. [DOI] [PubMed] [Google Scholar]
- 65.Kerman BE, Cheshire AM, Myat MM, Andrew DJ. Ribbon modulates apical membrane during tube elongation through Crumbs and Moesin. Developmental biology. 2008;320:278–288. doi: 10.1016/j.ydbio.2008.05.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheshire AM, Kerman BE, Zipfel WR, Spector AA, Andrew DJ. Kinetic and mechanical analysis of live tube morphogenesis. Developmental dynamics: an official publication of the American Association of Anatomists. 2008;237:2874–2888. doi: 10.1002/dvdy.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medina E, Williams J, Klipfell E, Zarnescu D, Thomas G, Le Bivic A. Crumbs interacts with moesin and beta(Heavy)-spectrin in the apical membrane skeleton of Drosophila. J Cell Biol. 2002;158:941–951. doi: 10.1083/jcb.200203080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polesello C, Delon I, Valenti P, Ferrer P, Payre F. Dmoesin controls actin-based cell shape and polarity during Drosophila melanogaster oogenesis. Nature cell biology. 2002;4:782–789. doi: 10.1038/ncb856. [DOI] [PubMed] [Google Scholar]
- 69.Xu N, Bagumian G, Galiano M, Myat MM. Rho GTPase controls Drosophila salivary gland lumen size through regulation of the actin cytoskeleton and Moesin. Development. 2011;138:5415–5427. doi: 10.1242/dev.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reed RA, Womble MA, Dush MK, Tull RR, Bloom SK, Morckel AR, Devlin EW, Nascone-Yoder NM. Morphogenesis of the primitive gut tube is generated by Rho/ROCK/myosin II-mediated endoderm rearrangements. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:3111–3125. doi: 10.1002/dvdy.22157. [DOI] [PubMed] [Google Scholar]
- 71.Tepass U, Hartenstein V. The development of cellular junctions in the Drosophila embryo. Developmental biology. 1994;161:563–596. doi: 10.1006/dbio.1994.1054. [DOI] [PubMed] [Google Scholar]
- 72.Pirraglia C, Jattani R, Myat MM. Rac function in epithelial tube morphogenesis. Developmental biology. 2006;290:435–446. doi: 10.1016/j.ydbio.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 73.Bokoch GM. Biology of the p21-activated kinases. Annual review of biochemistry. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 74.Arias-Romero LE, Chernoff J. A tale of two Paks. Biology of the cell/under the auspices of the European Cell Biology Organization. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- 75.Pirraglia C, Walters J, Myat MM. Pak1 control of E-cadherin endocytosis regulates salivary gland lumen size and shape. Development. 2010;137:4177–4189. doi: 10.1242/dev.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin VP, Thummel CS. Mechanisms of steroid-triggered programmed cell death in Drosophila. Seminars in cell & developmental biology. 2005;16:237–243. doi: 10.1016/j.semcdb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 77.Ismat A, Cheshire AM, Andrew DJ. The secreted AdamTS-A metalloprotease is required for collective cell migration. Development. 2013;140:1981–1993. doi: 10.1242/dev.087908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlichting K, Wilsch-Brauninger M, Demontis F, Dahmann C. Cadherin Cad99C is required for normal microvilli morphology in Drosophila follicle cells. Journal of cell science. 2006;119:1184–1195. doi: 10.1242/jcs.02831. [DOI] [PubMed] [Google Scholar]
- 79.D'Alterio C, Tran DD, Yeung MW, Hwang MS, Li MA, Arana CJ, Mulligan VK, Kubesh M, Sharma P, Chase M, Tepass U, Godt D. Drosophila melanogaster Cad99C, the orthologue of human Usher cadherin PCDH15, regulates the length of microvilli. J Cell Biol. 2005;171:549–558. doi: 10.1083/jcb.200507072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chung SY, Andrew DJ. Cadherin 99C regulates apical expansion and cell rearrangement during epithelial tube elongation. Development. 2014 doi: 10.1242/dev.104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bradley PL, Myat MM, Comeaux CA, Andrew DJ. Posterior migration of the salivary gland requires an intact visceral mesoderm and integrin function. Dev Biol. 2003;257:249–262. doi: 10.1016/s0012-1606(03)00103-9. [DOI] [PubMed] [Google Scholar]
