Abstract
Biologists have long observed that physiological and developmental processes are insensitive, or robust, to many genetic and environmental perturbations. A complete understanding of the evolutionary causes and consequences of this robustness is lacking. Recent progress has been made in uncovering the regulatory mechanisms that underlie environmental robustness in particular. Less is known about robustness to the effects of mutations, and indeed the evolution of mutational robustness remains a controversial topic. The controversy has spread to related topics, in particular the evolutionary relevance of cryptic genetic variation. This review aims to synthesize current understanding of robustness mechanisms and to cut through the controversy by shedding light on what is and is not known about mutational robustness. Some studies have confused mutational robustness with non-additive interactions between mutations (epistasis). We conclude that a profitable way forward is to focus investigations (and rhetoric) less on mutational robustness and more on epistasis.
Keywords: microenvironmental variation, macroenvironmental variation, phenotypic plasticity, canalization, mutational robustness, epistasis, genotype networks, cryptic genetic variation, conditional neutrality
1. INTRODUCTION
Physiological and developmental processes produce outcomes that are relatively insensitive, or robust, to many genetic and environmental perturbations (Wagner 2007, Masel & Siegal 2009). This observation is simultaneously trivial and highly controversial. It is trivial because no living system can persist without regulating its internal composition. Homeostatic mechanisms maintain the balance of metabolic products in the face of fluctuating environments, and pattern-formation mechanisms operate with high fidelity through feedback controls and switch-like fate decisions. Robustness to some changes in the internal or external environment is therefore virtually a given, although much remains to be discovered about how such robustness is achieved, how it relates to phenotypic plasticity (stereotyped phenotypic changes in response to environmental differences), how lowering its level might in some cases be adaptive, and how it constrains (or promotes) phenotypic divergence (Levy & Siegal 2012).
Unlike robustness to environmental perturbations, robustness to genetic perturbations attracts much debate. To be clear, there is no debate over whether some mutations have no phenotypic consequence. Indeed, it is a cornerstone of the neutral theory of molecular evolution that many genetic changes are inconsequential, and even the highest estimates of the proportion of genomic sites that are functional do not approach 100% (ENCODE Proj Consort 2012). Instead, debate surrounds whether robustness to mutations is something that evolved via natural selection to be greater than it otherwise would be. The argument favoring such selection is that would-be deleterious genotypes arise continually through mutation and recombination, at a rate sufficient to present a selective force to increase robustness against their effects (Waddington 1957, Wagner et al. 1997, Lauring et al. 2013). A common counterargument is that robustness to genetic perturbations is a by-product of robustness to environmental perturbations. That is, the feedbacks, thresholds and other non-linearities that evolved to confer greater robustness against the effects of environmental perturbations are expected to confer greater robustness against the effects of mutations as well (Waddington 1957, Meiklejohn & Hartl 2002).
Note that this counterargument does not challenge the notion that organisms are more robust to the phenotypic effects of mutations than they otherwise would be. Indeed, it makes the strong prediction that impairing a mechanism that confers greater environmental robustness will increase the phenotypic effects of mutations. However, little empirical evidence exists to corroborate this prediction (Masel & Siegal 2009). What is worse is that a number of studies touted as demonstrating higher mutational robustness in wild-type organisms have not in reality done so (Hermisson & Wagner 2004). It has been argued that failure to acknowledge the paucity of evidence has contributed to controversy and directed the field away from more useful directions (Hermisson & Wagner 2004, Richardson et al. 2013).
Further debate surrounds the claim that some gene products modulate the extent of mutational robustness in a way that is evolutionarily adaptive. Such gene products have been called evolutionary capacitors (Rutherford & Lindquist 1998). The standard definition states that a capacitor normally contributes to a high level of robustness to mutations. As a result, mutations with effects on gene function but no effects on phenotypes accumulate in the population. Under rare circumstances, perhaps tied to environmental stresses in which new phenotypes might be beneficial, the capacitor’s robustness-conferring capacity is lowered so that the potentially adaptive accumulated (“cryptic”) genetic variation is released (Rutherford 2000, Sangster et al. 2004). Response to this capacitor hypothesis has ranged from outright dismissal of any evolutionary mechanism that appears to anticipate the future to full embracing of the idea that stores of previously hidden genetic variation are a major contributor to key evolutionary transitions (Dickinson & Seger 1999, Rohner et al. 2013). Again, controversy has obscured questions — in this case about the evolutionary implications of cryptic genetic variation and genetic interactions in general — that are important whether the capacitor hypothesis is true or not (Hermisson & Wagner 2004, Richardson et al. 2013, Siegal 2013, Masel 2013).
This review aims to assess the current state of research on how robustness is achieved and what its evolutionary implications are. Areas of misunderstanding and controversy are addressed directly from both theoretical and empirical angles. It is hoped that this direct approach will help to cut through some of the controversy and reveal key areas that need greater investigation.
2. MECHANISMS OF ROBUSTNESS
Environmental or genetic perturbation can influence living cells at different levels, from individual genes or gene products to pathways to global cellular homeostasis. It is conceivable that cells may evolve different mechanisms to achieve robustness depending on the source of perturbation. However, there might not be a simple one-to-one mapping of perturbation types and robustness mechanisms: Multiple mechanisms may be used simultaneously to cope with a single type of perturbation or a single mechanism may handle multiple types of perturbations.
Uncovering the detailed mechanisms underlying robustness against various perturbations will advance the more general goals of understanding (a) how organisms integrate and regulate different systems in response to prevailing external and internal conditions and (b) how such integrated systems evolve through time in response to different selective pressures. Although these issues have long drawn biologists’ attention (e.g., Waddington 1942), progress in understanding robustness at the molecular and cellular levels has accelerated recently with advances in modeling and experimental tools. We discuss this recent progress in this section, considering different sources of perturbation separately then together.
2.1 Robustness against microenvironmental variation
Most cellular processes are executed by proteins. During the transcriptional, translational and posttranslational processes that produce functional proteins (as well as during the function of those proteins), variation will inevitably arise due to stochastic fluctuations, particularly in the steps involving small numbers of molecules. Variation due to fluctuations in the internal cellular environment or the local external environment is defined as microenvironmental variation. Microenvironmental variation is a pervasive obstacle to fidelity of cellular behaviors.
A simple solution to the problem of microenvironmental variation for those proteins without stoichiometric restrictions is to be produced in excess so that their performance will not be compromised by such variation (Hartl et al. 1985). This increased expression might explain why some duplicated genes are maintained for a long time without obvious functional diversification (Wilkins et al. 1997, Kellis et al. 2004). Consistent with the idea that higher average expression buffers against microenvironmental variation is the observation that diploid cells can tolerate losing one copy of the vast majority of genes without obvious growth defects (Deutschbauer et al. 2005, Springer et al. 2010). Protein abundance typically correlates well with gene copy number, so the unaltered fitness of hemizygous cells suggests that most proteins are expressed in excess (Springer et al. 2010, Torres et al. 2010). However, using such a strategy bears the cost of wasting energy on making extra materials and losing the sensitivity to respond to other perturbations.
For those proteins with stoichiometric restrictions, mechanisms that specifically reduce cell-to-cell variability in protein abundance might be beneficial. Although cell-to-cell variability in protein abundance scales inversely with average abundance, essential yeast proteins tend to have proportionally less variability than non-essential ones (Bar-Even et al. 2006). Nonetheless, high variability might in some cases be beneficial. Because some populations experience acute changes to stressful environments, heterogeneity in benign conditions can constitute a bet-hedging strategy (Fraser & Kaern 2009, Levy et al. 2012, Levy & Siegal 2012). Consistent with bet hedging, expression of stress-related genes in yeast cells is more variable than that of housekeeping genes (Fraser et al. 2004, Bar-Even et al. 2006, Newman et al. 2006).
Several mechanisms have been observed to control stochastic cell-to-cell variation (often called “noise”) in specific genes. Lower noise can be achieved by frequent promoter activation even if average transcription levels are the same (Blake et al. 2003, Raser & O’Shea 2004). In yeast, the promoter-activation frequency is mainly determined by the promoter architecture (Blake et al. 2006, Hornung et al. 2012, Carey et al. 2013). This finding suggests that natural selection is able to shape expression robustness in a gene-specific manner. Similarly, protein-expression noise can be reduced, without changing the average protein abundance, by increasing mRNA abundance while decreasing translation rate. Cells need to pay the cost of making more mRNA molecules when using this control mechanism. Nonetheless, this strategy appears to be common in the housekeeping genes of both prokaryotes and eukaryotes, indicating that it is a common mechanism selected to regulate noise (Ozbudak et al. 2002, Fraser et al. 2004).
