Abstract
We must reliably map the interactomes of cellular macromolecular complexes in order to fully explore and understand biological systems. However, there are no methods to accurately predict how to capture a given macromolecular complex with its physiological binding partners. Here, we present a screen that comprehensively explores the parameters affecting the stability of interactions in affinity-captured complexes, enabling the discovery of physiological binding partners and the elucidation of their functional interactions in unparalleled detail. We have implemented this screen on several macromolecular complexes from a variety of organisms, revealing novel profiles even for well-studied proteins. Our approach is robust, economical and automatable, providing an inroad to the rigorous, systematic dissection of cellular interactomes.
High-throughput DNA sequencing facilitates whole genome characterization within weeks1,2. Likewise, advances in mass spectrometry (MS)3,4 are enabling cellular proteomes to be defined. However, we have yet to exhaustively map any interactome – the cell’s comprehensive biomolecular interaction network5,6; we may have identified less than 20% of the protein interactions in humans, not counting dynamic, tissue- or disease-specific interactions7–9.
A main approach for interactomic exploration is affinity capture10,11. For this, cells are broken and their contents extracted into a solution that ideally preserves each target macromolecular complex. Complexes are then specifically enriched from the cell extract using affinity reagents – usually antibodies – that recognize the target, either directly or through an epitope tag, permitting subsequent characterization of the complex. However, one of the foremost challenges facing affinity capture studies is the precise optimization of the extraction conditions, because no single condition is optimal for the preservation of the many different types of interactions found in macromolecular complexes12–14. As a result, affinity capture experiments either require time-consuming optimization on a case-by-case basis, or a compromise must be made by using un-optimized conditions; the latter is a common strategy but often results in sparse coverage of protein-protein interactions and error-prone data15–17. A variety of advanced bioinformatics tools18 and databases of common contaminant proteomes19,20 have attempted to mitigate this problem21–24, but cannot fully substitute for optimized sample preparation15. Because any given extraction solution influences the complement of copurifying proteins, multiple extractant formulations are required if one intends to broadly sample the interactome, as underscored by a recent high-throughput study of membrane protein interactions in yeast25.
The problem of maintaining post-extraction protein complex stability is comparable to that which once hindered protein crystallographic efforts. Crystallography requires the empirical determination of conditions promoting interactions that permit efficient crystal growth. Similarly, affinity capture requires the empirical determination of conditions that support the retention of in vivo interactions and minimize the in vitro artifacts. For crystallography, the answer came with the development of massively parallel crystallization optimization screens26,27 that allow hundreds of conditions to be simultaneously explored28. Inspired by this, we have developed improved methods for the rapid processing of cellular material in conjunction with parallelized, multi-parameter searches of extraction conditions. Our approach is compatible with both standard lab scale investigations and high-throughput robotics, and facilitates the systematic exploration of the interactome of any given protein in a cell.
Results
Designing a large-scale interactomics screen
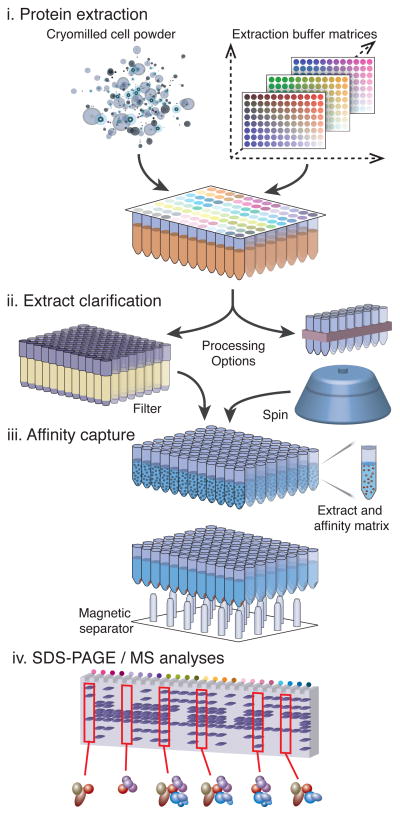
Our strategy (Fig. 1), starts with the distribution of cryomilled cell material29,30 to a multi-well plate. To enable the uniform delivery of frozen cell powder to each well in the plate, we designed dispensing manifolds (Fig. 2a,d and Supplementary Fig. 1). After dispensing, the powder in the wells is thawed by addition of an array of distinct extractants. The resulting extracts are clarified of insoluble material using a clog-resistant filtration device (Fig. 2b,d) that provides a filtrate matching the quality of centrifugally clarified cell extract (Fig. 2c). The remainder of the procedure implements commercially available supplies and equipment (Online Methods and Supplementary Protocol 1).
Figure 1.
Schematic representation of the parallelized affinity capture procedure. (i) cells expressing a tagged protein of interest are mechanically disrupted at cryogenic temperature to produce a micron-scale powder and precise aliquots of the frozen powder are deposited into the wells of a multi-well plate using a dedicated manifold. A diverse set of extraction solvents are rapidly added in parallel and complete re-suspension with concomitant extraction of the powder is ensured using immediate brief mechanical agitation and/or low-power sonication; (ii) rapid removal of insoluble material is achieved by either centrifugation or using a novel deep-bed filtration device (this study); (iii) affinity capture is then performed on the clarified extract using magnetic beads coupled with an affinity reagent for the tag used, and a multipronged magnet separator in register with the multiwell plate, allowing subsequent washing steps; (iv) elution of the complexes is followed by SDS-PAGE, Coomassie blue staining, and (as desired) MS analysis. The resulting copurification profiles are catalogued and cross-compared to infer interactomes and determine preparative conditions appropriate for further biochemical and analytical means.
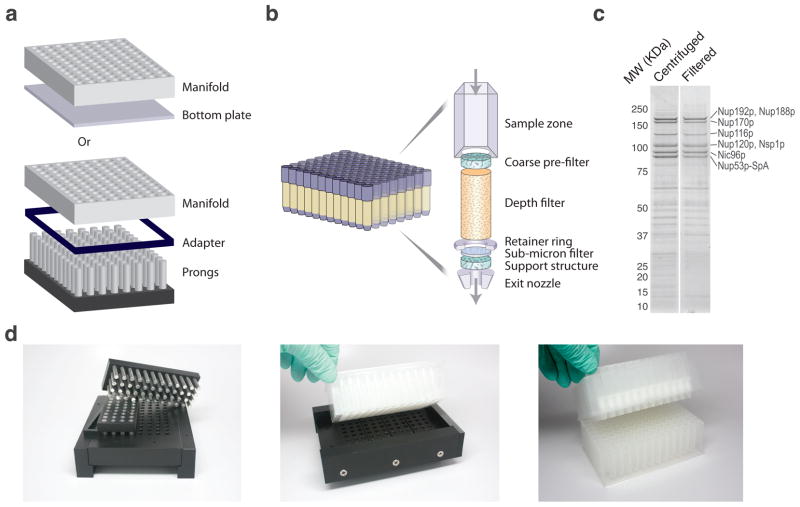
Figure 2.