- 82.Vining MS, Bradley PL, Comeaux CA, Andrew DJ. Organ positioning in Drosophila requires complex tissue-tissue interactions. Dev Biol. 2005;287:19–34. doi: 10.1016/j.ydbio.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 83.Harris KE, Schnittke N, Beckendorf SK. Two ligands signal through the Drosophila PDGF/VEGF receptor to ensure proper salivary gland positioning. Mechanisms of development. 2007;124:441–448. doi: 10.1016/j.mod.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kolesnikov T, Beckendorf SK. NETRIN and SLIT guide salivary gland migration. Dev Biol. 2005;284:102–111. doi: 10.1016/j.ydbio.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 85.Jones NA, Kuo YM, Sun YH, Beckendorf SK. The Drosophila Pax gene eye gone is required for embryonic salivary duct development. Development. 1998;125:4163–4174. doi: 10.1242/dev.125.21.4163. [DOI] [PubMed] [Google Scholar]
- 86.Kerman BE, Andrew DJ. Staying alive: dalmation mediated blocking of apoptosis is essential for tissue maintenance. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239:1609–1621. doi: 10.1002/dvdy.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Irvine KD, Wieschaus E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development. 1994;120:827–841. doi: 10.1242/dev.120.4.827. [DOI] [PubMed] [Google Scholar]
- 88.Iwaki DD, Johansen KA, Singer JB, Lengyel JA. drumstick, bowl, and lines are required for patterning and cell rearrangement in the Drosophila embryonic hindgut. Developmental biology. 2001;240:611–626. doi: 10.1006/dbio.2001.0483. [DOI] [PubMed] [Google Scholar]
- 89.Lengyel JA, Iwaki DD. It takes guts: the Drosophila hindgut as a model system for organogenesis. Developmental biology. 2002;243:1–19. doi: 10.1006/dbio.2002.0577. [DOI] [PubMed] [Google Scholar]
- 90.Jung AC, Denholm B, Skaer H, Affolter M. Renal tubule development in Drosophila: a closer look at the cellular level. Journal of the American Society of Nephrology: JASN. 2005;16:322–328. doi: 10.1681/ASN.2004090729. [DOI] [PubMed] [Google Scholar]
- 91.Caussinus E, Colombelli J, Affolter M. Tip-cell migration controls stalk-cell intercalation during Drosophila tracheal tube elongation. Current biology: CB. 2008;18:1727–1734. doi: 10.1016/j.cub.2008.10.062. [DOI] [PubMed] [Google Scholar]
- 92.Keller R, Davidson L, Edlund A, Elul T, Ezin M, Shook D, Skoglund P. Mechanisms of convergence and extension by cell intercalation. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2000;355:897–922. doi: 10.1098/rstb.2000.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 94.Moore AW, Barbel S, Jan LY, Jan YN. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133:495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- 96.Lin W, Metzakopian E, Mavromatakis YE, Gao N, Balaskas N, Sasaki H, Briscoe J, Whitsett JA, Goulding M, Kaestner KH, Ang SL. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Developmental biology. 2009;333:386–396. doi: 10.1016/j.ydbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 97.Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. The EMBO journal. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- 98.Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & development. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao N, Le Lay J, Qin W, Doliba N, Schug J, Fox AJ, Smirnova O, Matschinsky FM, Kaestner KH. Foxa1 and Foxa2 maintain the metabolic and secretory features of the mature beta-cell. Molecular endocrinology. 2010;24:1594–1604. doi: 10.1210/me.2009-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smit RB, Schnabel R, Gaudet J. The HLH-6 transcription factor regulates C. elegans pharyngeal gland development and function. PLoS genetics. 2008;4:e1000222. doi: 10.1371/journal.pgen.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barbosa S, Fasanella G, Carreira S, Llarena M, Fox R, Barreca C, Andrew D, O'Hare P. An orchestrated program regulating secretory pathway genes and cargos by the transmembrane transcription factor CREB-H. Traffic. 2013;14:382–398. doi: 10.1111/tra.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murakami T, Saito A, Hino S, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Wanaka A, Imaizumi K. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nature cell biology. 2009;11:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- 103.Seshaiah P, Miller B, Myat MM, Andrew DJ. pasilla, the Drosophila homologue of the human Nova-1 and Nova-2 proteins, is required for normal secretion in the salivary gland. Developmental biology. 2001;239:309–322. doi: 10.1006/dbio.2001.0429. [DOI] [PubMed] [Google Scholar]