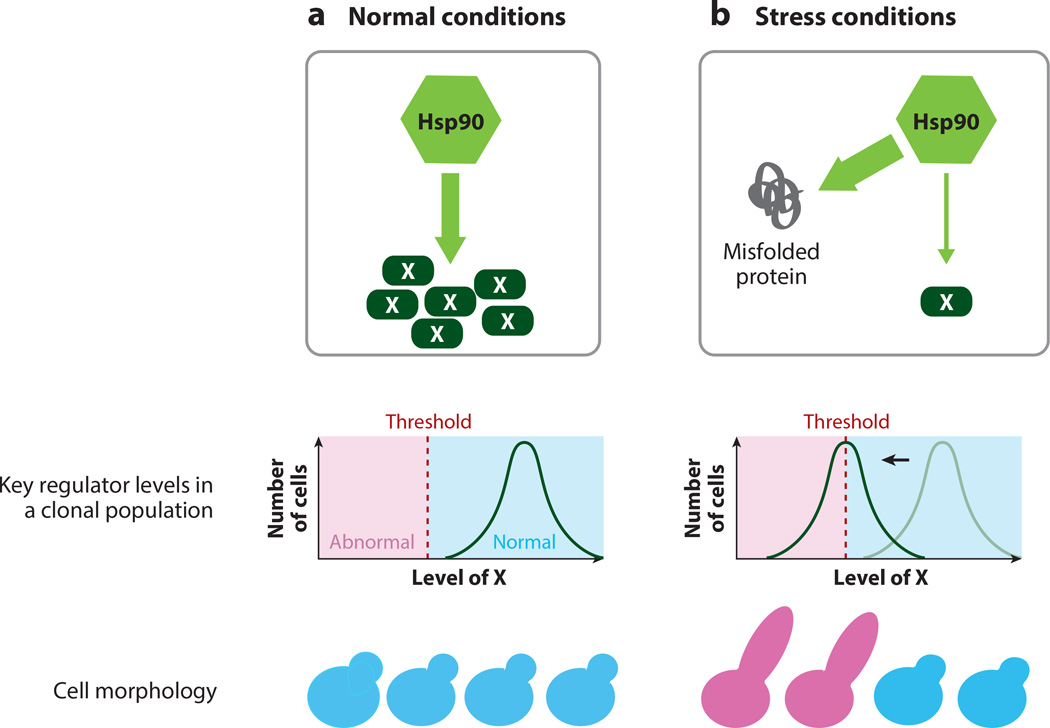
Specific noise control also occurs after translation. The heat-shock protein 90 (Hsp90) chaperone represents the best-studied example. In eukaryotic cells, Hsp90 promotes maturation and stability of many key regulators (Taipale et al. 2010). It is hypothesized that Hsp90 allows cells to maintain above-threshold levels of those regulators and thereby reduces the impact of stochastic fluctuations. Decreased Hsp90 activity leads to increased levels of within-strain variation in Arabidopsis seedling phenotypes and yeast morphology (Sangster et al. 2007, Hsieh et al. 2013). In the yeast study, the average abundances of two morphogenesis regulators were reduced to near the threshold level, resulting in the observed morphological variation in low-Hsp90 cells (Hsieh et al. 2013) (Figure 1). A potentially important feature of this mechanism is that, because Hsp90’s chaperone capacity can be overwhelmed by stress, Hsp90 can work as an environmental sensor and, thus, enable cells to fine-tune phenotypic diversity in response to environmental stress. In Drosophila, other molecular chaperones — Hsp22, Hsp67, and Hsp70 — were also observed to affect either within-individual variation (measured by asymmetry of bilateral traits) or among-individual variation in morphology, but the detailed molecular mechanisms are still unclear (Takahashi et al. 2010; 2011a).
Figure 1. Model for Hsp90 impairment causing increases to microenvironmental variation.
(a) Under normal conditions, a key morphogenesis regulator and client of Hsp90 (X) is present at sufficient levels in all cells, so all cells bud properly. (b) Under stress conditions, Hsp90 is diverted to other proteins, so fewer properly folded X molecules are present. The level of active X per cell therefore approaches the threshold required for proper bud morphology. Some cells have sufficient active X (blue) whereas others do not (pink), leading to phenotypic heterogeneity. (Figure based on Hsieh et al. 2013).
Gene-regulatory interactions have also been shown to be critical in maintaining robustness against microenvironmental variation (Ozbudak et al. 2002, Ramsey et al. 2006, Benazet et al. 2009, MacNeil & Walhout 2011, Paulsen et al. 2011, Denby et al. 2012). A simple negative feedback loop comprising a regulator that represses its own transcription is sufficient to increase output stability by twofold (Becskei & Serrano 2000). Because of their homeostatic nature, it is not surprising that negative feedback loops are frequently observed in eukaryotic signaling pathways (Freeman 2000, Tsang et al. 2007).
More complicated interactions between a few regulators (termed network motifs) may also confer robustness. The incoherent feedforward loop (IFFL) — in which an upstream regulator and its target jointly regulate a third gene but have opposing effects on that gene — can have similar dynamic properties to the negative feedback loop in that fluctuations in the upstream regulator lead to concerted changes in both activation and repression of the third gene (Tsang et al. 2007). A particularly important type of IFFL in this respect is one in which an upstream factor activates transcription of a target gene and a microRNA (miRNA) that represses translation of the target gene’s mRNA (Tsang et al. 2007). Some species-specific regulatory pathways involve novel IFFLs (Freeman 2000, Tsang et al. 2007), perhaps suggesting that they have arisen from existing feedback or feedforward loops and confer additional robustness on them. One caveat, however, is that kinetics are quite important to the motif behavior. For example, some IFFLs might indeed buffer fluctuations whereas others, depending on delays in the system, might generate pulses or accelerate responses (Alon 2007). Therefore the ability to make inferences merely from knowing whether activating or repressing interactions exist is limited. Moreover, the absence of a clear IFFL does not necessarily mean that a miRNA is not involved in buffering: miR-263a and miR-263b were shown to increase fidelity of apoptotic cell pruning during Drosophila eye development, but a putative IFFL was found not to be operating (Hilgers et al. 2010).
Beyond network motifs, complex regulatory networks with many interconnections are also thought to play a role in robustness. Computational simulations suggest that regulatory networks that produce biologically relevant outputs (e.g., yield stable steady states of gene expression or regular spatial patterns) are intrinsically robust to environmental and genetic variations (von Dassow et al. 2000, Siegal & Bergman 2002). Corroborating this notion, a genome-wide screen for gene deletions in yeast that increase morphological variation among genetically identical cells in the same environment yielded hundreds of genes, which tended to be highly connected in cellular networks (Levy & Siegal 2008).
2.2 Robustness against macroenvironmental variation versus phenotypic plasticity
It has been argued that environmental perturbations represent the major selective pressure for the evolution of buffering systems (Wagner et al. 1997, Meiklejohn & Hartl 2002). Variation due to changes in temperature, moisture, nutrient concentrations, and other variables is termed macroenvironmental variation to distinguish it from the stochastic fluctuations of microenvironmental variation. In simple terms, organisms might successfully suppress macroenvironmental variation in two ways: maintaining a single phenotype despite environmental changes (robustness) or altering developmental or physiological processes to produce a different phenotype suited to each environment (phenotypic plasticity). Plasticity implies a kind of robustness in that particular environments reliably induce particular phenotypes, so robustness and plasticity should not be thought of as opposites (Levy & Siegal 2012). The relative advantages of unitary versus plastic responses likely depend on life-history characteristics of the organism as well as on complicated ecological forces that include the dynamics of environmental change and a population’s geographic distribution (Moran 1992, Sultan & Spencer 2002, Liefting et al. 2009, Pfennig et al. 2010).
Theoretically, many of the aforementioned microenvironmental buffering systems would be expected to help suppress macroenvironmental variation as well. However, protecting against particularly acute perturbations might require specific mechanisms. One indication that mechanisms of macroenvironmental robustness differ from mechanisms of microenvironmental robustness comes from a Drosophila screen for genomic regions that affect variation in bristle traits (Takahashi et al. 2012). Deficiencies of 29 genomic regions were found to alter the environmental sensitivity of the bristle traits. However, the deficiencies’ effects on within-individual variation were not correlated with their effects on environmental sensitivity, suggesting that independent mechanisms confer robustness against macro- and microenvironmental variations (Takahashi et al. 2012). Likewise, although miR-263a and miR-263b were found to promote microenvironmental robustness of Drosophila eye development, they do not appear to have a role in robustness against macroenvironmental perturbations (Hilgers et al. 2010).
Despite the miR-263 result, regulatory interactions involving miRNAs retain particular interest in the study of macroenvironmental robustness. As described above, miRNAs are a common component of negative feedback and feedforward loops and have been specifically proposed to damp perturbations (Hornstein & Shomron 2006, Tsang et al. 2007). Computational simulations and mathematical analysis suggest that the buffering efficiency of miRNAs is often better than that of protein regulators (Osella et al. 2011). Regulatory motifs involving miRNAs can respond to perturbations more quickly because miRNAs regulate genes posttranscriptionally. In Drosophila, miR-7 maintains stability against environmental perturbations during development of sensory organs (Li et al. 2009). Another miRNA acting in the specification of sensory cell fate in Drosophila, miR-9a, also confers robustness against environmental perturbations (Cassidy et al. 2013). It remains to be determined how many miRNAs have evolved to play roles similar to those of miR-7 and miR-9a, but the number is potentially very large. New miRNA families continuously appear, and their target genes sometimes change quickly through evolution (Hertel et al. 2006, Li et al. 2009). Even conserved miRNAs can have few or no noticeable effects when removed under standard laboratory conditions (Pelaez & Carthew 2012). Moreover, individual miRNAs tend to target mRNAs that encode proteins that group into modules or occupy bottlenecks in regulatory networks, possibly indicating that the miRNAs’ primary role is to confer robustness rather than to alter regulatory programs (Pelaez & Carthew 2012).
The connection between robustness and plasticity might be especially important to evolution. High levels of phenotypic variation generated through plasticity might increase the chance of population survival in novel hostile environments, which in turn would buy time for the population to accumulate adaptive mutations (Baldwin 1896, Price et al. 2003). Temporary perturbations that reduce robustness might also turn unitary phenotypes into plastic ones that then give natural selection a substrate on which to act, either to favor a particular new phenotype (“genetic assimilation”) or to establish the plastic response even in the absence of the original perturbation (“genetic accommodation”) (Waddington 1953, Behera & Nanjundiah 2004, Suzuki & Nijhout 2006).
2.3 Robustness against mutations
Theory and computer simulations predict that mechanisms increasing robustness to the effects of mutations can evolve in a population with a high mutation rate or a large population size under stabilizing or fluctuating selection (Wagner et al. 1997, Kawecki 2000, Wilke et al. 2001). The effects of mutations might be manifest at multiple levels of cellular organization, from individual codons within protein-coding sequences to the stabilities and activities of individual gene products to networks of interacting genes to complex multicellular phenotypes. For example, greater genetic robustness of a protein-coding gene can be achieved by biased codon usage (Stephens & Waelbroeck 1999). Because synonymous codons can differ in the number of mutations that result in an amino acid substitution (termed codon volatility; Plotkin et al. 2004), by biasing to codons with lower volatility, a protein can tolerate more mutations or transcriptional errors without changing its amino acid sequence. Analyses of genes or genomes from a few viruses and pathogens concluded that protein domains recognized by host immune systems exhibit biases to more volatile codons, indicating that proteins experiencing different selective regimes adopt different codon usage (Stephens & Waelbroeck 1999, Plotkin & Dushoff 2003, Plotkin et al. 2004), consistent with a population-genetic model (Plotkin et al. 2006).