Dispensing manifold and filtration device. (a) Schematic representations of the manifold used to dispense a calibrated amount of frozen cell powder into a 96-well plate. A set of adapters and volume displacing prongs are used to deliver the required amount of cell material. (b) Schematic representation of the filtration device used to clarify crude yeast cell extracts. Each well contains a composite filter comprised of multiple filtration elements, including: a coarse pre-filter that retards the flow of highly aggregated material; a diatomaceous earth depth filter that permits the passage of soluble material, and traps the insoluble debris that can clog submicron filters; and a 0.2 micron membrane filter, which provides a uniform final clarification. (c) Coomassie blue stained SDS-PAGE analysis of Nup53p-SpA affinity capture (100mg cell material resuspended with 600uL extraction solvent: 40 mM TRIS-Cl, pH 8, 250 mM trisodium citrate, 150 mM NaCl, 1% v/v Triton X-100) comparing extract clarification by centrifugation at 14k rpm for 10 min (“Centrifuged”) and filtration at 3.5k rpm for 5 min (“Filtered”), MW – molecular weight standard. Duplicate experiments produced identical results (not shown). Proteins labeled in accordance with Figure 4a. (d) Pictures of the actual devices from left to right: adjustable volume dispensing manifold (as in a), shown bottom up; dispensing manifold with 96-well deep-well plate atop, cell material transfer is achieved upon inversion of this assembly; a 96-well filtration device (as in b) atop a 96-well, deep well collection plate.
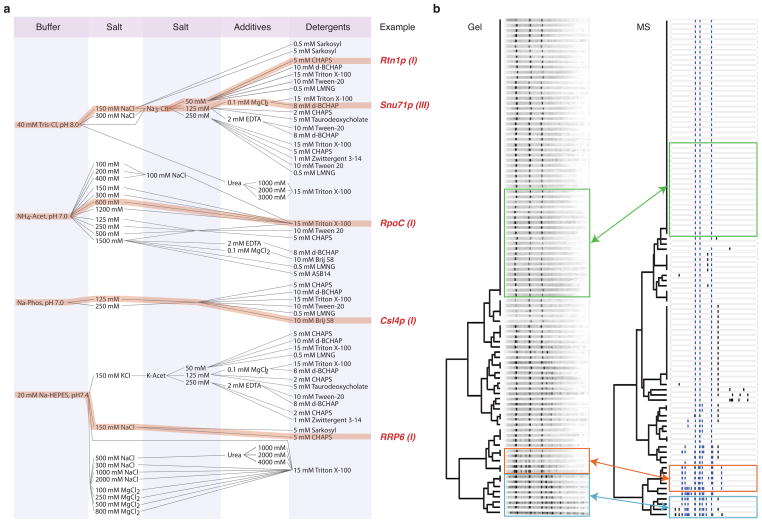
The bandwidth of our screen allowed us to thoroughly explore the most common reagents used in affinity capture experiments: salts, buffers, and detergents (Fig. 3a, Supplementary Table 1)12,31. Salts are frequently classed as kosmotropes (e.g. sodium citrate or ammonium acetate) or chaotropes (e.g. sodium perchlorate), in accordance with their tendency to respectively stabilize or disrupt protein structures in aqueous solution (the so-called Hofmeister series32). The mechanism of these effects is still largely uncharacterized33,34 and cannot be predicted a priori. The chemical character of the buffering agent can also contribute to the efficacy of affinity capture in unpredictable ways beyond simple pH control12. Detergents, used to extract membrane-anchored complexes and inhibit aggregation of all complexes, also exhibit unpredictable behaviors and require empirical optimization31.
Figure 3.
Extraction condition design and copurification pattern analysis. (a) Flow-chart representation of mixtures of components in select extraction solvent formulations. The main components are a pH buffer, 1 or 2 salts, an additive and a detergent. Some examples of useful formulations discovered through screening are indicated (refer to Fig. 5 to view the associated copurification patterns, indicated by Roman numerals). (b) Comparison of SDS-PAGE (“Gel”) and LC-MS/MS (“MS”) clustering analysis of Nup1p-SpA 96-well purification. For a visual comparison the MS data is represented as a pseudo-gel, where each band corresponds to a protein above a certain intensity threshold (see Methods). Known Nup1p interacting proteins are indicated with blue bands, the rest are labeled black. Co-clustering conditions with identical or highly similar components producing distinct copurification profiles are highlighted in blue (low ammonium acetate or low potassium acetate), orange (high sodium citrate/high ammonium acetate with Triton X-100) and green (sodium citrate/potassium acetate with CHAPS) boxes. See Supplementary Figure 3 for lane labels.
Evaluation of affinity capture profiles
There are two commonly used approaches for analyzing affinity captured samples: SDS-PAGE with dye-based visualization35,36 and subsequent MS of select conditions; or, direct MS of the samples10,11. We compared these two approaches for the 96-well purification of Nup1p, a component of the yeast nuclear pore complex (NPC), exploring a diverse set of extraction conditions. Two replicates were carried out for the comparison: one set resolved by SDS-PAGE and stained with Coomassie blue (Supplementary Fig. 2), the other set processed for LC-MS/MS (Supplementary Table 2, see Online Methods for details).
Gel images were segmented into lanes, aligned, and the intensity of bands ranked. The lanes were clustered based on the intensity rank of bands exhibiting similar apparent molecular mass (Fig. 3b, Supplementary Fig. 3). MS data were filtered of exogenous and endogenous contaminants (Supplementary Table 3), and proteins exhibiting intensities below 10% that of the most intense species detected were removed. The results for each set were clustered based on the presence of common proteins and represented as a dendrogram (Fig. 3b, Supplementary Fig. 3). The cophenetic correlation coefficient between the two dendrograms was found to be 0.53 with a p-value of < 1 × 10−7 (Supplementary Fig. 4), indicating that both analytical methods describe the data similarly to a high degree of significance – revealing that the various extractions yield complexes with a number of distinct protein compositions and a high degree of redundancy; and also confirming that individual well failures do not compromise the conclusions of the whole experiment. Three proteins were revealed in the majority of conditions: Kap95p, Kap60p and Nup1p-SpA (Fig. 3b, green box), and a larger number of proteins were observed in e.g. low acetate (Fig. 3b, blue box) or high citrate/acetate (Fig. 3b, orange box).
Our conclusions are twofold. First, SDS-PAGE provides a representative readout of the composition of affinity captured samples, faithfully revealing the effects of changing affinity capture conditions, in a rapid, robust and inexpensive fashion. Second, with sufficient resources, direct sample-to-MS analytical approaches may be utilized. Both conclusions bolster the prospects for automated, unsupervised high-throughput approaches for optimizing affinity capture and dissecting interactomes (more below).
Exploring of the molecular organization of a 50 MDa complex
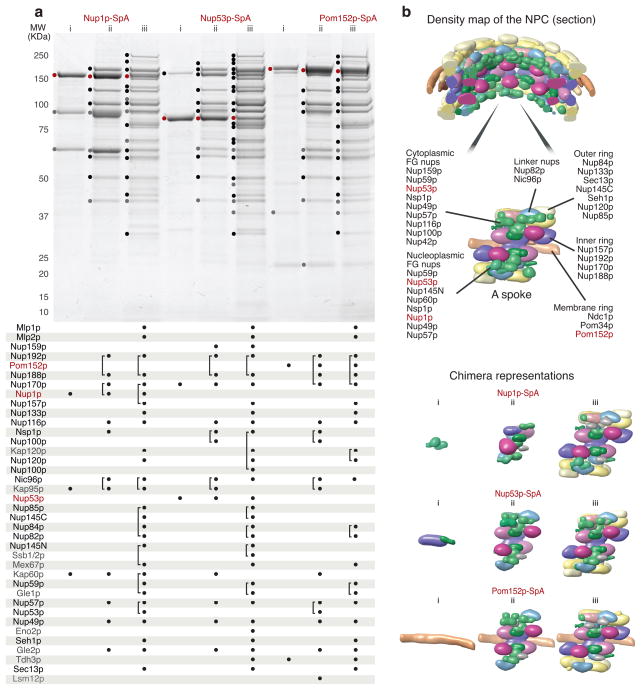
The yeast NPC is ~50 MDa in size and consists of multiple copies of ~30 different proteins. It presents an excellent test bed for our screen because it comprises a diverse physicochemical landscape and has a modular architecture consisting of subcomplexes of different sizes. Moreover, an extensive catalog of already existing affinity capture results37, enabled us to assess our findings and the quality of results produced by the screen. Based on the above-described initial results, we further modified our conditions matrix to test other reagents and applied it collectively to SpA-tagged NPC proteins Nup1p, Nup53p and Pom152p.