- 104.Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR. Conservation of an RNA regulatory map between Drosophila and mammals. Genome research. 2011;21:193–202. doi: 10.1101/gr.108662.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Massarwa R, Schejter ED, Shilo BZ. Apical secretion in epithelial tubes of the Drosophila embryo is directed by the Formin-family protein Diaphanous. Developmental cell. 2009;16:877–888. doi: 10.1016/j.devcel.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 106.Rousso T, Shewan AM, Mostov KE, Schejter ED, Shilo BZ. Apical targeting of the formin Diaphanous in Drosophila tubular epithelia. eLife. 2013;2:e00666. doi: 10.7554/eLife.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Geron E, Schejter ED, Shilo BZ. Directing exocrine secretory vesicles to the apical membrane by actin cables generated by the formin mDia1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10652–10657. doi: 10.1073/pnas.1303796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Redfern CPF. Ecdysteroid synthesis by the ring gland of Drosophila melanogaster during late-larval, prepupal and pupal development. Journal of insect physiology. 1983;29:65–71. [Google Scholar]
- 109.Warren JT, Yerushalmi Y, Shimell MJ, O'Connor MB, Restifo LL, Gilbert LI. Discrete pulses of molting hormone, 20-hydroxyecdysone, during late larval development of Drosophila melanogaster: correlations with changes in gene activity. Developmental dynamics: an official publication of the American Association of Anatomists. 2006;235:315–326. doi: 10.1002/dvdy.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clever U, Karlson P. Induction of puff changes in the salivary gland chromosomes of Chironomus tentans by ecdysone. Experimental cell research. 1960;20:623–626. doi: 10.1016/0014-4827(60)90141-5. [DOI] [PubMed] [Google Scholar]
- 111.Ashburner M, Lemeunier F. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VII. Homology of puffing patterns on chromosome arm 3L in D. melanogaster and D. yakuba, with notes on puffing in D. teissieri. Chromosoma. 1972;38:283–295. doi: 10.1007/BF00290926. [DOI] [PubMed] [Google Scholar]
- 112.Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VI. Induction by ecdysone in salivary glands of D. melanogaster cultured in vitro. Chromosoma. 1972;38:255–281. doi: 10.1007/BF00290925. [DOI] [PubMed] [Google Scholar]
- 113.Lehmann M, Korge G. Ecdysone regulation of the Drosophila Sgs-4 gene is mediated by the synergistic action of ecdysone receptor and SEBP 3. Embo J. 1995;14:716–726. doi: 10.1002/j.1460-2075.1995.tb07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crossgrove K, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complex early gene directly regulates late gene transcription during the ecdysone-induced puffing cascade. Developmental biology. 1996;180:745–758. doi: 10.1006/dbio.1996.0343. [DOI] [PubMed] [Google Scholar]
- 115.Fletcher JC, D'Avino PP, Thummel CS. A steroid-triggered switch in E74 transcription factor isoforms regulates the timing of secondary-response gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4582–4586. doi: 10.1073/pnas.94.9.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun G, Zhu J, Li C, Tu Z, Raikhel AS. Two isoforms of the early E74 gene, an Ets transcription factor homologue, are implicated in the ecdysteroid hierarchy governing vitellogenesis of the mosquito, Aedes aegypti. Molecular and cellular endocrinology. 2002;190:147–157. doi: 10.1016/s0303-7207(01)00726-2. [DOI] [PubMed] [Google Scholar]
- 117.Zhou B, Hiruma K, Jindra M, Shinoda T, Segraves WA, Malone F, Riddiford LM. Regulation of the transcription factor E75 by 20-hydroxyecdysone and juvenile hormone in the epidermis of the tobacco hornworm, Manduca sexta, during larval molting and metamorphosis. Developmental biology. 1998;193:127–138. doi: 10.1006/dbio.1997.8798. [DOI] [PubMed] [Google Scholar]
- 118.Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes & development. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- 119.Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- 120.Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis Developmental biology. 1974;39:141–157. doi: 10.1016/s0012-1606(74)80016-3. [DOI] [PubMed] [Google Scholar]
- 121.Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- 122.King-Jones K, Charles JP, Lam G, Thummel CS. The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell. 2005;121:773–784. doi: 10.1016/j.cell.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 123.Talbot WS, Swyryd EA, Hogness DS. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell. 1993;73:1323–1337. doi: 10.1016/0092-8674(93)90359-x. [DOI] [PubMed] [Google Scholar]
- 124.Biyasheva A, Do TV, Lu Y, Vaskova M, Andres AJ. Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Developmental biology. 2001;231:234–251. doi: 10.1006/dbio.2000.0126. [DOI] [PubMed] [Google Scholar]