Theoretical work also suggests that robustness to mutations emerges as a property of complex metabolic and regulatory networks. A classic example is that the dependence of overall flux in a metabolic pathway on any individual enzyme decreases as the number of enzymes in the pathway increases; this is most likely why wild-type alleles are typically dominant over loss-of-function alleles (Kacser & Burns 1981). Similarly, computationally simulated gene-regulatory networks of sufficient complexity evolve to become less sensitive to the effects of new mutations (Siegal & Bergman 2002). Moreover, deleting an arbitrary gene in such an evolved network tends to reveal cryptic variation that had accumulated in the other genes, suggesting that each member of the network, rather than a few dedicated factors, contributes to the higher robustness to mutations (Bergman & Siegal 2003).
The most direct evidence supporting the existence of mechanisms that alter the level of mutational robustness comes from laboratory evolution experiments. When cytochrome P450 proteins were evolved under constant selection pressure to maintain the same biochemical function, evolved proteins from larger populations of molecules became more robust to mutations than those from smaller populations of molecules (Bloom et al. 2007). Laboratory evolution of an RNA virus provides additional evidence. Populations of bacteriophage φ6 cultured serially at a high multiplicity of infection evolve lower robustness to the phenotypic effects of mutations than those cultured serially at a low multiplicity of infection, presumably because genetic complementation between co-infecting strains provides a form of robustness that reduces selection for individual-level robustness (Montville et al. 2005, McBride et al. 2008). Greater mutational robustness was favored during competition experiments with vesicular stomatitis virus populations as well (Sanjuan et al. 2007).
These results indicate that selection can directly enhance or relax mutational robustness. They also highlight the recurring finding that a high product of two population-genetic parameters, the population size and the mutation rate, favors greater mutational robustness. RNA viruses meet this criterion well, making them excellent for investigating the mechanisms of mutational robustness (Lauring et al. 2013). To date, most focus has been on viral genome structure, although the hope is that more precise molecular mechanisms will be identified (Lauring et al. 2013).
One proposed mechanism of increasing mutational robustness is via molecular chaperones. By aiding correct folding and enhancing stability, chaperones might allow mutated proteins to retain function and therefore not alter phenotypes (Sangster et al. 2004). Mutation-accumulation experiments in bacteria found that levels of the chaperones DnaK and GroEL were increased in cells evolved with high mutation loads. In addition, the fitnesses of these mutant cells, but not the ancestral wild-type cells, could be improved when GroEL was overexpressed (Fares et al. 2002, Maisnier-Patin et al. 2005). In endosymbiotic bacteria, which possess degenerated genomes, GroEL is also overexpressed, suggesting a role in stabilizing otherwise inferior proteins (Charles et al. 1997, Fares et al. 2004). However, these forms of evidence do not directly implicate chaperones in natively increasing mutational robustness. In particular, the results of overexpression do not necessarily reveal the endogenous function of a protein.
In eukaryotes, Hsp90 impairment has been found to reveal cryptic genetic variation in organisms ranging from yeast to flies to vertebrates to plants (Jarosz et al. 2010, Rohner et al. 2013). Other than a few cases in yeast, the molecular identities of the Hsp90-dependent cryptic variants remain elusive (Jarosz & Lindquist 2010). Here again, it is important to recognize the nature of the evidence and what it does or does not say about the role of Hsp90 in mutational robustness. Because the cryptic variants in the Hsp90 studies were not new mutations but mutations that had survived the filter of natural selection to exist in cryptic form, they do not measure mutational robustness (Hermisson & Wagner 2004). Similar considerations apply to the demonstration that deletions of chromatin regulators reveal cryptic differences between yeast species in gene expression (Tirosh et al. 2010). The distinction between impairing mutational robustness and revealing cryptic genetic variation is discussed in detail in Section 3, Evolutionary Consequences of Robustness, below. For now, the key point is that the cases many tout as showing mechanisms of mutational robustness are circumstantial at best. To be clear, the best evidence that mutational robustness can be increased comes from viruses and experimental evolution, in which the product of the population size and the mutation rate is high. This situation might not apply to many organisms.
2.4 Congruence of robustness mechanisms
How do organisms acquire different types of robustness mechanisms in the first place? The current level of robustness may have evolved directly under natural selection or may be an intrinsic property of a system. Two possible scenarios are speculated under the selection hypothesis. The first is that systems conferring robustness to environmental or mutational perturbations are selected independently. One weakness of this hypothesis is that the conditions under which mutational robustness is expected to evolve are quite restrictive. The adaptive benefit of greater robustness to environmental perturbations increases with the intensity of the perturbations. By contrast, only a sweet spot of intermediate selection intensity favors evolution of greater mutational robustness: Strong stabilizing selection quickly purges mutations from the population that would have been the selective pressure to evolve greater robustness, whereas weak stabilizing selection is not sufficient to drive the evolution of greater robustness because robustness is already a second-order effect (Wagner et al. 1997). This imbalance in adaptive benefits has led to the “congruence” hypothesis that mutational robustness evolves as a by-product of environmental robustness (Meiklejohn & Hartl 2002). Of course, the congruence hypothesis is only necessary if one is trying to explain why organisms are more robust to both environmental and mutational perturbations than one would otherwise expect them to be. However, as argued in the previous section and as is revisited in Section 3, Evolutionary Consequences of Robustness, the evidence that organisms are indeed more robust to mutations than they “should” be is limited, except perhaps for RNA viruses, which might very well live in that sweet spot. Perhaps this is why evidence bearing on the congruence hypothesis is so mixed.
Some work supports the congruence hypothesis, including in silico and laboratory evolution studies. For example, simulations and analyses of RNA secondary structures suggest that their mutational robustness and environmental robustness are inherently correlated (Ancel & Fontana 2000). Mutation-accumulation experiments in phage φ6 also suggest that mutational and environmental robustness have the same developmental basis (Burch & Chao 2004). In a bacterial evolution experiment an antibiotic resistance gene (TEM-1 β-lactamase) was selected to retain its enzyme activity under error-prone transcription (representing a type of environmental perturbation). The evolved proteins exhibited increased mutational robustness and protein stability (Goldsmith & Tawfik 2009). These results indicate that enhanced environmental and mutational robustness of a molecule may be achieved by selecting for more stable conformations, a conclusion drawn in the experimental evolution of cytochrome P450 as well (Bloom et al. 2007).
Congruence has also been observed in more complex systems. In an early Drosophila experiment, five fitness components were measured for their environmental robustness (the variation within inbred lines) and genetic robustness (the variation among lines). Robustness of the traits increased with their impact on fitness, and the strengths of environmental and genetic robustness were correlated (Stearns et al. 1995). A more recent genome-wide analysis of yeast showed that the effects of a gene deletion on environmental robustness (defined by the number of different environments in which the growth is affected by a mutation) and on genetic robustness (defined by the number of synthetic-lethal interactions made by a mutation) are significantly correlated (Lehner 2010). It is not clear if or how synthetic-lethal interactions can be extrapolated to the spontaneous mutation spectrum, which contains many mutations of much less extreme effect than gene deletions. In a similar way, although Hsp90 has been found in yeast to confer environmental robustness, it is not necessarily the case that its pervasive interactions with cryptic genetic variation are indicative of mutational robustness and, therefore, congruence (Jarosz & Lindquist 2010, Hsieh et al. 2013).
Examples against congruence can also be found, although they too provide indirect evidence at best because spontaneous mutations were not assayed. For example, a quantitative genetic analysis of gene-expression traits in mice found that polymorphisms buffering genetic (among-line) variation were different from those buffering environmental (within-line) variation (Fraser & Schadt 2010). Similarly, in Drosophila deficiency-mapping experiments, distinct genomic regions influenced between-strain variation and within-individual variation of wing-shape development (Takahashi et al. 2011b, Takahashi 2013) (Figure 2). With their analyses of within-strain and between-strain variation, these two studies by Takahashi and colleagues are analogous to the above-noted study by Stearns et al. (1995), but these researchers reached opposite conclusions. However, it should be noted that all polymorphisms in these experiments had been filtered by natural selection, so these experiments give information about cryptic genetic variation but not necessarily mutational robustness. An analogous study to that by Lehner (2010) is that by Cooper et al. (2006), who measured the effects of environmental perturbation and mutational perturbation on Escherichia coli cells after deletion of individual genes. Cooper and colleagues found environmental and mutational robustness to be not positively correlated but instead negatively correlated (Cooper et al. 2006). However, like the study by Lehner (2010), the mutational perturbations were not spontaneous mutations (in this case they were laboratory-generated transposon insertions) (Cooper et al. 2006).
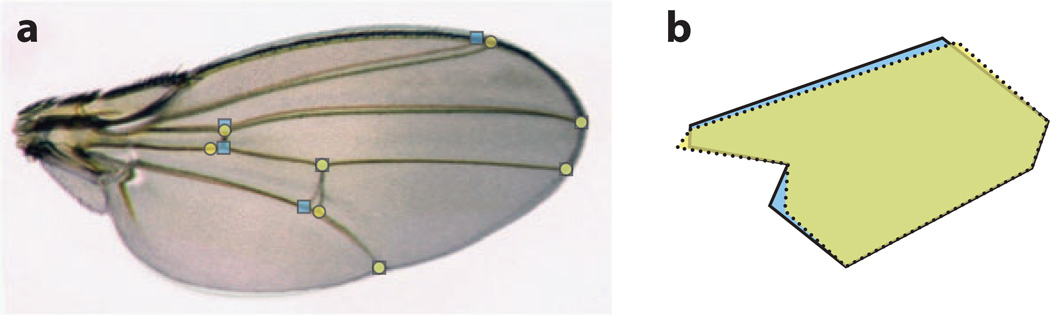
Figure 2. Morphometric analysis enables detection and characterization of loci affecting trait variation.