For all three proteins the screen revealed novel conditions that exhibited improvements in yield, background, and hierarchical coverage, compared to the best results previously obtained37 (Fig. 4a). Interestingly, all three proteins responded similarly to the extraction conditions presented (Fig. 4a): condition (i) gave small complexes with few interactors for all three, and no common components; condition (ii) gave more complicated profiles that partially overlapped in composition with each other; and condition (iii) gave the most complicated and highly similar profiles, representing almost the entirety of the NPC (Supplementary Data). These profiles were in agreement with the previously determined arrangement of proteins in the NPC, constituting interaction shells of increasing size and degree of overlap37 (Fig. 4b). These overlaps demonstrated our method’s ability to “walk” from protein to protein via common complex components through the entire NPC, showing how reagent screening can be a tool for comprehensive interactomic mapping of different macromolecular complexes.
Figure 4.
NPC purification – from single proteins to macromolecular assemblies. (a) A representative SDS-PAGE image and MS analysis of affinity capture of 3 nucleoporins – Nup1p, Nup53p and Pom152p. Composition of affinity isolation solvents: i – 50 mM trisodium citrate, 300 mM NaCl, 10 mM Tween 20, 2 mM EDTA, 40 mM TRIS, pH 8; ii – 250 mM trisodium citrate, 10 mM Brij58, 0.3 mM Sarkosyl, 40 mM TRIS, pH 8; iii - 1.5 M ammonium acetate, 15 mM Triton X-100. The protein bands identified by MS are marked on the gel. The table below contains the list of identified proteins. Affinity tagged nups are labeled red, the remaining NPC constituents are black and non-NPC proteins are gray. Each protein identified by MS is marked by a dot under the corresponding lane. The brackets indicate comigrating proteins identified in a single band. (b) Section through the density map of the NPC37 with one spoke enlarged and minimum Chimera representations of NPC subcomplexes in (a) for 1 spoke.
Simultaneous mapping of distinct interaction networks
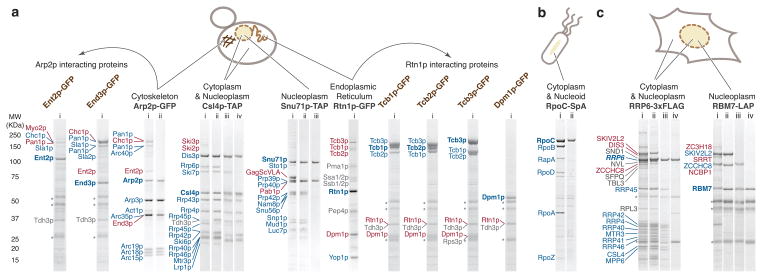
We tested whether the screen was equally applicable for many different types of macromolecular complexes. We examined four proteins with two different tags acquired from commercial collections. These proteins (Arp2p-GFP, Csl4p-TAP, Snu71p-TAP and Rtn1p-GFP) exhibit distinct subcellular localization patterns and functions. Each protein was subjected to a 32-condition screen (Supplementary Figs. 5–8), allowing us to assay multiple proteins within the same 96-well plate. High quality copurification profiles were obtained (Fig. 5a, Supplementary Data), including the observation of novel and distinctive copurification patterns for proteins already extensively subjected to affinity capture MS strategies.
Figure 5.
Affinity capture strategy implementation on different protein complexes, affinity tags and model organisms. Affinity capture profiles: (a) S. cerevisiae; (b) E. coli; (c) H. sapiens. Representative SDS-PAGE profiles are shown. The cell schematics indicate the localization of the tagged proteins; the different tagged proteins screened are indicated in black; each lane corresponds to a different purification and is assigned an arbitrary Roman numeral (see Supplementary Table 1 for extraction conditions, except when specified). Some of the newly identified putative interactors were subjected to affinity capture (labeled brown) and the resulting profiles are indicated by arced arrows originating from the profiles in which they were identified. Copurifying protein bands identified by MS are marked next to each profile (see Supplementary Data). Protein names marked in blue are previously characterized physical interactors, those in red are select novel physical interactors or proteins of interest discussed in the main text, and those in grey are contaminants or proteins of indeterminate specificity based on their high frequency of copurification20, or as determined by I-DIRT analysis (Fig. 6a). The bands labeled with an “*” indicate heavy and light chains of the antibody used for affinity capture. Ent2p and End3p extraction solvent – 40mM TRIS-Cl, pH 8, 150 mM NaCl, 250 mM trisodium citrate, 10 mM deoxy-BigCHAP, Tcb1p, Tcb2p, Tcb3p, and Dpm1p extraction solvent - 40 mM TRIS-Cl, pH 8, 150 mM NaCl, 50 mM trisodium citrate, 5 mM CHAPS.
The Arp2/3 complex is a conserved actin nucleator that participates in multiple actin-dependent processes, including endocytosis38,39. Screening revealed a putative novel macromolecular assembly comprised of clathrin (Chc1p) with its adaptor protein (Ent2p) and actin (Act1p) with its recruiting and activating proteins40 (Pan1p, End3p and Arp2/3 complex; Fig. 5a, Arp2p-GFP, i). Of these, however, only Act1p and Pan1p are known to physically interact with Arp2p41,42. As End3p is linked to Arp2p genetically38 and no direct links between Arp2p and Ent2p have been demonstrated, we chose to perform a secondary affinity capture for GFP-tagged versions of these proteins. The results (Fig. 5a, Ent2p-GFP, Ent3p-GFP and Arp2p-GFP, see also Supplementary Note 1), supported the role of Pan1p as a core scaffold, coupling actin and clathrin with the rest of the endocytic machinery42, in an interaction network involved in the early stages of actin-dependent clathrin-mediated endocytosis42–46.
Csl4p is a component of the eukaryotic exosome, a modular multi-protein ribonuclease with compartment-specific components47,48. Screening revealed conditions that selectively destabilized the compartment-specific components Rrp6p, Lrp1p, and Ski7p (Fig. 5a, Csl4p-TAP, iii and iv, compared with the canonical exosome, profile ii), while retaining the component Dis3p. Due to the relative stabilities of these different components in established purification conditions48–50, strains with genetic deletions have been necessary to obtain comparable complexes (e.g.50). In a separate profile, components of the cytoplasmic exosome cofactor Ski complex Ski2p/Ski3p were observed (Fig. 5a, Csl4p-TAP, i), which are considered recalcitrant to copurification51,52.