- 125.Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, Cherbas P, Evans RM. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 126.Costantino BF, Bricker DK, Alexandre K, Shen K, Merriam JR, Antoniewski C, Callender JL, Henrich VC, Presente A, Andres AJ. A novel ecdysone receptor mediates steroid-regulated developmental events during the mid-third instar of Drosophila. PLoS genetics. 2008;4:e1000102. doi: 10.1371/journal.pgen.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Costantino BF. UNLV Theses/Dissertations/Professional Papers/Capstones. 2008. The Larval salivary gland of Drosophila melanogaster: A model system for temporal and spatial steroid hormone regulation. [Google Scholar]
- 128.Mach V, Ohno K, Kokubo H, Suzuki Y. The Drosophila fork head factor directly controls larval salivary gland-specific expression of the glue protein gene Sgs3. Nucleic acids research. 1996;24:2387–2394. doi: 10.1093/nar/24.12.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roth GE, Wattler S, Bornschein H, Lehmann M, Korge G. Structure and regulation of the salivary gland secretion protein gene Sgs-1 of Drosophila melanogaster. Genetics. 1999;153:753–762. doi: 10.1093/genetics/153.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li TR, White KP. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Developmental cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 131.Muskavitch MA, Hogness DS. An expandable gene that encodes a Drosophila glue protein is not expressed in variants lacking remote upstream sequences. Cell. 1982;29:1041–1051. doi: 10.1016/0092-8674(82)90467-6. [DOI] [PubMed] [Google Scholar]
- 132.Garfinkel MD, Pruitt RE, Meyerowitz EM. DNA sequences, gene regulation and modular protein evolution in the Drosophila 68C glue gene cluster. Journal of molecular biology. 1983;168:765–789. doi: 10.1016/s0022-2836(83)80074-6. [DOI] [PubMed] [Google Scholar]
- 133.Farkas R, Suakova G. Developmental regulation of granule size and numbers in larval salivary glands of drosophila by steroid hormone ecdysone. Cell biology international. 1999;23:671–676. doi: 10.1006/cbir.1999.0433. [DOI] [PubMed] [Google Scholar]
- 134.Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Developmental biology. 1993;160:388–404. doi: 10.1006/dbio.1993.1315. [DOI] [PubMed] [Google Scholar]
- 135.Burgess J, Jauregui M, Tan J, Rollins J, Lallet S, Leventis PA, Boulianne GL, Chang HC, Le Borgne R, Kramer H, Brill JA. AP-1 and clathrin are essential for secretory granule biogenesis in Drosophila. Molecular biology of the cell. 2011;22:2094–2105. doi: 10.1091/mbc.E11-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thummel CS. Ecdysone-regulated puff genes 2000. Insect biochemistry and molecular biology. 2002;32:113–120. doi: 10.1016/s0965-1748(01)00112-6. [DOI] [PubMed] [Google Scholar]
- 137.Cao C, Liu Y, Lehmann M. Fork head controls the timing and tissue selectivity of steroid-induced developmental cell death. J Cell Biol. 2007;176:843–852. doi: 10.1083/jcb.200611155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Juhasz G, Sass M. Hid can induce, but is not required for autophagy in polyploid larval Drosophila tissues. European journal of cell biology. 2005;84:491–502. doi: 10.1016/j.ejcb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 139.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee CY, Simon CR, Woodard CT, Baehrecke EH. Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Developmental biology. 2002;252:138–148. doi: 10.1006/dbio.2002.0838. [DOI] [PubMed] [Google Scholar]
- 141.Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJ, Marra MA. A SAGE approach to discovery of genes involved in autophagic cell death. Current biology: CB. 2003;13:358–363. doi: 10.1016/s0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 142.Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Current biology: CB. 2003;13:350–357. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 143.Wang L, Lam G, Thummel CS. Med24 and Mdh2 are required for Drosophila larval salivary gland cell death. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239:954–964. doi: 10.1002/dvdy.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee HH, Frasch M. Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development. 2005;132:1429–1442. doi: 10.1242/dev.01687. [DOI] [PubMed] [Google Scholar]
- 145.Maruyama R, Andrew DJ. Drosophila as a model for epithelial tube formation. Developmental dynamics: an official publication of the American Association of Anatomists. 2012;241:119–135. doi: 10.1002/dvdy.22775. [DOI] [PMC free article] [PubMed] [Google Scholar]