(a) Two wings with subtle differences in vein positioning are overlaid, with positions of eight landmarks shown for each wing (yellow-filled circles and blue-filled squares, respectively). (b) The landmarks define polygons whose differences can be quantified using morphometric analysis of variation within and between strains. (Image courtesy of Kazuo Takahashi).
Analyses of miRNA precursor stem-loop structures highlight the disagreement in the field about what forms of robustness have been selected and why. An initial analysis of precursor structure did not find congruence between environmental and mutational robustness (Borenstein & Ruppin 2006), suggesting that the previous finding of congruence for RNA structure (Ancel & Fontana 2000) is not universal. However, another study showed that changing the measure of environmental robustness leads to a correlation with mutational robustness, arguing support for congruence in miRNA precursor structures (Szollosi & Derenyi 2009). Yet another study used phylogenetic analysis to reconstruct ancestral miRNA sequences, and these researchers concluded that the primary force acting on miRNA sequences is purifying selection on secondary structure and that neither direct selection of mutational robustness nor selection of mutational robustness as a by-product of environmental applies (Price et al. 2011). It is clear that analyses of robustness have far to go before any consensus is reached on the congruence hypothesis.
3. EVOLUTIONARY CONSEQUENCES OF ROBUSTNESS
A naïve view of the effect of robustness on evolution is that it would constrain divergence. If physiological and developmental processes are organized to reduce the influence of environmental and mutational perturbations, then generating a new phenotype should be less likely. Although true in the very short term, this statement is likely to be false over evolutionarily relevant timescales.
When cryptic genetic variation accumulates it causes diversification of genetic backgrounds on which new mutations may arise. To the extent that epistasis exists, the effect of any new mutation will be background-dependent: There will be some backgrounds on which the new mutation would have a particular phenotypic effect and others on which it would have a different or no effect. The diversity of genetic backgrounds would then mean that the population as a whole would have access to more new phenotypes than if it were isogenic (Wagner 2007, 2011, 2012). This conceptual argument for evolvability correlating positively with mutational robustness has been formalized in mathematical models of so-called neutral networks in genotype space (more recently termed genotype networks) and has some empirical support (McBride et al. 2008, Hayden et al. 2011, Lauring et al. 2013). These studies are discussed further below.
The idea of a population comprising many genetic backgrounds and thereby having greater adaptive potential is reminiscent of the notion of an evolutionary capacitor modulating the effects of cryptic genetic variation. If the cryptic genetic variation is enriched for mutations that are beneficial under circumstances in which the capacitor reveals them, then evolvability will be higher than if no modulation occurs (Masel 2006). Note that the key difference between the genotype-network formulation and the capacitance formulation is that the genotype-network formulation requires no special perturbation to reveal new phenotypes. One way to think about the distinction is that, in the genotype-network formulation, many new mutations could act as capacitor-like perturbations, interacting with one or more previously neutral mutations at other loci to produce a novel phenotype. Determining which formulation (or both or neither) is the best representation of reality therefore boils down to understanding the extent and nature of gene-by-environment (G×E) and gene-by-gene (G×G) interactions.
There are many fields of research, from human disease genetics to developmental genetics to breeding, where understanding G×E and G×G interactions is valuable. It can be fruitful, therefore, to think about the evolutionary consequences of robustness as part of larger conceptual issues in genetics and evolution. In particular, robustness is profitably viewed as a property of the mapping of genotype to phenotype. No evolutionary genetic account is complete without addressing this mapping (Landry & Rifkin 2012). Robustness to mutations would manifest as many-to-one relationships between genotypes and phenotypes (Ancel & Fontana 2000, Landry & Rifkin 2012). Because genotype-phenotype maps should also include environmental inputs, robustness to environmental variation would also be an element of the mapping, as would phenotypic plasticity. Thinking about robustness in this way highlights the importance of dissecting the physiological and developmental mechanisms that give the genotype-phenotype map its structure (Landry & Rifkin 2012). It also highlights the need for greater theoretical and empirical attention to epistatic interactions, as is discussed below.
3.1 Genotype networks and evolvability
Consider a set of genotypes that produce the same phenotype. Mutational robustness implies that such sets would exist and might be quite large. It also implies that “neighboring” genotypes — i.e., those connected by single mutational steps — would tend to belong to the same set. Because the mutational steps can be considered as links between genotypes, sets of genotypes connected by mutational steps that do not change the phenotype are termed genotype networks (Wagner 2012). “Mutational steps” are referred to, rather than single mutations, because some steps may involve multiple mutations, depending on population-genetic parameters. For example, in a very large population a non-negligible proportion of progeny will inherit two or more mutations simultaneously, allowing the population to avoid low-fitness neighboring genotypes by “tunneling” to a different region of genotype space (Wagner 2012). Moreover, comparative genomics has revealed many cases of compensatory mutations within macromolecules and even between interacting molecules (Clark et al. 2012), implying that weakly deleterious mutations occur, and then fitness is restored through mutations at other sites; a mutational step that includes a weakly deleterious mutation and the compensatory mutation would be included in the genotype network (Wagner 2012). It is because individual mutations need not be neutral that the term genotype network is now preferred over neutral network (Wagner 2012) (Figure 3).
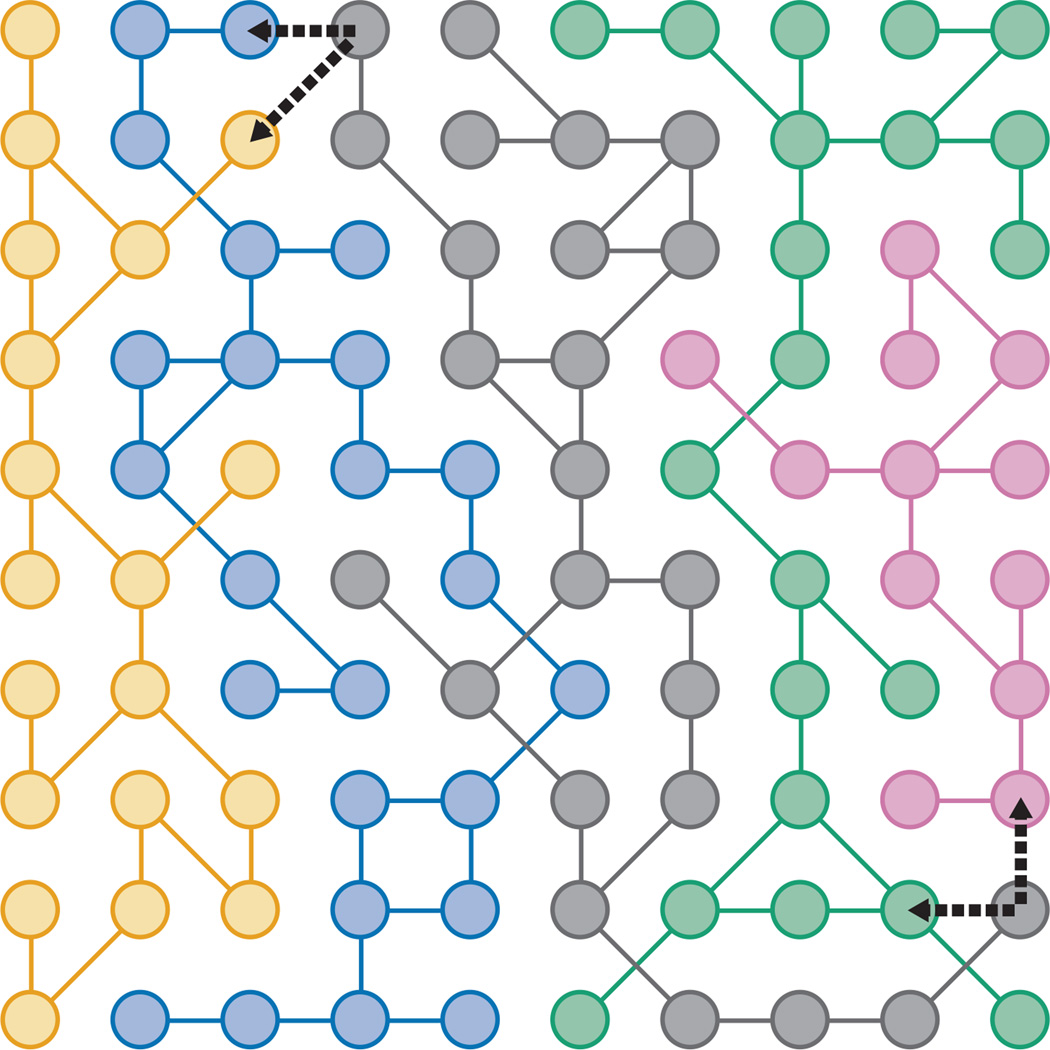
Figure 3. Populations spreading on genotype networks gain access to more novel phenotypes.
An abstract space of genotypes is depicted in two dimensions, although an actual genotype space would be of extremely high dimension. Genotypes (circles) connected by lines are those that are accessible by single mutational steps and that produce identical phenotypes. The collection of connected genotypes for a particular phenotype is a genotype network. Five genotype networks are depicted, each in a different color. Spreading out across a genotype network might increase evolvability by allowing a population access to more novel phenotypes. For example, at two distant genotypes on the gray network, single mutational steps (thick dashed arrows) can produce the blue or yellow phenotypes (upper left) or the green or pink phenotypes (lower right).