Snu71p is a component of the nuclear localized U1 snRNP complex, a constituent of the spliceosome53,54. Of interest, we purified U1 snRNP (Fig. 5a, Snu71p-TAP, ii) with nuclear mRNA associated proteins Sto1p and Pab1p and the major coat protein of ScVLA virus, which is known to covalently bind the mRNA cap55 (Fig. 5a, Snu71p-TAP, i). We also noted a profile demonstrating a direct interaction between Snu71p and Prp40p (Fig. 5a, Snu71p-TAP, iii). Despite more than a decade of research on the composition of this complex, this dimer was only unambiguously shown recently by the introduction of a deletion mutation to a third constituent of this RNP that contains 17 distinct components56. Here, it was obtained within a single screen.
Rtn1p, an integral endoplasmic reticulum (ER) membrane component, simultaneously embodies many of the challenges to affinity capture approaches: it is spread between multiple localizations and functionalities, is expected to form particularly dynamic or transient complexes57–59, and as a membrane protein is among a class of proteins often refractory to interactomics60. Although it is important in numerous cellular processes58,59,61 and has been subjected to an affinity capture screen intended for membrane proteins25, there are comparatively few validated physical interaction data available for Rtn1p. Lacking prior knowledge of the expected interactions, we selected a condition from our screen using our experience-based SDS-PAGE profile criteria (see Discussion). The copurifying proteins (Fig. 5a) included known and uncharacterized putative Rtn1p-interacting partners. Among these was the ER membrane-associated protein, Dpm1p, known to have a negative genetic interaction with Rtn1p62. We used secondary affinity capture to validate this interaction with Dpm1p, as well as with the ER membrane tricalbins (Tcb1-3p), which also copurified but have no previously demonstrated physical links to Rtn1p; all four GFP-tagged proteins copurified Rtn1p, the Tcb proteins each copurifed one another63 and also yielded Dpm1p.
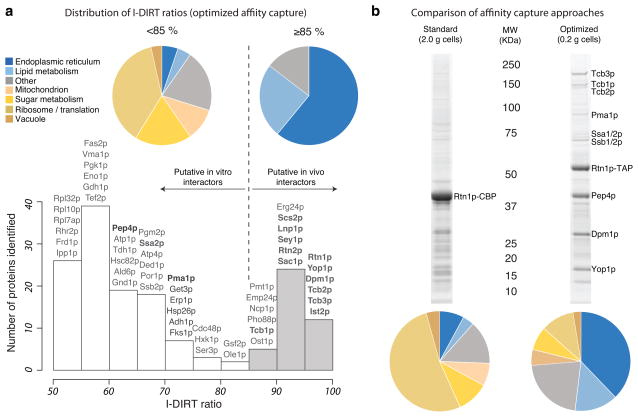
We repeated the Rtn1p-GFP affinity capture experiment, implementing isotopic differentiation of interactions as random or targeted (I-DIRT) analysis to distinguish interactions formed in vivo from those likely to be in vitro artifacts64. I-DIRT analysis (see Supplementary Table 4 for unprocessed and analyzed data) indeed confirmed that most of the strong bands in our optimized affinity capture represent protein interactions with Rtn1p that were present in vivo (Fig. 6a), with a few prominent bands corresponding to common contaminants (see also Supplementary Note 1). Among putative Rtn1p interactors were 5 out of the 6 proteins known to tether ER to plasma membrane, namely Tcb1-3p, Ist2p, and Scs2p. Notably, Rtn1p, Tcb1p, and Tcb3p were identified in Sac1p (lipid phosphatase) and Scs2p (ER-plasma membrane tether) purifications65. Given its proposed function to stabilize curved membranes58,66, we suggest that Rtn1p may help stabilize membrane curvature at ER-plasma membrane contact sites where lipid transfer/modification occurs.
Figure 6.
An in-depth analysis of Rtn1p affinity capture. (a) Frequency distribution of I-DIRT ratios – light protein intensity/total protein intensity, normalized to 100% – from Rtn1p-GFP affinity capture experiment (extraction condition as in Fig. 5a, Rtn1p-GFP). Putative in vivo interactors are represented with shaded bars (≥85%), likely post-extraction associations – with open bars (<85%). Representative proteins are shown above each bar, proteins labeled in Figure 5a (Rtn1p-GPF) and known interactors of Rtn1p are bolded. The proteins with <85% and ≥85% I-DIRT ratio were separately analyzed for the subcellular localization and molecular function. The corresponding pie charts are plotted above the I-DIRT ratio distribution (see Methods for details). (b) SDS-PAGE and MS comparison of standard and optimized affinity capture. 4g of Rtn1p-TAP powder was processed essentially as previously described25 using Triton X-100 as a detergent in a 2-step affinity capture experiment and 0.4g of Rtn1p-TAP was processed in a 1-step affinity capture experiment using conditions revealed in the present study (Fig. 5a, Rtn1p-GFP). Half of the elution was analyzed by SDS-PAGE followed by Coomassie staining and the other half was analyzed by LC-MS/MS. The distribution of subcellular localizations and molecular functions was analyzed as in (a) and is plotted bellow the corresponding gel lanes.
Could a standard affinity capture approach have reproduced these results? To address this question, we executed a side-by-side comparison of the popular tandem affinity purification procedure, recently tuned for membrane proteins25, with an optimized procedure emerging from our screen (see Supplementary Table 4 for MS analysis). The results illustrate that the classic approach cannot compete with our screening strategy, either in terms of quality or efficiency (Fig. 6b).
Adaptability of the screen for diverse model organisms
Different model organisms often exhibit idiosyncrasies associated with affinity capture experiments. Our screen allows alternatives to be explored at each step. For example, an issue found with E. coli was the high viscosity of cell extracts due to high concentrations of released genomic DNA. We therefore modified our procedure to include a short low energy sonication using a multi-tip probe, sufficient to re-suspend the frozen cell powder during extraction and reduce viscosity to levels compatible with affinity capture. Similarly, low quantities of starting material may present an issue when working with tissue culture cells. Thus, we also modified the screen for a 24-well format in order to economize on cell usage. In our procedure, the mass of yeast cell pellet required per purification was reduced from the typical order of grams25,67–69 to the range of tens to hundreds of milligrams; similarly, we consume only 50 mg cryomilled human cells per profile, an ~8-fold reduction over contemporary high-throughput studies21,24. These modifications therefore enable economical interactomic screens in diverse model organisms.
From E. coli we purified the RNA polymerase (RNAP)70 complex corresponding to the σ70 containing holoenzyme in complex with RapA71 and RpoC-SpA isolated away from RNAP (Supplementary Data, Fig. 5b, RpoC-SpA i and ii) demonstrating that the implemented modifications provide affinity capture results comparable in quality to those from yeast (Supplementary Fig. 9). Utilizing human cells, we revisited the RNA exosome, conducting purifications via a 3xFLAG-tagged hRRP6 (EXOSC10) – adding another common tag variety to those tested thus far (Supplementary Fig. 10). Among our observations (see also Supplementary Note 1), we noted the stable retention of SKIV2L2 (hMTR4) in numerous interaction profiles (see Supplementary Data). SKIV2L2 is also a member of the nuclear-specific human exosome cofactor NEXT complex72,73, along with ZCCHC8 and RBM7. We readily observed another member of this complex, ZCCHC872, in human exosome profiles (e.g. Fig. 5c RRP6-3xFLAG, i and ii), raising the question as to whether SKIV2L2 is single or multiple copy in the combined exosome/NEXT containing fractions. To extend our exploration we applied this screen to the NEXT complex itself, purified via LAP-tagged RBM7 (Supplementary Fig. 11). Among our findings, we observed NEXT in association with NCBP1 (CBP80), SRRT (ARS2), and ZC3H18 (NHN1) (Supplementary Data, Fig. 5c, RBM7-LAP, i; see also Supplementary Note 1) and made the novel observation of a direct interaction between RBM7 and ZCCHC8 (Fig. 5c, RBM7-LAP, compare ii to iii), demonstrating that these interactors form a stable dimer. We validated the above interactors extensively in a parallel study74 via both MS-based label free quantitation and a suite of functional assays – revealing a new pathway of RNA surveillance utilizing the mRNA cap-binding complex, the NEXT complex and the exosome complex.