The genotype-network perspective is very useful for thinking about robustness and the effects it has on evolutionary divergence, as well as about genotype-phenotype maps more generally. Early work focused on the folding properties of single RNA molecules (Schuster et al. 1994). The RNA sequence provides the genotype while the secondary structure of the folded RNA provides the phenotype. Although only a tiny part of the full genotype-phenotype map for any organism, this study system is still highly complex. Indeed the number of possible genotypes of an RNA molecule of length 100 is astronomical. Nevertheless, inferences about the genotype-phenotype map can be made because computational algorithms exist for predicting secondary structures from the primary sequence. This map was discovered to have three major properties. First, large genotype networks do exist; indeed a substantial fraction of these networks traverse vast swaths of genotype space to include sequences that have identity at no base position (Schuster et al. 1994). Second, a few folded structures constitute a disproportionately high fraction of phenotype space and can be reached from many places in genotype space; that is, any random sequence is likely to be a few mutational steps away from a sequence that folds into any common structure (Schuster et al. 1994). Third, the overlap between the set of structures neighboring one genotype and the set of structures neighboring another on the same network decreases rapidly as the number of mutational steps separating the two increases; that is, different regions of the genotype network tend to give access to different uncommon structures (Sumedha et al. 2007).
Investigations of other systems have yielded similar conclusions. For example, analysis of protein structure and function suggests that large genotype networks exist and that different regions of a genotype network contain different neighboring structures (Ferrada & Wagner 2010, Wagner 2011). A potentially important caveat, however, is that genotype networks for protein structure appear to be less extensive than those for RNA structure and appear to have more similar neighborhoods (Ferrada & Wagner 2012). Nevertheless, interactions between proteins might amplify robustness and diversity: Flux balance analysis of possible metabolic pathways suggests both that there are many routes to the same ability to grow on particular carbon sources (i.e., there are extensive genotype networks when genotypes are conceived as sets of enzyme-encoding genes) and that different regions of a genotype network tend to give access to different alternative metabolisms (Wagner 2011). Abstract models of gene-regulatory circuits suggest the same is true of regulatory systems: Extensive genotype networks exist that produce identical patterns of gene activity, and different regions of a genotype network tend to give access to different gene-activity patterns (Wagner 2011). One empirical manifestation of these genotype networks is the observation of so-called developmental system drift, whereby divergent organisms carry out similar functions using distinct regulatory pathways (True & Haag 2001, Wagner 2011).
The existence of extensive genotype networks begs an evolutionary question: How do populations “spread out” on genotype networks? That is, assuming that a population starts with a single genotype and that stabilizing selection acts to preserve its associated phenotype, what will be the steady-state distribution of genotypes? An important consideration is the topology of the genotype network. Some genotypes in the network might have a high proportion of neighbors that are also on the network, whereas others might have a low proportion. One might expect the population to concentrate in parts of the network that are highly connected (where genotypes’ neighbors tend to be on the network as well), because in the highly connected parts of the network mutations are less likely to create unfit genotypes. Mathematical modeling and simulations of evolving RNA structures support this expectation (van Nimwegen et al. 1999).
What consequence does the spreading out of a population on a genotype network have for the evolution of new phenotypes? Is evolvability increased because mutational robustness allows diverse genotypes to accumulate in a population? The studies of genotype networks for RNA structure, protein structure, metabolism, and gene regulation imply the answer should be yes. Those studies not only suggest that extensive genotype networks exist but that spreading out on a network should give access to more novel phenotypes because different regions of a network tend to have neighbors with different phenotypes. However, details might matter. For instance, if robustness is too low, there might be too little genotypic diversity in a population to take advantage of the different access points; if robustness is too high, there might be too few access points to exploit; if dissimilar genotypes have access to the same alternative phenotypes, there might be no advantage to spreading to more access points (Draghi et al. 2010). For this reason, the differences in the genotype networks inferred for RNA structure and protein structure might be more important than their similarities.
Quantitative insight into the non-trivial relationship between robustness and evolvability comes from a population-genetic model with three key parameters: (a) the probability that a genotypic neighbor has the same phenotype (a measure of robustness), (b) the number of possible phenotypes, and (c) the number of phenotypes accessible by single mutational steps from any one genotype (Draghi et al. 2010). If the number of accessible phenotypes from any genotype equals the number of possible phenotypes, then spreading out through the genotype network confers no advantage, and more robust populations take longer to adapt to a new selective pressure than less robust populations (Draghi et al. 2010). However, if the number of accessible phenotypes from any genotype is smaller than the number of possible phenotypes, meaning that different neighborhoods provide access to different phenotypes, then populations of intermediate robustness adapt fastest and, correspondingly, produce the greatest diversity of mutant phenotypes (Draghi et al. 2010). The relationship between robustness and evolvability is further dependent on the mutation rate and population size, again highlighting that details matter (Draghi et al. 2010).
Ultimately, whether mutational robustness speeds adaptation is an empirical question. The RNA virus φ6 provides an example of robustness promoting evolvability (McBride et al. 2008). High- and low-robustness strains of φ6 were compared for their abilities to adapt to an imposed selective pressure. Although high- and low-robustness strains did not differ in their initial abilities to tolerate heat shock, 50 generations of selection involving periodic heat shocks produced different outcomes: populations founded with high-robustness strains tended to evolve greater heat-shock tolerance than those founded with low-robustness strains (McBride et al. 2008). Studies of other viruses largely support a connection between robustness and evolvability, suggesting that clinical interventions that alter mutational robustness are worth pursuing (Lauring et al. 2013).
Laboratory evolution using a different system — a ribozyme (RNA enzyme) capable of cleaving an RNA oligonucleotide and joining part of the oligonucleotide to the 3' end of the ribozyme itself — addressed the same question from a different angle. Instead of a comparison between high robustness and low robustness, the comparison was between two populations that either did or did not accumulate neutral variation before selection for a new phenotype was imposed (Hayden et al. 2011). All steps in the evolutionary process were carried out ex vivo. Mutations were introduced through mutagenic amplification of template DNA sequences, and selection was imposed by creating the next generation’s template sequences by reverse transcription of ribozymes that had successfully catalyzed cleavage and joining (Hayden et al. 2011). A new selective pressure was imposed by challenging the ribozyme to catalyze cleavage of a phosphorothioate linkage in an otherwise identical oligonucleotide substrate (Hayden et al. 2011). DNA sequencing confirmed that a diversity of sequences encoding functional ribozymes had accumulated after 10 generations of random mutagenesis and purifying selection for cleavage of the native target (Hayden et al. 2011). Under selection for ability to cleave the phosphorothioate-containing substrate, populations that had accumulated neutral diversity adapted more rapidly than ones that had not (Hayden et al. 2011).
3.2 Cryptic genetic variation and capacitance
Evidence for the existence of cryptic genetic variation in nature is widespread (Paaby & Rockman 2014). By definition, cryptic genetic variation is invisible at first glance, so genetic or environmental perturbations are required to reveal it (Masel & Siegal 2009). To take one example, the nematode Caenorhabditis elegans shows a famously invariant pattern of cell divisions during development. The signaling pathways that pattern the C. elegans hermaphrodite’s vulva were well worked out through genetic analysis in the standard laboratory strain. However, mutating genes acting in vulval development or ablating key cells yields different vulval phenotypes in different genetic backgrounds (Milloz et al. 2008). The C. elegans population thus appears to have spread out on a genotype network. Indeed, different strains differ quantitatively in their signaling activities to the extent that, had a different strain been chosen initially for genetic analysis, different conclusions about the regulation of vulval development might have been reached (Milloz et al. 2008).
The most prominent example of a perturbation revealing cryptic genetic variation is that of impairment of the chaperone Hsp90 (Jarosz et al. 2010). In Drosophila melanogaster, reducing Hsp90 levels by mutation or pharmacological inhibition yields phenotypic differences between strains that otherwise are similar in phenotype (Rutherford & Lindquist 1998). Subsequent studies in other organisms, spanning animals, plants, and fungi, gave similar results (Jarosz et al. 2010). Hsp90 was the first gene product to be termed an evolutionary capacitor, suppressing the effects of cryptic genetic variation under normal conditions and revealing cryptic genetic variation when its activity is impaired (Rutherford & Lindquist 1998).
The ability of Hsp90 impairment to reveal cryptic genetic variation is largely undisputed. A recent study showed that reduced Hsp90 activity causes transposable elements to mobilize, raising the possibility that what was thought to be cryptic genetic variation is actually new mutations (Specchia et al. 2010). However, transposable-element mobilization cannot fully account for the effects of Hsp90 impairment on phenotypic variation, so revealing cryptic genetic variation remains a valid explanation (Gangaraju et al. 2011, Siegal & Masel 2012).
The more controversial claim about Hsp90 is that its ability to hide and reveal variation plays a role in evolution. There are two questions pertinent to this claim that are important to separate: (a) Does Hsp90-interacting genetic variation contribute to evolutionary divergence?, and (b) was Hsp90’s ability to modulate the phenotypic effects of genetic variation itself favored by natural selection? The first question had, until recently, not been directly addressed. All that was known about Hsp90 in natural populations was that there are two alleles of Hsp90 found in D. melanogaster populations that appear to reduce Hsp90 activity sufficiently to reveal cryptic genetic variation (Sgro et al. 2010, Chen & Wagner 2012). Recently, however, investigation of Hsp90 in blind cavefish suggested that Hsp90-interacting variation might be relevant to the adaptation of formerly surface-dwelling fish to the cave environment (Rohner et al. 2013). Experimental Hsp90 inhibition in fish from surface populations reveals cryptic genetic variation that affects eye size, and culturing surface fish in cave-like conditions not only induces a stress-like response similar to that induced by pharmacological Hsp90 inhibition but also increases eye-size variation (Rohner et al. 2013). Still to be determined is whether the actual alleles conferring reduced-eye phenotypes on the cavefish derive from Hsp90-interacting standing variation in the surface population. Nonetheless, this case is the most promising evidence to date of a role for Hsp90-mediated cryptic genetic variation release in a natural adaptive process. Also remaining to be determined is whether the cavefish case is special because the adaptive event involves loss of function (eye reduction). Other cave-adaptive traits, such as changes in body size and mechanosensory organs, did not show evidence of Hsp90-interacting cryptic genetic variation (Rohner et al. 2013).