Robotic automation and gel curation for higher throughput
Our pipeline is designed for easy bench-scale execution. However, translation to automation has several advantages, including increased throughput and reproducibility. Using a liquid handling robot, we developed a version of the screen that includes automated production of extractant matrices and sample handling from the addition of affinity medium to clarified extracts through to the final wash. Given the intriguing results observed through the course of this study using trisodium citrate during manual screening (e.g. Fig. 4a), we implemented automation to systematically explore the effect of this reagent on Nup53p-SpA affinity capture profiles over 48 increments from 50 to 300 mM. We observed three distinct profiles (Fig. 7, detailed in Supplementary Note 1). These results demonstrated that the automated procedure was precise and revealed systematic changes in the copurification pattern specific to trisodium citrate, which involved the loss of Kap121p and the increased retention of a large number of NPC components as the concentration increased.
Figure 7.
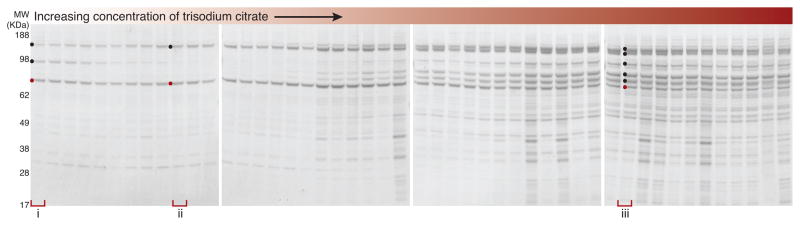
Robotic implementation. SDS-PAGE analysis of one Nup53p-SpA purification screen performed using a Hamilton STAR liquid handling workstation, testing 50 – 300 mM trisodium citrate (40mM TRIS-Cl, pH 8, 1% v/v Triton X-100 are common to all lanes). The purifications form bracketed lanes were manually repeated and analyzed by MALDI-TOF-MS (Supplementary Data). Three distinct profiles are observed: copurification with Nup170p and Kap121p (i); dimer with Nup170p (ii); and a larger subcomplex of the NPC: Nup192p, Nup188p, Nup170p, Nup116p, Nup120p, Nsp1p and Nic96p (iii).
Because large amounts of data were generated during screening, we developed a web portal to assist in affinity capture data management. Our software (described in Supplementary Note 2 and publicly accessible at www.copurification.org) accepts images of gels along with experimental metadata. Gel images are automatically sectored lane-by-lane and annotated with the conditions applied to each, respectively. The lanes are also clustered according to protein banding pattern similarity, to ease the discovery of lane-to-lane differences and trends (Fig. 3b). This database provides a platform for the work of different experimentalists to be compared side-by-side, with instantaneous access to the respective experimental conditions for ease of reproduction.
Discussion
The solvent environment of the extractant plays a crucial role in dictating the stability of both real and artifactual protein-protein interactions during affinity capture12–14. The difficulty in finding extractants that maximally explore the real interactome while minimizing artifacts15–17 has limited high-throughput screens21–24,68,69,75. Our approach addressed this limitation, providing a fast, efficient and cost-effective means to scan many conditions for their ability to preserve physiological interactions and minimize noise. This is particularly important for studies that hope to go beyond protein identifications and further obtain high quality protein preparations for biochemical and structural studies16.
Doubtless because of the huge diversity of interaction types, we have not yet found one set of conditions that works well for all the protein interactomes we have studied, underscoring the need for our screen. However, encouragingly, our results suggest that the optimal set of extraction conditions determined for a subset of constituents in a given complex, will suffice for the interactomic exploration of all the components in that complex (e.g., Fig. 4a). Moreover, we observed that during the secondary affinity capture of putative interactors identified in a primary screen (i.e. biochemical validation), copurification profiles containing both overlapping and distinct proteins were frequently obtained (see e.g. Fig. 4a, Fig. 5a, Arp2p-GFP and Rtn1p-GFP, and Fig. 5c RRP6-3xFLAG and RBM7-LAP) – highlighting the potential of this screen to uncover local (sub)complexes as well as the broader interactome. These combined attributes are particularly important given the current efforts to create a “human proteome encyclopedia76” – which will undoubtedly require rigorous attention by any investigator to the preparation of the highest quality samples for affinity capture MS analyses.
For general purposes, we favor SDS-PAGE with protein staining for sample quality assessment, followed by band excision and MS to determine protein identities. SDS-PAGE is a proven, fast, parallel and quantifiable assay giving the approximate number, size, and amount of each band on the gel35. Our findings, time and again, reinforced the notion that high quality affinity capture experiments are typified in SDS-PAGE profiles by a discrete pattern of sharp, abundant, and roughly stoichiometric bands as well as a paucity of background staining from other fainter bands (see e.g.37,54, Figs. 4a and 5). The existence of increasingly sensitive general protein stains provides gel-based visualization options even for very low abundance samples77. These criteria in turn allow for the judicious application of MS analyses to only the most potentially informative samples.
When tens to hundreds of SDS-PAGE protein copurification profiles are viewed in parallel, patterns of common and changing proteins and their solvent dependencies typically become readily apparent, and several promising conditions reveal themselves. A typical 96-well screen, as a consequence of being thorough, may yield many gel lanes with comparable banding profiles (Fig. 3b), and many that do not meet the criteria for further analysis. To modulate between throughput and screen breadth the total number of conditions can be adjusted (presented here at multiplicities of 96, 32, and 24).
While a promising protein copurification profile accompanied by high quality MS-based protein identifications provides the basis for a strong hypothesis regarding the existence of a physically associated protein complex that exists in vivo, such data should encourage the design of orthogonal experiments intended to rigorously test this hypothesis, including the importance of affinity capture optimization and the utility of I-DIRT in revealing high confidence targets for in vivo experimentation78. Hence, in one sense, the presented screen can be considered a rapid and efficient hypothesis generation machine for physical interactions.
Our procedure is also compatible with direct sample-to-MS analyses (Fig. 3b) and can be implemented using robotic automation (Fig. 7), greatly enabling throughput. Future data mining opportunities will include the development of unsupervised, machine-based classification schemes to further improve our ability to identify promising samples, greatly augmenting high-throughput interactomic studies.
Online Methods
Affinity capture
All cell lines/strains utilized in this study are listed in Supplementary Table 5. Yeast, E. coli and Human cell lines were cultured using standard procedures and cryomilled and affinity captured essentially as previously described29,30, except adapted for 96-well plates as described in text and elaborated step-wise in the Supplementary Protocol 1. Human cell lines have not been subjected to mycoplasma testing during the course of the study. Rabbit IgG used for purifying TAP and SpA tagged proteins was purchased from Innovative Research. Anti-GFP polyclonal antibodies were prepared and conjugated as previously described30, except the concentration of ammonium sulfate used during the conjugation was 1.5 M. In all cases, proteins were eluted from the affinity medium by the addition of 1x NuPAGE LDS sample loading solution (Life Technologies); elution of GFP-tagged proteins included incubation at 70°C for 10 min. Extraction solvent working solutions were mixed from concentrated stock solutions in 2.5 ml deep-well plates (VWR) manually, using a Formulator (Formulatrix), or using a Hamilton STAR liquid handling workstation (program files provided in Supplementary Protocol 2). Supplementary Figure 12 contains detailed engineering diagram of the powder dispensing manifold. Resuspension of powders in extraction solvents included sonication with an ice water chilled microplate horn (yeast) or 8-tip micro-probe (bacteria & human) (Qsonica). Yeast lysates were also vortexed with steel beads to aid rapid homogenization. Supplementary Figure 13 displays the bead dispensing manifold utilized in yeast affinity capture experiments. Custom manufactured filters (Fig. 2, Orochem Technologies) were used to clarify yeast cell extracts for screens, otherwise extracts were clarified by centrifugation at 14k RPM and 4°C, for 10 min in a bench top microfuge. To ensure reproducibility all purifications presented (and processed for MS) were repeated individually in microfuge tubes using an otherwise comparable procedure except that extracts were clarified via centrifugation. Polyacrylamide gels were stained with either a homemade colloidal Coomassie brilliant blue G-250 solution79 or with Imperial Protein Stain (Thermo Fisher Scientific). Gel images were recorded in TIFF using a Fujifilm LAS-3000 or an Epson Photo v700. In addition to the cited publications, detailed protocols for many preparatory procedures utilized in this study can be obtained at http://www.ncdir.org/public-resources/protocols/.
Standard mass spectrometric identification of proteins
The major bands observed in SDS-polyacrylamide gels were excised and analyzed either by MALDI-TOF-MS essentially as previously described30, or nanoLC-ESI-MS/MS on an LTQ Orbitrap XL, Orbitrap Velos, Q Exactive Plus or Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). For analysis of excised protein bands using the LTQ Orbitrap XL or Orbitrap Velos the dry peptides were resuspended in 0.5% v/v acetic acid and pressure loaded on a self-packed C18 column and subjected to a 10 min gradient: 8 min 0 – 43%, 2 min 43 – 100% solvent B (solvent A = 0.1M acetic acid, solvent B = 0.1M acetic acid, 70% acetonitrile, 100 nl/min). The eluted peptides were analyzed with the following settings: top 10 peaks were selected for fragmentation, without dynamic exclusion. For the analysis on Q Exactive Plus and Orbitrap Fusion the peptides were resuspended in 0.1% formic acid, and separated using a 10 minutes gradient (8 min 0 – 30%, 2 min 30 – 100%, 1 min 100% solvent B) on an EASY-Spray column (Thermo Fisher Scientific) using an EASY-nLC 1000 (Thermo Fisher Scientific; solvent A = 0.1 % v/v formic acid, solvent B = 0.1 % v/v formic acid in acetonitrile, flow rate 300 nl/min). The 3 most abundant ions were selected in each full scan and sequentially fragmented by HCD with Q Exactive Plus. With Orbitrap Fusion a fixed duty cycle of 3 seconds was used. The RAW files were converted to MZXML format with the MM File Conversion tool (http://www.massmatrix.net/mm-cgi/downloads.py) or MGF format by ProteoWizard80 and searched against the yeast protein database with X! Tandem81.
For the analysis of whole affinity captured fractions in Figures 3b and 6b, the protein samples were run ~4–6mm into an SDS-polyacrylamide gel (gel plug), and gels were Coomassie blue stained. Stained gel regions were excised, cut into 1 mm cubes, de-stained, and digested for 6 hours with 120 μl of 3.1 ng/μl trypsin (Promega) in 25 mM ammonium bicarbonate. An equal volume of 2.5 mg/ml POROS R2 20 μm beads (Life Technologies) in 5% v/v formic acid, 0.2% v/v trifluoroacetic acid was added, and the mixture incubated on a shaker at 4°C for 24 hr. Digests were desalted on C18 tips, eluted, dried by vacuum centrifugation, resuspended in 0.1% formic acid, and separated using a 10 minute gradient on an EASY-Spray column (as above). The 12 most abundant ions were selected in each full scan and sequentially fragmented by HCD (Q Exactive Plus); dynamic exclusion was enabled. RAW files were converted and searched as above. In order to determine the molecular functions of constituent proteins (Fig. 6b) we searched the gene descriptions for key words (as for I-DIRT analysis, see below).
Gel and MS data clustering and correlation analysis
The details of the gel image analysis are provided in the Supplementary Note 2; the source code is publicly available at https://github.com/FenyoLab/copurification. Once the lanes were sectored and bands identified, quantified and assigned an apparent molecular weight they were categorized as dark, light or, not observed (Fig. 3b, Supplementary Fig. 3).
Supplementary Table 2 contains the unfiltered search results of 96 LC-MS/MS runs. For each sample we extracted the intensities of all the hits, filtered out exogenous and endogenous contaminants (Supplementary Table 3) and considered the hits that were at least 10% as intense as the most intense hit (after initial contaminant filtering). We used a modified version of a source code available at the GPM repository ftp://ftp.thegpm.org to output the resulting protein sets as a pseudogel (Fig. 3b, Supplementary Fig. 3).
For both data sets, the Ward method was used for hierarchical clustering with distance between data points calculated as Euclidean distance82. To perform the correlation, the cophenetic distance was calculated for the gel and MS dendrograms83. The cophenetic correlation was then calculated, which is defined as the Pearson correlation between the cophenetic distance matrices of the dendrograms83. A p-value was obtained by a permutation test: the labels of the MS dendrogram were shuffled 10 million times, and a correlation calculated between the MS and gel dendrograms for each random shuffle (see Supplementary Fig. 4 for frequency distribution). A p-value of < 1 × 10−7 was calculated as a proportion of the random distribution equal to or greater than the actual correlation (0.53).
Graphical representation of NPC subcomplexes
We used the density maps for individual nucleoporins available at http://salilab.org/npc/ and the UCSF Chimera package84 to graphically represent the NPC subcomplexes.