Additional support for a capacitor contributing to evolutionary divergence comes from the prion [PSI+] in the yeast Saccharomyces cerevisiae. [PSI+] is an aggregated form of the translation termination factor Sup35; [PSI+] cells spontaneously convert at a low rate to [psi−] (prion-lacking) cells and vice versa. Readthrough of stop codons is more frequent in [PSI+] cells than in [psi−] cells. Phenotypic diversity, as measured by growth on different media, is also greater among [PSI+] cells, presumably because different genetic backgrounds have different sequences following stop codons that are incorporated into proteins when termination is impaired (True & Lindquist 2000). Observation of increased phenotypic diversity in the laboratory does not necessarily imply that prion formation contributes to evolution. It could be, for instance, that all revealed variation is deleterious in the wild, even if a growth advantage is seen on particular media in the lab. The strongest, albeit indirect, evidence for the adaptive value of [PSI+]-revealed variation comes from comparative genomics. In the Saccharomyces lineage, DNA sequence changes that cause additions of formerly 3'-untranslated regions into coding sequences disproportionately preserve the reading frame, consistent with these additions being cases of selection of pre-existing cryptic variation (Giacomelli et al. 2007). No such bias is seen in rodents, where no [PSI+]-like mechanism is known to operate (Giacomelli et al. 2007). Mathematical modeling also suggests that [PSI+] is plausibly an evolutionary capacitor, given realistic rates of prion formation and outcrossing, and assuming that episodes of adaptation requiring more than one genetic change occur at a sufficient frequency (Masel & Bergman 2003, Griswold & Masel 2009, Lancaster et al. 2010).
The second question about Hsp90 and any other capacitor — whether or not they evolved to modulate the effects of cryptic genetic variation — is extremely difficult to test in any conclusive way. That Hsp90 appears to modulate the effects of cryptic genetic variation across a broad phylogenetic range does not necessarily imply selection to preserve its ability to do so, as this ability might merely be a by-product of selection for some other necessary function of Hsp90. The case for [PSI+] is similar. The ability to form [PSI+] is apparently conserved over hundreds of millions of years of yeast evolution (Santoso et al. 2000), and although the part of Sup35 required for prion propagation is dispensable, it likely has other functions besides prion formation (True & Lindquist 2000, Jarosz et al. 2010). Mathematical modeling does, however, suggest that it is plausible for natural selection to favor capacitor function (Masel 2005).
3.3 Robustness and epistasis
The revelation of cryptic genetic variation by a genetic or environmental perturbation has been taken as evidence that the perturbation decreases mutational robustness, but, as hinted at in Section 2, Mechanisms of Robustness, this conclusion is flawed. The flaw has been discussed in detail elsewhere (Hermisson & Wagner 2004, Richardson et al. 2013), so it is not to be belabored here. In brief the flaw is that, although decreasing mutational robustness is a sufficient condition for the release of cryptic genetic variation, it is not a necessary condition (Hermisson & Wagner 2004, Richardson et al. 2013). Revelation of cryptic genetic variation merely indicates that epistasis (or, in the case of cryptic genetic variation revealed by environmental perturbation, G×E interaction) exists.
Although the logical flaw was noted a decade ago (Hermisson & Wagner 2004), the view persists that release of cryptic genetic variation implies decrease of mutational robustness (e.g., Rohner et al. 2013). Of course, release of cryptic genetic variation means that mutations that formerly did not have phenotypic effects now do, so one might be justified in saying that robustness to the effects of those particular mutations has been compromised. However, such a statement would say nothing about the fate of the next mutation to arise, which is the relevant evolutionary consideration. Mutational robustness is only properly defined according to the distribution of effects of spontaneous mutations that have not already survived the filter of natural selection.
In other words, a proper test of whether a gene product increases mutational robustness is to measure, in the presence and absence of the gene product, the phenotypic effects of new mutations (Hermisson & Wagner 2004, Richardson et al. 2013). To date, this test has only been performed once (Richardson et al. 2013). Perhaps surprisingly, that test concluded that an excellent candidate in S. cerevisiae for conferring increased mutational robustness, the histone variant Htz1, does not do so: There was no difference in between-strain variation in cell morphology among yeast mutation-accumulation strains with HTZ1 versus without HTZ1 (Richardson et al. 2013). However, HTZ1 did show extensive epistasis with accumulated mutations. That is, the morphological effects of new mutations depended heavily on whether HTZ1 was present or not, even though the average morphological effect of a new mutation did not increase in the absence of HTZ1 (Richardson et al. 2013) (Figure 4). HTZ1 deletion also increases within-line variation in cell morphology (Levy & Siegal 2008, Richardson et al. 2013). HTZ1 therefore increases microenvironmental robustness but not mutational robustness, providing direct evidence against congruence.
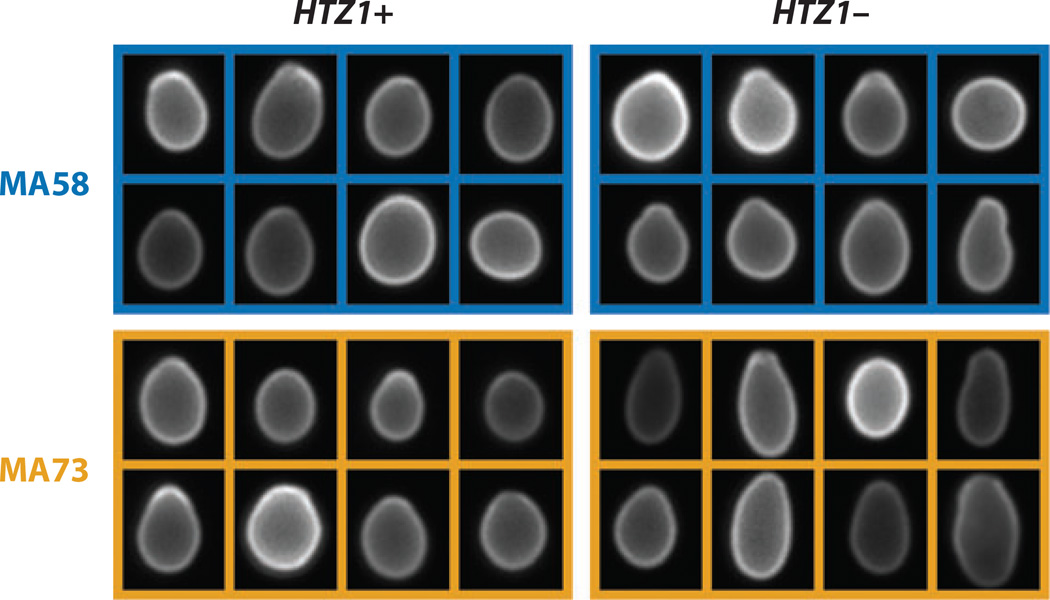
Figure 4. Deletion of HTZ1 in yeast interacts epistatically with accumulated mutations.
Yeast mutation-accumulation lines were assayed for cell morphology with and without a wild-type copy of the HTZ1 gene (Richardson et al. 2013). Two pairs of such lines are shown, illustrating epistasis. In line MA58, HTZ1 deletion has little effect on average cell shape (compare cells at upper left with cells at upper right). In contrast, in line MA73, HTZ1 deletion causes elongated cells (compare lower left with lower right). Although wild-type HTZ1 does not increase mutational robustness, it does increase robustness to microenvironmental variation: There is more cell-to-cell variation within mutation-accumulation lines in the absence of HTZ1 than in its presence. Figure modified from Richardson et al. (2013).
Much more investigation is necessary to determine whether the HTZ1 result is an exception or represents the rule. Meanwhile, the HTZ1 result clearly indicates that the logical flaw in connecting cryptic genetic variation and mutational robustness cannot be ignored. At the same time, the HTZ1 result (or any similar future result) should not be taken as an indication that cryptic genetic variation is irrelevant to evolution. Rather, the HTZ1 result strengthens the case that the focus should be on epistasis instead of mutational robustness. Indeed, as pointed out a decade ago, the question of the potential importance of cryptic genetic variation in evolution can — and should — be completely separated from questions about the existence of mutational-robustness mechanisms (Hermisson & Wagner 2004).
One way to begin thinking about epistasis rather than mutational-robustness mechanisms is to consider that a mutational-robustness mechanism implies a particular form of epistasis: In the context of a genetic background with the mutational-robustness mechanism in place, a new mutation will, on average, have less phenotypic effect than in the context of a genetic background with the mutational-robustness mechanism impaired. However, other forms of epistasis are also relevant to cryptic genetic variation. Indeed, any interaction that includes some amount of conditional neutrality is relevant. HTZ1 is an example: Some new mutations do not have a phenotypic effect on an HTZ1+ genetic background but do on an HTZ1− genetic background (Richardson et al. 2013). Therefore, if these mutations occurred in nature, they could accumulate as cryptic genetic variation and HTZ1 impairment would reveal them.
A similar result was seen for Hsp90 in S. cerevisiae (Jarosz & Lindquist 2010). Their study was not technically a test of mutational robustness conferred by Hsp90, as natural variation segregating in the progeny of divergent parental strains, rather than new mutations, was assayed. Nonetheless, results from mapping quantitative trait loci (QTLs) affecting growth under a variety of conditions, with and without pharmacological Hsp90 impairment, support the conclusion that conditional effects are symmetrical with respect to wild-type and impaired Hsp90 function. Nearly as many QTLs were discovered when Hsp90 was fully functional as when Hsp90 was impaired (Jarosz & Lindquist 2010). Because of this and related observations, Hsp90 has been called both a capacitor (suppressing phenotypic effects of mutations except when impaired) and a potentiator (enabling phenotypic effects of mutations except when impaired) (Jarosz et al. 2010). Both capacitance and potentiation are forms of epistasis involving conditional neutrality and are perhaps best thought of as such rather than as distinct phenomena (Richardson et al. 2013, Masel 2013).