I-DIRT data analysis
I-DIRT was carried out essentially as previously described64: the Rtn1-GFP strain was grown in synthetic complete minimal medium lacking lysine and supplemented with 50 mg/L of isotopically light lysine and a wild type DF5α strain was grown in the same medium but supplemented with 50 mg/L of isotopically heavy lysine (L-Lysine:2HCl 13C6, Cambridge Isotopes), both were frozen, mixed and cryomilled. Rtn1-GFP was affinity captured from the mixed powder extracted in 40 mM Tris-Cl pH 8, 50 mM trisodium citrate, 150 mM NaCl and 5 mM CHAPS. The eluted sample was reduced, alkylated and precipitated with methanol/chloroform85. The precipitate was resuspended in 50 mM ammonium bicarbonate, 0.1% w/v RapiGest (Waters) via bath sonication with heating (20 min at 70°C, followed by 2min at 95°C). The proteins were digested with trypsin (Promega) overnight. Rapigest was depleted following the manufacturer’s instructions and the digest was desalted over C18 Omix tips (Agilient Technologies). The eluted fractions were analyzed on LTQ Orbitrap XL as described before except with 1h gradient and dynamic exclusion enabled. The output data was processed with MaxQuant86 (http://maxquant.org/) using essentially the default parameters (for the light sample all amino acids were set to light, for the heavy sample 6 Da heavy lysine was selected; the yeast translated ORF sequences – http://www.yeastgenome.org/ – reversed sequences and contaminants database were searched) to identify and measure the intensity of heavy and light peptides. The “Evidence.txt” file containing all the peptide identifications and heavy/light measurements was used in the final analysis. Peptides mapped to contaminants, constituents of a reversed sequence database or those containing no lysine were excluded from the analysis. We further excluded non-unique peptides and peptides with a single MS/MS fragmentation event. For the remaining peptides I-DIRT ratios were calculated by dividing the intensity of the peptide with light lysine by the total intensity (light and heavy). To calculate the I-DIRT ratio of proteins the I-DIRT ratios of its constituent peptides were averaged. Proteins with a single peptide contributing to the I-DIRT ratio measurement were excluded from the analysis as unreliable. For proteins with 4 or more peptides contributing to the I-DIRT ratio measurement, those peptides with outlying I-DIRT ratios were filtered out (no more than 1 peptide removed per protein) using the following criteria: if the calculated I-DIRT ratio for a peptide was < Q1 (first quartile) - 1.5 x IQR (interquartile range) or > Q3 (third quartile) + (1.5 x IQR), that peptide was excluded. All statistical calculations and plotting were done with R (http://www.R-project.org)87. To assess the normality of protein I-DIRT ratio distribution a Q-Q plot was constructed, which revealed a notable deviation from the y = x line implying that the data was not normally distributed (Supplementary Fig. 14). To assess the shape of the distribution the data were binned in 5% intervals (Supplementary Fig. 15). Despite a low bimodality coefficient88,89 (0.2956), the distribution deviates significantly from unimodal by Hartigan’s dip test90 (p-value = 0) and has a positive Akaike’s information criterion difference91 (0.1061) – suggesting bimodality. We used Mixtools package in R92 to fit 2 normal distributions to the data (Supplementary Figs. 15). We accepted a cut off of mean ± 2 standard deviations of the second distribution as stable interactors of Rtn1p (≥85%). We considered anything below 85% to constitute interactions indistinguishable from those formed post-extraction. In order to determine the molecular functions of constituent proteins we searched the gene descriptions for key words. The following are the categories and key words searched: Endoplasmic reticulum – “ er”/“endoplasmic reticulum”; Sugar metabolism – “glycolysis”/“gluconeogenesis”/“glucose”/“glycolytic”/“pentose”; Vacuole – “vacuol”; Ribosome/Translation – “ribosom”/“ translat”; Lipid metabolism – “lipid”/“fatty acid”/“choline”/“sterol”/“ceramide”; Mitochondrion – “mitochond”; Other - everything that didn’t match. All searches were case insensitive. Once a gene description matched a keyword it was put into the corresponding category, allowing us to count the number of proteins in each category and construct a pie chart of the distribution of molecular functions/localization for a given protein set (Fig. 6).
Supplementary Material
Acknowledgments
We thank The Rockefeller University High Energy Physics Instrument Shop for diligence in custom apparatus design and fabrication, X. Wang for assistance with MS data analysis, and members of the Chait, Jensen and Rout laboratories for help and discussion. I. Poser and A. Hyman (Max Planck Institute of Molecular Biology and Genetics, Dresden DE) provided the RBM7-LAP cell line. This work was funded by the National Institutes of Health (NIH) grant no. U54 GM103511 and P41 GM109824 (J.D.A, B.T.C., and M.P.R.), P41 GM103314 (B.T.C), P50 GM076547 (J.D.A.), and The Lundbeck (to T.H.J. and J.L.) and Danish National Research Foundations (to T.H.J.).
Footnotes
Author Contribution
J.L. and M.P.R. conceived the screening strategy; J.L. carried out proof of concept, assisted by L.H.; L.H. and V.S. designed manifolds, fabricated by V.S., and tested by L.H., J.L. and Z.H.; filters were designed by A.A.O, A.R.O, and J.L., fabricated by A.A.O. and A.R.O., and tested by J.L. and Z.H.; J.L., Z.H., and M.D. designed experiments and executed screens and further developed procedures – with yeast work primarily carried out by Z.H. and human cell line work primarily carried out by M.D.; mass spectrometry analyses were carried out by J.L., Z.H., and K.M. – with I-DIRT done by Z.H.; transposing the procedure to robotic automation was carried about by D.J.D. assisted by J.L.; J.L., Z.H., and D.F. conceived the protein copurification gel database and software, which was built by S.K. and D.F. with testing and feedback from J.L. and Z.H.; J.D.A., D.F., B.T.C., T.H.J., M.P.R., and J.L. supervised the project; Z.H., B.T.C., M.P.R. and J.L. wrote the paper.
Competing interests
A.A.O., A.R.O., M.P.R., and J.L. are inventors on a US patent application encompassing the filter work described in this manuscript.
References
- 1.Brockhurst MA, Colegrave N, Rozen DE. Next-generation sequencing as a tool to study microbial evolution. Mol Ecol. 2011;20:972–980. doi: 10.1111/j.1365-294X.2010.04835.x. [DOI] [PubMed] [Google Scholar]
- 2.Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol. 2011;136:527–539. doi: 10.1309/AJCPR1SVT1VHUGXW. [DOI] [PubMed] [Google Scholar]
- 3.Charbonnier S, Gallego O, Gavin AC. The social network of a cell: recent advances in interactome mapping. Biotechnol Annu Rev. 2008;14:1–28. doi: 10.1016/S1387-2656(08)00001-X. [DOI] [PubMed] [Google Scholar]
- 4.Collins MO, Choudhary JS. Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr Opin Biotechnol. 2008;19:324–330. doi: 10.1016/j.copbio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Kiemer L, Cesareni G. Comparative interactomics: comparing apples and pears? Trends Biotechnol. 2007;25:448–454. doi: 10.1016/j.tibtech.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Williamson MP, Sutcliffe MJ. Protein-protein interactions. Biochem Soc T. 2010;38:875–878. doi: 10.1042/BST0380875. [DOI] [PubMed] [Google Scholar]
- 7.del Sol A, Balling R, Hood L, Galas D. Diseases as network perturbations. Curr Opin Biotechnol. 2010;21:566–571. doi: 10.1016/j.copbio.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Stumpf MPH, et al. Estimating the size of the human interactome. Proc Natl Acad Sci USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menche J, et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardo M, Choudhary JS. Assignment of protein interactions from affinity purification/mass spectrometry data. J Proteome Res. 2012;11:1462–1474. doi: 10.1021/pr2011632. [DOI] [PubMed] [Google Scholar]
- 11.Dunham WH, Mullin M, Gingras AC. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 12.Ugwu SO, Apte SP. The Effect of Buffers on Protein Conformational Stability. Pharmaceutical Technology. 2004;28:86–108. [Google Scholar]
- 13.Oeffinger M. Two steps forward-one step back: Advances in affinity purification mass spectrometry of macromolecular complexes. Proteomics. 2012;12:1591–1608. doi: 10.1002/pmic.201100509. [DOI] [PubMed] [Google Scholar]
- 14.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 15.Bell AW, Nilsson T, Kearney RE, Bergeron JJM. The protein microscope: Incorporating mass spectrometry into cell biology. Nat Methods. 2007;4:783–784. doi: 10.1038/nmeth1007-783. [DOI] [PubMed] [Google Scholar]