4. PERSPECTIVES AND FUTURE DIRECTIONS: OPENING THE DOOR TO EPISTASIS
Opening the door to thinking about epistasis naturally leads to new questions such as: (a) What is the probability that a pair of naturally occurring mutations will have non-additive effects on phenotypes?, (b) How common is conditional neutrality as a type of G×G or G×E interaction?, and (c) Is there widespread congruence between mechanisms that increase environmental robustness and those that interact with cryptic genetic variation (as is true for HTZ1) and, if so, what is the molecular basis of this congruence? The answers to these questions could have profound implications for our understanding of evolution. For example, if conditionally neutral G×G interactions are common, then each new mutation that produces a new phenotype can be thought of as a perturbation to a capacitor (or, from a flipped perspective, as having been enabled by a potentiator). As noted above, this would favor the perspective that views evolution as a process of populations spreading out over genotype networks and gaining access to more novel phenotypes as a result. In the end, particular genes, such as Hsp90, might be more valuable as illustrations of how such evolution might always proceed rather than as candidates for contributing to rare adaptive events that punctuate long periods of stasis (Siegal 2013).
There has been much recent interest in whether adaptive genotypes more commonly derive from new mutations or from so-called standing genetic variation that is present in the population before selective pressures change. Although most accounts of standing variation focus on simple reasons for its existence, such as mutation-selection balance and recessive alleles, conditionally neutral genetic variation could make up a very important component of standing variation not only with respect to evolution (Phillips 2008, Draghi et al 2010, Rohner et al 2013, Siegal 2013) but also with respect to human health and the increasingly prevalent “diseases of modernity” (Phillips 2008, Gibson 2009). Indeed it might be so important that it warrants its own name: “crouching variation” (Siegal 2013). In support of this possibility, computational simulations suggest that capacitor-like behavior might be a property of many or all genes in gene-regulatory networks (Bergman & Siegal 2003). Some empirical support also exists. In Drosophila a screen for deleted genomic regions that increase genetic-background-dependent effects on wing morphology revealed 10 such regions out of 61 tested (Takahashi 2013). Moreover, a number of trait-mapping studies have identified QTLs with weak or absent primary effects but strong epistatic effects, despite the methodological challenges and low power associated with identifying epistatic effects in such studies (Phillips 2008).
Further progress in understanding the evolutionary role of conditionally neutral genetic variation requires much more empirical work to determine its extent and ultimate fate. In turn, methodological advances that overcome the difficulties of identifying epistatic interactions between naturally occurring mutations will be required. Studies focused on molecular mechanisms need to be done to elucidate the causes of conditional neutrality and how they relate to suppression of the effects of microenvironmental and macroenvironmental variation. Theoretical advances are also necessary. Models of genotype-phenotype relationships necessarily make simplifying assumptions in the interest of tractability, because of the combinatorial explosion of parameters that accompanies any attempt to include interactions. Greater understanding of the forms epistasis and G×E interactions take could help guide and constrain parameterizations so tractability can be preserved while biological realism is increased.
ACKNOWLEDGMENTS
This work was supported by funding from the NIH (R01GM097415 and R01GM086673) and NSF (IOS-1258078) to M.L.S. J.-Y.L. was supported by Academia Sinica of Taiwan (100-CDA-L04) and the National Science Council of Taiwan (NSC102-2321-B-001-053). We thank members of the Siegal and Leu laboratories for helpful discussions.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Mark L. Siegal, Center for Genomics and Systems Biology, Department of Biology, New York University, New York, New York 10003; mark.siegal@nyu.edu
Jun-Yi Leu, Institute of Molecular Biology, Academia Sinica, Nankang, Taipei, Taiwan 11529; jleu@imb.sinica.edu.tw.
LITERATURE CITED
- Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Ancel LW, Fontana W. Plasticity, evolvability, and modularity in RNA. J. Exp. Zool. 2000;288:242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Baldwin JM. A new factor in evolution. Am. Nat. 1896;30:441–451. [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, et al. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Behera N, Nanjundiah V. Phenotypic plasticity can potentiate rapid evolutionary change. J. Theor. Biol. 2004;226:177–184. doi: 10.1016/j.jtbi.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Benazet JD, Bischofberger M, Tiecke E, Goncalves A, Martin JF, et al. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science. 2009;323:1050–1053. doi: 10.1126/science.1168755. [DOI] [PubMed] [Google Scholar]
- Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Kærn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Bloom JD, Lu Z, Chen D, Raval A, Venturelli OS, Arnold FH. Evolution favors protein mutational robustness in sufficiently large populations. BMC Biol. 2007;5:29. doi: 10.1186/1741-7007-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein E, Ruppin E. Direct evolution of genetic robustness in microRNA. Proc. Natl. Acad. Sci. USA. 2006;103:6593–6598. doi: 10.1073/pnas.0510600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch CL, Chao L. Epistasis and its relationship to canalization in the RNA virus φ6. Genetics. 2004;167:559–567. doi: 10.1534/genetics.103.021196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey LB, van Dijk D, Sloot PM, Kaandorp JA, Segal E. Promoter sequence determines the relationship between expression level and noise. PLOS Biol. 2013;11:e1001528. doi: 10.1371/journal.pbio.1001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJ, et al. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155:1556–1567. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles H, Heddi A, Guillaud J, Nardon C, Nardon P. A molecular aspect of symbiotic interactions between the weevil Sitophilus oryzae and its endosymbiotic bacteria: over-expression of a chaperonin. Biochem. Biophys. Res. Commun. 1997;239:769–774. doi: 10.1006/bbrc.1997.7552. [DOI] [PubMed] [Google Scholar]
- Chen B, Wagner A. Hsp90 is important for fecundity, longevity, and buffering of cryptic deleterious variation in wild fly populations. BMC Evol. Biol. 2012;12:25. doi: 10.1186/1471-2148-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Alani E, Aquadro CF. Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 2012;22:714–720. doi: 10.1101/gr.132647.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF, Morby AP, Gunn A, Schneider D. Effect of random and hub gene disruptions on environmental and mutational robustness in Escherichia coli. BMC Genomics. 2006;7:237. doi: 10.1186/1471-2164-7-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby CM, Im JH, Yu RC, Pesce CG, Brem RB. Negative feedback confers mutational robustness in yeast transcription factor regulation. Proc. Natl. Acad. Sci. USA. 2012;109:3874–3878. doi: 10.1073/pnas.1116360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson WJ, Seger J. Cause and effect in evolution. Nature. 1999;399:30. doi: 10.1038/19894. [DOI] [PubMed] [Google Scholar]
- Draghi JA, Parsons TL, Wagner GP, Plotkin JB. Mutational robustness can facilitate adaptation. Nature. 2010;463:353–355. doi: 10.1038/nature08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Proj. Consort. Bernstein BE, Birney E, Dunham I, Green ED, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares MA, Moya A, Barrio E. GroEL and the maintenance of bacterial endosymbiosis. Trends Genet. 2004;20:413–416. doi: 10.1016/j.tig.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-Gonzalez MX, Moya A, Elena SF, Barrio E. Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature. 2002;417:398. doi: 10.1038/417398a. [DOI] [PubMed] [Google Scholar]
- Ferrada E, Wagner A. Evolutionary innovations and the organization of protein functions in genotype space. PLOS ONE. 2010;5:e14172. doi: 10.1371/journal.pone.0014172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrada E, Wagner A. A comparison of genotype-phenotype maps for RNA and proteins. Biophys. J. 2012;102:1916–1925. doi: 10.1016/j.bpj.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser D, Kærn M. A chance at survival: gene expression noise and phenotypic diversification strategies. Mol. Microbiol. 2009;71:1333–1340. doi: 10.1111/j.1365-2958.2009.06605.x. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLOS Biol. 2004;2:E137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Schadt EE. The quantitative genetics of phenotypic robustness. PLOS ONE. 2010;5:e8635. doi: 10.1371/journal.pone.0008635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–319. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 2011;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli MG, Hancock AS, Masel J. The conversion of 3’ UTRs into coding regions. Mol. Biol. Evol. 2007;24:457–464. doi: 10.1093/molbev/msl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Decanalization and the origin of complex disease. Nat. Rev. Genet. 2009;10:134–140. doi: 10.1038/nrg2502. [DOI] [PubMed] [Google Scholar]
- Goldsmith M, Tawfik DS. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc. Natl. Acad. Sci. USA. 2009;106:6197–6202. doi: 10.1073/pnas.0809506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold CK, Masel J. Complex adaptations can drive the evolution of the capacitor [PSI+], even with realistic rates of yeast sex. PLOS Genet. 2009;5:e1000517. doi: 10.1371/journal.pgen.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Dykhuizen DE, Dean AM. Limits of adaptation: the evolution of selective neutrality. Genetics. 1985;111:655–674. doi: 10.1093/genetics/111.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Wagner GP. The population genetic theory of hidden variation and genetic robustness. Genetics. 2004;168:2271–2284. doi: 10.1534/genetics.104.029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, et al. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Bushati N, Cohen SM. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLOS Biol. 2010;8:e1000396. doi: 10.1371/journal.pbio.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38(Suppl):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Hornung G, Bar-Ziv R, Rosin D, Tokuriki N, Tawfik DS, et al. Noise-mean relationship in mutated promoters. Genome Res. 2012;22:2409–2417. doi: 10.1101/gr.139378.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YY, Hung PH, Leu JY. Hsp90 regulates nongenetic variation in response to environmental stress. Mol. Cell. 2013;50:82–92. doi: 10.1016/j.molcel.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu. Rev. Genet. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ. The evolution of genetic canalization under fluctuating selection. Evolution. 2000;54:1–12. doi: 10.1111/j.0014-3820.2000.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Rifkin SA. The genotype-phenotype maps of systems biology and quantitative genetics: distinct and complementary. Adv. Exp. Med. Biol. 2012;751:371–398. doi: 10.1007/978-1-4614-3567-9_17. [DOI] [PubMed] [Google Scholar]