- 16.Devos D, Russell RB. A more complete, complexed and structured interactome. Curr Opin Struct Biol. 2007;17:370–377. doi: 10.1016/j.sbi.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Breitkreutz B-J, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armean IM, Lilley KS, Trotter MWB. Popular computational methods to assess multiprotein complexes derived from label-free affinity purification and mass spectrometry (AP-MS) experiments. Mol Cell Proteomics. 2013;12:1–13. doi: 10.1074/mcp.R112.019554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trinkle-Mulcahy L, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013 doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jäger S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babu M, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- 26.Jancarik J, Kim SH. Sparse matrix sampling. A screening method for crystallization of proteins. J Appl Crystallogr. 1991;24:409–411. [Google Scholar]
- 27.Chayen NE. High-throughput protein crystallization. Adv Protein Chem Struct Biol. 2009;77:1–22. doi: 10.1016/S1876-1623(09)77001-4. [DOI] [PubMed] [Google Scholar]
- 28.Montelione GT. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol Rep. 2012;4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oeffinger M, et al. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 30.Domanski M, et al. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. BioTechniques. 2012;0:1–6. doi: 10.2144/000113864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: Not just a soap opera. Biochimica et Biophysica Acta - Biomembranes. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Kunz W, Nostro, Lo P, Ninham B. The present state of affairs with Hofffieister effects. Curr Opin Colloid In. 2004;9:1–18. [Google Scholar]
- 33.Nostro, Lo P, Ninham BW. Hofmeister phenomena: an update on ion specificity in biology. Chem Rev. 2012;112:2286–2322. doi: 10.1021/cr200271j. [DOI] [PubMed] [Google Scholar]
- 34.Wilson EK. HOFMEISTER STILL MYSTIFIES. Chemical & Engineering News. 2012;90:42–43. [Google Scholar]
- 35.Gauci VJ, Wright EP, Coorssen JR. Quantitative proteomics: assessing the spectrum of in-gel protein detection methods. J Chem Biol. 2011;4:3–29. doi: 10.1007/s12154-010-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller I, Crawford J, Gianazza E. Protein stains for proteomic applications: Which, when, why? Proteomics. 2006;6:5385–5408. doi: 10.1002/pmic.200600323. [DOI] [PubMed] [Google Scholar]
- 37.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 38.Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999;11:117–121. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 40.Moseley JB, Goode BL. The yeast actin cytoskeleton: From cellular function to biochemical mechanism. Microbiol Mol Biol R. 2006;70:605. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu SL, Needham KM, May JR, Nolen BJ. Mechanism of a Concentration-dependent Switch between Activation and Inhibition of Arp2/3 Complex by Coronin. J Biol Chem. 2011;286:17039–17046. doi: 10.1074/jbc.M111.219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- 43.Tang HY, Munn A, Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. Embo J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aguilar RC, Watson HA, Wendland B. The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J Biol Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell P, et al. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wasmuth EV, Lima CD. Structure and Activities of the Eukaryotic RNA Exosome. Enzymes. 2012;31:53–75. doi: 10.1016/B978-0-12-404740-2.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allmang C, et al. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 51.Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Synowsky SA, Van Den Heuvel RHH, Mohammed S, Pijnappel PWWM, Heck AJR. Probing genuine strong interactions and post-translational modifications in the heterogeneous yeast exosome protein complex. Mol Cell Proteomics. 2006;5:1581–1592. doi: 10.1074/mcp.M600043-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Gottschalk A, et al. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 54.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 55.Blanc A, Goyer C, Sonenberg N. The Coat Protein of the Yeast Double-Stranded-Rna Virus L-a Attaches Covalently to the Cap Structure of Eukaryotic Messenger-Rna. Mol Cell Biol. 1992;12:3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Görnemann J, et al. Cotranscriptional spliceosome assembly and splicing are independent of the Prp40p WW domain. RNA. 2011;17:2119–2129. doi: 10.1261/rna.02646811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geng J, Shin ME, Gilbert PM, Collins RN, Burd CG. Saccharomyces cerevisiae Rab-GDI displacement factor ortholog Yip3p forms distinct complexes with the Ypt1 Rab GTPase and the reticulon Rtn1p. Eukaryotic Cell. 2005;4:1166–1174. doi: 10.1128/EC.4.7.1166-1174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Craene JO, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helbig AO, Heck AJR, Slijper M. Exploring the membrane proteome-Challenges and analytical strategies. J Proteomics. 2010;73:868–878. doi: 10.1016/j.jprot.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 61.Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 63.Creutz CE, Snyder SL, Schulz TA. Characterization of the yeast tricalbins: membrane-bound multi-C2-domain proteins that form complexes involved in membrane trafficking. Cell Mol Life Sci. 2004;61:1208–1220. doi: 10.1007/s00018-004-4029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tackett AJ, et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 65.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 67.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 68.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 69.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nat Cell Biol. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 70.Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 71.Sukhodolets MV, Cabrera JE, Zhi HJ, Jin DJ. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes Dev. 2001;15:3330–3341. doi: 10.1101/gad.936701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lubas M, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 73.Sloan KE, Schneider C, Watkins NJ. Comparison of the yeast and human nuclear exosome complexes. Biochem Soc T. 2012;40:850–855. doi: 10.1042/BST20120061. [DOI] [PubMed] [Google Scholar]
- 74.Andersen PR, et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol. 2013;20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 76.The call of the human proteome. Nat Methods. 2010;7:661. doi: 10.1038/nmeth0910-661. [DOI] [PubMed] [Google Scholar]
- 77.Mackintosh JA, et al. A fluorescent natural product for ultra sensitive detection of proteins in one-dimensional and two-dimensional gel electrophoresis. Proteomics. 2003;3:2273–2288. doi: 10.1002/pmic.200300578. [DOI] [PubMed] [Google Scholar]
- 78.Taylor MS, et al. Affinity Proteomics Reveals Human Host Factors Implicated in Discrete Stages of LINE-1 Retrotransposition. Cell. 2013;155:1034–1048. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Candiano G, et al. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- 80.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 82.Ward JH., Jr Hierarchical Grouping to Optimize an Objective Function. Journal of the American Statistical Association. 1963;58:236–244. [Google Scholar]
- 83.Sokal RR, Rohlf FJ. The Comparison of Dendrograms by Objective Methods. Taxon. 1962;11:33–40. [Google Scholar]
- 84.Pettersen EF, et al. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 85.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 86.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 87.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2012. [Google Scholar]
- 88.Ellison AM. Effect of Seed Dimorphism on the Density-Dependent Dynamics of Experimental Populations of Atriplex Triangularis (Chenopodiaceae) American Journal of Botany. 1987;74:1280–1288. [Google Scholar]
- 89.Statistical Analysis System Institute (Cary, NC) SAS/STAT User’s Guide. 1990 [Google Scholar]
- 90.Hartigan JA, Hartigan PM. The Dip Test of Unimodality. Annals of Statistics. 1985;13:70–84. [Google Scholar]
- 91.Akaike H. New look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;AC-19:716–723. [Google Scholar]
- 92.Benaglia T, Chauveau D, Hunter DR, Young DS. mixtools: An R Package for Analyzing Finite Mixture Models. Journal of Statistical Software. 2009;32:1–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.