- Lauring AS, Frydman J, Andino R. The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 2013;11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. Genes confer similar robustness to environmental, stochastic, and genetic perturbations in yeast. PLOS ONE. 2010;5:e9035. doi: 10.1371/journal.pone.0009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLOS Biol. 2008;6:e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. The robustness continuum. Adv. Exp. Med. Biol. 2012;751:431–452. doi: 10.1007/978-1-4614-3567-9_20. [DOI] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLOS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefting M, Hoffmann AA, Ellers J. Plasticity versus environmental canalization: population differences in thermal responses along a latitudinal gradient in Drosophila serrata. Evolution. 2009;63:1954–1963. doi: 10.1111/j.1558-5646.2009.00683.x. [DOI] [PubMed] [Google Scholar]
- Macneil LT, Walhout AJ. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 2011;21:645–657. doi: 10.1101/gr.097378.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S, Roth JR, Fredriksson A, Nystrom T, Berg OG, Andersson DI. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat. Genet. 2005;37:1376–1379. doi: 10.1038/ng1676. [DOI] [PubMed] [Google Scholar]
- Masel J. Evolutionary capacitance may be favored by natural selection. Genetics. 2005;170:1359–1371. doi: 10.1534/genetics.105.040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2006;172:1985–1991. doi: 10.1534/genetics.105.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J. Q&A: evolutionary capacitance. BMC Biol. 2013;11:103. doi: 10.1186/1741-7007-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Bergman A. The evolution of the evolvability properties of the yeast prion [PSI+] Evolution. 2003;57:1498–1512. doi: 10.1111/j.0014-3820.2003.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride RC, Ogbunugafor CB, Turner PE. Robustness promotes evolvability of thermotolerance in an RNA virus. BMC Evol. Biol. 2008;8:231. doi: 10.1186/1471-2148-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL. A single mode of canalization. Trends Ecol. Evol. 2002;17:468–473. [Google Scholar]
- Milloz J, Duveau F, Nuez I, Felix MA. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 2008;22:3064–3075. doi: 10.1101/gad.495308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. Evolution of mutational robustness in an RNA virus. PLOS Biol. 2005;3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. The evolutionary maintenance of alternative phenotypes. Am. Nat. 1992;139:971–989. [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Osella M, Bosia C, Cora D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLOS Comput. Biol. 2011;7:e1001101. doi: 10.1371/journal.pcbi.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat. Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Rockman MV. Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 2014;15:247–258. doi: 10.1038/nrg3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M, Legewie S, Eils R, Karaulanov E, Niehrs C. Negative feedback in the bone morphogenetic protein 4 (BMP4) synexpression group governs its dynamic signaling range and canalizes development. Proc. Natl. Acad. Sci. USA. 2011;108:10202–10207. doi: 10.1073/pnas.1100179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaez N, Carthew RW. Biological robustness and the role of microRNAs: a network perspective. Curr. Top. Dev. Biol. 2012;99:237–255. doi: 10.1016/B978-0-12-387038-4.00009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP. Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol. Evol. 2010;25:459–467. doi: 10.1016/j.tree.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc. Natl. Acad. Sci. USA. 2003;100:7152–7157. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Dushoff J, Desai MM, Fraser HB. Codon usage and selection on proteins. J. Mol. Evol. 2006;63:635–653. doi: 10.1007/s00239-005-0233-x. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Dushoff J, Fraser HB. Detecting selection using a single genome sequence of M. tuberculosis and P. falciparum. Nature. 2004;428:942–945. doi: 10.1038/nature02458. [DOI] [PubMed] [Google Scholar]
- Price N, Cartwright RA, Sabath N, Graur D, Azevedo RB. Neutral evolution of robustness in Drosophila microRNA precursors. Mol. Biol. Evol. 2011;28:2115–2123. doi: 10.1093/molbev/msr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TD, Qvarnstrom A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. B-Biol. Sci. 2003;270:1433–1440. doi: 10.1098/rspb.2003.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey SA, Smith JJ, Orrell D, Marelli M, Petersen TW, et al. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat. Genet. 2006;38:1082–1087. doi: 10.1038/ng1869. [DOI] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JB, Uppendahl LD, Traficante MK, Levy SF, Siegal ML. Histone variant HTZ1 shows extensive epistasis with, but does not increase robustness to**new mutations. PLOS Genet. 2013;9:e1003733. doi: 10.1371/journal.pgen.1003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, et al. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL. From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays. 2000;22:1095–1105. doi: 10.1002/1521-1878(200012)22:12<1095::AID-BIES7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Bahrami A, Wilczek A, Watanabe E, Schellenberg K, et al. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLOS ONE. 2007;2:e648. doi: 10.1371/journal.pone.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster TA, Lindquist S, Queitsch C. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- Sanjuan R, Cuevas JM, Furio V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLOS Genet. 2007;3:e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proc. R. Soc. B-Biol. Sci. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- Sgrò CM, Wegener B, Hoffmann AA. A naturally occurring variant of Hsp90 that is associated with decanalization. Proc. R. Soc. B-Biol. Sci. 2010;277:2049–2057. doi: 10.1098/rspb.2010.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal ML. Crouching variation revealed. Mol. Ecol. 2013;22:1187–1189. doi: 10.1111/mec.12195. [DOI] [PubMed] [Google Scholar]
- Siegal ML, Bergman A. Waddington’s canalization revisited: developmental stability and evolution. Proc. Natl. Acad. Sci. USA. 2002;99:10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal ML, Masel J. Hsp90 depletion goes wild. BMC Biol. 2012;10:14. doi: 10.1186/1741-7007-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D’Alessandro R, et al. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- Springer M, Weissman JS, Kirschner MW. A general lack of compensation for gene dosage in yeast. Mol. Syst. Biol. 2010;6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC, Kaiser M, Kawecki TJ. The differential genetic and environmental canalization of fitness components in Drosophila melanogaster. J. Evol. Biol. 1995;8:539–557. [Google Scholar]
- Stephens CR, Waelbroeck H. Codon bias and mutability in HIV sequences. J. Mol. Evol. 1999;48:390–97. doi: 10.1007/pl00006483. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Spencer HG. Metapopulation structure favors plasticity over local adaptation. Am. Nat. 2002;160:271–283. doi: 10.1086/341015. [DOI] [PubMed] [Google Scholar]
- Sumedha, Martin OC, Wagner A. New structural variation in evolutionary searches of RNA neutral networks. Biosystems. 2007;90:475–485. doi: 10.1016/j.biosystems.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nijhout HF. Evolution of a polyphenism by genetic accommodation. Science. 2006;311:650–652. doi: 10.1126/science.1118888. [DOI] [PubMed] [Google Scholar]
- Szollosi GJ, Derenyi I. Congruent evolution of genetic and environmental robustness in micro-RNA. Mol. Biol. Evol. 2009;26:867–874. doi: 10.1093/molbev/msp008. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Takahashi KH. Multiple capacitors for natural genetic variation in Drosophila melanogaster. Mol. Ecol. 2013;22:1356–1365. doi: 10.1111/mec.12091. [DOI] [PubMed] [Google Scholar]
- Takahashi KH, Daborn PJ, Hoffmann AA, Takano-Shimizu T. Environmental stress-dependent effects of deletions encompassing Hsp70Ba on canalization and quantitative trait asymmetry in Drosophila melanogaster. PLOS ONE. 2011a;6:e17295. doi: 10.1371/journal.pone.0017295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi KH, Okada Y, Teramura K. Genome-wide deficiency mapping of the regions responsible for temporal canalization of the developmental processes of Drosophila melanogaster. J. Hered. 2011b;102:448–457. doi: 10.1093/jhered/esr026. [DOI] [PubMed] [Google Scholar]
- Takahashi KH, Okada Y, Teramura K. Deficiency screening for genomic regions with effects on environmental sensitivity of the sensory bristles of Drosophila melanogaster. Evolution. 2012;66:2878–2890. doi: 10.1111/j.1558-5646.2012.01636.x. [DOI] [PubMed] [Google Scholar]
- Takahashi KH, Rako L, Takano-Shimizu T, Hoffmann AA, Lee SF. Effects of small Hsp genes on developmental stability and microenvironmental canalization. BMC Evol. Biol. 2010;10:284. doi: 10.1186/1471-2148-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Sigal N, Assia Y, Barkai N. Chromatin regulators as capacitors of interspecies variations in gene expression. Mol. Syst. Biol. 2010;6:435. doi: 10.1038/msb.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen E, Crutchfield JP, Huynen M. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA. 1999;96:9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:118–126. [Google Scholar]
- Waddington CH. The Strategy of the Genes. London: George Allen & Unwin Ltd.; 1957. p. 62. [Google Scholar]
- Wagner A. Robustness and Evolvability in Living Systems. Princeton: Princeton Univ. Press; 2007. p. 367. [Google Scholar]
- Wagner A. The molecular origins of evolutionary innovations. Trends Genet. 2011;27:397–410. doi: 10.1016/j.tig.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Wagner A. The role of robustness in phenotypic adaptation and innovation. Proc. Biol. Sci. 2012;279:1249–1258. doi: 10.1098/rspb.2011.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Booth G, Bagheri-Chaichian H. A population genetic theory of canalization. Evolution. 1997;51:329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412:331–333. doi: 10.1038/35085569. [DOI] [PubMed] [Google Scholar]
- Wilkins AS. Canalization: a molecular genetic perspective. Bioessays. 1997;19:257–262. doi: 10.1002/bies.950190312. [DOI] [PubMed] [Google Scholar]